Abstract
Broadly neutralizing antibodies have been isolated that bind the glycan shield of the HIV-1 envelope spike. One such antibody, PGT135, contacts the intrinsic mannose patch of gp120 at the Asn332, Asn392, and Asn386 glycosylation sites. Here, site-specific glycosylation analysis of recombinant gp120 revealed glycan microheterogeneity sufficient to explain the existence of a minor population of virions resistant to PGT135 neutralization. Target microheterogeneity and antibody glycan specificity are therefore important parameters in HIV-1 vaccine design.
TEXT
The isolation of potent, broadly neutralizing antibodies (bnAbs) from infected HIV-1 individuals has focused efforts toward the molecular characterization of their epitopes (1–5). These antibodies have potential value in a therapeutic context, and their epitopes represent key vaccine leads (6–12). Structural and biochemical studies have revealed that a number of the recently isolated bnAbs penetrate the heavily glycosylated surface of the HIV-1 envelope spike, making contacts with both the glycans and the protein underneath (1–3, 13–22) Characterization of the glycan-containing epitopes has revealed that much of the glycan shield is vulnerable to antibody recognition (5). Many glycans within the outer domain of gp120 are protected from normal glycan processing and do not form complex-type glycans, instead remaining as immature oligomannose-type glycans. This region is known as the “intrinsic mannose patch” since it contains oligomannose-type glycans, regardless of whether presented in the context of isolated gp120 monomers or functional virions (23–25).
The intrinsic mannose patch is targeted by the so-called “mannose patch-dependent” antibodies, which include PGT121 to -124, 10-1074, PGT125 to -128, PGT130 and -131, PGT135 to -137, and 2G12 (14–16, 26–29). These antibodies display remarkable potencies against a diverse panel of HIV-1 strains, although their breadth varies both between and within families (2, 30). PGT135 was found to neutralize 33% of viruses from a 162-cross-clade-pseudovirus panel. This neutralization is equivalent to the breadth of b12, which has a protein-based epitope at the CD4 binding site, but is lower than those of other Asn332-dependent bnAbs, such as PGT128 and PGT121, which neutralized 72% and 70% of the panel, respectively (2). This lower breadth of neutralization has been attributed to the limited prevalence of the larger number of critical contact residues (Asn332, Asn392, and His330) across different isolates (15) compared to PGT121 and PGT128. In addition to these properties, inspection of neutralization profiles reveals that, despite containing the required target residues, for some strains of HIV-1, neutralization is incomplete, with plateaus that do not reach 100% (15). A crystal structure of a PGT135 Fab domain in complex with the gp120 core revealed that the majority of the interactions were mediated through contact with the glycans at the Asn332, Asn392, and Asn386 sites, with 1,010 Å2 and 438 Å2 of buried surface area contacting gp120 glycans and protein, respectively (15). Given the extensive contribution of glycans to the binding interaction, we hypothesized that the incomplete neutralization of some isolates by PGT135 could partially derive from microheterogeneity at the target glycan sites, whereby the presence of certain glycoforms precludes the binding of PGT135.
To investigate this, we performed site-specific glycosylation analysis of the glycan sites targeted by PGT135, as observed in the crystal structure (15): Asn332, Asn386, and Asn392 (Fig. 1). The BaL isolate was chosen as this has been demonstrated to exhibit some resistance to neutralization by PGT135, with only about 80% of wild-type virus neutralized (15). Recombinant monomeric gp120BaL was expressed in HEK 293T cells and purified by immobilized metal affinity chromatography followed by size exclusion chromatography. We previously observed that recombinant gp120 expressed in this way reproduces the intrinsic population of the oligomannose-type glycans present on virus produced in peripheral blood mononuclear cells (PBMCs), providing a good model for analyzing this component of Env glycosylation (24, 25). Glycopeptides containing a target glycan site were generated by in-solution protease digestions of reduced and alkylated gp120BaL and isolated by reverse-phase high-performance liquid chromatography (RP-HPLC).
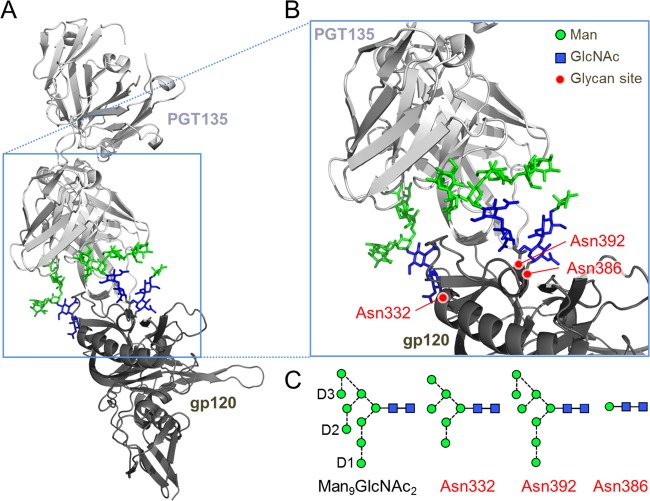
FIG 1.
The glycan epitope of PGT135 encompasses the Asn332, Asn392, and Asn386 sites. (A) A previously reported crystal structure reveals the interaction of a PGT135 Fab domain with the Asn332 (Man6GlcNAc2), Asn392 (Man8GlcNAc2), and Asn386 (Man1GlcNAc2) glycans from a gp120JR-FL core (15). The protein moiety is depicted in a ribbon diagram, and glycans are depicted as sticks. Mannose (Man) residues are colored in green, and N-acetlyglucosamine (GlcNAc) residues are colored in blue. (B) Enlarged view of the PGT135 glycan epitope. (C) Schematic representation of a Man9GlcNAc2 glycan, with the D1 to D3 arms annotated and the glycans resolvable in the crystal structure. Glycan structures are shown according to the proposed method of Harvey et al. (40), with residues colored according to panels A and B. Images were made in PyMol using PDB code 4JM2.
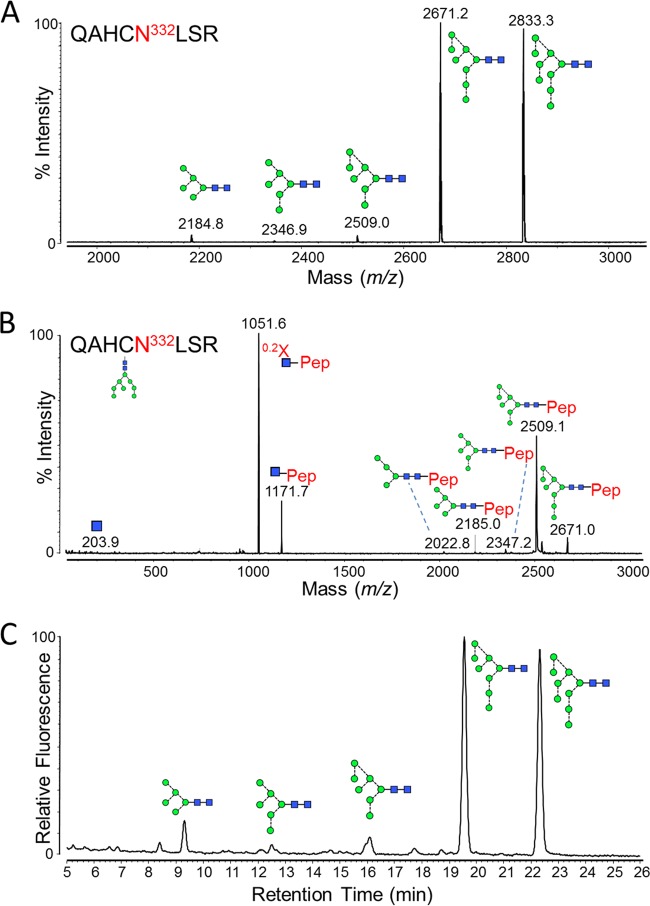
Asn332-containing glycopeptides (sequence QAHCN332LSR) were isolated in a fraction from a tryptic digest, performed according to the manufacturer's instructions (Promega), and were analyzed by matrix-assisted laser desorption ionization mass spectrometry (MALDI MS) (Fig. 2A). This revealed the glycoforms at the Asn332 site to be overwhelmingly dominated by Man8GlcNAc2 and Man9GlcNAc2 glycans, with trace levels of Man5–7GlcNAc2 (Table 1). Confirmation of the glycopeptide identity was performed by tandem MS (MS/MS) fragmentation (Fig. 2B). Since the ionization of molecules can be influenced by their chemical composition, the measured abundances were validated by quantitating the glycans directly. This was achieved by PNGase F digestion of the glycopeptide fractions to release the glycans, which were then labeled with 2-aminobenzamide (2-AB) and analyzed by hydrophilic interaction liquid chromatography-ultraperformance liquid chromatography (HILIC-UPLC) (Fig. 2C). Comparison of the MALDI-MS glycopeptide spectrum and the HILIC-UPLC glycan profile revealed a very high degree of similarity, validating the use of MALDI-MS for the analysis of glycopeptides containing only oligomannose-type glycans.
FIG 2.
Glycans present at the Asn332 glycosylation site. Recombinant, monomeric gp120BaL was expressed in HEK 293T cells from the pHLsec vector (41) and purified by metal-affinity and size exclusion chromatography, as previously described (42). gp120 was reduced, alkylated, and digested with trypsin (Promega), before fractionation using a Jupiter C18 5-μm 250- by 4.5-mm column (300-Å pore size) and a Dionex U3000 liquid chromatography system. Fractions were collected every minute, at a flow rate of 1 ml/min, for 90 min, and then analyzed on an Autoflex Speed MALDI tandem time of flight (TOF/TOF) instrument (Bruker), operated in positive-ion mode. (A) MALDI MS of pooled fractions containing the Asn332-containing glycopeptide, QAHCNLSR. The glutamine carried a pyro-Glu modification (−17), and the cysteine was modified due to treatment with iodoacetamide (carbamidomethyl, +57). Glycan structures corresponding to the observed glycopeptide masses are indicated. (B) MALDI MS/MS fragmentation spectrum of the 2,671.2 peak, corresponding to a Man8GlcNAc2 glycopeptide. Fragment ions were observed that were characteristic of glycopeptide MALDI MS/MS fragmentation (43), including [Mpep+H + 83]+ (corresponding to 0.2X-ring cleavage of the innermost GlcNAc) and [Mpep+H + 203]+ (corresponding to Y-type cleavage of the di-N-acetylchitobiose core). Y-type fragmentation of glycosidic bonds was also observed. (C) HILIC-UPLC profile of Asn332 glycans. Glycans were released from the QAHCNLSR glycopeptides by in-solution PNGase F (QA-Bio) digestion, according to the manufacturer's instructions, and then labeled using a LudgerTag 2-AB labeling kit (Ludger Ltd., Abingdon, United Kingdom). Chromatography was performed on a Waters Acquity UPLC instrument. Glycans were assigned by comparison with known oligomannose-type glycan standards (Ludger Ltd., Abingdon, United Kingdom).
TABLE 1.
Summary of glycoforms and their abundances found at the glycan epitopes of PGT135 by MALDI MS
| Asn glycan site | Peptidea | Peptide mass [M+H]+ |
m/z |
Glycoforme | % of total | |
|---|---|---|---|---|---|---|
| Observed | Calculated | |||||
| N332 | QAHCNLSRf,g | 968.4 | 2,184.8b | 2,184.9 | M5 | 2 |
| 2,346.9b | 2,346.9 | M6 | <1 | |||
| 2,509.0b | 2,509.0 | M7 | 1 | |||
| 2,671.2b | 2,671.0 | M8 | 49 | |||
| 2,833.3b | 2,833.1 | M9 | 47 | |||
| N392 | NSTW | 507.2 | 1,583.4c | 1,583.6 | M4 | 1 |
| 1,745.4c | 1,745.6 | M5 | 19 | |||
| 1,907.4c | 1,907.7 | M6 | <1 | |||
| 2,069.5c | 2,069.7 | M7 | 2 | |||
| 2,231.5c | 2,231.8 | M8 | 62 | |||
| 2,247.4d | 2,247.9 | |||||
| 2,393.5c | 2,393.8 | M9 | 16 | |||
| 2,409.4d | 2,409.9 | |||||
| N386 | YCNSTQLFg | 1,032.4 | 2,086.9b | 2,086.8 | M4 | 2 |
| 2,249.0b | 2,248.8 | M5 | 7 | |||
| 2,270.9c | 2,270.8 | |||||
| 2,286.9d | 2,286.9 | |||||
| 2,411.0b | 2,410.9 | M6 | 7 | |||
| 2,433.0c | 2,432.9 | |||||
| 2,449.0d | 2,449.0 | |||||
| 2,573.1b | 2,573.0 | M7 | 33 | |||
| 2,595.1c | 2,594.9 | |||||
| 2,611.1d | 2,611.0 | |||||
| 2,735.2b | 2,735.0 | M8 | 42 | |||
| 2,757.1c | 2,757.0 | |||||
| 2,773.1d | 2,773.1 | |||||
| 2,897.3b | 2,897.1 | M9 | 9 | |||
| 2,919.2c | 2,919.0 | |||||
| 2,935.2d | 2,935.1 | |||||
The Asn glycan sites are underlined.
[M+H]+.
[M+Na]+.
[M+K]+.
Mx, ManxGlcNAc2.
Pyro-glu modification of glutamine (−17).
Carbamidomethyl modification of cysteine (+57).
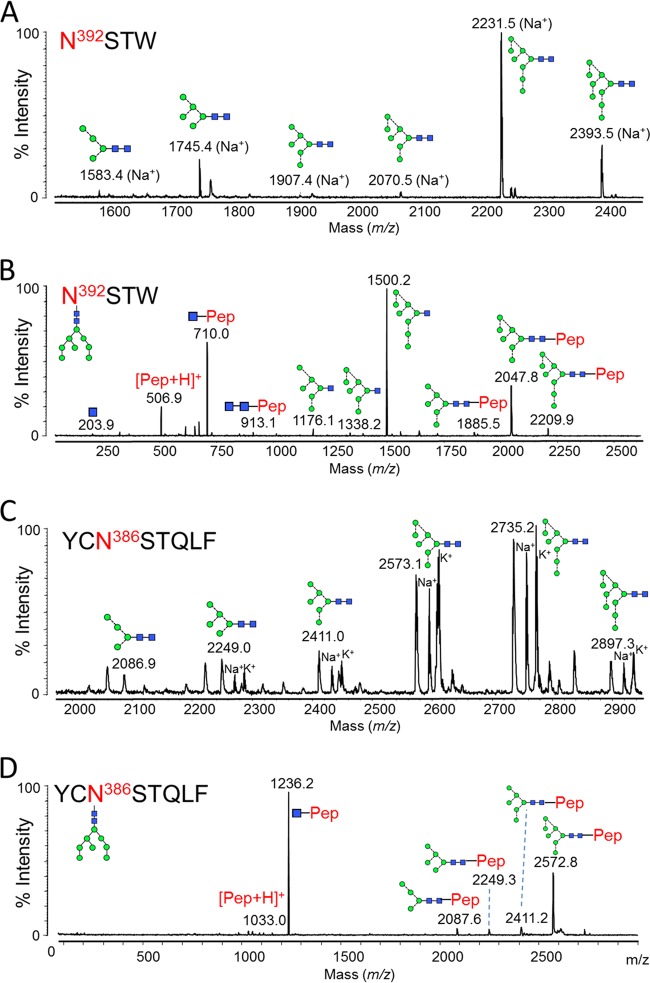
In the crystal structure, PGT135 contacts an area of 365 Å2 across the Asn332 glycan of gp120, with a further 547 Å2 of contacts with Asn392 and a minor contribution of 99 Å2 from Asn386 (15). The Asn392 and Asn386 sites were not resolvable using a tryptic digestion; instead chymotrypsin (Promega) was used to produce N392STW and YCN386STQLF glycopeptides (Fig. 3). These glycosylation sites displayed different patterns of microheterogeneity to the Asn332 site (Table 1). The Asn392 site was primarily populated by Man8GlcNAc2 glycans (62%), with lower levels of Man5GlcNAc2 and Man9GlcNAc2 and trace amounts of Man4GlcNAc2, Man6GlcNAc2, and Man7GlcNAc2 (Fig. 3A; Table 1). Meanwhile the Asn386 site displayed a more even distribution of oligomannose-type glycans, with Man7GlcNAc2 and Man8GlcNAc2 predominating (Fig. 3C; Table 1).
FIG 3.
Glycans present at the Asn392 and Asn386 glycosylation sites. gp120 was digested with chymotrypsin (Promega) before RP-HPLC and MALDI analysis. (A) MALDI MS of Asn392 glycopeptides (NSTW). Sodium, [M + 23]+, and potassium, [M + 39]+, adducts were observed: both peaks were used for measuring abundances (Table 1). (B) MALDI MS/MS fragmentation spectrum of the peak corresponding to the Man8GlcNAc2 glycopeptide. Fragmentation peaks corresponded to the protonated masses. (C) MALDI MS of Asn386 glycopeptides (YCNSTQLF). The cysteine was modified due to treatment with iodoacetamide (carbamidomethyl, +57). Protonated glycopeptides, as well as sodium and potassium adducts, were detected: all were used for calculation of abundances (Table 1). (D) MALDI MS/MS fragmentation spectrum of the peak corresponding to the Man8GlcNAc2 glycopeptide. Fragmentation peaks corresponded to the protonated masses.
The oligomannose-type nature of the glycans at the Asn332, Asn392, and Asn386 sites is consistent with their location on the densely glycosylated outer domain: the crowded environment helps restrict access by glycosidases and glycosyltransferases, which would otherwise lead to the formation of an array of complex- and hybrid-type glycan structures, as observed at more peripheral glycan sites (24, 31–33). However, despite the more restricted microheterogeneity at these sites, there remains a distribution of the individual oligomannose-type glycans, which may affect the capacity for PGT135 binding given its glycan specificity. Glycan array data revealed a preference for large oligomannose-type glycans (Man7–9GlcNAc2) (15). Such a preference, at least at the Asn392 site, can be rationalized by the extensive interactions of PGT135 along the full length of the D1 arm of a Man8GlcNAc2 glycan present in the crystal structure (15). While the population of smaller oligomannose glycans at the Asn392 site might not capitalize on all possible binding interactions with PGT135, their presence does not necessarily preclude binding. However, the plateau in neutralization observed by us (Fig. 4A) and others (15) at around 80 to 85% for BaL pseudovirus suggests there's a population of virus that cannot be bound effectively. Modeling of a Man9GlcNAc2 glycan at the Asn392 site revealed that the presence of a terminal D2 mannose residue would induce a steric clash with the complementarity determining region (CDR) H3 loop of PGT135 (15).
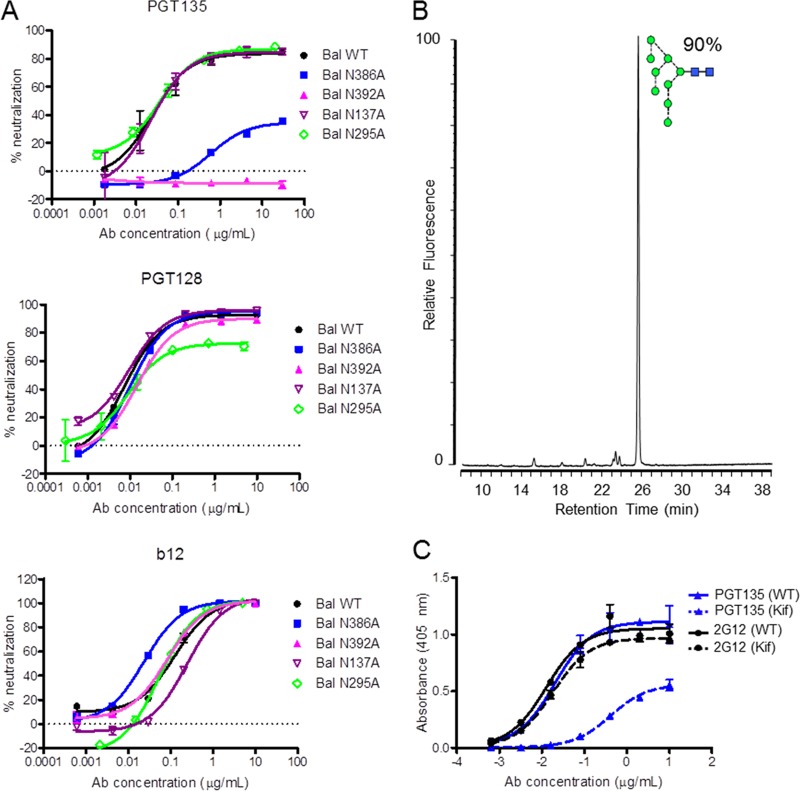
FIG 4.
Influences of gp120 glycans on bnAb recognition. (A) Neutralization sensitivity of PGT135, PGT128, and b12 against BaL wild-type pseudovirus and glycan site mutants. bnAbs PGT128 and b12 are used as controls. To produce pseudoviruses, plasmids encoding Env were cotransfected with an Env-deficient genomic backbone plasmid (pSG3ΔEnv) in a 1:2 ratio with the transfection reagent polyethyleneimine (PEI) (1 mg/ml, 1:3 PEI-total DNA [Polysciences]) into HEK 293T cells. Pseudoviruses were harvested 72 h posttransfection. Neutralizing activity was assessed using a single-round replication pseudovirus assay with TZM-bl target cells, as described previously (2). Glycan sites N137, N295, N386, and N392 were removed using site-directed mutagenesis through an Asn-to-Ala mutation. Mutations were verified by DNA sequencing (MWG Eurofins, Germany). (B) Glycan profile of recombinant gp120 expressed in HEK 293T cells in the presence of kifunensine, analyzed as described in the legend to Fig. 2C. (C) Enzyme-linked immunosorbent assay (ELISA) data of PGT135 (blue) and 2G12 (black) binding to wild-type (continuous line) and kifunensine-treated (dashed line) gp120. ELISAs were performed as previously described (34).
To assess the compatibility of Man9GlcNAc2 glycans with PGT135 binding, we expressed gp120 in the presence of kifunensine, which produced a glycan profile containing over 90% Man9GlcNAc2 (Fig. 4B). 2G12 binding, which has a known tolerance for Man9GlcNAc2 glycans, was mostly unchanged upon kifunensine treatment (Fig. 4C), consistent with previous observations (34). In contrast, PGT135 binding was dramatically reduced compared to that of the wild type. Our site-specific glycosylation analysis showed that 16% of the glycans at the Asn392 site of recombinant gp120BaL were Man9GlcNAc2, which corresponds well with the observed maximal neutralization of BaL pseudovirus (Fig. 4A). In contrast the magnitude of the plateau is not consistent with the much larger population of Man9GlcNAc2 glycans (almost 50%) observed at the N332 site. This constraint, excluding Man9GlcNAc2 at the Asn392 site, is further supported by the significant decrease in neutralization observed upon kifunensine treatment (15). In contrast, modeling of a Man9GlcNAc2 glycan at the Asn332 site suggested that a D2 terminal mannose could be accommodated with a minor conformational change, and thus the microheterogeneity observed at this site is likely to be better tolerated. Similarly, we note that PGT128 could achieve almost complete neutralization despite the observed microheterogeneity at the Asn332 site (Fig. 4A). Incompatible glycoforms at the secondary binding site, N301, could account for the incomplete neutralization. The interaction between PGT135 and the glycan at Asn386 was observed to make a much smaller contribution to binding (15), suggesting that an imperfect glycan match would not be as detrimental to binding. Indeed, removal of the Asn392 glycan site completely abolished neutralization by PGT135, whereas a degree of neutralization was retained upon removal of the N386 glycan site, with the neutralization plateau reduced to approximately 30% (Fig. 4A). Removal of Asn137, which has been shown to enhance neutralization of some viruses by other N332-directed antibodies (35, 36), had no effect on PGT135 or PGT128 neutralization. Similarly the N295 glycan, which has been observed to be important in a strain-dependent manner (15), had no effect on PGT135 binding. In contrast to PGT135 and PGT128, b12 neutralization of wild-type virus reached 100%. However, neutralization potency was increased by removal of the Asn386 glycan (Fig. 4A). This phenomenon has been previously reported and is thought to arise from improved accessibility to the b12 epitope at the CD4 binding site following removal of this proximal glycan (37, 38). Thus, Env glycans can influence neutralization by both glycan-directed and protein-directed bnAbs via distinct mechanisms.
Neutralization plateaus below 100% have previously been observed with glycan-reactive bnAbs (1, 3, 15, 22, 39). The corresponding resistant populations could arise from either incomplete occupancy or the presence of unfavorable glycan structures at target glycosylation sites. A recent study suggested that most sites were 100% occupied (33), while other work has suggested occupancy may be incomplete for some sites (32). We did not detect unglycosylated peptides at the N392 site (data not shown) but cannot formally exclude the possibility of such a small population existing below our detection limits. While nonoccupancy of a target glycosylation site may offer one mechanism of neutralization resistance, the ability of glycosylation inhibitors such as kifunensine or swainsonine to modulate the plateau of neutralization strongly suggests that the presence of tolerable glycoforms is an important parameter for the efficacy of glycan-reactive bnAbs. This is an important consideration for vaccine design, since any elicited bnAb response will likely require a degree of plasticity in glycan recognition in order to counteract the intrinsic microheterogeneity of gp120 glycosylation. The precise degree of microheterogeneity will likely differ between isolates, as the various distributions of glycan sites may alter the local accessibility of processing enzymes. While this could be considered a potential barrier to targeting the HIV-1 glycan shield for vaccine purposes, recent investigations into the specificity of glycan-reactive bnAbs have suggested that they possess considerable plasticity in their mode of recognition. This has been demonstrated in particular for the Asn332-targeted antibodies, many of which can still neutralize diverse viruses when the Asn332 glycan is shifted or completely removed (30). Thus, while greater insight is needed into the microheterogeneity of key antibody epitopes and the flexibility of bnAb recognition, the elicitation of glycan-targeting bnAbs remains a promising strategy for vaccine design.
ACKNOWLEDGMENTS
We thank Li Phing Liew and Richard Gardner for technical assistance and David J. Harvey and Holger Kramer for helpful discussions.
L.K.P. is supported by a scholarship from the Department of Biochemistry, University of Oxford. M.C. is the Against Breast Cancer Fellow of Oriel College, Oxford. This work was supported by an International AIDS Vaccine Initiative Neutralizing Antibody Center and NAC CAVD grant (M.C.), CHAVI-ID grant 1UM1AI100663 (M.C.), and Medical Research Council grant MR/K024426/1 (K.J.D).
REFERENCES
- 1.Walker LM, Phogat SK, Chan-Hui P-Y, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien J-P, Wang S-K, Ramos A, Chan-Hui P-Y, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong C-H, Phogat S, Wrin T, Simek MD, Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, Ramirez A, Derking R, van Gils MJ, Liang C-H, Mcbride R, von Bredow B, Shivatare SS, Wu C-Y, Chan-Hui P-Y, Liu Y, Feizi T, Zwick MB, Koff WC, Seaman MS, Swiderek K, Moore JP, Evans D, Paulson JC, Wong C-H, Ward AB, Wilson IA, Sanders RW, Poignard P, Burton DR. 2014. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity 40:657–668. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burton DR, Ahmed R, Barouch DH, Butera ST, Crotty S, Godzik A, Kaufmann DE, McElrath MJ, Nussenzweig MC, Pulendran B, Scanlan CN, Schief WR, Silvestri G, Streeck H, Walker BD, Walker LM, Ward AB, Wilson IA, Wyatt R. 2012. A blueprint for HIV vaccine discovery. Cell Host Microbe 12:396–407. doi: 10.1016/j.chom.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crispin M, Bowden TA. 2013. Antibodies expose multiple weaknesses in the glycan shield of HIV. Nat Struct Mol Biol 20:771–772. doi: 10.1038/nsmb.2627. [DOI] [PubMed] [Google Scholar]
- 6.Barouch DH, Whitney JB, Moldt B, Klein F, Oliveira TY, Liu J, Stephenson KE, Chang H-W, Shekhar K, Gupta S, Nkolola JP, Seaman MS, Smith KM, Borducchi EN, Cabral C, Smith JY, Blackmore S, Sanisetty S, Perry JR, Beck M, Lewis MG, Rinaldi W, Chakraborty AK, Poignard P, Nussenzweig MC, Burton DR. 2013. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature 503:224–228. doi: 10.1038/nature12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hessell AJ, Rakasz EG, Poignard P, Hangartner L, Landucci G, Forthal DN, Koff WC, Watkins DI, Burton DR. 2009. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog 5:e1000433. doi: 10.1371/journal.ppat.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mascola JR, Stiegler G, VanCott TC, Katinger H, Carpenter CB, Hanson CE, Beary H, Hayes D, Frankel SS, Birx DL, Lewis MG. 2000. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med 6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 9.Moldt B, Rakasz EG, Schultz N, Chan-Hui P-Y, Swiderek K, Weisgrau KL, Piaskowski SM, Bergman Z, Watkins DI, Poignard P, Burton DR. 2012. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc Natl Acad Sci U S A 109:18921–18925. doi: 10.1073/pnas.1214785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shingai M, Nishimura Y, Klein F, Mouquet H, Donau OK, Plishka R, Buckler-White A, Seaman M, Piatak M, Lifson JD, Dimitrov DS, Nussenzweig MC, Martin MA. 2013. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature 503:277–280. doi: 10.1038/nature12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu X, Yang Z-Y, Li Y, Hogerkorp C-M, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O'Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao H-X, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia S-M, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BTM, Kwong PD, Mascola JR, Haynes BF. 2013. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pancera M, Shahzad-Ul-Hussan S, Doria-Rose NA, McLellan JS, Bailer RT, Dai K, Loesgen S, Louder MK, Staupe RP, Yang Y, Zhang B, Parks R, Eudailey J, Lloyd KE, Blinn J, Alam SM, Haynes BF, Amin MN, Wang L-X, Burton DR, Koff WC, Nabel GJ, Mascola JR, Bewley CA, Kwong PD. 2013. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16. Nat Struct Mol Biol 20:804–813. doi: 10.1038/nsmb.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pejchal R, Doores KJ, Walker LM, Khayat R, Huang P-S, Wang S-K, Stanfield RL, Julien J-P, Ramos A, Crispin M, Depetris R, Katpally U, Marozsan A, Cupo A, Maloveste S, Liu Y, McBride R, Ito Y, Sanders RW, Ogohara C, Paulson JC, Feizi T, Scanlan CN, Wong C-H, Moore JP, Olson WC, Ward AB, Poignard P, Schief WR, Burton DR, Wilson IA. 2011. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong L, Lee JH, Doores KJ, Murin CD, Julien J-P, McBride R, Liu Y, Marozsan A, Cupo A, Klasse P-J, Hoffenberg S, Caulfield M, King CR, Hua Y, Le KM, Khayat R, Deller MC, Clayton T, Tien H, Feizi T, Sanders RW, Paulson JC, Moore JP, Stanfield RL, Burton DR, Ward AB, Wilson IA. 2013. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol 20:796–803. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PNP, Spencer DIR, Seaman MS, Schuitemaker H, Feizi T, Nussenzweig MC, Bjorkman PJ. 2012. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A 109:E3268-77. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Julien J-P, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse P-J, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. 2013. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLellan JS, Pancera M, Carrico C, Gorman J, Julien J-P, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O'Dell S, Patel N, Shahzad-ul Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang G-Y, Diwanji D, Georgiev I, Kwon Do Y, Lee D, Louder MK, Moquin S, Schmidt SD, Yang Z-Y, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang L-X, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Julien J-P, Lee JH, Cupo A, Murin CD, Derking R, Hoffenberg S, Caulfield MJ, King CR, Marozsan AJ, Klasse PJ, Sanders RW, Moore JP, Wilson IA, Ward AB. 2013. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc Natl Acad Sci U S A 110:4351–4356. doi: 10.1073/pnas.1217537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blattner C, Lee JH, Sliepen K, Derking R, Falkowska E, de la Peña AT, Cupo A, Julien J-P, van Gils M, Lee PS, Peng W, Paulson JC, Poignard P, Burton DR, Moore JP, Sanders RW, Wilson IA, Ward AB. 2014. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity 40:669–680. doi: 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scharf L, Scheid JF, Lee JH, West AP, Chen C, Gao H, Gnanapragasam PNP, Mares R, Seaman MS, Ward AB, Nussenzweig MC, Bjorkman PJ. 2014. Antibody 8ANC195 reveals a site of broad vulnerability on the HIV-1 envelope spike. Cell Rep 7:785–795. doi: 10.1016/j.celrep.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang J, Kang BH, Pancera M, Lee JH, Tong T, Feng Y, Georgiev IS, Chuang G-Y, Druz A, Doria-Rose NA, Laub L, Sliepen K, van Gils MJ, de la Peña AT, Derking R, Klasse P-J, Migueles SA, Bailer RT, Alam M, Pugach P, Haynes BF, Wyatt RT, Sanders RW, Binley JM, Ward AB, Mascola JR, Kwong PD, Connors M. 2014. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41–gp120 interface. Nature 515:138–142. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leonard CK, Spellman MW, Riddle L, Harris RJ, Thomas JN, Gregory TJ. 1990. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem 265:10373–10382. [PubMed] [Google Scholar]
- 24.Doores KJ, Bonomelli C, Harvey DJ, Vasiljevic S, Dwek RA, Burton DR. 2010. Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc Natl Acad Sci U S A 107:13800–13805. doi: 10.1073/pnas.1006498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonomelli C, Doores KJ, Dunlop DC, Thaney V, Dwek RA, Burton DR, Crispin M, Scanlan CN. 2011. The glycan shield of HIV is predominantly oligomannose independently of production system or viral clade. PLoS One 6:e23521. doi: 10.1371/journal.pone.0023521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Julien J-P, Sok D, Khayat R, Lee JH, Doores KJ, Walker LM, Ramos A, Diwanji DC, Pejchal R, Cupo A, Katpally U, Depetris RS, Stanfield RL, McBride R, Marozsan AJ, Paulson JC, Sanders RW, Moore JP, Burton DR, Poignard P, Ward AB, Wilson IA. 2013. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog 9:e1003342. doi: 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scanlan CN, Pantophlet R, Wormald MR, Saphire EO, Stanfield R, Wilson IA, Katinger H, Dwek RA, Rudd PM, Burton DR. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J Virol 76:7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanders RW, Venturi M, Schiffner L, Kalyanaraman R, Katinger H, Lloyd KO, Kwong PD, Moore JP. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J Virol 76:7293–7305. doi: 10.1128/JVI.76.14.7293-7305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murin CD, Julien J-P, Sok D, Stanfield RL, Khayat R, Cupo A, Moore JP, Burton DR, Wilson IA, Ward AB. 2014. Structure of 2G12 Fab2 in complex with soluble and fully glycosylated HIV-1 Env by negative-stain single particle electron microscopy. J Virol 88:10177–10188. doi: 10.1128/JVI.01229-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sok D, Doores KJ, Briney B, Le KM, Saye-Francisco KL, Ramos A, Kulp DW, Julien J-P, Menis S, Wickramasinghe L, Seaman MS, Schief WR, Wilson IA, Poignard P, Burton DR. 2014. Promiscuous glycan site recognition by antibodies to the high-mannose patch of gp120 broadens neutralization of HIV. Sci Transl Med 6:236ra63. doi: 10.1126/scitranslmed.3008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu X, Borchers C, Bienstock RJ, Tomer KB. 2000. Mass spectrometric characterization of the glycosylation pattern of HIV-gp120 expressed in CHO cells. Biochemistry 39:11194–11204. doi: 10.1021/bi000432m. [DOI] [PubMed] [Google Scholar]
- 32.Go EP, Irungu J, Zhang Y, Dalpathado DS, Liao H, Sutherland LL, Alam SM, Haynes BF, Desaire H. 2008. Glycosylation site-specific analysis of HIV envelope proteins (JR-FL and CON-S) reveals major differences in glycosylation site occupancy, glycoform profiles, and antigenic epitopes' accessibility. J Proteome Res 7:1660–1674. doi: 10.1021/pr7006957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pabst M, Chang M, Stadlmann J, Altmann F. 2012. Glycan profiles of the 27 N-glycosylation sites of the HIV envelope protein CN54gp140. Biol Chem 393:719–730. doi: 10.1515/hsz-2012-0148. [DOI] [PubMed] [Google Scholar]
- 34.Doores KJ, Fulton Z, Huber M, Wilson IA, Burton DR. 2010. Antibody 2G12 recognizes di-mannose equivalently in domain- and nondomain-exchanged forms but only binds the HIV-1 glycan shield if domain exchanged. J Virol 84:10690–10699. doi: 10.1128/JVI.01110-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garces F, Sok D, Kong L, McBride R, Kim HJ, Saye-Francisco KF, Julien J-P, Hua Y, Cupo A, Moore JP, Paulson JC, Ward AB, Burton DR, Wilson IA. 2014. Structural evolution of glycan recognition by a family of potent HIV antibodies. Cell 159:69–79. doi: 10.1016/j.cell.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doores KJ, Kong L, Krumm SA, Le KM, Sok D, Laserson U, Garces F, Poignard P, Wilson IA, Burton DR. 2015. Two classes of broadly neutralizing antibodies within a single lineage directed to the high-mannose patch of HIV envelope. J Virol 89:1105–1118. doi: 10.1128/JVI.02905-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koch M, Pancera M, Kwong PD, Kolchinsky P, Grundner C, Wang L, Hendrickson WA, Sodroski J, Wyatt R. 2003. Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology 313:387–400. doi: 10.1016/S0042-6822(03)00294-0. [DOI] [PubMed] [Google Scholar]
- 38.Duenas-Decamp MJ, Peters P, Burton D, Clapham PR. 2008. Natural resistance of human immunodeficiency virus type 1 to the CD4bs antibody b12 conferred by a glycan and an arginine residue close to the CD4 binding loop. J Virol 82:5807–5814. doi: 10.1128/JVI.02585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doores KJ, Burton DR. 2010. Variable loop glycan dependency of the broad and potent HIV-1-neutralizing antibodies PG9 and PG16. J Virol 84:10510–10521. doi: 10.1128/JVI.00552-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harvey DJ, Merry AH, Royle L, Campbell MP, Dwek RA, Rudd PM. 2009. Proposal for a standard system for drawing structural diagrams of N- and O-linked carbohydrates and related compounds. Proteomics 9:3796–3801. doi: 10.1002/pmic.200900096. [DOI] [PubMed] [Google Scholar]
- 41.Aricescu AR, Lu W, Jones EY. 2006. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr 62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 42.Harvey DJ, Sobott F, Crispin M, Wrobel A, Bonomelli C, Vasiljevic S, Scanlan CN, Scarff CA, Thalassinos K, Scrivens JH. 2011. Ion mobility mass spectrometry for extracting spectra of N-glycans directly from incubation mixtures following glycan release: application to glycans from engineered glycoforms of intact, folded HIV gp120. J Am Soc Mass Spectrom 22:568–581. doi: 10.1007/s13361-010-0053-0. [DOI] [PubMed] [Google Scholar]
- 43.Wuhrer M, Catalina MI, Deelder AM, Hokke CH. 2007. Glycoproteomics based on tandem mass spectrometry of glycopeptides. J Chromatogr B Analyt Technol Biomed Life Sci 849:115–128. doi: 10.1016/j.jchromb.2006.09.041. [DOI] [PubMed] [Google Scholar]