ABSTRACT
Endothelial cells (ECs) are a critical target of viruses, and infection of the endothelium represents a defining point in viral pathogenesis. Human cytomegalovirus (HCMV), the prototypical betaherpesvirus, encodes proteins specialized for entry into ECs and delivery of the genome to the nuclei of ECs. Virus strains competent to enter ECs replicate with differing efficiencies, suggesting that the virus encodes genes for postentry tropism in ECs. We previously reported a specific requirement for the UL133/8 locus of HCMV for replication in ECs. The UL133/8 locus harbors four genes: UL133, UL135, UL136, and UL138. In this study, we find that while UL133 and UL138 are dispensable for replication in ECs, both UL135 and UL136 are important. These genes are not required for virus entry or the expression of viral genes. The phenotypes associated with disruption of either gene reflect phenotypes observed for the UL133/8NULL virus, which lacks the entire UL133/8 locus, but are largely distinct from one another. Viruses lacking UL135 fail to properly envelop capsids in the cytoplasm, produce fewer dense bodies (DB) than the wild-type (WT) virus, and are unable to incorporate viral products into multivesicular bodies (MVB). Viruses lacking UL136 also fail to properly envelop virions and produce larger dense bodies than the WT virus. Our results indicate roles for the UL135 and UL136 proteins in commandeering host membrane-trafficking pathways for virus maturation. UL135 and UL136 represent the first HCMV genes crucial for early- to late-stage tropism in ECs.
IMPORTANCE Human cytomegalovirus (HCMV) persists in the majority of the world's population. While typically asymptomatic in healthy hosts, HCMV can cause significant morbidity and mortality in immunocompromised or naïve individuals, particularly transplant patients and patients with congenital infections, respectively. Lifelong persistence of the virus may also contribute to age-related pathologies, such as vascular disease. One aspect of HCMV infection contributing to complex and varied pathogenesis is the diverse array of cell types that this virus infects in the host. The vascular endothelium is a particularly important target of infection, contributing to viral dissemination and likely leading to CMV complications following transplantation. In this work, we identify two viral gene products required for postentry tropism in endothelial cells. Identifying tropism factors required for replication in critical cell targets of infection is important for the development of strategies to restrict virus replication.
INTRODUCTION
Human cytomegalovirus (HCMV) is a ubiquitous herpesvirus with 50 to 99% seroprevalence in the global population. Like all herpesviruses, HCMV persists for the lifetime of the host by way of latent infection (1–3). The persistence of HCMV is asymptomatic in immunocompetent individuals and is characterized by states of subclinical reactivation from latency and low-level virus shedding (1). However, in immunocompromised hosts, HCMV causes significant morbidity and mortality. Of particular concern are stem cell and solid-organ transplant patients, HIV/AIDS patients, and cancer patients undergoing chemotherapy or radiation regimens (4, 5). Additionally, HCMV is the leading cause of infectious disease-related birth defects in the United States, affecting 1 in 150 children born in the United States and most commonly resulting in mild to severe hearing loss (6). While CMV infection of seronegative women during pregnancy poses the most significant risk for severe sequelae in infants (microcephaly, cerebral palsy, and severe hearing loss or cognitive deficits), as many as 75% of congenital infections occur in infants whose mothers were seropositive at the time of conception, indicating that these infections result from reinfection or reactivation (7). Finally, the persistence of HCMV is increasingly associated with age-related pathologies, even when overt clinical symptoms are absent. Age-related pathologies include vascular disease, immune dysfunction, and frailty (8–13). In spite of the pressing health significance of HCMV infection, the mechanisms underlying infection in multiple tissues and the persistence of the virus in the host are poorly understood.
While highly restricted in host tropism, HCMV infects a wide variety of cell types within the human host. Each cell type offers a unique context that may profoundly affect the outcome of infection: latency or replicative states of infection ranging from smoldering, chronic virus shedding to high-titer virus production (5, 14, 15). The human vascular endothelium is a key target of infection for a myriad of viruses, including HCMV, as a primary site for both hematogenous dissemination and dissemination to organ tissues (1, 16). Infected vascular endothelial cells (ECs) can detach and circulate to infect distal tissues (17, 18). Further, ECs are thought to factor importantly into viral persistence (19). Infection and persistence at the vascular endothelium also have the potential to impact tissue integrity. It has been shown that HCMV infection of ECs increases inflammation at the endothelium, promoting monocyte recruitment and extravasation, and increasing vascular permeability (1, 20–25). Further, proinflammatory signaling elicited by the infection of vascular ECs contributes to angiogenesis and vascular disease (8, 26, 27).
HCMV infection of the vascular endothelium and its impact on vascular biology would undoubtedly factor importantly into HCMV-related vascular pathologies. HCMV seropositivity is an important risk factor for graft and solid-organ transplant failure (28). In the case of heart transplantation, HCMV nearly doubles the 5-year rate of cardiac graft rejections as a consequence of transplant vascular arteriosclerosis (29). Additionally, HCMV persistence in healthy individuals is associated with an increased risk of age-related vascular pathologies, including atherosclerosis and restenosis, and correlates positively with greater risks for frailty and cardiac mortality (27, 30–36). In spite of the marked importance of ECs to HCMV infection, little is known about tropism barriers in these cells or the viral factors important for replication specifically in ECs.
Low-passage or clinical strains of HCMV readily infect ECs, whereas laboratory-adapted strains do not (37, 38). The restriction to replication of laboratory strains in ECs may be due to genome rearrangements that occur during passage in cell culture. For example, the ULb′ region of the genome is lost upon serial passage of the virus in cultured fibroblasts (39–41). The UL128 to UL131A genes are required for HCMV entry into ECs, epithelial cells, leukocytes, and dendritic cells (21, 42–48), but not fibroblasts. However, low-passage-number HCMV strains or laboratory strains repaired for their ability to enter ECs replicate with various efficiencies suggesting the existence of postentry viral factors important to replication specifically in these cell types (21, 46). Accordingly, US16 has been reported to be specifically required for delivery of the viral genome to the nucleus in ECs and epithelial cells following viral entry (49). Viral genes required for later stages of replication specifically in ECs likely exist but have been more elusive.
We recently demonstrated a requirement for the ULb′ (UL133/8) polycistronic locus for replication in ECs (50, 51). The UL133/8 locus encodes four genes, UL133, UL135, UL136, and UL138. We originally demonstrated that the entire locus and the individual genes encoded play important and antagonistic roles in the establishment or maintenance of latency and in reactivation (50, 52, 53). While UL133/8 locus genes, particularly UL133, UL138, and one of the isoforms of UL136, are suppressive to viral replication in fibroblasts and CD34+ hematopoietic progenitor cells (HPCs) (50, 52–55), disruption of the UL133/8 locus resulted in a defect in virus replication in ECs, suggesting that UL133/8 genes are required for efficient replication in this cell type (51). Electron microscopic analysis of ECs infected with a virus lacking the UL133/8 locus indicated a postentry, late-stage defect in virus maturation that might be due to increased vesiculation of cytoplasmic membranes and failure to form the viral assembly compartment (VAC), a distinct restructuring of endocytic compartments into a concentric perinuclear organelle in which viral progeny is assembled during HCMV infection (51). UL133/8 mutant virus-infected ECs produced virions that lacked envelopes and had decreased numbers of dense bodies (DBs), host-derived vesicles containing viral tegument proteins. Another intriguing observation is that multivesicular bodies (MVB) induced by HCMV infection become packed with virions and DBs and that the recruitment of these viral products into MVB requires the UL133/8 locus. In contrast, fibroblasts infected with a UL133/8NULL virus produce essentially wild-type (WT) yields of virus with no defects in virion maturation and no alteration in DBs or intracellular membranes, suggesting that these functions of the UL133/8 locus are EC specific (51). Cell type-specific requirements for the UL133/8 locus illustrate the importance of unique cellular environments to the outcome of infection.
In the present study, we have identified pUL135 and pUL136 as the critical viral gene products encoded within the UL133/8 locus for postentry viral replication in ECs. Viruses lacking UL135 or UL136 exhibited intermediate defects in viral replication relative to that of the UL133/8NULL virus. The phenotypes associated with the loss of the UL133/8 locus in EC infection are complex and largely segregate between UL135 and UL136 mutant viruses. Both UL135 and UL136 are required for the maintenance of intracellular membrane organization and for efficient formation of the VAC, but distinct defects in virion maturation were observed with UL135 or UL136 mutant virus infection. Our findings demonstrate a cooperative requirement for UL135 and UL136 for efficient viral replication in ECs and indicate roles for pUL135 and pUL136 in commandeering membrane-trafficking pathways at early/late stages during infection in ECs. To our knowledge, these are the first HCMV proteins to be identified as required for post-immediate early (post-IE) stages of infection in ECs. These findings suggest novel mechanisms involving membrane organization or trafficking in mediating tropism specifically in ECs.
MATERIALS AND METHODS
Cells.
Primary human microvascular lung endothelial cells (HMVEC) (Lonza, Walkersville, MD) were cultured using EGM-2MV BulletKit medium (microvasular endothelial cell growth medium 2; Lonza) with 5% fetal bovine serum (FBS) and 100 U/ml penicillin. HMVEC were cultured at 37°C under 5% CO2 and were passaged as described previously (51).
Viruses.
Bacterial artificial chromosome (BAC) clones of the HCMV TB40/E wild-type (WT) and all variant strains express green fluorescent protein (GFP) as a marker of infection. The construction of the UL135STOP variant has been described previously (53). The TB40/E-UL136GalK construct was generated by replacing the entire UL136 open reading frame (ORF) with a GalK cassette. Briefly, the GalK cassette was amplified by PCR using primers flanked by homologous viral sequences (UL136 GalK Fwd [5′ gcagcacacgccttccctctttttcaccgcagctaagagagagaaagagagtCCTGTTGACAATTAATCATCGGCA 3′] and UL136 GalK Rev [5′ gtctctctcttttgtacagcactcgcgcgggaacggccccctcaaccctcTCAGCACTGTCCTGCTCCTT 3′] [lowercase letters stand for HCMV homology arms]) and was recombined into the WT BAC by homologous recombination. GalK-positive recombinants were selected on M63 minimal medium plates followed by MacConkey medium plates containing galactose as the sole carbon source. Multiple independent recombinant viruses were screened by BAC digestion and PCR to confirm the replacement of UL136 with the GalK cassette. Construction of the UL135STOP variant and the UL136NULL (54) and UL135STOP/UL138STOP (53) viruses have been described previously.
Virus stocks were propagated by transfection of 25 μg of BAC DNA plus 3 μg of a plasmid expressing HCMV UL82 (encoding the pp71 protein) into 5 × 106 MRC-5 fibroblasts by electroporation to obtain primary (P0) virus stocks. The secondary virus stocks (P1) were generated by infecting MRC-5 fibroblasts with P0 stocks. The P1 stocks were purified by density gradient centrifugation through a 20% d-sorbitol cushion at 20,000 rpm in an SW28 rotor (Beckman Coulter, CA) for 80 min at 22°C. Virions were resuspended in IMDM (Iscove's modified DMEM [Dulbecco's modified Eagle medium]) containing 10% BIT (bovine serum albumin, insulin, and transferrin; Stem Cell Technologies) and were stored at −80°C. Virus titers were determined by the 50% tissue culture infective dose (TCID50) method on MRC-5 fibroblasts. Multistep virus growth curves were determined by TCID50 analysis of cell lysates harvested over a time course.
Quantitative PCR.
Total viral DNA and viral RNA were isolated from infected cells by using the Zymo Duet kit with on-column DNase digestion for RNA samples according to the manufacturers' instructions (Zymo Research, Irvine, CA, and Macherey-Nagel, Inc., Bethlehem, PA). cDNA was synthesized using a Transcriptor First Strand cDNA synthesis kit (Roche, Indianapolis, IN). Both viral DNA and viral cDNA were analyzed using advanced relative quantitation on a Roche LightCycler 480 instrument according to the manufacturer's instructions (Roche, Indianapolis, IN). Viral DNA and cDNA levels were normalized to those of the cellular housekeeping genes RNaseP and β-actin, respectively. The primers and corresponding Universal ProbeLibrary probes are listed in Table 1.
TABLE 1.
Real-time PCR primers
| Gene | Directiona | Sequence | UPLb probe no. |
|---|---|---|---|
| UL123 | Fwd | CAGTGCACCCCCTAACTTGT | 73 |
| Rev | ATAAGCGGGAGATGTGGATG | ||
| UL99 | Fwd | TTGTCCTCGCGAAACGTC | 83 |
| Rev | GGGGGAAACGACAGTAGTAGC | ||
| UL69 | Fwd | CCTACGACTTTCGGTTCTTCTC | 30 |
| Rev | CGTCCAGTTCGTCGTCAATAA | ||
| UL32 | Fwd | TCTTGACGGGGGAACCTAC | 58 |
| Rev | CCAAAAGCAGCGTATCGAAT | ||
| RNaseP | Fwd | GACGGACTGCGCAGGTTA | 24 |
| Rev | CCATGCTGAAGTCCCATGA |
Fwd, forward; Rev, reverse.
UPL, Universal ProbeLibrary.
Immunoblotting.
Immunoblotting was performed as described previously (52). Briefly, protein lysates (15 to 20 μg) were separated on 12% Bis-Tris gels through electrophoresis. Proteins were transferred to 0.45-μm-pore-size polyvinylidene difluoride membranes (Immobilon-FL; Millipore, MA). The membranes were then immunoblotted using mouse monoclonal or rabbit polyclonal antibodies directed against each protein. Monoclonal and polyclonal antibodies were detected using fluorescently conjugated secondary antibodies and the Odyssey infrared imaging system (Li-Cor, NE). The mouse monoclonal antibodies used in this study include antibodies against IE1/IE2 (clone 3H4; dilution, 1:100; a generous gift from Tom Shenk), UL44 (clone 10D8; dilution, 1:2,500; Virusys), pp28 (clone 10B4-29; dilution, 1:50), pp150 (clone 36-14; dilution, 1:20; a generous gift from William Britt), and pp65 (clone 3A12; dilution, 1:16,000; Virusys). Anti-β-actin is a rabbit monoclonal antibody (clone ACTN05-C4; Abcam) used at a 1:1,000 dilution.
TEM.
HMVEC were either mock infected or infected at a multiplicity of infection (MOI) of 4 with the WT, UL135STOP, or UL136GalK virus. Infection was aided by using centrifugal enhancement. For HMVEC, the infection medium was replaced at 24 h postinfection (hpi) with normal EGM-2MV medium. Infected cells were harvested 5 days postinfection (dpi) and were fixed in 2.5% glutaraldehyde and 0.1 M PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] for 20 min. Fixed cell pellets were first postfixed with osmium tetroxide in 0.1 M PIPES and then dehydrated in a graded series of alcohol. Pellets were infiltrated with resin and were cut into 100-nm sections, which were floated onto copper grids. Images were acquired using a Philips CM-12s transmission electron microscope (TEM). Infected cells were embedded and sectioned by the Arizona Research Laboratories, Arizona Health Sciences Center Core Facility.
Indirect immunofluorescence.
Indirect immunofluorescence was performed as described previously (51, 52). Briefly, HMVEC were plated onto 24-well dishes containing 12-mm coverslips 24 h prior to infection. Cells were either mock infected or infected at an MOI of 2 with the WT, UL135STOP, or UL136GalK virus. Cells were processed for indirect immunofluorescence at 144 hpi for HMVEC. A mouse monoclonal antibody to pp28 was used at a 1:20 dilution. The Golgi apparatus was stained with a rabbit monoclonal antibody specific to GM130 (clone EP892Y; Abcam) at a 1:250 dilution. Cells from each infection condition were fixed in 2% formaldehyde in phosphate-buffered saline (PBS) and were then permeabilized. Primary and secondary antibodies were incubated in PBS-Tween 20 (PBS-T) for 1 h. DNA from infected cells was stained with 4′,6-diamidino-2-phenylindole (DAPI) (Molecular Probes) after primary staining. Coverslips were mounted onto slides using Aqua Poly/Mount medium (Polysciences, Inc.). Cells were imaged as lambda stacks by using a Zeiss 510 Meta confocal microscope. Images were unmixed using the Zeiss 510 Meta software, version 4.2, and were processed using Zeiss software.
RESULTS
UL135 and UL136 are required for efficient viral replication in endothelial cells.
We demonstrated previously that the UL133/8 locus was required for postentry viral tropism across various micro- and macrovascular EC types (50, 51). Infection with a recombinant virus lacking the UL133/8 locus resulted in as much as 1,000-fold-reduced yields of progeny virus compared to that for WT infection. Moreover, the reduction in viral titers was not due to defects in viral entry or in immediate early (IE), early, or late gene expression. Rather, we observed disorganization of host intracellular membranes, failure to form a VAC, and defects in the maturation of progeny virus. The requirement for this locus for virus replication was specific to endothelial cells, since the UL133/8 locus was dispensable for replication in primary fibroblasts and primary epithelial cells. The phenotypes associated with UL133/8NULL infection in ECs are summarized in Table 2. These findings suggested that genes within the UL133/8 locus are required for late-stage tropism in ECs (51).
TABLE 2.
Segregation of UL133/8NULL phenotypes between UL135 and UL136
| Virus | Feature of infection in ECsa |
||||
|---|---|---|---|---|---|
| Fold reduction in virus yieldb | Formation of viral assembly compartment | Dense bodies | MVB containing viral products | Enveloped virions | |
| WT | + | + | + | + | |
| UL133/8NULL | 1,000 | − | − | − | − |
| UL135STOP | 100 | +/− | − | − | −c |
| UL136GalK | 50 | +/− | ++ | + | − |
+, WT feature; +/−, reduction of the feature; −, absence of the feature.
Relative to that of the WT.
Phenotype also observed in fibroblasts.
To define the UL133/8 genes important for replication in ECs, we examined the replication of a series of mutant viruses, each containing a disruption in one of the four viral genes encoded within the locus. UL133, UL135, and UL138 were disrupted by the substitution of stop codons for translation initiation codons, resulting in the UL133STOP, UL135STOP, and UL138STOP recombinant viruses, which fail to express the respective proteins (50, 52, 53). Because UL136 encodes a number of protein isoforms (54), the entire UL136 gene was disrupted by the insertion of a cassette encoding galactosidase K (UL136GalK). We examined the replication of each of these viruses in primary human microvascular lung ECs (HMVEC) infected at a low MOI (0.05). Virus yield was measured over a time course (Fig. 1). While viruses containing disruptions in UL133 or UL138 replicated with kinetics and yields similar to those of the WT virus, the UL135STOP and UL136GalK viruses exhibited 100- and 50-fold reductions in progeny virus production. The defect associated with replication in the absence of UL135 or UL136 has been observed with multiple independent clones of each virus. Based on these results, we conclude that UL135 and UL136 are important for efficient viral replication in ECs.
FIG 1.
UL135 and UL136 are required for replication in endothelial cells. (A) HMVEC were infected with the TB40/E-WT, -UL133/8NULL, -UL133STOP, -UL135STOP, -UL136GalK, or -UL138STOP virus at an MOI of 0.05. (B) HMVEC were infected with the TB40/E-WT, -UL136NULL, or -UL135STOP/UL138STOP virus at an MOI of 0.05. Virus yields were measured in lysates harvested over a time course (measured in days postinfection) by TCID50 on fibroblasts. The values plotted are averages for 3 to 4 independent experiments. The standard deviation for each time point is indicated by error bars.
To confirm that the defects in replication are specific to the disruption of UL135 and UL136, we examined the replication of independent mutant viruses containing disruptions in these genes. UL136 is expressed as 5 protein isoforms. We recently mapped the origins of these isoforms, and we have disrupted the expression of all protein isoforms by converting each ATG in the UL136 coding sequence to a stop or alanine codon (54). The resulting UL136NULL virus replicates with a defect equal to or greater than that observed for the UL136GalK virus (Fig. 1B). We previously demonstrated a requirement for UL135 for replication following transfection of infectious genomic clones in fibroblasts or for reactivation in CD34+ HPCs (53). The requirement for UL135 for replication from BAC transfection in fibroblasts could be overcome by subsequent disruption of UL138. Therefore, we analyzed the replication of a virus containing disruptions in both UL135 and UL138 (UL135STOP/UL138STOP) in order to confirm the importance of UL135 to replication in ECs and to determine if disruption of UL138 could compensate for the loss of UL135 in ECs. The UL135STOP/UL138STOP virus replicated with a defect equal to or greater than that of the UL135STOP virus (Fig. 1B). Further, disruption of UL138 did not compensate for the defect in replication associated with the loss of UL135. Taken together, these results confirm the requirement for UL135 and UL136 for replication in ECs and indicate that the requirement for UL135 in ECs is independent of any effects of UL138.
Distribution of pUL135 and pUL136 during infection in ECs.
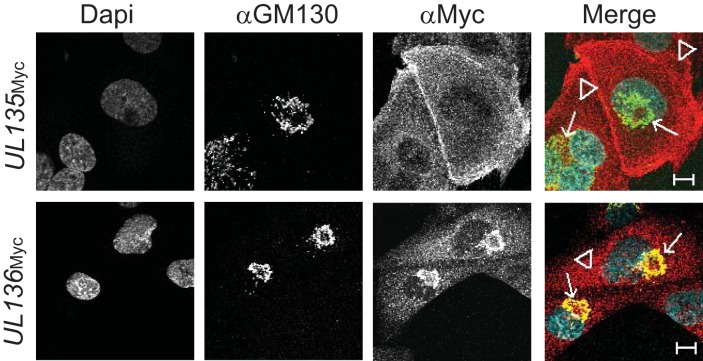
We have previously analyzed the localization of pUL135 and pUL136 in fibroblasts (50, 54). While all proteins encoded in the UL133/8 locus are associated with Golgi membranes and localize to the VAC at late times in infection, the protein encoded by UL135 exhibits cell surface localization early in infection (50) and associates with the cytoskeleton (56). UL136 is expressed as multiple protein isoforms, some of which localize with the Golgi apparatus while others are more diffusely distributed throughout the cytoplasm (54). To determine if the localization of the UL135 or UL136 protein is distinct in ECs, we examined the subcellular distribution of these proteins in ECs infected with recombinant viruses expressing a variant of UL135 or UL136 fused with a C-terminal Myc epitope tag (UL135Myc or UL136Myc). pUL135 localized throughout the cytoplasm and at the cell surface (Fig. 2). Interestingly, we did not observe pUL135 colocalization with Golgi membranes or the VAC, which is the predominant localization in fibroblasts. The distinct localization of pUL135 in ECs may reflect cell type-specific protein functions or differential trafficking of the protein. pUL136 colocalized with the Golgi marker GM130, with some punctate staining throughout the cytoplasm (Fig. 2). The distribution of pUL136 in infected ECs is similar to that during infection in fibroblasts. Further studies are required to define the subcellular distribution of individual pUL136 isoforms during infection in ECs.
FIG 2.
UL135 and UL136 exhibit unique distributions throughout the cytoplasm of infected cells. HMVEC were infected at an MOI of 2 with one of two TB40/E recombinant viruses (UL135Myc or UL136Myc), each expressing the indicated protein with a C-terminal Myc epitope tag. At 144 hpi, cells were processed for indirect immunofluorescence microscopy using monoclonal antibodies specific to the Golgi marker GM130 and the Myc epitope tag. Nuclei were stained by DAPI. UL135 is located primarily at the plasma membrane (arrowheads) and is dispersed in the cytoplasm, whereas UL136 colocalizes extensively with the Golgi marker (arrows). Bars, 10 μm.
UL135 and UL136 are not required for viral gene expression.
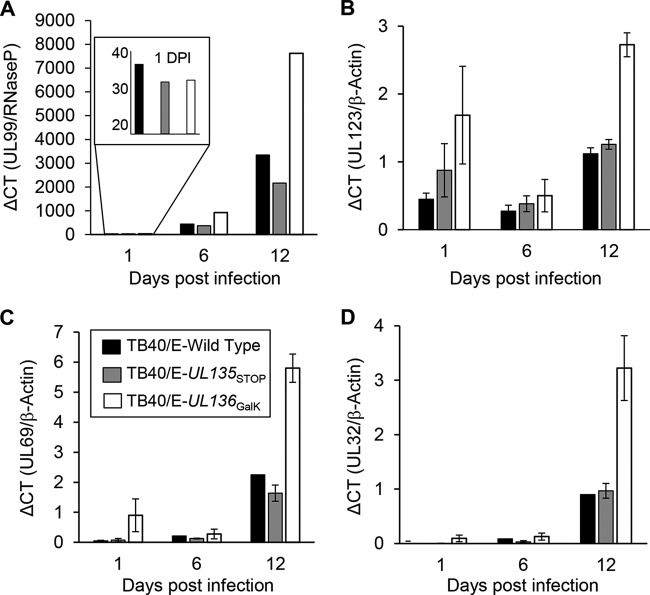
ECs infected with the UL133/8NULL virus exhibited no statistically significant difference in gene expression at the immediate early, early, or late stage of viral infection from ECs infected with the WT virus (51). However, because we have observed requirements for individual UL133/8 locus genes that are not realized in the context of deletion of the entire UL133/8 locus (53), we analyzed the accumulation of viral genomes, transcripts, and proteins in HMVEC infected at an MOI of 0.1 with the WT, UL135STOP, or UL136GalK virus. The accumulation of viral genomes and transcripts was measured at 1, 6, and 12 dpi by quantitative PCR (Fig. 3). The 1-dpi genome data are graphed alone in the Fig. 3A inset in order to better represent input genomes. HCMV genomes, measured using a primer pair specific to the UL99 gene, accumulated similarly over time in cells infected with the WT or UL135STOP virus, while cells infected with the UL136GalK virus accumulated slightly higher levels of genomes by 12 dpi. In agreement with the analysis of viral genomes, UL136GalK virus infection also resulted in greater accumulation of UL123/IE1 (Fig. 3B), UL69 (Fig. 3C), and UL32/pp150 (Fig. 3D) transcripts than infection with the WT or UL135STOP virus during the 12-day time course. These results suggest that the defect in replication observed in ECs infected with UL135 or UL136 mutant viruses is not due to a failure to replicate viral genomes or to express viral genes. Indeed, viral gene expression was enhanced during infection with the UL136 mutant virus.
FIG 3.
Accumulation of viral genomes and transcripts in infected HMVEC. (A) Total DNA was isolated from HMVEC infected with TB40/E-WT (filled bars), -UL135STOP (shaded bars), or -UL136GalK (open bars) (MOI, 0.1) at the time points indicated. Genomes were quantified by real-time PCR using primers specific to UL99, and values were normalized to those for the cellular gene RNaseP. The results of one experiment representative of three total experiments are shown. (B to D) Transcripts for UL123/IE1 (B), UL69 (C), and UL32/pp150 (D) were quantified from cDNA synthesized from total RNA isolated from infected HMVEC over a time course. Transcript levels were normalized to those for β-actin. The bars represent average values from three independent experiments. ΔCT, change in threshold cycle.
We then examined representative IE (IE1 [72 kDa] and IE2 [86 kDa]), early (UL44), and late (pp28) viral proteins over a time course by immunoblotting (Fig. 4A). In each infection, proteins at each stage of infection accumulated with similar kinetics and to levels similar to those of WT virus infection. Quantification of IE1 and pp28 bands over 3 independent experiments indicated that there was no statistically significant difference in the accumulation of IE or late proteins between WT virus infection and infection with either mutant virus (Fig. 4B and C). These data are consistent with the defect associated with UL133/8NULL virus infection in ECs. The absence of a substantial defect in viral gene expression in HMVEC infected with these mutant viruses indicates a requirement for UL135 and UL136 in the maturation of progeny virus, as was observed with the UL133/8NULL virus.
FIG 4.
Accumulation of IE, early, and late viral proteins in infected HMVEC. (A) HMVEC were infected at an MOI of 2 with TB40/E-WT, -UL135STOP, or -UL136GalK. Protein lysates were harvested over the time course indicated (in hours postinfection [hpi]) and were analyzed by immunoblotting using antibodies specific to the 72-kDa IE1 and 86-kDa IE2 proteins, the early protein UL44, the late protein pp28, and β-actin. β-Actin served as a loading control. (B and C) Quantification of the average IE1 (B) and pp28 (C) protein levels (normalized to those for actin) from three independent experiments. The standard deviations are indicated. Student's t test indicates that there is no significant difference between WT and mutant viruses at each time point.
UL135 and UL136 are important for efficient formation of the viral assembly compartment.
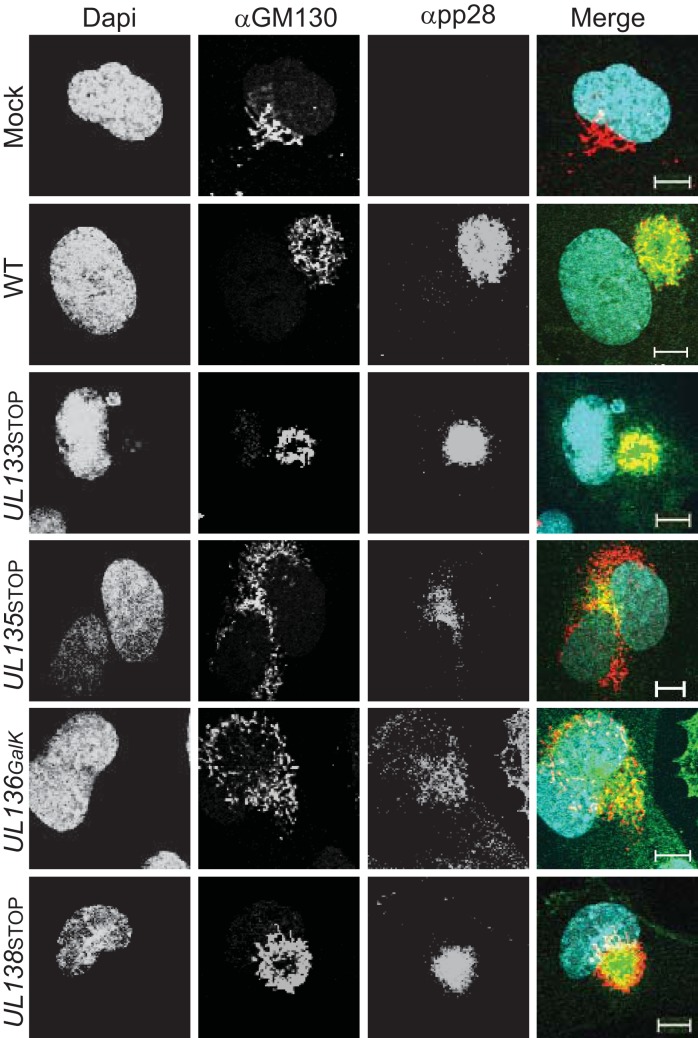
The VAC is a virus-induced reorganization of intercellular cytoplasmic membranes into a concentric perinuclear compartment important for virus maturation (57–62). Our previous work demonstrated a requirement of the UL133/8 locus for maintaining intracellular membrane structures and for the formation of the VAC (51). In the absence of the UL133/8 locus, a number of cellular trafficking markers associated with the VAC are dispersed throughout the cell (51, 58, 59). In the present study, we examined the formation of the VAC in HMVEC infected with a WT, UL135 mutant, or UL136 mutant virus. Infected cells were labeled with antibodies specific to the Golgi marker GM130 or the late protein pp28, both known to mark the VAC (59). Both pUL135 and pUL136 were required for efficient VAC formation (Fig. 5). Mutant viruses lacking either UL135 or UL136 resulted in the dispersal of Golgi membranes and the VAC in approximately 50% of infected cells. We also analyzed VAC formation in cells infected with UL133 or UL138 mutant viruses to ensure that the requirement for VAC formation was limited to UL135 and UL136. UL133 and UL138 mutant viruses formed the VAC similarly to the WT virus. These results indicate that UL135 and UL136, but not UL133 and UL138, are important for membrane organization during HCMV infection of ECs.
FIG 5.
UL135 and UL136 are required for efficient formation of the viral assembly compartment. HMVEC were infected at an MOI of 2 with TB40/E-WT, -UL133STOP, -UL135STOP, UL136GalK, or UL138STOP. At 144 h postinfection, cells were processed for indirect immunofluorescence microscopy using monoclonal antibodies specific to the Golgi marker GM130 and the late viral protein pp28. Nuclei were stained by DAPI. Both the UL135STOP and UL136GalK viruses failed to reorganize the intracellular membranes into the VAC. A merge of all three images is shown at the right. Bars, 10 μM.
UL135 and UL136 are required for the maturation of progeny virions.
We previously characterized late-stage defects in virion maturation and egress in ECs infected with the UL133/8NULL virus by using transmission electron microscopy (TEM) (51). To explore the roles of UL135 and UL136 in the maturation of virions, we infected HMVEC with TB40/E-WT, -UL135STOP, or UL136GalK at an MOI of 4 and processed the cells for TEM at 5 dpi, a time point at which the level of infectious virus production is high.
We observed striking defects in the maturation of virus particles in ECs infected with the UL135STOP or UL136GalK virus relative to that with the WT (Fig. 6). Interestingly, the phenotypes associated with UL133/8NULL infection segregated between those for UL135STOP and UL136GalK infections (Table 2). WT infection results in the production of virus particles containing an encapsidated viral genome, tegument, and envelope (Fig. 6A). HMVEC infected with the UL135STOP virus accumulated two types of cytoplasmic virus particles in addition to a minority of virions that appeared morphologically normal: nonenveloped capsids and particles with abnormal envelopes that appeared to wrap the capsid loosely (Fig. 6B). Only 27% of the virus particles in cells infected with the UL135STOP virus morphologically resembled WT virions. We previously reported a similar envelopment defect for UL135STOP infection in fibroblasts and a corresponding increase in the number of genome-containing particles required to form an infectious unit (53). Figure 7B shows representative UL135STOP virus particles in infected fibroblasts, where the envelopment defect is marked. Further, in fibroblasts infected with the UL135STOP virus, we had noted an accumulation of noninfectious enveloped particles (NIEPs); however, NIEPs were far less pronounced in ECs infected with the UL135STOP virus (data not shown). Sixty-five percent of the capsids that accumulated in the cytoplasm of ECs infected with the UL136GalK virus lacked envelopes entirely, a phenotype similar to the UL133/8NULL infection phenotype in ECs (Fig. 6C). Nonenveloped capsids were not observed in fibroblasts infected with the UL136GalK virus (Fig. 7C). In ECs, nonenveloped or aberrantly enveloped virus particles in UL136GalK and UL135STOP infection, respectively, did not appear to be fully tegumented (Fig. 6).
FIG 6.
UL135 and UL136 are required for virion maturation. HMVEC were infected at an MOI of 4 with the TB40/E-WT (A), -UL135STOP (B), or -UL136GalK (C) virus. At 5 days postinfection, cells were fixed, embedded, and sectioned for TEM. Representative micrographs are shown to illustrate the accumulation and maturation of virus particles in the cytoplasm. Three distinct virus particle morphologies are observed: normal virions with characteristic HCMV morphology (filled arrowheads), aberrantly enveloped virions (open arrowheads), and nonenveloped capsids (filled arrows). DB, dense body. Bars, 500 nm. (D) Seven hundred to 1,300 total virions were counted in 30 to 45 cells. The percentages of normal virions (open portions of bars), aberrantly enveloped virions (shaded portions of bars), and nonenveloped capsids (filled portions of bars) in each infection are illustrated.
FIG 7.

UL135 and UL136 mutant virus progeny in fibroblasts. MRC-5 fibroblasts were infected with the TB40/E-WT (A), -UL135STOP (B), or –UL136GalK (C) virus at an MOI of 4. At 5 days postinfection, cells were fixed, embedded, and sectioned for TEM. Representative micrographs are shown to illustrate the accumulation and maturation of virus particles in the cytoplasm. Three distinct virus particle morphologies are observed: normal virions with characteristic HCMV morphology (filled arrowheads), aberrantly enveloped virions (open arrowheads), and nonenveloped capsids (filled arrows). Bars, 2 μm.
We observed striking differences between DBs accumulating in cells infected with the UL135 or UL136 mutant virus and those in WT-infected cells (Fig. 8A to C). DBs are vesicles containing viral tegument proteins. Infection of HMVEC with the UL135STOP virus resulted in an average of 2-fold fewer DBs than WT infection (Fig. 8B), a finding similar to the reduced presence of DBs in UL133/8NULL infection. In UL136GalK virus-infected ECs, DBs accumulated to the same numbers as in WT infection but were strikingly larger (Fig. 6C and 8C). Measurements of >200 DBs indicated that DBs in UL136GalK infection were on average 2.5 times larger than DBs in WT infection. We have not observed any alterations in DB formation in fibroblasts infected with the UL135STOP or UL136GalK virus (Fig. 7) (53). To determine if the accumulation of dense bodies is the result of altered accumulation of proteins that form dense bodies, we analyzed the levels of two DB/tegument proteins, pp65 and pp150, in the context of infection (Fig. 9A). The levels of pp65 and pp150 in multiple experiments were quantified and are shown in Fig. 9B and C, respectively. Infection with the UL135STOP virus resulted in pp65 and pp150 levels 2- to 3-fold lower than those in WT infection. In contrast, UL136GalK virus-infected cells accumulated viral proteins to levels similar to those for WT infection despite the higher level of accumulation of viral transcripts in UL136GalK infection (Fig. 3B to D). Therefore, while reductions in pp65 and pp150 levels may contribute to the reduction in the number of DBs in UL135STOP infection, the enlarged DBs in UL136GalK infection do not result simply from increased protein levels. These results suggest that there is a defect in the expression of at least some late genes during infection with UL135STOP virus that become apparent after 96 hpi (comparing time courses in Fig. 4 and 9). The late stage defects associated with UL135STOP infection may be related to a defect in IE2 expression associated with UL135STOP infection in fibroblasts (53).
FIG 8.
Dense body formation impacted by the disruption of UL135 or UL136. HMVEC infected with the TB40/E-WT (A), -UL135STOP (B), or -UL136GalK (C) virus were fixed, embedded, and sectioned for TEM at 5 dpi. Representative micrographs are shown to illustrate the accumulation of DBs (arrows). Bars, 2 μm.
FIG 9.
Enlarged DBs are not due to increased accumulation of DB proteins. (A) HMVEC were infected at an MOI of 2 with the TB40/E-WT, -UL135STOP, or -UL136GalK virus. Protein lysates were harvested over the time course indicated (in hours postinfection [hpi]) and were analyzed by immunoblotting using antibodies specific to the pp150 and pp65 proteins. β-Actin served as a loading control. (B and C) Quantification of the average pp65 (B) and pp150 (C) protein levels (normalized to actin) in three independent experiments. The standard deviations are indicated. Student's t test indicated that there was no significant difference between WT and mutant viruses at each time point.
MVB are large vesicles formed by the fusion of endosomes and other secretory vesicles. MVB have important functions in cells: (i) the fusion of MVB with lysosomes results in the degradation of MVB contents; (ii) MVB serve as a platform for intracellular signaling; and (iii) MVB trafficking to and fusion at the cell surface results in the secretion of exosomes that may affect intracellular signaling (63–67). MVB are induced during HCMV infection of ECs. The MVB in infected ECs typically contain virus progeny and DBs in addition to intraluminal vesicles (51). The UL133/8 locus is required for the incorporation of viral progeny into MVB. This observation supports the intriguing possibility that MVB are vehicles for viral egress. MVB were dramatically altered during infection with the UL135STOP virus relative to WT or UL136GalK infection. While MVB were present at equivalent frequencies in ECs infected with the WT (Fig. 10A), UL135STOP (Fig. 10B), and UL136GalK (Fig. 10C) viruses, virions and DBs were excluded from MVB in UL135STOP infection (Fig. 10B). These results suggest that UL135 recruits viral products into MVB. Taken together, these results suggest that UL135 and UL136 play roles in membrane reorganization and structure that are particularly important for virus maturation at late stages of infection in ECs.
FIG 10.

pUL135 is required for the incorporation of viral products into MVB. HMVEC were infected at an MOI of 4 with the TB40/E-WT (A), -UL135STOP (B), or UL136GalK (C) virus. At 5 days postinfection, cells were fixed, embedded, and sectioned for TEM. Representative micrographs are shown to illustrate the accumulation of MVB (arrows) and the incorporation of virions (arrowheads) and DBs (open arrowheads) into MVB. Bars, 500 nm.
DISCUSSION
The vascular endothelium represents a key target for myriad viruses as a gateway for viral dissemination throughout the human host (68). HCMV is no exception; vascular ECs represent a site of infection critical to viral dissemination, persistence, and pathogenesis (1, 18, 20, 21, 24, 25, 69, 70). Understanding the mechanisms underlying HCMV replication in key targets of infection such as ECs is crucial for the development of new therapeutics to control HCMV spread and the associated disease in the host. Given the importance of ECs as a target of infection, cytomegaloviruses encode a number of genes to ensure access to and tropism in ECs. HCMV mediates entry into ECs using a pentameric complex comprising the glycoproteins gH and gL in complex with UL128, UL130, and UL131A (42, 44, 47, 71, 72). In addition to the genes involved in entry, two genes that function in postentry steps in the replication of cytomegaloviruses in ECs have been identified previously. The HCMV US16 gene is specifically required for the delivery of viral genomes and tegument proteins to the nucleus following entry into ECs (49), and the murine CMV M45 gene contributes to viral replication specifically in ECs and macrophages by inhibiting apoptosis (73). These studies suggest that HCMV has evolved functions to overcome specific tropism barriers and to navigate unique cell type environments. Our present study identifies two additional gene products, UL135 and UL136, and their roles in mediating intracellular membrane organization as important for the later stages of replication in ECs. To our knowledge, these are the first post-immediate early stage viral determinants of tropism for ECs to be identified.
We reported previously that EC infection with a virus lacking the entire UL133/8 locus, the UL133/8NULL virus, resulted in dramatic disruption of intracellular membranes and abnormal virion morphology (51). The defect in virion morphology might be secondary to the disruption of intracellular membrane organization. The phenotypes observed during infection with the UL133/8NULL virus in ECs segregated between the phenotypes for infection with UL135 or UL136 mutant viruses (Table 2)—implying that the functions of UL135 and UL136 are distinct, albeit cooperative. Both UL135 and UL136 were required for efficient formation of the VAC during infection in ECs (Fig. 5). Dispersal of Golgi membranes and the VAC was observed in ∼50% of ECs infected with either the UL135STOP or the UL136GalK virus. The dispersal of the VAC is more pronounced in ECs infected with the UL133/8NULL virus, lacking both UL135 and UL136, and further work is required to understand how these proteins function cooperatively to maintain membrane organization for VAC formation. It is possible that the dispersal of intracellular membranes observed during UL133/8NULL, UL135STOP, or UL136GalK infection in ECs is an intermediate step in normal VAC formation and that UL135 and UL136 are required for the reorganization of membranes for VAC formation. Studies at higher resolution will be required to determine if the VACs formed in a portion of the mutant virus-infected ECs are truly equivalent to those formed in WT infection. This requirement for pUL135 and pUL136 in restructuring host membranes to form the VAC is consistent with a role for these proteins in membrane trafficking and reorganization. Neither the UL133/8 locus nor UL135 and UL136 are required for VAC formation in fibroblasts, indicating unique membrane-trafficking requirements in ECs. The HCMV tegument proteins pUL48, pUL94, and pUL103 have been shown to be required for VAC formation in fibroblasts (74); however, it is not known if these proteins are required in ECs. Defining the mechanisms by which viral proteins function to mediate VAC formation will contribute to our understanding of cellular trafficking pathways in ECs.
Distinct defects in virion maturation, possibly secondary to the defects in membrane organization, were observed in UL135 and UL136 mutant virus-infected cells. For both mutant infections, as many as 75% of the virions in infected ECs deviated from the WT in their morphology. The majority of virions lack a secondary envelope altogether in both UL135STOP virus- and UL136GalK virus-infected ECs (Fig. 6), a phenotype commonly observed in UL133/8NULL infection of ECs. Nearly half of the virions that acquired an envelope in UL135STOP virus-infected ECs were abnormal; the envelope was loosely wrapped, and the virions appeared to lack teguments (Fig. 6B). Not surprisingly, UL135STOP virions could not be banded on glycerol tartrate gradients (data not shown). One intriguing possibility is that these mutant viruses have allowed us to capture an intermediate in virion maturation due to the stalling of the process at a point following the egress of the nucleocapsid into the cytoplasm and prior to full tegumentation and envelopment of the capsid. The formation of DBs was also altered in cells infected with UL135 or UL136 mutant viruses: UL135STOP infection produced fewer DBs, and UL136GalK produced enlarged DBs (Fig. 8). While little is known about the formation of DBs and their contribution to infection, they are thought to be an intermediate in the maturation process and may contribute to secondary rounds of infection by delivering a payload of tegument proteins (75). No alteration in DB formation was seen for either UL135 or UL136 mutant viruses in fibroblasts (Fig. 7). A similar phenotype of enlarged DBs was observed in fibroblasts infected with a mutant virus containing a disruption in UL97, a viral kinase important for early and late stages of infection (76), but it is not known if that phenotype exists in EC infection.
While there are important distinctions, some of the defects associated with the disruption of UL135 in ECs are reminiscent of phenotypes we have observed during infection of fibroblasts with the UL135STOP virus (53). The UL135STOP bacterial artificial chromosome clone is defective in initiating infection following its electroporation into fibroblasts (53). Once produced, virus stocks have an increased particle-to-PFU ratio (a greater number of genome-containing particles is required to form an infectious unit) but replicate with little defect relative to the WT virus when fibroblasts are infected with equivalent multiplicities. In contrast, infection with the UL135STOP virus stock results in a 100-fold defect in replication in ECs relative to WT infection (Fig. 1A), indicating a greater requirement for UL135 for replication in ECs than in fibroblasts. In agreement with the defect in ECs, the UL135STOP virus is defective for reactivation from latency in CD34+ HPCs even at MOIs that result in WT virus yields in fibroblasts (53). The UL135STOP defect in reconstituting viral replication from BAC transfection in fibroblasts and much of the defect in virion maturation can be overcome by the additional disruption of UL138 (53). However, the defect in UL135STOP replication in ECs was not reduced by the additional disruption of UL138 (Fig. 1B). Aberrant envelopment of UL135STOP virions is observed in both fibroblasts and ECs. Taken together, these findings suggest that while UL135 is important for viral replication across different cell types, unique, EC-specific functions exist that are independent of the antagonistic relationship between UL135 and UL138.
The fate of virus particles found within MVB is not yet entirely clear. While MVB could represent a dead-end pathway for the virus if they were to fuse with the lysosome, MVB may be a vehicle for transport out of the cell by exocytosis (77). Viral hijacking of MVB is an important route of entry and egress for enveloped viruses and has recently been shown to be a means by which viruses may alter intercellular communication and inflammation by redesigning exosomes (65, 77–87). HIV proteins interact with a number of components of ESCRT-I and ESCRT-III complexes to facilitate budding at the plasma membrane (88), yet MVB appear to be the site of assembly for HIV in monocyte-derived macrophages (89, 90). Within the herpesvirus family, ESCRT components and MVB are a means of maturation and egress for herpes simplex virus 1 (HSV-1) (64, 91, 92). Specifically, secondary envelopment of HSV-1 requires VPS-4, a component of the ESCRT-III machinery required for the generation of intraluminal vesicles in MVB (92). Similarly, HCMV and human herpesvirus 6 (HHV-6) virions have been observed within MVB (51, 93, 94), and HCMV replication depends on the ESCRT proteins VPS-4 and CHIMP-1 (95). MVB and recycling endosomal markers, including Rab11 and transferrin, localize within the VAC, further suggesting a role for these structures in assembly and egress (58). Intriguingly, ESCRT and MVB have also been implicated in the envelopment and egress of viruses traditionally classified as nonenveloped. The picornavirus hepatitis A virus hijacks an envelope from cellular membranes in a VPS-4B- and Alix-dependent fashion (78).
In agreement with a role for MVB in virus maturation and egress, our studies demonstrate that MVB are induced during HCMV infection in ECs and become densely packed with virions and DBs (Fig. 10). In contrast to those in WT infection, MVB in ECs infected with the UL135STOP virus were devoid of virions and DBs (Fig. 10B). This finding suggests that virions and DBs are actively recruited into MVB by the virus, a process that may require UL135. Further evidence that HCMV encodes mechanisms to modulate MVB biogenesis comes from the disruption of the gene encoding the UL71 tegument protein, which results in enlarged vesicles resembling MVB with virions clustered on the limiting membrane (96). In work from two groups, UL71 mutant viruses exhibit dramatic alteration of intracellular membranes, altered VAC formation, and defects in virus egress (96, 97). In addition to their role in egress, MVB are increasingly being recognized as important organelles for intra- and intercellular signaling (63, 98) through their ability to attenuate signaling or secrete signaling molecules in exosomes, respectively. MVB have been shown to be important for intercellular signaling during Epstein-Barr virus infection of B cells (67, 79, 80). Along these lines, it is intriguing to consider how UL135 may alter exosome composition in HCMV-infected ECs and to speculate on the potential impact of this manipulation on virus dissemination and inflammation of the endothelium. It was reported recently that pUL135 remodels the actin cytoskeleton through an interaction with ABI1 and ABI2 to evade natural killer cell recognition (56), and these interactions have also been observed in our laboratory (M. Rak and F. Goodrum, unpublished results). These host interactions might underlie pUL135-mediated remodeling of the cytoskeleton for proper formation of the VAC, incorporation of virus particles into MVB, and virus maturation in ECs.
UL136 is unique in that it is expressed as five protein isoforms derived from multiple transcription initiation sites throughout the UL136 ORF in combination with alternative translation initiation (54). Small, soluble isoforms of UL136 appear to suppress virus replication in both fibroblasts and CD34+ HPCs. Studies are under way to understand the relative contributions of each UL136 isoform to replication in ECs and to reveal how disruption of individual isoforms or combinations of isoforms may contribute to the phenotypes described in these studies. While the soluble isoforms also suppress virus replication in ECs (K. Caviness and F. Goodrum, unpublished results), further work is required to distinguish complex roles emerging for the other isoforms that were not apparent during infection in fibroblasts.
The number and diversity of cell types infected by HCMV in the human host underlie the complex nature of HCMV persistence and pathogenesis. While we have long appreciated the complexity of HCMV infection in the host, we have little knowledge of how the microenvironment inherent to specialized cell types infected by the virus influences viral persistence. Defining the roles of UL135 and UL136 and the mechanisms by which they ensure tropism in ECs is critical to understanding how viruses target and modulate infection in key cell types in the host. These questions have important implications for understanding how infection of the vascular endothelium impacts inflammation of the endothelium and vascular pathologies such as atherosclerosis, as well as how we might control HCMV persistence and chronic virus shedding by limiting infection of ECs.
ACKNOWLEDGMENTS
We thank William Day of the Arizona Research Laboratories and Patricia Jansma of the Molecular and Cellular Biology Imaging Facility for their expertise and assistance with transmission electron microscopy and confocal microscopy, respectively. We thank Tom Shenk and Bill Britt for the generous gifts of antibodies.
This work was supported by Public Health Service grant AI079059 from the National Institute of Allergy and Infectious Diseases.
The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
REFERENCES
- 1.Britt W. 2008. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr Top Microbiol Immunol 325:417–470. doi: 10.1007/978-3-540-77349-8_23. [DOI] [PubMed] [Google Scholar]
- 2.Goodrum F, Caviness K, Zagallo P. 2012. Human cytomegalovirus persistence. Cell Microbiol 14:644–655. doi: 10.1111/j.1462-5822.2012.01774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jarvis MA, Nelson JA. 2007. Molecular basis of persistence and latency, chapter 42 In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed] [Google Scholar]
- 4.Boeckh M, Geballe AP. 2011. Cytomegalovirus: pathogen, paradigm, and puzzle. J Clin Invest 121:1673–1680. doi: 10.1172/JCI45449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mocarski ES, Shenk T, Pass RF. 2007. Cytomegaloviruses, p 2701–2673. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 6.Demmler GJ. 1996. Congenital cytomegalovirus infection and disease. Adv Pediatr Infect Dis 11:135–162. [PubMed] [Google Scholar]
- 7.Bate SL, Dollard SC, Cannon MJ. 2010. Cytomegalovirus seroprevalence in the United States: the national health and nutrition examination surveys, 1988–2004. Clin Infect Dis 50:1439–1447. doi: 10.1086/652438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caposio P, Orloff SL, Streblow DN. 2011. The role of cytomegalovirus in angiogenesis. Virus Res 157:204–211. doi: 10.1016/j.virusres.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horváth R, Cerny J, Benedik J Jr, Hokl J, Jelinkova I, Benedik J. 2000. The possible role of human cytomegalovirus (HCMV) in the origin of atherosclerosis. J Clin Virol 16:17–24. doi: 10.1016/S1386-6532(99)00064-5. [DOI] [PubMed] [Google Scholar]
- 10.Pawelec G, Derhovanessian E. 2011. Role of CMV in immune senescence. Virus Res 157:175–179. doi: 10.1016/j.virusres.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 11.Pourgheysari B, Khan N, Best D, Bruton R, Nayak L, Moss PA. 2007. The cytomegalovirus-specific CD4+ T-cell response expands with age and markedly alters the CD4+ T-cell repertoire. J Virol 81:7759–7765. doi: 10.1128/JVI.01262-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vasto S, Colonna-Romano G, Larbi A, Wikby A, Caruso C, Pawelec G. 2007. Role of persistent CMV infection in configuring T cell immunity in the elderly. Immun Ageing 4:2. doi: 10.1186/1742-4933-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang GC, Kao WH, Murakami P, Xue QL, Chiou RB, Detrick B, McDyer JF, Semba RD, Casolaro V, Walston JD, Fried LP. 2010. Cytomegalovirus infection and the risk of mortality and frailty in older women: a prospective observational cohort study. Am J Epidemiol 171:1144–1152. doi: 10.1093/aje/kwq062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sinzger C, Grefte A, Plachter B, Gouw AS, The TH, Jahn G. 1995. Fibroblasts, epithelial cells, endothelial cells and smooth muscle cells are major targets of human cytomegalovirus infection in lung and gastrointestinal tissues. J Gen Virol 76:741–750. doi: 10.1099/0022-1317-76-4-741. [DOI] [PubMed] [Google Scholar]
- 15.Sinzger C, Hahn G, Digel M, Katona R, Sampaio KL, Messerle M, Hengel H, Koszinowski U, Brune W, Adler B. 2008. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J Gen Virol 89:359–368. doi: 10.1099/vir.0.83286-0. [DOI] [PubMed] [Google Scholar]
- 16.Adler B, Sinzger C. 2009. Endothelial cells in human cytomegalovirus infection: one host cell out of many or a crucial target for virus spread? Thromb Haemost 102:1057–1063. doi: 10.1160/TH09-04-0213. [DOI] [PubMed] [Google Scholar]
- 17.Grefte A, van der Giessen M, van Son W, The TH. 1993. Circulating cytomegalovirus (CMV)-infected endothelial cells in patients with an active CMV infection. J Infect Dis 167:270–277. doi: 10.1093/infdis/167.2.270. [DOI] [PubMed] [Google Scholar]
- 18.Grefte A, Blom N, van der Giessen M, van Son W, The TH. 1993. Ultrastructural analysis of circulating cytomegalic cells in patients with active cytomegalovirus infection: evidence for virus production and endothelial origin. J Infect Dis 168:1110–1118. doi: 10.1093/infdis/168.5.1110. [DOI] [PubMed] [Google Scholar]
- 19.Jarvis MA, Nelson JA. 2007. Human cytomegalovirus tropism for endothelial cells: not all endothelial cells are created equal. J Virol 81:2095–2101. doi: 10.1128/JVI.01422-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grundy JE, Lawson KM, MacCormac LP, Fletcher JM, Yong KL. 1998. Cytomegalovirus-infected endothelial cells recruit neutrophils by the secretion of C-X-C chemokines and transmit virus by direct neutrophil-endothelial cell contact and during neutrophil transendothelial migration. J Infect Dis 177:1465–1474. doi: 10.1086/515300. [DOI] [PubMed] [Google Scholar]
- 21.Hahn G, Revello MG, Patrone M, Percivalle E, Campanini G, Sarasini A, Wagner M, Gallina A, Milanesi G, Koszinowski U, Baldanti F, Gerna G. 2004. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J Virol 78:10023–10033. doi: 10.1128/JVI.78.18.10023-10033.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Revello MG, Gerna G. 2010. Human cytomegalovirus tropism for endothelial/epithelial cells: scientific background and clinical implications. Rev Med Virol 20:136–155. doi: 10.1002/rmv.645. [DOI] [PubMed] [Google Scholar]
- 23.Sinzger C, Digel M, Jahn G. 2008. Cytomegalovirus cell tropism. Curr Top Microbiol Immunol 325:63–83. doi: 10.1007/978-3-540-77349-8_4. [DOI] [PubMed] [Google Scholar]
- 24.Waldman WJ, Knight DA, Huang EH, Sedmak DD. 1995. Bidirectional transmission of infectious cytomegalovirus between monocytes and vascular endothelial cells: an in vitro model. J Infect Dis 171:263–272. doi: 10.1093/infdis/171.2.263. [DOI] [PubMed] [Google Scholar]
- 25.Bentz GL, Jarquin-Pardo M, Chan G, Smith MS, Sinzger C, Yurochko AD. 2006. Human cytomegalovirus (HCMV) infection of endothelial cells promotes naïve monocyte extravasation and transfer of productive virus to enhance hematogenous dissemination of HCMV. J Virol 80:11539–11555. doi: 10.1128/JVI.01016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Streblow DN, Dumortier J, Moses AV, Orloff SL, Nelson JA. 2008. Mechanisms of cytomegalovirus-accelerated vascular disease: induction of paracrine factors that promote angiogenesis and wound healing. Curr Top Microbiol Immunol 325:397–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Botto S, Streblow DN, DeFilippis V, White L, Kreklywich CN, Smith PP, Caposio P. 2011. IL-6 in human cytomegalovirus secretome promotes angiogenesis and survival of endothelial cells through the stimulation of survivin. Blood 117:352–361. doi: 10.1182/blood-2010-06-291245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Almond PS, Matas A, Gillingham K, Dunn DL, Payne WD, Gores P, Gruessner R, Najarian JS. 1993. Risk factors for chronic rejection in renal allograft recipients. Transplantation 55:752–757. doi: 10.1097/00007890-199304000-00013. [DOI] [PubMed] [Google Scholar]
- 29.Grattan MT, Moreno-Cabral CE, Starnes VA, Oyer PE, Stinson EB, Shumway NE. 1989. Cytomegalovirus infection is associated with cardiac allograft rejection and atherosclerosis. JAMA 261:3561–3566. doi: 10.1001/jama.1989.03420240075030. [DOI] [PubMed] [Google Scholar]
- 30.Bentz GL, Yurochko AD. 2008. Human CMV infection of endothelial cells induces an angiogenic response through viral binding to EGF receptor and β1 and β3 integrins. Proc Natl Acad Sci U S A 105:5531–5536. doi: 10.1073/pnas.0800037105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsich E, Zhou YF, Paigen B, Johnson TM, Burnett MS, Epstein SE. 2001. Cytomegalovirus infection increases development of atherosclerosis in Apolipoprotein-E knockout mice. Atherosclerosis 156:23–28. doi: 10.1016/S0021-9150(00)00608-0. [DOI] [PubMed] [Google Scholar]
- 32.Koskinen PK, Nieminen MS, Krogerus LA, Lemstrom KB, Mattila SP, Hayry PJ, Lautenschlager IT. 1993. Cytomegalovirus infection and accelerated cardiac allograft vasculopathy in human cardiac allografts. J Heart Lung Transplant 12:724–729. [PubMed] [Google Scholar]
- 33.Popović M, Smiljanic K, Dobutovic B, Syrovets T, Simmet T, Isenovic ER. 2012. Human cytomegalovirus infection and atherothrombosis. J Thromb Thrombolysis 33:160–172. doi: 10.1007/s11239-011-0662-x. [DOI] [PubMed] [Google Scholar]
- 34.Streblow DN, Orloff SL, Nelson JA. 2001. Do pathogens accelerate atherosclerosis? J Nutr 131:2798S–2804S. [DOI] [PubMed] [Google Scholar]
- 35.Vliegen I, Stassen F, Grauls G, Blok R, Bruggeman C. 2002. MCMV infection increases early T-lymphocyte influx in atherosclerotic lesions in apoE knockout mice. J Clin Virol 25(Suppl 2):S159–S171. doi: 10.1016/S1386-6532(02)00095-1. [DOI] [PubMed] [Google Scholar]
- 36.Zhou YF, Leon MB, Waclawiw MA, Popma JJ, Yu ZX, Finkel T, Epstein SE. 1996. Association between prior cytomegalovirus infection and the risk of restenosis after coronary atherectomy. N Engl J Med 335:624–630. doi: 10.1056/NEJM199608293350903. [DOI] [PubMed] [Google Scholar]
- 37.Bolovan-Fritts CA, Wiedeman JA. 2002. Mapping the viral genetic determinants of endothelial cell tropism in human cytomegalovirus. J Clin Virol 25(Suppl 2):S97–S109. doi: 10.1016/S1386-6532(02)00089-6. [DOI] [PubMed] [Google Scholar]
- 38.Sinzger C, Schmidt K, Knapp J, Kahl M, Beck R, Waldman J, Hebart H, Einsele H, Jahn G. 1999. Modification of human cytomegalovirus tropism through propagation in vitro is associated with changes in the viral genome. J Gen Virol 80(Part 11):2867–2877. [DOI] [PubMed] [Google Scholar]
- 39.Cha TA, Tom E, Kemble GW, Duke GM, Mocarski ES, Spaete RR. 1996. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol 70:78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davison AJ. 2002. Evolution of the herpesviruses. Vet Microbiol 86:69–88. doi: 10.1016/S0378-1135(01)00492-8. [DOI] [PubMed] [Google Scholar]
- 41.Murphy E, Yu D, Grimwood J, Schmutz J, Dickson M, Jarvis MA, Hahn G, Nelson JA, Myers RM, Shenk TE. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc Natl Acad Sci U S A 100:14976–14981. doi: 10.1073/pnas.2136652100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adler B, Scrivano L, Ruzcics Z, Rupp B, Sinzger C, Koszinowski U. 2006. Role of human cytomegalovirus UL131A in cell type-specific virus entry and release. J Gen Virol 87:2451–2460. doi: 10.1099/vir.0.81921-0. [DOI] [PubMed] [Google Scholar]
- 43.Ryckman BJ, Chase MC, Johnson DC. 2008. HCMV gH/gL/UL128-131 interferes with virus entry into epithelial cells: evidence for cell type-specific receptors. Proc Natl Acad Sci U S A 105:14118–14123. doi: 10.1073/pnas.0804365105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryckman BJ, Jarvis MA, Drummond DD, Nelson JA, Johnson DC. 2006. Human cytomegalovirus entry into epithelial and endothelial cells depends on genes UL128 to UL150 and occurs by endocytosis and low-pH fusion. J Virol 80:710–722. doi: 10.1128/JVI.80.2.710-722.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scrivano L, Sinzger C, Nitschko H, Koszinowski UH, Adler B. 2011. HCMV spread and cell tropism are determined by distinct virus populations. PLoS Pathog 7:e1001256. doi: 10.1371/journal.ppat.1001256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang D, Shenk T. 2005. Human cytomegalovirus UL131 open reading frame is required for epithelial cell tropism. J Virol 79:10330–10338. doi: 10.1128/JVI.79.16.10330-10338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang D, Shenk T. 2005. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc Natl Acad Sci U S A 102:18153–18158. doi: 10.1073/pnas.0509201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nogalski MT, Chan GC, Stevenson EV, Collins-McMillen DK, Yurochko AD. 2013. The HCMV gH/gL/UL128-131 complex triggers the specific cellular activation required for efficient viral internalization into target monocytes. PLoS Pathog 9:e1003463. doi: 10.1371/journal.ppat.1003463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bronzini M, Luganini A, Dell'Oste V, De Andrea M, Landolfo S, Gribaudo G. 2012. The US16 gene of human cytomegalovirus is required for efficient viral infection of endothelial and epithelial cells. J Virol 86:6875–6888. doi: 10.1128/JVI.06310-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Umashankar M, Petrucelli A, Cicchini L, Caposio P, Kreklywich CN, Rak M, Bughio F, Goldman DC, Hamlin KL, Nelson JA, Fleming WH, Streblow DN, Goodrum F. 2011. A novel human cytomegalovirus locus modulates cell type-specific outcomes of infection. PLoS Pathog 7:e1002444. doi: 10.1371/journal.ppat.1002444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bughio F, Elliott DA, Goodrum F. 2013. An endothelial cell-specific requirement for the UL133-UL138 locus of human cytomegalovirus for efficient virus maturation. J Virol 87:3062–3075. doi: 10.1128/JVI.02510-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petrucelli A, Rak M, Grainger L, Goodrum F. 2009. Characterization of a novel Golgi apparatus-localized latency determinant encoded by human cytomegalovirus. J Virol 83:5615–5629. doi: 10.1128/JVI.01989-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Umashankar M, Rak M, Bughio F, Zagallo P, Caviness K, Goodrum FD. 2014. Antagonistic determinants controlling replicative and latent states of human cytomegalovirus infection. J Virol 88:5987–6002. doi: 10.1128/JVI.03506-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caviness K, Cicchini L, Rak M, Umashankar M, Goodrum F. 2014. Complex expression of the UL136 gene of human cytomegalovirus results in multiple protein isoforms with unique roles in replication. J Virol 88:14412–14425. doi: 10.1128/JVI.02711-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petrucelli A, Umashankar M, Zagallo P, Rak M, Goodrum F. 2012. Interactions between proteins encoded within the human cytomegalovirus UL133-UL138 locus. J Virol 86:8653–8662. doi: 10.1128/JVI.00465-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stanton RJ, Prod'homme V, Purbhoo MA, Moore M, Aicheler RJ, Heinzmann M, Bailer SM, Haas J, Antrobus R, Weekes MP, Lehner PJ, Vojtesek B, Miners KL, Man S, Wilkie GS, Davison AJ, Wang EC, Tomasec P, Wilkinson GW. 2014. HCMV pUL135 remodels the actin cytoskeleton to impair immune recognition of infected cells. Cell Host Microbe 16:201–214. doi: 10.1016/j.chom.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Britt B. 2007. Maturation and egress, chapter 20 In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed] [Google Scholar]
- 58.Das S, Pellett PE. 2011. Spatial relationships between markers for secretory and endosomal machinery in human cytomegalovirus-infected cells versus those in uninfected cells. J Virol 85:5864–5879. doi: 10.1128/JVI.00155-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Das S, Vasanji A, Pellett PE. 2007. Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J Virol 81:11861–11869. doi: 10.1128/JVI.01077-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Homman-Loudiyi M, Hultenby K, Britt W, Soderberg-Naucler C. 2003. Envelopment of human cytomegalovirus occurs by budding into Golgi-derived vacuole compartments positive for gB, Rab 3, trans-Golgi network 46, and mannosidase II. J Virol 77:3191–3203. doi: 10.1128/JVI.77.5.3191-3203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanchez V, Greis KD, Sztul E, Britt WJ. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J Virol 74:975–986. doi: 10.1128/JVI.74.2.975-986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sanchez V, Sztul E, Britt WJ. 2000. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-Golgi-intermediate compartment. J Virol 74:3842–3851. doi: 10.1128/JVI.74.8.3842-3851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hanson PI, Cashikar A. 2012. Multivesicular body morphogenesis. Annu Rev Cell Dev Biol 28:337–362. doi: 10.1146/annurev-cellbio-092910-154152. [DOI] [PubMed] [Google Scholar]
- 64.Calistri A, Sette P, Salata C, Cancellotti E, Forghieri C, Comin A, Gottlinger H, Campadelli-Fiume G, Palu G, Parolin C. 2007. Intracellular trafficking and maturation of herpes simplex virus type 1 gB and virus egress require functional biogenesis of multivesicular bodies. J Virol 81:11468–11478. doi: 10.1128/JVI.01364-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Calistri A, Salata C, Parolin C, Palu G. 2009. Role of multivesicular bodies and their components in the egress of enveloped RNA viruses. Rev Med Virol 19:31–45. doi: 10.1002/rmv.588. [DOI] [PubMed] [Google Scholar]
- 66.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Wurdinger T, Middeldorp JM. 2010. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A 107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pegtel DM, van de Garde MD, Middeldorp JM. 2011. Viral miRNAs exploiting the endosomal-exosomal pathway for intercellular cross-talk and immune evasion. Biochim Biophys Acta 1809:715–721. doi: 10.1016/j.bbagrm.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 68.Sahni SK. 2007. Endothelial cell infection and hemostasis. Thromb Res 119:531–549. doi: 10.1016/j.thromres.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 69.Grefte A, Harmsen MC, van der Giessen M, Knollema S, van Son WJ, The TH. 1994. Presence of human cytomegalovirus (HCMV) immediate early mRNA but not ppUL83 (lower matrix protein pp65) mRNA in polymorphonuclear and mononuclear leukocytes during active HCMV infection. J Gen Virol 75(Part 8):1989–1998. doi: 10.1099/0022-1317-75-8-1989. [DOI] [PubMed] [Google Scholar]
- 70.Sacher T, Andrassy J, Kalnins A, Dolken L, Jordan S, Podlech J, Ruzsics Z, Jauch KW, Reddehase MJ, Koszinowski UH. 2011. Shedding light on the elusive role of endothelial cells in cytomegalovirus dissemination. PLoS Pathog 7:e1002366. doi: 10.1371/journal.ppat.1002366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huber MT, Compton T. 1997. Characterization of a novel third member of the human cytomegalovirus glycoprotein H–glycoprotein L complex. J Virol 71:5391–5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li L, Nelson JA, Britt WJ. 1997. Glycoprotein H-related complexes of human cytomegalovirus: identification of a third protein in the gCIII complex. J Virol 71:3090–3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brune W, Menard C, Heesemann J, Koszinowski UH. 2001. A ribonucleotide reductase homolog of cytomegalovirus and endothelial cell tropism. Science 291:303–305. doi: 10.1126/science.291.5502.303. [DOI] [PubMed] [Google Scholar]
- 74.Das S, Ortiz DA, Gurczynski SJ, Khan F, Pellett PE. 2014. Identification of human cytomegalovirus genes important for biogenesis of the cytoplasmic virion assembly complex. J Virol 88:9086–9099. doi: 10.1128/JVI.01141-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kalejta RF. 2008. Tegument proteins of human cytomegalovirus. Microbiol Mol Biol Rev 72:249–265. doi: 10.1128/MMBR.00040-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goldberg MD, Honigman A, Weinstein J, Chou S, Taraboulos A, Rouvinski A, Shinder V, Wolf DG. 2011. Human cytomegalovirus UL97 kinase and nonkinase functions mediate viral cytoplasmic secondary envelopment. J Virol 85:3375–3384. doi: 10.1128/JVI.01952-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pelchen-Matthews A, Raposo G, Marsh M. 2004. Endosomes, exosomes and Trojan viruses. Trends Microbiol 12:310–316. doi: 10.1016/j.tim.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 78.Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong SH, Walker C, Lanford RE, Lemon SM. 2013. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496:367–371. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Meckes DG Jr, Gunawardena HP, Dekroon RM, Heaton PR, Edwards RH, Ozgur S, Griffith JD, Damania B, Raab-Traub N. 2013. Modulation of B-cell exosome proteins by gamma herpesvirus infection. Proc Natl Acad Sci U S A 110:E2925–E2933. doi: 10.1073/pnas.1303906110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Meckes DG Jr, Shair KH, Marquitz AR, Kung CP, Edwards RH, Raab-Traub N. 2010. Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci U S A 107:20370–20375. doi: 10.1073/pnas.1014194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fleming A, Sampey G, Chung MC, Bailey C, van Hoek ML, Kashanchi F, Hakami RM. 2014. The carrying pigeons of the cell: exosomes and their role in infectious diseases caused by human pathogens. Pathog Dis 71:109–120. doi: 10.1111/2049-632X.12135. [DOI] [PubMed] [Google Scholar]
- 82.Sampey GC, Meyering SS, Asad Zadeh M, Saifuddin M, Hakami RM, Kashanchi F. 2014. Exosomes and their role in CNS viral infections. J Neurovirol 20:199–208. doi: 10.1007/s13365-014-0238-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Watanabe T, Sorensen EM, Naito A, Schott M, Kim S, Ahlquist P. 2007. Involvement of host cellular multivesicular body functions in hepatitis B virus budding. Proc Natl Acad Sci U S A 104:10205–10210. doi: 10.1073/pnas.0704000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Morita E, Sandrin V, Alam SL, Eckert DM, Gygi SP, Sundquist WI. 2007. Identification of human MVB12 proteins as ESCRT-I subunits that function in HIV budding. Cell Host Microbe 2:41–53. doi: 10.1016/j.chom.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Prange R. 2012. Host factors involved in hepatitis B virus maturation, assembly, and egress. Med Microbiol Immunol 201:449–461. doi: 10.1007/s00430-012-0267-9. [DOI] [PubMed] [Google Scholar]
- 86.Kolesnikova L, Berghofer B, Bamberg S, Becker S. 2004. Multivesicular bodies as a platform for formation of the Marburg virus envelope. J Virol 78:12277–12287. doi: 10.1128/JVI.78.22.12277-12287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stuchell MD, Garrus JE, Muller B, Stray KM, Ghaffarian S, McKinnon R, Krausslich HG, Morham SG, Sundquist WI. 2004. The human endosomal sorting complex required for transport (ESCRT-I) and its role in HIV-1 budding. J Biol Chem 279:36059–36071. doi: 10.1074/jbc.M405226200. [DOI] [PubMed] [Google Scholar]
- 88.Martin-Serrano J, Neil SJ. 2011. Host factors involved in retroviral budding and release. Nat Rev Microbiol 9:519–531. doi: 10.1038/nrmicro2596. [DOI] [PubMed] [Google Scholar]
- 89.Pelchen-Matthews A, Kramer B, Marsh M. 2003. Infectious HIV-1 assembles in late endosomes in primary macrophages. J Cell Biol 162:443–455. doi: 10.1083/jcb.200304008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Raposo G, Moore M, Innes D, Leijendekker R, Leigh-Brown A, Benaroch P, Geuze H. 2002. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic 3:718–729. doi: 10.1034/j.1600-0854.2002.31004.x. [DOI] [PubMed] [Google Scholar]
- 91.Crump CM, Yates C, Minson T. 2007. Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. J Virol 81:7380–7387. doi: 10.1128/JVI.00222-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pawliczek T, Crump CM. 2009. Herpes simplex virus type 1 production requires a functional ESCRT-III complex but is independent of TSG101 and ALIX expression. J Virol 83:11254–11264. doi: 10.1128/JVI.00574-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fraile-Ramos A, Pelchen-Matthews A, Risco C, Rejas MT, Emery VC, Hassan-Walker AF, Esteban M, Marsh M. 2007. The ESCRT machinery is not required for human cytomegalovirus envelopment. Cell Microbiol 9:2955–2967. doi: 10.1111/j.1462-5822.2007.01024.x. [DOI] [PubMed] [Google Scholar]
- 94.Mori Y, Koike M, Moriishi E, Kawabata A, Tang H, Oyaizu H, Uchiyama Y, Yamanishi K. 2008. Human herpesvirus-6 induces MVB formation, and virus egress occurs by an exosomal release pathway. Traffic 9:1728–1742. doi: 10.1111/j.1600-0854.2008.00796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tandon R, AuCoin DP, Mocarski ES. 2009. Human cytomegalovirus exploits ESCRT machinery in the process of virion maturation. J Virol 83:10797–10807. doi: 10.1128/JVI.01093-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schauflinger M, Fischer D, Schreiber A, Chevillotte M, Walther P, Mertens T, von Einem J. 2011. The tegument protein UL71 of human cytomegalovirus is involved in late envelopment and affects multivesicular bodies. J Virol 85:3821–3832. doi: 10.1128/JVI.01540-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Womack A, Shenk T. 2010. Human cytomegalovirus tegument protein pUL71 is required for efficient virion egress. mBio 1(5):e00282-10. doi: 10.1128/mBio.00282-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Piper RC, Katzmann DJ. 2007. Biogenesis and function of multivesicular bodies. Annu Rev Cell Dev Biol 23:519–547. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]