ABSTRACT
Dengue virus (DENV) nonstructural protein 4B (NS4B) is an endoplasmic reticulum (ER) membrane-associated protein, and mutagenesis studies have revealed its significance in viral genome replication. In this work, we demonstrated that NS4B is an N-glycosylated protein in virus-infected cells as well as in recombinant protein expression. NS4B is N glycosylated at residues 58 and 62 and exists in two forms, glycosylated and unglycosylated. We manipulated full-length infectious RNA clones and subgenomic replicons to generate N58Q, N62Q, and N58QN62Q mutants. Each of the single mutants had distinct effects, but the N58QN62Q mutation resulted in dramatic reduction of viral production efficiency without affecting secretion or infectivity of the virion in mammalian and mosquito C6/36 hosts. Real-time quantitative PCR (qPCR), subgenomic replicon, and trans-complementation assays indicated that the N58QN62Q mutation affected RNA replication possibly by the loss of glycans. In addition, four intragenic mutations (S59Y, S59F, T66A, and A137T) were obtained from mammalian and/or mosquito C6/36 cell culture systems. All of these second-site mutations compensated for the replication defect of the N58QN62Q mutant without creating novel glycosylation sites. In vivo protein stability analyses revealed that the N58QN62Q mutation alone or plus a compensatory mutation did not affect the stability of NS4B. Overall, our findings indicated that mutation of putative N-glycosylation sites affected the biological function of NS4B in the viral replication complex.
IMPORTANCE This is the first report to identify and reveal the biological significance of dengue virus (DENV) nonstructural protein 4B (NS4B) posttranslation N-glycosylation to the virus life cycle. The study demonstrated that NS4B is N glycosylated in virus-infected cells and in recombinant protein expression. NS4B is modified by glycans at Asn-58 and Asn-62. Functional characterization implied that DENV NS4B utilizes the glycosylation machinery in both mammalian and mosquito hosts. Four intragenic mutations were found to compensate for replication and subsequent viral production deficiencies without creating novel N-glycosylation sites or modulating the stabilities of the protein, suggesting that glycans may be involved in maintaining the NS4B protein conformation. NS4B glycans may be necessary elements of the viral life cycle, but compensatory mutations can circumvent their requirement. This novel finding may have broader implications in flaviviral biology as the most likely glycan at Asn-62 of NS4B is conserved in DENV serotypes and in some related flaviviruses.
INTRODUCTION
Dengue virus (DENV) belongs to the Flaviviridae family of the Flavivirus genus, exists in four serotypes (DENV1 to -4), and is transmitted to humans by Aedes mosquitoes. Incidences of dengue have grown dramatically around the globe in recent decades and are endemic to tropical and subtropical countries. According to the latest report of the World Health Organization (WHO) (February 2015), approximately 50% of the world's population are now at risk from dengue. It is estimated that there are around 50 to 100 million new infections annually, resulting in around 22,000 deaths (1).
The DENV genome is a positive-sense, single-stranded RNA molecule 10.7 kb in length. The genome contains a single open reading frame (ORF) encoding a polyprotein that is co- and posttranslationally processed by cellular and viral proteases into three structural proteins (C, prM, and E) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). The structural proteins are components of mature virions, and most of the nonstructural (NS) proteins are believed to be involved in viral genome replication (2). DENV replication occurs in specialized, endoplasmic reticulum (ER)-derived membranous compartments (3). The ER-anchored DENV nonstructural proteins (NS2A, NS2B, NS4A, and NS4B) may rearrange the intracellular membranes, and NS4A has been shown to induce membrane alterations, possibly to serve as a platform for the formation of the viral replicase complex (3).
DENV NS4B is the largest of the four transmembrane proteins, consisting of 248 amino acids (aa) with an apparent molecular mass of 28 kDa. Previous studies reported that mutations in DENV NS4B adversely affect viral genome replication, strongly supporting the fact that NS4B is a component of the replication complex (4–6). Additionally, NS4B was found to interact with other NS proteins, such as NS3 and NS4A in DENV (7–9). Recently, NS4B was shown to have genetic and physical interactions with NS4A and NS1 in the related flaviviruses Japanese encephalitis virus (JEV) and West Nile virus (WNV), and these interactions were essential for viral genome replication (10, 11). Further, NS4B was also shown to have other biological functions, including evasion of host immune response (12) and suppression of host RNA interference (RNAi) response (13).
The membrane topology of NS4B revealed two membrane-associated domains, pTMD1 and pTMD2, in the ER lumen and three transmembrane domains, TMD3, TMD4, and TMD5 (14). Further, NS4B was suggested to be an N-glycosylated protein based on the results of Western blotting examining recombinant protein expression (14). However, another report showed that NS4B is not N glycosylated in virus-infected cells (15). In spite of these controversies, we were interested in investigating whether DENV NS4B is an N-glycosylated protein. Many viruses depend on N-linked glycosylation for vital biological functions. Viruses use this host cell process to modify their structural proteins, and it is ultimately beneficial in multiple ways, such as for maintaining viral protein folding and stability and for conducting efficient virion morphogenesis, egress, and infection (16). For example, the DENV envelope (E) protein is N glycosylated at Asn-67 and Asn-153, and the loss of N-glycan at residue 67 abolishes infectious particle production while the lack of N-glycan at residue 153 reduces infectivity (17, 18). In the related flavivirus Japanese encephalitis virus (JEV), the loss of Asn-15 glycan from precursor membrane protein (prM) affects the efficiency of infectious virus production (19). However, there has been almost no similar evidence for the NS proteins. Only DENV NS1 is known to be N glycosylated at two positions, Asn-130 and Asn-207, and several groups have reported the defective function of these NS1 N-glycans in viral genome replication and virion production (20–22). N-linked glycans on DENV NS1 are believed to maintain the protein structure and function in the replication complex (22).
According to the DENV NS4B protein topology (14), glycosylation site prediction, and NS4B sequence alignment among DENV serotypes and related flaviviruses, DENV NS4B is suspected to be an N-glycosylated protein. Flavivirus prM, E, and NS1 are known N-glycosylated proteins, and this study provides evidence that NS4B is also an N-glycosylated protein. NS1 and NS4B are components of the replication complex but differ in their cellular localization. NS1 is a luminal and a secretory protein, whereas NS4B is an integral ER membrane protein. The elucidation of the effects of NS4B modifications by glycans would improve our understanding of its biological function in the DENV life cycle. In this work, we demonstrated that NS4B is an N-glycosylated protein in virus-infected cells as well as in recombinant protein expression. We used a full-length (FL) infectious clone, subgenomic replicon, and recombinant virus to characterize the biological significance of NS4B glycosylation in various stages of the virus life cycle. We found that mutation of putative NS4B N-glycosylation sites affected the replication step in both mammalian and mosquito C6/36 cell culture systems. Four compensatory NS4B intragenic mutations were obtained from our screening experiments that mitigated the defective function associated with the N58QN62Q double mutant, possibly by restoring protein folding and conformation. Loss of glycans did not affect the NS4B protein stability. Collectively, these findings indicate that NS4B glycans may be required for normal DENV viral replication but are not critically important in the presence of compensatory mutations.
MATERIALS AND METHODS
Cells and virus.
Baby hamster kidney cell line BHK-21 and human embryonic kidney cell line HEK-293T were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS) and 2% penicillin-streptomycin at 37°C in a 5% CO2 humidified incubator. Aedes albopictus cell clone C6/36 was maintained in RPMI 1640 medium (Invitrogen) supplemented with 10% FBS, 2 mM l-glutamine, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, and 2% penicillin-streptomycin at 28°C in a 5% CO2 humidified incubator. The human hepatocellular carcinoma Huh7 cell line was maintained in DMEM supplemented with 10% FBS, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, and 2% penicillin-streptomycin at 37°C in a 5% CO2 humidified incubator. DENV2 strain PL046 (GenBank sequence accession number AJ968413.1) and a constructed recombinant full-length infectious clone of DENV2 strain PL046 (GenBank sequence accession number KJ734727) (23), amplified from C6/36 mosquito hosts, are used for high-multiplicity-of-infection (MOI) experiments.
Construct for NS4B protein expression.
The p2K4B-V5 (4X) plasmid was derived from the p2K-NS4B-HA (2X)-EGFP plasmid. The construct p2K-NS4B-HA (2X)-EGFP was generated by modifying the parental pEGFP-N3 plasmid (Clontech). First, two copies of the hemagglutinin (HA) tag were inserted between BamHI and the enhanced green fluorescent protein (EGFP) coding region in pEGFP-N3 by linker DNA. Then, the PCR-amplified NS4B with its 2K signal peptide at the N terminus was subcloned between XhoI and BamHI and referred to as p2K-NS4B-HA (2X)-EGFP plasmid. At the C terminus of EGFP, the NotI restriction site is present, and these plasmids for wild-type (WT) and mutant NS4B were digested with BamHI and NotI to remove the HA (2X)-EGFP segment. A PCR-amplified V5 tag (four copies) was subcloned between BamHI and NotI, retaining the intact open reading frame of 2K-NS4B and V5 tag. The 2K signal sequence at the N terminus of NS4B was used for measuring the integration of the protein into the ER membrane, and the V5 tag fused at the C terminus was used for Western detection. Asparagine (N) residues of the canonical NXS/T motif at amino acid positions 58 and 62 of the NS4B protein were individually mutated to glutamine (Q) by site-directed mutagenesis (SDM). The SDM DNA primers changed the two nucleotides at the respective sites (AAT→CAG and AAC→CAG).
Recombinant NS4B protein expression and cell lysate preparation.
HEK-293T (2 × 105) cells were plated in 12-well culture dishes, and the next day, cells were transfected using calcium phosphate with 2 μg of 2K-NS4B-V5 (4X) expression plasmid DNA encoding WT or glycosylation mutations as indicated in the figures. For tunicamycin treatment, after 6 h posttransfection (hpt), cells were grown in the presence of 0.5% dimethyl sulfoxide (DMSO) or 0-, 1-, 5-, and 10-μg/ml doses of tunicamycin for 12 h. For protein stability analysis, at 24 hpt, cells were treated with 1 mg/ml cycloheximide (CHX) or 0.5% DMSO and chased for 0 to 6 h. For MG132 treatment, at 24 hpt, cells were treated with either 0.4% DMSO or 40 μM MG132 (Sigma-Aldrich) for 12 h and then treated with 1 mg/ml cycloheximide (CHX) or 0.5% DMSO for 0 to 3 h. For all of the above conditions, cell lysate was harvested in 1× passive lysis buffer (PLB; Promega) and 10 μg of total protein was mixed with 5× sample buffer (250 mM Tris-HCl, pH 6.8, 50% glycerol, 0.5 M β-mercaptoethanol, 10% SDS, and 0.1% bromophenol blue) and heated at 95°C for 10 min, and proteins were resolved by 10% SDS-PAGE and subjected to Western detection. In the case of E protein detection, protein sample was mixed with 4× sample buffer without reducing agent (250 mM Tris-HCl, pH 6.8, 40% glycerol, 8% SDS, and 0.02% bromophenol blue) and proteins were resolved by 10% SDS-PAGE, followed by Western blotting.
Western blot analysis.
Western transfer was performed on nitrocellulose membranes. After blocking with 5% skim milk, the blot was incubated with rabbit polyclonal anti-NS4B antibody (1:1,000; GeneTex), mouse monoclonal anti-E (4G2) antibody (1:1,000), rabbit polyclonal anti-NS3 antibody (1:5,000), rabbit polyclonal anti-NS5 antibody (1:5,000), mouse monoclonal anti-V5 antibody (1:2,000; Serotec), mouse monoclonal antiubiquitin antibody (1:2,000; Santa Cruz), mouse monoclonal anti-alpha-tubulin antibody (1:5,000; GeneTex), or mouse monoclonal anti-glyceraldehyde3-phosphate dehydrogenase (anti-GAPDH) antibody (1:5,000; GeneTex), followed by horseradish peroxidase (HRP)-conjugated goat anti-rabbit (PerkinElmer) or mouse (Merck Millipore) IgG (1:5,000). The blot was processed for detection with a commercial enhanced chemiluminescence (ECL) detection system (Advantsa).
Enrichment of glycoproteins using ConA agarose.
A total of 8 × 106 BHK-21 or Huh7 cells in one 10-cm cell culture dish were infected (IF) with DENV2 or heat-inactivated (HI) virus at a multiplicity of infection (MOI) of 10. At 48 h postinfection (hpi), cells were washed with 1× PBS and lysed in 1.5 ml of 1× passive lysis buffer (PLB; Promega), and then the protein concentration was measured by the Bradford assay. One-third of cell lysate corresponding to 3 mg of total protein in a volume of 500 to 600 μl was dissolved in 1× binding buffer (20 mM Tris-HCl, pH 7.4, and 0.5 M NaCl) containing 1% Triton X-100 and incubated at room temperature (RT) for 10 min. Then, cell lysate was centrifuged at 13,000 rpm for 30 min at RT to remove the insoluble particles. The protocol for the purification of glycoproteins by concanavalin A-coupled agarose (ConA agarose 6B; GB Biosciences) was followed according to the manufacturer's instructions with some modifications, and all buffers were supplemented with 0.01% NP-40 for better elution efficiency. Two hundred microliters of 50% ConA agarose slurry was centrifuged in a 1.5-ml Eppendorf tube to remove the storage buffer. The ConA agarose bed was washed with 200 μl of 0.01% NP-40–equilibration buffer (1 M NaCl, 5 mM MgCl2, 5 mM MnCl2, and 5 mM CaCl2, pH 7.0) three times to equilibrate the ConA agarose with metal ions and then washed once with 0.01% NP-40–1× binding buffer. One-tenth of the clarified cell lysate was saved to serve as the total protein control. The remaining clarified cell lysate was incubated with ConA agarose at RT with end-to-end mixing for 1 h and then centrifuged at 3,000 rpm for 1 min to remove the unbound proteins. After washing with 200 μl of 0.01% NP-40–1× binding buffer at least 5 times to remove the loosely bound proteins, ConA agarose beads were then incubated with 100 μl 0.01% NP-40–elution buffer (0.5 M α-methyl-α-d-mannopyranoside [Sigma-Aldrich] in 1× binding buffer) at RT for 2 h. Glycoprotein eluate (the E1 fraction) was collected by centrifugation at 3,000 rpm for 1 min. The ConA agarose beads were incubated with 100 μl 0.01% NP-40–elution buffer for the second time, overnight, and the eluate was collected (the E2 fraction). Proteins from the various fractions were resolved by 12% SDS-PAGE and analyzed by Western blotting.
For the purification of recombinant NS4B glycosylated form and NS4B N58QN62Q variant by ConA agarose, 8 × 106 HEK-293T cells in one 10-cm cell culture dish were transfected using calcium phosphate with 10 μg p2K-NS4B-V5 (4X) expression plasmid encoding WT or N58QN62Q mutant NS4B. At 36 hpt, the cells were washed with 1× PBS and lysed in 1.5 ml of 1× PLB. The above-described protocol was followed except that 1/10 cell lysate (100 to 150 μl) corresponding to 0.5 mg of total protein was applied to ConA agarose. Proteins from the various fractions were resolved by 10% SDS-PAGE and analyzed by Western blotting.
Glycosidase treatment.
The peptide N-glycosidase F (PNGase F; NEB) treatment was carried out according to the manufacturer's instructions. In brief, the protein samples in 1× glycoprotein denaturing buffer were heated at 95°C for 10 min and then cooled at RT for 5 min. After a short spin, the PNGase F reaction was carried out in the presence of 1× G7 reaction buffer and 1% NP-40 for 18 h at 37°C. Subsequently, the samples were mixed with 5× sample buffer and heated at 95°C for 5 min, and proteins were resolved by SDS-PAGE followed by Western blotting. For the ConA agarose-eluted samples; approximately 1 mg protein from the eluted fraction was digested with 1,000 units of PNGase F in 60 μl of total reaction volume. In the case of recombinant NS4B mutant proteins; 10 μg of total proteins was digested with 500 units of PNGase F in 20 μl of total reaction volume.
In vitro RNA synthesis and RNA transfection.
Our laboratory has several SP6 promoter-driven plasmids, namely, DENV2 full-length (FL) infectious clone (23) and subgenomic replicon pD2-SGR-Rlu, in which DENV prM and E genes were deleted in frame and a Renilla luciferase reporter gene was inserted in the 3′ untranslated region (UTR). To facilitate the replacement of NS4B mutation into the full-length infectious clone and the subgenomic replicon, the intermediate plasmid pRS424ΔCE, which contains the coding sequence from NS3 to NS5, was used. Site-directed mutagenesis (SDM) was performed to create N58Q, N62Q, and N58QN62Q mutations in NS4B. The digested product from this intermediate plasmid was subcloned into a full-length infectious clone and subgenomic replicon between the XmaI and MluI restriction sites.
The full-length infectious clone construct and the subgenomic replicon construct of DENV2 were linearized by ClaI digestion. The linearized plasmid was phenol-chloroform extracted, ethanol precipitated, and resuspended in RNase-free water at a concentration of 1 μg/μl. FL infectious clone RNA (FL RNA) and SGR-Rluc replicon RNA (SGR-Rluc RNA) were in vitro synthesized using the Amplicap SP6 High-Yield Message Maker kit (Cellscript) according to the manufacturer's protocol. The transcription reaction mixture was incubated at 37°C for 2 h. The DNA templates were removed from the reaction mixture by DNase I digestion, and the RNAs were purified using lithium chloride precipitation and finally resuspended in RNase-free water. The transcribed RNA was quantified with a spectrophotometer, and the quality was checked on a 2% formaldehyde agarose gel. For RNA transfection, a total of 10 μg FL RNA or SGR-Rluc RNA was electroporated into 3 × 106 BHK-21 cells or 4 × 106 mosquito C6/36 cells using the Gene Pulser Xcell electroporation system (Bio-Rad) according to the manufacturer's instructions (140 V, 25 ms, 1 pulse).
Preparation of culture supernatants and cell lysates.
A total of 1 × 106 FL RNA-electroporated BHK-21 cells were seeded in 6-cm dishes with 5 ml of 2% FBS-DMEM and incubated at 37°C. After 24 h (24 h postelectroporation [hpe]), culture dishes were moved to 30°C and kept for up to several days, as indicated in the figures. At various time points, culture fluid was collected for extracellular viral titer determination. For intracellular virus titer, electroporated cells were trypsinized and washed with 1× PBS twice at 144 hpe. Cell pellets were resuspended in 1 ml 10% FBS-DMEM and subjected to multiple rounds of freeze-thaw treatment. Cell lysates and cell debris were separated by centrifugation at 5,000 × g for 5 min. Then, the clarified cell lysate was subjected to intracellular viral titer determination.
Focus-forming assay and plaque assay for viral titer determination.
The BHK-21 cell monolayer in a 12-well plate was infected with virus inoculum for 2 h at 37°C. Cells were washed with 1× PBS to remove the unattached virus, followed by overlaying the cell monolayer with 2% FBS-DMEM and 1.2% Avicel RC-591 (FMC Biopolymer). After 72 h of incubation at 37°C, the overlay medium was removed and cells were fixed for 15 min in 4% formaldehyde. Immunofluorescence assay was carried out overnight at 4°C with rabbit polyclonal anti-NS3 antibody (1:300). The cells were then counterstained with Cy3-conjugated goat anti-rabbit IgG (1:300). After several washes with 1× PBS, immunostained foci were counted. For plaque assay, the confluent cell monolayers in 6-well plates were infected with serial dilutions of virus inoculum. After virus adsorption for 2 h at 37°C, cells were washed with 1× PBS followed by overlaying with 2% FBS-DMEM and 1% low-melting-point agarose and incubation at 30°C. Twelve or 14 days postinfection (dpi), cells were fixed with 10% formaldehyde for 2 h and the agarose layer was removed. Cells were stained with crystal violet solution (8.5 mg/ml NaCl and 2.5 mg/ml crystal violet in 50% ethanol).
Extraction of viral RNA from transfected cells and real-time qPCR.
Viral RNA was extracted from cells using TRIzol reagent, and reverse transcription was performed on 1 μg of total cellular RNA. Real-time quantitative PCR (qPCR) was performed in a 96-well plate in which each well contained 2 μl of cDNA, 0.4 μl of 5 μM specific primers (final concentration, 0.1 nM), 7.2 μl H2O, and 10 μl 2× SYBR green master mix (ABI). Samples were run in triplicate. Real-time PCR was conducted with the ABI 7500 Fast system for over 40 cycles with an annealing temperature of 60°C. Assessment of the expression of each target gene was based on relative quantification (RQ) using the comparative critical threshold cycle (CT) value method. The RQ of a specific gene was evaluated in each reaction by normalization to the CT obtained for the endogenous control gene (GAPDH gene). Data from all the later time points were normalized with 4-hpe data that indicated the transfection efficiency. At least two or three independent transfection experiments were conducted, and data presented are representative results from one independent transfection.
The primers for quantitative real-time PCR used in this study were as follows: random hexamer for reverse transcription; primers for viral RNA amplification, NS3 forward (5′-CATTT CAGAA ATGGG TGCC-3′) and NS3 reverse (5′-GGGTC ACTGG CATGG GTCC-3′); primers for endogenous internal control, GADPH forward (5′-GTGGA GTCTA CTGGC GTCTT-3′) and GADPH reverse (5′-AGAAG GTGCG GAGAT GATGA-3′); and primers for Renilla luciferase reporter, forward (5′-CATGG GATGA ATGGC CTGAT-3′) and reverse (5′-ATGGT TTCCA CGAAG AAGTT ATTCTC-3′).
Transient replicon assay and trans-complementation analysis.
For transient replicon assay, 10 μg of luciferase reporter subgenomic replicon SGR-Rlu RNA encoding wild-type or mutant NS4B or the replication-defective NS5GDDm mutant was electroporated into 3 × 106 BHK-21 cells. For trans-complementation of an NS4B N-glycosylation mutant, 3 × 106 BHK-21 cells or D2-Flu-SGR-Neo stable cells (24) were electroporated with 10 μg of SGR-Rlu RNA encoding wild-type or N58QN62Q mutant NS4B or a helicase-defective mutant, NS3 K199AT200A. The transfected cells were resuspended in 10% FBS-DMEM at a concentration of 1 × 105 cells/ml and seeded in 24-well plates at a density of 1 × 105 cells/well in triplicate. At various time points, the cells were washed with 1× PBS, 60 μl of 1× PLB was added to each well, and the plate containing the cell lysate was stored at −20°C. Once samples had been collected for all time points, 10-μl portions of the cell lysate were transferred to a 96-well plate and assayed for Rlu and/or Flu luciferase activity. Alternatively, total cellular RNAs of cells harvested 4 hpe and 96 hpe were isolated, and RT-qPCR was performed for trans-complementation analysis.
Statistics.
Data were analyzed by Student's t test, and a two-tailed P value of <0.05 was considered significant.
RESULTS
DENV NS4B is N glycosylated in virus-infected cells.
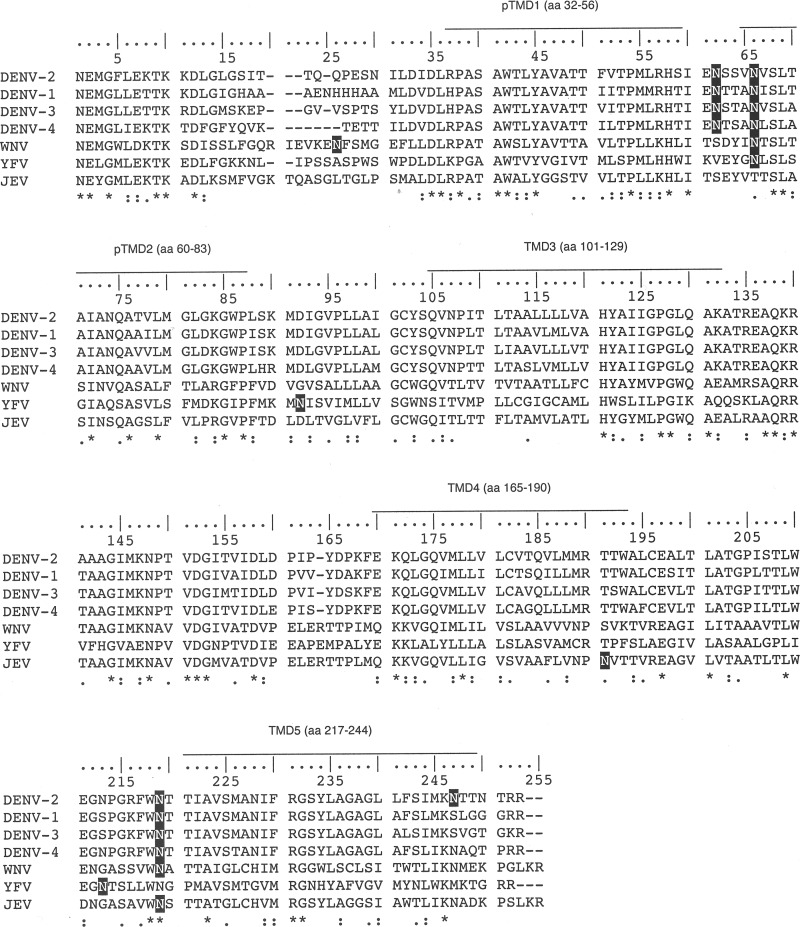
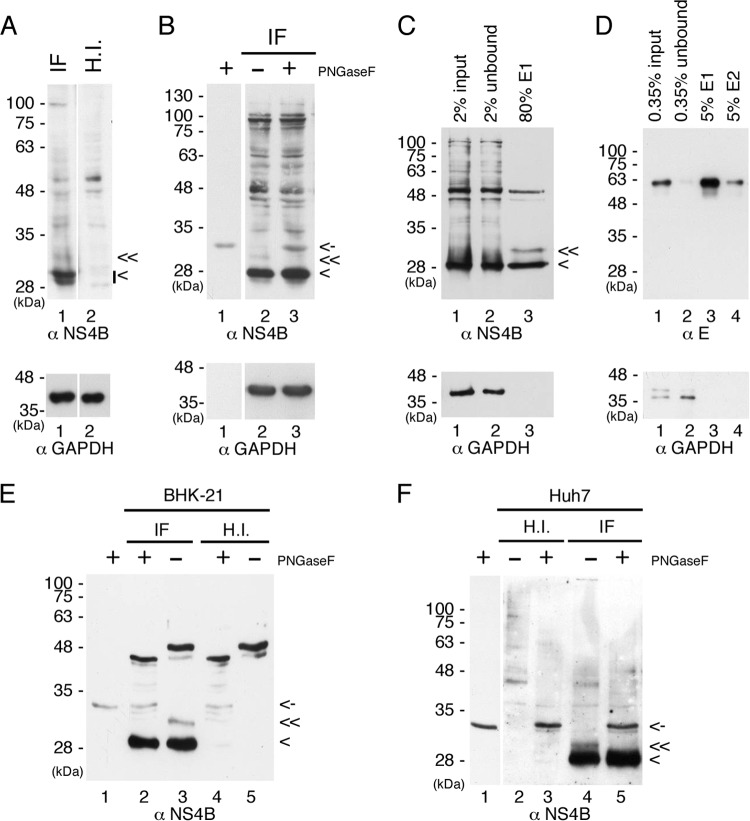
Miller et al. (14) showed that the C-terminally truncated versions of DENV 2K-NS4B protein gave rise to two bands in Western analyses, and the upper band was digested by PNGase F treatment, indicating the possibility that NS4B could be a glycosylated protein. NS4B amino acid sequence alignment was performed for DENV serotypes 1 to 4 and closely related flaviviruses WNV, JEV, and yellow fever virus (YFV). We used the NetNGlyc 1.0 server to predict the potential N-linked glycosylation sites in NS4B. The various putative N-glycosylation sites in flaviviral NS4B (Fig. 1) led us to speculate that N-glycosylation may be occurring on DENV NS4B. To this end, BHK-21 cells were infected with DENV at a multiplicity of infection (MOI) of 10 for 48 h followed by Western analysis. The commercial antibody detected NS4B at 28 kDa as a doublet band, which could be the mature NS4B and the unprocessed 2K-NS4B, a faint band above the 28-kDa doublet, and a band above 100 kDa in virus-infected BHK-21 cells (Fig. 2A, lane 1). The antibody also had nonspecific interaction with some BHK-21 cellular proteins (Fig. 2A, lanes 1 and 2). We were interested to determine whether the band immediately above the 28-kDa doublet band is the putative N-glycosylated form of NS4B. The DENV-infected cell lysate was treated with PNGase F (an enzyme to remove N-linked glycans), and we found that the band right above the 28-kDa doublet was digested after PNGase F treatment (Fig. 2B), implying that NS4B is likely an N-glycosylated protein in the virus-infected cells. However, the reason for the consistent observation of the low intensity of the NS4B glycosylated form compared to that of the unglycosylated form in Western analyses is not known. We noticed that the NS4B doublet band was not clearly resolved in the presence of PNGase F enzyme reaction buffer (Fig. 2A versus B), and we also observed that the PNGase F enzyme was recognized by the NS4B antibody (Fig. 2B, lanes 1 and 3).
FIG 1.
Alignment of mosquito-borne flavivirus NS4B. Multiple-sequence alignment of flaviviral NS4B sequences was performed using Clustal Omega software. The corresponding amino acids of transmembrane helices of DENV2 are indicated above the DENV2 sequence. An asterisk represents a fully conserved residue, a colon represents strong similarity, and a period represents weak similarity among the residues according to the parameters of Clustal Omega. The putative N-glycosylation sites from NetNGlyc prediction software are shown in white on a black background. DENV2, strain PL046, Asn-58 (NSSV), Asn-62 (NVSL), Asn-214 (NTTI), and Asn-242 (NTTN). DENV1, strain 45AZ5 vaccine, Asn-59 (NTTA), Asn-63 (NISL), and Asn-215 (NTTI). DENV3, strain H87, Asn-58 (NSTA), Asn-62 (NVSL), and Asn-214 (NTTI). DENV4, GenBank accession number AF326825.1, Asn-55 (NTSA), Asn-59 (NLSL), and Asn-211(NTTI). WNV, West Nile virus, strain NY99, Asn-26 (NFSM), Asn-66 (NTSL), and Asn-219 (NATT). YFV, yellow fever virus, strain 17D vaccine, Asn-64 (NLSL), Asn-90 (NISV), and Asn-211(NTSL). JEV, Japanese encephalitis virus, strain JaOArS982, Asn-191 (NVTT) and Asn-219 (NSTT).
FIG 2.
DENV NS4B is an N-glycosylated protein in virus-infected cells. (A and B) NS4B is modified by N-glycosylation in DENV-infected cells. BHK-21 cells (4 × 105 cells in 6-well culture dish) were infected with DENV2 (IF) or heat-inactivated virus (H.I.) at an MOI of 10 for 48 h. For panel A, 50 μg of total cellular proteins was resolved by 12% SDS-PAGE. For panel B, 50 μg of total cellular proteins from virus-infected cells (IF) was digested with PNGase F or not and resolved by 12% SDS-PAGE. (C to F) Enrichment of glycoproteins by ConA agarose and PNGase F treatment of putative NS4B glycosylated form. BHK-21 cells for panels C, D, and E and Huh7 cells for panel F (8 × 106 cells in 10-cm cell culture dish) were infected (IF) with DENV2 or heat-inactivated virus (H.I.) at an MOI of 10 for 48 h. Cell lysates were harvested, and 3 mg of total cellular proteins was applied to ConA agarose for glycoprotein enrichment according to the procedures described in Materials and Methods. For panel C, 2% of the starting material (input), 2% of the unbound fraction (unbound), and 80% of the E1 elution fraction were resolved by 10% SDS-PAGE for Western detection of NS4B protein. For panel D, 0.35% of the starting material (input), 0.35% of the unbound fraction (unbound), and 5% of the elution fractions E1 and E2 were resolved by nonreducing 10% SDS-PAGE for Western detection of envelope (E) protein. For panels E and F, 30% of the E1 elution fraction from IF or H.I. cell lysates was digested with PNGase F or not and resolved by 12% SDS-PAGE. Lanes 1 of panels B, E, and F contain the PNGase F enzyme only. DENV NS4B protein was detected by polyclonal anti-rabbit NS4B antibody, DENV E protein was detected by monoclonal anti-E (4G2) antibody, and GAPDH served as a loading control. “<” indicates the signals derived from NS4B protein, “≪” indicates the signal derived from the putative glycosylated form of NS4B protein, and “<-” indicates the signal derived from PNGase F. All experiments were performed three times, and the data shown are from one representative infection experiment.
The concanavalin A (ConA) agarose resin was used to enrich glycoproteins in virus-infected BHK-21 cell lysate before Western analysis. In theory, the unglycosylated proteins that did not bind to ConA agarose were removed in unbound fraction and the glycoproteins that bound to the ConA agarose were retrieved by elution with the competing sugar molecule of methyl-α-d-mannopyranoside. The putative glycosylated form of NS4B, which migrated right above the 28-kDa marker, became clearly detectable in the elution fraction (Fig. 2C, lane 3). The unglycosylated form of NS4B was not removed completely during washing steps, although more than 90% was observed in the unbound fraction (Fig. 2C, lanes 1 and 2), and therefore, we have confirmed the specificity of ConA agarose and the enrichment of viral glycoprotein (envelope [E]) in virus-infected cell lysates (Fig. 2D). Further, the putative glycosylated form of NS4B was completely digested by PNGase F (Fig. 2E, lanes 2 and 3), indicating that NS4B is an N-glycosylated protein. Some glycoprotein(s) of 48 kDa from BHK-21 cells showed a mobility shift after PNGase F treatment (Fig. 2E, lanes 2 to 5).
To further examine whether the modification of N-glycosylation on NS4B occurred in other cell types, we performed the same ConA agarose affinity procedure for the Huh7 cells infected with DENV or heat-inactivated virus. We observed relatively less nonspecific cellular glycoproteins in Huh7 cells than in BHK-21 cells (Fig. 2F versus E). The NS4B glycosylated form that was purified by ConA agarose from DENV-infected Huh7 cells was digested with PNGase F, suggesting that NS4B is an N-glycosylated protein and that the modification is not cell type dependent. (Fig. 2F, lanes 4 and 5). The small difference in the mobility of the NS4B glycosylated form between BHK-21 and Huh7 cells may be due to differences in oligosaccharyl transferase efficiency in adding glycans to NS4B. Overall, these results demonstrated that NS4B is modified by N-glycans in virus-infected cells. However, the ratio of the glycosylated form to the unglycosylated form remains to be investigated.
DENV NS4B is N glycosylated at both residue Asn-58 and residue Asn-62.
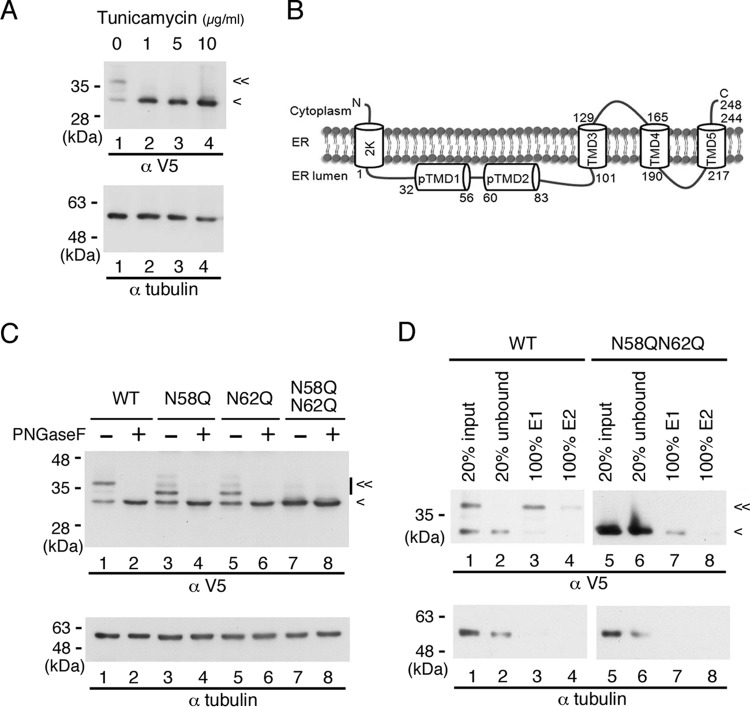
To assess whether DENV NS4B is N glycosylated outside the context of viral infection, we expressed the recombinant p2K-NS4B-V5 (4X) plasmid DNA construct in HEK-293T cells. At 6 h posttransfection (hpt), transfected cells were treated with a spectrum of tunicamycin concentrations for 12 h to block N-linked glycosylation, following which cell lysates were prepared and subjected to Western analysis. As indicated in Fig. 3A, we observed two major bands of this protein (lane 1); the upper band disappeared when treated with as little as 1 μg/ml tunicamycin, and this was accompanied with increasing intensity of the lower band (lanes 2 to 4). These data imply that NS4B may be modified by N-glycosylation in the absence of other viral proteins. The same Western blot was used to probe with tubulin antibody for internal loading control because of the similar electrophoretic mobilities of the recombinant NS4B glycosylated form and GAPDH.
FIG 3.
Recombinant NS4B is an N-glycosylated protein. (A) Effect of tunicamycin on recombinant DENV NS4B. HEK-293T cells (2 × 105 cells in a 12-well culture dish) were transfected with p2K-NS4B-V5 (4X) plasmid DNA construct (2 μg) encoding wild-type (WT) sequence. After 6 h posttransfection (hpt), cells were grown in the presence or absence of 0-, 1-, 5-, and 10-μg/ml doses of tunicamycin for 12 h. Cell lysates were harvested, and 10 μg of total protein was resolved by 10% SDS-PAGE. (B) Topology of DENV 2K-NS4B protein. Two domains, pTMD1 and pTMD2, in the ER lumen at the N terminus and three transmembrane domains, TMD3, TMD4, and TMD5, at the C terminus were previously published (14). (C) PNGase F treatment of recombinant NS4B proteins. HEK-293T cells (2 × 105 cells in a 12-well culture dish) were transfected with p2K-NS4B-V5 (4X) plasmid DNA constructs (2 μg) encoding WT, N58Q, N62Q, or N58QN62Q NS4B. Cell lysates were harvested 24 h posttransfection (hpt). Ten micrograms of total protein was treated with or without PNGase F and resolved by 10% SDS-PAGE. (D) Enrichment of recombinant NS4B proteins by ConA agarose. HEK-293T cells (8 × 106 cells in 10-cm culture dish) were transfected with p2K-NS4B-V5 (4X) plasmid DNA construct (10 μg) encoding WT or N58QN62Q NS4B. At 36 hpt, the cells were washed with 1× PBS and lysed in 1.5 ml of 1× PLB. Five hundred micrograms of total cellular protein was applied to ConA agarose for glycoprotein enrichment as described in Materials and Methods. Twenty percent of the starting material (input), 20% of the unbound fraction (unbound), and 100% of the E1 or E2 elution fraction were resolved by 10% SDS-PAGE. In panels A, B, and D, recombinant NS4B protein expression was detected with monoclonal anti-V5 antibody followed by HRP-conjugated anti-mouse antibody. Alpha-tubulin expression served as a loading control. “<” indicates the signal derived from NS4B protein, and “≪” indicates the signal derived from the N-glycosylated form of NS4B protein. All experiments were performed three times independently, and the data shown are from one representative transfection experiment.
The NetNGlyc 1.0 server revealed four putative N-glycosylation sites at asparagine residues 58, 62, 214, and 242 of NS4B, with higher scores for Asn-58 and Asn-62. According to the topology information (14), Asn-58 and Asn-62 localize at the ER lumen, Asn-214 resides in the ER luminal loop between transmembrane domains TMD4 and TMD5, and Asn-242 faces toward the cytosol (Fig. 3B). Miller et al. (14) showed that C-terminally truncated versions of recombinant DENV 2K-NS4B protein that contain residues 1 to 93 of NS4B gave rise to two bands in Western analyses, and the upper band was digested by PNGase F, ruling out the possibility of N-glycan modification on Asn-214 and Asn-242 residues. Considering the scores given by the NetNGlyc 1.0 server and the topology of the NS4B protein, it was speculated that NS4B could be modified by N-glycans at Asn-58 and Asn-62, as the oligosaccharyl transferase required for N-glycosylation resides in the ER lumen.
Asparagine (N) residues of the canonical NXS/T motif at residues 58 and 62 of NS4B were individually mutated to glutamine (Q), which represents an amino acid substitution differing by only the addition of a methylene group. We transfected recombinant p2K-NS4B-V5 (4X) plasmids encoding WT, N58Q, N62Q, and N58QN62Q into HEK-293T cells. We observed two major bands for WT, single mutant N58Q, and N62Q proteins in Western analysis; however, a single band was detected for double mutant N58QN62Q protein (Fig. 3C, odd-numbered lanes). Moreover, the electrophoretic mobility of the upper bands for single mutant proteins was higher than that for WT protein, and the single band of double mutant protein shared similar electrophoretic mobility with the lower band of WT and single mutant proteins (Fig. 3C, odd-numbered lanes). We also observed a similar mobility shift pattern of WT and single mutant proteins when these NS4B proteins were expressed in BHK-21 cells (data not shown), suggesting that the modification of NS4B protein is not cell type dependent. We suspected that the upper, lower-mobility bands were from glycosylated NS4B protein. To confirm this speculation, the cell lysates from p2K-NS4B-V5 (4X)-transfected HEK-293T cells were examined for PNGase F sensitivity. We found that the slower-migrating forms of WT and single mutant proteins were sensitive, whereas the single form of double mutant protein was resistant, to PNGase F treatment (Fig. 3C, even-numbered lanes). These results revealed that NS4B protein exists in at least two forms: the N-glycosylated and unglycosylated forms of low and high electrophoretic mobility, respectively. Furthermore, NS4B is N glycosylated at both N58 and N62 positions, and the other two possible N-glycosylation sites at amino acid positions 214 and 242 may not be N glycosylated, as the glycanase-treated N58QN62Q mutant protein did not show any difference in mobility from the PNGase F-untreated sample (Fig. 3C, lanes 7 and 8).
Next, we investigated whether the recombinant NS4B glycosylated form, which is free from other viral proteins, binds to ConA agarose resin. For this purpose, we used HEK-293T cell lysates that were transfected with p2K-NS4B-V5 (4X) plasmid DNA encoding WT or N58QN62Q mutant NS4B in the glycoprotein enrichment procedure. As shown in Fig. 3D, most of the unglycosylated form of WT NS4B as well as unglycosylated N58QN62Q mutant NS4B was in the unbound fraction (lanes 1 and 2 and lanes 5 and 6), and the NS4B glycosylated form was purified by ConA agarose resin (lanes 3 and 4). Hence, the NS4B glycosylated form bound to ConA agarose resin in the absence of other viral proteins. These results revealed that NS4B is an N-glycosylated protein and the N-to-Q mutation at residues 58 and 62 completely blocks the N-glycosylation modification.
Mutation of NS4B N-glycosylation residues reduced infectious DENV production in mammalian BHK-21 cells and mosquito C6/36 cells.
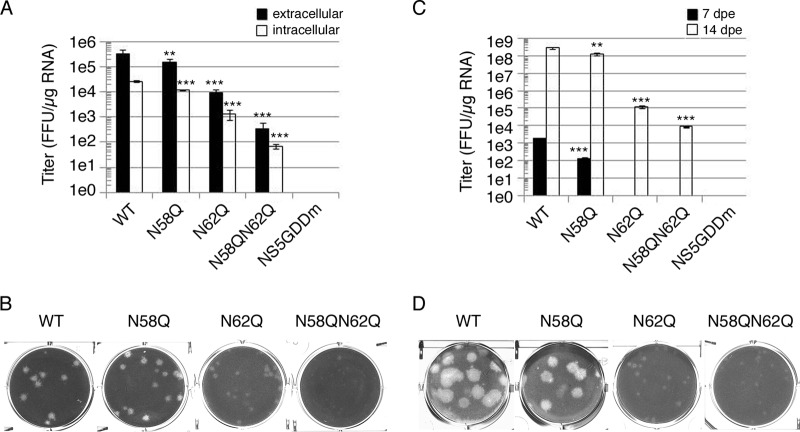
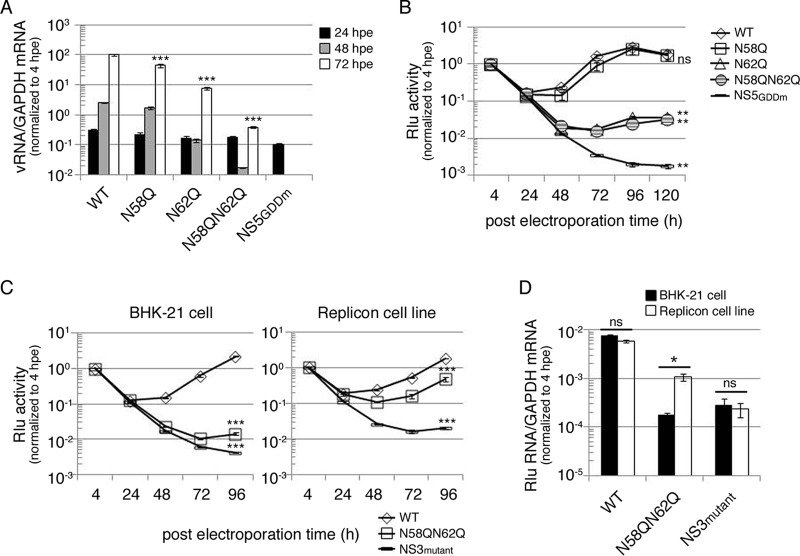
To investigate the biological role of NS4B N-glycosylation residues N58 and N62, several full-length infectious clones of DENV2 strain PL046 (named FL RNA) containing WT, N58Q, N62Q, or N58QN62Q in NS4B or the RNA-dependent RNA polymerase (RDRP)-defective NS5 GDD mutant (NS5GDDm) were generated. The in vitro-transcribed FL RNA was electroporated into BHK-21 cells followed by a systematic characterization of these FL RNAs. At 4 h postelectroporation (hpe), similar expression levels of DENV NS proteins suggested similar transfection efficiencies, and the Asn-to-Gln (N-to-Q) mutation at residues 58 and 62 of NS4B did not affect viral polyprotein translation and processing (data not shown). The extracellular viral titers from cell culture fluid at 144 hpe showed that the N58Q mutation alone exhibited a milder effect but the N62Q mutation dramatically decreased viral production efficiency, and further reduction in the virus titer was observed for the N58QN62Q double mutant (Fig. 4A). For WT and NS4B mutant FL RNAs, the intracellular viral titer from cell lysate and the extracellular viral titer exhibited similar patterns (Fig. 4A), suggesting that the NS4B N-glycosylation residue mutation does not affect the maturation, egress, or infectivity of the NS4B mutant viruses. Plaque morphology of BHK-21 cell-derived virus examined at 12 days postinfection (dpi) showed that the plaque was small and turbid for the N62Q mutant and much smaller and also turbid for N58QN62Q double mutant (Fig. 4B). These data revealed the distinct abilities of Asn-58 and Asn-62 N-glycosylation residues; mutation of these residues affects virion production, and this effect may be mediated by loss of NS4B glycans.
FIG 4.
Mutation of putative N-glycosylation sites on DENV NS4B affected infectious virus production. (A and C) Detection of viral production efficiency in mammalian BHK-21 cells (A) and mosquito C6/36 cells (C). FL RNA (10 μg for BHK-21 cells or 15 μg for C6/36) encoding WT, N58Q, N62Q, or N58QN62Q NS4B was electroporated into BHK-21 cells (A) or C6/36 cells (C). For panel A, culture fluid (extracellular fraction) and cell lysate (intracellular fraction) were collected at 144 hpe. For panel C, culture fluid was collected at 7 dpe and 14 dpe (days postelectroporation). Viral titer was determined by focus-forming assay in naive BHK-21 cells. Focus-forming assays were performed in triplicate with one dilution to determine the mean and standard deviation. The experiment was performed independently in triplicate, and the data shown are representative of one independent electroporation experiment. Asterisks (**, P < 0.005; ***, P < 0.0005) indicate results significantly different from the WT. (B and D) Characterization of NS4B N-glycosylation mutant viruses derived from WT or NS4B mutant FL RNA-electroporated BHK-21 cells (B) and C6/36 cells (D). The culture fluids from 144 hpe of BHK-21 cells (B) and 14 dpe of C6/36 cells (D) were subjected to plaque assay in naive BHK-21 cells, and plaques were fixed after 12 days postinfection (dpi).
Mosquitoes and humans are hosts of DENV. Hence, we also examined the effect of NS4B N58Q and N62Q mutations in mosquito C6/36 cells. Only the WT and N58Q mutant FL RNAs produced virus at 7 dpe (Fig. 4C). All FL RNAs could produce virus at 14 dpe, but the N58QN62Q double mutant showed dramatic reduction in the infectious viral titer compared to the WT and single mutants (Fig. 4C), suggesting that NS4B N-glycans may be essential for the DENV life cycle in the mosquito host system. Plaque morphology of mosquito C6/36 cell-derived virus showed small and turbid plaques for N58QN62Q mutant virus similar to those seen for BHK-21 cell-derived N58QN62Q mutant virus (Fig. 4B and D). Therefore, we concluded that NS4B may be N glycosylated in mammalian and mosquito hosts, and the NS4B N58QN62Q mutation appeared to hinder the generation of infectious viruses in cultured cells.
DENV RNA replication is decreased by mutation of NS4B N-glycosylation sites.
To examine the effect of the NS4B N-glycosylation site mutation in the replication step of the virus life cycle, BHK-21 cells were electroporated with FL RNAs, and viral RNA synthesis as a function of time was analyzed using real-time qPCR. The input RNA amounts at 4 hpe were nearly identical, indicating similar transfection efficiencies (data not shown). At 48 hpe and 72 hpe, viral RNA synthesis for the replication-defective NS5GDDm was below the detection limit, and N62Q single mutant and N58QN62Q double mutant showed significant decreases in viral RNA synthesis compared to the WT (Fig. 5A).
FIG 5.
Mutation of putative N-glycosylation sites on DENV NS4B decreased viral RNA replication. (A) Replication analysis of NS4B mutants using full-length (FL) infectious clone RNA. FL RNA (10 μg) encoding WT, N58Q, N62Q, or N58QN62Q NS4B was electroporated into BHK-21 cells (3 × 106 cells). The NS5GDDm FL RNA was a replication-deficient control. At the indicated time points postelectroporation, total cellular RNA was isolated. The viral RNA NS3 protein coding region was quantitated using real-time qPCR, and host GAPDH mRNA quantification served as a control. All the later time points were normalized with the 4-hpe value. Asterisks (***, P < 0.0005) indicate results significantly different from the WT. The data for the last time point in the experiment were chosen for statistical analysis. (B) Replication analysis of NS4B mutants using DENV subgenomic replicon with Renilla luciferase reporter. DENV SGR-Rlu replicon RNA (10 μg) encoding WT, N58Q, N62Q, or N58QN62Q NS4B was electroporated into BHK-21 cells (3 × 106 cells). The NS5GDDm SGR-Rlu replicon was a replication-deficient control. Cell lysate was prepared at 4, 24, 48, 72, 96, and 120 hpe to determine the Renilla luciferase activity at each time point. All the later time points were normalized against 4-h Renilla luciferase activity that indicated the electroporation efficiency. The normalized Renilla luciferase activity at different time points is expressed as the mean ± standard deviation of triplicates. The experiment was performed in triplicate, and the data shown are from one representative electroporation experiment. Asterisks (**, P < 0.005) indicate results significantly different from the WT, and “ns” represents no significant difference. The data for the last time point in the experiment were chosen for statistical analysis. (C and D) trans-complementation analyses of NS4B N58QN62Q mutant by Renilla luciferase reporter (C) and real-time qPCR (D). For panel C, naive BHK-21 cells or the D2-Flu-SGR-Neo replicon cell line was electroporated with DENV SGR-Rlu replicon RNA (10 μg) encoding WT NS4B, N58QN62Q mutant NS4B, or NS3 K199AT200A mutant. The normalized Renilla luciferase activity at different time points postelectroporation is expressed as mean ± standard deviation for triplicate experiments. The experiment was performed in triplicate, and the data shown are from one representative experiment. Asterisks (***, P < 0.0005) indicate results significantly different from the WT. The data from the last time point in the experiment were chosen for statistical analysis. For panel D, at 4 hpe and 96 hpe, total RNA was isolated from both host cells. The Renilla luciferase region was amplified with qPCR primers, GAPDH mRNA quantification served as a control, and 96-hpe data were normalized to the 4-hpe value. An asterisk (*, P < 0.05) indicates a significant difference from the naive BHK-21 cells electroporated with DENV SGR-Rlu replicon NS4B N58QN62Q mutant RNA, and “ns” represents no significant difference. The experiment was performed twice, and the data shown are from one representative electroporation experiment.
From real-time qPCR results, it was noticed that the N58QN62Q double mutant affected RNA replication of the DENV genome. However, reinfection by virus would increase the RNA level. Therefore, to rule out this possibility, a DENV subgenomic replicon, SGR-Rlu, was used for the replication study, wherein the prM and E genes were deleted in frame to prevent virion formation and an internal ribosome entry site (IRES)-driven Renilla luciferase reporter gene was introduced at the 3′ UTR. SGR-Rlu replicon RNA containing WT, N58Q, N62Q, or N58QN62Q NS4B or NS5GDDm was electroporated into BHK-21 cells to analyze the efficiency of replication-dependent reporter expression as a function of time in BHK-21 cells. The luciferase signals for N62Q and N58QN62Q replicons were 1 order of magnitude lower at 48 hpe and 2 orders of magnitude lower at 72 hpe than those for WT and N58Q replicons (Fig. 5B). The replication-defective NS5GDDm replicon showed very low levels of luciferase signal throughout the course of the experiment (Fig. 5B). These results indicate that the NS4B N62Q and N58QN62Q mutations reduced the replication of DENV replicon.
NS4B N58QN62Q mutant can be trans-complemented by functional replication complex.
Previously, the lethal P104R mutation of DENV NS4B was shown to be trans-complemented by a WT NS4B harboring helper replicon (4). Recently, the same group reported that the lethal NS4B mutant replicon cannot be trans-complemented by WT NS4B protein alone but requires NS4B from the replication complex (15). To examine whether the replication-incompetent replicon SGR-Rlu-NS4B N58QN62Q could be rescued by a functionally active replicon in trans, we used the D2-Flu-SGR-Neo replicon cell line, which was originally established for an anti-DENV inhibitor study and harbors an autonomously replicating WT replicon and firefly luciferase gene (24). The NS3 helicase-defective K199AT200A mutant replicon was used as a negative control in trans-complementation assays. Electroporation of different replicons into D2-Flu-SGR-Neo cells did not affect its replication efficiency as determined by analysis of firefly luciferase activity as a function of time (data not shown). The SGR-Rlu-NS4B N58QN62Q replicon showed low levels of Renilla luciferase activity in naive BHK-21 cells (Fig. 5C, left), indicating its inability to replicate. However, its Renilla luciferase activity was significantly increased in D2-Flu-SGR-Neo cells, which provide a functionally active replication complex (Fig. 5C, right). In contrast, Renilla luciferase activity steadily declined for the negative-control SGR-Rlu-NS3 K199AT200A replicon from 4 hpe onward to 96 hpe in BHK-21 cells and D2-Flu-SGR-Neo cells (Fig. 5C). These data revealed that the replication deficiency of NS4B N58QN62Q mutant replicon could be trans-complemented.
Furthermore, to confirm this observation at the RNA level, real-time qPCR analysis at 96 hpe was performed. RNA levels in both naive BHK-21 cells and D2-Flu-SGR-Neo cells harboring the WT replicon were comparable (Fig. 5D), indicating similar replication efficiencies in the two cell lines. The negative-control NS3 K199AT200A replicon exhibited reduced RNA synthesis in both cell lines. As expected, NS4B N58QN62Q replicon synthesized significantly higher levels of RNA in D2-Flu-SGR-Neo cells than in BHK-21 cells (Fig. 5D). These results reveal that the defective replication complex caused by the NS4B N58QN62Q mutation may be trans-complemented by helper replication complex containing N-glycosylated NS4B.
Strategy for screening mutations capable of compensating for NS4B N58QN62Q.
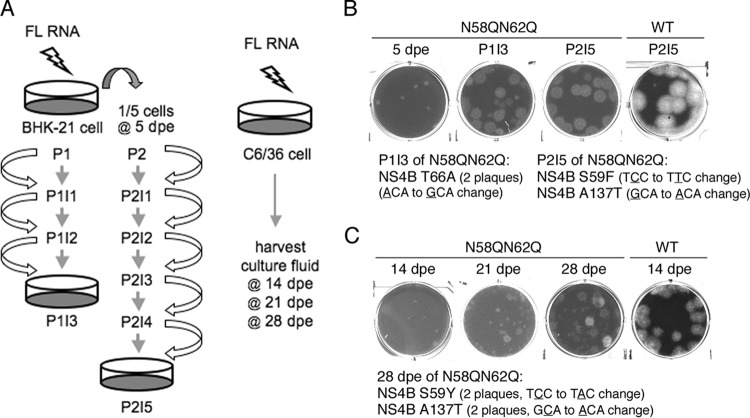
Since it was observed that the NS4B N58QN62Q mutation affected genome replication in mammalian and mosquito host systems, it was hypothesized that the glycans on NS4B might function through interacting with other components or modulating NS4B structure and function in the replicase complex. To gain insight into how the NS4B N58QN62Q mutation affects viral genome replication, we aimed to encourage the emergence of compensatory mutations that might correct the defect in replication. FL RNA of the WT or NS4B N58QN62Q mutant was electroporated in BHK-21 cells. As shown in the left panel of Fig. 6A, cells were trypsinized at 5 days postelectroporation (dpe) (referred to as P1) and one-fifth of the cells were subcultured into a new culture dish for another 3 days (referred to as P2), with the expectation that the electroporated cells would undergo replication stress and increase the viral progeny yield. Naive BHK-21 cells were infected with the culture fluid from both passages P1 and P2 for 3 days, referred to as P1I1 and P2I1, respectively. Subsequently, naive BHK-21 cells were infected with the culture fluid from passages P1I2 and P2I2 for 3 days and are indicated as P1I2 and P2I2, respectively. Likewise, this procedure was followed until the number of NS3-positive cells significantly increased and the virus in the culture fluid gave a focus size similar to that of WT virus. As expected, the focus size generated by N58QN62Q mutant virus at P1I1 or P2I1 was smaller, but surprisingly, focus sizes were increased by P1I2 or P2I4 and reached sizes comparable to those of the WT by P1I3 or P2I5 (data not shown), indicating the likely emergence of rescued virus that had corrected the defects in replication and virus production.
FIG 6.
Screening strategy for compensatory mutations of NS4B NS58QN62Q. (A) Schematic representation of procedures used to screen compensatory mutations of NS4B N58QNS62Q mutant virus from mammalian BHK-21 cells (left panel) and mosquito C6/36 cells (right panel). BHK-21 cells were electroporated with WT or N58QN62Q mutant FL RNA (10 μg). At 5 dpe, one-fifth of the electroporated cells from P1 were subcultured to a new culture dish and named P2. The culture fluid was transferred to naive BHK-21 cells at 3-day intervals until P1I3 and P2I5, thus enriching any rescued virus. C6/36 cells were electroporated with WT or N58QN62Q mutant FL RNA (10 μg), and the culture fluids were harvested at 14, 21, and 28 dpe. (B and C) Analysis of rescued virus from BHK-21 cells (B) and C6/36 cells (C). For panel B, culture fluids of 5 dpe, P1I3, and P2I5 for NS4B N58QN62Q FL RNA were used for plaque assays. The plaque assay of the culture fluid of P2I5 for WT FL RNA was used for comparison. Two isolated plaques from P1I3 contained an NS4B Y66A second-site mutation, and independent plaques from P2I5 contained NS4B S59F or A137T second-site mutations. For panel C, culture fluids of 14, 21, and 28 dpe for NS4B N58QN62Q FL RNA were used for the plaque assay. For comparison, a culture fluid of 14 dpe for WT FL RNA was used for the plaque assay. NS4B S59Y and A137 second-site mutations were identified in isolated plaques from 28 dpe, with two clones for each mutation.
Plaque assay was performed with the culture fluids of P1I3 and P2I5, using the culture fluid from 5 dpe as a control. It was observed that the NS4B N58QN62Q virus from P1I3 and P2I5 gave rise to significantly bigger plaques than that of 5 dpe, although the plaques were turbid compared to those of WT virus (Fig. 6B). At this point, viruses from larger plaques were amplified in mosquito C6/36 cells and the viral RNA was extracted for cDNA sequencing. We sequenced the majority of the nonstructural protein-encoding region, including NS1, NS3, NS4A, NS4B, and NS5 genes, of the viral genome. Interestingly, the plaques from both P1I3 and P2I5 revealed the presence of intragenic mutations in NS4B, i.e., S59F, T66A, and A137T, while viruses from all large plaques retained the original N58QN62Q mutation (Fig. 6B).
Simultaneously, experiments were carried out to obtain the compensatory mutation for the NS58QN62Q mutation from mosquito C6/36 cells, as this host is believed to provide a different environment for virus survival. Previously, it was observed that culture fluid of 14 dpe obtained from NS4B N58QN62Q FL RNA-electroporated C6/36 cells still yielded small plaques, implying that the genome needs more time to generate compensatory mutations in this host. Hence, the incubation period was increased to at least 4 weeks and the culture fluid was collected at 7-day intervals for plaque morphological analysis (Fig. 6A, right). Culture fluid of 21-dpe cells yielded few large plaques, and the number of large plaques was increased in culture fluid of 28-dpe cells (Fig. 6C), indicating an enrichment of rescued viruses. Samples from large plaques were selected, amplified in C6/36 cells, and subjected to viral genome cDNA sequencing. Notably, two NS4B intragenic second-site mutations were found, i.e., S59Y and A137T, wherein the amino acid positions were identical to the mutations obtained from screening with BHK-21 cells (Fig. 6C), implying the significance of the amino acid position in NS4B structure and function in mammalian and mosquito hosts.
Intragenic suppressors rescue the defective function of the NS4B N58QN62Q mutation.
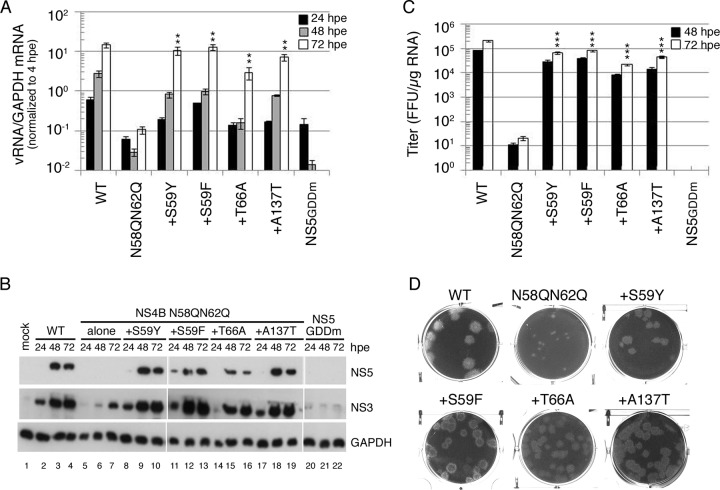
The screening of the suppressor for NS4B N58QN62Q mutant FL RNA in both mammalian cells and mosquito cells revealed the occurrence of four intragenic second-site mutations: S59Y and S59F within the glycosylation acceptor motif, T66A in the membrane-associated pTMD2 domain, and A137T in the cytosolic loop (Fig. 3B). To determine whether these mutations were responsible for restoring the replication defect and the subsequent infectious virus production, they were cloned into the FL RNA containing the original NS4B N58QN62Q mutation. We found that WT FL RNA synthesized RNA efficiently as a function of time, whereas no RNA synthesis was detected from 48-hpe and 72-hpe cells for replication-defective NS5GDDm FL RNA. NS4B N58QN62Q mutant FL RNA was defective in RNA synthesis (Fig. 7A). Notably, all of the second-site mutations rescued the RNA replication defect of the N58QN62Q mutation that might be associated with the loss of N-glycans from NS4B (Fig. 7A). Additionally, Western analyses revealed that all NS4B intragenic second-site mutations restored the viral NS protein expression defect caused by the N58QN62Q mutation, while for NS5GDDm FL RNA, NS proteins were not detectable throughout the course of the experiment (Fig. 7B).
FIG 7.
Rescuing effect of compensatory mutations on NS4B N58QN62Q mutation. (A) Viral RNA synthesis. BHK-21 cells were electroporated with FL RNA (10 μg) encoding WT, N58QN62Q, and N58QN62Q+suppressor NS4B. The levels of viral RNA and cellular GAPDH mRNA at 4, 24, 48, and 72 hpe were quantified by real-time qPCR. The data were normalized to the 4-hpe value, which reflects the FL RNA electroporation efficiency. The electroporation experiment was performed three times, and real-time qPCR was done in triplicate. The mean ± standard deviation shown here is from one representative electroporation experiment. Asterisks (**, P < 0.005) indicate results significantly different from the NS4B N58QN62Q mutation. The data from the last time point in the experiment were chosen for statistical analysis. (B) Replication-dependent translation of FL RNA. At 24, 48, and 72 hpe, the cells were lysed, and NS3 and NS5 expression was analyzed by Western blotting. GAPDH expression served as a protein loading control. (C) Virus production. Culture fluid of FL RNA electroporated BHK-21 cells collected at 48 hpe and 72 hpe was used to infect naive BHK-21 cells, and virus titer was determined by focus-forming assay. Results are expressed as mean ± standard deviation of triplicates. The experiment was performed three times, and the data shown are from one representative electroporation experiment. Asterisks (***, P < 0.0005) indicate results significantly different from the NS4B N58QN62Q mutation. The data from the last time point in the experiment were chosen for statistical analysis. (D) Plaque morphology. Culture fluid from 144 hpe was subjected to plaque assay in BHK-21 cells. Plaques were fixed at 12 dpi.
To investigate whether the suppressor mutation corrected all the steps in the virus life cycle, we examined the virus production efficiencies at 48 hpe and 72 hpe. As expected, NS4B N58QN62Q FL RNA produced nearly 4-orders-of-magnitude-fewer infectious virus particles than did WT FL RNA. In contrast, NS4B N58QN62Q+suppressor FL RNAs yielded high viral titers nearly comparable to those for the WT FL RNA (Fig. 7C). Additionally, viruses collected from 72 hpe were used for the plaque assay. We found that the NS4B N58QN62Q mutant virus yielded smaller, turbid plaques while all four NS4B N58QN62Q+suppressor viruses resulted in homogeneous plaques larger than the original N58QN62Q mutant virus but with a more turbid phenotype than that of the WT virus (Fig. 7D). These results implied that NS4B intragenic second-site mutations rescued the defects in viral RNA replication and the subsequent infectious virion production that might be associated with loss of N-glycans from NS4B.
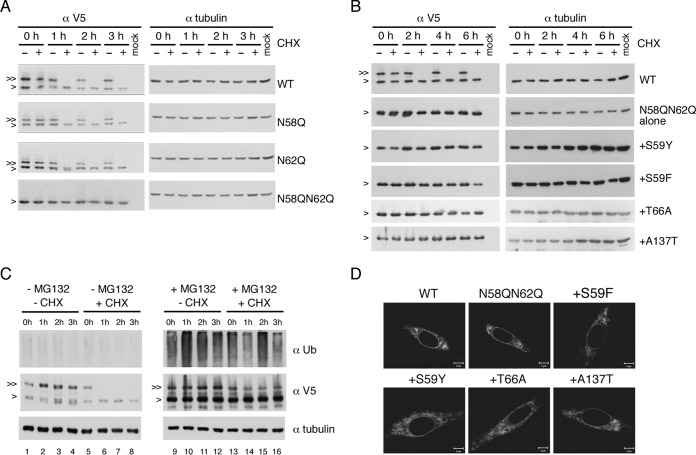
Defective N-glycosylation did not affect the stability of NS4B mutant proteins.
It is reported elsewhere that removal of N-glycosylation from glycoproteins leads to folding defects (25) and that subsequently the stability of the improperly folded protein is affected (26, 27). Therefore, we wished to examine whether NS4B N-glycosylation site mutations have any effect on the stability of NS4B and whether suppressor mutations of the N58QN62Q variant modulate the stability of unglycosylated NS4B, which may provide some hints at the biological role of NS4B N-glycosylation. For this purpose, p2K-NS4B-V5 (4X) plasmids containing NS4B N-glycosylation mutations and NS4B N58QN62Q+suppressor mutations were transfected into HEK-293T cells and at 24 h posttransfection were treated with DMSO or cycloheximide (CHX) for up to 6 h. We found that the unglycosylated form was more stable than the glycosylated form for WT NS4B protein as well as N58Q and N62Q single mutant proteins (Fig. 8A). Additionally, the stability of the unglycosylated protein by N58QN62Q mutation was comparable to the unglycosylated form of WT and single mutant proteins (Fig. 8A). Expression of NS4B N58QN62Q+suppressor proteins did not produce any form other than the unglycosylated form of NS4B (Fig. 8B), suggesting that the suppressor mutation did not create a novel glycosylation site. Similar stabilities among all N58QN62Q+suppressor mutant proteins were observed up to 6 h post-CHX treatment, and over the time course, these proteins were indistinguishable from the original NS4B N58QN62Q mutant protein and the unglycosylated form of WT NS4B (Fig. 8B). These data suggest that suppressor mutations rescue the replication and subsequent virus production deficiencies of NS4B N58QN62Q, possibly by correcting the NS4B protein conformation that might have been altered by the loss of N-glycans rather than by altering the stabilities of NS4B protein.
FIG 8.
Ablation of glycosylation did not affect NS4B stability. (A to C) Stability analyses of NS4B proteins. For panels A and B, HEK-293T cells (2 × 105 cells) were transfected with p2K-NS4B-V5 (4X) plasmid DNA constructs (2 μg) encoding WT NS4B or NS4B with N58Q, N62Q, N58QN62Q, or N58QN62Q+suppressor mutations. Cells were treated with 0.5% DMSO or cycloheximide (CHX) (1 mg/ml in 0.5% DMSO) at 24 hpt for 0 to 6 h. For panel C, HEK-293T cells were transfected with p2K-NS4B-V5 (4X) plasmid DNA construct (2 μg) encoding WT NS4B. After 24 h, cells were treated with or without 40 μM MG132 for 12 h and then treated with or without CHX (1 mg/ml) for 0 to 3 h. Cell lysates were harvested, and proteins were resolved by 10% SDS-PAGE. 2K-NS4B-V5 (4X) fusion protein was probed with monoclonal anti-V5 antibody followed by HRP-conjugated anti-mouse antibody. For panel C, the detection of a ubiquitination smear under MG132 treatment indicated the successful inhibition of 26S proteasome machinery and alpha-tubulin expression served as a loading control. “>” indicates the NS4B protein; “≫” indicates the N-glycosylated form of NS4B protein. All experiments were performed twice independently, and the data shown are from one representative transfection experiment. (D) Cellular distribution of NS4B mutant proteins. BHK-21 cells were transfected with p2K-NS4B-V5 (4X) plasmid DNA constructs containing WT, N58QN62Q, or N58QN62Q+suppressor mutations. After 24 h, cells were fixed with 4% paraformaldehyde and the immunofluorescence assay was carried out with anti-V5 monoclonal primary antibody and anti-mouse-Cy3 secondary antibody.
Further, we analyzed the fate of glycosylated NS4B under treatment with a 26S proteasome inhibitor, MG132. HEK-293T cells were transfected with p2K-NS4B-V5 (4X) plasmid for 24 h, treated with MG132 for 12 h, and then treated with or without CHX for 0 to 3 h. Detection of a ubiquitination smear under MG132 treatment indicated the successful inhibition of 26S proteasome machinery (Fig. 8C, top panel, lanes 9 to 16 versus 1 to 8). The glycosylated form of NS4B was almost completely degraded at as early as 1 h post-CHX treatment (Fig. 8C, middle panel, lanes 5 to 8) but remained almost intact in the presence of MG132 (Fig. 8C, middle panel, lanes 13 to 16). These results suggest that the NS4B glycosylated form degrades through the 26S proteasome machinery.
Furthermore, the cellular distribution of N58QN62Q+suppressor mutant proteins was examined to detect any changes in the staining pattern at the cellular level. We found that WT, NS58QN62Q, and NS58QN62Q+suppressor proteins had similar reticular staining patterns (Fig. 8D).
DISCUSSION
In this study, we demonstrated that DENV NS4B is N glycosylated in virus-infected cells and in cells expressing recombinant NS4B protein. Mutation of putative N-glycosylation sites on NS4B affected viral genome replication and the subsequent viral production efficiencies, whereas intragenic mutations compensated for these deficiencies without creating novel glycosylation sites, suggesting that the glycans could be involved in maintenance of NS4B folding.
We showed that recombinant NS4B protein could be N glycosylated at the Asn-58 and Asn-62 residues, and this modification may not be cell type specific. Zou et al. showed that DENV NS4B forms a dimer or multimer but is not N glycosylated in virus-infected cells (15). Moreover, the same report showed a single species (15) and a later report of the same research group showed two species from recombinant NS4B protein expression which were assumed to be 2K-4B unprocessed and processed forms (8). To some extent, their data are not in agreement with our data and others' findings (14). These discrepancies in the results could possibly be due to different antibodies used in the experiments. We detected glycosylated and unglycosylated forms of NS4B of similar intensities in recombinant NS4B protein expression experiments using V5 tag antibody, which detects C-terminus-fused V5, but unfortunately, recombinant NS4B protein was not detected by the commercial NS4B antibody in Western analyses. This commercial NS4B antibody specifically detects NS4B protein in the virus-infected cell lysates, whereas it may have limitations in the detection of the NS4B glycosylated form as the antibody was raised against an N terminus peptide, making it unable to recognize the NS4B protein containing N-glycans.
In addition to the antibody issue, there could be other reasons for the huge difference in Western detection of glycosylated versus unglycosylated forms of NS4B in virus-infected cells. First is low solubility; the overall yield of NS4B protein from our membrane solubilization attempts was lower than the yield of another membrane-associated envelope (E) glycoprotein (data not shown). This may also account for the better enrichment of E glycoprotein than of the glycosylated form of NS4B in the ConA agarose affinity approach (Fig. 1C and D). Second, the process of N-glycosylation onto NS4B may be transient or the glycosylated form of NS4B may have degraded. Third is the low recovery of glycosylated NS4B from ConA agarose resin. We could improve the recovery efficiency by heating the ConA agarose-bound glycoproteins in the presence of SDS reducing sample buffer (data not shown). Nevertheless, this heating approach was not possible for better enrichment of the NS4B glycosylated form from virus-infected cell lysates because ConA agarose beads were damaged and released the ConA lectin, which was around the size of the NS4B glycosylated form, and unfortunately, ConA was recognized nonspecifically by the NS4B antibody. Because of the lower abundance, it was technically difficult to further demonstrate the NS4B glycosylation, for example, by mass spectrometry. The de novo N-glycosylation occurring on NS4B in the cells was demonstrated in cells transfected with the recombinant NS4B protein expression construct by treatment with tunicamycin. However, it was not possible to show in virus-infected cells as tunicamycin severely reduced viral genome replication under the optimized conditions of a 1-μg/ml concentration for 1 h of treatment (data not shown). Regardless of the commercial NS4B antibody detection limit and other possibilities, we have demonstrated the affinity of the NS4B glycosylated form for ConA agarose and further confirmed that it is an N-glycosylated form by PNGase F treatment in at least two mammalian cell lines, BHK-21 and Huh7, infected with DENV. Furthermore, the observation of glycosylated and unglycosylated forms of NS4B was corroborated by the results of recombinant protein expression, strongly indicating that DENV NS4B is an N-glycosylated protein.
NS4B protein physically interacts with NS3 and NS4A (7–9) and with NS1 in WNV (11) and exhibits genetic interaction with NS4A in JEV (10), indicating that NS4B may have the ability to tightly associate with viral replicase proteins and also with host proteins in the replication complex. Given that NS4B plays a significant role in the establishment of protein-protein interaction in the replication complex, maintenance of its structure and function would be of the utmost importance. N-linked glycans on NS4B may be essential to maintain its structural integrity and its biological function, as our subgenomic replicon, quantitative PCR, and trans-complementation studies revealed that putative NS4B glycans are necessary at the replication step in the DENV life cycle.
We have checked the virus pathogen database and resource (http://www.viprbrc.org). There are 4,427 DENV2 strains, and for 1,138 strains, the complete genomes are available in this database. We observed that Asn-58 and Asn-62 are well conserved in 1,138 DENV2 strains. Our studies showed that mutation of putative Asn-58 and Asn-62 glycosylation residues of NS4B to Gln (Q) residues exhibited distinct effects on viral RNA replication and subsequent virion production and the Asn (N)-to-Gln (Q) mutation of Asn-62 exhibited more severe effects, which could be mediated by the loss of glycans. Asn-58 is conserved in dengue viruses while it is absent in related flaviviruses, whereas Asn-62 is conserved in four DENV serotypes and also in related WNV and YFV but not JEV (Fig. 1). Thus, NS4B Asn-62 has some evolutionary significance, and it would be interesting to examine this potential N-glycan in other flaviviruses such as WNV and YFV.
Besides the functional characterizations of putative NS4B glycans in mammalian and mosquito hosts, the other key finding of this work is the screening and identification of NS4B intragenic compensatory mutations that rescue the viral RNA replication deficiency of the NS4B N58QN62Q mutation. When these NS4B intragenic mutations, including S59Y/F, T66A, and A137T, were introduced back into the N58QN62Q genotype background, viral RNA replication and virion production efficiencies were increased by 100- to 1,000-fold relative to that of the N58QN62Q variant, although they remained less efficient than that of WT FL RNA (Fig. 7A and C). We speculate that loss of glycans may affect the structure of NS4B, most probably in the N-terminal luminal domains, and the compensatory abilities of the aromatic amino acid (tyrosine or phenylalanine) within the Asn-58 glycosylation acceptor motif, the alanine at the amino acid (aa) 66 position, or the threonine at the aa 137 position play a significant role in correcting the structural defect of NS4B N58QN62Q. A recent study showed the presence of an interaction between the cytosolic loop of NS4B (aa 129 to 165) and NS3 protein (9). It is possible that NS4B N58QN62Q mutation alters the conformation of the cytosolic loop and disrupts NS4B-NS3 interaction, and different compensatory mutations might correct the conformation of the cytosolic loop, rebuilding the interaction with NS3. Previously, the mutation was observed in the NS4B gene that was screened from 1,284 chemically mutagenized virus clones of DENV4 (6), and in another report, the drug resistance mutations were mainly found in the NS4B gene of DENV2 (4). In this study, we found compensatory mutations only in the NS4B region. These observations hint that the NS4B protein may have some flexibility/plasticity in its structure due to which compensatory mutations arise.
In our stability analyses of recombinant NS4B protein, the glycosylated form of NS4B is found to be less stable than the unglycosylated form. Further, we also found that recombinant NS4B significantly degraded through 26S proteasome machinery (Fig. 8C). However, a block of proteasome machinery in virus-infected cells showed a mild but not significant increase in glycosylated NS4B detection (data not shown), giving no reason for the low level of glycosylated NS4B in virus-infected cells. All the N58QN62Q+suppressor mutant NS4B proteins that lacked the glycans were as stable as the unglycosylated form of WT NS4B protein and the original N58QN62Q mutant NS4B protein (Fig. 8B), suggesting that at least protein stability may not be the reason for the replication defect of the NS4B N58QN62Q mutant. Our experiments with full-length infectious clones were primarily performed in BHK-21 cells, which lack the interferon (IFN) production pathway (28), and hence, we believe that NS4B glycans may not be involved in the hallmark function of NS4B antagonism of interferon signaling (12). Moreover, one group demonstrated that NS4B has RNAi suppressor activity and expression of the NS4B construct containing residues 1 to 93 did not have any effect on the RNAi suppressor mechanism (13). Another report showed that NS4B can form a dimer or multimer and that residues 129 to 165 or 166 to 248 of NS4B are involved in the NS4B dimerization process (15). Thus, considering the location of N-glycosylation sites (aa 58 and aa 62) in NS4B, glycans may not regulate dengue virus replication through RNAi suppression mechanisms or modulation of the NS4B dimerization process.
Taking all these observations into account and irrespective of the possibilities whether the glycosylated form of NS4B is degraded or is due to the detection limits of commercial NS4B antibody in virus-infected cells, our data supported the fact that NS4B is an N-glycosylated protein. Mutation of putative N-glycosylation sites on NS4B dramatically reduced replication and subsequent virus production. We speculate that loss of glycans by N58QN62Q mutation may lead to the misfolding of NS4B, and as a result, NS4B loses the ability to interact with viral proteins or host proteins in the replication complex. Searching for the interacting partners of glycosylated and unglycosylated NS4B in the replication complex is required to better understand the molecular mechanism presented here. In summary, we have identified NS4B N-glycosylation sites and characterized the function of putative NS4B glycosylation in mammalian and mosquito hosts. NS4B glycans appeared to be required for efficient viral RNA replication and subsequent virion production but are dispensable in the presence of suppressor mutations. Our data also showed that a single point mutation converted the folding of NS4B from N-glycosylation dependent to N-glycosylation independent. Finally, this study added evidence that in addition to the other well-known flaviviral prM, E, and NS1 glycoproteins, NS4B is an N-glycosylated protein, and the glycosylated form of NS4B might potentially be utilized in the DENV replication complex. This novel finding may have broader implications in flaviviral biology as the most likely N-glycosylation site at Asn-62 of NS4B is well conserved in DENV serotypes and in some related flaviviruses (WNV and YFV). Moreover, this finding may also lead to deeper insights into the role of NS4B in the replication complex as NS4B has recently emerged as a major antiviral drug target (29).
ACKNOWLEDGMENTS
Some of the preliminary experiments were performed by Chiao-Ling Hung. We are grateful to AndreAna Pena for English editing.
This work was supported by a grant from the Ministry of Science and Technology, Taiwan, and Academia Sinica, Taiwan.
REFERENCES
- 1.Guzman A, Isturiz RE. 2010. Update on the global spread of dengue. Int J Antimicrob Agents 36(Suppl 1):S40–S42. doi: 10.1016/j.ijantimicag.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 2.Lindenbach BD, Thiel H-J, Rice CM. 2007. Flaviviridae: the viruses and their replication, p 1101–1152. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed),. Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 3.Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie X, Wang QY, Xu HY, Qing M, Kramer L, Yuan Z, Shi PY. 2011. Inhibition of dengue virus by targeting viral NS4B protein. J Virol 85:11183–11195. doi: 10.1128/JVI.05468-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grant D, Tan GK, Qing M, Ng JK, Yip A, Zou G, Xie X, Yuan Z, Schreiber MJ, Schul W, Shi PY, Alonso S. 2011. A single amino acid in nonstructural protein NS4B confers virulence to dengue virus in AG129 mice through enhancement of viral RNA synthesis. J Virol 85:7775–7787. doi: 10.1128/JVI.00665-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanley KA, Manlucu LR, Gilmore LE, Blaney JE Jr, Hanson CT, Murphy BR, Whitehead SS. 2003. A trade-off in replication in mosquito versus mammalian systems conferred by a point mutation in the NS4B protein of dengue virus type 4. Virology 312:222–232. doi: 10.1016/S0042-6822(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 7.Umareddy I, Chao A, Sampath A, Gu F, Vasudevan SG. 2006. Dengue virus NS4B interacts with NS3 and dissociates it from single-stranded RNA. J Gen Virol 87:2605–2614. doi: 10.1099/vir.0.81844-0. [DOI] [PubMed] [Google Scholar]
- 8.Zou J, Xie X, Wang QY, Dong H, Lee MY, Kang C, Yuan Z, Shi PY. 2015. Characterization of dengue virus NS4A and NS4B protein interaction. J Virol 89:3455–3470. doi: 10.1128/JVI.03453-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou J, Lee le, Wang TQY, Xie X, Lu S, Yau YH, Yuan Z, Geifman Shochat S, Kang C, Lescar J, Shi PY. 2015. Mapping the interactions between the NS4B and NS3 proteins of dengue virus. J Virol 89:3471–3483. doi: 10.1128/JVI.03454-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li XD, Ye HQ, Deng CL, Liu SQ, Zhang HL, Shang BD, Shi PY, Yuan ZM, Zhang B. 9 January 2015. Genetic interaction between NS4A and NS4B for replication of Japanese encephalitis virus. J Gen Virol doi: 10.1099/vir.0.000044. [DOI] [PubMed] [Google Scholar]
- 11.Youn S, Li T, McCune BT, Edeling MA, Fremont DH, Cristea IM, Diamond MS. 2012. Evidence for a genetic and physical interaction between nonstructural proteins NS1 and NS4B that modulates replication of West Nile virus. J Virol 86:7360–7371. doi: 10.1128/JVI.00157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munoz-Jordan JL, Laurent-Rolle M, Ashour J, Martinez-Sobrido L, Ashok M, Lipkin WI, Garcia-Sastre A. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J Virol 79:8004–8013. doi: 10.1128/JVI.79.13.8004-8013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kakumani PK, Ponia SS, Rajgokul KS, Sood V, Chinnappan M, Banerjea AC, Medigeshi GR, Malhotra P, Mukherjee SK, Bhatnagar RK. 2013. Role of RNA interference (RNAi) in dengue virus replication and identification of NS4B as an RNAi suppressor. J Virol 87:8870–8883. doi: 10.1128/JVI.02774-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller S, Sparacio S, Bartenschlager R. 2006. Subcellular localization and membrane topology of the dengue virus type 2 non-structural protein 4B. J Biol Chem 281:8854–8863. doi: 10.1074/jbc.M512697200. [DOI] [PubMed] [Google Scholar]
- 15.Zou J, Xie X, Lee LT, Chandrasekaran R, Reynaud A, Yap L, Wang QY, Dong H, Kang C, Yuan Z, Lescar J, Shi PY. 2014. Dimerization of flavivirus NS4B protein. J Virol 88:3379–3391. doi: 10.1128/JVI.02782-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vigerust DJ, Shepherd VL. 2007. Virus glycosylation: role in virulence and immune interactions. Trends Microbiol 15:211–218. doi: 10.1016/j.tim.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mondotte JA, Lozach PY, Amara A, Gamarnik AV. 2007. Essential role of dengue virus envelope protein N glycosylation at asparagine-67 during viral propagation. J Virol 81:7136–7148. doi: 10.1128/JVI.00116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee E, Leang SK, Davidson A, Lobigs M. 2010. Both E protein glycans adversely affect dengue virus infectivity but are beneficial for virion release. J Virol 84:5171–5180. doi: 10.1128/JVI.01900-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JM, Yun SI, Song BH, Hahn YS, Lee CH, Oh HW, Lee YM. 2008. A single N-linked glycosylation site in the Japanese encephalitis virus prM protein is critical for cell type-specific prM protein biogenesis, virus particle release, and pathogenicity in mice. J Virol 82:7846–7862. doi: 10.1128/JVI.00789-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crabtree MB, Kinney RM, Miller BR. 2005. Deglycosylation of the NS1 protein of dengue 2 virus, strain 16681: construction and characterization of mutant viruses. Arch Virol 150:771–786. doi: 10.1007/s00705-004-0430-8. [DOI] [PubMed] [Google Scholar]
- 21.Pryor MJ, Gualano RC, Lin B, Davidson AD, Wright PJ. 1998. Growth restriction of dengue virus type 2 by site-specific mutagenesis of virus-encoded glycoproteins. J Gen Virol 79:2631–2639. [DOI] [PubMed] [Google Scholar]
- 22.Tajima S, Takasaki T, Kurane I. 2008. Characterization of Asn130-to-Ala mutant of dengue type 1 virus NS1 protein. Virus Genes 36:323–329. doi: 10.1007/s11262-008-0211-7. [DOI] [PubMed] [Google Scholar]
- 23.Teoh PG, Huang ZS, Pong WL, Chen PC, Wu HN. 2014. Maintenance of dimer conformation by dengue virus core protein alpha4-alpha4′ helix pair is critical for nucleocapsid formation and virus production. J Virol 88:7998–8015. doi: 10.1128/JVI.00940-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu YC, Chen NC, Chen PC, Wang CC, Cheng WC, Wu HN. 2012. Identification of a small-molecule inhibitor of dengue virus using a replicon system. Arch Virol 157:681–688. doi: 10.1007/s00705-012-1224-z. [DOI] [PubMed] [Google Scholar]
- 25.Helenius A. 1994. How N-linked oligosaccharides affect glycoprotein folding in the endoplasmic reticulum. Mol Biol Cell 5:253–265. doi: 10.1091/mbc.5.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roth J, Zuber C, Park S, Jang I, Lee Y, Kysela KG, Le Fourn V, Santimaria R, Guhl B, Cho JW. 2010. Protein N-glycosylation, protein folding, and protein quality control. Mol Cells 30:497–506. doi: 10.1007/s10059-010-0159-z. [DOI] [PubMed] [Google Scholar]
- 27.Parodi AJ. 2000. Role of N-oligosaccharide endoplasmic reticulum processing reactions in glycoprotein folding and degradation. Biochem J 348:1–13. doi: 10.1042/0264-6021:3480001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otsuki K, Maeda J, Yamamoto H, Tsubokura M. 1979. Studies on avian infectious bronchitis virus (IBV). III. Interferon induction by and sensitivity to interferon of IBV. Arch Virol 60:249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie X, Zou J, Wang QY, Shi PY. 19 March 2015. Targeting dengue virus NS4B protein for drug discovery. Antiviral Res doi: 10.1016/j.antiviral.2015.03.007. [DOI] [PubMed] [Google Scholar]