ABSTRACT
Gene-engineered CD34+ hematopoietic stem and progenitor cells (HSPCs) can be used to generate an HIV-1-resistant immune system. However, a certain threshold of transduced HSPCs might be required for transplantation into mice for creating an HIV-resistant immune system. In this study, we combined CCR5 knockdown by a highly efficient microRNA (miRNA) lentivector with pretransplantation selection of transduced HSPCs to obtain a rather pure population of gene engineered CD34+ cells. Low-level transduction of HSPCs and subsequent sorting by flow cytometry yielded >70% transduced cells. Mice transplanted with these cells showed functional and persistent resistance to a CCR5-tropic HIV strain: viral load was significantly decreased over months, and human CD4+ T cells were preserved. In one mouse, viral mutations, resulting presumably in a CXCR4-tropic strain, overcame HIV resistance. Our results suggest that HSPC-based CCR5 knockdown may lead to efficient control of HIV in vivo. We overcame a major limitation of previous HIV gene therapy in humanized mice in which only a proportion of the cells in chimeric mice in vivo are anti-HIV engineered. Our strategy underlines the promising future of gene engineering HIV-resistant CD34+ cells that produce a constant supply of HIV-resistant progeny.
IMPORTANCE Major issues in experimental long-term in vivo HIV gene therapy have been (i) low efficacy of cell transduction at the time of transplantation and (ii) transduction resulting in multiple copies of heterologous DNA in target cells. In this study, we demonstrated the efficacy of a transplantation approach with a selection step for transduced cells that allows transplantation of an enriched population of HSPCs expressing a single (low) copy of a CCR5 miRNA. Efficient maintenance of CD4+ T cells and a low viral titer resulted only when at least 70% of the HIV target cells were genetically modified. These findings imply that clinical protocols of HIV gene therapy require a selective enrichment of genetically targeted cells because positive selection of modified cells is likely to be insufficient below this threshold. This selection approach may be beneficial not only for HIV patients but also for other patients requiring transplantation of genetically modified cells.
INTRODUCTION
Combined antiretroviral therapy (cART) changed the face of HIV medicine: patients have a life expectancy close to that of uninfected people (1). However, cART has major disadvantages, including adverse events, emergence of drug-resistant strains in patients with poor adherence, a need for lifelong intake, psychological dependence, and cost. Thus, cART has not halted the pandemic (http://www.who.int/hiv/en/), and alternative therapies are needed to cure HIV.
Gene therapy has been widely discussed as a possible strategy to cure HIV and has been tested in phase I and II clinical trials. Autologous CD4+ T cells (2, 3) or CD34+ cells (4, 5) were gene engineered to express various anti-HIV moieties, including a combination of three RNA-based anti-HIV moieties (tat/rev short hairpin RNA [shRNA], TAR decoy, and CCR5 ribozyme) (4), a tat-vpr-specific anti-HIV ribozyme (5), and a conditionally replicating lentiviral vector expressing a long antisense to HIV (3), or were gene edited by zinc finger nucleases for CCR5 knockout (2). Gene engineering also generated HIV-specific CD4+ or CD8+ T cells (6, 7). Overall, the effects on HIV infection were modest, but importantly, gene engineering proved to be safe in humans.
The concept of engineering an HIV-resistant immune system received new impetus from the “Berlin patient,” who was infected with HIV and was treated with hematopoietic stem cell transplantation for acute myeloid leukemia. He received bone marrow from a donor homozygous for the Δ32 CCR5 mutation, and thus, the progeny cells did not express CCR5. His case was the first in which a cure for HIV was documented (8) and provided hope that eliminating CCR5 from the cell surface would be the “Holy Grail” for the cure of HIV. However, another HIV-infected patient suffering from anaplastic large-cell lymphoma also received a stem cell transplant from a homozygous CCR5-null donor. Unfortunately, in that case, X4-tropic HIV strains emerged that necessitated the reinitiation of cART (9).
In view of the modest success of phase I and II clinical trials and the data from stem cell transplantation, preclinical studies are needed to define the best anti-HIV moieties and the minimal number of gene-engineered cells required to advance gene therapy in HIV. Humanized (hu) mice, which are generated by the transplantation of CD34+ cells, are of particular value in this context. These mice excel in their multilineage hematopoiesis (10), are highly permissive to HIV (11), and allow for the gene engineering of human CD34+ cells before transplantation (12). Indeed, various anti-HIV moieties have been investigated in hu mice as gene therapy options, including cellular factors, boosting the anti-HIV immune response, and the HIV genome itself (12). These mice were used in extenso to investigate the effects of targeting CCR5 by shRNA (13–15) or zinc finger nucleases (ZNF) (16). All these studies reported a decrease in CCR5 expression in circulating and tissue leukocytes, which were not permissive to HIV ex vivo, but only the study by Holt et al. reported a significant decrease of HIV RNA copy number in vivo (16). The other studies either did not analyze the effects on HIV infection in vivo (14) or demonstrated no effect on viral load (15). The results of Holt et al. revealed disruption of CCR5 in only ∼20% of all CD4+ T cells. The follow-up in that study was only 8 weeks. Gene engineering of hematopoietic stem and progenitor cells (HSPCs) with a lentiviral vector encoding the broadly neutralizing anti-HIV human antibody 2G12 showed suppression of HIV RNA but was studied only 7 days after a challenge with a virus containing the corresponding epitope (17); gene engineering of an HIV-specific T-cell receptor also lowered the HIV RNA but only modestly (18). A very elegant study with CD34+ cells edited with an HIV-1 long terminal repeat (LTR)-specific Tre recombinase showed a potent lowering of HIV RNA activity after HIV challenge in hu mice (19). All these data are promising; however, we lack a long-term follow-up of the effects of anti-HIV gene therapy in hu mice, the number of gene-engineered CD34+ cells needed in the various studies to obtain an HIV RNA lowering effect, and a detailed characterization of the hematopoietic system. A major advance was recently presented by Barclay et al., who purified gene-engineered CD34+ cells via the expression of a truncated version of CD25 (20).
Various means are available for gene engineering of CD34+ cells; each has its pros and cons. A great deal of experience exists with shRNA (21); potential cons may be its potential to trigger the innate immune system (22) and its less-than-absolute downregulation of the target gene. Targeted gene disruption by ZFN, Talen, or Crispr/cas has the advantage of complete disruption of the gene of interest (23–25); however, the modest rate of gene engineering of CD34+ cells (26), the potential of off-target effects (27–29), and the lack of clinical experience represent substantial hurdles for wider use in vivo.
We recently reported a novel microRNA (miRNA)-based gene knockdown (KD) strategy with improved knockdown relative to that obtained by methods conventionally used (30). A triple-hairpin cassette targeting CCR5 resulted in >90% CCR5 knockdown upon single-copy transduction in HeLa cells. The aim of the present study was to assess whether gene engineering of CD34+ cells with this vector construct results in downregulation of CCR5 in progeny cells in hu mice and whether it could protect against HIV challenge ex vivo as well as in vivo. Since there is some evidence in the literature that the number of gene-engineered CD34+ cells is a major determinant of the success of the anti-HIV moieties (15, 16), we also made a major effort to generate hu mice with a very high number of gene-engineered CD34+ cells. Finally, we characterized in extenso the hematopoietic system subsequent to HIV infection in hu mice with gene-engineered CD34+ cells.
MATERIALS AND METHODS
Ethics statement.
The procurement and use of CD34+ cells from human cord blood were approved by the Cantonal Ethical Committee of Zurich (EK-1103). All adult subjects provided written informed consent. Animal care and experimental protocols were in accordance with the “Swiss Ethical Principles and Guidelines for Experiments on Animals” (http://www.akademien-schweiz.ch/en/index/Portrait/Kommissionen-AG/Kommission-fuer-Tierversuchsethik.html) and approved by the Veterinary Office of the Canton of Zurich, permit 26/2011. Manipulations of mice were in accordance with the regulations of the Veterinary Office of the Canton of Zurich (http://www.veta.zh.ch/internet/gesundheitsdirektion/veta/de/home.html).
Lentiviral vector production and titration.
Lentiviral vector stocks were generated using transient transfection of HEK 293T/17 cells with the self-inactivating vector pCLX-R4-DEST-R2 encoding the microRNA to CCR5 (30), the psPAX2 plasmid encoding gag/pol, and the pCAG-VSVG envelope plasmid, as described previously (31). Lentivector titration was performed using transduction of HeLa cells, followed by quantification of green fluorescent protein (GFP)-positive cells 5 days after infection by flow cytometry as described previously (31).
Generation of humanized mice.
NOD.scid.IL2R−/− (NSG) mice were bred and maintained in individual ventilated cages and were fed autoclaved food and water. Mice with a human immune system (humanized [hu] mice) were generated as described previously (32). Briefly, newborn (<5 days old) NSG mice received sublethal (1 Gy) total-body irradiation with a Cs source and then received 2 × 105 transduced or untransduced CD34+ human HSPCs with a 50-μl Hamilton syringe via the intrahepatic (i.h.) route. For the fluorescence-activated cell sorter (FACS)-sorted R5 knockdown animals, CD34+ cells were sorted postransduction into GFP-positive and -negative fractions, and then 2 × 105 CD34+ GFP-positive or GFP-negative cells were injected i.h. into respective cohorts. All manipulations of hu mice were performed under laminar flow. Cell suspensions of the hu mouse organs were prepared in RPMI 1640 medium supplemented with 2% fetal calf serum.
HIV stock and infection of mice.
Viral stocks were obtained by polyethylenimine (PEI)-mediated transfection (Polysciences) of 293T cells with either pYU-2 (R5 tropic) or JRCSF (R5 tropic) (provided through the NIH AIDS Research and Reference Reagent Program). At 48 h after transfection, virus was harvested, filtered (0.45 μm), and frozen at −80°C until use. Viral titers were determined as described previously (33). Briefly, 50% tissue culture infectious dose (TCID50) was determined by infecting human CD8+ T-cell-depleted peripheral blood mononuclear cells (PBMCs) from three donors that were stimulated by adding interleukin-2 (IL-2), phytohemagglutinin (PHA), and anti-CD3 beads (Dynal 11131D; Life Technologies). Mice were infected intraperitoneally (i.p.) with either HIV YU-2 or JRCSF at 2 × 105 TCID50s per mouse. Plasma HIV RNA levels were measured by reverse transcription-PCR (RT-PCR) (AmpliPrep/COBAS TaqMan HIV-1 test; Roche) at various times after infection.
Flow cytometry.
The cells in whole blood were counted in a Beckman cell counter. Cell suspensions were labeled with anti-human monoclonal antibodies (MAb) targeting the following cell surface markers: CD45-peridinin chlorophyll protein (PerCP), CD3-allophycocyanin (APC), CD4-Pe Cy7, CD8-BVa, CCR5-phycoerythrin (PE), CD34-APC, CD45RA-APC, and CCR7-PE (all from BD Biosciences or Biolegend). Washing and reagent dilutions were done with FACS buffer (phosphate-buffered saline [PBS] containing 2% fetal calf serum and 0.05% sodium azide). All acquisitions were performed on a Cyan ADP (Beckman Coulter) flow cytometer. Data were analyzed with FlowJo software (Ashland, OR). Cellular debris and dead cells were excluded by their light-scattering characteristics. Transduced CD34+ cells were sorted according to intrinsic GFP expression as measured by a BD FACSARIA III cell sorter.
HIV challenge ex vivo.
Spleens of five hu mice transplanted with HSPCs gene engineered with the microRNA to CCR5 were dissociated through a nylon mesh, and red blood cells were lysed with ACK buffer (Lonza) for 3 min. Cells were sorted (≥99% pure) with an ARIA sorter (BD Bioscience) into GFP-positive and -negative cells and were subsequently activated for 24 h with PHA (4 mg/ml) in RPMI 1640 culture medium supplemented with IL-2 (100 U/ml) and 10% fetal calf serum. Thereafter, cells were infected with a TCID50 of 3.3 ×105/ml of YU-2 for 6 h and washed three times. The supernatant of the last wash was used as the baseline p24 antigen level measured by an in-house enzyme-linked immunosorbent assay (ELISA) (34). Virus spread was then monitored at days 1, 4, 6, 8, and 10 postinfection.
Analysis of HIV envelope sequences. (i) Nucleic acid extraction.
For viral envelope sequencing, total nucleic acid was extracted from 60 μl of mouse plasma on an EasyMag extractor (bioMérieux, Switzerland) according to the manufacturer's instructions. The elution volume was 50 μl.
(ii) Reverse transcription and PCR.
For cDNA synthesis, 9 μl of extracted nucleic acid was reverse transcribed in a total reaction volume of 20 μl at 42°C for 30 min using a sequence-specific primer, MSR5 (35), and the PrimeScript One Step RT-PCR kit (TaKaRa Bio Europe/SAS, France). After heat inactivation at 96°C for 5 min, amplification primers were added to the reaction mixture, and DNA corresponding to positions 5956 to 8535 in isolate HXB2 (GenBank accession numbers K03455 and M38432) was amplified for 20 cycles. Nested PCR in a total reaction volume of 40 μl with Phusion Hot Start II DNA polymerase (Thermo Scientific, Switzerland) was carried out to amplify gp120 (positions 6817 to 7812) and gp41 (positions 7789 to 8382).
(iii) DNA sequencing.
Before sequencing, amplified DNA was treated with Illustra ExoStar 1-step reagent (Fisher Scientific, Switzerland). For cycle sequencing, the BigDye Terminator v3.1 cycle sequencing kit (LifeTechnologies, Switzerland) and specific sequencing primers were used. Twenty-five cycles of heat denaturation at 96°C for 10 s, annealing at 50°C for 5 s, and synthesis at 60°C for 4 min were carried out on a 2720 thermal cycler (Life Technologies). Samples were further processed by ethanol precipitation, followed by capillary electrophoresis on a model 3130xl genetic analyzer (Applied Biosystems, Switzerland). The sequences were assembled and edited with SeqMan Pro from the Lasergene 11 package (DNAStar).
Statistical analyses.
Statistical analyses were performed using GraphPad Prism 5.04 (GraphPad Software). Data were subjected to either unpaired t tests or paired t tests, as indicated in the figure legends. The P values obtained were considered significant when <0.05. Statistical outlier analysis was performed using the GraphPad Outlier calculator with an alpha of 0.01 (http://graphpad.com/quickcalcs/Grubbs1.cfm).
RESULTS
Transplanting CD34+ cells with partial CCR5 knockdown does not hinder HIV replication.
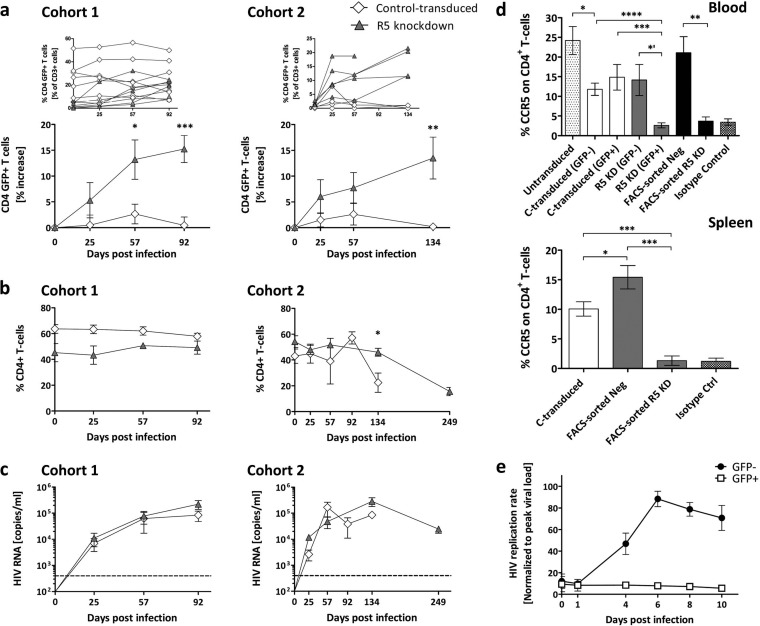
We previously developed a highly efficient microRNA called mirGE that allows efficient knockdown by single-copy transduction (30). In this study, we explored the potential of a mirGE lentivector targeting CCR5 to produce an HIV-resistant immune system in hu mice. The construct consists of a triple hairpin, and the vector cassette contains GFP driven by the same promoter as the miRNA that allows transduced cells to be identified directly. To minimize possible cellular perturbations from multiple vector inserts, we established a protocol that gave us a transduction rate of 20 to 30%. This transduction rate was based on previous work and should correlate with single-copy integration (36). In the first series of experiments, mice were transplanted with mirGE-transduced CD34+ cells without further manipulation (R5 knockdown mice); the CD34+ cells were a mixture of transduced (20 to 30%) and untransduced (70 to 80%) cells (data not shown). As controls, we used mice transplanted with CD34+ cells transduced either with a control GFP lentivector (control-transduced mice) (30) or with untransduced CD34+ cells (untransduced mice). Upon infection with an R5-tropic HIV (YU-2), the percentage and absolute number of GFP-positive CD4+ T cells were increased in R5 knockdown cohorts and not in control-transduced cohorts (Fig. 1a; see also Fig. 3g and h). The CD4+ cell population, however, remained the same over the observation period of 92 days (cohort 1) or showed a CD4+ T-cell loss only at day 134 days (cohort 2) (Fig. 1b). We explain this increase in GFP+ HIV-resistant CD4+ cells as the result of preferential expansion at the cost of untransduced CD4+ T cells, while the lymphoid system tries to keep the lymphoid T-cell number constant. However, the rates of HIV replication were similar in R5 knockdown mice and control-transduced mice at 92 and 134 days (Fig. 1c). Analysis of absolute numbers of CD4+ T cells (cohort 2) indicated that the control-transduced mice lost CD4+ T cells, whereas CD4+ T cells were more or less maintained in the R5 knockdown mice (data not shown).
FIG 1.
Homeostatic expansion of GFP-positive CD4+ T cells in R5 knockdown mice despite sustained R5-tropic (YU-2) HIV infection. (a) Percent change of GFP-positive CD4+ T cells in the peripheral blood of control-transduced mice (cohort 1, n = 8; cohort 2, n = 4) and R5 knockdown mice (cohort 1, n = 6; cohort 2, n = 5). Values are means ± SEMs. *, P = 0.0152; **, P = 0.0034; ***, P = 0.0002. P values were determined by two-tailed unpaired t test. Insets show the results for individual mice. (b) Frequency of total CD4+ T cells (percentage of total CD3 T cells) for the control-transduced and R5 knockdown cohorts 1 and 2. Values are means ± SEMs. *, P = 0.0146. P values were determined by two-tailed unpaired t test. (c) HIV RNA copies per milliliter of blood plasma collected for the control-transduced and R5 knockdown cohort 1 and cohort 2 over 92 and 134 days, respectively. Time of termination was chosen at random for the various groups. The dashed line indicates 400 copies/ml, the detection limit of the HIV RNA assay. Values are means ± SEMs. (d) Percentage CCR5 expression on total CD4+ T cells in peripheral blood and spleens of various cohorts of mice. Values are means ± SEMs. For blood, asterisks indicate P values as follows: *, P = 0.0237; *′, P = 0.0121; **, P = 0.0035; ***, P = 0.0007; ****, P = 0.0001. For spleens, asterisks indicate P values as follows: *, P = 0.041; ***, P = 0.0003. P values were determined by two-tailed unpaired t test. (e) HIV replication is inhibited ex vivo in GFP-positive sorted splenocytes from R5 knockdown mice. Splenocytes were isolated from R5 knockdown mice 20 weeks after CD34+ cell injection. Splenocytes were sorted into GFP-positive (n = 5) and GFP-negative (n = 5) fractions at 99% purity and challenged with R5-tropic HIV (YU-2). The amount of HIV production in the culture supernatant was monitored by HIV p24 ELISA. Viral loads were normalized to the peak viral load under each condition (means ± SEMs).
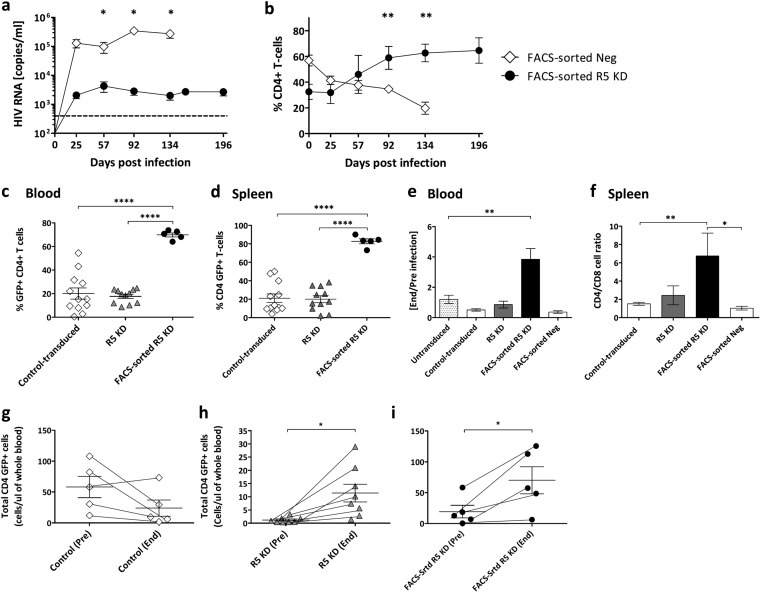
FIG 3.
Sustained HIV load inhibition in FACS-sorted R5 knockdown mice. (a) HIV RNA copies per milliliter in blood plasma from FACS-sorted R5 knockdown (YU-2 [n = 5]) and FACS-sorted negative (n = 15 [YU-2, n = 9, and JRCSF, n = 6]) mice collected over 134 and 196 days, respectively. The viral load detection limit is indicated by the dashed line (400 copies/ml). Values are means ± SEMs. Single asterisks indicate P values of 0.0486, 0.0188, and 0.0391 for days 57, 92, and 134, respectively. P values were determined by a two-tailed unpaired t test. (b) Percent CD4+ T cells of FACS-sorted R5 knockdown (n = 5) and FACS-sorted negative (n = 15) mice. Values are means ± SEMs. Double asterisks indicate P values of 0.0017 and 0.0018 for days 92 and 134, respectively. P values were determined by two-tailed unpaired t test. (c) Percent GFP-positive CD4+ T cells in the peripheral blood at termination. Values are means ± SEMs. ****, P = 0.0001. P values were determined by two-tailed unpaired t test. (d) Percent GFP-positive CD4+ T cells in the spleen at termination. Values are means ± SEMs. ****, P = 0.0001. P values were determined by two-tailed unpaired t test. (e) Change of CD4+/CD8+ T-cell ratio in the peripheral blood, comparing the CD4+/CD8+ T-cell ratio of each cohort end and preinfection. Values are means ± SEMs. **, P = 0.0026. P values were determined by two-tailed unpaired t test. (f) CD4+/CD8+ T-cell ratio at termination in the spleens of various cohorts of mice. Values are means ± SEMs. **, P = 0.0059; *, P = 0.039. P values were determined by two-tailed unpaired t test. (g to i) Absolute numbers of GFP+ CD4+ T cells or total CD4+ T cells/microliter of blood of representative mice from the control-transduced (n = 5), R5 knockdown (n = 8), and FACS-sorted R5 knockdown (n = 5) cohorts are shown. Values are means ± SEMs. Single asterisks in panels h and i indicate P values of 0.0194 and 0.0369, respectively. P values were determined by paired t test.
In the R5 knockdown mice, CCR5 was downregulated in the GFP-positive CD4+ T cells, but CCR5 was detected on the GFP-negative CD4+ T cells in blood and spleen (Fig. 1d). In the control-transduced mice, CCR5 was detected on GFP-negative and -positive CD4+ T cells. We verified the efficacy of our gene engineering approach by separating transduced from untransduced splenocytes (R5 knockdown mice) by FACS and infected the populations ex vivo with R5-tropic HIV. GFP-positive splenocytes had no HIV replication (Fig. 1e).
These results suggest that CCR5 knockdown efficiently protects CD4+ T cells from HIV infection, while CCR5-expressing CD4+ T cells are eradicated. In our mice, despite HIV challenge, at least a proportion of the ∼70 to 80% untransduced hematopoietic stem cells survived and continued to produce HIV-permissive CD4+ T cells, which sustained high HIV titers. It is still not entirely clear what effect HIV infection has on CD34+ cells and to what extent they are depleted, if at all (37).
Transplantation of purified CCR5 knockdown CD34+ cells results in mice with “pure” populations of transduced cells in vivo.
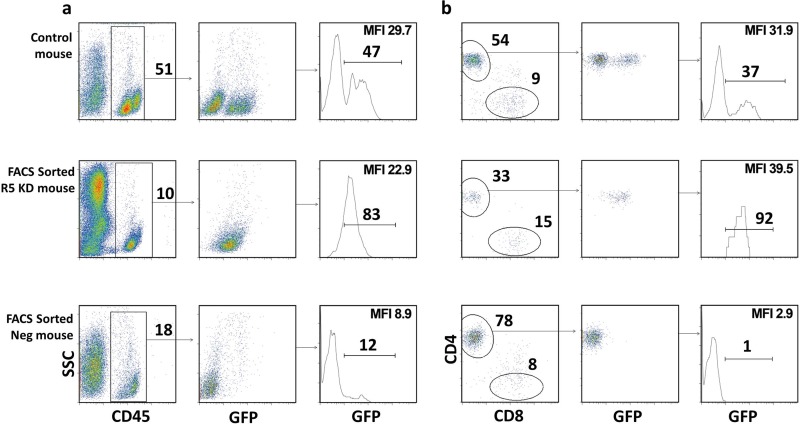
The lack of resistance to HIV infection was likely due to the chimerism of transduced and untransduced CD34+ cells in our initial experiments. Therefore, we sorted the CD34+ cells after transduction into CCR5 knockdown, GFP-positive and -negative fractions obtaining a >90% pure population of GFP-positive CD34+ cells. Mice transplanted with GFP-sorted cells were called FACS-sorted R5 knockdown mice. Analysis of the peripheral blood in six of the FACS-sorted R5 knockdown animals showed a single GFP-positive peak for human CD45+ and CD4+ T cells, suggesting that only transduced CD34+ cells were engrafted in these mice (Fig. 2); the level of GFP-positive cells was a major criterion for successful gene engineering and engraftment. Mice transplanted with the GFP-negative fraction were called FACS-sorted negative mice. They developed CD4+ and CD8+ T-cell populations with no GFP expression (Fig. 2). In contrast, the control-transduced mice had two distinct GFP-negative and -positive populations for CD45+ and CD4+ T cells (Fig. 2). A summary of mice used in this study is provided in Table 1. Mice with less than 5% human engraftment in the peripheral blood before HIV infection were excluded.
FIG 2.
GFP-positive CCR5 knockdown sorted CD34+ HSPCs produced mice with “pure” populations of transduced cells in vivo. (a) FACS plots showing the percent human engraftment of CD45+ (percentage of live cells) and GFP-positive CD45+ (percentage of human CD45) cells for representative control-transduced, FACS-sorted R5 knockdown, and FACS-sorted negative mice before HIV infection. (b) FACS plots showing the percentage CD4+ T cells (percent CD3+), CD8+ (percent CD3+), and GFP-positive CD4+ T cells (percent CD4+) in the peripheral blood before HIV infection of the same mice as in panel a. For all cell subset analyses, the subgating was done as follows: total live cells and CD45+, CD3+, CD4+, and CD8+ cells. Mean fluorescence intensity (MFI) for GFP is indicated.
TABLE 1.
Mice used in this study
| Trait | % GFP+ CD34+ cells pretransplantation | Total no. of mice | No. of mice with: |
|||
|---|---|---|---|---|---|---|
| % engraftment (>5% hCD45+) | % GFP+ CD4+ T cells (>70%) | % GFP+ CD4+ T cells (20–70%) | % GFP+ CD4+ T cells (<20%) | |||
| Untransduced | NAa | 15 | 11 | NA | NA | NA |
| Control transduced | 15–40 | 17 | 12 | 0 | 5 | 7 |
| R5 knockdown | 17–20 | 15 | 11 | 0 | 0 | 11 |
| FACS GFP positiveb | 12–32 | 20 | 10 | 6 | 4 | 0 |
| FACS GFP negative | NA | 26 | 15 | NA | NA | NA |
NA, not applicable.
FACS GFP-positive cells prior to FACS cell sorting for GFP-positive cells.
Transplanting purified CCR5 knockdown CD34+ cells dramatically lowered viral load and protected HIV target cells in vivo.
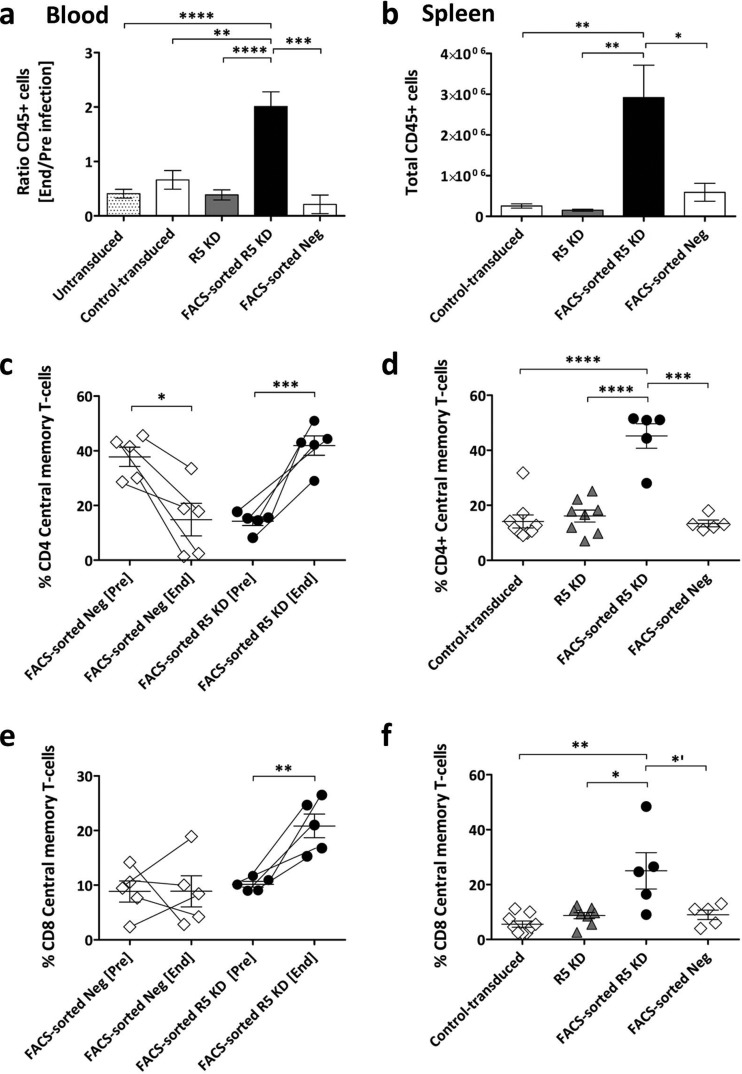
The FACS-sorted R5 knockdown mice had markedly lower viral loads than the FACS-sorted negative mice over 28 weeks (Fig. 3a). Peak viremia for the FACS-sorted R5 knockdown mice was, on average, 4.2 × 103 copies/ml, and FACS-sorted negative mice had 3.5 × 105 copies/ml. Viral loads for the untransduced, control-transduced, R5 knockdown, and FACS-sorted negative mice were similar. The FACS-sorted R5 knockdown mice had lower viral loads than all cohorts.
CD4+ T cells (percentage of total CD3+ T cells) from the FACS-sorted negative mice declined steadily upon infection (day 0, 55%; day 134, 20%) (Fig. 3b). In contrast, the FACS-sorted R5 knockdown mice showed a steady increase in CD4+ T cells (day 0, 33%; day 196, 65%). Furthermore, the absolute numbers of CD4+ T cells increased for the FACS-sorted R5 knockdown mice but decreased for the FACS-sorted negative mice (data not shown).
At euthanasia, for the FACS-sorted R5 knockdown mice, 70% of CD4+ T cells in the blood and 83% of CD4+ T cells in the spleen were GFP positive (Fig. 3c and d). In the control-transduced and R5 knockdown groups, the values were, on average, 20% and 18% in the blood and 21% and 20% in the spleen, respectively (Fig. 3c and d). Similarly, the CD4+/CD8+ T-cell ratios in the blood (end/preinfection) and spleen were very low in the various groups except for the FACS-sorted R5 knockdown group (Fig. 3e and f). Absolute numbers of GFP-positive CD4+ T cells expanded significantly upon HIV challenge in the FACS-sorted R5 KD mice (Fig. 3i), and CCR5 expression was downregulated in the blood and spleens of the FACS-sorted R5 knockdown mice (Fig. 1d).
In summary, transplantation of GFP-positive sorted CD34+ cells produced mice with high levels of gene-engineered CCR5 knockdown CD4+ T cells in vivo. This resulted in long-term inhibition of HIV replication in vivo and preservation of HIV target cells in the blood and spleen.
Outlier analysis and follow-up of mice that did not meet acceptance criteria.
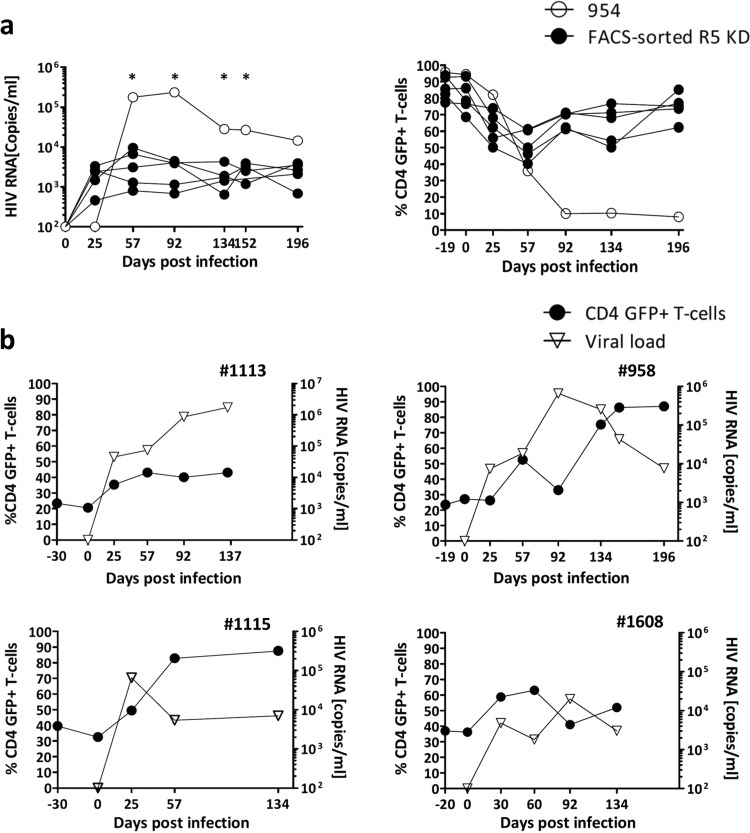
The FACS-sorted R5 knockdown mice typically controlled the virus long term (titers < 104 copies/ml) while maintaining a high level of GFP-positive CD4+ T cells in the blood (Fig. 4a). In contrast, a single FACS-sorted R5 knockdown mouse (number 954) showed an unexpected decline in GFP-positive CD4+ T cells with high HIV copy numbers, >105 copies/ml, on days 57 and 92 (Fig. 4a). We performed an outlier analysis (see Materials and Methods), and at four different times, viral loads detected in mouse 954 were statistical outliers (Fig. 4a). Based on this, mouse 954 was not included in the mean values and was analyzed separately (see below). At termination, this mouse had much lower splenic engraftment of total human splenocytes, with 31% human cells, than did the FACS-sorted R5 knockdown mice, which had 54% (standard error of the mean [SEM], ±11%). GFP-positive CD4+ T cells were barely detected in the spleen of mouse 954 (6%), while the FACS-sorted R5 knockdown mice had, on average, 83% (SEM, ±2%) GFP-positive CD4+ T cells in the spleen (Fig. 3d).
FIG 4.
Outlier analysis and examples of viral load control due to homeostatic expansion of transduced cells. (a) Left graph, viral load of the FACS-sorted R5 knockdown mice (YU-2) and viral load of outlier FACS-sorted R5 knockdown mouse 954 (YU-2). Right graph, percent GFP-positive CD4+ T cells of the FACS-sorted R5 knockdown mice and mouse 954. Percent GFP-positive CD4+ T cells is shown as a percentage of total CD4+ T cells. P values were determined by GraphPad Outlier calculator. *, P < 0.001. (b) Viral load and percent GFP-positive CD4+ T cells of four individual FACS-sorted R5 knockdown mice (mouse 1113, JRCSF; mouse 958, YU-2; mouse 1115, JRCSF; and mouse 1608, JRCSF) that were excluded due to not reaching the inclusion criteria of >70% GFP-positive CD4+ T cells before HIV infection.
The inclusion criterion we defined as successful reconstitution for FACS-sorted R5 knockdown mice was 70% GFP+ CD4+ T cells in the peripheral blood before infection. Four mice did not meet this criterion despite being transplanted with GFP-positive sorted CD34+ cells (Fig. 4b). Mouse 1113 had no protection against HIV and had a limited expansion of GFP-positive CD4+ T cells (23% on day 0 to 43% on day 137). Mice 958 and 1115 had massive expansions of GFP-positive cells, reaching close to 100% of the total CD4+ T-cell population, which went in parallel with a decrease in the viral load (Fig. 4b). For mouse 1608, the dynamics of recovery of GFP-positive CD4+ T cells and viral load were slower, and recovery of GFP-positive CD4+ T cells on day 134 was less extensive (Fig. 4b). These three mice (958, 1115, and 1608) maintained a high CD4/CD8 ratio, and the percentage of GFP-positive CD4+ T cells in the blood increased upon infection from 24 to 80, 39 to 82, and 37 to 52%, respectively. These animals also had high frequencies of GFP-positive CD4+ T cells in the spleens, i.e., 59, 60, and 83%, respectively.
Strikingly, we observed this level of expansion (Fig. 4b) of HIV-resistant GFP-positive CD4+ T cells and a concomitant inhibitory effect on HIV only in mice transplanted with CCR5 knockdown GFP-sorted CD34+ cells (these animals had 30% [±4% SEM] GFP+ CD4+ T cells on day 0). This degree of expansion was not seen in R5 knockdown mice (which had 5% ± 2% GFP+ CD4+ T cells before HIV infection). Based on these results, we estimate that at least 20% of CD4+ T cells need to be CCR5 repressed to observe homeostatic expansion and relevant effects on viremia.
Preserved engraftment and preferential expansion of central memory T cells in FACS-sorted R5 knockdown mice upon HIV infection.
Engraftment as reflected in peripheral blood decreased in all control cohorts but increased in the FACS-sorted R5 knockdown mice (Fig. 5a). This effect on total engraftment was even more impressive in the spleen. FACS-sorted R5 knockdown mice had 10 times more human cells than control cohorts (Fig. 5b).
FIG 5.
Increased engraftment and central memory T cells in blood and spleens of FACS-sorted R5 knockdown mice. (a) Change in level of peripheral blood engraftment expressed as the ratio of total CD45+ at the end and preinfection. Values are means ± SEMs. **, P = 0.0031; ***, P = 0.0006; ****, P < 0.0001. P values were determined by two-tailed unpaired t test. (b) Absolute numbers of human cells (CD45+) in the spleen at termination. Values are means ± SEMs. *, P = 0.0229; **, P < 0.0019. P values were determined by two-tailed unpaired t test. (c and e) Percent CD4+ and CD8+ central memory T cells in the peripheral blood, preinfection and at the end time point. Values are means and SEMs. *, P = 0.0153; **, P = 0.0042; ***, P = 0.0004. P values were determined by paired t test. (d and f) Percent CD4+ and CD8+ central memory T cells in the spleen at termination. Values are means ± SEMs. *′, P = 0.0472; *, P = 0.0106; **, P = 0.0022; ***, P = 0.0001; ****, P < 0.0001. P values were determined by two-tailed unpaired t test.
We evaluated the CD4+ and CD8+ effector (CD45RApos CCR7neg), effector memory (CD45RAneg CCR7neg), naive (CD45RApos CCR7pos), and central memory (CD45RAneg CCR7pos) T-cell subsets in the blood and spleens of the FACS-sorted R5 knockdown and representative FACS-sorted negative mice (Fig. 5c to f). In the peripheral blood of the FACS-sorted negative mice, the frequency of central memory CD4+ T cells was significantly decreased, and the CD8+ central memory T-cell subset was unchanged (Fig. 5c and e). In contrast, central memory CD4+ and CD8+ T cells were increased in the FACS-sorted R5 knockdown mice. Similarly, more CD4+ and CD8+ central memory T cells were present in the spleens of the FACS-sorted R5 knockdown mice (Fig. 5d and f). We observed no differences between the cohorts for effector and effector memory cells (data not shown). Notably, however, there was a trend toward a decrease in naive CD4+ and CD8+ T cells in all cohorts (data not shown).
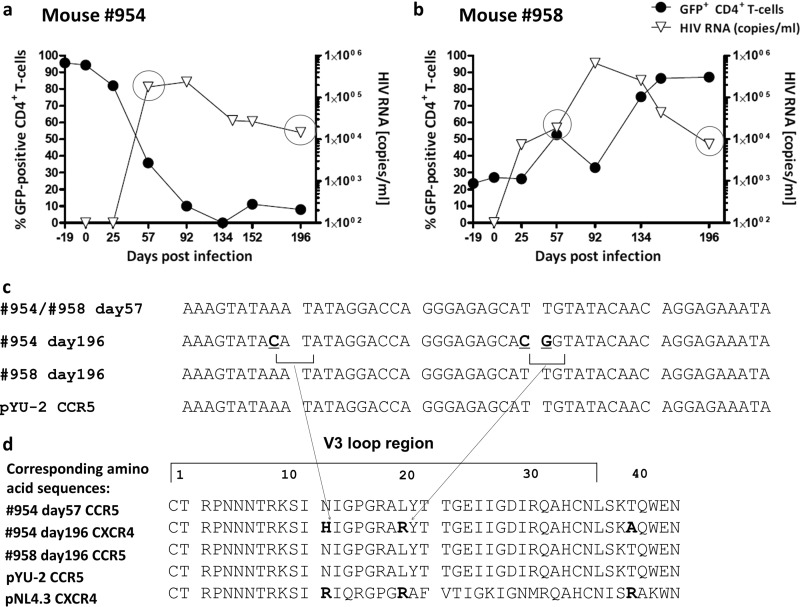
Evidence of shift from R5- to X4-tropic strain in one mouse.
As described above (Fig. 4), FACS-sorted R5 knockdown mice had a low viral load and maintained high levels of CCR5 knockdown CD4+ T cells. Mouse 954 was clearly an outlier: it had high viral titers and lost GFP-positive CD4+ T cells (Fig. 6a). We hypothesized that this mouse might have had a tropism shift of the virus from R5 to X4. We performed HIV population sequencing (from plasma) on days 57 and 196. As a control, we analyzed mouse 958 (Fig. 6b). Sequencing revealed no mutations in mouse 958. Mouse 954 had mutations in the V3 loop of the HIV envelope sequence (Fig. 6c), resulting in amino acid substitutions to basic amino acids as indicated (Fig. 6d). Substitutions in the V3 loop to basic amino acids have been reported to result in a switch from R5 to X4 tropism (38).
FIG 6.
Population sequencing of HIV in plasma: evidence for gene therapy failure. (a and b) Plots showing the percent GFP-positive CD4+ T cells in peripheral blood (as a percentage of CD3+ T cells) on the left y axis and HIV load on the right y axis for mice 954 and 958, respectively. Mouse 954 had a high and sustained viral load over time, with a complete loss of GFP-positive CD4+ T cells. Mouse 958 experienced an expansion of GFP-positive CD4+ T cells from less than 30% on day 19 to more than 80% on day 196. (c) On day 57, both animals had a homogenous HIV population in the peripheral blood; sequencing data of HIV envelope V3 loop are consistent with an R5-tropic HIV strain. On day 196, mouse 954, which experienced a complete loss of GFP-positive CD4+ T cells (blood and spleen), had detectable mutations within the V3 loop of HIV. For mouse 958, no mutations were detected in the V3 loop on day 196, indicating the presence of a homogenous HIV population. (d) Changes to basic amino acids H and R. According to a Geno2Pheno (http://www.geno2pheno.org/) analysis of the obtained sequence in panel c, there is only 18% confidence that the virus at day 196 of mouse 954 was not an X4 variant.
DISCUSSION
In this study, we investigated a lentiviral vector-based, CCR5-targeting miRNA as a tool for engineering an HIV-resistant human immune system. We show that (i) the miRNA-based vector was very efficient in downregulating CCR5 on T cells and prevented their infection by HIV ex vivo, (ii) only mice that were transplanted with a preselected population of transduced CD34+ cells and maintained gene-engineered CD4+ T cells had a dramatically reduced viral load (functional cure), and (iii) the HIV-infected mice transplanted with miRNA CCR5 gene-engineered CD34+ cells showed a dramatic expansion of memory T cells (i.e., the miRNA-edited T cells were mainly of this phenotype). Thus, we provide here preclinical proof of concept for gene engineering of an HIV-resistant immune system through the use of vector-mediated miRNA expression and the need for a certain threshold of gene-engineered CD34+ cells for functional cure of HIV.
While gene engineering of HIV-resistant cells is a viable option for cure of HIV, major issues remain to be solved. These include finding the best antiviral moiety or combination, the most efficacious way to gene engineer the CD34+ cells, and the threshold of gene-engineered CD34+ cells needed for functional cure.
Lentivirus-based transduction has been supplemented by gene-targeting methods, such as ZNF or Talen nucleases, or the Crispr/Cas system for gene editing (23–25). However, off-target effects of these methods are still unknown, and gene engineering in primary cells is only modestly effective (26). And even though no adverse events have been reported, there is less experience in clinical trials with gene-targeting methods than with lentivirus-based transduction. Thus, we opted for lentivirus-based gene engineering (39–41). Furthermore, we are the first to engage in miRNA technology for knocking down the HIV coreceptor CCR5 in CD4+ T cells via gene engineering of CD34+ cells. miRNAs closely mimic naturally occurring pri-miRNAs and thus are less likely to cause oversaturation of the RNA interference pathway and to affect cellular homeostasis than the widely used shRNAs (42, 43). However, miRNAs are thought to have a lower capacity to downregulate target genes than shRNAs. In this study, we used a miRNA we developed with optimized features that efficiently knock down target genes upon single-copy vector transduction (30). Ex vivo-sorted cells were indeed resistant to a challenge with CCR5-tropic strains. However, bulk transplantation of transduced CD34+ cells into mice resulted in a human lymphoid system that replicated HIV similarly to untransduced hu mice but preserved CD4+ T-cell counts. Similar data have been reported previously (15).
We hypothesized that the majority of HSPCs needs to be gene engineered to see an effect on the HIV load; otherwise, the HIV-permissive CD4+ T cells that originated from the untransduced CD34+ cells would “outnumber” the HIV-resistant cells. Thus, we used GFP to allow for efficient sorting of transduced CD34+ cells before transplantation. By doing so, we found that to achieve long-term suppression of viral load, more than 70% of CD34+ transplanted should be gene engineered. Walker et al. obtained an average engraftment (± standard deviation [STD]) level of anti-HIV vector-transduced cells of 17.5% ± 8% in the peripheral blood and argued that these numbers of cells were insufficient to see any decrease in plasma viremia (15). Furthermore, a very recent publication from the same group sorted the gene-engineered CD34+ cells with a truncated version of CD25 (tCD25) before their transplantation into 2- to 5-day-old NRG mice (20). They found that mice transplanted with tCD25-purified CD34+ cells had normal multilineage hematopoiesis, similar to mice transplanted with untransduced CD34+ cells. Upon HIV challenge, tCD25-transplanted mice did not suffer from HIV-induced CD4+ T-cell depletion as did the untransduced mice, and tCD25 mice had a 1.5-log inhibition in plasma viremia compared to that of mice with untransduced CD34+ cells. Our data nicely complement the data provided by Walker et al. and Barclay et al. and underline the importance of the number of transduced cells that are required for efficient HIV gene therapy. Notably, three hu mice transplanted with purified gene-engineered CD34+ cells showed at baseline ∼30% GFP+ cells which expanded substantially upon HIV infection; the expansion went along with a decrease in viral load. The data for these three mice were reminiscent of the data reported by Holt et al. showing that disruption of CCR5 by zinc finger nucleases was achieved in ∼20% of CD34+ cells and resulted in viral repression over time (16).
Obviously, in humans, GFP-based sorting would not be an option, given the xenogeneic nature of the protein. However, novel strategies for sorting of transduced CD34+ cells based on the expression of truncated cellular surface receptors, such as CD25 (20), the epidermal growth factor receptor (44), or the nerve growth factor receptor (45), are very promising for achieving high numbers of engrafted gene-engineered cells. An alternative approach to pretransplantation sorting would be in vivo selection of transduced cells (46, 47). Regrettably, current in vivo selection methods use potentially carcinogenic compounds, such as mycophenolate, methotrexate, or alkylating agents (i.e., O6-benzylguanine/bis-chloroethylnitrosourea), that offset their use in a disease, such as HIV, that is amenable to an efficient and well-tolerated cART. We want to emphasize that in our gene-engineering efforts, we aimed for single lentiviral copy integration. The two recent phase I clinical trials used gene engineering protocols that resulted in vector copy numbers ranging from 2 to 4 per genome of bone marrow cells prior to transplantation without documenting any adverse events over an observation period of >20 months (39, 40). Thus, ensuring CD34+ transduction might present another alternative for increasing the number of gene-engineered CD34+ cells. These protocols appeared not to affect the long-term engraftment negatively in these phase I clinical trials.
In fact, we do not know the number of gene-engineered HSPCs needed to render the immune system resistant to HIV. As outlined above, we aimed for a rather pure population of gene-engineered HSPCs as proof of preclinical concept. However, we observed HIV-lowering effects in some mice with 20 to 40% engraftment of transduced cells, data similar to those reported by Holt et al. (16). HIV, certainly by killing untransduced cells via its cytopathic effects, will promote the expansion of HIV-resistant cells. To what extent the HIV-resistant cells will foster an efficient HIV-specific immune response and thereby constrain HIV remains unknown. Whether additional factors contribute to HIV-lowering effects remains unknown.
White blood cell counts from HIV-infected mice generated with FACS-sorted R5 KD cells showed an expansion of central memory CD4+ and CD8+ T cells, while all other groups showed a progressive loss of these CD4+ memory T cells and no change in CD8+ T cells. This pattern was also evident when looking at the splenocytes. These memory CD8+ T cells might have contributed to the control of HIV. There was a decrease in the frequency of naive CD4+ and CD8+ T cells in the peripheral blood in both FACS-sorted negative and FACS-sorted R5 KD mice, whereas in the spleen, the naive CD4+ and CD8+ T cells in the FACS-sorted R5 KD mice tended to be higher (data not shown). The expansion of central memory T cells is reminiscent of the immune reconstitution seen in HIV-infected patients on ART (48).
HIV is known for its high mutational rate. In this respect, we observed one mouse (mouse 954) with an escape mutation. Despite high levels (day 0) of engraftment of CCR5 knockdown cells, this mouse had a high viral load and a complete loss of circulating CD4+ T cells. Population sequencing of the V3 loop indicated a likely shift to X4 tropism, which might explain the uncontrolled infection. The mutations were detectable in the blood only at relatively late time points. This might be due to an initial compartmentalized replication of the X4-tropic strains in the thymus. We previously showed that X4-HIV NL4-3 severely depleted the thymus, whereas YU-2 had only minor effects (49). However, we do not know whether the potential emergence was due to the CCR5 knockdown or was just a coincidence. Indeed, emergence of CXCR4-tropic strains may occur without any immune or drug pressure in hu mice infected with CCR5-tropic strains (50). In any case, CCR5 knockdown should be done in concert with another strategy to constrain HIV (i.e., including another anti-HIV moiety, combining with efficient antiretrovirals, or boosting the immune response in parallel to transplantation). Indeed, the solidness of successful gene engineering by the expression of more than one antiviral moiety may prevent HIV evolution (51). Gene engineering could be combined with conventional ART: combining two treatment modalities was efficient in cell lines (52), as induction therapy (53) or with anti-PD1 antibodies that decrease viral load and increase the level of CD4+ T cells in HIV-infected mice (54). In any case, gene engineering efforts cannot promote more virulent HIV strains, either for the individual patient or for the general population.
In summary, our results provide the first preclinical proof of concept that transplantation of miRNA CCR5 knockdown CD34+ cells can lead to long-term control of HIV viremia. Translation of our results to the clinical setting is relatively straightforward but will require the implementation of existing strategies for pre- and posttransplantation selection compatible with human use. At this point, our strategy demonstrates long-term viral control but not yet a cure. However, while a cure remains the ultimate goal, long-term viral control independent of antiretrovirals is a relevant intermediate step, worth translating to the clinical setting. We believe that a definitive cure of HIV might indeed come from a combination of different approaches such as CCR5 knockdown combined with drug therapy, vaccination, or a second gene therapy target.
ACKNOWLEDGMENTS
We thank Astrid Koster, Xenia Rüfenacht, and Fatbardha Dautaj (Clinic of Immunology, University Hospital Zurich, Zurich, Switzerland) for their work in obtaining the HIV loads of the mice. We thank Jürg Böni, Caroline Käser, and Cyril Shah (Institute of Medical Virology, University of Zurich) for performing the sequencing of the HIV genome and analyzing the data.
This work was supported by the clinical research focus program Human Hemato-Lymphatic Diseases of the University of Zurich and the Swiss South African Research Joint Program, the Swiss National Science Foundation, a University of Zurich Candoc grant, the South African National Research Foundation, the Medical Research Council of South Africa, and the Wolfermann Nägeli Stiftung.
R.M. designed and performed experiments, analyzed data, and wrote the paper. R.F.S. and K.-H.K. designed the experiments, supervised the work, analyzed the data, and wrote the paper. M.S.P. was involved in the initial conceptualization of the study, supervised aspects of the work, and helped to write the paper. A.A. helped to coordinate the work with the mice and transplantation of newborn mice, process cord blood, and write the paper. P.S. helped supervise aspects of the work and designed certain experiments. V.J. helped to write the paper and analyze data. G.G.-H., D.L., and M.-A.R. helped to terminate experiments and process cord blood samples. S.I. helped in processing of cord blood samples. S.R. provided the expertise and obtained the viral loads for HIV-infected mice. M.G.M. gave highly valuable input in the entire study.
We declare no conflicts of interest.
REFERENCES
- 1.Lohse N, Gerstoft J, Kronborg G, Larsen CS, Pedersen C, Pedersen G, Nielsen L, Sorensen HT, Obel N. 2011. Comorbidity acquired before HIV diagnosis and mortality in persons infected and uninfected with HIV: a Danish population-based cohort study. J Acquir Immune Defic Syndr 57:334–339. doi: 10.1097/QAI.0b013e31821d34ed. [DOI] [PubMed] [Google Scholar]
- 2.Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, Spratt SK, Surosky RT, Giedlin MA, Nichol G, Holmes MC, Gregory PD, Ando DG, Kalos M, Collman RG, Binder-Scholl G, Plesa G, Hwang WT, Levine BL, June CH. 2014. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med 370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tebas P, Stein D, Binder-Scholl G, Mukherjee R, Brady T, Rebello T, Humeau L, Kalos M, Papasavvas E, Montaner LJ, Schullery D, Shaheen F, Brennan AL, Zheng Z, Cotte J, Slepushkin V, Veloso E, Mackley A, Hwang WT, Aberra F, Zhan J, Boyer J, Collman RG, Bushman FD, Levine BL, June CH. 2013. Antiviral effects of autologous CD4 T cells genetically modified with a conditionally replicating lentiviral vector expressing long antisense to HIV. Blood 121:1524–1533. doi: 10.1182/blood-2012-07-447250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DiGiusto DL, Krishnan A, Li L, Li H, Li S, Rao A, Mi S, Yam P, Stinson S, Kalos M, Alvarnas J, Lacey SF, Yee JK, Li M, Couture L, Hsu D, Forman SJ, Rossi JJ, Zaia JA. 2010. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci Transl Med 2:36ra43. doi: 10.1126/scitranslmed.3000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitsuyasu RT, Merigan TC, Carr A, Zack JA, Winters MA, Workman C, Bloch M, Lalezari J, Becker S, Thornton L, Akil B, Khanlou H, Finlayson R, McFarlane R, Smith DE, Garsia R, Ma D, Law M, Murray JM, von Kalle C, Ely JA, Patino SM, Knop AE, Wong P, Todd AV, Haughton M, Fuery C, Macpherson JL, Symonds GP, Evans LA, Pond SM, Cooper DA. 2009. Phase 2 gene therapy trial of an anti-HIV ribozyme in autologous CD34+ cells. Nat Med 15:285–292. doi: 10.1038/nm.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deeks SG, Wagner B, Anton PA, Mitsuyasu RT, Scadden DT, Huang C, Macken C, Richman DD, Christopherson C, June CH, Lazar R, Broad DF, Jalali S, Hege KM. 2002. A phase II randomized study of HIV-specific T-cell gene therapy in subjects with undetectable plasma viremia on combination antiretroviral therapy. Mol Ther 5:788–797. doi: 10.1006/mthe.2002.0611. [DOI] [PubMed] [Google Scholar]
- 7.Riddell SR, Elliott M, Lewinsohn DA, Gilbert MJ, Wilson L, Manley SA, Lupton SD, Overell RW, Reynolds TC, Corey L, Greenberg PD. 1996. T-cell mediated rejection of gene-modified HIV-specific cytotoxic T lymphocytes in HIV-infected patients. Nat Med 2:216–223. doi: 10.1038/nm0296-216. [DOI] [PubMed] [Google Scholar]
- 8.Hütter G, Nowak D, Mossner M, Ganepola S, Mussig A, Allers K, Schneider T, Hofmann J, Kucherer C, Blau O, Blau IW, Hofmann WK, Thiel E. 2009. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med 360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 9.Kordelas L, Verheyen J, Beelen DW, Horn PA, Heinold A, Kaiser R, Trenschel R, Schadendorf D, Dittmer U, Esser S, Essen HIVAG. 2014. Shift of HIV tropism in stem-cell transplantation with CCR5 Delta32 mutation. N Engl J Med 371:880–882. doi: 10.1056/NEJMc1405805. [DOI] [PubMed] [Google Scholar]
- 10.Rongvaux A, Takizawa H, Strowig T, Willinger T, Eynon EE, Flavell RA, Manz MG. 2013. Human hemato-lymphoid system mice: current use and future potential for medicine. Annu Rev Immunol 31:635–674. doi: 10.1146/annurev-immunol-032712-095921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nischang M, Gers-Huber G, Audige A, Akkina R, Speck RF. 2012. Modeling HIV infection and therapies in humanized mice. Swiss Med Wkly 142:w13618. doi: 10.4414/smw.2012.13618. [DOI] [PubMed] [Google Scholar]
- 12.Bennett MS, Akkina R. 2013. Gene therapy strategies for HIV/AIDS: preclinical modeling in humanized mice. Viruses 5:3119–3141. doi: 10.3390/v5123119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimizu S, Hong P, Arumugam B, Pokomo L, Boyer J, Koizumi N, Kittipongdaja P, Chen A, Bristol G, Galic Z, Zack JA, Yang O, Chen IS, Lee B, An DS. 2010. A highly efficient short hairpin RNA potently down-regulates CCR5 expression in systemic lymphoid organs in the hu-BLT mouse model. Blood 115:1534–1544. doi: 10.1182/blood-2009-04-215855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ringpis GE, Shimizu S, Arokium H, Camba-Colon J, Carroll MV, Cortado R, Xie Y, Kim PY, Sahakyan A, Lowe EL, Narukawa M, Kandarian FN, Burke BP, Symonds GP, An DS, Chen IS, Kamata M. 2012. Engineering HIV-1-resistant T-cells from short-hairpin RNA-expressing hematopoietic stem/progenitor cells in humanized BLT mice. PLoS One 7:e53492. doi: 10.1371/journal.pone.0053492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker JE, Chen RX, McGee J, Nacey C, Pollard RB, Abedi M, Bauer G, Nolta JA, Anderson JS. 2012. Generation of an HIV-1-resistant immune system with CD34(+) hematopoietic stem cells transduced with a triple-combination anti-HIV lentiviral vector. J Virol 86:5719–5729. doi: 10.1128/JVI.06300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, Crooks GM, Kohn DB, Gregory PD, Holmes MC, Cannon PM. 2010. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol 28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joseph A, Zheng JH, Chen K, Dutta M, Chen C, Stiegler G, Kunert R, Follenzi A, Goldstein H. 2010. Inhibition of in vivo HIV infection in humanized mice by gene therapy of human hematopoietic stem cells with a lentiviral vector encoding a broadly neutralizing anti-HIV antibody. J Virol 84:6645–6653. doi: 10.1128/JVI.02339-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitchen SG, Levin BR, Bristol G, Rezek V, Kim S, Aguilera-Sandoval C, Balamurugan A, Yang OO, Zack JA. 2012. In vivo suppression of HIV by antigen specific T cells derived from engineered hematopoietic stem cells. PLoS Pathog 8:e1002649. doi: 10.1371/journal.ppat.1002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hauber I, Hofmann-Sieber H, Chemnitz J, Dubrau D, Chusainow J, Stucka R, Hartjen P, Schambach A, Ziegler P, Hackmann K, Schrock E, Schumacher U, Lindner C, Grundhoff A, Baum C, Manz MG, Buchholz F, Hauber J. 2013. Highly significant antiviral activity of HIV-1 LTR-specific tre-recombinase in humanized mice. PLoS Pathog 9:e1003587. doi: 10.1371/journal.ppat.1003587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barclay SL, Yang Y, Zhang S, Fong R, Barraza A, Nolta JA, Torbett BE, Abedi M, Bauer G, Anderson JS. 2015. Safety and efficacy of a tCD25 pre-selective combination anti-HIV lentiviral vector in human hematopoietic stem and progenitor cells. Stem Cells 33:870–879. doi: 10.1002/stem.1919. [DOI] [PubMed] [Google Scholar]
- 21.Berkhout B, Liu YP. 2014. Towards improved shRNA and miRNA reagents as inhibitors of HIV-1 replication. Future Microbiol 9:561–571. doi: 10.2217/fmb.14.5. [DOI] [PubMed] [Google Scholar]
- 22.Jackson AL, Linsley PS. 2004. Noise amidst the silence: off-target effects of siRNAs? Trends Genet 20:521–524. doi: 10.1016/j.tig.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 23.Bibikova M, Golic M, Golic KG, Carroll D. 2002. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161:1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho SW, Kim S, Kim JM, Kim JS. 2013. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 25.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ. 2011. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 26.Genovese P, Schiroli G, Escobar G, Di Tomaso T, Firrito C, Calabria A, Moi D, Mazzieri R, Bonini C, Holmes MC, Gregory PD, van der Burg M, Gentner B, Montini E, Lombardo A, Naldini L. 2014. Targeted genome editing in human repopulating haematopoietic stem cells. Nature 510:235–240. doi: 10.1038/nature13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sander JD, Ramirez CL, Linder SJ, Pattanayak V, Shoresh N, Ku M, Foden JA, Reyon D, Bernstein BE, Liu DR, Joung JK. 2013. In silico abstraction of zinc finger nuclease cleavage profiles reveals an expanded landscape of off-target sites. Nucleic Acids Res 41:e181. doi: 10.1093/nar/gkt716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gabriel R, Lombardo A, Arens A, Miller JC, Genovese P, Kaeppel C, Nowrouzi A, Bartholomae CC, Wang J, Friedman G, Holmes MC, Gregory PD, Glimm H, Schmidt M, Naldini L, von Kalle C. 2011. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol 29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- 29.Cradick TJ, Ambrosini G, Iseli C, Bucher P, McCaffrey AP. 2011. ZFN-site searches genomes for zinc finger nuclease target sites and off-target sites. BMC Bioinformatics 12:152. doi: 10.1186/1471-2105-12-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myburgh R, Cherpin O, Schlaepfer E, Rehrauer H, Speck RF, Krause KH, Salmon P. 2014. Optimization of critical hairpin features allows miRNA-based gene knockdown upon single-copy transduction. Mol Ther Nucleic Acids 3:e207. doi: 10.1038/mtna.2014.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giry-Laterrière M, Verhoeyen E, Salmon P. 2011. Lentiviral vectors. Methods Mol Biol 737:183–209. doi: 10.1007/978-1-61779-095-9_8. [DOI] [PubMed] [Google Scholar]
- 32.Nischang M, Sutmuller R, Gers-Huber G, Audige A, Li D, Rochat MA, Baenziger S, Hofer U, Schlaepfer E, Regenass S, Amssoms K, Stoops B, Van Cauwenberge A, Boden D, Kraus G, Speck RF. 2012. Humanized mice recapitulate key features of HIV-1 infection: a novel concept using long-acting anti-retroviral drugs for treating HIV-1. PLoS One 7:e38853. doi: 10.1371/journal.pone.0038853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDougal JS, Cort SP, Kennedy MS, Cabridilla CD, Feorino PM, Francis DP, Hicks D, Kalyanaraman VS, Martin LS. 1985. Immunoassay for the detection and quantitation of infectious human retrovirus, lymphadenopathy-associated virus (LAV). J Immunol Methods 76:171–183. doi: 10.1016/0022-1759(85)90489-2. [DOI] [PubMed] [Google Scholar]
- 34.Moore JP, Sattentau QJ, Clapham PR. 1990. Enhancement of soluble CD4-mediated HIV neutralization and gp 120 binding by CD4 autoantibodies and monoclonal antibodies. AIDS Res Hum Retroviruses 6:1273–1279. [DOI] [PubMed] [Google Scholar]
- 35.Salminen MO, Koch C, Sanders-Buell E, Ehrenberg PK, Michael NL, Carr JK, Burke DS, McCutchan FE. 1995. Recovery of virtually full-length HIV-1 provirus of diverse subtypes from primary virus cultures using the polymerase chain reaction. Virology 213:80–86. doi: 10.1006/viro.1995.1548. [DOI] [PubMed] [Google Scholar]
- 36.Fehse B, Kustikova OS, Bubenheim M, Baum C. 2004. Pois(s)on—it's a question of dose. Gene Ther 11:879–881. doi: 10.1038/sj.gt.3302270. [DOI] [PubMed] [Google Scholar]
- 37.De Luca A, Teofili L, Antinori A, Iovino MS, Mencarini P, Visconti E, Tamburrini E, Leone G, Ortona L. 1993. Haemopoietic CD34+ progenitor cells are not infected by HIV-1 in vivo but show impaired clonogenesis. Br J Haematol 85:20–24. doi: 10.1111/j.1365-2141.1993.tb08640.x. [DOI] [PubMed] [Google Scholar]
- 38.Milich L, Margolin B, Swanstrom R. 1993. V3 loop of the human immunodeficiency virus type 1 Env protein: interpreting sequence variability. J Virol 67:5623–5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biffi A, Montini E, Lorioli L, Cesani M, Fumagalli F, Plati T, Baldoli C, Martino S, Calabria A, Canale S, Benedicenti F, Vallanti G, Biasco L, Leo S, Kabbara N, Zanetti G, Rizzo WB, Mehta NA, Cicalese MP, Casiraghi M, Boelens JJ, Del Carro U, Dow DJ, Schmidt M, Assanelli A, Neduva V, Di Serio C, Stupka E, Gardner J, von Kalle C, Bordignon C, Ciceri F, Rovelli A, Roncarolo MG, Aiuti A, Sessa M, Naldini L. 2013. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science 341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- 40.Aiuti A, Biasco L, Scaramuzza S, Ferrua F, Cicalese MP, Baricordi C, Dionisio F, Calabria A, Giannelli S, Castiello MC, Bosticardo M, Evangelio C, Assanelli A, Casiraghi M, Di Nunzio S, Callegaro L, Benati C, Rizzardi P, Pellin D, Di Serio C, Schmidt M, Von Kalle C, Gardner J, Mehta N, Neduva V, Dow DJ, Galy A, Miniero R, Finocchi A, Metin A, Banerjee PP, Orange JS, Galimberti S, Valsecchi MG, Biffi A, Montini E, Villa A, Ciceri F, Roncarolo MG, Naldini L. 2013. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science 341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGarrity GJ, Hoyah G, Winemiller A, Andre K, Stein D, Blick G, Greenberg RN, Kinder C, Zolopa A, Binder-Scholl G, Tebas P, June CH, Humeau LM, Rebello T. 2013. Patient monitoring and follow-up in lentiviral clinical trials. J Gene Med 15:78–82. doi: 10.1002/jgm.2691. [DOI] [PubMed] [Google Scholar]
- 42.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. 2006. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 43.An DS, Qin FX, Auyeung VC, Mao SH, Kung SK, Baltimore D, Chen IS. 2006. Optimization and functional effects of stable short hairpin RNA expression in primary human lymphocytes via lentiviral vectors. Mol Ther 14:494–504. doi: 10.1016/j.ymthe.2006.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X, Chang WC, Wong CW, Colcher D, Sherman M, Ostberg JR, Forman SJ, Riddell SR, Jensen MC. 2011. A transgene-encoded cell surface polypeptide for selection, in vivo tracking, and ablation of engineered cells. Blood 118:1255–1263. doi: 10.1182/blood-2011-02-337360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dupré L, Trifari S, Follenzi A, Marangoni F, Lain de Lera T, Bernad A, Martino S, Tsuchiya S, Bordignon C, Naldini L, Aiuti A, Roncarolo MG. 2004. Lentiviral vector-mediated gene transfer in T cells from Wiskott-Aldrich syndrome patients leads to functional correction. Mol Ther 10:903–915. doi: 10.1016/j.ymthe.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 46.Phaltane R, Lachmann N, Brennig S, Ackermann M, Modlich U, Moritz T. 2014. Lentiviral MGMT(P140K)-mediated in vivo selection employing a ubiquitous chromatin opening element (A2UCOE) linked to a cellular promoter. Biomaterials 35:7204–7213. doi: 10.1016/j.biomaterials.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 47.Jonnalagadda M, Brown CE, Chang WC, Ostberg JR, Forman SJ, Jensen MC. 2013. Engineering human T cells for resistance to methotrexate and mycophenolate mofetil as an in vivo cell selection strategy. PLoS One 8:e65519. doi: 10.1371/journal.pone.0065519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muñoz-Calleja C, Costantini A, Silvestri G, Butini L, Regnery CM, Mancini S, Montroni M. 2001. Highly active antiretroviral therapy induces specific changes in effector and central memory T cell sub-populations. AIDS 15:1887–1890. doi: 10.1097/00002030-200109280-00022. [DOI] [PubMed] [Google Scholar]
- 49.Baenziger S, Tussiwand R, Schlaepfer E, Mazzucchelli L, Heikenwalder M, Kurrer MO, Behnke S, Frey J, Oxenius A, Joller H, Aguzzi A, Manz MG, Speck RF. 2006. Disseminated and sustained HIV infection in CD34+ cord blood cell-transplanted Rag2−/−γc−/− mice. Proc Natl Acad Sci U S A 103:15951–15956. doi: 10.1073/pnas.0604493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ince WL, Zhang L, Jiang Q, Arrildt K, Su L, Swanstrom R. 2010. Evolution of the HIV-1 env gene in the Rag2−/− γC−/− humanized mouse model. J Virol 84:2740–2752. doi: 10.1128/JVI.02180-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Herrera-Carrillo E, Liu YP, Berkhout B. 2014. The impact of unprotected T cells in RNAi-based gene therapy for HIV-AIDS. Mol Ther 22:596–606. doi: 10.1038/mt.2013.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boutimah F, Eekels JJ, Liu YP, Berkhout B. 2013. Antiviral strategies combining antiretroviral drugs with RNAi-mediated attack on HIV-1 and cellular co-factors. Antiviral Res 98:121–129. doi: 10.1016/j.antiviral.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 53.Horwitz JA, Halper-Stromberg A, Mouquet H, Gitlin AD, Tretiakova A, Eisenreich TR, Malbec M, Gravemann S, Billerbeck E, Dorner M, Buning H, Schwartz O, Knops E, Kaiser R, Seaman MS, Wilson JM, Rice CM, Ploss A, Bjorkman PJ, Klein F, Nussenzweig MC. 2013. HIV-1 suppression and durable control by combining single broadly neutralizing antibodies and antiretroviral drugs in humanized mice. Proc Natl Acad Sci U S A 110:16538–16543. doi: 10.1073/pnas.1315295110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palmer BE, Neff CP, Lecureux J, Ehler A, Dsouza M, Remling-Mulder L, Korman AJ, Fontenot AP, Akkina R. 2013. In vivo blockade of the PD-1 receptor suppresses HIV-1 viral loads and improves CD4+ T cell levels in humanized mice. J Immunol 190:211–219. doi: 10.4049/jimmunol.1201108. [DOI] [PMC free article] [PubMed] [Google Scholar]