Abstract
Epstein-Barr virus latent membrane protein 2A (LMP2A) induces many characteristics of carcinoma, including transformation, migration, invasion, and impaired differentiation. The MCF10A cell line differentiates to form hollow acini when grown in Matrigel, and expression of LMP2A inhibited differentiation and anoikis induced by loss of matrix attachment. LMP2A-infected cells formed large, lobular structures rather than hollow acini. Autophagy inhibitors impaired this abnormal growth and induced caspase 3 activation and acinus formation. LMP2A also increased autophagosome formation and expression of proteins in the autophagosome pathway. These findings suggest that LMP2A may inhibit anoikis and luminal clearance in acini through induction of autophagy.
TEXT
Epstein-Barr virus (EBV) latent membrane protein 2A (LMP2A) is consistently detected at the RNA and protein levels in the epithelial cancer nasopharyngeal carcinoma (1–4). LMP2A has been shown to impair differentiation and induce migration, which likely contributes to the development of epithelial cancers. Human MCF10A cells are nonmalignant breast epithelial cells that undergo distinct differentiation when grown in three-dimensional culture and form spherical acini with a characteristic hollow lumen. Many oncogenes affect this process and block acinus formation, a process that is dependent on anoikis or apoptosis induced by lack of attachment to the extracellular matrix. Previously, LMP2A has been shown to block anoikis in MCF10A cells and to induce the formation of large, irregular growths with a filled lumen (5). The N-terminal src binding domain and the syc-binding immunoreceptor tyrosine-based activation motif were required for inhibition of anoikis (5). In contrast, mutation of the PY domain known to bind NEDD4 ubiquitin ligases, including Itch, did not block anoikis but rather resulted in the formation of acini that were hollow, spherical, and larger than those formed in control cells. This finding suggested that PY interactions with NEDD4-like ubiquitin ligases are important for impairment of anoikis (5).
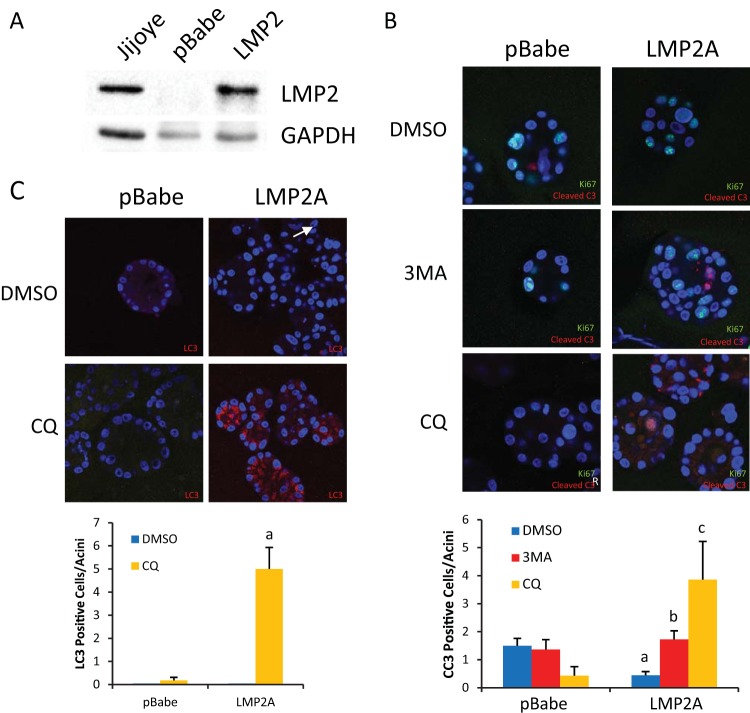
Autophagy is a process that provides energy conservation to cells undergoing stress and is thought to inhibit anoikis by prolonging cell viability until matrix reattachment occurs (6–11). Interestingly, a high-throughput screen for ubiquitin ligase inhibitors recently identified clomipramine as an inhibitor of Itch and identified a role for Itch in the regulation of autophagy (12). To test the hypothesis that LMP2A blocked anoikis through induction of autophagy, MCF10A cell lines that contained the pBabe vector control or expressed wild-type LMP2A were established. The LMP2A expression levels were comparable to those of the Jijoye lymphocyte line (Fig. 1A). The cells were seeded in three-dimensional culture to induce acinus formation in the presence of autophagy inhibitors. Treatment with the type III phosphoinositol 3-kinase inhibitor 3-methyladenine (3-MA; 10 mM, days 6 to 8), which blocks the activation of the autophagosome (AP) initiation complex, induced increased cleaved caspase 3 in the LMP2A spheres with evidence of luminal clearing, suggesting that the autophagy initiation complex is required for resistance to anoikis and impaired lumen formation (Fig. 1B). Some of the LMP2A-expressing cells within the lumen remained positive for staining with Ki67 as an indicator of DNA synthesis. Treatment of LMP2A acini with the late-stage autophagy inhibitor chloroquine (CQ; 30 μg/ml) also induced luminal clearing and significantly higher levels of cleaved caspase 3 (Fig. 1B). Treatment with either inhibitor did not affect the formation of acini in vector control cells.
FIG 1.
Autophagy inhibitors induce the formation of acini and inhibit luminal filling in LMP2A-expressing cells. (A) LMP2A expression in pBabe vector control or LMP2A-expressing MCF10A cells compared to that in EBV+ Jijoye lymphoid cells. (B) Vector control and LMP2A-expressing MCF10A cells were grown for 6 days in Matrigel and treated with the DMSO vehicle control or the autophagy inhibitor 3-MA or CQ from day 6 to day 8. On day 8, acini were fixed and stained for Ki67 (green), a marker of proliferation, cleaved caspase 3 (CC3, red), or 4′,6-diamidino-2-phenylindole (DAPI) (blue) to visualize nuclei. The numbers of CC3-positive cells per acinus were determined, and means were calculated from seven images and are presented graphically. Bars: a, P = 0.00033 for LMP2A versus pBabe DMSO; b, P = 0.000094 for LMP2A 3-MA treated cells versus LMP2A DMSO; c, P = 0.000024 for LMP2A CQ versus LMP2A DMSO. (C) pBabe vector control and LMP2A-expressing cells grown in Matrigel were treated with CQ from day 6 to day 8 and stained with LC3 (red) to identify APs and with DAPI (blue) to visualize nuclei. The mean values of pBabe LC3-positive cells versus those of LMP2A LC3-positive cells are presented graphically (a, P = 0.00029). Acinus images were acquired with a 63× oil objective on a Zeiss 710 confocal laser scanning microscope and are representative of three experiments.
Treatment of acini with CQ blocks autophagy through inhibition of enzymes that require an acidic environment. Thus, CQ treatment prevents the degradation of AP components and stabilizes APs. APs can be identified by the binding of LC3II, which is produced by lipidation of LC3 in a process that resembles ubiquitination. Treatment with CQ induces AP accumulation and enables the detection of immunofluorescent LC3II. LMP2A MCF10A cells treated with dimethyl sulfoxide (DMSO) during growth in Matrigel (days 6 to 8) formed characteristic multilobular structures, whereas CQ treatment inhibited autophagy, resulting in spherical acini, some evidence of hollowing, and significantly increased detection of LC3II, indicating that autophagy had been activated by LMP2A (Fig. 1C).
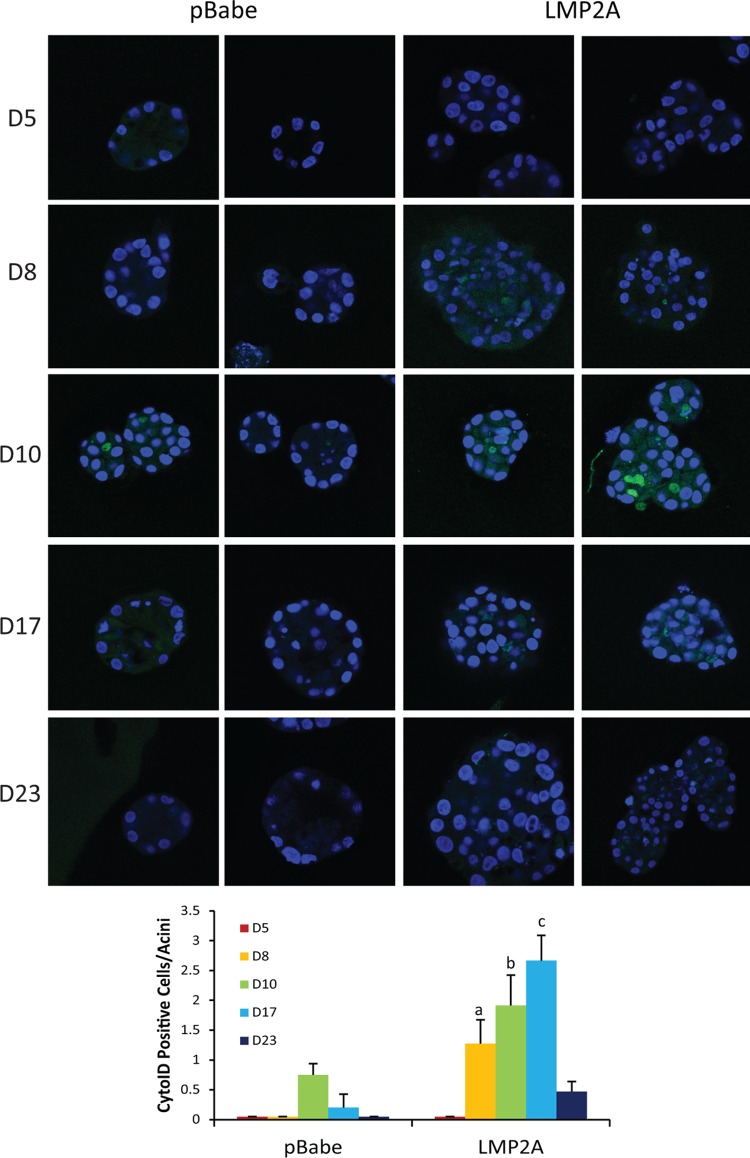
To determine when LMP2A induced autophagy, APs were detected with a stain specific for APs (Enzo Life Sciences Cyto-ID staining kit) in vector control and LMP2A-expressing MCF10A cells from day 5 to day 23 during growth in Matrigel (Fig. 2). Positive staining for APs was detected in LMP2A acini at low levels by day 8, with strong detection on days 10 and 17. In contrast, pBabe acini had low AP levels detectable only on day 10. These findings indicate that LMP2A produced AP formation greater than that in vector control cells.
FIG 2.
LMP2A induces autophagy in MCF10A acini. MCF10A acini were grown from pBabe- and LMP2A-expressing cells for 23 days. Acini were stained with the Cyto-ID autophagy detection reagent and Hoechst 33342 to visualize nuclei on days 5, 8, 10, 17, and 23. Two representative images from each time point are shown. Images were acquired with a 63× oil objective on a Zeiss 710 confocal laser scanning microscope and are representative of two experiments. The numbers of Cyto-ID-positive cells per acinus were determined, and mean values are presented graphically. Bars: a, P = 0.019 versus pBabe; b, P = 0.034 versus pBabe; c, P = 0.00081 versus pBabe. Mean values were calculated from five images.
To test the hypothesis that LMP2A induced autophagy to promote anoikis resistance following detachment, pBabe vector control and wild-type LMP2A- and PY mutant LMP2A-expressing MCF10A cells were cultured for 24 h in dishes coated with poly(2-hydroxyethyl methacrylate) (polyHEMA) at 10 mg/ml with or without CQ treatment. After 24 h, suspension cells were stained with the Cyto-ID staining to kit to label APs. Hoechst 33342 nuclear staining indicated that LMP2A-expressing cells formed large clumps when cultured on HEMA plates, in contrast to single cells in the pBabe vector control or small colonies formed by PY mutant cells (Fig. 3). Trace levels of APs were detected in pBabe vector control cells, with higher AP levels in LMP2A- and PY-expressing cells. Treatment with CQ enhanced AP detection. These findings reveal that LMP2A induced AP formation but this induction did not require the PY domain (Fig. 3).
FIG 3.
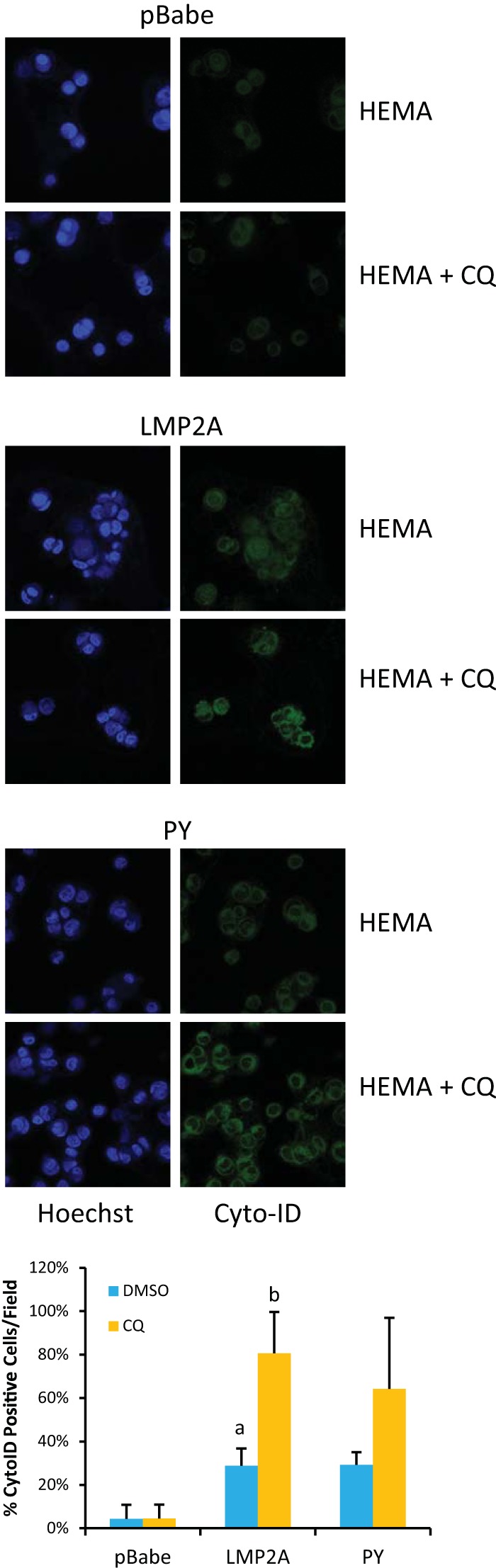
LMP2A potentiates autophagy induced by suspension culture. MCF10A cells expressing the pBabe vector control, wild-type LMP2A, or PY mutant LMP2A were seeded onto polyHEMA-coated cell culture dishes for 24 h. Autophagy was detected by visualization of autophagic vesicles following treatment with the Cyto-ID autophagy detection reagent (green). Nuclei were visualized with Hoechst 33342 (blue). Images were acquired with a 63× oil objective on a Zeiss 710 confocal laser scanning microscope. The number of Cyto-ID positive cells was determined from three experiments, and mean values are expressed graphically as percentages of positive cells per field. Bars: a, P = 0.00067 for LMP2A versus pBabe DMSO; b, P = 0.00014 for LMP2A CQ versus LMP2A DMSO. The percentage of Cyto-ID-positive LMP2A PY cells was increased, although the P value of the mean change was not significant.
To confirm AP formation in HEMA cultures, whole-cell lysates were analyzed by immunoblotting 24 h after seeding (Fig. 4A). Slightly lower levels of cleaved poly(ADP-ribose) polymerase 1 (PARP), as an indicator of cell death, were detected in LMP2A-expressing cells than in pBabe vector control cells. LMP2A-expressing cells also had slightly greater expression of Atg7, which is activated early in the induction of autophagy. LC3B expression was increased in LMP2A-expressing cells, and treatment with protease inhibitors enhanced its detection (Fig. 4A). To further assess the activation of autophagy, the formation of the Atg5/Atg12 E3-like ligase complex, which is required for LC3 lipidation, was assessed by determining the association of Atg5 with Atg12. Immunoprecipitation of Atg5 from whole-cell lysates, followed by Western blotting for Atg12, indicated increased complex formation in LMP2A-expressing cells, while the levels in PY mutant LMP2A-expressing cells were comparable to those in vector control cells (Fig. 4B). These data indicate that following matrix detachment, LMP2A increases the expression of AP proteins and the formation of ATG complexes.
FIG 4.
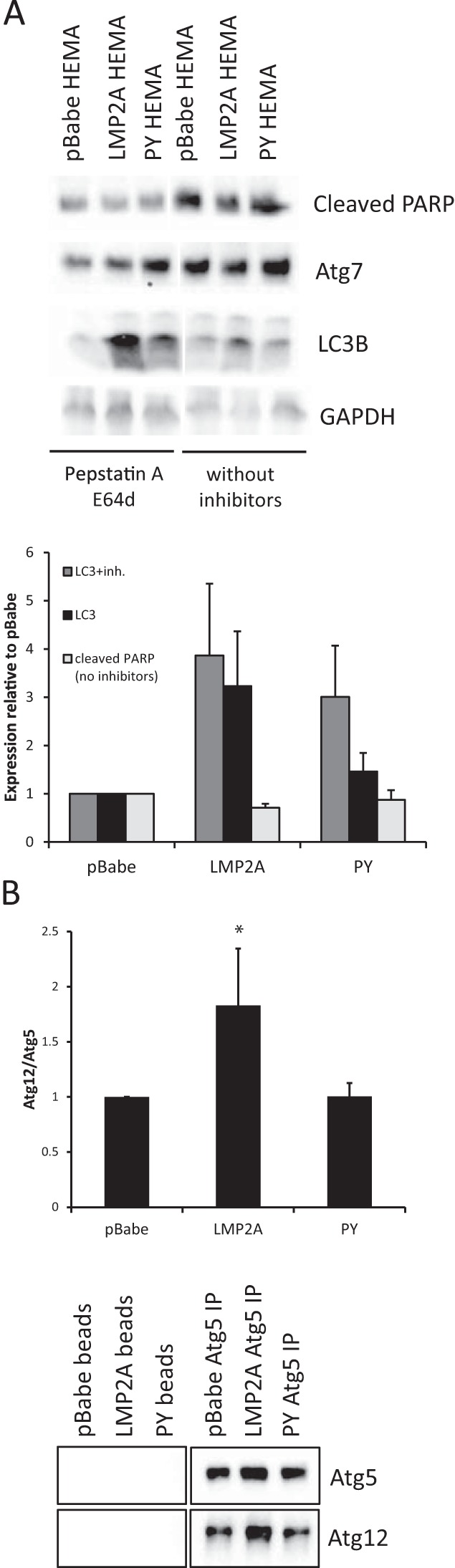
LMP2A promotes expression of LC3A/B-II following autophagy induction. (A) MCF10A cells expressing the pBabe vector control, wild-type LMP2A, or PY mutant LMP2A were cultured on polyHEMA-coated tissue culture plates for 24 h to induce autophagy. Cleaved PARP, LC3A/B, Atg7, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) protein expression was measured by Western blotting of whole-cell lysates. The protease inhibitors (inh.) pepstatin A and E64d were added where indicated for the last 4 h of culture to enhance LC3 detection. Western blot assays were quantitated and normalized to GAPDH, and the results are expressed relative to those obtained with pBabe (n = 4 for LC3; n = 3 for PARP). (B) Atg5 protein complexes were immunoprecipitated from whole-cell lysates, and Atg5 association with Atg12 was detected by Western blotting. Atg12 expression was normalized to Atg5 and expressed relative to that of pBabe controls (n = 3; *, P = 0.049).
These findings reveal that inhibition of autophagy impaired LMP2A-mediated anoikis resistance in MCF10A cells, resulting in the formation of hollow lumens and the activation of caspase 3. Additionally, induction of detachment-induced cell death following culture on polyHEMA-coated culture plates revealed that LMP2A increased the expression of proteins required for AP formation, including the formation of the Atg5/Atg12 complex and LC3. These findings indicate that by increasing the expression of proteins required for AP formation, LMP2A increases autophagy to maintain viability after loss of matrix attachment. The LMP2A-mediated induction of autophagy in detached luminal cells likely contributes to the impaired development of acini and indicates that LMP2A uses autophagy to evade anoikis. Enhanced autophagy is likely a critical factor in the inhibition of acinus formation and continued growth of luminal cells induced by LMP2A (6). Evasion of anoikis is thought to be a critical ability for the metastasis of malignant cells. The ability of LMP2A to induce migration is also thought to potentially contribute to metastasis. Thus, the properties of LMP2A that inhibit differentiation, induce migration, and block anoikis through induction of autophagy are likely to be critical to the contribution of LMP2A to the metastasis of EBV-positive carcinomas.
ACKNOWLEDGMENTS
Aron Marquitz assisted with figure preparation and critically read the manuscript.
This study was supported by NCI CA 32979 and CA 19014 to N.R.-T.
REFERENCES
- 1.Brooks L, Yao QY, Rickinson AB, Young LS. 1992. Epstein-Barr virus latent gene transcription in nasopharyngeal carcinoma cells: coexpression of EBNA1, LMP1, and LMP2 transcripts. J Virol 66:2689–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Busson P, McCoy R, Sadler R, Gilligan K, Tursz T, Raab-Traub N. 1992. Consistent transcription of the Epstein-Barr virus LMP2 gene in nasopharyngeal carcinoma. J Virol 66:3257–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lan YY, Hsiao JR, Chang KC, Chang JS, Chen CW, Lai HC, Wu SY, Yeh TH, Chang FH, Lin WH, Su IJ, Chang Y. 2012. Epstein-Barr virus latent membrane protein 2A promotes invasion of nasopharyngeal carcinoma cells through ERK/Fra-1-mediated induction of matrix metalloproteinase 9. J Virol 86:6656–6667. doi: 10.1128/JVI.00174-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heussinger N, Buttner M, Ott G, Brachtel E, Pilch BZ, Kremmer E, Niedobitek G. 2004. Expression of the Epstein-Barr virus (EBV)-encoded latent membrane protein 2A (LMP2A) in EBV-associated nasopharyngeal carcinoma. J Pathol 203:696–699. doi: 10.1002/path.1569. [DOI] [PubMed] [Google Scholar]
- 5.Fotheringham JA, Raab-Traub N. 2013. Epstein-Barr virus latent membrane protein 2 effects on epithelial acinus development reveal distinct requirements for the PY and YEEA motifs. J Virol 87:13803–13815. doi: 10.1128/JVI.02203-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avivar-Valderas A, Salas E, Bobrovnikova-Marjon E, Diehl JA, Nagi C, Debnath J, Aguirre-Ghiso JA. 2011. PERK integrates autophagy and oxidative stress responses to promote survival during extracellular matrix detachment. Mol Cell Biol 31:3616–3629. doi: 10.1128/MCB.05164-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Debnath J. 2008. Detachment-induced autophagy during anoikis and lumen formation in epithelial acini. Autophagy 4:351–353. doi: 10.4161/auto.5523. [DOI] [PubMed] [Google Scholar]
- 8.Debnath J. 2009. Detachment-induced autophagy in three-dimensional epithelial cell cultures. Methods Enzymol 452:423–439. doi: 10.1016/S0076-6879(08)03625-2. [DOI] [PubMed] [Google Scholar]
- 9.Fung C, Lock R, Gao S, Salas E, Debnath J. 2008. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell 19:797–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green DR, Levine B. 2014. To be or not to be? How selective autophagy and cell death govern cell fate. Cell 157:65–75. doi: 10.1016/j.cell.2014.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levine B, Kroemer G. 2008. Autophagy in the pathogenesis of disease. Cell 132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossi M, Rotblat B, Ansell K, Amelio I, Caraglia M, Misso G, Bernassola F, Cavasotto CN, Knight RA, Ciechanover A, Melino G. 2014. High throughput screening for inhibitors of the HECT ubiquitin E3 ligase ITCH identifies antidepressant drugs as regulators of autophagy. Cell Death Dis 5:e1203. doi: 10.1038/cddis.2014.113. [DOI] [PMC free article] [PubMed] [Google Scholar]