Abstract
Dental implants are commonly used to replace missing teeth. However, the dysbiotic polymicrobial communities of peri-implant sites are responsible for peri-implant diseases, such as peri-implant mucositis and peri-implantitis. In this study, we analyzed the microbial characteristics of oral plaque from peri-implant pockets or sulci of healthy implants (n = 10), peri-implant mucositis (n = 8) and peri-implantitis (n = 6) sites using pyrosequencing of the 16S rRNA gene. An increase in microbial diversity was observed in subgingival sites of ailing implants, compared with healthy implants. Microbial co-occurrence analysis revealed that periodontal pathogens, such as Porphyromonas gingivalis, Tannerella forsythia, and Prevotella intermedia, were clustered into modules in the peri-implant mucositis network. Putative pathogens associated with peri-implantitis were present at a moderate relative abundance in peri-implant mucositis, suggesting that peri-implant mucositis an important early transitional phase during the development of peri-implantitis. Furthermore, the relative abundance of Eubacterium was increased at peri-implantitis locations, and co-occurrence analysis revealed that Eubacterium minutum was correlated with Prevotella intermedia in peri-implantitis sites, which suggests the association of Eubacterium with peri-implantitis. This study indicates that periodontal pathogens may play important roles in the shifting of healthy implant status to peri-implant disease.
Implants have revolutionized dental rehabilitation, prosthetic dentistry, and maxillary reconstruction1,2. Marketing estimates show that over 2 million dental implants were inserted annually in the United States at the turn of the millennium3. Although dental implants survive well, infections at peri-implant sites have been widely reported4,5,6. Peri-implant diseases present in two forms: peri-implant mucositis (PM) and peri-implantitis (PI). In PM, inflammation is confined to the soft tissues surrounding a dental implant, with no sign of any loss of supporting bone after the initial bone remodeling that takes place during healing5. PI is characterized by inflammation around the implant, involving both soft tissues and a progressive loss of supporting bone to an extent greater than occurs upon biological remodeling, and may eventually lead to loss of the implant (implant failure)7. Peri-implant diseases have become emerging problems as the number of implants placed increases. The prevalence of mucositis is ~80% in implant patients and ~50% in the implants per se, whereas peri-implantitis has been diagnosed in 28–56% of implant patients and 12–43% of implants8,9. Bacteria colonize the peri-implant crevice soon after implant placement to establish polymicrobial communities9,10, and the failure of dental implants is commonly ascribed to inflammation of the supporting bone and related soft tissues caused by microbiota in peri-implant biofilms11,12.
PM and PI correspond in basic terms to gingivitis and periodontitis. Persistent gingivitis may lead to chronic periodontitis in susceptible individuals13. From the viewpoint of microbial ecology, red and orange complexes are more prevalent and more numerous in the lesions of established gingivitis, and this is even more apparent in periodontitis14. The microbial compositions of gingivitis have been compared with those of PM15, and those of periodontitis with PI16,17,18. However, PM, regarded as the precursor of PI, has seldom been investigated separately, and the relationships between the microbial communities of PM and PI remain unclear.
Traditionally, studies on the pathogenesis of peri-implant microbiota have analyzed individual bacterial species in complex microbial communities. More recent work has shown that peri-implant diseases may be polymicrobial in etiology, caused by a shift in the microbial community, rather than a single pathogen16. Previous studies, using culture-based methods, 16S rRNA gene PCR, or DNA-DNA hybridization techniques, commonly addressed roles played by individual bacterial species and afforded limited information on the overall diversity of the peri-implant environment. Sequencing of 16S ribosomal genes has yielded deeper insights into the composition of the oral microbiome in health and disease, creating a paradigm shift in our understanding of such microbial communities19. Pyrosequencing of PCR-amplified 16S rRNA is a next-generation sequencing method that simultaneously generates thousands of sequences from individual samples. Such an unprecedented amount of information allows comprehensive examination of a taxonomically heterogeneous community and has revealed ever-greater levels of microbial diversity20,21. A recent study on peri-implant bacterial communities using 16S pyrosequencing revealed that the microbial profile of healthy implants was significantly more diverse than that of PI sites16. However, when the prevalence of individual species was evaluated using DNA-DNA hybridization methods, Renvert S. et al.6 found no difference in microbial diversity between PI and healthy sites, whereas others detected fewer species in healthy sites compared to PI sites22,23.
In the present study, we analyzed subgingival plaque samples from healthy implants, PM and PI, using the 16S rRNA pyrosequencing method. This study was performed to compare the differences of the microbial communities of healthy implants, PM and PI, aiming to reveal the potential pathogens associated with peri-implant diseases.
Methods
Subject recruitment
Ten individuals with healthy peri-implant sites (n = 10), eight cases with PM (n = 8), and six cases with PI (n = 6), participated in the study. The project was approved by the Peking University Biomedical Ethics Committee (Beijing, China). Subjects gave written informed consent with the approval of the Ethics Committee of the Peking University School and Hospital of Stomatology. The methods were carried out in accordance with the approved guidelines. All patients had received dental implants in our hospital using the Straumann Dental Implant System (Straumann, California, USA).
Diagnosis and sample collection
The diagnostic criteria for peri-implant diseases were in accordance with the recognized definitions of PM and PI8. Plaque samples were collected from peri-implant sulci or pockets, at the maximum possible probing depth, using a sterile periodontal probe.
Microbial DNA extraction, 16S rRNA gene library preparation, and pyrosequencing
DNA from plaque samples was extracted and the v1–v3 hypervariable regions of bacterial 16S ribosomal RNA genes were amplified via PCR. The libraries were pyrosequenced on a 454-GS-FLX sequencing platform (454 Life Sciences, Branford, USA) at the BGI Institute (BGI Institute, Shenzhen, China). These sequence data have been submitted to the Short Reads Archive (Accession number SRP043555).
16S data processing and statistical analysis
The raw sequencing data were analyzed using (principally) the pipeline tools MOTHUR24 and QIIME25, as described in supplementary methods. Student’s t-test was used to compare alpha and beta diversities. Differences in the relative abundances of taxa in healthy implant, PM, and PI samples were analyzed using the Wilcoxon rank-sum test. Differences in prevalence were compared using Fisher’s exact test. P values <0.05 were considered to indicate statistical significance. P values have not been corrected for multiple comparisons. We calculated the Pearson correlation coefficients (PCC) for each pair of operational taxonomic units (OTUs) and used the permutation test to compute the statistical significance of the PCC value. Edges were set between pairs of OTUs for which the PCC was significant (P < 0.01).
Quantification of bacterial loads of the Eubacterium brachy subgroup
Bacterial loads of members of the Eubacterium brachy subgroup were determined via real-time PCR using modified genus-specific primers26.
Detailed methods were provided as Supplementary data.
Results
Peri-implant diseases were associated with increased microbial diversity
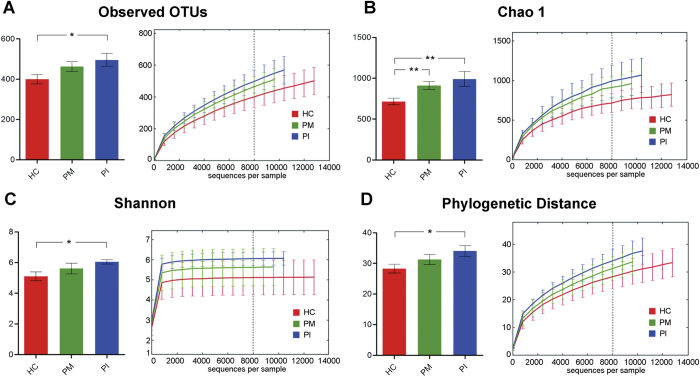
Schematic diagrams of healthy implant, PM and PI are shown in Fig. 1A. The demographic and clinical parameters of all subjects are shown in Table 1. In total, 424,579 final reads were generated after processing, with a mean of 17,692 ± 6,236 (range 9,720–38,763) per sample. We finally detected 15,766 OTUs, with 311–1,028 OTUs in individual specimens, using a 97% similarity cutoff (for details please see Table S1).
Figure 1. Sample collection and microbial community variation within groups.
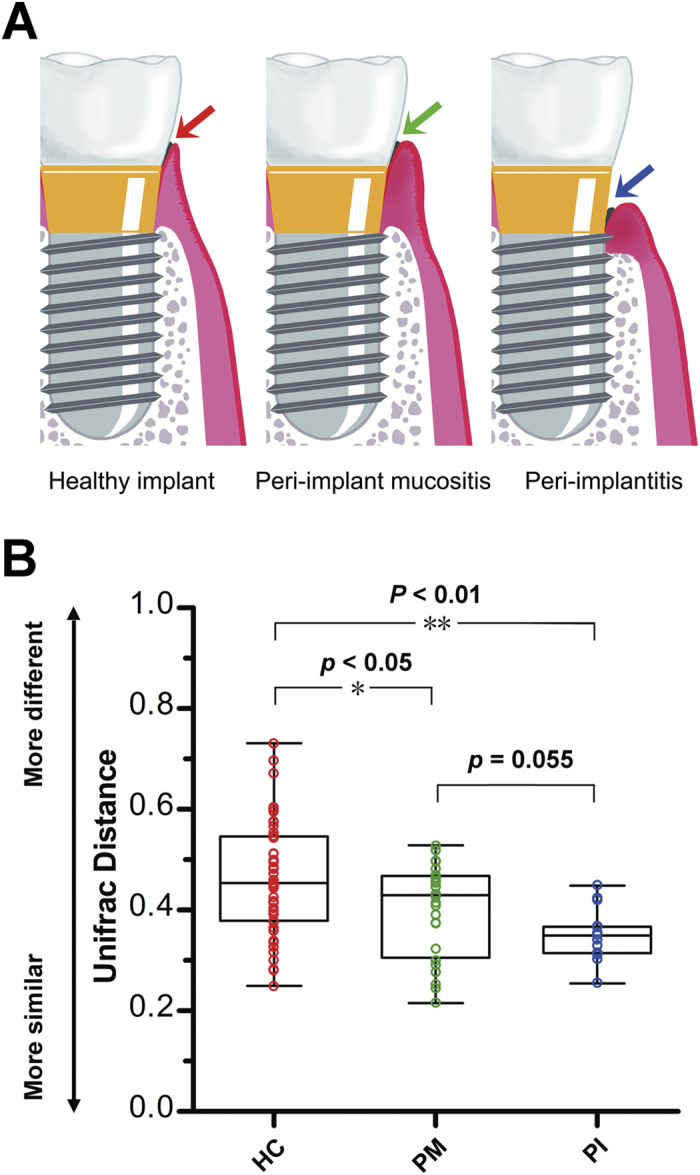
(A) A diagrammatic representation of our sample collection procedure. Plaque from healthy implant, peri-implant mucositis, and peri-implantitis sites was sampled from the deepest pockets or sulci. (B) The average weighted UniFrac distance values (the beta diversities) of healthy implant (HC), peri-implant mucositis (PM), and peri-implantitis (PI) sites. Healthy implant sites tended to host diverse bacterial communities, whereas peri-implantitis sites showed the greatest similarity in microbial communities. *P < 0.05, **P < 0.01 by two-tailed t-test.
Table 1. Demographic and clinical characteristics of all subjects.
| Characteristic | Healthy subjects (n=10) | Peri-implant mucositis patients (n=8) | Peri-implantitis patients (n=6) |
|---|---|---|---|
| Male/female | 3/7 | 6/2 | 3/3 |
| Age (years ± s.d.) | 42.6 ± 3.6 | 46.0 ± 3.5 | 48.2 ± 7.8 |
| Years of functional loading (years ± s.d.) | 3.9 ± 0.7 | 4.2 ± 0.7 | 4.5 ± 1.4 |
| Plaque index (mean ± s.d.) | 1.3 ± 0.5 | 1.4 ± 0.5 | 1.16 ± 0.4 |
| Peri-implant probing depth (mm ± s.d.) | 2.0 ± 0.9 | 2.0 ± 1.6 | 4.0 ± 2.7 |
| Bone loss (mm ± s.d.) | 0 | 0 | 2 ± 3.6 |
| Bleeding on probing (+/–) | 0/10 | 5/3 | 6/0 |
The variation in overall bacterial community composition based on weighted UniFrac distance measurements was compared among the three groups (Fig. 1B). The variations of the microbial characteristics were similar for the PI sites, but greater for among healthy implants; the difference was statistically significant. Microbial diversity within each sample (the alpha diversity) was calculated at given numbers of reads (n = 8000). Microbial diversity differed significantly between healthy implants and infected sites: 1) OTU richness was higher in plaque samples from peri-implant diseases, compared with samples from healthy implant sites (observed OTUs and Chao 1 index values, Fig. 2A,B); 2) The microbial diversity estimator (the Shannon diversity index) showed that PI sites harbored statistically significantly more diverse bacterial communities than did healthy sites (Fig. 2C); and, 3) the phylogenetic diversity (PD) measure also revealed that the microbial communities of PI sites were the most diverse, and those of healthy implant sites were the least diverse (Fig. 2D).
Figure 2. Calculation of alpha diversity values for comparison of the total microbial diversity of healthy implant (HC), peri-implant mucositis (PM), and peri-implantitis (PI) sites.
Alpha diversity values were calculated based on a subsample of 8000 sequences from each dataset. *P < 0.05, **P < 0.01 by two-tailed t-test. (A) The numbers of observed OTUs increased in both PM and PI. (B) The estimated OTU numbers (Chao1) of PM and PI were significantly greater than that of HC. (C) Microbial community diversity analysis (Shannon index) showed that the PI microbial community exhibited the greatest diversity. (D) Phylogenetic diversity (PD) measures of community diversity.
Healthy implant sites and peri-implantitis sites harbor distinct bacterial communities
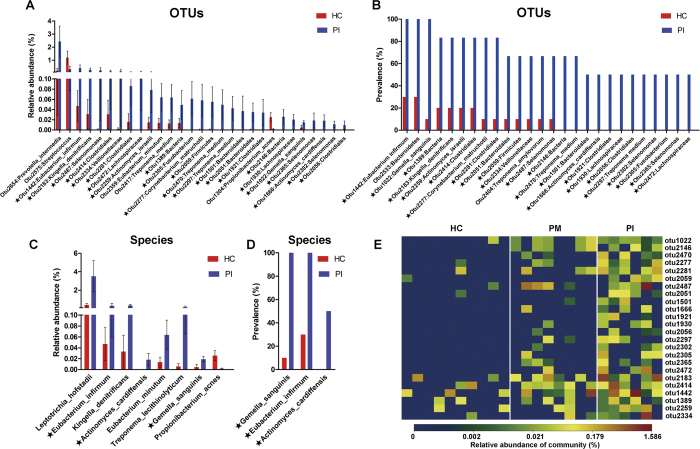
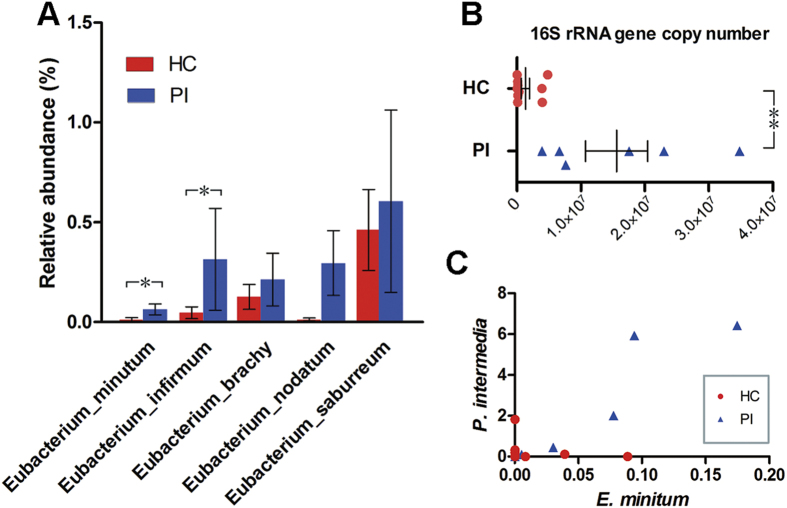
Analysis of the relative abundance of microbial taxonomic groups showed that bacterial compositions differed between healthy and PI sites. Generally, the dominant phyla at implant sites were Firmicutes, Bacteroidetes, Fusobacteria, Actinobacteria, and Proteobacteria. Other dominant taxa are described in Figure S1-S2. The relative abundance levels of 29 OTUs differed significantly between healthy implants and PI sites, of which 27 were over-represented in the latter sites (P < 0.05, Wilcoxon rank-sum test; Fig. 3A). A total of 26 OTUs showed higher prevalence in PI sites than in healthy implants (P < 0.05, Fisher’s exact test; Fig. 3B). A total of 24 OTUs showed higher relative abundance and prevalence in PI sites compared with in healthy implants. Analysis at the species level showed that the relative abundances of Leptotrichia hofstadii, Eubacterium infirmum, Kingella denitrificans, Actinomyces cardiffensis, Eubacterium minutum, Treponema lecithinolyticum, and Gemella sanguinis were higher in PI sites, whereas Propionibacterium acnes showed lower proportion. Gemella sanguinis, Eubacterium minutum, and Actinomyces cardiffensis were more prevalent in PI sites (Fig. 3C,D). The proportions of other members of Eubacterium, including Eubacterium brachy, Eubacterium nodatum, and Eubacterium saburreum, were also higher, although the differences were not significant (Fig. 4A). Real-time PCR using genus-specific primers showed that the bacterial load of the Eubacterium brachy subgroup (as measured by 16S rRNA gene copy number) was significantly higher in PI sites compared with healthy implants. The real-time PCR results were in agreement with OTU-based analysis, confirming that members of Eubacterium were more abundant in PI sites (Fig. 4B). Analysis of the co-occurrence revealed a positive correlation between Eubacterium minutum and Prevotella intermedia, a periodontal pathogen27 (Fig. 4C).
Figure 3. OTUs and taxa differing between healthy implant (HC) and peri-implantitis (PI) sites.
(A) A total of 29 OTUs exhibited significant differences in mean relative abundances between HC and PI sites (Wilcoxon rank-sum test, P < 0.05). The bars show mean ± SEM relative abundances. In total, levels of 27 OTUs were higher in PI. (B) OTUs differing in terms of detection frequency between HC and PI sites (Fisher’s exact test, P < 0.05). (C) Species differing in terms of relative abundance between HC and PI sites (Wilcoxon rank-sum test, P < 0.05). The bars show mean ± SEM relative abundances. (D) Species differing in terms of detection frequency between HC and PI sites (Fisher’s exact test, P < 0.05). OTUs or species marked with stars (★) differed significantly in terms of both relative abundance and detection frequency. (E) A heat map of the relative abundances of OTUs that differed significantly between health and disease. The diagram shows OTUs that differed both in relative abundance (Wilcoxon rank-sum test, P < 0.05) and frequency of detection (Fisher’s exact test, P < 0.05) in HC and PI sites. Peri-implant mucositis sites were intermediate in terms of both relative abundance and prevalence.
Figure 4. Members of the genus Eubacterium in healthy implant (HC) and peri-implantitis (PI) sites.
(A) The relative abundances of Eubacterium species were compared. Bars represent the means ± SEMs of the relative abundances of detected species. *P < 0.05 by the Wilcoxon rank-sum test. (B) Total abundances, measured via real-time qPCR, of the Eubacterium brachy subgroup (including E. brachy, E. infirmum, E. nodatum, and E. tardum). **P < 0.01 by Wilcoxon rank-sum test. (C) Positive correlation between Eubacterium minutum and Prevotella intermedia.
The microbial communities of PM sites were intermediate in nature between those of healthy implants and PI sites
From a microbial viewpoint, PM appears to be a transitional phase on the course to PI. It is likely that the microbial characteristics of PM are intermediate between those of healthy implants and PI site. 1) The extent of variation in the microbial community of PM was intermediate between that of healthy implants and PI site (Fig. 1B); 2) PM was also intermediate in terms of alpha diversity (Fig. 2). PM was associated with greater bacterial diversity than were healthy implant sites, but lower than that of PI sites; 3) PI-associated OTUs were of moderate relative abundance (Fig. 3E).
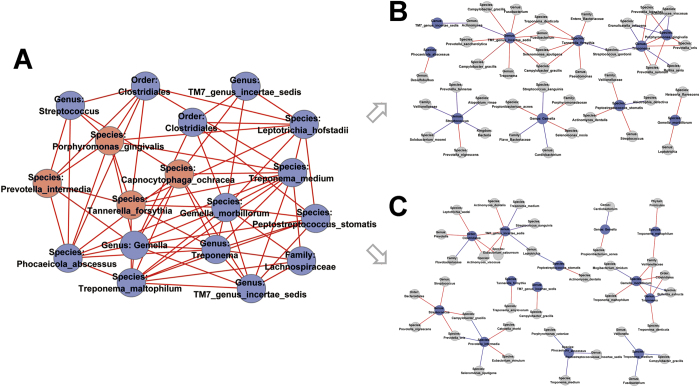
We further analyzed the co-occurrence network of microbiota in peri-implant sites, and the results revealed that the microbial components were strongly connected (Figure S3–S5). Interestingly, we found unique and clearly delimited modules for PM with18 nodes (Fig. 5A). Modules of PM networks consisted of nodes with at least five degrees. Periodontal pathogens, such as Porphyromonas gingivalis, Tannerella forsythia, Prevotella intermedia and Capnocytophaga ochracea were clustered as part of the module. However, corresponding nodes did not form any pairwise modules in HC and PI sites (Fig. 5B,C).
Figure 5. Co-occurring network modules in PM site and corresponding OTUs in HC and PI sites.
Edges between each pair of OTUs indicate significant correlations (P < 0.01 by permutation test). Red and blue edges indicate positive and negative correlations, respectively. (A) Module in PM network consisted of OTUs with at least five degrees. Periodontal pathogens were marked red. (B, C) Corresponding OTUs did not cluster into pairwise modules in HC and PI sites.
The core subgingival microbiome of healthy implant and peri-implant diseases
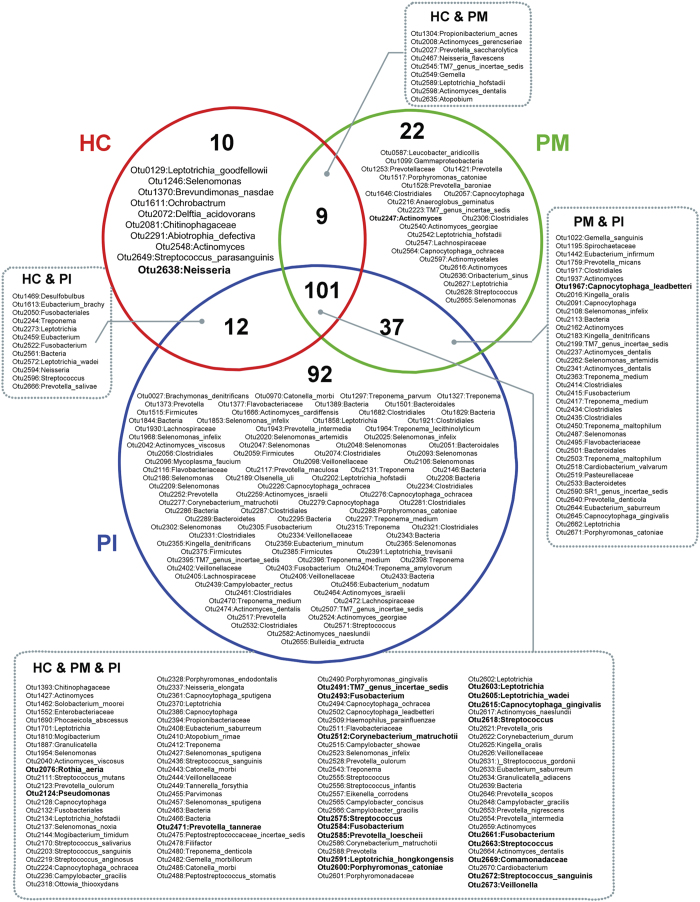
Several taxa differed between healthy implant and PM sites (Figure S6) and between PM and PI sites (Figure S7). We defined the core peri-implant microbiome. Generally, healthy implant, PM, and PI sites shared most OTUs. Of the 383 OTUs present in at least 50% of all subjects, 101 were common to all subjects; these included members of Streptococcus, Leptotrichia, Actinomyces, Capnocytophaga, Prevotella, Fusobacterium, Neisseria, and TM7. These taxa predominated in all subjects (Fig. 6). Healthy implant sites shared few OTUs with PM or PI. Ten OTUs were more prevalent in healthy implant sites, suggesting associations between such bacteria and implant health. PM and PI sites shared 37 OTUs, and 151 OTUs were more prevalent in peri-implant diseases. In total, 22 OTUs were unique to PM sites and 92 to PI sites.
Figure 6. Venn diagram of the core microbiome of peri-implant sites.
Each circle (red, green or blue) contains OTUs present in at least 50% of subjects within a group. OTUs in the overlapping regions were shared by two or three groups. Numerically dominant OTUs with mean relative abundances >0.5% are shown in bold.
Discussion
We have described the complexity of microbial communities in peri-implant sites, and we found that peri-implant diseases were associated with dysbiotic subgingival microbial communities. Our study indicates the important role played by the microbiota in peri-implant diseases.
We measured the complexity of microbial communities in PI sites, which contained larger numbers of OTUs and had higher Shannon index values than did healthy implant sites. These results were confirmed by showing that the presence of non-abundant OTUs explained the increase in microbial diversity. The results are consistent with previous findings of microbial enrichment in ailing implant sites23,28,29. However, our results contradict some earlier report, which claimed that PI was attributable to a simple infection of relatively low microbial diversity16. This may be explained by differences in sampling methods (a periodontal probe vs. a pointed piece of paper). The paper point sampling method may collect only the superficial region of a submucosal biofilm30, thus underestimating the richness and diversity of the microbial community around a dental implant. In our present study, we obtained plaque from the deepest pockets of PI sites using periodontal probe. To allow results to be comparable, healthy implants, which lack deep peri-implant pockets, were also sampled from shallow peri-implant sulci using periodontal probe instead of curette. In recent work using the 16S rRNA gene pyrosequencing method31, no significant difference in the microbial diversity of healthy and ailing implants was found (although the Shannon index of ailing implants was higher), which is in part explained by the fact that PM and PI patients were both assigned to the diseased group. In summary, our results indicate that peri-implant diseases are associated with changes in the microbial enrichment (healthy implant < peri-implant mucositis < peri-implantitis). Implants and teeth share histopathological and ecological similarities, and it has thus been proposed that the microbial communities around these structures should be similar6,22,32,33,34. Evidence that the host responses to microbiota differ at implants and teeth is lacking. However, recent studies have shown that microbial peri-implant communities differ markedly from those of periodontal sites16,31; the former sites exhibited lower microbial diversity and a simpler microbial composition. In the present study, we did not seek to address the similarity (or otherwise) of the microbiota of teeth and dental implants. However, the dominant bacterial taxa in the plaque in peri-implant sites were similar to those of teeth (Figure S2)35,36. We assumed that progression of a healthy implant to PI was similar to the development of periodontitis, and, in this preliminary study, we aimed to identify potential “key pathogens” of peri-implant diseases, which may be less abundant in health. At the present level of sequencing depth, we found that the most significant difference between PI and healthy implant sites was associated with the levels of poorly abundant OTUs, or taxonomic “species.”
Periodontal pathogens including Porphyromonas gingivalis, Treponema denticola, Tannerella forsythia, Aggregatibacter actinomycetemcomitans, Prevotella intermedia, Fusobacterium, and Campylobacter have been reported to be associated with implant diseases22,37,38,39. The relative abundances of Treponema denticola, Prevotella intermedia, Fusobacterium, and Campylobacter were markedly increased in the lesion sites in our study, but the differences were not significant upon Wilcoxon rank-sum testing (data not shown). The relative abundances of Porphyromonas gingivalis and Tannerella forsythia were similar in healthy implant and PI sites, but were reduced in PM site (data not shown). Taken together, our data were largely consistent with those of previous studies23,40. Moreover, analysis at the OTU level identified several suspected periodontal pathogens, which increased in relative abundance, and prevalence, in PI sites. Such suspects included members of Eubacterium, Treponema, and Selenomonas. Particularly, Eubacterium spp. appear to be promising candidate peri-implant pathogens. Our results agree with those of previous reports that Eubacterium spp. were of higher relative abundance, and present in greater numbers, at peri-implantitis sites41. Also, Eubacterium species such as Eubacterium minutum and Eubacterium nodatum, the numbers of which were increased significantly at PI sites, have been associated with periodontitis36,42,43. Moreover, Eubacterium minutum were likely to co-exist with Prevotella intermedia, a well-known periodontal pathogen27. Our study highlights the urgency of conducting further research on the role played by Eubacterium in peri-implant diseases.
Although lacking sufficiently large samples, we used network analysis to clarify the co-occurrence patterns of microbial communities in peri-implant sites. In this study, microbial networks were constructed based on the inter-taxa correlations of closely related bacteria co-existing in subgingival sites of implants. Networks of co-occurring microbial taxa in implant sites consisted mainly of periodontal bacteria, implying a niche similarity between periodontal and peri-implant sites36. Positively correlated microbial taxa may have interactions, such as habitat affinities and symbiotic relationships44. As random associations between the taxa in the networks are expected, a module network was constructed using strongly linked taxa that may play core roles in the interactions with other taxa. We found that periodontal pathogens such as Porphyromonas gingivalis, Tannerella forsythia, and Prevotella intermedia were among members of the module in the PM co-occurring network, indicative of their potential biotic interaction with other microbes during the early stages of peri-implant diseases. The “keystone pathogen” hypothesis suggests that pathogens of low abundance can cause inflammatory diseases by rendering a symbiotic microbial community dysbiotic45. Based on network analysis, it is possible that periodontal pathogens play key roles and contribute to an overall shift in the microbial community, despite their low abundance.
We defined the core microbiome of healthy peri-implant, PM, and PI sites, defined as the “most commonly detected OTUs across samples”. Overall, we found that the OTUs were similar in the three groups. Most OTUs were found in all individuals, indicating that microbial ecosystems of peri-implant sites are similar. Certain OTUs—such as the Streptococcus, Leptotrichia, Capnocytophaga, Prevotella, Fusobacterium, Neisseria, and Rothia genera—were dominant in all samples. We also aimed to describe the core microbiota associated with health and disease. For example, OTUs identified as Neisseria spp. were found in most healthy implant samples, and at relatively high abundances (>0.5%). Although this taxon remains unclassified at the species level, these bacteria may maintain health. Many OTUs present in the majority of PI samples were rare in the other two groups. Such PI-associated OTUs contributed to the complexity of the submucosal microbial community.
We also investigated the microbial diversity in PM sites, the clinical parameters of which were intermediate between those of healthy implants and PI. These sites appeared to not exhibit a distinct microbial pattern, rather hosting candidate pathogens causing PI. This reflects the fact that, from a microbiological viewpoint, PM precedes PI, and should be considered an early event in the development of PI. Our results are in accordance with the consensus of the Seventh European Workshop on Periodontology46. We acknowledge, however, that we cannot presently conclude that PM definitely progresses to PI, although inflammation generally develops rapidly around dental implants47. There is a causal relationship between gingivitis and periodontitis, but further work is needed to evaluate the relationship between PM and PI48. Periodontal pathogens such as Porphyromonas gingivalis, Tannerella forsythia, Prevotella intermedia and Capnocytophaga ochracea, clustered together in PM sites, suggesting that periodontal pathogens may play important roles in the pathogenesis of peri-implant diseases.
In conclusion, we have described and compared the microbial communities of healthy implant, PM, and PI sites, affording deep insight into the dysbiosis in the microbial community of ailing implants. Our work adds to the current knowledge that periodontal pathogens may play important roles in oral peri-implant diseases.
Additional Information
How to cite this article: Zheng, H. et al. Subgingival microbiome in patients with healthy and ailing dental implants. Sci. Rep. 5, 10948; doi: 10.1038/srep10948 (2015).
Supplementary Material
Acknowledgments
This work was supported by funding from the Peking University School of Stomatology (PKUSS20130210), and by the Scientific Research Foundation for Returned Overseas Chinese Scholars, State Education Ministry.
Footnotes
Author Contributions H.Z. conducted experiments, processed sequence data and wrote the main part of the manuscript. L.X. recruited volunteers and collected samples. Z.W. and L.L performed analysis of microbial co-occurrence network and processed statistical analysis. J.Z. and Q.Z. participated in sample collection and revision of the manuscript. T.C. provided algorithms and revised the manuscript. J.L. and F.C. conceived the study and revised the manuscript.
References
- Barber A. J., Butterworth C. J. & Rogers S. N. Systematic review of primary osseointegrated dental implants in head and neck oncology. Br J Oral Maxillofac Surg 49, 29–36, 10.1016/j.bjoms.2009.12.007 (2011). [DOI] [PubMed] [Google Scholar]
- Demarosi F., Leghissa G. C., Sardella A., Lodi G. & Carrassi A. Localised maxillary ridge expansion with simultaneous implant placement: a case series. Br J Oral Maxillofac Surg 47, 535–540, 10.1016/j.bjoms.2008.11.012 (2009). [DOI] [PubMed] [Google Scholar]
- Klinge B., Hultin M. & Berglundh T. Peri-implantitis. Dent Clin North Am 49, 661–676, vii–viii, 10.1016/j.cden.2005.03.007 (2005). [DOI] [PubMed] [Google Scholar]
- Jepsen S., Ruhling A., Jepsen K., Ohlenbusch B. & Albers H. K. Progressive peri-implantitis. Incidence and prediction of peri-implant attachment loss. Clin Oral Implants Res 7, 133–142 (1996). [DOI] [PubMed] [Google Scholar]
- Lindhe J., Meyle J. & Group D. o. E. W. o. P. Peri-implant diseases: Consensus Report of the Sixth European Workshop on Periodontology. J Clin Periodontol 35, 282–285, 10.1111/j.1600-051X.2008.01283.x (2008). [DOI] [PubMed] [Google Scholar]
- Renvert S., Roos-Jansaker A. M., Lindahl C., Renvert H. & Rutger Persson G. Infection at titanium implants with or without a clinical diagnosis of inflammation. Clin Oral Implants Res 18, 509–516, 10.1111/j.1600-0501.2007.01378.x (2007). [DOI] [PubMed] [Google Scholar]
- Sanz M., Chapple I. L. & Working Group 4 of the, V. E. W. o. P. Clinical research on peri-implant diseases: consensus report of Working Group 4. J Clin Periodontol 39 (Suppl 12), 202–206, 10.1111/j.1600-051X.2011.01837.x (2012). [DOI] [PubMed] [Google Scholar]
- Zitzmann N. U. & Berglundh T. Definition and prevalence of peri-implant diseases. J Clin Periodontol 35, 286–291, 10.1111/j.1600-051X.2008.01274.x (2008). [DOI] [PubMed] [Google Scholar]
- Sanz M. et al. Characterization of the subgingival microbial flora around endosteal sapphire dental implants in partially edentulous patients. Int J Oral Maxillofac Implants 5, 247–253 (1990). [PubMed] [Google Scholar]
- Quirynen M. et al. Dynamics of initial subgingival colonization of ‘pristine’ peri-implant pockets. Clin Oral Implants Res 17, 25–37, 10.1111/j.1600-0501.2005.01194.x (2006). [DOI] [PubMed] [Google Scholar]
- Esposito M., Hirsch J. M., Lekholm U. & Thomsen P. Biological factors contributing to failures of osseointegrated oral implants. (II). Etiopathogenesis. Eur J Oral Sci 106, 721–764 (1998). [DOI] [PubMed] [Google Scholar]
- Berglundh T., Persson L. & Klinge B. A systematic review of the incidence of biological and technical complications in implant dentistry reported in prospective longitudinal studies of at least 5 years. J Clin Periodontol 29 (Suppl 3), 197–212; discussion 232-193 (2002). [DOI] [PubMed] [Google Scholar]
- Kistler J. O., Booth V., Bradshaw D. J. & Wade W. G. Bacterial community development in experimental gingivitis. PLoS One 8, e71227, 10.1371/journal.pone.0071227 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky S. S. & Haffajee A. D. Periodontal microbial ecology. Periodontol 2000 38, 135–187, 10.1111/j.1600-0757.2005.00107.x (2005). [DOI] [PubMed] [Google Scholar]
- Salvi G. E. et al. Reversibility of experimental peri-implant mucositis compared with experimental gingivitis in humans. Clin Oral Implants Res 23, 182–190, 10.1111/j.1600-0501.2011.02220.x (2012). [DOI] [PubMed] [Google Scholar]
- Kumar P. S., Mason M. R., Brooker M. R. & O’Brien K. Pyrosequencing reveals unique microbial signatures associated with healthy and failing dental implants. J Clin Periodontol 39, 425–433, 10.1111/j.1600-051X.2012.01856.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meffert R. M. Periodontitis vs. peri-implantitis: the same disease? The same treatment? Crit Rev Oral Biol Med 7, 278–291 (1996). [DOI] [PubMed] [Google Scholar]
- Algraffee H., Borumandi F. & Cascarini L. Peri-implantitis. Br J Oral Maxillofac Surg 50, 689–694, 10.1016/j.bjoms.2011.11.020 (2012). [DOI] [PubMed] [Google Scholar]
- Shchipkova A. Y., Nagaraja H. N. & Kumar P. S. Subgingival microbial profiles of smokers with periodontitis. J Dent Res 89, 1247–1253, 10.1177/0022034510377203 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlson E. S. et al. Disordered microbial communities in the upper respiratory tract of cigarette smokers. PLoS One 5, e15216, 10.1371/journal.pone.0015216 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keijser B. J. et al. Pyrosequencing analysis of the oral microflora of healthy adults. J Dent Res 87, 1016–1020 (2008). [DOI] [PubMed] [Google Scholar]
- Shibli J. A. et al. Composition of supra- and subgingival biofilm of subjects with healthy and diseased implants. Clin Oral Implants Res 19, 975–982, 10.1111/j.1600-0501.2008.01566.x (2008). [DOI] [PubMed] [Google Scholar]
- Maximo M. B. et al. Short-term clinical and microbiological evaluations of peri-implant diseases before and after mechanical anti-infective therapies. Clin Oral Implants Res 20, 99–108, 10.1111/j.1600-0501.2008.01618.x (2009). [DOI] [PubMed] [Google Scholar]
- Schloss P. D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75, 7537–7541, 10.1128/AEM.01541-09 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336, 10.1038/nmeth.f.303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt D. A., Weightman A. J. & Wade W. G. Diversity of oral asaccharolytic Eubacterium species in periodontitis--identification of novel phylotypes representing uncultivated taxa. Oral Microbiol Immunol 14, 56–59 (1999). [DOI] [PubMed] [Google Scholar]
- Socransky S. S., Haffajee A. D., Cugini M. A., Smith C. & Kent R. L. Jr. Microbial complexes in subgingival plaque. J Clin Periodontol 25, 134–144 (1998). [DOI] [PubMed] [Google Scholar]
- Tabanella G., Nowzari H. & Slots J. Clinical and microbiological determinants of ailing dental implants. Clin Implant Dent Relat Res 11, 24–36, 10.1111/j.1708-8208.2008.00088.x (2009). [DOI] [PubMed] [Google Scholar]
- Koyanagi T., Sakamoto M., Takeuchi Y., Ohkuma M. & Izumi Y. Analysis of microbiota associated with peri-implantitis using 16S rRNA gene clone library. J Oral Microbiol 2, 10.3402/jom.v2i0.5104 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijnge V. et al. Oral biofilm architecture on natural teeth. PLoS One 5, e9321, 10.1371/journal.pone.0009321 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabdoub S. M., Tsigarida A. A. & Kumar P. S. Patient-specific analysis of periodontal and peri-implant microbiomes. J Dent Res 92, 168S–175S, 10.1177/0022034513504950 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botero J. E., Gonzalez A. M., Mercado R. A., Olave G. & Contreras A. Subgingival microbiota in peri-implant mucosa lesions and adjacent teeth in partially edentulous patients. J Periodontol 76, 1490–1495, 10.1902/jop.2005.76.9.1490 (2005). [DOI] [PubMed] [Google Scholar]
- Mombelli A. & Decaillet F. The characteristics of biofilms in peri-implant disease. J Clin Periodontol 38 (Suppl 11), 203–213, 10.1111/j.1600-051X.2010.01666.x (2011). [DOI] [PubMed] [Google Scholar]
- Heitz-Mayfield L. J. & Lang N. P. Comparative biology of chronic and aggressive periodontitis vs. peri-implantitis. Periodontol 2000 53, 167–181, 10.1111/j.1600-0757.2010.00348.x (2010). [DOI] [PubMed] [Google Scholar]
- Li Y. et al. Phylogenetic and functional gene structure shifts of the oral microbiomes in periodontitis patients. ISME J, 10.1038/ismej.2014.28 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abusleme L. et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J 7, 1016–1025, 10.1038/ismej.2012.174 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultin M. et al. Microbiological findings and host response in patients with peri-implantitis. Clin Oral Implants Res 13, 349–358 (2002). [DOI] [PubMed] [Google Scholar]
- Alcoforado G. A., Rams T. E., Feik D. & Slots J. Microbial aspects of failing osseointegrated dental implants in humans. J Parodontol 10, 11–18 (1991). [PubMed] [Google Scholar]
- Persson G. R. & Renvert S. Cluster of Bacteria Associated with Peri-Implantitis. Clin Implant Dent Relat Res, 10.1111/cid.12052 (2013). [DOI] [PubMed] [Google Scholar]
- Leonhardt A., Renvert S. & Dahlen G. Microbial findings at failing implants. Clin Oral Implants Res 10, 339–345 (1999). [DOI] [PubMed] [Google Scholar]
- Koyanagi T. et al. Comprehensive microbiological findings in peri-implantitis and periodontitis. J Clin Periodontol 40, 218–226, 10.1111/jcpe.12047 (2013). [DOI] [PubMed] [Google Scholar]
- Haffajee A. D., Teles R. P. & Socransky S. S. Association of Eubacterium nodatum and Treponema denticola with human periodontitis lesions. Oral Microbiol Immunol 21, 269–282, 10.1111/j.1399-302X.2006.00287.x (2006). [DOI] [PubMed] [Google Scholar]
- Booth V., Downes J., Van den Berg J. & Wade W. G. Gram-positive anaerobic bacilli in human periodontal disease. J Periodontal Res 39, 213–220, 10.1111/j.1600-0765.2004.00726.x (2004). [DOI] [PubMed] [Google Scholar]
- Barberan A., Bates S. T., Casamayor E. O. & Fierer N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J 6, 343–351, 10.1038/ismej.2011.119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G., Darveau R. P. & Curtis M. A. The keystone-pathogen hypothesis. Nat Rev Microbiol 10, 717–725, 10.1038/nrmicro2873 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang N. P., Berglundh T. & Working Group 4 of Seventh European Workshop on, P. Periimplant diseases: where are we now?–Consensus of the Seventh European Workshop on Periodontology. J Clin Periodontol 38 (Suppl 11), 178–181, 10.1111/j.1600-051X.2010.01674.x (2011). [DOI] [PubMed] [Google Scholar]
- Berglundh T., Zitzmann N. U. & Donati M. Are peri-implantitis lesions different from periodontitis lesions? J Clin Periodontol 38 (Suppl 11), 188–202, 10.1111/j.1600-051X.2010.01672.x (2011). [DOI] [PubMed] [Google Scholar]
- Lang N. P., Schatzle M. A. & Loe H. Gingivitis as a risk factor in periodontal disease. J Clin Periodontol 36 (Suppl 10), 3–8, 10.1111/j.1600-051X.2009.01415.x (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.