ABSTRACT
Human immunodeficiency virus (HIV) in humans and simian immunodeficiency virus (SIV) in macaques (MAC) lead to chronic inflammation and AIDS. Natural hosts, such as African green monkeys (AGM) and sooty mangabeys (SM), are protected against SIV-induced chronic inflammation and AIDS. Here, we report that AGM plasmacytoid dendritic cells (pDC) express extremely low levels of CD4, unlike MAC and human pDC. Despite this, AGM pDC efficiently sensed SIVagm, but not heterologous HIV/SIV isolates, indicating a virus-host adaptation. Moreover, both AGM and SM pDC were found to be, in contrast to MAC pDC, predominantly negative for CCR5. Despite such limited CD4 and CCR5 expression, lymphoid tissue pDC were infected to a degree similar to that seen with CD4+ T cells in both MAC and AGM. Altogether, our finding of efficient pDC infection by SIV in vivo identifies pDC as a potential viral reservoir in lymphoid tissues. We discovered low expression of CD4 on AGM pDC, which did not preclude efficient sensing of host-adapted viruses. Therefore, pDC infection and efficient sensing are not prerequisites for chronic inflammation. The high level of pDC infection by SIVagm suggests that if CCR5 paucity on immune cells is important for nonpathogenesis of natural hosts, it is possibly not due to its role as a coreceptor.
IMPORTANCE The ability of certain key immune cell subsets to resist infection might contribute to the asymptomatic nature of simian immunodeficiency virus (SIV) infection in its natural hosts, such as African green monkeys (AGM) and sooty mangabeys (SM). This relative resistance to infection has been correlated with reduced expression of CD4 and/or CCR5. We show that plasmacytoid dendritic cells (pDC) of natural hosts display reduced CD4 and/or CCR5 expression, unlike macaque pDC. Surprisingly, this did not protect AGM pDC, as infection levels were similar to those found in MAC pDC. Furthermore, we show that AGM pDC did not consistently produce type I interferon (IFN-I) upon heterologous SIVmac/HIV type 1 (HIV-1) encounter, while they sensed autologous SIVagm isolates. Pseudotyping SIVmac/HIV-1 overcame this deficiency, suggesting that reduced uptake of heterologous viral strains underlays this lack of sensing. The distinct IFN-I responses depending on host species and HIV/SIV isolates reveal the host/virus species specificity of pDC sensing.
INTRODUCTION
Chronic inflammation and immune activation in human immunodeficiency virus (HIV)-infected humans and in simian immunodeficiency virus (SIV)-infected macaques (MAC) lead to depletion of CD4+ T cells and progression to AIDS. Natural hosts of SIV, such as African green monkeys (AGM) and sooty mangabeys (SM), do not display chronic inflammation or AIDS (1). This is due to resolution of inflammation before the end of acute infection rather than to a lack of SIV recognition by the innate immune system (2). Natural hosts further differ from pathogenic HIV/SIV infections by exhibiting reduced infection rates in certain cell subsets, such as central memory CD4+ T cells (Tcm) (3, 4). This relative resistance has been linked to reduced expression of the HIV/SIV CCR5 coreceptor on natural host CD4+ T cells and to downmodulation of CD4 on activated CD4+ T cells in AGM (3–5).
Plasmacytoid dendritic cells (pDC) form a rare cell population that is responsible for the vast majority of type I interferon (IFN-I) production after HIV encounter (6). This is also true for AGM pDC, as the depletion of pDC from AGM peripheral blood mononuclear cells (PBMCs) completely abrogates the IFN-I response to SIVagm stimulation (7). HIV/SIV sensing by pDC is mediated through endocytosis followed by TLR7 and TLR9 (TLR7/9) engagement. It requires CD4 but is independent of coreceptor expression (6). Data on the infection rates of pDC in vivo are scarce. One study reported the presence of HIV DNA in circulating pDC of chronically HIV-infected patients (8). Another study reported high infection levels in lymph node (LN) pDC during acute SIVmac infection (9).
Here, we discovered restricted CD4 and/or CCR5 expression on pDC in natural hosts. We evaluated the effect of low CD4 expression on the capacity of AGM pDC to efficiently sense distinct forms of SIVagm (free virus, noninfectious particles, and SIVagm-infected cells). Furthermore, we examined the frequency of infection of pDC during pathogenic and nonpathogenic SIV infection.
MATERIALS AND METHODS
Study approval.
All animal experimental protocols were approved either by the Ethical Committee of Animal Experimentation (CETEA-DSV, IDF, France) (notification no. 10-051b and 12-006) or by the Institutional Animal Care and Use Committees (IACUC) of Emory University (IACUC protocol no. 2000793, entitled “Comparative AIDS Program”). Animals were housed in the facilities of the CEA (Commissariat à l'Energie Atomique, Fontenay-aux-Roses, France; permit no. A 92-032-02), Institut Pasteur (Paris, France, permit no. A 78-100-3), or Yerkes National Primate Research Center (Atlanta, GA, USA). All experimental procedures were conducted in strict accordance with the international European guidelines (2010/63/UE) on protection of animals used for experimentation and other scientific purposes (French decree 2013-118) and with the recommendations of the Weatherall report or in strict accordance with USDA regulations and the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The CEA complies with the U.S. Standards for Human Care and Use of Laboratory of the Office for Laboratory Animal Welfare (OLAW) under OLAW Assurance no. A5826-01. The monitoring of the animals was under the supervision of the veterinarians in charge of the animal facilities.
In vivo infections and sample collection.
Twenty-six African green monkeys (Chlorocebus sabaeus) of the sabaeus species with a Caribbean origin, 18 Chinese rhesus macaques (Macaca mulatta), two cynomolgus macaques (Macaca fascularis), 16 Indian rhesus macaques (Macaca mulatta), and 16 sooty mangabeys (Cercocebus atys) were used in this study. Eleven AGMs were infected via intravenous (i.v.) inoculation with 250 50% tissue culture infective doses (TCID50) of SIVagm.sab92018, as described previously (10, 11). Four Chinese rhesus macaques were i.v. infected with 50 50% animal infectious doses (AID50) of SIVmac251, and two others were infected with 5,000 AID50 SIVmac251, as described previously (10, 12). Eight Indian rhesus macaques had been previously infected with SIVmac239 or SIVmac251, and eight sooty mangabeys were either naturally infected or infected experimentally with SIVsmE041 (13, 14). Blood was collected by venipuncture on sodium heparin tubes and shipped to Institut Pasteur or used on site at Yerkes National Primate Research Center. Bone marrow mononuclear cells were isolated on Ficoll, and tissue cells were put in suspension before staining. LNs and spleens were disrupted mechanically, and rectal tissues were enzymatically degraded as described previously (12, 15).
Viral stimulations.
Freshly isolated PBMCs were cultured at 0.5 × 106 cells/well in 24-well plates (Costar) at 37°C in 5% CO2 for 18 h with or without virus. Cell viability was measured using trypan blue and a Countess (Life Technologies) cell counter. The SIVagm strains used have been previously described: SIVagm.sab92018 (11), SIVagm.sabD46 and SIVagm.tanB14 (16), SIVagm.sab1c (11, 17), and SIVgri1 (18, 19). Free SIVagm was added to PBMCs at a concentration of 1,500 ng/ml p27, unless indicated otherwise. Herpes simplex virus 1 (HSV-1) was added at a TCID50 of 2 × 105. HIV-1.Bal-VSV, which is endocytosed independently of CD4 (20), was kindly provided by A. David and AT2-inactivated SIVagm as well as control microvesicles by Jeff Lifson (National Cancer Institute, Frederick, MD). SIVmac251-VSV was produced by cotransfecting 293T cells with SIVmac251Δenv and vesicular stomatitis virus G (VSV-G) expression vector using SuperFect (Qiagen), as described previously (21). SIVagm isolates were grown on SupT1 cells, as these cells express Bonzo and are susceptible to SIVagm, whereas SIVmac isolates were grown on CEMx174, as these cells express Bob and are susceptible to SIVmac (22).
Production of infected cells.
Cells were infected as previously described (23). Briefly, SupT1 cells were exposed for 1 h at 37°C to 3.3 × 104 TCID50 per 106 cells under conditions of constant agitation. Infection levels were assessed by measuring SIV Gag-positive (Gag+) cells using flow cytometry (see below).
Functional alpha interferon assay.
Bioactive IFN-I levels were quantified as described earlier (7). In short, Mardin-Darby bovine kidney (MDBK) cells were incubated with UV-inactivated supernatants for 18 h, after which the cytopathic effect of vesicular stomatitis virus was determined using a CellTiter 96 AQueous nonradioactive cell proliferation assay (Promega). Alternatively, a cell line stably transfected with a luciferase gene under the control of an IFN-I-inducible promoter was used to measure IFN-I levels, as described previously (23). The limit of detection threshold was set at 2 IU/ml.
Flow cytometry.
SM flow cytometric data were acquired at Yerkes National Primate Center, and data for AGM were acquired at Institut Pasteur. MAC data were acquired at both Yerkes and Pasteur, with similar results. The following antibodies were used to identify and characterize pDC phenotype and function in whole blood or isolated tissue cells: CD3 (SP34-2), CD4 (L200), HLA-DR (L243), CCR5 (3A9), CD123 (7G3) (all from BD Biosciences), CD20 (2H7; eBioscience), CD4 (M-T466), BDCA-2 (AC144) (both from Miltenyi), and CD4 (SFCI12T4D11; Beckman Coulter). FcR blocking reagent (Miltenyi) was used to block nonspecific antibody binding; in experiments using tissues other than blood, a LIVE/DEAD cell viability assay (Invitrogen) was used to exclude dead cells. For intracellular staining, cells were labeled with surface-binding antibodies and fixed with 4% paraformaldehyde and permeabilized using saponin prior to incubation with anti-CD4. For intracellular SIVagm detection, anti-p24 staining (KC57; Beckman Coulter) was used after cell permeabilization performed with an IntraPrep permeabilization reagent kit (Beckman Coulter) according to the manufacturer's instructions (23). Events with counts ranging from 100 to 1,000 pDC were collected on a BD LSR-II flow cytometer, running BD fluorescence-activated cell sorter (FACS) Diva 6.0 software, and analyzed with FlowJo 9.4.10 (TreeStar). Anti-mouse compensation beads (BD Biosciences) and an Arc Amine reactive compensation bead kit (Life Technologies) were used to define compensation levels. An isotype control antibody was used to define CCR5+ cells.
Cell sorting.
Splenocytes were frozen in 10% dimethyl sulfoxide (DMSO)–liquid nitrogen until use. Cells were thawed in the presence of DNase I (Roche) (10 IU/ml) and washed in fetal calf serum (FCS). Cells were labeled with antibodies against CD3, CD20, HLA-DR, CD4, and CD123 and with LIVE/DEAD reagent in the presence of FcR blocking reagent. Cells were sorted using a FACSAria II sorter (BD), running on BD FACS Diva 6.0 software. Sorted pDC and CD4+ T cells were purified a second time to increase the purity of the two fractions. CD4+ T cells were also isolated from LN cells using anti-CD4 beads and magnetic stands (Miltenyi) following the manufacturer's protocol, after which purity was verified by flow cytometry.
SIV DNA quantification.
DNA was extracted as follows: samples were lysed in NaCl (3 M), EDTA (0.5 M, pH 8), SDS (Bio-Rad) (10%), and proteinase K (Qiagen) (1 mg/ml) in a 45-min incubation at 55°C. Then, NaCl (5 M) was added and the reaction mixture was incubated at 4°C for between 15 and 60 min followed by centrifugation for 15 min at 3,000 rpm and 4°C. DNA was then precipitated from the supernatant in phenol:chloroform:isoamyl alcohol (Sigma-Aldrich) (25:24:1) (pH 8). After DNA extraction, viral DNA was measured by quantitative PCR (qPCR) in duplicate, using primers and probes designed specifically for SIVagm.sab and SIVmac (11, 15). SIVagm and SIVmac plasmids were used as standards to calculate SIV DNA copy numbers. CCR5 DNA quantification was used to normalize the viral levels to the number of cells (24). Sample preparation, enzyme mix preparation, and PCR setup were performed in three separate rooms to avoid PCR contamination. Positive and negative controls were used to exclude sample contamination.
Fluorescence microscopy.
Fluorescence microscopy was done as follows with markers against NKp30 (AF29-4D12, Miltenyi), CD123 (5B11, Biolegend), DAPI (4′,6-diamidino-2-phenylindole), and SIVagm env RNA (made from the region amplified by primers 5′-GAG GCT TGT GAT AAA ACT TAT TGG GAT-3′ and 5′-AGA GCA GTG ACG CGG GCA TTG AGG-3′ and labeled with fluorochrome Alexa Fluor 488 [Life Technologies]). Briefly, cryopreserved sections were permeabilized by incubation in 0.5% (vol/vol) Triton X-100. This was followed by hybridization of the probe and antibodies mounted on tissue. Secondary antibodies were used to visualize the bound antibodies. Donkey anti-mouse IgG-CFL 594 (Santa Cruz) was used to detect CD123 and a Zenon Alexa Fluor 647 mouse IgG1 labeling kit (Life technologies) to reveal the NKP30 antibody. As negative controls for SIV RNA in situ hybridization, we used an RNase degraded probe, taken up in hybridization buffer, as well as lymph nodes of uninfected animals. Images were acquired on a Leica TCS SP8 confocal laser scanning microscope, running LAS AF 3 (Leica Application Suite Advanced Fluorescence).
Statistics.
Statistical inference analyses were performed using Prism 5.0 (GraphPad). The nonparametric Wilcoxon signed-rank test and Mann-Whitney test were used to test paired and nonpaired observations, respectively. In cases of multiple testing of unpaired data, a nonparametric Kruskal-Wallis test, followed by a Dunn's multiple-comparison test, was used. Multiple testing of paired data without missing values was done by a Friedman test, followed by a Dunn's multiple-comparison test.
RESULTS
Low CD4 expression on AGM pDC.
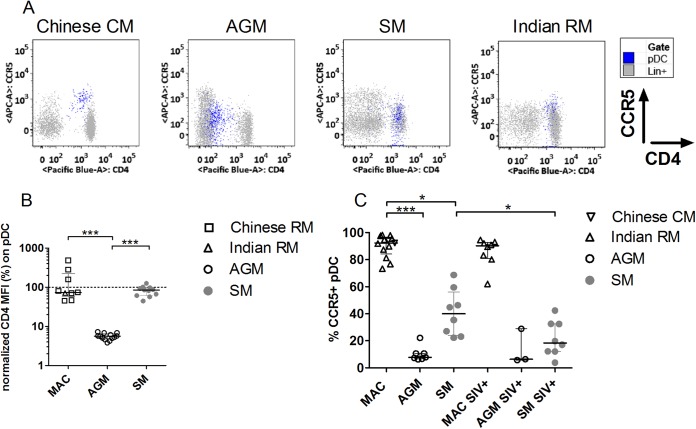
Given the importance of CD4 for HIV/SIV sensing by pDC, we measured CD4 on pDC from uninfected AGM, SM, and MAC (Fig. 1). pDC were defined as CD3− CD20− HLA-DR+ CD123hi cells (Fig. 2A) (2). MAC and SM pDC displayed CD4 levels similar to those seen with the CD4+ T cells (Fig. 1A and B). Surprisingly, AGM pDC expressed CD4 at levels >1 log lower than the levels seen with the CD4+ T cells (P < 0.001).
FIG 1.
CD4 and CCR5 expression on blood plasmacytoid dendritic cells. (A) CD4 and CCR5 expression on pDC (blue) and CD3/CD20+ (Lin+) cells (gray) of one (each) representative Chinese cynomolgus macaque (CM), African green monkey, sooty mangabey, and Indian rhesus macaque (RM). (B) CD4 expression on pDC from SIV-negative Chinese rhesus macaques (n = 8, open squares), Indian rhesus macaques (n = 1, open upward triangles), AGM (n = 13, open circles), and SM (n = 11, filled gray circles). CD4 mean fluorescent intensity (MFI) on pDC was normalized (%) to the CD4 MFI on CD4+ T cells. The horizontal, dashed line designates expression equal to that seen with CD4+ T cells. (C) The percentage of CCR5+ pDC was determined for SIV-negative Indian rhesus macaques (n = 11, open upward triangles), Chinese cynomolgus macaques (n = 2, open downward triangles), AGM (n = 7, open circles), and SM (n = 8, gray filled circles) and chronically SIV-infected Indian rhesus macaques (n = 8, open upward triangles), AGM (n = 3, open circles), and SM (n = 8, gray filled circles). Symbols represent individual animals, and lines and bars represent medians and interquartile ranges. *, Kruskal-Wallis, P < 0.05; ***, Kruskal-Wallis, P < 0.001.
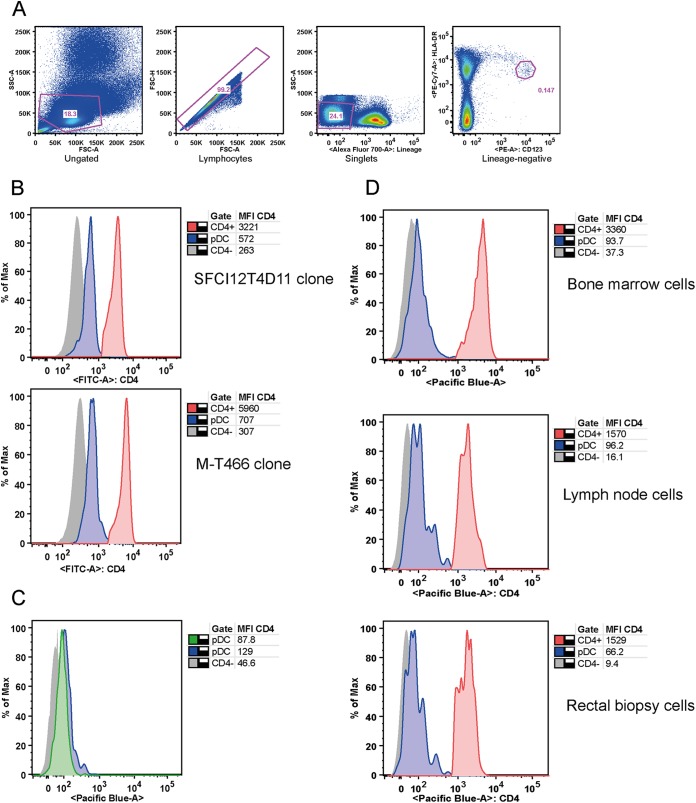
FIG 2.
Low CD4 expression on AGM pDC. (A) Dot plots showing the gating strategy used to identify pDC [forward-scatter and side-scatter (FSC/SSC) gated, singlets, Lineage (CD3/CD20), and HLA-DR CD123hi cells] for one representative animal. PE, phycoerythrin. (B) Low CD4 expression on AGM pDC (n = 3) was confirmed using two additional monoclonal anti-CD4 antibodies (M-T466 and SFCI12T4D11). Histograms of representative animals are shown, depicting CD4 expression on CD4− cells (gray, solid), pDC (blue, solid with contour), and Lineage+ CD4+ cells (red, solid with contour). MFI values of CD4 for the three cell populations are shown in the table. FITC, fluorescein isothiocyanate; max, maximum. (C) Intracellular and extracellular staining of AGM pDC (n = 3). Histograms for a representative animal are shown, cell surface CD4 expression is depicted for CD4− cells (gray, solid) and for pDC, and intracellular (blue, solid with contour) and extracellular (green, solid with contour) expression is shown, indicating the absence of an intracellular CD4 pool in AGM pDC. (D) Low CD4 expression was observed on pDC from bone marrow samples (n = 1 AGM), LN samples (n = 1 AGM), and rectal biopsy specimens (n = 1 AGM). Histograms depict CD4 expression on CD4− cells (gray, solid), pDC (blue, solid with contour), and Lineage+ CD4+ cells (red, solid with contour). MFI values of CD4 are listed in the tables.
As (i) AGM and MAC CD4+ T cell CD4 mean fluorescent intensity levels (MFI) were similar and (ii) analysis of two additional CD4 antibody clones confirmed low CD4 expression on AGM pDC (Fig. 2B), it is unlikely that low AGM pDC CD4 expression levels were due to species-specific antibody issues. The absence of intracellular CD4 in AGM pDC indicated that recycling from the cell surface was not the underlying mechanism of this low expression (Fig. 2C). AGM pDC from primary, secondary, and tertiary lymphoid tissues all expressed low levels of CD4 (Fig. 2D).
Efficient SIV sensing by AGM pDC.
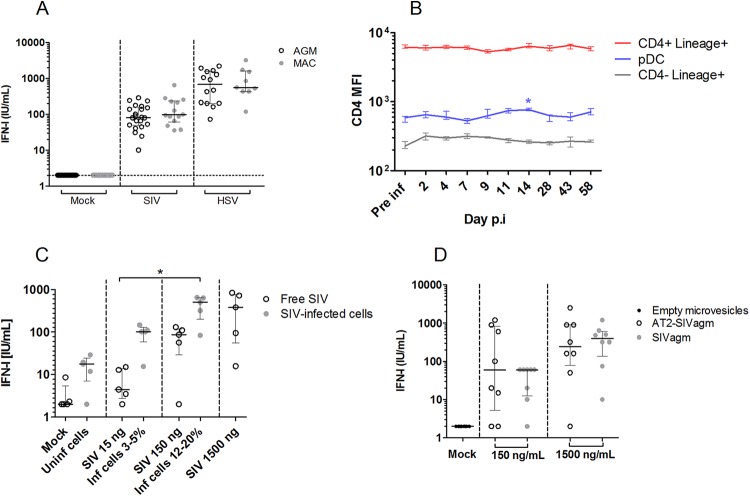
The finding of low AGM pDC CD4 expression is paradoxical, since CD4 is essential for HIV/SIV sensing by pDC and AGM pDC have been shown to efficiently sense SIVagm (7, 12). Nonetheless, a lower level of production of IFN-I has been previously described during SIVagm infection in vivo (12, 25). We wondered whether low pDC CD4 levels could have subtle effects on SIV sensing in vivo. For instance, only sensing of infectious SIVagm particles has been previously investigated (12, 25), while (i) most virions produced in vivo are noninfectious and (ii) sensing of virus-infected cells is more efficient in human and MAC (23). We stimulated peripheral blood mononuclear cells (PBMCs) of a large number of uninfected AGM and MAC with infectious SIV, using HSV as a control for CD4-independent sensing. No quantitative differences between AGM and MAC in IFN-I production were observed (Fig. 3A). Both SIV-infected cells and AT2-inactivated SIVagm also induced normal IFN-I responses (Fig. 3B and C). Longitudinal measurement showed that CD4 levels on pDC did not further decrease during SIV infection (Fig. 3D). Altogether, these data demonstrate that low AGM pDC CD4 levels did not impair their capacity to sense SIVagm.
FIG 3.
SIV sensing capacity of AGM pDC. (A) SIVagm.sab92018 was used to stimulate AGM (n = 22) and SIVmac251 was used to stimulate Chinese rhesus MAC (n = 14) PBMCs. Alternatively, PBMCs were stimulated with HSV-1. (B) CD4 MFI was followed throughout SIVagm infection of four AGMs. Median and interquartile ranges are shown for CD4+ Lineage+ cells, pDC, and CD4− Lineage+ cells. Pre inf, preinfection time point; Day p.i, day postinfection. (C) SIVagm-infected SupT1 cells and free virions were used to stimulate AGM PBMCs (n = 5). Inf, infected; Uninf, uninfected. (D) SIVagm or AT2-inactivated SIVagm was used to stimulate AGM PBMCs (n = 8). Individual symbols represent distinct animals. Medians and interquartile ranges are shown. Vertical dashes separate the different viral preparations used to test pDC sensing. *, Friedman, P < 0.05.
SIV isolate-dependent sensing of AGM pDC.
We wondered how SIVagm is sensed by pDC despite low CD4 expression. CD4 is a highly polymorphic molecule among primates. We considered the hypothesis that, given the long circulation of SIVagm in the AGM population, SIVagm is well adapted to AGM CD4 (26). This could entail the possibility that SIVagm, but not heterologous viruses, can elicit robust IFN-I responses by AGM pDC. We tested this hypothesis by stimulating AGM (sabaeus) and MAC PBMC with nine SIV/HIV isolates (Fig. 4A and B). The three SIVagm.sab isolates and the SIVagm.tan isolate induced robust IFN-I production by AGM pDC. SIVagm.gri did not induce an efficient response, which corresponds with the observation that the CD4 genes of the sabaeus and tantalus AGM species are more closely related to each other than to the CD4 gene of the grivet AGM species (26).
FIG 4.
Viral and host determinants of SIV sensing by pDC. (A and B) PBMCs from AGM (n = 5 to 23) (A) and Chinese rhesus MAC (n = 2 to 15) (B) were stimulated with medium (Mock), five SIVagm strains, three SIVmac strains, or HIV-1 (HIV-1.bal) at three concentrations: 1,500 ng/ml, 150 ng/ml, and 15 ng/ml p24/p27. Median values are represented by a horizontal line (blue and red for SIVagm and HIV/SIVmac, respectively). *, Wilcoxon, P < 0.05; **, Wilcoxon, P < 0.01 (compared to stimulation with an equal dose of SIVagm.sab92018). nd = not determined. tox = cytotoxic in 18-h culture. (C) AGM PBMCs (n = 10) were stimulated with medium (gray circle) or with 15, 150, or 1,500 ng/ml p27 SIVmac251 (red square) or SIVmac239 (blue triangle) for 18 h, after which viability was measured. Median and interquartile ranges are depicted. *, Wilcoxon, P < 0.05. Neg. Ctrl, negative control. (D) AGM PBMCs were stimulated with HIV (n = 14), VSV-G-pseudotyped HIV (n = 10), SIVmac251 (n = 10), VSV-G-pseudotyped SIVmac251 (n = 9), or SIVagm.sab92018 (n = 23) at 150 ng/ml p24 or p27. Median values and interquartile ranges are represented by a line and bars, respectively. *, Kruskal-Wallis, P < 0.05; **, Kruskal-Wallis, P < 0.01; ***, Kruskal-Wallis, P < 0.001. Symbols represent individual animals; horizontal dashed lines represent the limit of detection.
In contrast, HIV-1 and two (SIVmac.251 and SIVmac.STM) of three SIVmac isolates did not induce robust IFN-I production by AGM pDC (P < 0.05 or P < 0.01), even at a high viral dose. Only SIVmac239 induced IFN-I production from AGM PBMC, already at a low viral dose. A low dose of SIVmac239 also induced high levels of IFN-I in MAC PBMCs. At the highest dose, SIVmac239 was more cytotoxic than SIVmac251 for AGM PBMC (P < 0.05) (Fig. 4C). However, at a dose of 150 ng/ml p27, SIVmac239 did not influence viability, so it is unlikely that the strong response to SIVmac239 was related to dying cells, which are known to induce IFN-I responses (Fig. 4C). It should be noted that SIVmac239 has been shown to be highly virulent in vivo compared to SIVmac251 (27). In contrast to AGM pDC, MAC pDC produced IFN-I upon stimulation with all SIV/HIV isolates, including SIVagm (Fig. 4B).
If the lack of SIVmac/HIV sensing by AGM pDC is indeed due to low CD4 levels, then forcing viral endocytosis should overcome this sensing deficiency. In line with this, HIV-1, or SIVmac251, pseudotyped with VSV-G was sensed similarly to SIVagm (Fig. 4D). Indeed, pseudotyped HIV-1 and SIVmac251 induced higher levels of IFN-I than the corresponding wild-type isolates (P < 0.05 and P < 0.01, respectively) (Fig. 4D). Altogether, these findings indicate a virus-host coadaptation in AGM for viral sensing by pDC.
Predominance of CCR5-negative pDC in natural hosts.
We next examined CCR5 expression on AGM pDC, since coreceptor expression, in addition to CD4, is essential for infection. MAC pDC were predominantly CCR5+ (median, 92.3%), while the majority of AGM and SM pDC did not express detectable levels of CCR5 (7.7% and 40% CCR5+ pDC, respectively) (Fig. 1A and C). In SM, the percentage of CCR5+ pDC was lower after SIV infection (18.4%, P = 0.02) (Fig. 1C).
pDC are highly infected during pathogenic and nonpathogenic SIV infection.
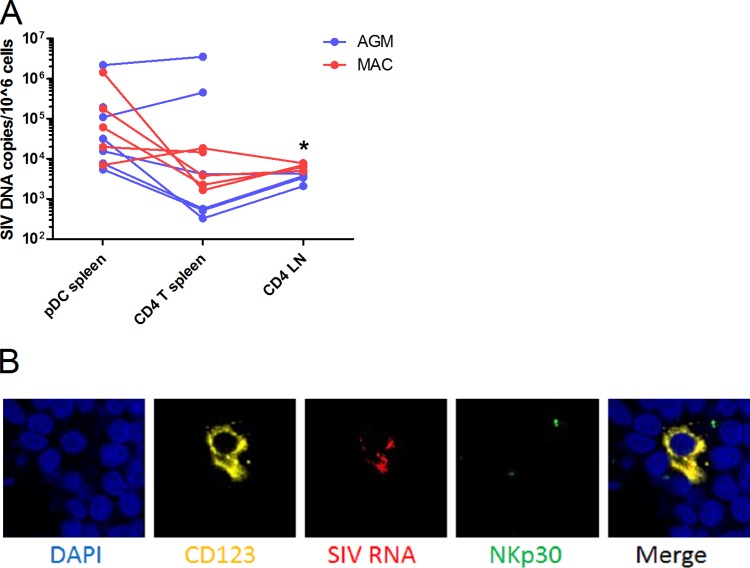
We then addressed the issue of whether these reduced levels of CD4 and CCR5 expression are associated with low levels of infection of AGM pDC. To test this, we purified splenic pDC and CD4+ T cells of chronically SIV-infected AGM and MAC and measured cell-associated viral DNA levels. Cells from uninfected animals were never positive for viral DNA (data not shown). Spleen pDC and CD4+ T cells from MAC harbored medians of 8.1 × 104 and 9.2 × 103 copies per million cells, respectively (P = 0.37) (Fig. 5A). AGM harbored 3.2 × 104 and 4.1 × 103 copies per million splenic pDC and CD4+ T cells, respectively (P = 0.44) (Fig. 5A). AGM and MAC pDC were thus infected to similar extents in vivo. As the infection levels of pDC and CD4+ T cells were similar, the presence of potentially contaminating CD4+ T cells in the pDC fraction cannot explain the levels of SIV DNA detected in pDC. As pDC have limited phagocytic capacities, the high levels of viral DNA in these cells are also unlikely to be associated with engulfed infected T cells.
FIG 5.
In vivo pDC infection. (A) pDC and CD4+ T cells of chronically SIV-infected AGM (n = 7) and Chinese rhesus MAC (n = 5) were sorted from 2 × 108 to 4 × 108 splenocytes, yielding a median of 6,000 pDC after two subsequent sorts (purity, 91%). CD4+ T cells were also purified from lymph node (LN) cells (purity, 97%). SIV DNA was normalized to CCR5 and is represented as copies per million cells. Symbols represent individual animals. CD4+ T cells in LNs were infected to a higher extent in MAC than in AGM (*, Mann-Whitney, P = 0.016). SIV DNA copy numbers of spleen and LN CD4+ T cells were similar in the two species. (B) Fluorescence microscopy was performed on a LN of one chronically infected AGM. DAPI (blue) staining shows nuclei, CD123 (yellow) is expressed on pDC, SIV RNA (red) shows infected cells, and NKp30 (green) is a marker not expressed on pDC. The merge shows an overlap of CD123 and SIV signals.
To further demonstrate the infection status of AGM pDC, we immunohistochemically examined a LN of a chronically infected AGM (Fig. 5B). CD123 and SIVagm RNA signals overlapped, while NKp30, which is not expressed on pDC, did not costain with CD123 or SIVagm RNA.
We then measured CD4 and CCR5 expression on splenic pDC of chronically infected AGM and MAC to exclude phenotypic differences compared to pDC from blood or from lymphoid tissues of uninfected animals. Splenic pDC of SIV-infected AGM, but not MAC, expressed low levels of CD4 (Fig. 6). Only 4.7% of AGM splenic pDC had detectable CCR5 expression, while 92.9% of MAC splenic pDC were CCR5+ (P = 0.0079). In conclusion, pDC of AGM and MAC were infected at high levels despite restricted CD4 and CCR5 expression on AGM pDC.
FIG 6.
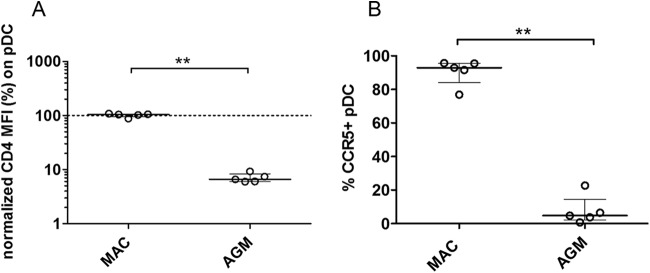
CD4 and CCR5 expression on splenic AGM and MAC pDC. (A) CD4 expression was measured on pDC from spleen of chronically SIV-infected Chinese rhesus MACs (n = 5) and AGMs (n = 5). The mean fluorescent intensity (MFI) of CD4 on splenic pDC was normalized (%) to the CD4 MFI on splenic CD4+ T cells of the same animal. The horizontal, dashed line designates CD4 expression equal to that seen with CD4+ Lineage+ cells. Symbols represent individual animals, and the line and bars represent the median and interquartile ranges, respectively. (B) The percentage of CCR5+ splenic pDC was determined for SIV-infected Chinese rhesus MACs (n = 5) and AGMs (n = 5). Symbols represent individual animals, and the line and bars represent the median and interquartile ranges. **, Mann-Whitney, P < 0.01.
DISCUSSION
The low levels of CD4 expression such as the level we discovered on AGM pDC are remarkable given the evolutionarily conserved high level of expression of CD4 on mammalian pDC, including primate, murine, cattle, and swine pDC (6, 28, 29). The role of CD4 in pDC biology is currently unknown, but this low expression raises questions on the physiological impact for AGM. The low CD4 levels on pDC were sufficient to allow SIVagm endocytosis and subsequent sensing, and we demonstrated for the first time that natural host pDC can sense SIV-infected cells. In contrast, most SIVmac and HIV-1 isolates tested were not sensed by AGM pDC. Pseudotyping increased the efficiency of HIV-1/SIVmac251 sensing, which indicates that the inefficient sensing of heterologous virus was due to restricted viral uptake. Altogether, this suggests an adaptation between SIVagm and its host-specific CD4. Similarly to our findings, it has been shown that HIV-1 interacts poorly with MAC CD4 compared to human CD4 and that HIV-1 could thus infect cells expressing low levels of human CD4 but not cells expressing low levels of MAC CD4 (30). Low CD4 expression also did not prevent AGM pDC from being infected, as they were infected at high levels in vivo. Since the pDC sensing capacities and infection levels of AGM and MAC were similar, these factors do not determine the level of chronic inflammation.
Our study revealed a high rate of infection of pDC in secondary lymphoid tissue during chronic SIV infection. These data resemble those determined in analysis of acute SIVmac infection, where pDC were found to be infected at levels similar to those seen with CD4+ T cells in LN (9). Since only very few data on pDC infection in vivo are available, this is important and underlines the potential role of pDC as a viral reservoir. Of note, the ratio of pDC to CD4+ T cells is low, at approximately 1:300 in lymph nodes (2). Therefore, the majority of the viral burden in SIV infections is still associated with CD4+ T cells. However, the contribution of pDC should not be underestimated, given their presence in mucosae, their capacity to migrate to lymph nodes, and their ability to efficiently transmit HIV/SIV to CD4+ T cells.
The lack of correlation between the frequency of CCR5+ pDC and SIV infection status does not support the hypothesis that absence of CCR5 expression protects against target cell infection (4). In line with this, a mutation in the SM CCR5 allele, disrupting functional CCR5 expression, does not diminish SIVsm infection prevalence (31). This can be explained by the fact that SIVs, including SIVagm, efficiently use alternative coreceptors, such as CXCR6 (Bonzo) and GPR15 (Bob) (22, 31). It is, however, possible that the low percentage of CCR5 expression in natural hosts is related indirectly to their resistance to disease. Indeed, while CCR5 is not a specific gut-homing receptor, it can induce migration to inflamed tissues (14). Such extremely low levels of CCR5+ pDC in natural hosts could therefore be related to the lack of pDC accumulation in the gut after infection (32). Our results therefore suggest that evaluation of the function of CCR5 in the seeding of viral reservoirs outside its role as an HIV coreceptor is warranted. Teasing out such mechanisms could be helpful for curative approaches.
ACKNOWLEDGMENTS
This work was supported by Fondation AREVA, the French National Agency for Research on AIDS and Viral Hepatitis (ANRS), Sidaction, Fondation TOTAL (to F.B.-S.), the French Ministry of Higher Education and Research, and Institut Pasteur. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We are grateful to Christophe Joubert, Benoît Delache, Jean-Marie Héliès, Patrick Flament, and the staff of the CEA animal facilities for outstanding work in animal care. We also sincerely thank the staff of the Institut Pasteur animal facility. We thank Kathryn Faulkner for performing flow cytometric analyses on SM cells. We appreciate the excellent technical assistance of Claire Torres, Julie Morin, Aurélien Corneau, and the TIPIV staff of the CEA. We thank the Centre d'Immunologie Humaine (CIH) at Institut Pasteur for access to the FACSAria II cell sorter. We also acknowledge the state-of-the-art National Center for Infectious Disease Models and Innovative Therapies (IDMIT). Viruses were kindly provided by A. David, D. Negre, and F.-L. Cosset as well as by J. Lifson and J. Bess, Jr. (AIDS & Cancer Virus Program, NCI at Frederick). SIVagm(gri-1) was obtained through the AIDS Research and Reference Program, Division of AIDS, NIAID, NIH, from J. Allan. We thank A. Isaacs for reading of the manuscript and are grateful to J-P. Herbeuval and N. Smith for helpful discussions.
REFERENCES
- 1.Sodora DL, Allan JS, Apetrei C, Brenchley JM, Douek DC, Else JG, Estes JD, Hahn BH, Hirsch VM, Kaur A, Kirchhoff F, Muller-Trutwin M, Pandrea I, Schmitz JE, Silvestri G. 2009. Toward an AIDS vaccine: lessons from natural simian immunodeficiency virus infections of African nonhuman primate hosts. Nat Med 15:861–865. doi: 10.1038/nm.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diop OM, Ploquin MJ, Mortara L, Faye A, Jacquelin B, Kunkel D, Lebon P, Butor C, Hosmalin A, Barre-Sinoussi F, Muller-Trutwin MC. 2008. Plasmacytoid dendritic cell dynamics and alpha interferon production during simian immunodeficiency virus infection with a nonpathogenic outcome. J Virol 82:5145–5152. doi: 10.1128/JVI.02433-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaumier CM, Harris LD, Goldstein S, Klatt NR, Whitted S, McGinty J, Apetrei C, Pandrea I, Hirsch VM, Brenchley JM. 2009. CD4 downregulation by memory CD4+ T cells in vivo renders African green monkeys resistant to progressive SIVagm infection. Nat Med 15:879–885. doi: 10.1038/nm.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paiardini M, Cervasi B, Reyes-Aviles E, Micci L, Ortiz AM, Chahroudi A, Vinton C, Gordon SN, Bosinger SE, Francella N, Hallberg PL, Cramer E, Schlub T, Chan ML, Riddick NE, Collman RG, Apetrei C, Pandrea I, Else J, Munch J, Kirchhoff F, Davenport MP, Brenchley JM, Silvestri G. 2011. Low levels of SIV infection in sooty mangabey central memory CD4+ T cells are associated with limited CCR5 expression. Nat Med 17:830. doi: 10.1038/nm.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pandrea I, Onanga R, Souquiere S, Mouinga-Ondeme A, Bourry O, Makuwa M, Rouquet P, Silvestri G, Simon F, Roques P, Apetrei C. 2008. Paucity of CD4+ CCR5+ T cells may prevent transmission of simian immunodeficiency virus in natural nonhuman primate hosts by breast-feeding. J Virol 82:5501–5509. doi: 10.1128/JVI.02555-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beignon AS, McKenna K, Skoberne M, Manches O, DaSilva I, Kavanagh DG, Larsson M, Gorelick RJ, Lifson JD, Bhardwaj N. 2005. Endocytosis of HIV-1 activates plasmacytoid dendritic cells via Toll-like receptor-viral RNA interactions. J Clin Invest 115:3265–3275. doi: 10.1172/JCI26032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jochems SP, Petitjean G, Kunkel D, Liovat AS, Ploquin MJ, Barre-Sinoussi F, Lebon P, Jacquelin B, Muller-Trutwin MC. 2015. Modulation of type I interferon-associated viral sensing during acute simian immunodeficiency virus infection in African green monkeys. J Virol 89:751–762. doi: 10.1128/JVI.02430-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donaghy H, Gazzard B, Gotch F, Patterson S. 2003. Dysfunction and infection of freshly isolated blood myeloid and plasmacytoid dendritic cells in patients infected with HIV-1. Blood 101:4505–4511. doi: 10.1182/blood-2002-10-3189. [DOI] [PubMed] [Google Scholar]
- 9.Brown KN, Wijewardana V, Liu X, Barratt-Boyes SM. 2009. Rapid influx and death of plasmacytoid dendritic cells in lymph nodes mediate depletion in acute simian immunodeficiency virus infection. PLoS Pathog 5:e1000413. doi: 10.1371/journal.ppat.1000413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacquelin B, Petitjean G, Kunkel D, Liovat AS, Jochems SP, Rogers KA, Ploquin MJ, Madec Y, Barre-Sinoussi F, Dereuddre-Bosquet N, Lebon P, Le Grand R, Villinger F, Muller-Trutwin M. 2014. Innate immune responses and rapid control of inflammation in African green monkeys treated or not with interferon-alpha during primary SIVagm infection. PLoS Pathog 10:e1004241. doi: 10.1371/journal.ppat.1004241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diop OM, Gueye A, Dias-Tavares M, Kornfeld C, Faye A, Ave P, Huerre M, Corbet S, Barre-Sinoussi F, Muller-Trutwin MC. 2000. High levels of viral replication during primary simian immunodeficiency virus SIVagm infection are rapidly and strongly controlled in African green monkeys. J Virol 74:7538–7547. doi: 10.1128/JVI.74.16.7538-7547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacquelin B, Mayau V, Targat B, Liovat AS, Kunkel D, Petitjean G, Dillies MA, Roques P, Butor C, Silvestri G, Giavedoni LD, Lebon P, Barre-Sinoussi F, Benecke A, Muller-Trutwin MC. 2009. Nonpathogenic SIV infection of African green monkeys induces a strong but rapidly controlled type I IFN response. J Clin Invest 119:3544–3555. doi: 10.1172/JCI40093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taaffe J, Chahroudi A, Engram J, Sumpter B, Meeker T, Ratcliffe S, Paiardini M, Else J, Silvestri G. 2010. A five-year longitudinal analysis of sooty mangabeys naturally infected with simian immunodeficiency virus reveals a slow but progressive decline in CD4+ T-cell count whose magnitude is not predicted by viral load or immune activation. J Virol 84:5476–5484. doi: 10.1128/JVI.00039-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meythaler M, Martinot A, Wang Z, Pryputniewicz S, Kasheta M, Ling B, Marx PA, O'Neil S, Kaur A. 2009. Differential CD4+ T-lymphocyte apoptosis and bystander T-cell activation in rhesus macaques and sooty mangabeys during acute simian immunodeficiency virus infection. J Virol 83:572–583. doi: 10.1128/JVI.01715-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gueye A, Diop OM, Ploquin MJ, Kornfeld C, Faye A, Cumont MC, Hurtrel B, Barre-Sinoussi F, Muller-Trutwin MC. 2004. Viral load in tissues during the early and chronic phase of non-pathogenic SIVagm infection. J Med Primatol 33:83–97. doi: 10.1111/j.1600-0684.2004.00057.x. [DOI] [PubMed] [Google Scholar]
- 16.Müller MC, Saksena NK, Nerrienet E, Chappey C, Hervé VM, Durand JP, Legal-Campodonico P, Lang MC, Digoutte JP, Georges AJ, Georges-Courbot M-C, Sonigo P, Barre-Sinouss F. 1993. Simian immunodeficiency viruses from central and western Africa: evidence for a new species-specific lentivirus in tantalus monkeys. J Virol 67:1227–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin MJ, Hui H, Robertson DL, Muller MC, Barre-Sinoussi F, Hirsch VM, Allan JS, Shaw GM, Sharp PM, Hahn BH. 1994. Mosaic genome structure of simian immunodeficiency virus from West African green monkeys. EMBO J 13:2935–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allan JS, Kanda P, Kennedy RC, Cobb EK, Anthony M, Eichberg JW. 1990. Isolation and characterization of simian immunodeficiency viruses from two subspecies of African green monkeys. AIDS Res Hum Retroviruses 6:275–285. doi: 10.1089/aid.1990.6.275. [DOI] [PubMed] [Google Scholar]
- 19.Fomsgaard A, Hirsch VM, Allan JS, Johnson PR. 1991. A highly divergent proviral DNA clone of SIV from a distinct species of African green monkey. Virology 182:397–402. doi: 10.1016/0042-6822(91)90689-9. [DOI] [PubMed] [Google Scholar]
- 20.Aiken C. 1997. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J Virol 71:5871–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.David A, Saez-Cirion A, Versmisse P, Malbec O, Iannascoli B, Herschke F, Lucas M, Barre-Sinoussi F, Mouscadet JF, Daeron M, Pancino G. 2006. The engagement of activating FcgammaRs inhibits primate lentivirus replication in human macrophages. J Immunol 177:6291–6300. doi: 10.4049/jimmunol.177.9.6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller MC, Barre-Sinoussi F. 2003. SIVagm: genetic and biological features associated with replication. Front Biosci 8:d1170–d1185. doi: 10.2741/1130. [DOI] [PubMed] [Google Scholar]
- 23.Lepelley A, Louis S, Sourisseau M, Law HK, Pothlichet J, Schilte C, Chaperot L, Plumas J, Randall RE, Si-Tahar M, Mammano F, Albert ML, Schwartz O. 2011. Innate sensing of HIV-infected cells. PLoS Pathog 7:e1001284. doi: 10.1371/journal.ppat.1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kornfeld C, Ploquin MJ, Pandrea I, Faye A, Onanga R, Apetrei C, Poaty-Mavoungou V, Rouquet P, Estaquier J, Mortara L, Desoutter JF, Butor C, Le Grand R, Roques P, Simon F, Barre-Sinoussi F, Diop OM, Muller-Trutwin MC. 2005. Antiinflammatory profiles during primary SIV infection in African green monkeys are associated with protection against AIDS. J Clin Invest 115:1082–1091. doi: 10.1172/JCI23006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campillo-Gimenez L, Laforge M, Fay M, Brussel A, Cumont MC, Monceaux V, Diop O, Levy Y, Hurtrel B, Zaunders J, Corbeil J, Elbim C, Estaquier J. 2010. Nonpathogenesis of simian immunodeficiency virus infection is associated with reduced inflammation and recruitment of plasmacytoid dendritic cells to lymph nodes, not to lack of an interferon type I response, during the acute phase. J Virol 84:1838–1846. doi: 10.1128/JVI.01496-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fomsgaard A, Muller-Trutwin MC, Diop O, Hansen J, Mathiot C, Corbet S, Barre-Sinoussi F, Allan JS. 1997. Relation between phylogeny of African green monkey CD4 genes and their respective simian immunodeficiency virus genes. J Med Primatol 26:120–128. doi: 10.1111/j.1600-0684.1997.tb00043.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Y, Bao R, Haigwood NL, Persidsky Y, Ho WZ. 2013. SIV infection of rhesus macaques of Chinese origin: a suitable model for HIV infection in humans. Retrovirology 10:89. doi: 10.1186/1742-4690-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Summerfield A, Auray G, Ricklin M. 2015. Comparative dendritic cell biology of veterinary mammals. Annu Rev Anim Biosci 3:533–557. doi: 10.1146/annurev-animal-022114-111009. [DOI] [PubMed] [Google Scholar]
- 29.Ferrero I, Held W, Wilson A, Tacchini-Cottier F, Radtke F, MacDonald HR. 2002. Mouse CD11c(+) B220(+) Gr1(+) plasmacytoid dendritic cells develop independently of the T-cell lineage. Blood 100:2852–2857. doi: 10.1182/blood-2002-01-0214. [DOI] [PubMed] [Google Scholar]
- 30.Humes D, Emery S, Laws E, Overbaugh J. 2012. A species-specific amino acid difference in the macaque CD4 receptor restricts replication by global circulating HIV-1 variants representing viruses from recent infection. J Virol 86:12472–12483. doi: 10.1128/JVI.02176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riddick NE, Hermann EA, Loftin LM, Elliott ST, Wey WC, Cervasi B, Taaffe J, Engram JC, Li B, Else JG, Li Y, Hahn BH, Derdeyn CA, Sodora DL, Apetrei C, Paiardini M, Silvestri G, Collman RG. 2010. A novel CCR5 mutation common in sooty mangabeys reveals SIVsmm infection of CCR5-null natural hosts and efficient alternative coreceptor use in vivo. PLoS Pathog 6:e1001064. doi: 10.1371/journal.ppat.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwa S, Kannanganat S, Nigam P, Siddiqui M, Shetty RD, Armstrong W, Ansari A, Bosinger SE, Silvestri G, Amara RR. 2011. Plasmacytoid dendritic cells are recruited to the colorectum and contribute to immune activation during pathogenic SIV infection in rhesus macaques. Blood 118:2763–2773. doi: 10.1182/blood-2011-02-339515. [DOI] [PMC free article] [PubMed] [Google Scholar]