Abstract
Methadone is a long-acting opioid with considerable unexplained interindividual variability in clearance. Cytochrome P450 2B6 (CYP2B6) mediates clinical methadone clearance and metabolic inactivation via N-demethylation to 2-ethyl-1,5-dimethyl-3,3-diphenylpyrrolidine (EDDP). Retrospective studies suggest that individuals with the CYP2B6*6 allelic variant have higher methadone plasma concentrations. Catalytic activities of CYP2B6 variants are highly substrate- and expression-system dependent. This investigation evaluated methadone N-demethylation by expressed human CYP2B6 allelic variants in an insect cell coexpression system containing P450 reductase. Additionally, the influence of coexpressing cytochrome b5, whose role in metabolism can be inhibitory or stimulatory depending on the P450 isoform and substrate, on methadone metabolism, was evaluated. EDDP formation from therapeutic (0.25–1 μM) R- and S-methadone concentrations was CYP2B6.4 ≥ CYP2B6.1 ≥ CYP2B6.5 >> CYP2B6.9 ≈ CYP2B6.6, and undetectable from CYP2B6.18. Coexpression of b5 had small and variant-specific effects at therapeutic methadone concentrations but at higher concentrations stimulated EDDP formation by CYP2B6.1, CYP2B6.4, CYP2B6.5, and CYP2B6.9 but not CYP2B6.6. In vitro intrinsic clearances were generally CYP2B6.4 ≥ CYP2B6.1 > CYP2B6.5 > CYP2B6.9 ≥ CYP2B6.6. Stereoselective methadone metabolism (S>R) was maintained with all CYP2B6 variants. These results show that methadone N-demethylation by CYP2B6.4 is greater compared with CYP2B6.1, whereas CYP2B6.9 and CYP2B6.6 (which both contain the 516G>T, Q172H polymorphism), are catalytically deficient. The presence or absence of b5 in expression systems may explain previously reported disparate catalytic activities of CYP2B6 variants for specific substrates. Differences in methadone metabolism by CYP2B6 allelic variants provide a mechanistic understanding of pharmacogenetic variability in clinical methadone metabolism and clearance.
Introduction
Methadone is a long-duration opioid used (primarily as a racemate) to treat multiple types of acute, chronic, and cancer pain, as well as opiate addiction. Methadone use, particularly for pain, has grown exponentially over the past decades. However, the incidence of unanticipated methadone toxicity, and related fatalities, has grown disproportionately, even more so than the increase in methadone use (Paulozzi et al., 2012). There is considerable inter- and intraindividual variability in constitutive methadone metabolism and clearance, and also susceptibility to drug interactions, with the greatest risk related to unanticipated accumulation (Ferrari et al., 2004; Bruce et al., 2013). Variable disposition complicates the clinical use of methadone, and, despite considerable research, mechanisms of variability remain insufficiently understood.
Hepatic methadone N-demethylation to the inactive metabolite 2-ethyl-1,5-dimethyl-3,3-diphenylpyrrolidine (EDDP) is the major route of systemic clearance. Both methadone clearance and N-demethylation are stereoselective. Methadone N-demethylation in vitro, by human liver microsomes and by expressed cytochrome P450s (P450s), is catalyzed most efficiently by CYP2B6 and CYP3A4, and only CYP2B6 N-demethylates methadone stereoselectively (S>R) (Gerber et al., 2004; Kharasch et al., 2004; Totah et al., 2007, 2008; Chang et al., 2011; Gadel et al., 2013).
Although both P450s metabolize methadone in vitro, it has become clear that CYP2B6, rather than CYP3A4, is the predominant P450 responsible for clinical methadone disposition. Evidence derives from drug interaction and genetic studies (Greenblatt, 2014). CYP2B6 induction or inhibition correspondingly modulated methadone metabolism, clearance, and plasma concentrations (Kharasch et al., 2004, 2008a,b; Kharasch and Stubbert, 2013b). In contrast, strong CYP3A inhibitors (Kharasch et al., 2004, 2008a, 2012; van Heeswijk et al., 2013; Kharasch and Stubbert, 2013a) failed to diminish (and sometimes increased) methadone N-demethylation and clearance, and CYP3A induction also had no effect (Vourvahis et al., 2012). CYP2B6 polymorphisms may influence clinical methadone disposition. Gene-association studies suggested that the CYP2B6*6 polymorphism was associated with higher dose-adjusted steady-state plasma methadone concentrations (Crettol et al., 2005, 2006; Eap et al., 2007; Wang et al., 2011) or use of lower methadone doses (Hung et al., 2011; Levran et al., 2013). Formal determination of methadone N-demethylation and clearance showed that both were greater and lesser than wild-types, respectively, in CYP2B6*4 and CYP2B6*6 carriers (Kharasch et al., 2014).
Whereas the fraction of total hepatic P450 represented by CYP2B6 is small, it nonetheless metabolizes a disproportionately greater percentage of drugs (Wang and Tompkins, 2008; Mo et al., 2009). The CYP2B6 gene is highly polymorphic (Zanger and Klein, 2013), with thirty-eight CYP2B6 protein variants identified to date (http://www.cypalleles.ki.se/cyp2b6.htm). The functional consequences of CYP2B6 allelic variants on catalytic activity in vitro are allele-, substrate-, and expression system–dependent (Turpeinen and Zanger, 2012; Zanger and Klein, 2013). P450 function can also be influenced (or not) by coexpression of cytochrome b5 (Xu et al., 2012), and allelic variant effects in vivo are additionally influenced by quantitative differences in CYP2B6 protein expression (Turpeinen and Zanger, 2012; Zanger and Klein, 2013). Among the more well studied variants, CYP2B6*4 (785A>G, K262R) is described as causing increased expression and variably increased or decreased enzymatic activity, CYP2B6*5 (1459C>T, R487C) causing decreased expression and increased specific activity, CYP2B6*6 (516G>T, Q172H; 785A>G, K262R) causing markedly reduced expression and substrate-dependent changes in activity, and CYP2B6*18 (983T>C, I328T) having reduced expression and activity (Turpeinen and Zanger, 2012; Zanger and Klein, 2013). The CYP2B6*6 allele is of particular interest, owing to its frequent occurrence (particularly in African, Asian, and Hispanic populations) and therapeutic significance for the metabolism, pharmacokinetics, and clinical effects of efavirenz, cyclophosphamide, and bupropion (Turpeinen and Zanger, 2012; Zanger and Klein, 2013). We recently reported that methadone N-demethylation catalyzed by CYP2B6.6, the CYP2B6 variant encoded by the CYP2B6*6 polymorphism, is catalytically deficient compared with wild-type CYP2B6.1, and that human liver microsomes with diminished CYP2B6 content owing to a CYP2B6*6 allele had lower rates of methadone N-demethylation (Gadel et al., 2013).
The purpose of the present investigation was to further characterize methadone N-demethylation by CYP2B6 allelic variants, including CYP2B6.1, CYP2B6.4, CYP2B6.5, CYP2B6.6, CYP2B6.9, and CYP2B6.18, coexpressed with NADPH cytochrome P450 reductase in an insect cell system. The second purpose was to evaluate the influence of coexpressed cytochrome b5 on the methadone N-demethylase activities of wild-type and variant CYP2B6 proteins.
Materials and Methods
Chemicals and Reagents.
EDDP and EDDP-d3 were purchased from Cerilliant (Round Rock, TX). R- and S-methadone were from the National Institute on Drug Abuse (Bethesda, MD). All other reagents were from Sigma-Aldrich (St. Louis, MO).
Construction of Plasmids.
Human CYP2B6, P450 reductase (POR), and cytochrome b5 were polymerase chain reaction–amplified from the Human Liver QUICK-Clone cDNA library (Clontech, Mountain View, CA). CYP2B6 variants (2B6.4, 2B6.5, 2B6.6, 2B6.9, 2B6.18) were made from CYP2B6.1 DNA using the QuikChange XL Site-Directed Mutagenesis Kit (Agilent Technologies, Inc., Santa Clara, CA). CYP2B6, POR, and b5 DNA were individually inserted into the pVL1393 vector using the In-Fusion HD Cloning system (Clontech, Mountain View, CA). All sequences were verified by the Protein and Nucleic Acid Chemistry Laboratory (PNACL) at Washington University in St. Louis.
Generation of Virus.
Spodoptera frugiperda (SF9) cells (ATCC, Manassas, VA) were maintained in 500 ml polycarbonate Erlenmeyer flasks with vented caps (Corning, Corning, NY) shaken at 115 rpm and 27°C in Sf-900 III SFM (Life Technologies, Carlsbad, CA). The pVL1393/CYP2B6, pVL1393/POR, and pVL1393/b5 vectors were each individually cotransfected in SF9 cells using the BestBac 2.0 Baculovirus Cotransfection Kit (Expression Systems, Davis, CA). Briefly, 2 ml of SF9 cells were plated at 4,6 × 105 cells/ml in a six-well tissue culture plate and allowed to adhere for 30 minutes. pVL1393/CYP2B6, pVL1393/POR, or pVL1393/b5 DNA was then combined with linearized viral DNA, Expression Systems transfection media, and Expression Systems transfection reagent. The media was removed from the cells, the transfection solution was added, and plates were incubated at 27°C for 4.5 hours. After 4.5 hours, 3 ml of media was added to each well and the plates were incubated for 5 days at 27°C. The resulting cells and supernatant were collected as the p0 viral generation. Subsequent viral generations, up to p3, were performed in SF9 cells grown in suspension. All viral titers were determined using the BacPAK Baculovirus Rapid Titer Kit (Clontech).
Recombinant Protein Expression.
Trichoplusia ni (High Five) cells (Life Technologies) were maintained in Express Five serum-free medium (Life Technologies) supplemented with 16 mM l-glutamine, in 500 ml polycarbonate Erlenmeyer flasks with vented caps (Corning) at 27°C with shaking at 115 rpm. CYP2B6 and POR with or without b5 were coexpressed in High Five cells. On day 0, 100 ml of High Five cells were seeded at a density of 1 × 106 cells/ml. On day 1, cells were counted and infected with the following multiplicities of infection: 4:2:1 (CYP2B6/P450 reductase/cytochrome b5) or 4:2 (CYP2B6/P450 reductase) plaque-forming units per milliliter (pfu/ml) in the presence of 100 μM δ-aminolevulinic acid and 100 μM ferric citrate (Lee et al., 1995). After 72 hours, infected cells were harvested by centrifugation for 15 minutes at 3000g and washed two times with phosphate-buffered saline and pelleted between each wash. The cell pellet was resuspended in 100 mM potassium phosphate buffer (pH 7.4) and homogenized for 2 minutes on ice using a TissueRuptor (Qiagen, Hilden, Germany). Aliquots (500 μl) were stored at –70°C. Cytochrome P450 content, P450 reductase activity, and cytochrome b5 content of the various expression systems are provided in Table 1.
TABLE 1.
Characteristics of the CYP2B6 variant constructs
| P450 contenta | Cytochrome c reductase activity | Cytochrome b5 content | P450/reductase/b5 ratio | |
|---|---|---|---|---|
| pmol/mg | μmol/min per milligram protein | nmol/mg protein | ||
| CYP2B6.1 + P450 reductase + cytochrome b5 | 77 | 4.5 | 0.25 | 1:18:3 |
| CYP2B6.4 + P450 reductase + cytochrome b5 | 107 | 3.3 | 0.28 | 1:10:3 |
| CYP2B6.5 + P450 reductase + cytochrome b5 | 172 | 4.1 | 0.49 | 1:7:3 |
| CYP2B6.6 + P450 reductase + cytochrome b5 | 458 | 4.0 | 0.28 | 1:2:1 |
| CYP2B6.9 + P450 reductase + cytochrome b5 | 198 | 5.9 | 0.32 | 1:9:2 |
| CYP2B6.18 + P450 reductase + cytochrome b5 | 0 | 2.1 | 0.34 | |
| CYP2B6.1 + P450 reductase | 64 | 3.6 | 1:18 | |
| CYP2B6.4 + P450 reductase | 28 | 2.6 | 1:30 | |
| CYP2B6.5 + P450 reductase | 29 | 2.6 | 1:28 | |
| CYP2B6.6 + P450 reductase | 37 | 2.9 | 1:24 | |
| CYP2B6.9 + P450 reductase | 28 | 3.0 | 1:34 | |
| CYP2B6.18 + P450 reductase | 0 | 1.9 |
Determined spectrophotometrically from CO difference spectra, as described in Materials and Methods.
Characterization of Expressed Protein.
Protein concentrations of all infections were determined using Protein Assay Dye Reagent Concentrate (Bio-Rad, Hercules, CA), following manufacturer’s instructions. For electrophoresis, 20 μg of protein from whole cell lysate was mixed with 4× Protein Loading Buffer (LI-COR Biosciences, Lincoln, NE) and subjected to electrophoresis on a precast NuPAGE 4–12% Bis-Tris Gel (Life Technologies). Precision Plus Protein Dual Color Prestained Standard (10 μl; Bio-Rad, Hercules, CA) was used as a molecular weight standard. Proteins were transferred to a nitrocellulose membrane using the iBlot Transfer System (Life Technologies), per manufacturer’s instructions. Membranes were blocked (1 hour at room temperature) in Blocking Buffer (LI-COR). Blocked membranes were incubated overnight at 4°C with rabbit anti-CYP2B6 (H-110) antibody (1:1000 dilution), rabbit anti-CYPOR (H-300) antibody (1:2500 dilution), and mouse anti-cytochrome b5 (36) antibody (1:1000 dilution) (Santa Cruz Biotechnology, Dallas, TX) in blocking buffer containing 0.1% Tween-20. After washing, membranes were incubated with goat anti-rabbit IRDye 680 (1:10000 dilution) and goat anti-mouse IRDye 800CW (1:10000 dilution) (LI-COR) for 30 minutes at room temperature in blocking buffer containing 0.1% Tween-20. CYP2B6, POR, and b5 were all visualized using an Odyssey Infrared Imager (LI-COR). P450 concentration was determined as previously described with minor modifications (Matsubara et al., 1976). Briefly, cell lysate was diluted to 1 mg/ml, bubbled with CO, and the baseline absorbance reading was taken from 400–500 nm using a Synergy MX Microplate Reader (Biotek, Winooski, VT). Sodium dithionite (final concentration 25 mM) was added immediately after the reference baseline was obtained, and the final absorbance reading was taken after 2 minutes. Cytochrome c reductase activity was determined using 0.3 M potassium phosphate buffer (pH 7.7) at 37°C (Dignam and Strobel, 1977) and the Synergy MX Microplate Reader. Cytochrome b5 content was determined as previously described (McLaughlin et al., 2010). The P450/reductase ratio was calculated on the basis of an assumed specific activity of 3200 nmol of cytochrome c reduced/min per nanomole reductase (Parikh et al., 1997).
Methadone Metabolism.
Incubations (200 μl) with R- or S-methadone were performed as previously described (Gadel et al., 2013), with minor modifications. Preliminary experiments showed that N-demethylation was linear for up to 45 minutes; routine incubations were 10 minutes. Reactions (10 minutes) were quenched with 40 μl 20% trichloroacetic acid containing internal standard (d3-EDDP, final concentration 1.6 ng/ml), and centrifuged for 5 minutes at 2500g; the supernatant (150 μl) was processed immediately by solid-phase extraction as described previously (Kharasch et al., 2004), except that Strata-X-C 33-μm, 30 mg/well plates (Phenomenex, Torrance, CA) were used. EDDP and methadone achiral analysis was performed on an Agilent 6140 single quadrupole mass spectrometer with an electrospray ionization source, Agilent 1100 series high-pressure liquid chromatography system equipped with a 96-well plate autosampler (Agilent Technologies, Inc., Santa Clara, CA), and a Sunfire C18 column (2.1 × 50 mm, 3.5-μm) (Waters, Milford, MA) with a 2-μm column filter guard (Supleco Analytical, Bellefonte, PA). Sample injections were 25 μl and the column oven was held at 30°C. Mobile phase A was 4.5 mM ammonium acetate in Milli-Q water, pH 4.5, and mobile phase B was 4.5 mM ammonium acetate in acetonitrile. The mobile phase gradient (0.4 ml/min) was 25% B for 0.5 minutes, linear gradient to 75% B between 0.5 and 4.2 minutes, held at 75% B until 5.0 minutes, immediately decreased back to 25% B and re-equilibrated at initial conditions for 3.0 minutes. Under these conditions, EDDP retention time was 4.6 minutes. Mass spectrometer parameters were: positive ion mode, nitrogen drying gas at 12 l/min and 300°C, nebulizer pressure of 35 psig, capillary voltage 3000 V, and fragmentor voltage of 80 V for EDDP 70 V for d3-EDDP. All ions were monitored in the same ion group: m/z of 278.2 and 281.2 for EDDP and d3-EDDP, respectively. Analytes were quantified using peak area ratios and standard curves prepared using calibration standards in buffer. Control incubations lacking enzyme were included for all reactions to determine background EDDP content, which was subtracted from all results.
Data and Statistical Analysis.
Results are the mean ± S.D. (3–6 replicates) unless otherwise indicated. EDDP formation by CYP2B6 variants was compared by analysis of variance. EDDP formation versus substrate concentration data were analyzed by nonlinear regression analysis (SigmaPlot 12.5; Systat, San Jose, CA) evaluating a single-enzyme Michaelis-Menten, Adair-Pauling, or substrate (or product inhibition) model as described previously (Totah et al., 2007, 2008; Gadel et al., 2013). The choice of model was based on whether the Eadie-Hofstee plots were linear or nonlinear and the goodness of fit regression diagnostics. Modeling results are the parameter estimate plus or minus standard error of the estimate. Parameter estimates for CYP2B6 variants and CYP2B1.1 were compared using an unpaired t test. Significance was assigned at P < 0.05.
Results
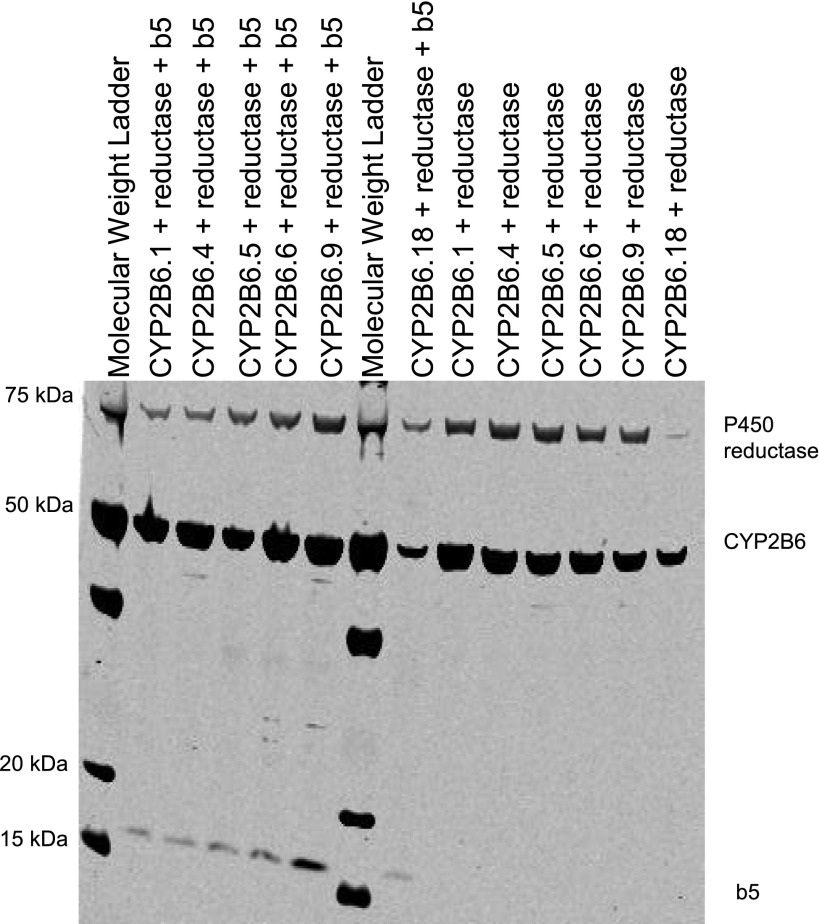
CYP2 2B6.1, 2B6.4, 2B6.6, 2B6.9, and 2B6.18 were successfully coexpressed with cytochrome P450 reductase, with or without coexpressed cytochrome b5. Expression of all proteins was confirmed by Western blot (Fig. 1). CYP2B6.18 protein was expressed but did not generate a CO-difference spectrum. P450 content, P450 reductase activity, and b5 content of the various constructs are provided in Table 1.
Fig. 1.
Western blot showing expression of CYP2B6, P450 reductase, and cytochrome b5.
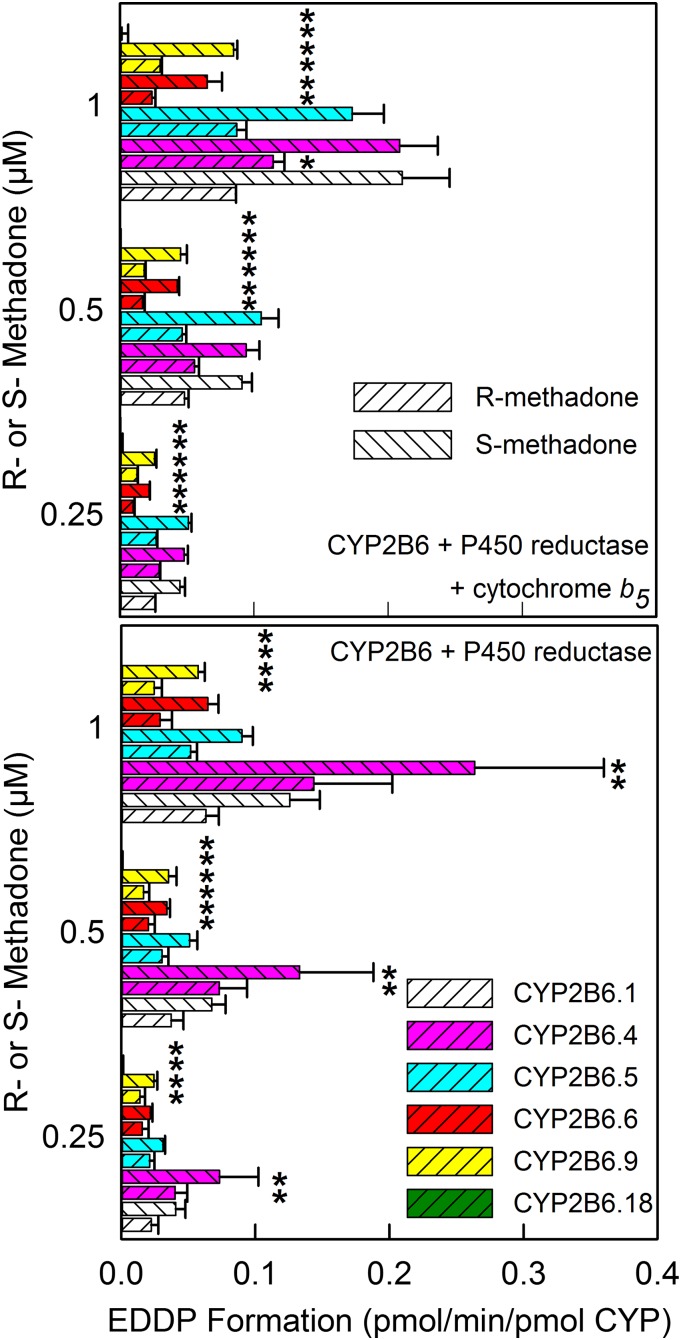
Methadone enantiomer N-demethylation was evaluated at plasma concentrations (0.25–1 μM) typically occurring in patients receiving low and high doses of methadone for treatment of pain or substance abuse, respectively (Fig. 2). EDDP formation from therapeutic (0.25–1 μM) R- and S-methadone concentrations was CYP2B6.4 = CYP2B6.1 = CYP2B6.5 > CYP2B6.9 ≈ CYP2B6.6, and undetectable from CYP2B6.18 in expression systems with P450, P450 reductase, and coexpressed cytochrome b5, and CYP2B6.4 > CYP2B6.1 ≥ CYP2B6.5 > CYP2B6.9 ≈ CYP2B6.6, and undetectable from CYP2B6.18 in expression systems with P450 and P450 reductase but without cytochrome b5. EDDP formation by CYP2B6.6 and CYP2B6.9 was one-half to one-third that by CYP2B6.1. Coexpression of b5 had little effect at therapeutic methadone concentrations. EDDP formation ratios with/without b5 were 1.3, 0.7, 1.7, 0.9, and 1.2 for CYP2Bs 6.1, 6.4, 6.5, 6.6, and 6.9, respectively. With all CYP2B6 variants and expression systems, methadone N-demethylation was stereoselective (S>R). With coexpressed cytochrome b5, the S/R ratio averaged 2.1, whereas it was slightly less (1.8) without b5.
Fig. 2.
Recombinant CYP2B6-catalyzed N-demethylation of methadone enantiomers at therapeutic concentrations. Results are shown for CYP2B6 and cytochrome P450 reductase, with (top) or without (bottom) coexpressed cytochrome b5. Results are the mean ± S.D. of 5–6 determinations. *Significantly different from CYP2B6.1 (P < 0.05).
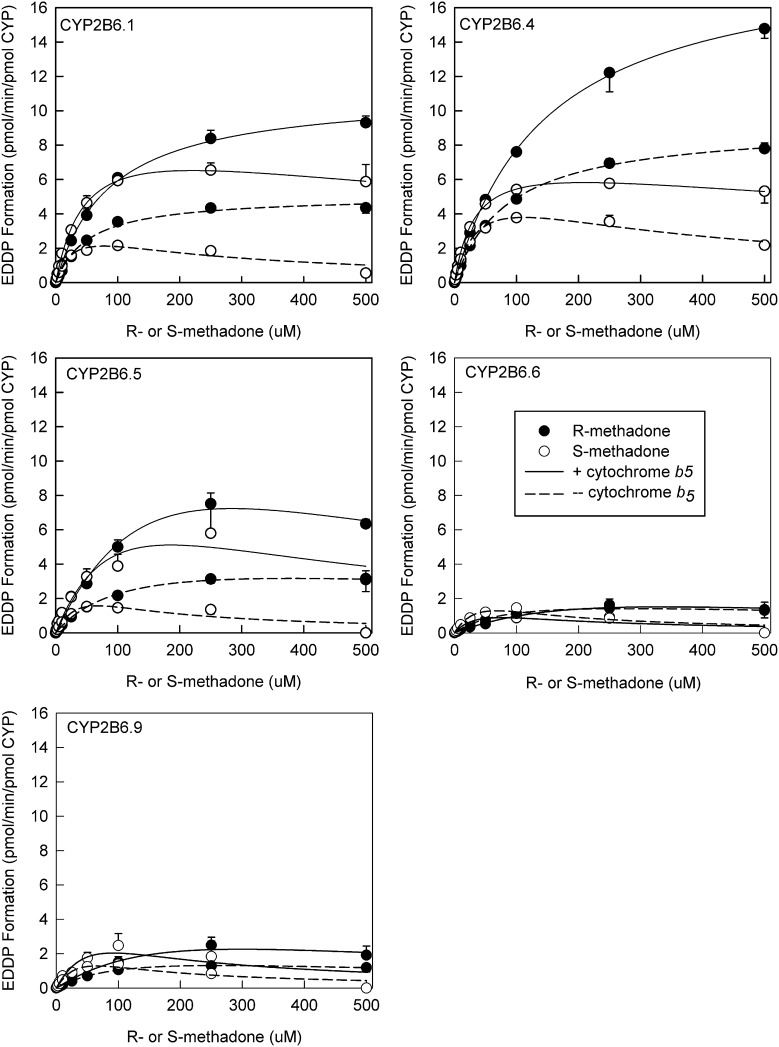
The concentration-dependence of methadone enantiomers N-demethylation was determined both in the presence and absence of coexpressed cytochrome b5 for CYP2Bs 6.1, 6.4, 6.5, 6.6, and 6.9 (Fig. 3). There was evidence for substrate or product inhibition at the highest concentration of S-methadone (but not R-methadone), as observed previously (Gadel et al., 2013). Eadie-Hosfstee plots were generally linear, with or without coexpressed b5 (not shown). Linear plots were interpreted as methadone binding to a single site, and analyzed with the Michaelis-Menten model, with substrate inhibition where apparent. Kinetic parameters are provided in Table 2. In vitro intrinsic clearances (Clint) were generally greater for S- versus R-methadone N-demethylation, regardless of CYP2B6 variant or the absence or presence of b5. Clint was generally of the order CYP2B6.4 ≥ CYP2B6.1 > CYP2B6.5 > CYP2B6.9 ≥ CYP2B6.6. Figure 3 also shows the influence of cytochrome b5 coexpression on N-demethylase activity for each CYP2B6 variant. For CYP2B6.1, 6.4, and 6.5, cytochrome b5 was stimulatory, and Clint was increased 20–80% compared with expression systems omitting b5. Coexpression of b5 had little effect on methadone N-demethylation by CYP2B6.6 and CYP2B6.9.
Fig. 3.
Concentration-dependence of recombinant CYP2B6-catalyzed N-demethylation of methadone enantiomers to EDDP. Each data point is the mean ± S.D. of 3–6 determinations. Solid and open symbols show R-EDDP and S-EDDP, respectively. Solid and open lines show the presence and absence of cytochrome b5. Lines represent rates predicted from nonlinear regression analysis of observed metabolite formation. Kinetic parameters are summarized in Table 2.
TABLE 2.
Kinetic parameters for methadone N-demethylation
Results are the parameter estimate ± standard error of the estimate.
| Parameter | CYP2B6.1 |
CYP2B6.4 |
CYP2B6.5 |
CYP2B6.6 |
CYP2B6.9 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| R-EDDP formation | S-EDDP formation | R-EDDP formation | S-EDDP formation | R-EDDP formation | S-EDDP formation | R-EDDP formation | S-EDDP formation | R-EDDP formation | S-EDDP formation | |
| CYP2B6 + P450 reductase + cytochrome b5 | ||||||||||
| Vmax (pmol/min per picomole P450) | 11 ± 1 | 9 ± 1 | 19 ± 1* | 8 ± 1* | 22 ± 4* | 13 ± 5 | 4 ± 3* | 2 ± 1* | 7 ± 4 | 6 ± 4 |
| Km (μM) | 87 ± 4 | 48 ± 6 | 148 ± 6* | 34 ± 3* | 284 ± 73* | 138 ± 78 | 337 ± 290 | 54 ± 46 | 298 ± 204 | 92 ± 91 |
| Clint (ml/min per nanomole) | 0.13 ± 0.01 | 0.19 ± 0.03 | 0.13 ± 0.01 | 0.23 ± 0.02 | 0.08 ± 0.03 | 0.09 ± 0.06 | 0.01 ± 0.01* | 0.04 ± 0.04* | 0.02 ± 0.02* | 0.07 ± 0.08 |
| CYP2B6 + P450 reductase | ||||||||||
| Vmax (pmol/min per picomole P450) | 5 ± 1 | 4 ± 1 | 9 ± 1* | 7 ± 1* | 5 ± 1 | 5 ± 1 | 2 ± 1* | 4 ± 2 | 3 ± 1* | 4 ± 2 |
| Km (μM) | 52 ± 3 | 33 ± 8 | 87 ± 3* | 43 ± 5 | 119 ± 33* | 67 ± 45 | 96 ± 32 | 67 ± 44 | 132 ± 32* | 65 ± 36 |
| Clint (ml/min per nanomole) | 0.10 ± 0.01 | 0.12 ± 0.04 | 0.11 ± 0.01 | 0.16 ± 0.02 | 0.04 ± 0.01* | 0.07 ± 0.06 | 0.02 ± 0.01* | 0.06 ± 0.05 | 0.02 ± 0.01* | 0.06 ± 0.04 |
Significantly different versus CYP2B6.1 (P < 0.05)
Discussion
The major finding was that at therapeutic concentrations, N-demethylation of R- and S-methadone by the variant CYP2B6 isoform CYP2B6.4 was greater than that by wild-type CYP2B6.1, whereas EDDP formation by CYP2B6.6 and CYP2B6.9 was less than CYP2B6.1, and CYP2B6.18 was catalytically incompetent. Assessed over broader methadone concentrations, in vitro intrinsic clearances for CYP2B6.6 and CYP2B6.9 were significantly lower than for CYP2B6.1. Stereoselectivity of methadone metabolism was maintained in all CYP2B6 variants, with S-methadone N-demethylation 2-fold greater than that of R-methadone, similar to that with CYP2B6.1 (Gerber et al., 2004; Totah et al., 2007, 2008; Chang et al., 2011; Gadel et al., 2013). Lower rates of CYP2B6.6-catalyzed methadone N-demethylation in the present investigation, in which insect cells were transfected with individual virus constructs for CYP2B6, P450 reductase, and cytochrome b5, were also seen in an insect cell system transfected with a single virus containing all three proteins (Gadel et al., 2013).
The kinetics of methadone N-demethylation by CYP2B6 are complex (Totah et al., 2007; Gadel et al., 2013). Previous experiments with expressed CYP2B6.1 and CYP2B6.6 suggested apparent multisite or multiple-affinity methadone binding with complex allosteric kinetics or homotropic cooperativity, modeled best using the Adair-Pauling equation (Totah et al., 2007; Gadel et al., 2013). Although there appeared to be substrate or product inhibition with CYP2B6.6 at the highest S-methadone concentration, this could not be modeled (Gadel et al., 2013). In the present investigation, substrate (or product) inhibition with S-methadone was more apparent, and more amenable to modeling, and Michaelis-Menten kinetics with substrate inhibition provided better fits to the data. Nonetheless, differences between kinetic models did not materially affect the intrinsic clearances or conclusions of this investigation that metabolism by CYP2B6.4 was greater than by CYP2B6.1, CYP2B6.5 was lower, and CYP2B6.6 and CYP2B6.9 were substantially less.
CYP2B6 expression levels in the triple protein constructs differed between allelic variants, resulting in varying P450/P450 reductase molar ratios. Nonetheless, this did not explain the differences between variants in methadone metabolism. Such varying ratios were also seen previously, with CYP2B6 systems using a single virus for the three proteins (P450, reductase, and b5) (Xu et al., 2012; Gadel et al., 2013). When reductase levels are sufficient for catalytic activity, overexpression of reductase may not be a factor (Nakajima et al., 2002).
The influence of CYP2B6 allelic mutations on catalytic activity, in general, is CYP2B6 variant- and substrate-dependent (Mo et al., 2009; Zanger and Klein, 2013). In an insect cell system with coexpressed P450 reductase and b5, CYP2B6.6 Clint for bupropion 4-hydroxylation was lower, whereas efavirenz 8-hydroxylation was less but not significantly different from CYP2B6.1 (Xu et al., 2012). CYP2B6.6 activity in other systems (E. coli, Cos-1, Cos-7) was lower toward bupropion and efavirenz (Zhang et al., 2011), and ketamine (Li et al., 2013), but greater for cyclophosphamide (Xie et al., 2003; Ariyoshi et al., 2011; Raccor et al., 2012) and artemether (Honda et al., 2011), and unchanged for selegiline (Watanabe et al., 2010). The present and previous (Gadel et al., 2013) experiments show that methadone is one of the most catalytically diminished substrates for CYP2B6.6. A novel finding herein is that CYP2B6.9 is also catalytically deficient toward methadone. Data regarding the functional activity of CYP2B6.9 are said to be rare (Zanger and Klein, 2013). In a COS-1 system, the activities of CYP2B6.6 and CYP2B6.9, in which 516G>T is the common polymorphism, were both extremely low, but this was attributed to minimal expression (Hofmann et al., 2008). CYP2B6.9 activity toward artemether was somewhat lower than CYP2B6.1 in COS-7 cells (Honda et al., 2011), and toward 7-ethoxy-4-trifluoromethylcoumarin, bupropion, and efavirenz in a reconstituted bacterial expression system (Zhang et al., 2011). The present results suggest that methadone is also one of the more catalytically diminished substrates with CYP2B6.9. Although CYP2B6.5 has been reported to have shown increased catalytic activity that compensates for lower expression (Zanger and Klein, 2013), this was not observed with methadone. CYP2B6.5 activities are variable with other substrates. CYP2B6.5 cyclophosphamide 4-hydroxylation in E. coli, Cos-1, and Cos-7 systems was half that of CYP2B6.1 (Raccor et al., 2012), and artemether and selegiline metabolism was diminished (Watanabe et al., 2010; Honda et al., 2011), whereas 7-ethoxy-4-trifluoromethylcoumarin, bupropion, and efavirenz metabolism was decreased by one-third, half, and increased, respectively, compared with CYP2B6.1 (Zhang et al., 2011; Radloff et al., 2013). CYP2B6.4 results are even more substrate-dependent. In E. coli, Cos-1, and Cos-7 systems, CYP2B6.4 cyclophosphamide 4-hydroxylation was 25% lower than CYP2B6.1 (Ariyoshi et al., 2011; Raccor et al., 2012), the catalytic efficiency for 7-ethoxy-4-trifluoromethylcoumarin, bupropion, and efavirenz was decreased to half, one-third, and unchanged, respectively (Zhang et al., 2011), and artemether and selegiline metabolism was almost doubled (Watanabe et al., 2010; Honda et al., 2011). Methadone metabolism by CYP2B6.4 in the present investigation was greater than by CYP2B6.1. Thus, whereas 785A>G (K262R) alone (CYP2B6.4) caused increased methadone N-demethylation, 785A>G together with 516G>T (Q172H) (CYP2B6.6), and 516G>T alone (CYP2B6.9) diminished methadone metabolism. Catalytic incompetence of CYP2B6.18 toward methadone is consistent with other CYP2B6 substrates (Zanger and Klein, 2013).
The influence of CYP2B6 allelic mutations on catalytic activity appears dependent on the enzyme system used. In addition to differences noted above between various expression systems, most have also not included cytochrome b5, and the absence or presence of b5 may influence results. Efavirenz metabolism by both CYP2B6.1 and CYP2B6.6 was unchanged by b5, whereas bupropion metabolism by CYP2B6.1 was unchanged by b5 but metabolism by CYP2B6.6 was diminished (Xu et al., 2012). The present results showed that b5 increased methadone Clint by CYP2B6.1 and CYP2B6.4 but not metabolism at therapeutic concentrations. Effects of b5 on CYP2B6-catalyzed metabolism may underlie, at least in part, different results with various expression systems, and the kinetic consequences of CYP2B6 polymorphisms (Zanger and Klein, 2013). This investigation appears to be one of the first to use an insect cell system with a panel of CYP2B6 variants and coexpressed P450 reductase and b5. Use of fully competent CYP2B6 systems, containing both P450 reductase and cytochrome b5, may be advantageous.
Although the present investigation evaluated only catalytic activity, some clinical implications can be inferred regarding the influence of CYP2B6 variants on methadone disposition. Clinical consequences of CYP2B6 allelic variants will depend on both the intrinsic activity of the mutant protein and its level of expression. The CYP2B6*6 allele causes low human hepatic CYP2B expression (Lang et al., 2001; Desta et al., 2007; Hofmann et al., 2008), thus both deficient catalytic efficiency and decreased P450 content combine to cause a diminished CYP2B6 metabolizer phenotype in CYP2B6*6 carriers. Liver microsomes from CYP2B6*6 carriers had diminished methadone N-demethylation (Gadel et al., 2013). The lower CYP2B6.9 and CYP2B6.18 activities toward methadone suggest that similar results might be expected with CYP2B6*9 and CYP2B6*18 carriers; however, microsomal methadone metabolism by livers from carriers of variants other than CYP2B6*6 have not been reported. Clinical genetic association studies of methadone plasma concentrations are consistent with diminished methadone N-demethylation by CYP2B6.6 and liver microsomes from CYP2B6*6 carriers. In patients, methadone doses were lower (Hung et al., 2011; Levran et al., 2013), and dose-adjusted steady-state S-methadone concentrations were greater (Crettol et al., 2005, 2006; Eap et al., 2007; Wang et al., 2011) in CYP2B6*6 homozygotes compared with heterozygotes and noncarriers. While these observations suggested that CYP2B6 allelic variants might influence clinical methadone pharmacokinetics, only recently has this been confirmed. Methadone enantiomer N-demethylation and clearance were increased and decreased, respectively, in CYP2B6*4 and CYP2B6*6 carriers (Kharasch et al., 2014). Thus in vitro methadone metabolism by CYP2B6 variants does predict clinical methadone metabolism and clearance.
An interesting finding is that b5 coexpression influenced methadone metabolism, and this was CYP2B6 variant–specific. Coexpression of b5 stimulated R- and S-methadone metabolism by CYP2B6.1, CYP2B6.4, and CYP2B6.5, with less or minimal effects on CYP2B6.6 and CYP2B6.9. In the P450 catalytic cycle, involving two sequential one-electron transfers from NADPH to P450 and substrate, P450 reductase transfers the first electron, and the second electron may be transferred by P450 reductase or by b5 (Hildebrandt and Estabrook, 1971; Schenkman and Jansson, 2003). Effects of b5 are however variable, stimulating, inhibiting, or not affecting metabolism, depending on the P450 isoform, substrate, and experimental conditions (Schenkman and Jansson, 2003; Finn et al., 2008; Im and Waskell, 2011). Recently this variability was shown for the first time to extend to P450 variants, with some CYP1A2 mutants quite affected by b5 (Palma et al., 2013). On the basis of several kinetic, mutagenesis, and NMR binding studies of wild-type rabbit CYP2B4, Waskell et al. proposed a model for an electron transfer complex between the acidic convex surface of b5 and the concave basic proximal surface of CYP2B4 (Im and Waskell, 2011; Ahuja et al., 2013). More specifically, the b5 binding site is on the CYP2B4 C-helix and β-bulge, with Asp65 and Val66 of b5 in contact with Arg122, Arg126, and Lys433 of CYP2B4, with Arg133 also critical for interaction of the two proteins. Other sites, specifically Met137 and Lys139, which were important for CYP2B4 binding to b5, are thought to perturb the C-helix, and R422 also affected CYP2B4 binding (Ahuja et al., 2013). On the basis of sequence alignments, the CYP2B4 residues Arg122, Arg126, Met137, Lys139, and Arg422, which influence binding to b5, correspond to Lys122, Arg126, Met137, Lys139, and Lys422 of CYP2B6. The mutated residues in CYP2B6.6 (Gln172His, Lys262Arg), however, do not correspond to the b5 binding site identified on CYP2B4. With CYP1A2, the variant Gly299Ser (CYP1A2.13) had the most altered b5 response compared with wild-type, with a lesser influence of the variants Thr83Met (CYP1A2.9), Ile386Phe (CYP1A2.4), and Cys406Tyr (CYP1A2.5) (Palma et al., 2013). Gly299Ser was reported to be on the surface of the heme domain near the interaction site with b5, and close to the CYP1A2 C-helix, which is also thought to be important in interaction with b5 (Palma et al., 2013). On the basis of sequence alignment of CYP2B6 with CYP1A2, Thr83, Gly299, Ile386, and Cys406 of CYP1A2 correspond to CYP2B6 Arg73, His280, Val367, and Ile387. These residues are not mutated in the CYP2B6 variants with altered response to b5. Thus there appears no consistent pattern to explain P450 mutant–specific effects of b5. Another suggested explanation is that b5 stimulates metabolism of poorer substrates but not those more avidly metabolized (Im and Waskell, 2011). The present results show that b5 enhanced metabolism by the more active CYP2B6 variants. As stated previously, the role of cytochrome b5 remains enigmatic (Schenkman and Jansson, 2003).
In summary, methadone N-demethylation by CYP2B6 variants was in the order CYP2B6.4 ≥ CYP2B6.1 > CYP2B6.5 > CYP2B6.9 ≥ CYP2B6.6 >> CYP2B6.18. Differences in methadone metabolism by CYP2B6 allelic variants provide a mechanistic understanding for pharmacogenetic variability in clinical methadone metabolism and clearance.
Abbreviations
- EDDP
2-ethyl-1,5-dimethyl-3,3-diphenylpyrrolidine
- P450
cytochrome P450
- POR
P450 reductase
Authorship Contributions
Participated in research design: Gadel, Kharasch.
Conducted experiments: Gadel, Friedel.
Contributed new reagents or analytic tools: Gadel.
Performed data analysis: Gadel, Friedel, Kharasch.
Wrote or contributed to the writing of the manuscript: Gadel, Kharasch.
Footnotes
This work was supported by the National Institutes of Health [Grants R01-DA14211 and K24-DA00412]
References
- Ahuja S, Jahr N, Im SC, Vivekanandan S, Popovych N, Le Clair SV, Huang R, Soong R, Xu J, Yamamoto K, et al. (2013) A model of the membrane-bound cytochrome b5-cytochrome P450 complex from NMR and mutagenesis data. J Biol Chem 288:22080–22095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariyoshi N, Ohara M, Kaneko M, Afuso S, Kumamoto T, Nakamura H, Ishii I, Ishikawa T, Kitada M. (2011) Q172H replacement overcomes effects on the metabolism of cyclophosphamide and efavirenz caused by CYP2B6 variant with Arg262. Drug Metab Dispos 39:2045–2048. [DOI] [PubMed] [Google Scholar]
- Bruce RD, Moody DE, Altice FL, Gourevitch MN, Friedland GH. (2013) A review of pharmacological interactions between HIV or hepatitis C virus medications and opioid agonist therapy: implications and management for clinical practice. Expert Rev Clin Pharmacol 6:249–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Fang WB, Lin SN, Moody DE. (2011) Stereo-selective metabolism of methadone by human liver microsomes and cDNA-expressed cytochrome P450s: a reconciliation. Basic Clin Pharmacol Toxicol 108:55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crettol S, Déglon JJ, Besson J, Croquette-Krokkar M, Gothuey I, Hämmig R, Monnat M, Hüttemann H, Baumann P, Eap CB. (2005) Methadone enantiomer plasma levels, CYP2B6, CYP2C19, and CYP2C9 genotypes, and response to treatment. Clin Pharmacol Ther 78:593–604. [DOI] [PubMed] [Google Scholar]
- Crettol S, Déglon JJ, Besson J, Croquette-Krokar M, Hämmig R, Gothuey I, Monnat M, Eap CB. (2006) ABCB1 and cytochrome P450 genotypes and phenotypes: influence on methadone plasma levels and response to treatment. Clin Pharmacol Ther 80:668–681. [DOI] [PubMed] [Google Scholar]
- Desta Z, Saussele T, Ward B, Blievernicht J, Li L, Klein K, Flockhart DA, Zanger UM. (2007) Impact of CYP2B6 polymorphism on hepatic efavirenz metabolism in vitro. Pharmacogenomics 8:547–558. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Strobel HW. (1977) NADPH-cytochrome P-450 reductase from rat liver: purification by affinity chromatography and characterization. Biochemistry 16:1116–1123. [DOI] [PubMed] [Google Scholar]
- Eap CB, Crettol S, Rougier JS, Schläpfer J, Sintra Grilo L, Déglon JJ, Besson J, Croquette-Krokar M, Carrupt PA, Abriel H. (2007) Stereoselective block of hERG channel by (S)-methadone and QT interval prolongation in CYP2B6 slow metabolizers. Clin Pharmacol Ther 81:719–728. [DOI] [PubMed] [Google Scholar]
- Ferrari A, Coccia CP, Bertolini A, Sternieri E. (2004) Methadone—metabolism, pharmacokinetics and interactions. Pharmacol Res 50:551–559. [DOI] [PubMed] [Google Scholar]
- Finn RD, McLaughlin LA, Ronseaux S, Rosewell I, Houston JB, Henderson CJ, Wolf CR. (2008) Defining the in vivo role for cytochrome b5 in cytochrome P450 function through the conditional hepatic deletion of microsomal cytochrome b5. J Biol Chem 283:31385–31393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadel S, Crafford A, Regina K, Kharasch ED. (2013) Methadone N-demethylation by the common CYP2B6 allelic variant CYP2B6.6. Drug Metab Dispos 41:709–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber JG, Rhodes RJ, Gal J. (2004) Stereoselective metabolism of methadone N-demethylation by cytochrome P4502B6 and 2C19. Chirality 16:36–44. [DOI] [PubMed] [Google Scholar]
- Greenblatt DJ. (2014) Drug interactions with methadone: Time to revise the product label. Clin Pharmacol Drug Dev 3:249–251. [DOI] [PubMed] [Google Scholar]
- Hildebrandt A, Estabrook RW. (1971) Evidence for the participation of cytochrome b 5 in hepatic microsomal mixed-function oxidation reactions. Arch Biochem Biophys 143:66–79. [DOI] [PubMed] [Google Scholar]
- Hofmann MH, Blievernicht JK, Klein K, Saussele T, Schaeffeler E, Schwab M, Zanger UM. (2008) Aberrant splicing caused by single nucleotide polymorphism c.516G>T [Q172H], a marker of CYP2B6*6, is responsible for decreased expression and activity of CYP2B6 in liver. J Pharmacol Exp Ther 325:284–292. [DOI] [PubMed] [Google Scholar]
- Honda M, Muroi Y, Tamaki Y, Saigusa D, Suzuki N, Tomioka Y, Matsubara Y, Oda A, Hirasawa N, Hiratsuka M. (2011) Functional characterization of CYP2B6 allelic variants in demethylation of antimalarial artemether. Drug Metab Dispos 39:1860–1865. [DOI] [PubMed] [Google Scholar]
- Hung CC, Chiou MH, Huang BH, Hsieh YW, Hsieh TJ, Huang CL, Lane HY. (2011) Impact of genetic polymorphisms in ABCB1, CYP2B6, OPRM1, ANKK1 and DRD2 genes on methadone therapy in Han Chinese patients. Pharmacogenomics 12:1525–1533. [DOI] [PubMed] [Google Scholar]
- Im SC, Waskell L. (2011) The interaction of microsomal cytochrome P450 2B4 with its redox partners, cytochrome P450 reductase and cytochrome b(5). Arch Biochem Biophys 507:144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharasch ED, Stubbert K. (2013a) Cytochrome P4503A does not mediate the interaction between methadone and ritonavir-lopinavir. Drug Metab Dispos 41:2166–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharasch ED, Stubbert K. (2013b) Role of cytochrome P4502B6 in methadone metabolism and clearance. J Clin Pharmacol 53:305–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharasch ED, Hoffer C, Whittington D, Sheffels P. (2004) Role of hepatic and intestinal cytochrome P450 3A and 2B6 in the metabolism, disposition, and miotic effects of methadone. Clin Pharmacol Ther 76:250–269. [DOI] [PubMed] [Google Scholar]
- Kharasch ED, Bedynek PS, Park S, Whittington D, Walker A, Hoffer C. (2008a) Mechanism of ritonavir changes in methadone pharmacokinetics and pharmacodynamics: I. Evidence against CYP3A mediation of methadone clearance. Clin Pharmacol Ther 84:497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharasch ED, Mitchell D, Coles R, Blanco R. (2008b) Rapid clinical induction of hepatic cytochrome P4502B6 activity by ritonavir. Antimicrob Agents Chemother 52:1663–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharasch ED, Bedynek PS, Hoffer C, Walker A, Whittington D. (2012) Lack of indinavir effects on methadone disposition despite inhibition of hepatic and intestinal cytochrome P4503A (CYP3A). Anesthesiology 116:432–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharasch ED, Parchomski J, Regina K, Blood J, Yang Y. (2014) Methadone enantiomers metabolism and clearance are impaired in individuals with CYP2B6*6 genotype. Clin Pharmacol Ther 95:S53. [Google Scholar]
- Lang T, Klein K, Fischer J, Nüssler AK, Neuhaus P, Hofmann U, Eichelbaum M, Schwab M, Zanger UM. (2001) Extensive genetic polymorphism in the human CYP2B6 gene with impact on expression and function in human liver. Pharmacogenetics 11:399–415. [DOI] [PubMed] [Google Scholar]
- Lee CA, Kadwell SH, Kost TA, Serabjit-Singh CJ. (1995) CYP3A4 expressed by insect cells infected with a recombinant baculovirus containing both CYP3A4 and human NADPH-cytochrome P450 reductase is catalytically similar to human liver microsomal CYP3A4. Arch Biochem Biophys 319:157–167. [DOI] [PubMed] [Google Scholar]
- Levran O, Peles E, Hamon S, Randesi M, Adelson M, Kreek MJ. (2013) CYP2B6 SNPs are associated with methadone dose required for effective treatment of opioid addiction. Addict Biol 18:709–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Coller JK, Hutchinson MR, Klein K, Zanger UM, Stanley NJ, Abell AD, Somogyi AA. (2013) The CYP2B6*6 allele significantly alters the N-demethylation of ketamine enantiomers in vitro. Drug Metab Dispos 41:1264–1272. [DOI] [PubMed] [Google Scholar]
- Matsubara T, Koike M, Touchi A, Tochino Y, Sugeno K. (1976) Quantitative determination of cytochrome P-450 in rat liver homogenate. Anal Biochem 75:596–603. [DOI] [PubMed] [Google Scholar]
- McLaughlin LA, Ronseaux S, Finn RD, Henderson CJ, Roland Wolf C. (2010) Deletion of microsomal cytochrome b5 profoundly affects hepatic and extrahepatic drug metabolism. Mol Pharmacol 78:269–278. [DOI] [PubMed] [Google Scholar]
- Mo SL, Liu YH, Duan W, Wei MQ, Kanwar JR, Zhou SF. (2009) Substrate specificity, regulation, and polymorphism of human cytochrome P450 2B6. Curr Drug Metab 10:730–753. [DOI] [PubMed] [Google Scholar]
- Nakajima M, Tane K, Nakamura S, Shimada N, Yamazaki H, Yokoi T. (2002) Evaluation of approach to predict the contribution of multiple cytochrome P450s in drug metabolism using relative activity factor: effects of the differences in expression levels of NADPH-cytochrome P450 reductase and cytochrome b(5) in the expression system and the differences in the marker activities. J Pharm Sci 91:952–963. [DOI] [PubMed] [Google Scholar]
- Palma BB, Silva E Sousa M, Urban P, Rueff J, Kranendonk M. (2013) Functional characterization of eight human CYP1A2 variants: the role of cytochrome b5. Pharmacogenet Genomics 23:41–52. [DOI] [PubMed] [Google Scholar]
- Parikh A, Gillam EM, Guengerich FP. (1997) Drug metabolism by Escherichia coli expressing human cytochromes P450. Nat Biotechnol 15:784–788. [DOI] [PubMed] [Google Scholar]
- Paulozzi LJ, Mack KA, Jones CM, Centers for Disease Control and Prevention (CDC) (2012) Vital signs: risk for overdose from methadone used for pain relief - United States, 1999-2010. MMWR Morb Mortal Wkly Rep 61:493–497. [PubMed] [Google Scholar]
- Raccor BS, Claessens AJ, Dinh JC, Park JR, Hawkins DS, Thomas SS, Makar KW, McCune JS, Totah RA. (2012) Potential contribution of cytochrome P450 2B6 to hepatic 4-hydroxycyclophosphamide formation in vitro and in vivo. Drug Metab Dispos 40:54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff R, Gras A, Zanger UM, Masquelier C, Arumugam K, Karasi JC, Arendt V, Seguin-Devaux C, Klein K. (2013) Novel CYP2B6 enzyme variants in a Rwandese population: functional characterization and assessment of in silico prediction tools. Hum Mutat 34:725–734. [DOI] [PubMed] [Google Scholar]
- Schenkman JB, Jansson I. (2003) The many roles of cytochrome b5. Pharmacol Ther 97:139–152. [DOI] [PubMed] [Google Scholar]
- Totah RA, Allen KE, Sheffels P, Whittington D, Kharasch ED. (2007) Enantiomeric metabolic interactions and stereoselective human methadone metabolism. J Pharmacol Exp Ther 321:389–399. [DOI] [PubMed] [Google Scholar]
- Totah RA, Sheffels P, Roberts T, Whittington D, Thummel K, Kharasch ED. (2008) Role of CYP2B6 in stereoselective human methadone metabolism. Anesthesiology 108:363–374. [DOI] [PubMed] [Google Scholar]
- Turpeinen M, Zanger UM. (2012) Cytochrome P450 2B6: function, genetics, and clinical relevance. Drug Metabol Drug Interact 27:185–197. [DOI] [PubMed] [Google Scholar]
- van Heeswijk R, Verboven P, Vandevoorde A, Vinck P, Snoeys J, Boogaerts G, De Paepe E, Van Solingen-Ristea R, Witek J, Garg V. (2013) Pharmacokinetic interaction between telaprevir and methadone. Antimicrob Agents Chemother 57:2304–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vourvahis M, Wang R, Gruener DM, Bruce RD, Haider S, Tawadrous M. (2012) Effect of lersivirine co-administration on pharmacokinetics of methadone in healthy volunteers. Drug Alcohol Depend 126:183–188. [DOI] [PubMed] [Google Scholar]
- Wang H, Tompkins LM. (2008) CYP2B6: new insights into a historically overlooked cytochrome P450 isozyme. Curr Drug Metab 9:598–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SC, Ho IK, Tsou HH, Tian JN, Hsiao CF, Chen CH, Tan HK, Lin L, Wu CS, Su LW, et al. (2011) CYP2B6 polymorphisms influence the plasma concentration and clearance of the methadone S-enantiomer. J Clin Psychopharmacol 31:463–469. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Sakuyama K, Sasaki T, Ishii Y, Ishikawa M, Hirasawa N, Hiratsuka M. (2010) Functional characterization of 26 CYP2B6 allelic variants (CYP2B6.2-CYP2B6.28, except CYP2B6.22). Pharmacogenet Genomics 20:459–462. [DOI] [PubMed] [Google Scholar]
- Xie HJ, Yasar U, Lundgren S, Griskevicius L, Terelius Y, Hassan M, Rane A. (2003) Role of polymorphic human CYP2B6 in cyclophosphamide bioactivation. Pharmacogenomics J 3:53–61. [DOI] [PubMed] [Google Scholar]
- Xu C, Ogburn ET, Guo Y, Desta Z. (2012) Effects of the CYP2B6*6 allele on catalytic properties and inhibition of CYP2B6 in vitro: implication for the mechanism of reduced efavirenz metabolism and other CYP2B6 substrates in vivo. Drug Metab Dispos 40:717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanger UM, Klein K. (2013) Pharmacogenetics of cytochrome P450 2B6 (CYP2B6): advances on polymorphisms, mechanisms, and clinical relevance. Front Genet 4:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Sridar C, Kenaan C, Amunugama H, Ballou DP, Hollenberg PF. (2011) Polymorphic variants of cytochrome P450 2B6 (CYP2B6.4-CYP2B6.9) exhibit altered rates of metabolism for bupropion and efavirenz: a charge-reversal mutation in the K139E variant (CYP2B6.8) impairs formation of a functional cytochrome p450-reductase complex. J Pharmacol Exp Ther 338:803–809. [DOI] [PMC free article] [PubMed] [Google Scholar]