ABSTRACT
The A/H3N8 canine influenza virus (CIV) emerged from A/H3N8 equine influenza virus (EIV) around the year 2000 through the transfer of a single virus from horses to dogs. We defined and compared the biological properties of EIV and CIV by examining their genetic variation, infection, and growth in different cell cultures, receptor specificity, hemagglutinin (HA) cleavage, and infection and growth in horse and dog tracheal explant cultures. Comparison of sequences of viruses from horses and dogs revealed mutations that may be linked to host adaptation and tropism. We prepared infectious clones of representative EIV and CIV strains that were similar to the consensus sequences of viruses from each host. The rescued viruses, including HA and neuraminidase (NA) double reassortants, exhibited similar degrees of long-term growth in MDCK cells. Different host cells showed various levels of susceptibility to infection, but no differences in infectivity were seen when comparing viruses. All viruses preferred α2-3- over α2-6-linked sialic acids for infections, and glycan microarray analysis showed that EIV and CIV HA-Fc fusion proteins bound only to α2-3-linked sialic acids. Cleavage assays showed that EIV and CIV HA proteins required trypsin for efficient cleavage, and no differences in cleavage efficiency were seen. Inoculation of the viruses into tracheal explants revealed similar levels of infection and replication by each virus in dog trachea, although EIV was more infectious in horse trachea than CIV.
IMPORTANCE Influenza A viruses can cross species barriers and cause severe disease in their new hosts. Infections with highly pathogenic avian H5N1 virus and, more recently, avian H7N9 virus have resulted in high rates of lethality in humans. Unfortunately, our current understanding of how influenza viruses jump species barriers is limited. Our aim was to provide an overview and biological characterization of H3N8 equine and canine influenza viruses using various experimental approaches, since the canine virus emerged from horses approximately 15 years ago. We showed that although there were numerous genetic differences between the equine and canine viruses, this variation did not result in dramatic biological differences between the viruses from the two hosts, and the viruses appeared phenotypically equivalent in most assays we conducted. These findings suggest that the cross-species transmission and adaptation of influenza viruses may be mediated by subtle changes in virus biology.
INTRODUCTION
Influenza A viruses are maintained in aquatic birds as intestinal infections, occasionally transfer to and become established as respiratory infections in mammals, including humans, and sometimes spread from one mammal to another (1, 2). Mammalian hosts that have been commonly seen to maintain avian-derived viruses include swine, horses, humans, mink, seals, and, recently, dogs (2–5). Host transfers between different birds, from birds to mammals, or between different mammalian hosts are relatively common but mostly result in single infections or limited outbreaks. On rare occasions, the host-transferred viruses go on to cause sustained epidemics or pandemics in their new hosts. Influenza viruses causing epidemics in new hosts often have mutations that appear to be specific to the new hosts in several gene segments, and in some cases these have been shown to control host adaptation (6–8). In many cases, the transferred virus was observed to be a reassortant with segments from a number of different ancestors, or it soon reassorted with another influenza virus infecting that host (9, 10).
A number of different viral functions have been associated with host adaptation of influenza viruses. Specific sialic acid binding and/or cleavage is often a key factor in host adaptation because sialic acids are primary influenza virus receptors, and mutations in the receptor-interacting proteins, the hemagglutinin (HA) and neuraminidase (NA), often appear upon host transfer. Key traits include HA recognition of α2-3- or α2-6-linked sialic acids; avian viruses are generally specific for α2-3-linked receptors, and human viruses are generally specific for α2-6-linked receptors (11–13). There is often a coordination of the NA activity and specificity that correlates with HA binding and the sialic acid linkages that are present in the host (14). Importantly, mutations in other gene segments, including PB2, PA, NP, M, and NS, are often seen (15–18). In particular, polymerase subunits PB2 and PA control replication in different host cells and at different temperatures (19). Some mutations in the M gene segment have been associated with transmission (20), while NP mutations control the interactions with MxA, a host-derived antiviral molecule (21). Mutations in the NS1 gene control a variety of host-specific functions and innate immune responses (22, 23). Despite the identification of these mutations, we lack a complete understanding of the factors that control specific virus host range, particularly in nature, or of the host barriers that regulate the transfer of viruses to new hosts.
In this study, we examined the host tropism associated with the transfer to and continuing replication of the A/H3N8 equine influenza virus (EIV) in dogs to create the phylogenetically distinct lineage of A/H3N8 canine influenza viruses (CIVs) (Fig. 1) (3, 24–26). CIV was first identified in Florida in 2004, when it caused an outbreak in greyhounds in a training facility, and it was soon recognized to be closely related to EIV (3). Infected greyhounds carried the virus to different regions of the United States, and many other breeds of dogs have since been infected (24, 27, 28). CIV has continued to circulate in some regions of the United States, and for the past several years it appears to have been primarily maintained in several hot spots where there are high-density and high-turnover dog populations, including animal shelters in New York, NY, Philadelphia, PA, Colorado Springs, CO, and Denver, CO (26, 28, 29). Dogs appear to be a naturally receptive host of influenza virus, because in addition to the equine-origin A/H3N8 virus, an avian-origin A/H3N2 subtype has spread among dogs in Korea and China since 2006 (30, 31).
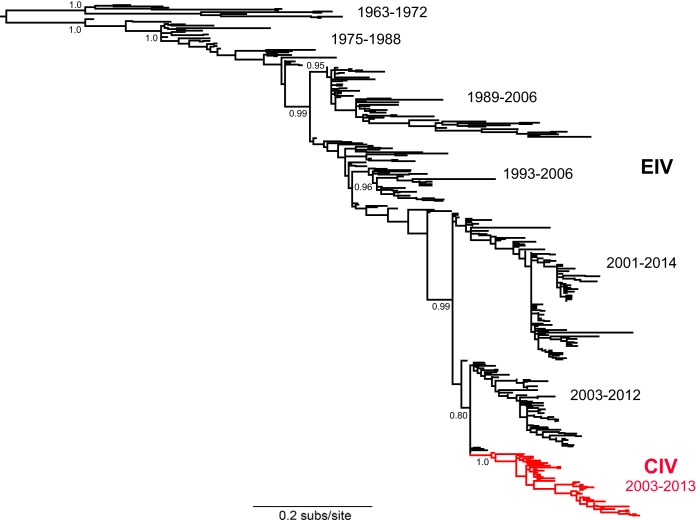
FIG 1.
Maximum likelihood phylogenetic tree of 400 EIV and CIV HA sequences, with the latter shown in red. All tip labels were removed for clarity of representation. SH-like branch supports are shown for key nodes, as are time ranges of sampling for the main clusters of viral sequences. The CIV sequences clearly form a single monophyletic group, indicative of a single viral emergence event in dogs.
Examination of A/H3N8 isolates collected from dogs has shown that all segments contained CIV-specific mutations not seen in equine viruses, or seen only at very low frequencies (26, 28). However, it is not known how these CIV-specific mutations alter host range and tropism or whether there has been ongoing selection of canine-adaptive mutations during the extended passage in dogs. To determine whether any of the CIV-specific mutations play a role in virus host switching and adaptation, we examined protein sequences of all available CIV sequences deposited in GenBank (NCBI). We also prepared reverse-genetics plasmid sets of EIV and CIV and used these to derive viruses that we tested for host tropism and infectivity in cells. Additionally, we examined receptor specificity and HA cleavage, and lastly, we looked at growth and infectivity of these viruses in horse and dog tracheal explants.
MATERIALS AND METHODS
Cells and cell culture.
All mammalian cells were grown at 37°C under 5% CO2 and included Marbin-Darby canine kidney (MDCK) cells, canine tumor (A72) fibroblasts, Norden laboratory feline kidney (NLFK) cells, human lung cancer (A549) cells, human kidney embryonic kidney (HEK293T) cells, ferret (Mpf) fibroblasts, Chinese hamster ovary (CHO) cells, and equine kidney (EQKD) cells. Cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Insect cells (Sf9 and High Five) were grown in Grace's insect medium supplemented with 10% FBS at 23°C. CHO cells express only α2-3-linked sialic acids (32). To generate CHO cells with various levels of α2-6-linked sialic acids, cells were transfected with a plasmid expressing α2-6 sialyltransferase (33) using Lipofectamine 2000 (Life Technologies, Carlsbad, CA) and selected with hygromycin B (Life Technologies) at 250 μg/ml. Stably transfected cells were double stained to detect the levels of expression of α2-3- or α2-6-linked sialic acids. Biotinylated Maackia amurensis agglutinin type 1 (MAA1) (Vector Laboratories, Burlingame, CA) was used to detect α2-3-linked sialic acids, and fluorescein isothiocyanate (FITC)-conjugated Sambucus nigra agglutinin (SNA) (Vector Laboratories) was used to detect α2-6-linked sialic acids. Cells were incubated with biotinylated MAA1 for 1 h on ice and then with FITC-conjugated SNA and phycoerythrin (PE)-conjugated streptavidin (Life Technologies). Cells were assayed by flow cytometry by following the commercial protocol using the Millipore Guava EasyCyte Plus flow cytometer (EMD Millipore, Billerica, MA), and expression levels were analyzed by FlowJo software (TreeStar, Ashland, OR).
Viruses, plasmids, and virus rescue.
A/equine/NY/61191/2003 and A/canine/NY/dog23/2009 were plaque purified and then passaged in MDCK cells. The eight gene segments of each virus were cloned into modified pDZ plasmids (34). Cocultures of HEK293T and MDCK cells (2:1 ratio) in 6-well plates were transfected with 300 ng of each of the eight influenza virus plasmids using TransIT-LT1 (Mirus Bio, Madison, WI). At 24 h posttransfection, medium was changed to DMEM with 0.3% bovine serum albumin (BSA) containing 1 μg/ml of tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK) trypsin from bovine pancreas (Sigma-Aldrich, St. Louis, MO). After 48 h, the supernatant was harvested and used to inoculate MDCK cells supplemented with 1 μg/ml of trypsin in 6-well plates to grow P2 virus. The supernatant was harvested 72 h later and clarified by low-speed centrifugation. Standard hemagglutination assays using 0.5% chicken erythrocytes (Lampire Biological Laboratories, Pipersville, PA) confirmed the presence of P2 virus (35). The virus was used to infect MDCK cells supplemented with 1 μg/ml of trypsin in 75-cm2 flasks to grow up working stocks of P3 virus. At 72 h postinfection, supernatant was harvested and clarified, and virus was frozen down at −80°C in 500-μl aliquots.
Virus titration.
Virus stocks were quantified by 50% tissue culture infectious dose (TCID50), HA assays, and genome copies (by real-time quantitative reverse transcription-PCR [real time qRT-PCR]). TCID50 was determined using 96-well plates seeded with MDCK cells. Briefly, virus stocks were 10-fold serially diluted in DMEM and 50-μl volumes were inoculated into each well across 8 rows. After 48 h of incubation, cells were fixed with 4% paraformaldehyde (PFA) for 10 min. Cells were then washed with phosphate-buffered saline (PBS), and mouse IgG anti-nucleoprotein (anti-NP) antibody (Creative Diagnostics, Shirley, NY) was added to each well in permeabilization buffer (PBS with 0.5% saponin). After 1 h of incubation, cells were washed with PBS and then incubated for 1 h with goat IgG anti-mouse Alexa Fluor 488-conjugated antibody (Life Technologies) in permeabilization buffer. Cells were washed with PBS and viewed by a Nikon TE300 fluorescence microscope. Each well was scored as positive for infection as long as there was a single infected cell, and TCID50 was calculated using the Reed and Muench method. Standard hemagglutination assays were performed using 0.5% chicken erythrocytes as mentioned above. Virus genome copies were calculated by real-time qRT-PCR targeting the influenza virus M gene segment (36). First, viral RNA was extracted using a QIAmp viral RNA minikit (Qiagen, Venlo, Netherlands). Next, the viral RNA, two outside primers specific for influenza virus M, and a TaqMan probe were used to set up standard reaction cocktails using the QuantiTect Probe PCR kit (Qiagen) supplemented with ImProm-II reverse transcriptase (Promega, Madison, WI). Samples were exposed to a reverse transcription step and subsequent 40-cycle amplification step in an AB StepOnePlus real-time qRT-PCR machine (Applied Biosciences, Foster City, CA), and genome copies per microliter were calculated based on the threshold cycle (CT) values of a standard curve generated using the influenza virus M gene plasmid.
Phylogenetic analysis.
A total of 400 representative HA sequences of EIV and CIV sequences were downloaded from GenBank. A minimum sequence length comprising at least the HA1 domain was set, and identical sequences were excluded. The sequences were easily aligned using the MAFFT method available in Geneious (37), resulting in a total alignment length of 1,710 bp. The phylogenetic relationships among these sequences were then determined using the maximum likelihood (ML) approach available in the PhyML program (38). This analysis utilized the generalized time-reversible plus (GTR+) model of nucleotide substitution and a combination of subtree pruning and regrafting (SPR) and nearest neighbor interchange (NNI) branch swapping. The robustness of individual nodes on the phylogeny was assessed using Shimodaira-Hasegawa-like (SH-like) branch supports.
Virus sequencing.
Virus RNA was extracted using a QIAmp viral RNA minikit. The cDNA was synthesized using avian myeloblastosis virus (AMV) reverse transcriptase (Promega) and universal primer Uni12 (5′-AGCAAAAGCAGG-3′). Influenza HA and NA gene segments were amplified by PCR with EIV and CIV gene-specific primers. The PCR products were purified using the E.Z.N.A. Cycle-Pure kit (Omega Bio-Tek, Norcross, GA). Purified DNA was sequenced using an Applied Biosystems 3730xl DNA analyzer (Life Technologies) at the Cornell University Institute of Biotechnology, and full-length genes were assembled using Lasergene software (DNASTAR, Madison, WI).
Virus sequence analysis.
Consensus protein sequences of EIV and CIV representing differences in host and time of sampling were generated and compared. In this context, a consensus sequence is used to define the most common amino acid in all available EIVs and CIVs (i.e., epidemiological scale) and not simply those from a single host. All available EIV and CIV sequences in GenBank were used. Sequence alignments were performed using MEGA (Arizona State University, Phoenix, AZ), and Clustal Omega (EMBL-EBI, Cambridge, United Kingdom) was used to find the most common amino acid at each position. Three groups of consensus EIV and CIV protein sequences (HA, NA, M1, NP, NS1, PA, PB1, and PB2) were generated. The first group represented EIV isolates sampled close to the ancestor of the CIVs, starting from 1990 to 2011, the second group represented CIV isolates sampled soon after the emergence in dogs (between 2003 and 2007), and the third group represented CIV isolates sampled after the virus had been circulating in dogs for at least 8 years (since 2000), from 2008 to 2013. Both EIV and CIV plasmid sets were compared with their respective consensus sequences to ensure that they were good representatives of EIV and CIV.
Virus growth curves.
MDCK cells were seeded in 12-well plates. Upon reaching confluence, cells were washed with DMEM and incubated with virus diluted in DMEM containing 0.3% BSA and 1 μg/ml of trypsin at a multiplicity of infection (MOI) of 0.0006 based on TCID50 for 1 h at 37°C. Cells were then washed with and replenished with fresh DMEM containing 0.3% BSA and 1 μg/ml of trypsin. Supernatants were harvested from each well every 24 h for 5 days and stored at −80°C. TCID50 and genome copies for all time points were determined as described above.
Virus infection of cell lines derived from different hosts.
Different host cells were seeded in 48-well plates. Upon reaching confluence, cells were washed and incubated with virus diluted in DMEM containing 0.3% BSA and 0.5 μg/ml of trypsin at an MOI of 0.05 based on TCID50 for 1 h at 37°C. Cells were then washed and replenished with fresh DMEM containing 0.3% BSA and 0.5 μg/ml of trypsin. Cells were fixed with 4% PFA for 10 min at 24 h postinfection and stained for NP expression as described above and with 4′,6-diamidino-2-phenylindole (DAPI; Life Technologies) for 5 min following the commercial protocol. Stained cells were visualized by fluorescence microscopy. Infections of different host cells were also quantified by flow cytometry. Briefly, cells were grown in 48-well plates and, upon reaching confluence, inoculated with virus as described above at an MOI of 0.1 based on TCID50. Cells were harvested and fixed with 4% PFA for 10 min at 24 and 48 h postinfection. Cells were stained for NP and quantified by flow cytometry as described above, and results were analyzed by FlowJo.
Lectin staining.
Cells were grown in 48-well plates, and upon reaching confluence, they were collected and fixed using 4% PFA. Cells were incubated with either FITC-conjugated MAA1 or FITC-conjugated SNA to detect α2-3- or α2-6-linked sialic acids, respectively. After 1 h of incubation, cells were washed with PBS with 1% BSA. Cells were assayed by flow cytometry as described above, and results were analyzed using FlowJo. In addition to flow cytometry, fluorescence microscopy was also used to look at lectin-stained cells.
Construction and purification of HA-Fc fusion proteins.
The EIV and CIV HA ectodomains (the same sequences as in the reverse-genetics plasmids) were fused to human IgG1 Fc at the C terminus, followed by a hexahistidine tag (39, 40). The baculovirus gp64 secretion peptide was fused to the constructs at the N terminus. The genes were synthesized by GeneScript (Piscataway, NJ) and cloned into pFastBac-1 (Life Technologies) to generate recombinant bacmids by following the commercial protocol. Recombinant baculoviruses were recovered by bacmid transfection into Sf9 insect cells using Cellfectin II (Life Technologies). Viruses were then used to infect suspension High Five cells, and 2 days postinfection, the proteins were purified by binding to a HiTrap ProteinG HP 5-ml column (GE Healthcare Life Sciences, Piscataway, NJ) and eluted with 0.1 M citrate, pH 3.0 (pH neutralization to 7.8 with 1 M Tris, pH 9.0) using an ÄKTA fast protein liquid chromatography (FPLC) system (GE Healthcare Life Sciences). The HA-Fc containing fractions were dialyzed in PBS and concentrated using 30-kDa Amicon Ultra-15 centrifugal filter tubes (EMD Millipore). The proteins were stored at −80°C in aliquots. Concentration was measured using Beer-Lambert law calculation based on the A280 reading.
Testing virus receptor specificity.
CHO cells and CHO cells expressing α2-6-linked sialic acids (6H4 cells) were grown in 48-well plates. When confluent, cells were inoculated with viruses as described above at an MOI of 1 based on TCID50. After 24 h postinfection, cells were collected and fixed by 4% PFA. Cells were stained for NP and quantified by flow cytometry as described above, and results were analyzed using FlowJo. Purified EIV and CIV HA-Fc proteins were used in glycan binding microarrays. The microarrays were fabricated using epoxide-derivatized glass slides, and the high-throughput protein binding screening was carried out as previously described (41, 42). Briefly, freshly printed glycan microarray slides were blocked by ethanolamine, washed and dried, and then fitted in a multiwell microarray hybridization cassette (ArrayIt, CA) to divide them into subarrays. The subarrays were blocked with ovalbumin (1%, wt/vol) in PBS (pH 7.4) for 1 h at room temperature in a humid chamber with gentle shaking. Subsequently, the diluted HA-Fc samples were added to the subarrays and incubated for 2 h at room temperature with gentle shaking, and lastly, the slides were extensively washed. Fluorescently labeled antibody (Cy3-labeled goat anti-human IgG; Jackson ImmunoResearch Laboratories) was then applied and incubated for 1 h. Following final washes and drying, the developed glycan microarray slides were scanned with a Genepix 4000B microarray scanner (Molecular Devices Corp., Union City, CA). Data analysis was done using Genepix Pro 7.0 analysis software (Molecular Devices Corp., Union City, CA).
HA cleavage assays.
HEK293T cells were seeded in 24-well plates coated with poly-d-lysine. Upon reaching 70% confluence, 500 ng of each respective HA plasmid was transfected using Lipofectamine 2000 (Life Technologies) by following the manufacturer's protocol. After 18 h posttransfection, cells were washed with PBS and incubated with trypsin at 3 μg/ml for 15 min. Cells were kept at 4°C and surface biotinylated using sulfo-NHS-SS-biotin (Thermo Scientific) at 250 μg/ml for 30 min by following the manufacturer's protocol. Excess biotin was quenched using 50 mM glycine for 10 min. Cells were lysed using radioimmunoprecipitation assay (RIPA) buffer (EMD Millipore) with complete protease inhibitor cocktail tablets (Roche, Nutley, NJ) for 10 min. Lysed cells were high-speed centrifuged for 20 min at 4°C. Supernatant was collected and incubated with a 50% suspension of streptavidin agarose beads (Thermo Scientific) for 18 h with rotation at 4°C. Beads were then washed with RIPA buffer and resuspended in 2× Laemmli sample buffer containing 10% beta-mercaptoethanol for Western blotting (43). HA bands were detected using goat IgG polyclonal anti-H3 HA antibody (BEI Resources, Manassas, VA) followed by rabbit IgG anti-goat antibody conjugated to horseradish peroxidase (Thermo Scientific). Western blot images were taken using a FujiFilm LAS-3000 imaging system. The pixel density of HA bands was measured by ImageJ (National Institutes of Health, Bethesda, MD), and relative cleavage efficiencies were calculated based on the following formula: (HA1/HA0 + HA1) × 100 (43).
Infection of horse and dog tracheal explants.
Tracheal explant cultures were acquired, prepared, maintained, and tested for viability as described previously (44–46). Viruses were used to infect explants by inoculating 400 TCID50 units of each virus directly on the epithelium layer. Virus growth was assayed by plaque assays in MDCK cells every 24 h as described previously (44). Explant sections were used for hematoxylin and eosin staining and also for virus antigen NP staining on days 1, 3, and 5 postinfection (44). Due to the difficulty of obtaining fresh horse trachea from healthy animals, only one experimental replicate was done using the horse tracheal explants.
Statistics.
Statistical significance was measured by the Student t test using GraphPad Prism when appropriate.
RESULTS
EIV and CIV genetic analysis.
Three sets of epidemiological-scale consensus protein sequences were generated: EIV (1990 to 2011), early CIV (2003 to 2007), and more recent CIV (2008 to 2013) (Fig. 2). Sequence alignments revealed consensus amino acid mutations in the eight major proteins. Some mutations were seen only between EIV and early CIV sequences, while others appeared only between the early CIV and more recent CIV sequences. HA exhibited the greatest number of mutations, while M1 showed the least. In addition, a phylogenetic analysis of 400 EIV and CIV HA sequences clearly showed that the CIV sequences formed a single monophyletic group distinct from EIV (Fig. 1).
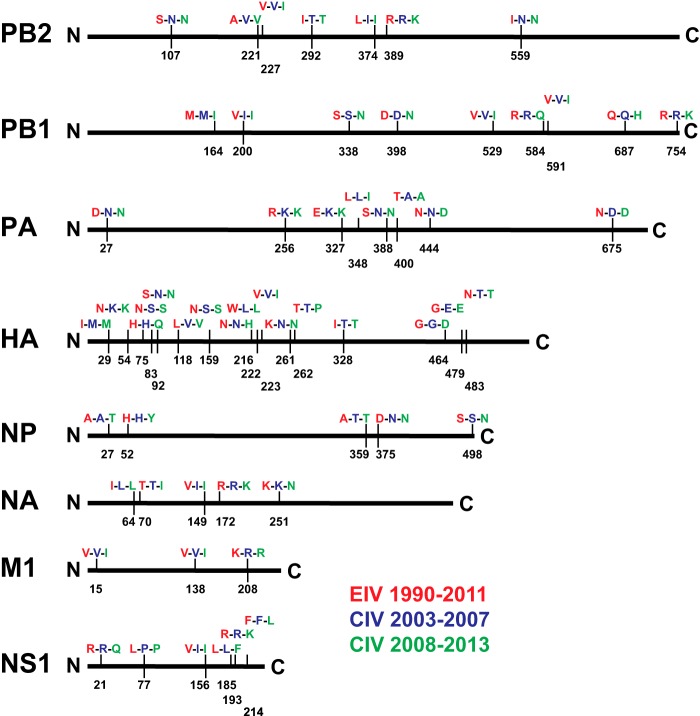
FIG 2.
Comparison of EIV and CIV epidemiological-scale consensus protein sequences. The consensus sequences of eight major influenza virus proteins were generated for EIV (1990 to 2011), for early CIV (2003 to 2007), and for more recent CIV (2008 to 2013). The three consensus sequences were aligned, and differences are indicated by vertical bars along each protein. Each vertical bar indicates the amino acid position (H3 and N2 numbering) and specific amino acids differences for EIV (red), early CIV (blue), and more recent CIV (green) sequences. A full vertical bar indicates a change comparing EIV to early CIV and no change between early CIV and more recent CIV. Half of a vertical bar indicates no change between EIV and early CIV but a change between early CIV and more recent CIV. Additionally, EIV NS1 can be truncated by 11 amino acids at the C terminus, while CIV NS1 is never truncated except for the first reported CIV sequence in 2003 (A/canine/Florida/242/2003 [H3N8]).
Our analysis revealed substitutions at putative HA antigenic sites (residues 54, 75, 83, 92, 159, and 216), the receptor binding pocket (residues 222 and 223), and sites that may influence HA cleavage (residues 328 and 483) (47–50). Although there were no NA mutations in the active site, changes at position 149 may affect sialidase activity because the 150 loop can incorporate sialic acid derivatives to inhibit NA enzymatic activity (51). Mutations at the N terminus of NP (residues 27 and 52) were located in RNA and PB2 binding domains, and mutations at the C terminus (residues 359, 375, and 498) may influence NP polymerization and binding to host actin (52). The M1 138 mutation was in a domain that is responsible for polymerization and binding to NP (53). Mutations in NS1 could potentially change a number of interactions with host proteins, including poly(A)-binding proteins I and II, importin-α, nucleolin, and translation initiation factors (54). Structural insights into the polymerase proteins have revealed functional domains, and our analysis revealed mutations in those regions. For example, mutations in PB2 (residues 374 and 389) were located in the host RNA cap binding domain, C-terminal mutations in PB1 (residues 687 and 754) may affect binding to PB2, and mutations in PA (residues 327, 348, 388, 400, 444, and 675) may alter interactions with PB1 (55). The mutation at position 27 in PA was located in the endonuclease domain, and with the recent discovery of PA-X, the mutation may affect PA-X-specific host gene suppression (56–59). Additionally, positions 400 in PA and 292 in PB2 may be host determinants and play roles in virus adaptation (60, 61).
Rescued viruses and virus growth.
An EIV sampled close to the ancestral sequence of CIV (A/equine/NY/61191/2003) and a CIV sampled from 9 years after the virus transferred into dogs (A/canine/NY/dog23/2009) were chosen as representatives of these viruses based on comparing their sequences to the consensus, and they were rescued by reverse genetics. Additionally, two reassortant viruses were recovered: EIV with CIV HA and NA and the reciprocal virus, CIV with EIV HA and NA. All viruses reached high infectious titers after minimal passages in MDCK cells and were able to hemagglutinate chicken erythrocytes (Table 1). The HA and NA genes of the virus stocks were sequenced, and there were no mutations determined by comparing the sequences to the reverse-genetics plasmids. Five-day growth curves for the viruses were determined in MDCK cells, and there was no significant difference (P > 0.05) in yields of infectious particles (Fig. 3A) or RNA copies (Fig. 3B). Infectious titers peaked for all viruses between 48 and 72 h and then steadily declined. RNA copies reached their peaks at similar time points and then plateaued.
TABLE 1.
Reverse-genetics rescue of EIV, CIV, and reassortant virusesa
| Virus | No. of log TCID50/ml | No. of log genome copies/μl | No. of HA units |
|---|---|---|---|
| EIV | 6.82 | 7.99 | 64 |
| CIV | 6.63 | 7.28 | 32 |
| EIV/CIV HA+NA | 6.40 | 7.11 | 32 |
| CIV/EIV HA+NA | 6.47 | 7.26 | 32 |
A representative EIV (A/equine/NY61191/2003) and CIV (A/canine/NY/dog23/2009) were recovered from 8 reverse-genetics plasmids along with HA and NA double reassortant viruses. Virus rescue was confirmed by TCID50, virus genome copies, and hemagglutination assays using chicken erythrocytes.
FIG 3.
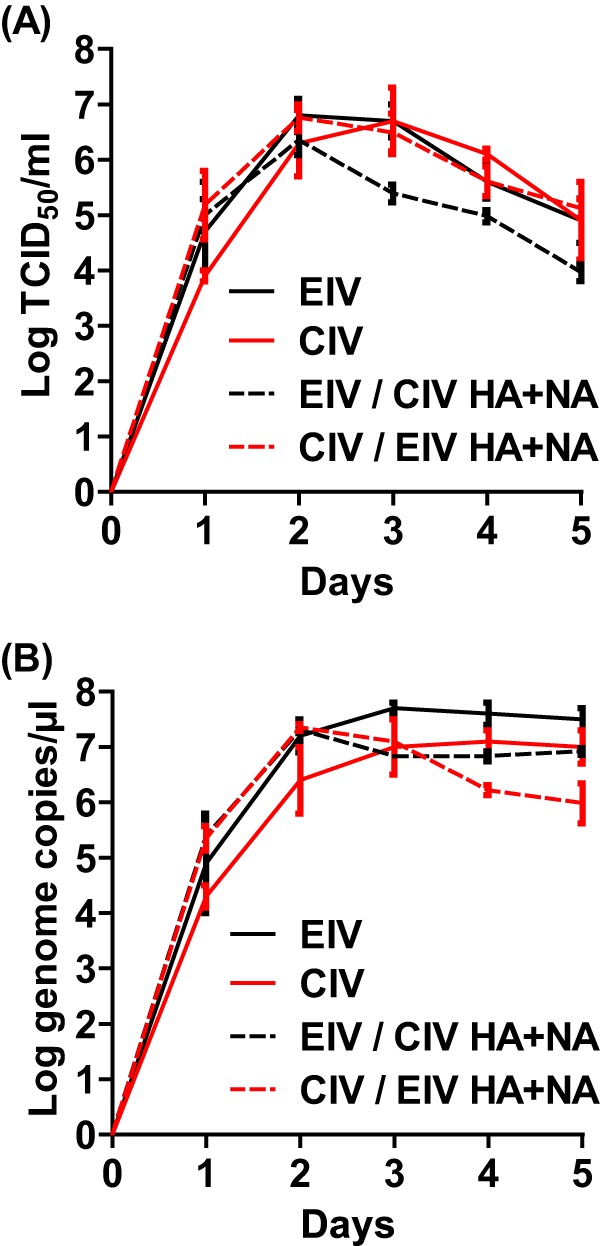
EIV, CIV, and reassortant virus growth curves in MDCK cells. Viruses were used to inoculate MDCK cells at an MOI of 0.0006 in the presence of 1 μg/ml of trypsin. Infectious titer (A) and genome copies per microliter (B) were determined every 24 h postinfection. Error bars represent standard deviations from three independent experiments.
EIV and CIV infections in different host cells.
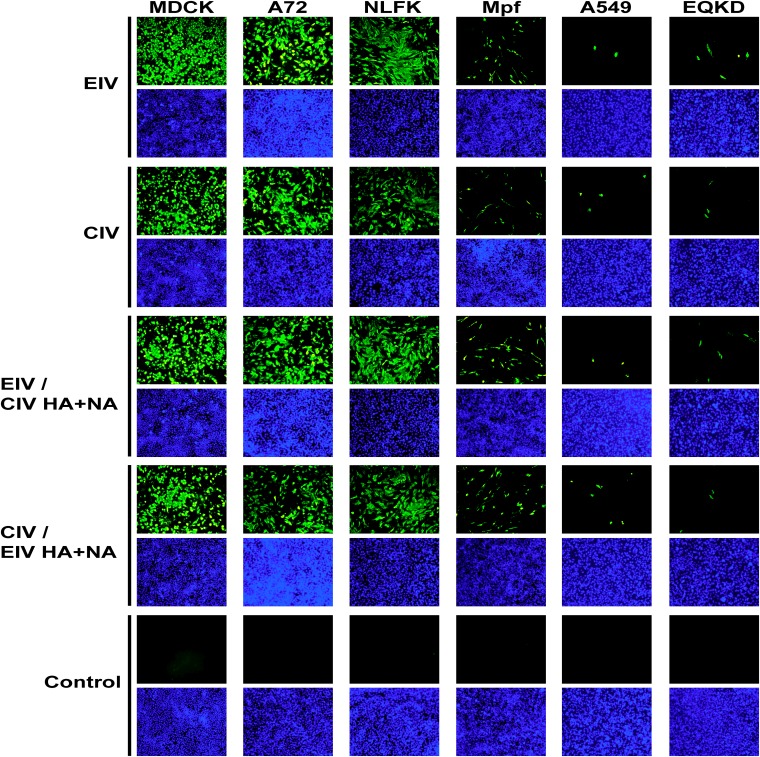
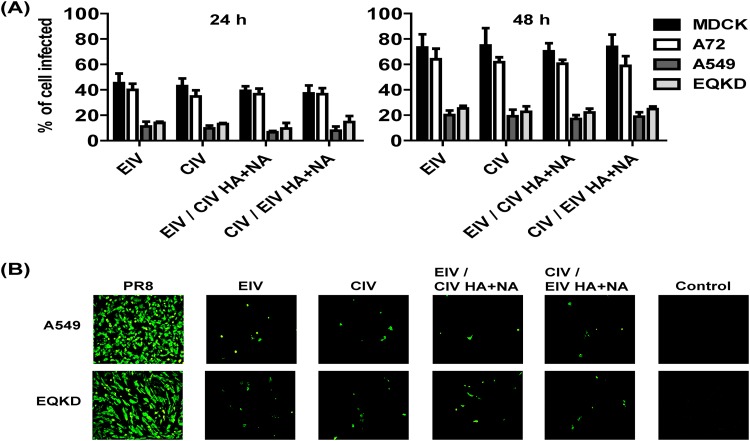
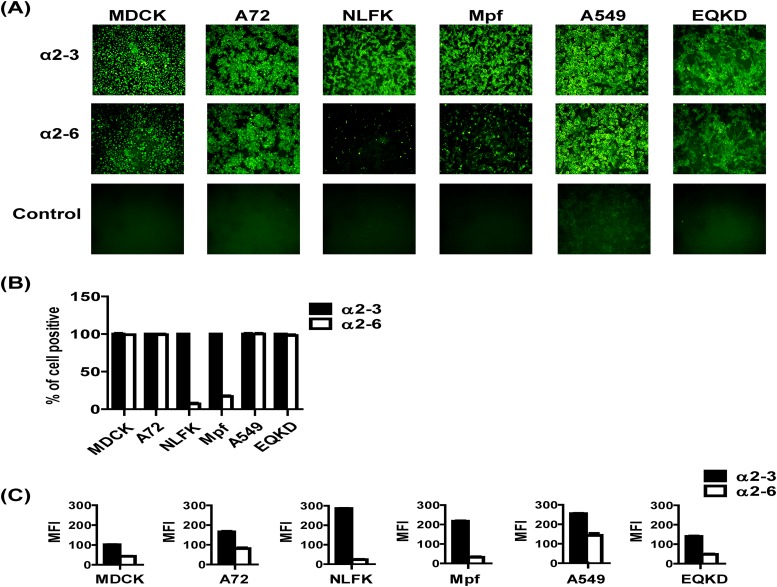
Viruses were used to inoculate cells (MOI = 0.05) from several hosts: MDCK (dog), A72 (dog), NLFK (cat), Mpf (ferret), A549 (human), and EQKD (horse). Cells were stained for virus NP at 24 h postinfection, and visually, the infectivities of the viruses looked similar; however, different host cells exhibited various levels of susceptibility to infection (Fig. 4). MDCK, A72, and NLFK cells were all heavily infected. Mpf cells were moderately infected, and A549 and EQKD cells were poorly infected. To further analyze infectivity in different host cells, viruses were used to inoculate four types of host cells (two that were permissive to infection, MDCK and A72 cells, and the two that were least permissive to infection, A549 and EQKD cells) (MOI = 0.1), and the percentage of infected cells was quantified by flow cytometry at 24 and 48 h postinfection (Fig. 5A). There was no significant difference (P > 0.05) in infectivity between the viruses. At 24 h postinfection, around 40% of MDCK and A72 cells were infected by all viruses. In contrast, about 15% of A549 and EQKD cells stained positive for infection. At 48 h postinfection, the percentage of MDCK and A72 infected cells rose to around 70 to 80%, while the percent increase of infected A549 and EQKD cells was much more modest, around 20% (Fig. 5A). To ensure that A549 and EQKD cells can be infected by influenza virus, the laboratory-adapted human virus A/Puerto Rico/8/1934 H1N1 (PR8) was used to infect A549 and EQKD cells (MOI = 0.05), and its infectivity was compared to those of EIV and CIV at 24 h postinfection (Fig. 5B). PR8 infected high proportions of A549 and EQKD cells, in contrast to EIV and CIV.
FIG 4.
Immunofluorescence images of EIV, CIV, and reassortant virus infections in different host cells. Viruses were used to inoculate different cells at an MOI of 0.05 in the presence of 0.5 μg/ml of trypsin. Anti-NP staining (green) was used to detect virus at 24 h postinfection, and DAPI (blue) was used for nuclear staining. Images were taken at a magnification of ×100 as overlays using a fluorescence microscope. The overlays were kept as separate panels for clarity.
FIG 5.
Quantification of EIV, CIV, and reassortant virus infections in different host cells, and PR8 infections of cells least permissive to EIV and CIV. Viruses were used to inoculate different host cells at an MOI of 0.1 in the presence of 0.5 μg/ml of trypsin and quantified by flow cytometry at 24 h and 48 h postinfection (A). Similarly, PR8, EIV, CIV, and reassortant viruses were used to inoculate A549 and EQKD cells at an MOI of 0.05 in the in the presence of 0.5 μg/ml of trypsin and visualized by fluorescence microscopy (anti-NP staining) 24 h postinfection (B). Error bars represent standard deviations from three independent experiments.
EIV and CIV receptor specificity.
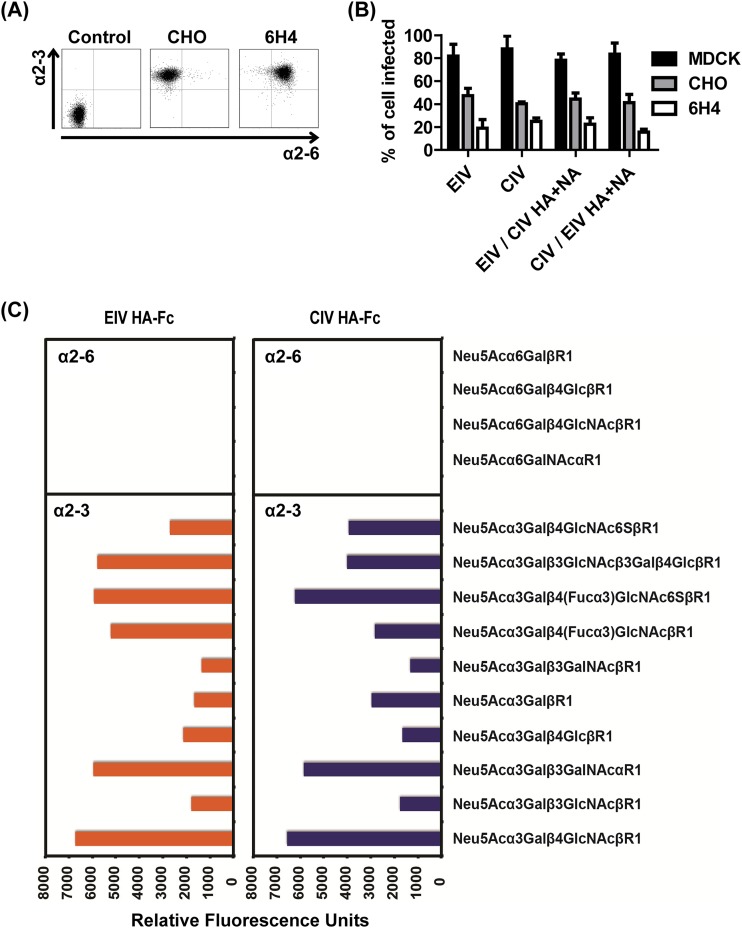
To determine if the difference in infectivity of various host cells was due to virus receptor availability and/or differences, cells were lectin stained and visualized by microscopy and fluorescence was measured by flow cytometry (Fig. 6). MDCK, A72, A549, and EQKD cells all stained positive for both α2-3- and α2-6-linked sialic acids, while NLFK and Mpf cells were stained predominately for α2-3-linked sialic acids (Fig. 6A and B). Overall, there was a higher relative concentration of α2-3-linked sialic acids compared to α2-6-linked sialic acids in all cell lines (Fig. 6C). CHO cells expressed only α2-3-linked sialic acids, while CHO cells stably transfected with α2-6 sialyltransferase (6H4) expressed both α2-3- and α2-6-linked sialic acids (Fig. 7A). Viruses were used to inoculate these cells (MOI = 1), and infectivity was assayed by flow cytometry 24 h postinfection. There was no significant difference (P > 0.05) in infectivity among the viruses, but all viruses showed around a 50% reduction in their ability to infect 6H4 cells compared to CHO cells (Fig. 7B). To further analyze EIV and CIV receptor specificity, HA-Fc fusion proteins were generated and used to bind different glycans on microarrays. The binding specificities for the EIV and CIV HA-Fc proteins were similar; both bound to α2-3-linked sialic acids, and neither bound to any α2-6-linked sialic acids in the array (Fig. 7C).
FIG 6.
Lectin staining of different host cells. Different host cells were lectin stained using FITC-conjugated MAA1 to detect α2-3-linked sialic acids and FITC-conjugated SNA to detect α2-6-linked sialic acids and visualized by fluorescence microscopy (A) or assayed by flow cytometry (B). Relative abundance of the sialic acids was measured by flow cytometry as well (C). Error bars represent standard deviations from three independent experiments.
FIG 7.
EIV and CIV receptor specificity. Lectin staining showed that CHO cells expressed only α2-3-linked sialic acids, while 6H4 cells expressed both α2-3- and α2-6-linked sialic acids (A). EIV, CIV, and reassortant viruses were used to inoculate MDCK (control), CHO, and 6H4 cells at an MOI of 1 in the presence of 0.5 μg/ml of trypsin. After 24 h postinfection, cells were stained for anti-NP and quantified by flow cytometry (B). EIV and CIV HA-Fc fusion proteins were used to bind α2-3- and α2-6-linked sialic acids on glycan microarrays (C). Error bars represent standard deviations from three independent experiments.
EIV and CIV HA cleavage efficiency.
HA cleavage assays showed that both wild-type proteins and CIV HA mutants (T328I and T483N) were efficiently cleaved by trypsin under the conditions of this trial (Fig. 8A). There was an extra band detected of around 38 kDa, and this was most likely degraded HA product detected by the polyclonal antibody. CIV HA T483N had a lower percentage of cleavage than did EIV and CIV HA T328I (Fig. 8B). However, the difference was minor (<20%), and overall, there was no significant difference (P > 0.05) in cleavage efficiency among the four proteins when tested for trypsin cleavage. All four HA proteins had minimal cleavage (<10%) without trypsin.
FIG 8.
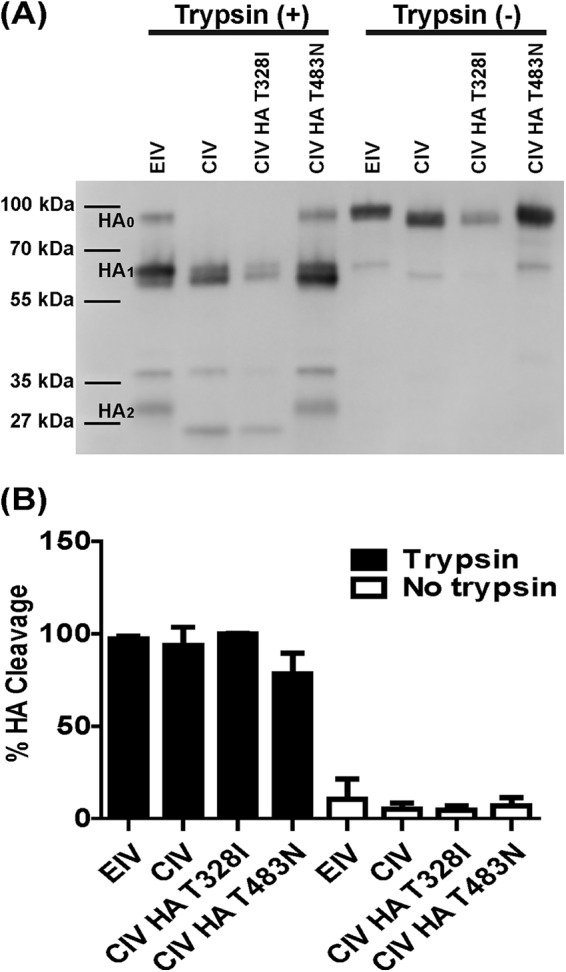
EIV and CIV HA cleavage assays. Both wild-type HA proteins, CIV HA T328I, and CIV HA T483N were surface expressed and biotinylated. Western blotting was used to look at HA0 cleavage into HA1 and HA2 (A). Cleavage efficiency was determined by band density of the Western blots and the following formula: (HA1/HA0 + HA1) × 100 (B). Error bars represent standard deviations from three independent experiments.
EIV and CIV infections of dog and horse tracheal explants.
Inoculation of dog tracheal explants with EIV, CIV, and the reassortants showed that all of the viruses infected explants to similar levels. Histology showed that canine tracheal explants that were infected with wild-type and reassortant viruses progressively had their epithelium layer thinned out (Fig. 9A), and this was most noticeable on day 5 compared to the control. Additionally, the number of ciliated cells gradually decreased in infected explants and ciliated cells completely disappeared on day 5 for all viruses, and these changes in the tracheal architecture were due to the presence of virus as confirmed by antigen detection (Fig. 9B). Virus was detected in the epithelium and did not infect the basal cells to a large degree. Levels of growth for all viruses were similar, reaching maximum titers between 48 and 72 h postinfection (Fig. 9C). Interestingly, at 24 h postinfection, the wild-type viruses' titers were around 2 logs lower (P < 0.05) than the reassortant viruses' titers.
FIG 9.
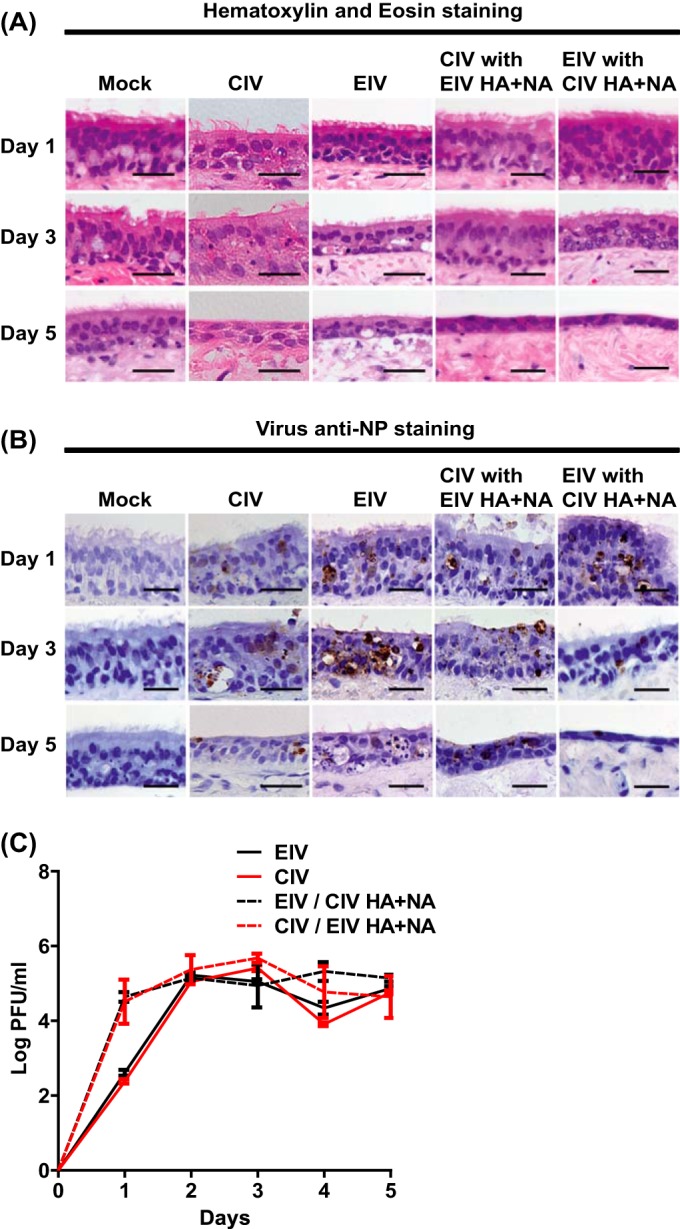
EIV, CIV, and reassortant virus infections of dog tracheal explants. Viruses (400 TCID50 units) were used to inoculate dog tracheal explants, and tissues were collected for hematoxylin and eosin staining (A) and for anti-NP staining (B) at days 1, 3, and 5 postinfection. Virus growth was assayed every 24 h for 5 days by plaque assays in MDCK cells (C). Error bars represent standard deviations from three independent experiments. Black scale bars represent 50 μm.
Inoculation of horse tracheal explants with the same viruses showed that EIV reached titers several logs higher than those of CIV on each day postinfection. CIV with EIV HA and NA grew better than CIV without EIV HA and NA, although its growth was still poor compared to that of the two EIVs. Conversely, EIV with CIV HA and NA reached lower titers (1- to 2-log difference) than EIV without CIV HA and NA, but it still grew to higher titers than the two CIVs (Fig. 10C). Histology showed that the epithelium layer of the equine tracheal cultures did not have an obvious decrease in thickness compared to the control, and the number of ciliated cells also was not greatly reduced on day 5 (Fig. 10A). Indeed, the ciliated cells decreased in number only when virus was detected in the epithelium (Fig. 10B), and overall virus detection was not as consistent compared to that with the infected dog tracheal explants. CIV was not detected in the horse trachea on days 1, 3, and 5 postinfection, and CIV with EIV HA and NA was not detected on day 1 postinfection (Fig. 10B and C). Interestingly, EIV with CIV HA and NA was detected only after screening several sections by NP staining on day 5, but growth was observed on each day postinfection.
FIG 10.
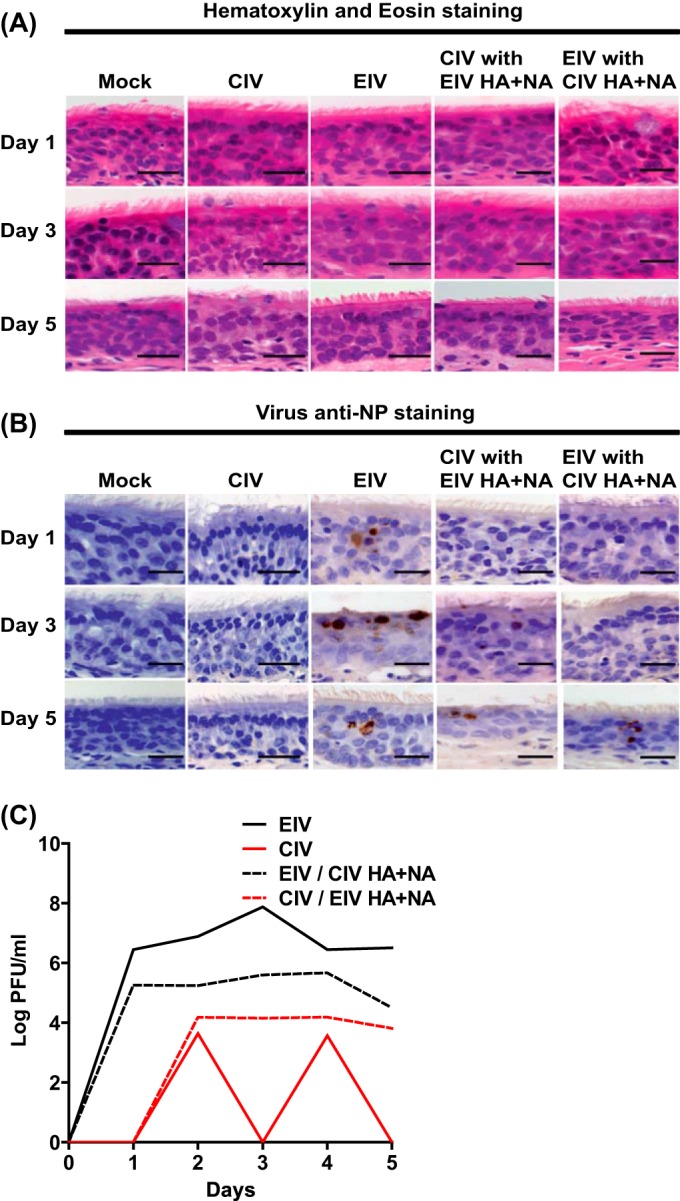
EIV, CIV, and reassortant virus infections of horse tracheal explants. Viruses (400 TCID50 units) were used to inoculate dog tracheal explants, and tissues were collected for hematoxylin and eosin staining (A) and for anti-NP staining (B) at days 1, 3, and 5 postinfection. Virus growth was assayed every 24 h for 5 days by plaque assays in MDCK cells (C). Black scale bars represent 50 μm.
DISCUSSION
Genetic differences in the EIV and CIV HA showed subtle differences in receptor specificity and cleavage efficiency.
There are five “signature” mutations that distinguish CIV HA from EIV HA: N54K, N83S, W222L, I328T, and N483T (Fig. 2) (25, 62). The W222L mutation is located in the sialic acid receptor binding pocket, and studies have shown that HA mutations in the 220 loop can alter binding to α2-3- and α2-6-linked sialic acids (35, 49, 63). There is also evidence that the W222L mutation in CIV H3N2 allowed the virus to infect dogs more efficiently (64), and so the species jump and subsequent adaptation of EIV H3N8 in dogs may have been mediated by changes in receptor recognition as well. However, no infectivity differences were seen between the viruses (or the reassortants) when those were inoculated into CHO or into 6H4 cells. However, all viruses exhibited a 50% reduction in their ability to infect 6H4 cells relative to CHO cells (Fig. 7B). This suggests that the viruses did not differ in their recognition of α2-3- and α2-6-linked sialic acids and in fact may prefer α2-3-linked sialic acids for infections. Furthermore, there were no significant differences in infectivity among viruses based on the infection of various host cells, which further suggests minimal differences in receptor specificity (Fig. 4 and 5A). Furthermore, a recent study showed EIV and CIV bound strongly to α2-3- and not α2-6-linked sialic acids using sialic acid glycopolymers (62), consistent with our conclusion.
Binding of EIV and CIV HA-Fc proteins to glycan microarrays showed support for the CHO and 6H4 cell infection results. Indeed, the two HA-Fc proteins bound strongly to α2-3-linked sialic acids in similar patterns, and neither protein bound to any α2-6-linked sialic acids (Fig. 7C). This preference toward binding to α2-3-linked (classical avian receptors) over α2-6-linked (classical human receptors) sialic acids suggests a potential human host restriction barrier for both viruses. Interestingly, there were differences in the relative preferences for binding to the various α2-3-linked glycans. For example, EIV HA-Fc showed higher binding to Neu5Acα3Galβ3GlcNAcβ3Galβ4GlcβR1 and Neu5Acα3Galβ4(Fucα3)GlcNAcβR1, while CIV HA-Fc showed higher binding to Neu5Acα3Galβ4GlcNAc6SβR1 and Neu5Acα3GalβR1. These findings suggest that there are subtle differences between the abilities of EIV and CIV HAs to recognize specific sialic acids. Indeed, a recent study revealed the atomic structures of EIV and CIV HA to be nearly identical, and both proteins preferred binding to α2-3- over α2-6-linked sialic acids, but there were subtle differences: CIV HA bound better to sulfated sialic acids than did EIV HA (65). Small differences such as these may be important for understanding virus host adaptation and tropism.
Previous research has suggested that the signature HA I328T mutation, which is at the P2 position of the cleavage site, may influence influenza virus HA cleavage (25, 62). However, we mutated the CIV HA to the EIV background, and it did not influence protein cleavage efficiency with and without trypsin compared to that of the wild type under the conditions of our assay (Fig. 8B). We also tested whether the signature HA N483T mutation, a glycosylation site that is close to the cleavage site in the HA structure, might influence cleavage by sterically hindering protease activity due to the presence of glycans. Changing the site in CIV to the EIV codon did not dramatically influence HA cleavage efficiency with and without trypsin (Fig. 8B). Overall, both EIV and CIV HA required trypsin activation, and there were no major differences in efficiency of cleavage between the wild type and the mutant proteins. Different hosts have been shown to express different proteases that can cleave HA (66), and it is therefore possible that there are dog-specific proteases that have selected the I328T and N483T mutations in CIV HA which allow for better replication. Testing dog-specific proteases might provide further insight into any differences in cleavage efficiency.
Other genetic differences between EIV and CIV may play roles in host adaptation and tropism.
Although many mutations documented here reside in known functional domains (Fig. 2), it is still unclear whether the specific mutations actually affect influenza virus host adaptation and replication or contribute to any host-specific functions. Interestingly, most (8 of 9) of the mutations in PB1 occurred between early and more recent CIV sequences (Fig. 2), whereas in the other proteins most mutations occurred between EIV and the earliest (2003 or 2004) CIV isolates (PB2, PA, and HA), or there was no difference between EIV and the two CIV groups (NP, NA, M1, and NS1) (Fig. 2). Whether mutations that occurred between early and more recent CIV isolates indicate adaptive changes to its canine host remains to be elucidated. Of the PB2 mutations, one (I292T) has been suggested to facilitate human H1N1 and H3N2 adaptation (60). Consequently, its possible role in dog adaptation needs to be considered. One mutation in PA (T400A) has also been reported to distinguish between human and avian influenza viruses. Human viruses almost always have a leucine at this site, while avian viruses show either serine or proline (61), such that it may represent another host range determinant in CIV. In addition, we found that EIV NS1 can be naturally truncated by 11 amino acids at the C terminus, while CIV NS1 is never truncated, except for the first CIV isolate. A previous study showed the 2009 human pandemic H1N1 virus also had an 11-amino-acid truncation which resulted in inefficient suppression of host genes, and extension of NS1 to full length restored its binding to host poly(A)-binding protein II, which increased NS1 host gene suppression activity (67). As mentioned above, the mutation at the N terminus of PA could affect PA-X host gene suppression. Furthermore, both CIVs (H3N8 and H3N2) have a 20-amino-acid truncation at the C terminus of PA-X, implying that the truncation may be a host determinant in dogs (57). In fact, a comparison of human full-length and truncated PA-X revealed a difference in gene suppression activity (58). Taken together, these genetic differences between EIV and CIV may be a consequence of dog-specific host pressures, including those generated by the innate and adaptive immune responses.
EIV and CIV did not show infectivity differences in various host cells.
Despite the relatively high number of mutations observed in the virus genomes, wild-type and reassortant viruses grew similarly in dog (MDCK) cells, which are generally considered the most susceptible cells for influenza viruses (Fig. 3). Other mammalian cells tested showed differences in susceptibility to infection (Fig. 4). The reassortant viruses appeared similar to the wild types, which was not surprising since the wild-type viruses did not show any infectivity differences. Interestingly, the viruses infected human (A549) and horse (EQKD) cells poorly (Fig. 4 and 5A). Previous results have indicated there is a host barrier for respiratory infection of CIV in horses (36, 62, 68, 69), so a low infectivity for CIV in EQKD cells may reflect a host difference that is also seen in kidney cells. However, EIV also infected EQKD cells poorly, so the block in infection may be due to biological differences between horse kidney and airway respiratory cells. The poor infection of CIV in A549 cells was interesting because there have been no reported cases of CIV (or EIV) naturally transmitting to humans, including people who were regularly exposed to CIV-infected dogs (70); whether there is a correlation between the poor infectivity in A549 cells and virus transmission to humans is unknown. Interestingly, the highly laboratory-adapted strain PR8 was able to infect both A549 and EQKD cells to high levels compared to those of the horse and dog viruses (Fig. 5B). This suggests that both cell types were permissive to influenza virus infection and that the limited infection from EIV and CIV was attributable to those specific viruses' biology. The poor infectivity in these cells could not be explained by sialic acid linkages, because both A549 and EQKD cells expressed high levels of both α2-3- and α2-6-linked sialic acids (Fig. 6), similar to cells that were permissive to infection. Indeed, the infectivity differences between host cells were likely not due to differences in sialic acid binding, because EIV and CIV HAs bound to sialic acid similarly, as described above. The viruses were able to infect cat (NLFK) and ferret (Mpf) cells, although the infection in ferret cells was poorer (Fig. 4). Both cells stained positive predominately for α2-3-linked sialic acids (Fig. 6), so this suggests that the infectivity difference between NLFK and Mpf cells was likely not due to receptor differences. These results were interesting because ferrets are susceptible to influenza and are used as a model for human virus infection and transmission (71), and there is already evidence that CIV H3N2 can infect cats (72, 73). Additionally, a previous study showed that EIV replicated in the upper respiratory tract in live ferrets but was restricted in the lungs (74). There are no extended analysis or reports of CIV H3N8 infections in ferrets, but seroconversion has been observed after inoculating ferrets (25). Interestingly, there is evidence of limited CIV H3N2 infection and transmission between ferrets in laboratory settings, but the virus could not transmit from dogs to ferrets (75, 76). Taken together, these findings show that there is potential for EIV and CIV to infect other mammalian hosts and subsequently adapt.
EIV and CIV infection of tracheal explants revealed a host-specific barrier.
To simulate the natural environment in which EIV and CIV cause infections, dog and horse tracheal explant cultures were prepared and used for virus infections. Overall, the results showed that the viruses could infect dog trachea, and there were no major differences among the viruses with respect to damaging the epithelium layer or the location of virus antigens in the tracheal explants (Fig. 9). The reassortant viruses reached a greater titer (around 2 logs) than did the wild types at 24 h postinfection, and this suggests that the protein mismatching altered viral biology (Fig. 9C). However, the change was not dramatic overall, because growth levels were very similar after 24 h postinfection, reaching peak titer between 48 and 72 h postinfection, similar to growth in MDCK cells (Fig. 3). In horse trachea all viruses caused some level of infection, but the physical damage of the epithelium was less pronounced than with the infections of the dog trachea, which may be due to the horse trachea being more structurally robust (Fig. 10). Interestingly, EIV grew much better than CIV, which provides further evidence of a host-specific barrier (Fig. 10C). Replacing the CIV HA and NA with EIV HA and NA allowed the virus to grow slightly better and stabilized (detected virus on days 3 and 5 postinfection) the virus compared to the wild-type CIV. Furthermore, replacing the EIV HA and NA with CIV HA and NA attenuated virus growth by around 1 to 2 logs across 5 days compared to that of the wild-type EIV. These findings suggest that the difference in growth between EIV and CIV in horse trachea may be attributed to the mutations in the glycoproteins. Overall, the reassortant viruses grew similarly with respect to their wild-type counterparts, and the experiment was carried out in one experimental replicate; thus, further experimentation using horse trachea and reassortant viruses is needed for validation. Also, the apparent attenuation of growth of CIV and EIV with CIV glycoproteins in horse trachea compared to dog trachea may not have been caused by differences in α2-3- and α2-6-linked sialic acids, as past studies have shown the existence of both in dog and horse tracheal tissues (77, 78). However, the distributions were shown to be different. While horse trachea epithelial cells stained positive for both sialic acids, dog trachea epithelial cells showed stronger staining for α2-3-linked sialic acids. Interestingly, the lamina propria of dog trachea stained positive for both. However, it is important to note that sialic acid distribution, variety, and presence can vary depending on individual animals and their age (79).
Overall, these results showed that despite 6 years of continuous evolution in dogs that separated the two viruses tested in this study, and the accumulation of mutations in all of the genomic segments such that EIV and CIV are clearly phylogenetically distinct (Fig. 1), there appeared to be minimal biological differences between them. We showed that the viruses infected various host cells with no infectivity differences among the viruses, although different host cells exhibited various degrees of permissiveness. We also showed that the viruses preferred α2-3- over α2-6-linked sialic acid receptors, and there may be subtle differences in receptor recognition. Virus inoculation in tracheal explants revealed limited CIV infectivity in horse trachea, and the restriction factors may reside in the receptor binding proteins. Notably, although EIV has circulated in many parts of the world since it emerged in 1963, CIV has not spread in a sustained fashion beyond North America, where it has been maintained mainly in large, high-turnover animal shelters. Given the scarce phenotypic differences between EIV and CIV, the relatively limited spread of CIV among the domestic dog population may reflect a lack of epidemiological contacts rather than constraints on viral fitness. This suggests that interspecies transmission and adaptation of influenza virus in this case are mediated by subtle factors.
ACKNOWLEDGMENTS
This work was supported by NIH/NIGMS grant R01 GM080533 to C.R.P. and E.C.H. and by grant R01GM32373 to A.V. E.C.H. is supported by an NHMRC Australia Fellowship (AF30). K.H.F. is supported by an NSF Graduate Research Fellowship. P.R.M. and G.G. are supported by the Medical Research Council of the United Kingdom (grant number G0801822).
REFERENCES
- 1.Parrish CR, Kawaoka Y. 2005. The origins of new pandemic viruses: the acquisition of new host ranges by canine parvovirus and influenza A viruses. Annu Rev Microbiol 59:553–586. doi: 10.1146/annurev.micro.59.030804.121059. [DOI] [PubMed] [Google Scholar]
- 2.Morens DM, Taubenberger JK. 2010. Historical thoughts on influenza viral ecosystems, or behold a pale horse, dead dogs, failing fowl, and sick swine. Influenza Other Respir Viruses 4:327–337. doi: 10.1111/j.1750-2659.2010.00148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crawford PC, Dubovi EJ, Castleman WL, Stephenson I, Gibbs EP, Chen L, Smith C, Hill RC, Ferro P, Pompey J, Bright RA, Medina MJ, Johnson CM, Olsen CW, Cox NJ, Klimov AI, Katz JM, Donis RO. 2005. Transmission of equine influenza virus to dogs. Science 310:482–485. doi: 10.1126/science.1117950. [DOI] [PubMed] [Google Scholar]
- 4.Anthony SJ, St Leger JA, Pugliares K, Ip HS, Chan JM, Carpenter ZW, Navarrete-Macias I, Sanchez-Leon M, Saliki JT, Pedersen J, Karesh W, Daszak P, Rabadan R, Rowles T, Lipkin WI. 2012. Emergence of fatal avian influenza in New England harbor seals. mBio 3(4):e00166–12. doi: 10.1128/mBio.00166-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoon KJ, Schwartz K, Sun D, Zhang J, Hildebrandt H. 2012. Naturally occurring influenza A virus subtype H1N2 infection in a Midwest United States mink (Mustela vison) ranch. J Vet Diagn Invest 24:388–391. doi: 10.1177/1040638711428349. [DOI] [PubMed] [Google Scholar]
- 6.Ince WL, Gueye-Mbaye A, Bennink JR, Yewdell JW. 2013. Reassortment complements spontaneous mutation in influenza A virus NP and M1 genes to accelerate adaptation to a new host. J Virol 87:4330–4338. doi: 10.1128/JVI.02749-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mänz B, Schwemmle M, Brunotte L. 2013. Adaptation of avian influenza A virus polymerase in mammals to overcome the host species barrier. J Virol 87:7200–7209. doi: 10.1128/JVI.00980-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Resa-Infante P, Gabriel G. 2013. The nuclear import machinery is a determinant of influenza virus host adaptation. Bioessays 35:23–27. doi: 10.1002/bies.201200138. [DOI] [PubMed] [Google Scholar]
- 9.Rose N, Herve S, Eveno E, Barbier N, Eono F, Dorenlor V, Andraud M, Camsusou C, Madec F, Simon G. 2013. Dynamics of influenza A virus infections in permanently infected pig farms: evidence of recurrent infections, circulation of several swine influenza viruses and reassortment events. Vet Res 44:72. doi: 10.1186/1297-9716-44-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trebbien R, Bragstad K, Larsen LE, Nielsen J, Botner A, Heegaard PM, Fomsgaard A, Viuff B, Hjulsager CK. 2013. Genetic and biological characterisation of an avian-like H1N2 swine influenza virus generated by reassortment of circulating avian-like H1N1 and H3N2 subtypes in Denmark. Virol J 10:290. doi: 10.1186/1743-422X-10-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imai M, Kawaoka Y. 2012. The role of receptor binding specificity in interspecies transmission of influenza viruses. Curr Opin Virol 2:160–167. doi: 10.1016/j.coviro.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sassaki GL, Elli S, Rudd TR, Macchi E, Yates EA, Naggi A, Shriver Z, Raman R, Sasisekharan R, Torri G, Guerrini M. 2013. Human (alpha2→6) and avian (alpha2→3) sialylated receptors of influenza A virus show distinct conformations and dynamics in solution. Biochemistry 52:7217–7230. doi: 10.1021/bi400677n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shichinohe S, Okamatsu M, Sakoda Y, Kida H. 2013. Selection of H3 avian influenza viruses with SAalpha2,6Gal receptor specificity in pigs. Virology 444:404–408. doi: 10.1016/j.virol.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 14.Chen Q, Huang S, Chen J, Zhang S, Chen Z. 2013. NA proteins of influenza A viruses H1N1/2009, H5N1, and H9N2 show differential effects on infection initiation, virus release, and cell-cell fusion. PLoS One 8:e54334. doi: 10.1371/journal.pone.0054334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le QM, Sakai-Tagawa Y, Ozawa M, Ito M, Kawaoka Y. 2009. Selection of H5N1 influenza virus PB2 during replication in humans. J Virol 83:5278–5281. doi: 10.1128/JVI.00063-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan S, Wu G. 2010. Evidence for cross-species infections and cross-subtype mutations in influenza A matrix proteins. Viral Immunol 23:105–111. doi: 10.1089/vim.2009.0080. [DOI] [PubMed] [Google Scholar]
- 17.Dankar SK, Wang S, Ping J, Forbes NE, Keleta L, Li Y, Brown EG. 2011. Influenza A virus NS1 gene mutations F103L and M106I increase replication and virulence. Virol J 8:13. doi: 10.1186/1743-422X-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakabe S, Ozawa M, Takano R, Iwastuki-Horimoto K, Kawaoka Y. 2011. Mutations in PA, NP, and HA of a pandemic (H1N1) 2009 influenza virus contribute to its adaptation to mice. Virus Res 158:124–129. doi: 10.1016/j.virusres.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiseleva IV, Voeten JT, Teley LC, Larionova NV, Drieszen-van der Cruijsen SK, Basten SM, Heldens JG, van den Bosch H, Rudenko LG. 2010. PB2 and PA genes control the expression of the temperature-sensitive phenotype of cold-adapted B/USSR/60/69 influenza master donor virus. J Gen Virol 91:931–937. doi: 10.1099/vir.0.017996-0. [DOI] [PubMed] [Google Scholar]
- 20.Chou YY, Albrecht RA, Pica N, Lowen AC, Richt JA, Garcia-Sastre A, Palese P, Hai R. 2011. The M segment of the 2009 new pandemic H1N1 influenza virus is critical for its high transmission efficiency in the guinea pig model. J Virol 85:11235–11241. doi: 10.1128/JVI.05794-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turan K, Mibayashi M, Sugiyama K, Saito S, Numajiri A, Nagata K. 2004. Nuclear MxA proteins form a complex with influenza virus NP and inhibit the transcription of the engineered influenza virus genome. Nucleic Acids Res 32:643–652. doi: 10.1093/nar/gkh192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim SH, Samal SK. 2010. Inhibition of host innate immune responses and pathogenicity of recombinant Newcastle disease viruses expressing NS1 genes of influenza A viruses. J Gen Virol 91:1996–2001. doi: 10.1099/vir.0.021766-0. [DOI] [PubMed] [Google Scholar]
- 23.Gao S, Song L, Li J, Zhang Z, Peng H, Jiang W, Wang Q, Kang T, Chen S, Huang W. 2012. Influenza A virus-encoded NS1 virulence factor protein inhibits innate immune response by targeting IKK. Cell Microbiol 14:1849–1866. doi: 10.1111/cmi.12005. [DOI] [PubMed] [Google Scholar]
- 24.Gibbs EP, Anderson TC. 2010. Equine and canine influenza: a review of current events. Anim Health Res Rev 11:43–51. doi: 10.1017/S1466252310000046. [DOI] [PubMed] [Google Scholar]
- 25.Payungporn S, Crawford PC, Kouo TS, Chen LM, Pompey J, Castleman WL, Dubovi EJ, Katz JM, Donis RO. 2008. Influenza A virus (H3N8) in dogs with respiratory disease, Florida. Emerg Infect Dis 14:902–908. doi: 10.3201/eid1406.071270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rivailler P, Perry IA, Jang Y, Davis CT, Chen LM, Dubovi EJ, Donis RO. 2010. Evolution of canine and equine influenza (H3N8) viruses co-circulating between 2005 and 2008. Virology 408:71–79. doi: 10.1016/j.virol.2010.08.022. [DOI] [PubMed] [Google Scholar]
- 27.Jirjis FF, Deshpande MS, Tubbs AL, Jayappa H, Lakshmanan N, Wasmoen TL. 2010. Transmission of canine influenza virus (H3N8) among susceptible dogs. Vet Microbiol 144:303–309. doi: 10.1016/j.vetmic.2010.02.029. [DOI] [PubMed] [Google Scholar]
- 28.Hayward JJ, Dubovi EJ, Scarlett JM, Janeczko S, Holmes EC, Parrish CR. 2010. Microevolution of canine influenza virus in shelters and its molecular epidemiology in the United States. J Virol 84:12636–12645. doi: 10.1128/JVI.01350-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holt DE, Mover MR, Brown DC. 2010. Serologic prevalence of antibodies against canine influenza virus (H3N8) in dogs in a metropolitan animal shelter. J Am Vet Med Assoc 237:71–73. doi: 10.2460/javma.237.1.71. [DOI] [PubMed] [Google Scholar]
- 30.Li S, Shi Z, Jiao P, Zhang G, Zhong Z, Tian W, Long LP, Cai Z, Zhu X, Liao M, Wan XF. 2010. Avian-origin H3N2 canine influenza A viruses in Southern China. Infect Genet Evol 10:1286–1288. doi: 10.1016/j.meegid.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng XJ, Lin Y, Zhao YB, Lu CP, Liu YJ. 2013. Experimental infection of dogs with H3N2 canine influenza virus from China. Epidemiol Infect 141:2595–2603. doi: 10.1017/S0950268813000472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim SF, Lee MM, Zhang P, Song Z. 2008. The Golgi CMP-sialic acid transporter: a new CHO mutant provides functional insights. Glycobiology 18:851–860. doi: 10.1093/glycob/cwn080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bragonzi A, Distefano G, Buckberry LD, Acerbis G, Foglieni C, Lamotte D, Campi G, Marc A, Soria MR, Jenkins N, Monaco L. 2000. A new Chinese hamster ovary cell line expressing alpha2,6-sialyltransferase used as universal host for the production of human-like sialylated recombinant glycoproteins. Biochim Biophys Acta 1474:273–282. doi: 10.1016/S0304-4165(00)00023-4. [DOI] [PubMed] [Google Scholar]
- 34.Zhou B, Donnelly ME, Scholes DT, St George K, Hatta M, Kawaoka Y, Wentworth DE. 2009. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza A viruses. J Virol 83:10309–10313. doi: 10.1128/JVI.01109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pawar SD, Parkhi SS, Koratkar SS, Mishra AC. 2012. Receptor specificity and erythrocyte binding preferences of avian influenza viruses isolated from India. Virol J 9:251. doi: 10.1186/1743-422X-9-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quintana AM, Hussey SB, Burr EC, Pecoraro HL, Annis KM, Rao S, Landolt GA. 2011. Evaluation of infectivity of a canine lineage H3N8 influenza A virus in ponies and in primary equine respiratory epithelial cells. Am J Vet Res 72:1071–1078. doi: 10.2460/ajvr.72.8.1071. [DOI] [PubMed] [Google Scholar]
- 37.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 39.Ayora-Talavera G, Shelton H, Scull MA, Ren J, Jones IM, Pickles RJ, Barclay WS. 2009. Mutations in H5N1 influenza virus hemagglutinin that confer binding to human tracheal airway epithelium. PLoS One 4:e7836. doi: 10.1371/journal.pone.0007836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shelton H, Ayora-Talavera G, Ren J, Loureiro S, Pickles RJ, Barclay WS, Jones IM. 2011. Receptor binding profiles of avian influenza virus hemagglutinin subtypes on human cells as a predictor of pandemic potential. J Virol 85:1875–1880. doi: 10.1128/JVI.01822-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deng L, Song J, Gao X, Wang J, Yu H, Chen X, Varki N, Naito-Matsui Y, Galan JE, Varki A. 2014. Host adaptation of a bacterial toxin from the human pathogen Salmonella Typhi. Cell 159:1290–1299. doi: 10.1016/j.cell.2014.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng L, Bensing BA, Thamadilok S, Yu H, Lau K, Chen X, Ruhl S, Sullam PM, Varki A. 2014. Oral streptococci utilize a Siglec-like domain of serine-rich repeat adhesins to preferentially target platelet sialoglycans in human blood. PLoS Pathog 10:e1004540. doi: 10.1371/journal.ppat.1004540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tse LV, Marcano VC, Huang W, Pocwierz MS, Whittaker GR. 2013. Plasmin-mediated activation of pandemic H1N1 influenza virus hemagglutinin is independent of the viral neuraminidase. J Virol 87:5161–5169. doi: 10.1128/JVI.00210-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gonzalez G, Marshall JF, Morrell J, Robb D, McCauley JW, Perez DR, Parrish CR, Murcia PR. 2014. Infection and pathogenesis of canine, equine, and human influenza viruses in canine tracheas. J Virol 88:9208–9219. doi: 10.1128/JVI.00887-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nunes SF, Murcia PR, Tiley LS, Brown IH, Tucker AW, Maskell DJ, Wood JL. 2010. An ex vivo swine tracheal organ culture for the study of influenza infection. Influenza Other Respir Viruses 4:7–15. doi: 10.1111/j.1750-2659.2009.00119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderton TL, Maskell DJ, Preston A. 2004. Ciliostasis is a key early event during colonization of canine tracheal tissue by Bordetella bronchiseptica. Microbiology 150:2843–2855. doi: 10.1099/mic.0.27283-0. [DOI] [PubMed] [Google Scholar]
- 47.Wiley DC, Wilson IA, Skehel JJ. 1981. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 48.Stray SJ, Pittman LB. 2012. Subtype- and antigenic site-specific differences in biophysical influences on evolution of influenza virus hemagglutinin. Virol J 9:91. doi: 10.1186/1743-422X-9-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin YP, Xiong X, Wharton SA, Martin SR, Coombs PJ, Vachieri SG, Christodoulou E, Walker PA, Liu J, Skehel JJ, Gamblin SJ, Hay AJ, Daniels RS, McCauley JW. 2012. Evolution of the receptor binding properties of the influenza A(H3N2) hemagglutinin. Proc Natl Acad Sci U S A 109:21474–21479. doi: 10.1073/pnas.1218841110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun X, Tse LV, Ferguson AD, Whittaker GR. 2010. Modifications to the hemagglutinin cleavage site control the virulence of a neurotropic H1N1 influenza virus. J Virol 84:8683–8690. doi: 10.1128/JVI.00797-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu Y, Qin G, Gao F, Liu Y, Vavricka CJ, Qi J, Jiang H, Yu K, Gao GF. 2013. Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding. Sci Rep 3:1551. doi: 10.1038/srep01551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Portela A, Digard P. 2002. The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J Gen Virol 83:723–734. [DOI] [PubMed] [Google Scholar]
- 53.Noton SL, Medcalf E, Fisher D, Mullin AE, Elton D, Digard P. 2007. Identification of the domains of the influenza A virus M1 matrix protein required for NP binding, oligomerization and incorporation into virions. J Gen Virol 88:2280–2290. doi: 10.1099/vir.0.82809-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hale BG, Randall RE, Ortin J, Jackson D. 2008. The multifunctional NS1 protein of influenza A viruses. J Gen Virol 89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- 55.Boivin S, Cusack S, Ruigrok RW, Hart DJ. 2010. Influenza A virus polymerase: structural insights into replication and host adaptation mechanisms. J Biol Chem 285:28411–28417. doi: 10.1074/jbc.R110.117531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yewdell JW, Ince WL. 2012. Virology frameshifting to PA-X influenza. Science 337:164–165. doi: 10.1126/science.1225539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi M, Jagger BW, Wise HM, Digard P, Holmes EC, Taubenberger JK. 2012. Evolutionary conservation of the PA-X open reading frame in segment 3 of influenza A virus. J Virol 86:12411–12413. doi: 10.1128/JVI.01677-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Desmet EA, Bussey KA, Stone R, Takimoto T. 2013. Identification of the N-terminal domain of the influenza virus PA responsible for the suppression of host protein synthesis. J Virol 87:3108–3118. doi: 10.1128/JVI.02826-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khaperskyy DA, Emara MM, Johnston BP, Anderson P, Hatchette TF, McCormick C. 2014. Influenza A virus host shutoff disables antiviral stress-induced translation arrest. PLoS Pathog 10:e1004217. doi: 10.1371/journal.ppat.1004217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miotto O, Heiny A, Tan TW, August JT, Brusic V. 2008. Identification of human-to-human transmissibility factors in PB2 proteins of influenza A by large-scale mutual information analysis. BMC Bioinformatics 9(Suppl 1):S18. doi: 10.1186/1471-2105-9-S1-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Allen JE, Gardner SN, Vitalis EA, Slezak TR. 2009. Conserved amino acid markers from past influenza pandemic strains. BMC Microbiol 9:77. doi: 10.1186/1471-2180-9-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pecoraro HL, Bennett S, Garretson K, Quintana AM, Lunn KF, Landolt GA. 2013. Comparison of the infectivity and transmission of contemporary canine and equine H3N8 influenza viruses in dogs. Vet Med Int 2013:874521. doi: 10.1155/2013/874521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu C, Cheng X, Wang X, Lv X, Yang F, Liu T, Fang S, Zhang R, Jinquan C. 2013. Clinical and molecular characteristics of the 2009 pandemic influenza H1N1 infection with severe or fatal disease from 2009 to 2011 in Shenzhen, China. J Med Virol 85:405–412. doi: 10.1002/jmv.23295. [DOI] [PubMed] [Google Scholar]
- 64.Yang G, Li S, Blackmon S, Ye J, Bradley KC, Cooley J, Smith D, Hanson L, Cardona C, Steinhauer DA, Webby R, Liao M, Wan XF. 2013. Mutation tryptophan to leucine at position 222 of haemagglutinin could facilitate H3N2 influenza A virus infection in dogs. J Gen Virol 94:2599–2608. doi: 10.1099/vir.0.054692-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Collins PJ, Vachieri SG, Haire LF, Ogrodowicz RW, Martin SR, Walker PA, Xiong X, Gamblin SJ, Skehel JJ. 2014. Recent evolution of equine influenza and the origin of canine influenza. Proc Natl Acad Sci U S A 111:11175–11180. doi: 10.1073/pnas.1406606111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peitsch C, Klenk HD, Garten W, Bottcher-Friebertshauser E. 2014. Activation of influenza A viruses by host proteases from swine airway epithelium. J Virol 88:282–291. doi: 10.1128/JVI.01635-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tu J, Guo J, Zhang A, Zhang W, Zhao Z, Zhou H, Liu C, Chen H, Jin M. 2011. Effects of the C-terminal truncation in NS1 protein of the 2009 pandemic H1N1 influenza virus on host gene expression. PLoS One 6:e26175. doi: 10.1371/journal.pone.0026175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamanaka T, Tsujimura K, Kondo T, Matsumura T, Ishida H, Kiso M, Hidari KI, Suzuki T. 2010. Infectivity and pathogenicity of canine H3N8 influenza A virus in horses. Influenza Other Respir Viruses 4:345–351. doi: 10.1111/j.1750-2659.2010.00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamanaka T, Nemoto M, Bannai H, Tsujimura K, Kondo T, Matsumura T, Muranaka M, Ueno T, Kinoshita Y, Niwa H, Hidari KI, Suzuki T. 2012. No evidence of horizontal infection in horses kept in close contact with dogs experimentally infected with canine influenza A virus (H3N8). Acta Vet Scand 54:25. doi: 10.1186/1751-0147-54-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krueger WS, Heil GL, Yoon KJ, Gray GC. 2014. No evidence for zoonotic transmission of H3N8 canine influenza virus among US adults occupationally exposed to dogs. Influenza Other Respir Viruses 8:99–106. doi: 10.1111/irv.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thangavel RR, Bouvier NM. 2014. Animal models for influenza virus pathogenesis, transmission, and immunology. J Immunol Methods 410:60–79. doi: 10.1016/j.jim.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Park SJ, Kang BK, Jeoung HY, Moon HJ, Hong M, Na W, Park BK, Poo H, Kim JK, An DJ, Song DS. 2013. Complete genome sequence of a canine-origin H3N2 feline influenza virus isolated from domestic cats in South Korea. Genome Announc 1(2):e00253–12. doi: 10.1128/genomeA.00253-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jeoung HY, Lim SI, Shin BH, Lim JA, Song JY, Song DS, Kang BK, Moon HJ, An DJ. 2013. A novel canine influenza H3N2 virus isolated from cats in an animal shelter. Vet Microbiol 165:281–286. doi: 10.1016/j.vetmic.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 74.Baz M, Paskel M, Matsuoka Y, Zengel J, Cheng X, Jin H, Subbarao K. 2013. Replication and immunogenicity of swine, equine, and avian H3 subtype influenza viruses in mice and ferrets. J Virol 87:6901–6910. doi: 10.1128/JVI.03520-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim H, Song D, Moon H, Yeom M, Park S, Hong M, Na W, Webby RJ, Webster RG, Park B, Kim JK, Kang B. 2013. Inter- and intraspecies transmission of canine influenza virus (H3N2) in dogs, cats, and ferrets. Influenza Other Respir Viruses 7:265–270. doi: 10.1111/j.1750-2659.2012.00379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee YN, Lee DH, Park JK, Yuk SS, Kwon JH, Nahm SS, Lee JB, Park SY, Choi IS, Song CS. 2013. Experimental infection and natural contact exposure of ferrets with canine influenza virus (H3N2). J Gen Virol 94(Part 2):293–297. doi: 10.1099/vir.0.042473-0. [DOI] [PubMed] [Google Scholar]
- 77.Ning ZY, Wu XT, Cheng YF, Qi WB, An YF, Wang H, Zhang GH, Li SJ. 2012. Tissue distribution of sialic acid-linked influenza virus receptors in beagle dogs. J Vet Sci 13:219–222. doi: 10.4142/jvs.2012.13.3.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Scocco P, Pedini V. 2008. Localization of influenza virus sialoreceptors in equine respiratory tract. Histol Histopathol 23:973–978. [DOI] [PubMed] [Google Scholar]
- 79.Varki A, Schauer R. 2009. Chapter 14. Sialic acids. In Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME (ed), Essentials of glycobiology, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]