ABSTRACT
Giant viruses are protist-associated viruses belonging to the proposed order Megavirales; almost all have been isolated from Acanthamoeba spp. Their isolation in humans suggests that they are part of the human virome. Using a high-throughput strategy to isolate new giant viruses from their original protozoan hosts, we obtained eight isolates of a new giant viral lineage from Vermamoeba vermiformis, the most common free-living protist found in human environments. This new lineage was proposed to be the faustovirus lineage. The prototype member, faustovirus E12, forms icosahedral virions of ≈200 nm that are devoid of fibrils and that encapsidate a 466-kbp genome encoding 451 predicted proteins. Of these, 164 are found in the virion. Phylogenetic analysis of the core viral genes showed that faustovirus is distantly related to the mammalian pathogen African swine fever virus, but it encodes ≈3 times more mosaic gene complements. About two-thirds of these genes do not show significant similarity to genes encoding any known proteins. These findings show that expanding the panel of protists to discover new giant viruses is a fruitful strategy.
IMPORTANCE By using Vermamoeba, a protist living in humans and their environment, we isolated eight strains of a new giant virus that we named faustovirus. The genomes of these strains were sequenced, and their sequences showed that faustoviruses are related to but different from the vertebrate pathogen African swine fever virus (ASFV), which belongs to the family Asfarviridae. Moreover, the faustovirus gene repertoire is ≈3 times larger than that of ASFV and comprises approximately two-thirds ORFans (open reading frames [ORFs] with no detectable homology to other ORFs in a database).
INTRODUCTION
Giant viruses were first described in 2003, with the discovery of Acanthamoeba polyphaga mimivirus (1, 2). They are protist-associated viruses that belong to a major monophyletic group of double-stranded DNA (dsDNA) viruses known as nucleocytoplasmic large DNA viruses (NCLDVs), and they have been classified under the proposed order Megavirales (3). Since the first description of Acanthamoeba polyphaga mimivirus, giant viruses have been isolated from other phagocytic protists, primarily Acanthamoeba spp. (4–6). Because these giant viruses are resistant to killing by phagocytic protists, we hypothesized that they may also reproduce in macrophages and might therefore infect humans. This proposition was validated experimentally by the isolation of mimivirus from atypical pneumonia patients and by the detection of marseilleviruses in blood donors and in human lymph nodes (7–9). Moreover, we and others identified sequences associated with giant viruses in metagenomes generated from human tissues, suggesting that giant viruses are a component of the human virome (10). Because the investigation of a virome typically starts with a filtration procedure that eliminates giant viruses (11), we developed a new culture approach that does not prevent the detection of these viruses.
In the present study, we developed a high-throughput strategy to isolate new giant viruses from 102 environmental samples. In addition to the commonly studied species A. polyphaga, we also assessed five other protists that were never previously used to isolate giant viruses, including Vermamoeba vermiformis, the most common free-living protist found in human environments (12, 13).
MATERIALS AND METHODS
Culture procedures.
As the support for the coculture, we used Vermamoeba vermiformis (strain CDC19). V. vermiformis strain CDC19 was maintained in a 75-cm2 cell culture flask with 30 ml of peptone-yeast extract-glucose (PYG) medium at 32°C as previously described for Acanthamoeba sp. (14, 15). After 48 h, cells were harvested and pelleted by centrifugation. The supernatant was removed, and the amoebae were resuspended in sterile Page's amoebal saline (PAS). Centrifugation and resuspension in PAS were repeated twice. After the last centrifugation step, the amoebae were resuspended in 30 ml of starvation medium with an antibiotic mix at a concentration of approximately 106 amoebae/ml. Samples (100 μl) were inoculated onto amoebae (500 μl in a 24-well plate) and incubated at 32°C in a humid environment. The starvation medium was composed of 1 liter of distilled water with 120 mg NaCl, 4 mg MgSO4 · 7H2O, 4 mg CaCl2 · 2H2O, 142 mg Na2HPO4 · 7H2O, 136 mg KH2PO4, 0.02 g (NH4)2Fe(SO4)2 · 6H2O, 2 g yeast extract, 18 g glucose, and an antimicrobial agent mix containing 10 μg/ml vancomycin (Meylan, Saint-Priest, France), 10 μg/ml imipenem, 20 μg/ml ciprofloxacin (Panpharma, Z.I. du Clairay, France), and 30 μg/ml thiabendazole (Sigma-Aldrich). These cocultures were incubated for 2 days and then subcultured as described above on fresh amoebae without any antibiotics. Sewage samples (24 from Marseille, France, and 7 from Dakar, Senegal) and 71 seawater/sediment samples were prepared as described previously (4).
Electron microscopy.
For preparation for transmission electron microscopy (TEM) observation, V. vermiformis-infected cells were recovered and pelleted for 10 min at 5,000 × g. The pellet was resuspended in 1 ml of phosphate-buffered saline (PBS) with 2% glutaraldehyde–0.1 M cacodylate and incubated for at least 1 h at 4°C. Each pellet was then washed three times with 0.1 M cacodylate–saccharose and resuspended in the same buffer. After repelleting, each sample was then embedded in Epon resin by using a standard method, as follows: 1 h of fixation in 1% osmium tetroxide, two washes in distilled water, dehydration in increasing ethanol concentrations (30%, 50%, 70%, 96%, and 100% ethanol), and embedding in Epon-812. Ultrathin (70 nm) sections were poststained with 5% uranyl acetate and lead citrate according to the Reynolds method (16) and were observed using a Morgagni 268 D TEM (Philips) operating at 60 keV and a Tecnai G2 TEM operating at 200 keV. Negative staining of faustovirus particles was performed using a 5% solution of ammonium molybdate and 1% trehalose.
Flow cytometric analyses. (i) Detection of amoeba lysis by fluorescence-activated cell sorting (FACS).
For the detection of amoeba lysis, 250 μl of each specimen was transferred to an adapted tube for analysis in a BD LSR Fortessa cytometer (BD Biosciences). The data acquisition and analysis were performed with BD FACSDiva software. Data acquisition was realized according to the parameters of size and structure (forward scatter [FSC] and side scatter [SSC]). Protozoal lysis associated with virus infection was detected by cytometer analysis with an arbitrarily designated threshold of 50% lysis. Detection of mimivirus and marseillevirus DNAs was performed by PCR as previously described (17).
(ii) Analyses for viral quantification.
For flow cytometry analyses for viral quantification, we used a BD LSR Fortessa cell analyzer (Becton Dickinson) equipped with 3 lasers (purple [405 nm], blue [488 nm], and red [633 nm]) (18). We performed faustovirus particle quantification using a suspension of fluorescent microspheres (Cytocount) as a reference population. The absolute number of viral cells (cells per microliter) in each sample was calculated using the following equation: (number of cells counted/number of Cytocount beads counted) × Cytocount bead concentration (1,100 beads/μl) × dilution factor. The parameters were adjusted using purified diluted concentrations of faustovirus with and without beads. Data acquisition was completed according to the parameters of size and structure (FSC and SSC), using Pacific Blue to visualize the DAPI (4′,6-diamidino-2-phenylindole) stain and fluorescein isothiocyanate (FITC) to visualize the Sybr green stain. We fixed thresholds for each parameter, and the logarithmic mode was used for each. Flow cytometry was performed on different dilutions (from 10−1 to 10−14). To 500 μl of each dilution, 500 μl of 0.1% Triton in PBS was added to permeabilize the viral cell wall. Cells were pelleted at 13,000 × g for 5 min in a microcentrifuge tube and resuspended in 1 ml of PBS. Each sample was stained with 1 μl of 1-μg/μl DAPI dye (Invitrogen). Samples were incubated for a minimum of 30 min at room temperature in the dark, and 25 μl of Cytocount beads was added to each sample before processing. The total number of recorded events was 10,000 for cell counting using a BD LSR Fortessa cell analyzer. Data analysis was performed using BD FACSDiva 6.2 software, and we created one-dimensional gates in the histogram for cells stained with DAPI, cells stained with Sybr green, and the liquid-containing fluorescent beads. Data acquisition and analysis were performed with BD FACSDiva software, according to the size and structure parameters (FSC and SSC). The number of events using each gate was calculated, and the viral load in 1 ml of sample was determined using the above equation. The results of this technique were compared to those of the routine endpoint dilution technique used by our lab to estimate viral concentrations (4).
Developmental cycle study.
V. vermiformis seeded at 106 cells/ml in starvation medium was infected with titrated faustovirus at a concentration of 107 particles/ml, with an amoeba cell:virus ratio of 1:10. The viral concentration was quantified by a flow cytometric technique used for microorganism enumeration, using fluorescent beads as a reference population. Amoeba viability was estimated by counting the cells on Kovaslides (Kova Glasstic slides; Hycor Biomedical Inc., Garden Grove, CA) immediately after centrifugation and every 2 h for the next 24 h and by flow cytometric quantification using beads after fixation in 3% paraformaldehyde.
DNA extractions and real-time PCR were performed using 200 μl of each coculture taken at every infection time point of the cycle (0 h and then every 2 h for 24 h). Automated extraction by use of an EZ1 DNA virus minikit (Qiagen, Hilden, Germany) was used for DNA extraction on a CFX96 thermocycler according to the manufacturer's instructions (Bio-Rad Laboratories Inc.). The following primers targeting the DNA-directed RNA polymerase subunit 1 gene were used: Fstv_S2F, 5′-CCA GGA CAT GAT GGT CAC ATA G-3′ (forward); and Fstv_S2R, 5′-TTG CAC CTC CGC AGT TAA A-3′ (reverse). Fstv_S2P (6-carboxyfluorescein [FAM]-TATGCTCCAATGGCCTTCAACGACA-6-carboxytetramethylrhodamine [TAMRA]) was used as a probe.
Quantification of the developmental cycle study results was also accomplished using flow cytometry (SI). V. vermiformis-infected cell cultures corresponding to each time point of infection were fixed by adding an equal volume of PBS to 2% glutaraldehyde and incubating them for 20 min at 4°C. The cells were then prepared for observation (SI).
Cryo-EM.
Samples for cryo-electron microscopy (cryo-EM) were prepared following published protocols. Purified particles were frozen in liquid ethane by using a Cryoplunge 3 instrument (Gatan, CA) on UC-A holey carbon grids (Ted Pella, CA). The virus was imaged using a Titan Krios electron microscope at a nominal magnification of ×47,000, with a 4,000-by-4,000 charge-coupled device (CCD) camera (Gatan, CA). A total of 660 particles were boxed using e2boxer from the EMAN2 software package (19). Due to the large size of the particles and their limited number, no attempts were made to correct the contrast transfer function (CTF). The images were instead low-pass filtered to a resolution below the first node in the CTF. An initial model was obtained by identifying the icosahedral 2-fold, 3-fold, and 5-fold axes in a subset of the images and combining them. A reconstruction assuming icosahedral symmetry was carried out using the EMAN program (20) and converged within 8 cycles.
Genomics. (i) Genome sequencing and assembly.
The genomic DNA of faustovirus was sequenced using MiSeq technology (Illumina Inc., San Diego, CA) with paired-end and mate pair applications. The paired-end and mate pair samples were bar coded and prepared with a Nextera XT DNA sample prep kit (Illumina) and a Nextera mate pair sample prep kit (Illumina). The faustovirus DNA was quantified as 10.3 ng/μl by use of a Qubit assay with a high-sensitivity kit (Life Technologies, Carlsbad, CA). Dilution was performed to obtain the required 1 ng for input to prepare the paired-end library. At the “tagmentation” step, DNA was fragmented, with an optimal size distribution at 1.2 kb, and tagged. Limited-cycle PCR amplification (12 cycles) was then completed to tag the adapters and introduce the dual-index bar codes. After purification with AMPure XP beads (Beckman Coulter Inc., Fullerton, CA), the libraries were normalized on specific beads according to the Nextera XT protocol (Illumina). They were then pooled into a single library for MiSeq sequencing. The pooled single-stranded library was loaded onto the reagent cartridge and then onto the instrument along with the flow cell. Automated cluster generation and paired-end sequencing with the dual-index reads were performed in a single 39-h run, providing 2 × 250-bp fragments. A total of 8.7 Gb of information was obtained from a cluster density of 1,006,000/mm2, with a cluster-passing quality control filter of 79.2% (21,480,000 clusters). Within this run, the index representation of the faustovirus was determined to be 6.25%. The 1,063,427 reads were filtered according to their quality. The mate pair library was prepared with 1 μg of genomic DNA, using an Illumina Nextera mate pair guide. The genomic DNA sample was simultaneously fragmented and tagged with a mate pair junction adapter. The profile of the fragmentation was validated on an Agilent 2100 BioAnalyzer with a DNA 7500 LabChip (Agilent Technologies Inc., Santa Clara, CA). The resulting DNA fragments ranged in size from 1.3 kb up to 11 kb, with an optimal size of 6 kb. No size selection was performed, and 600 ng of the tagged fragments was circularized. The circularized DNA was mechanically sheared on a Covaris S2 device in microtubes (Covaris, Woburn, MA) to obtain small fragments with an optimal size of 780 bp. The library profile was visualized on a high-sensitivity LabChip bioanalyzer (Agilent). The libraries were then normalized at 2 nM and pooled. After a denaturation step and dilution to 10 pM, the pooled libraries were loaded onto the reagent cartridge and into the instrument along with the flow cell. Automated cluster generation and sequencing runs were performed in a single 42-h run that provided 2 × 250-bp fragments. A total of 3.9 Gb of information was obtained from a cluster density of 399,000/mm2, with a cluster-passing quality control filter of 97.92% (7,840,000 clusters). Within this run, the index representation of faustovirus was determined to be 10.34%. The 793,201 reads were filtered according to their quality.
(ii) Genome assembly.
The whole set of reads was trimmed using Trimmomatic (21) and then preassembled with Anytag software v2.5 (22) to produce pseudoreads. Spades assembler (23, 24) was then used to assemble these reads, and the contigs obtained were combined using SSPACE v2.0 (25) and Opera software v1.4 (26), assisted by GapFiller v1.10 (27), to reduce the set. Some manual refinements using CLC Genomics v6 software (CLC Bio, Aarhus, Denmark) and homemade tools improved the genome. The final draft genome of faustovirus E12 consisted of a single molecule, without gaps, containing 466,265 bp and having a G+C content of 36.22%.
(iii) Genome annotation.
Open reading frame (ORF) prediction was performed for the genome of the faustovirus E12 prototype isolate by using GeneMarkS, using previously described strategies (14, 28–31) and the Prodigal tool (32). tRNAs were identified using the tRNAscan-SE search server (33). Intergenic regions of >1 kbp were translated into 6 frames, and BLASTp searches were done against the NCBI GenBank nonredundant (nr) protein sequence database (34) to identify any additional ORFs that may have been missed by GeneMarkS and Prodigal, as well as any frameshift mutations. Predicted ORFs were searched against the nr database, the Reference Sequence (Refseq) collection, and the NCVOGs database (35). Paralogous genes were detected by BLASTp analysis, using 1e−5 as the E value threshold. For delineation of the core genes and pan-genomes, a database of all the predicted proteins from the whole faustovirus genome was created. Protein clusters were built using COG triangles (36) and OrthoMCL (37) clustering algorithms (38), and the core genes and pan-genome were defined using GET_HOMOLOGUES software (38) with the following parameters: 75% minimum coverage and 30% minimum identity for the pairwise sequence alignments, with 1e−05 as the maximum E value.
(iv) Phylogenetic analyses.
Protein sequences were aligned using the MUSCLE program (39) with the default parameters. Phylogenetic reconstructions were performed for the 8 isolates of faustovirus (including the faustovirus E12 strain) and the other Megavirales members by the maximum likelihood method, using FastTree with the default parameters (JTT evolutionary model; discrete gamma model with 20 rate categories) (40), based on the conserved genes. FigTree software was used for visualization of the phylogenetic trees (http://tree.bio.ed.ac.uk/software/figtree/).
(v) Nucleotide composition, codon usage, and amino acid usage.
Nucleotide composition, codon usage, and amino acid usage were calculated using the CAIcal server (http://genomes.urv.es/CAIcal/) as described previously (41, 42). The resulting codon and amino acid usages are expressed as percentages and reflect the contributions made by each codon and amino acid, respectively. Giant viral sets of predicted genes that were analyzed were recovered from the NCBI GenBank nucleotide sequence database and were from representative members of each putative family of the proposed order. Megavirales species included mimivirus (accession no. NC_014649.1), marseillevirus (accession no. NC_013756.1), Pandoravirus salinus (accession no. NC_022098.1), Paramecium bursaria chlorella virus NY2A (accession no. NC_009898.1), African swine fever virus (accession no. NC_001659.1), faustovirus E12, invertebrate iridescent virus 6 (accession no. NC_003038.1), Heliothis virescens ascovirus 3e (accession no. NC_009233.1), vaccinia virus (accession no. NC_006998.1), and Pithovirus sibericum (accession no. NC_023423.1).
(vi) Sanger sequencing of the genome fragment harboring capsid genes.
The following 14 pairs of primers were designed to check the sequence of the faustovirus E12 genomic region encoding three capsid protein fragments (bp 199,950 to 213,525): C1Fwd, CCCGGGATATTTAGGCAATGA; C1Rev, GTAGGTGTGGGATCAGAGAAAC; C2Fwd, GACGACAGGTGACTGTCTTAAA; C2Rev, CCATAACGACTACGCTGACTAC; C3Fwd, GGCGTATTCGGGTATCAAAGT; C3Rev, GCGTCGTAGGCTGTATAATGAG; C4Fwd, GCACCTCTGTGAAAGCAGATA; C4Rev, TGGTCATCAGCACCGATAAAG; C5Fwd, CTACCTCGGGTGTGTATACTTTG; C5Rev, ACTACCGATCCATTGCGTATTAG; C6Fwd, GCCCAACAACCTCGGTATTA; C6Rev, GAACAAGAGTTTCGCAAGGTATG; C7Fwd, TCGGCATCAATCGCCTTATAG; C7Rev, GGCCAGAAGGGTCATTAACA; C8Fwd, GTCGCAAATCGCTTCGTAATC; C8Rev, AAACCCTATCCACACCTCATAAA; C9Fwd, GGGCTTTATGAGGTGTGGATAG; C9Rev, CTAGGCGTTAACGGTTGATAGG; C10Fwd, TGTATCCCGGGACCTATCAA; C10Rev, CGGCAGAACCGTCAGAAATA; C11Fwd, GTCGGTGATGCGTTGTTAATC; C11Rev, AGCGTTGACCATAGGGAATC; C12Fwd, CCTTGCTATTGCATCCGTTTC; C12Rev, AGATCATTTCACGACCTGCATA; C13Fwd, GTTCTGCCGTTCTCAGATATAG; C13Rev, GCCAATGAGTTTATAGTTTCCTATG; C14Fwd, CCGTTCTAGGTTCAGAGACTAAAG; and C14Rev, TGCGATACAACGCAGTATAAGG).
Proteomics.
To prepare samples for the proteomic analysis, the faustovirus pellet was suspended in 50 mM ammonium bicarbonate in the presence or absence of 1% N-lauroylsarcosine and disrupted by sonication (three times for 60 s at power 20 without pulsing) (Q700 sonicator; QSonica, Newtown, Connecticut, USA). Viral debris was removed by centrifugation (12,000 × g, 4°C, 10 min), and soluble proteins were diluted 1:1 with 50 mM ammonium bicarbonate. They were then subjected to a standard trypsin digestion protocol with reduction/carboxymethylation. Prior to mass spectrometry (MS) analysis, each digested sample (30 μg) containing detergent was processed through a 125-μl detergent removal spin column (Thermo Fisher Scientific) according to the manufacturer's protocol. The protein digest was analyzed using a nanoAcquity two-dimensional liquid chromatography (2D-LC) system connected to a Synapt G2Si Q-TOF ion mobility hybrid spectrometer. The first chromatographic dimension consisted of a 300-μm by 50-mm C18 column (Nano Ease 5 μm XBridge BEH130; Waters). Peptides were eluted onto a second dimension by using a gradient of seven steps at 1.5 μl/min, with 20 mM ammonium formate, pH 10, and 12, 15, 18, 20, 25, 35, and 65% acetonitrile. A trapping column (nanoAcquity UPLC 10K-2D-V/M Trap 5-μm Symmetry C18 column; 180 μm × 20 mm; Waters) was used to collect the first-dimension peptides for concentration and desalting, after dilution at 15 μl/min in 99.9% water–0.1% formic acid and 0.1% acetonitrile–0.1% formic acid. The second dimension consisted of a 75-μm by 250-mm C18 column (nanoAcquity UPLC 1.8-μm HSS T3; Waters). Peptides eluted from the first-dimension steps were separated using a 1-h gradient (275 nl/min; 5 to 40% acetonitrile–0.1% formic acid). Data-independent MS/MS analysis was performed with the ion mobility feature (HDMSe method). The capillary was set to 3 kV, the sampling cone to 40 V, and the source temperature to 90°C. The MS range was set to 50 to 2,000 m/z, the trap cell energy was 4 V, the transfer cell low energy was 5 V, and the high fragmentation energy was a ramp of 19 to 45 V. Sample loading was adjusted by injecting a known standard (Escherichia coli digestion standard; Waters MasPREP) by use of a pseudo-1D injection method (a unique first-dimension elution step in 50% acetonitrile followed by a 1.5-h analytical gradient). The typical on-column sample load was approximately 100 ng per fraction (700 ng injected). Raw MS data were processed using PLGS 3.0.1 software (low/high energy thresholds = 135/30 counts; intensity threshold = 750 counts). A lock mass correction was applied to all spectra (leucine enkephalin = 785.8426 m/z). An internal protein sequence database was used to identify the proteins in each fraction. The following workflow parameters were set: monoisotopic masses, minimum charge of +1, 1 missed cleavage, carbamidomethyl C as a fixed modification, deamination NQ and oxidation M as variable modifications, 4% false discovery rate, 1 minimum peptide per protein, and 3 minimum fragment ion matches per protein.
Accession numbers.
The complete genome sequence of faustovirus E12 was submitted to GenBank and assigned accession number KJ614390. The mass spectrometry proteomic data for faustovirus E12 were submitted to the Consortium Proteome Exchange database (43) and assigned accession number PXD001858.
RESULTS
Isolation and developmental cycle of faustovirus.
We documented the faustovirus replication strategy by following its propagation in axenic Vermamoeba vermiformis cultures over an entire multiplication cycle, beginning with purified viral particles at a multiplicity of infection of 10 (see Fig. S1 at http://www.mediterranee-infection.com/article.php?laref=373&titer=faustovirus). We processed the samples for TEM at different times postinfection, with 30 min after infection considered 0 h postinfection (p.i.). The replication cycle of faustovirus lasts 18 to 20 h.
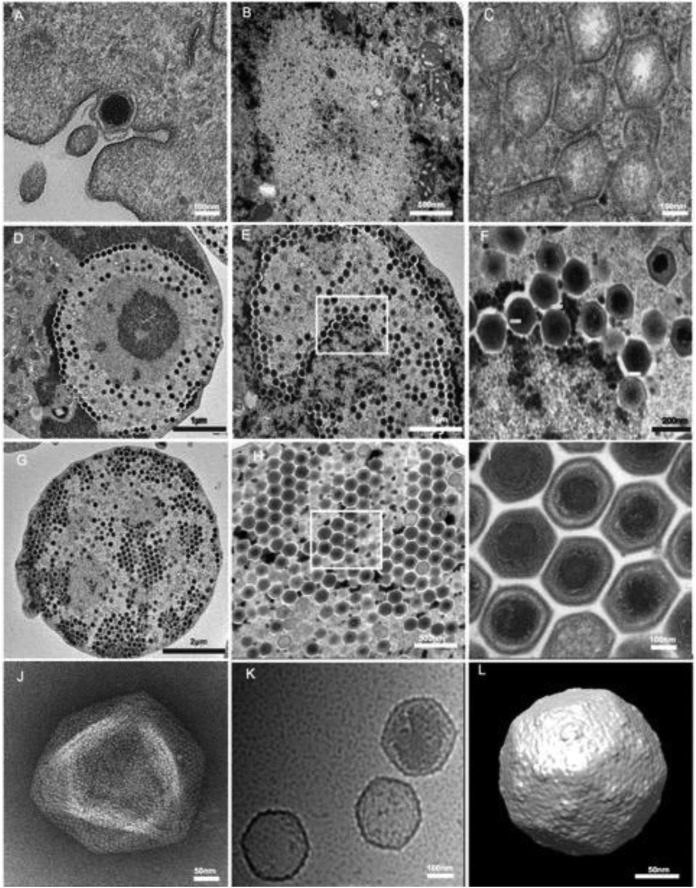
The phagocytosis of individual viral particles by the amoeba marked the beginning of the cycle (Fig. 1A). From 2 to 4 h p.i., faustovirus particles were detected within host phagosomes, demonstrating the internalization of the individual viral particles via phagocytic vacuoles. They could be seen near the host cell nucleus (data not shown). No evidence of interaction with the amoeba nucleus was observed; notably, no particles were seen within the nucleus or interacting with the nuclear membrane. The particles then emptied the contents of their internal compartment into the amoeba's cytoplasm. This process is similar to that of mimivirus, in which the internal lipid membrane delimiting the particle core fuses with the vacuole membrane, thereby creating a channel through which the particle proteins and DNA content can be delivered. This fusion process leads to an “eclipse” phase in which the content of the particles becomes invisible once delivered into the cytoplasm. It is remarkable that the “eclipse” phase of faustovirus seemed to be longer than that of mimivirus, taking place from 4 h p.i. until 6 h p.i. It is important that the host nucleus underwent some reorganization, which was initiated by the loss of its spherical appearance and a decrease of its surface area. Eight to 10 h after infection, the cells became rounded and lost their adherence, and new particles appeared at the center of a region forming a donut shape. This region was clearly distinct from the nucleus and represented the “virus factory” surrounded by mitochondria. At this time, the heterogeneous structure of the virus factory appeared near the cell nucleus. Some amoebae were observed to have a virus factory with only empty capsids (Fig. 1B and C), but others showed many newly synthesized viral particles or DNA-filled capsids that accumulated around the virus factory (Fig. 1D). At 12 and 14 h p.i., almost all the cytoplasmic space was occupied by the virus factory and was largely filled with new viral particles (Fig. 1E and F). These observations indicate that faustovirus replication and assembly take place in a very specific cytoplasmic structure composed of a dense central core from which newly formed particles appear like honeycomb stitches (Fig. 1H). At 16 and 18 h p.i., the virus factory still occupied the entire cell surface and was completely filled with viral particles ready to hatch (Fig. 1G). Complete viral particles were released through cell lysis at 18 to 20 h p.i., at which time the majority of amoebae were lysed.
FIG 1.
Electron microscopy imaging of the faustovirus replication cycle in V. vermiformis. (A) A faustovirus particle being phagocytosed by an amoeba at 0 h p.i. (B) Virus factory at 8 h p.i., with a dense replication center surrounded by empty capsids. (C) Higher-magnification view of a virus factory, showing the empty particles. (D) Virus factory at 8 h p.i., showing the donut-type morphology with both empty and DNA-filled particles. (E) Virus factory at 14 h p.i., showing the increased number of viral particles at different stages of morphogenesis. (F) Higher-magnification view of the boxed area in panel E. (G) Virus factory at 16 h p.i., with the new viral community occupying the entire cell cytoplasm area. (H and I) Higher-magnification views of panel G, demonstrating the honeycomb stitches. (J) Negative staining of a purified viral suspension showing a faustovirus in the typical aspect of Megavirales, with an icosahedral capsid and a size of 200 nm without fibrils. (K) Cryo-electron micrograph of faustovirus particles at different stages of maturation. The empty particles are 1,600 Å in diameter, whereas the full particles are 1,750 Å in diameter, vertex to vertex (n = 660). (L) Cryo-EM reconstruction of full faustovirus particles.
Genome analysis.
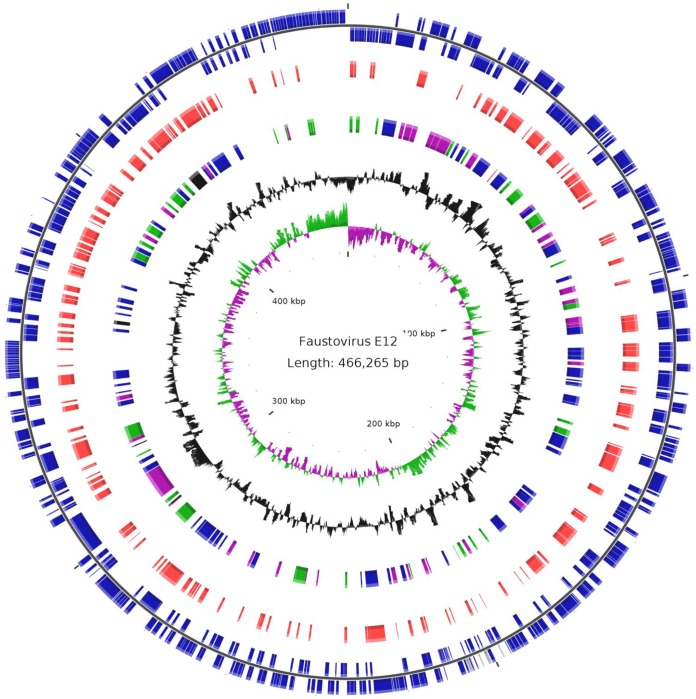
The genome of faustovirus E12 (GenBank accession no. KJ614390), the prototype isolate, is a 466,265-bp double-stranded DNA with a circular shape, as shown by paired-read assembly (Fig. 2). This size places the faustovirus lineage as the fourth largest viral genome, after pandoraviruses, Pithovirus sibericum, and mimiviruses (3, 5, 6). The G+C content of the faustovirus E12 genome is 36%, and it was predicted to encode 451 proteins, with a coding density of 85% (see Table S1 in the supplemental material). These proteins have a mean length (± standard deviation) of 295 ± 264 amino acids (ranging from 47 to 2,980 amino acids); 24 and 1 of them were found to be between 50 and 100 amino acids long and shorter than 50 amino acids, respectively. No tRNA genes were detected.
FIG 2.
Circular representation of the faustovirus E12 genome. The circles show the following, from the center to the outside: GC skew (green/purple), GC content (black), best-hit taxonomy (black, Archaea; green, Bacteria; purple, Eukarya; and blue, viruses), proteins in the virions as identified by proteomics (red), and ORFs on the plus and minus strands (blue).
Significant similarity to proteins from the NCBI GenBank nonredundant protein sequence database (with an E value threshold of 10−2) was detected for only 140 (31%) of the predicted proteins. For 42% of the faustovirus proteins with detectable homologs, the best matches were to proteins from other members of the Megavirales (n = 59 cases), mostly asfarviruses (39 cases; 28%) (see Table S1 in the Supplemental Material; see Fig. S2 at http://www.mediterranee-infection.com/article.php?laref=373&titer=faustovirus). The other best matches were to proteins from phycodnaviruses, mimiviruses, marseilleviruses, and an ascovirus, in 9, 8, 2, and 1 cases, respectively. In addition, 42 (30%), 31 (22%), 6 (4%), and 2 (1%) of the best hits were from bacteria, eukaryotes, archaea, and phages, respectively. Of the 317 ORFan genes (ORFs with no detectable homology to other ORFs in the database), 8 encoded putative proteins with significant matches against the NCBI GenBank environmental sequence database (making them meta-ORFans), corresponding to marine metagenomic sequences. Paralogous genes represented 19% of the gene complement, and their proportion was significantly higher in the one-fifth of the genomes harboring ORFs 0 to 43 and 399 to 457 (36% versus 14%; P < 1e−3) (see Fig. S3 at http://www.mediterranee-infection.com/article.php?laref=373&titer=faustovirus). Of these, 18% were predicted to encode membrane occupation and recognition nexus (MORN) repeat-containing proteins previously detected only in the marseilleviruses (14) and pandoraviruses (5) (among viruses) and were described to mediate membrane-membrane or membrane-cytoskeleton interactions (44). In contrast, only a few ankyrin repeat-containing proteins, another group of paralogs encountered in mimiviruses and pandoraviruses, were found. Altogether, genes encoding 98 predicted proteins of faustovirus fit into the clusters of orthologous genes of Megavirales (NCVOGs) (35). These included all 5 universal genes, 49 of the set common to ≥2 families, and 31 of the 47 genes that have been mapped to the common ancestor of the Megavirales. Some predicted proteins were of notable interest, including two polyproteins, of 220 kDa and 60 kDa (encoded by the adjacent ORFs 292 and 293), shared by faustovirus and African swine fever virus (ASFV), which are cleaved in ASFV-infected cells to yield several mature structural proteins. Other notable proteins included a ribosomal protein acetyltransferase (ORF 299), which is highly conserved in many bacteria and archaea and could modulate translation in faustovirus-infected cells, and a homolog of a bacteriophage tail fiber protein (ORF 46). A total of 162 proteins (36% of the predicted protein content) were detected in the faustovirus virions by nano-2D-LC–MS/MS, including 111 identified in at least two first-dimension LC fractions (Fig. 2; see Table S1 in the supplemental material). These included 76 proteins with homologs in the GenBank nr sequence database and 42 with functional annotations. Among the latter proteins are products of the Megavirales core genes, including homologs of the capsid protein, an A32-like packaging ATPase, and a possible T4-like proximal tail fiber from an uncultured phage.
The genomes of the seven other faustovirus isolates that were sequenced and mapped on the faustovirus E12 genome showed that they are closely related to the prototype isolate's genome, with a similar size and architecture (see Fig. S4 at http://www.mediterranee-infection.com/article.php?laref=373&titer=faustovirus). The mean size (± standard deviation) for these genomes was 467,340 ± 11,073 nucleotides. Overall, three groups could be delineated for the 8 faustovirus genomes, with viruses from different geographical origins (Senegal or France) falling between these groups (Fig. 3; see Fig. S5 at http://www.mediterranee-infection.com/article.php?laref=373&titer=faustovirus).
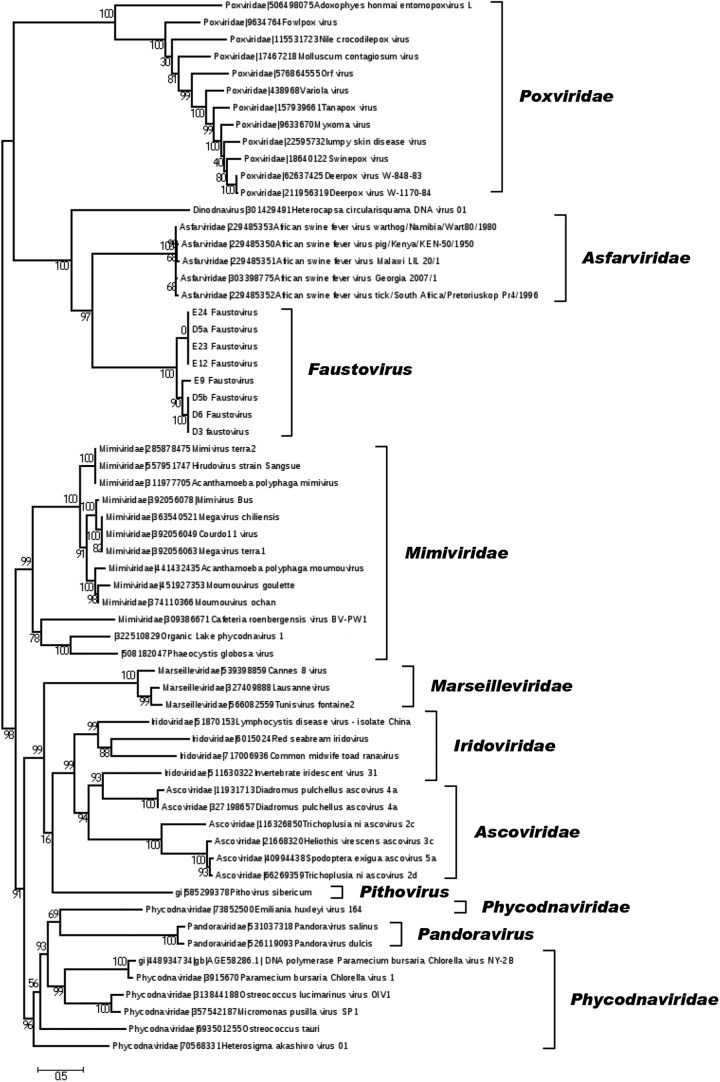
FIG 3.
Phylogeny reconstruction based on the family B DNA polymerase. Phylogenetic analyses were performed using the maximum likelihood method, based on the family B DNA polymerases from faustoviruses (including the faustovirus E12 strain) and representative members of the different families or new putative families of the proposed order Megavirales.
Of all the Megavirales members, faustovirus shared the largest number of orthologs, as defined by the bidirectional best-hit strategy (45), with ASFV. Thus, the faustovirus and ASFV protein sequences comprised 52 pairs of orthologs that shared 21 to 50% identity; 13 of these 52 genes were not found in any other members of the Megavirales. In addition, phylogenies of several conserved genes of Megavirales, including that encoding the family B DNA polymerase, showed that faustovirus E12 and other faustovirus isolates were distantly related to ASFV (Fig. 3; see Fig. S5 at http://www.mediterranee-infection.com/article.php?laref=373&titer=faustovirus). Nevertheless, this evolutionary relationship was supported only by analysis of a relatively small number of shared genes, constituting only ≈12% of the faustovirus gene complement. In addition, several features were found to differ significantly between faustovirus and ASFV. They included an ≈3 times larger genome in faustovirus and a G+C content and codon and amino acid usages that were closer to those of poxviruses than those of asfarviruses (see Fig. S6 and S7 at http://www.mediterranee-infection.com/article.php?laref=373&titer=faustovirus). Moreover, the size of the core genome decreased dramatically when the 8 faustovirus genomes and the 5 available ASFV genomes were included in the analysis, compared to the sizes estimated for the faustovirus genomes and the ASFV genomes taken separately (see Fig. S8 at http://www.mediterranee-infection.com/article.php?laref=373&titer=faustovirus). Indeed, the number of core genes dropped by factors of 10.6 and 5.8 for faustoviruses (from 231) and asfarviruses (from 116), respectively. In addition, the size of the pan-genome rose dramatically, from 752 and 212 for faustoviruses and ASFV, respectively, to 933 when both groups were taken together. Comparative genomics showed that these pan-genomes are strongly dissimilar. For comparison, 216 and 529 genes were estimated to comprise the core genome and the pan-genome of marseilleviruses, which have genomes that are ≈20% shorter (46). It is also worth noting that the mean amino acid identity between faustovirus/ASFV orthologous gene pairs is 30%. For comparison, the mean identity between orthologs from mimivirus and Megavirus chiliensis (47), which belong to different Mimiviridae lineages, is 50%, with 56 to 72% and 97% identities between orthologs from marseillevirus genomes of different lineages and the same lineage, respectively (46). Overall, these analyses indicate that the evolutionary distance between faustovirus and ASFV is comparable to that for pandoraviruses and phycodnaviruses (48). We therefore suggest that faustovirus is the first member of a new Megavirales family. In terms of the family B DNA polymerase, these two viruses were found to be distantly related to the Heterocapsa circularisquama DNA virus, a dsDNA virus that infects a marine dinoflagellate and has an ∼356,000-bp genome (49) (Fig. 3). Orthologs for 2 of the 6 proteins available for this virus were identified in faustovirus.
Apart from the substantial differences in the evolutionary and functional profiles of the unique parts of the gene repertoires, faustovirus and ASFV also unexpectedly differed in the architectures of their major capsid protein genes. Indeed, the capsid protein-encoding genes of faustovirus span an ≈17,000-kbp region that is interrupted by six group I self-splicing introns (see Table S1 in the supplemental material; see Fig. S9 at http://www.mediterranee-infection.com/article.php?laref=373&titer=faustovirus), whereas ASFV lacks introns entirely. Group I introns have previously been detected in other giant viruses isolated from protists, but not in members of the Megavirales that infect animals (30). The genome sequence of the regions encoding the capsid protein fragments was checked by Sanger sequencing, and the presence of group I introns was also observed in the other faustovirus genomes.
Finally, we analyzed sequences from two recent metagenomic studies that described ASFV-like sequences (50, 51), and we found 2 reads obtained from the serum samples of healthy Egyptian volunteers and 62 reads obtained from Mississippi ponds that had faustovirus sequences as the best hits, although until now these were considered ASFV-like sequences.
DISCUSSION
To date, the only protist used to culture giant viruses has been Acanthamoeba spp. Obviously, this reliance on a single host type has likely caused the research community to miss a substantial fraction of viruses. Our strategy for the isolation of giant viruses used the most common environmental protist combined with a new high-throughput procedure. This strategy proved fruitful and allowed for the opening of a new page in giant virus history. Using this new strategy, nearly 10% of all samples tested were found to be positive for faustovirus, using V. vermiformis as a support for coculture.
Faustovirus, with its 0.46-Mb genome, is larger than most members of the Megavirales, with the exception of mimiviruses, Pithovirus sibericum, and pandoraviruses (2, 3, 5), adding a sixth new viral family. Similar to the case for other giant viruses (52–54), a substantial majority (about two-thirds) of the predicted genes of faustovirus represent genomic “dark matter.” In addition, the faustovirus genome exhibits a substantial level of mosaicism, with genes from bacterial, archaeal, and eukaryotic origins, as previously noted for other giant viruses isolated from phagocytic protists, particularly marseillevirus (14). Phylogenetic analyses indicated that the evolutionary distance between faustovirus and the ASFVs is comparable to that between pandoraviruses and phycodnaviruses (48). We therefore suggest that faustovirus may be the first member of a new Megavirales family that is close to ASFV yet still distinct. Determining whether or not faustovirus should be merged with the Asfarviridae or should instead compose a new putative viral family will require a more comprehensive characterization of its morphology, host range, replicative cycle, and gene repertoire.
Among the six described giant virus species, four have been linked directly or indirectly to humans. The relationship reported here between faustovirus sequences and sequences from human and sewage metagenomes should prompt further studies to detect additional best matches with faustovirus sequences in environmental and human metagenomes retrieved worldwide. Time will show if this putative new virus family is associated with human diseases.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded in part by the IHU Méditerranée Infection Foundation. A part of the electron microcopy study in this paper was supported by National Institutes of Health grant AI011219, awarded to Michael G. Rossmann.
We are indebted to Fausto Strabato for sample collection and to Isabelle Pagnier, Olivier Croce, Catherine Robert, and Lina Barrassi for their technical help.
We have no conflicts of interest to report.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00115-15.
REFERENCES
- 1.La Scola B, Audic S, Robert C, Jungang L, de Lamballerie X, Drancourt M, Birtles R, Claverie JM, Raoult D. 2003. A giant virus in amoebae. Science 299:2033. doi: 10.1126/science.1081867. [DOI] [PubMed] [Google Scholar]
- 2.Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, La Scola B, Suzan M, Claverie JM. 2004. The 1.2-megabase genome sequence of Mimivirus. Science 306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- 3.Colson P, de Lamballerie X, Yutin N, Asgari S, Bigot Y, Bideshi DK, Cheng XW, Federici BA, Van Etten JL, Koonin EV, La Scola B, Raoult D. 2013. “Megavirales,” a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Arch Virol 158:2517–2521. doi: 10.1007/s00705-013-1768-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pagnier I, Reteno DG, Saadi H, Boughalmi M, Gaia M, Slimani M, Ngounga T, Bekliz M, Colson P, Raoult D, La Scola B. 2013. A decade of improvements in Mimiviridae and Marseilleviridae isolation from amoeba. Intervirology 56:354–363. doi: 10.1159/000354556. [DOI] [PubMed] [Google Scholar]
- 5.Philippe N, Legendre M, Doutre G, Coute Y, Poirot O, Lescot M, Arslan D, Seltzer V, Bertaux L, Bruley C, Garin J, Claverie JM, Abergel C. 2013. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 341:281–286. doi: 10.1126/science.1239181. [DOI] [PubMed] [Google Scholar]
- 6.Legendre M, Bartoli J, Shmakova L, Jeudy S, Labadie K, Adrait A, Lescot M, Poirot O, Bertaux L, Bruley C, Coute Y, Rivkina E, Abergel C, Claverie JM. 2014. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc Natl Acad Sci U S A 111:4274–4279. doi: 10.1073/pnas.1320670111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saadi H, Pagnier I, Colson P, Cherif JK, Beji M, Boughalmi M, Azza S, Armstrong N, Robert C, Fournous G, La Scola B, Raoult D. 2013. First isolation of Mimivirus in a patient with pneumonia. Clin Infect Dis 57:e127–e134. doi: 10.1093/cid/cit354. [DOI] [PubMed] [Google Scholar]
- 8.Popgeorgiev N, Boyer M, Fancello L, Monteil S, Robert C, Rivet R, Nappez C, Azza S, Chiaroni J, Raoult D, Desnues C. 2013. Marseillevirus-like virus recovered from blood donated by asymptomatic humans. J Infect Dis 208:1042–1050. doi: 10.1093/infdis/jit292. [DOI] [PubMed] [Google Scholar]
- 9.Popgeorgiev N, Michel G, Lepidi H, Raoult D, Desnues C. 2013. Marseillevirus adenitis in an 11-month-old child. J Clin Microbiol 51:4102–4105. doi: 10.1128/JCM.01918-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colson P, La Scola B, Raoult D. 2013. Giant viruses of amoebae as potential human pathogens. Intervirology 56:376–385. doi: 10.1159/000354558. [DOI] [PubMed] [Google Scholar]
- 11.Thurber RV, Haynes M, Breitbart M, Wegley L, Rohwer F. 2009. Laboratory procedures to generate viral metagenomes. Nat Protoc 4:470–483. doi: 10.1038/nprot.2009.10. [DOI] [PubMed] [Google Scholar]
- 12.Bradbury RS. 2014. Free-living amoebae recovered from human stool samples in Strongyloides agar culture. J Clin Microbiol 52:699–700. doi: 10.1128/JCM.02738-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coskun KA, Ozcelik S, Tutar L, Elaldi N, Tutar Y. 2013. Isolation and identification of free-living amoebae from tap water in Sivas, Turkey. Biomed Res Int 2013:675145. doi: 10.1155/2013/675145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyer M, Yutin N, Pagnier I, Barrassi L, Fournous G, Espinosa L, Robert C, Azza S, Sun S, Rossmann MG, Suzan-Monti M, La Scola B, Koonin EV, Raoult D. 2009. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc Natl Acad Sci U S A 106:21848–21853. doi: 10.1073/pnas.0911354106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.La Scola B, Campocasso A, N′Dong R, Fournous G, Barrassi L, Flaudrops C, Raoult D. 2010. Tentative characterization of new environmental giant viruses by MALDI-TOF mass spectrometry. Intervirology 53:344–353. doi: 10.1159/000312919. [DOI] [PubMed] [Google Scholar]
- 16.Reynolds ES. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol 17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ngounga T, Pagnier I, Reteno DG, Raoult D, La Scola B, Colson P. 2013. Real-time PCR systems targeting giant viruses of amoebae and their virophages. Intervirology 56:413–423. doi: 10.1159/000354563. [DOI] [PubMed] [Google Scholar]
- 18.van der Waaij LA, Mesander G, Limburg PC, van der Waaij D. 1994. Direct flow cytometry of anaerobic bacteria in human feces. Cytometry 16:270–279. doi: 10.1002/cyto.990160312. [DOI] [PubMed] [Google Scholar]
- 19.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 20.Ludtke SJ, Baldwin PR, Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 21.Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, Stitt M, Usadel B. 2012. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res 40:W622–W627. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruan J, Jiang L, Chong Z, Gong Q, Li H, Li C, Tao Y, Zheng C, Zhai W, Turissini D, Cannon CH, Lu X, Wu CI. 2013. Pseudo-Sanger sequencing: massively parallel production of long and near error-free reads using NGS technology. BMC Genomics 14:711–714. doi: 10.1186/1471-2164-14-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, Clingenpeel SR, Woyke T, McLean JS, Lasken R, Tesler G, Alekseyev MA, Pevzner PA. 2013. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol 20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 26.Gao S, Sung WK, Nagarajan N. 2011. Opera: reconstructing optimal genomic scaffolds with high-throughput paired-end sequences. J Comput Biol 18:1681–1691. doi: 10.1089/cmb.2011.0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boetzer M, Pirovano W. 2012. Toward almost closed genomes with GapFiller. Genome Biol 13:R56. doi: 10.1186/gb-2012-13-6-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.La Scola B, Desnues C, Pagnier I, Robert C, Barrassi L, Fournous G, Merchat M, Suzan-Monti M, Forterre P, Koonin E, Raoult D. 2008. The virophage as a unique parasite of the giant mimivirus. Nature 455:100–104. doi: 10.1038/nature07218. [DOI] [PubMed] [Google Scholar]
- 29.Colson P, Yutin N, Shabalina SA, Robert C, Fournous G, La Scola B, Raoult D, Koonin EV. 2011. Viruses with more than 1000 genes: Mamavirus, a new Acanthamoeba castellanii mimivirus strain, and reannotation of mimivirus genes. Genome Biol Evol 3:737–742. doi: 10.1093/gbe/evr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoosuf N, Yutin N, Colson P, Shabalina SA, Pagnier I, Robert C, Azza S, Klose T, Wong J, Rossmann MG, La Scola B, Raoult D, Koonin EV. 2012. Related giant viruses in distant locations and different habitats: Acanthamoeba polyphaga moumouvirus represents a third lineage of the Mimiviridae that is close to the Megavirus lineage. Genome Biol Evol 4:1324–1330. doi: 10.1093/gbe/evs109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Besemer J, Borodovsky M. 2005. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res 33:W451–W454. doi: 10.1093/nar/gki487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 35.Yutin N, Wolf YI, Raoult D, Koonin EV. 2009. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol J 17:223. doi: 10.1186/1743-422X-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kristensen DM, Kannan L, Coleman MK, Wolf YI, Sorokin A, Koonin EV, Mushegian A. 2010. A low-polynomial algorithm for assembling clusters of orthologous groups from intergenomic symmetric best matches. Bioinformatics 26:1481–1487. doi: 10.1093/bioinformatics/btq229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L, Stoeckert CJ Jr, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Contreras-Moreira B, Vinuesa P. 2013. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol 79:7696–7701. doi: 10.1128/AEM.02411-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Puigbo P, Bravo IG, Garcia-Vallve S. 2008. CAIcal: a combined set of tools to assess codon usage adaptation. Biol Direct 3:38. doi: 10.1186/1745-6150-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McInerney JO. 1998. GCUA: general codon usage analysis. Bioinformatics 14:372–373. doi: 10.1093/bioinformatics/14.4.372. [DOI] [PubMed] [Google Scholar]
- 43.Vizcaino JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Rios D, Dianes JA, Sun Z, Farrah T, Bandeira N, Binz PA, Xenarios I, Eisenacher M, Mayer G, Gatto L, Campos A, Chalkley RJ, Kraus HJ, Albar JP, Martinez-Bartolome S, Apweiler R, Omenn GS, Martens L, Jones AR, Hermjakob H. 2014. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol 32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gubbels M, Vaishnava S, Boot N, Dubremetz J, Striepen B. 2006. A MORN-repeat protein is a dynamic component of the Toxoplasma gondii cell division apparatus. J Cell Sci 119:2236–2245. doi: 10.1242/jcs.02949. [DOI] [PubMed] [Google Scholar]
- 45.Lechner M, Findeiss S, Steiner L, Marz M, Stadler PF, Prohaska SJ. 2011. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics 12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aherfi S, Boughalmi M, Pagnier I, Fournous G, La Scola B, Raoult D, Colson P. 2014. Complete genome sequence of Tunisvirus, a new member of the proposed family Marseilleviridae. Arch Virol 159:2349–2358. doi: 10.1007/s00705-014-2023-5. [DOI] [PubMed] [Google Scholar]
- 47.Arslan D, Legendre M, Seltzer V, Abergel C, Claverie JM. 2011. Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae. Proc Natl Acad Sci U S A 108:17486–17491. doi: 10.1073/pnas.1110889108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yutin N, Colson P, Raoult D, Koonin EV. 2013. Mimiviridae: clusters of orthologous genes, reconstruction of gene repertoire evolution and proposed expansion of the giant virus family. Virol J 10:106. doi: 10.1186/1743-422X-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ogata H, Toyoda K, Tomaru Y, Nakayama N, Shirai Y, Claverie JM, Nagasaki K. 2009. Remarkable sequence similarity between the dinoflagellate-infecting marine virus and the terrestrial pathogen African swine fever virus. Virol J 6:178. doi: 10.1186/1743-422X-6-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loh J, Zhao G, Presti RM, Holtz LR, Finkbeiner SR, Droit L, Villasana Z, Todd C, Pipas JM, Calgua B, Girones R, Wang D, Virgin HW. 2009. Detection of novel sequences related to African swine fever virus in human serum and sewage. J Virol 83:13019–13025. doi: 10.1128/JVI.00638-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wan XF, Barnett JL, Cunningham F, Chen S, Yang G, Nash S, Long LP, Ford L, Blackmon S, Zhang Y, Hanson L, He Q. 2013. Detection of African swine fever virus-like sequences in ponds in the Mississippi Delta through metagenomic sequencing. Virus Genes 46:441–446. doi: 10.1007/s11262-013-0878-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boyer M, Gimenez G, Suzan-Monti M, Raoult D. 2010. Classification and determination of possible origins of ORFans through analysis of nucleocytoplasmic large DNA viruses. Intervirology 53:310–320. doi: 10.1159/000312916. [DOI] [PubMed] [Google Scholar]
- 53.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 54.Sharma V, Colson P, Giorgi R, Pontarotti P, Raoult D. 2014. DNA-dependent RNA polymerase detects hidden giant viruses in published databanks. Genome Biol Evol 6:1603–1610. doi: 10.1093/gbe/evu128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.