LETTER
Modifications of RNA sequences by nucleotide insertions, deletions, or substitutions can result in the expression of multiple proteins in overlapping open reading frames (ORFs). In the case of viruses, polymerase slippage results in the alteration of newly synthesized RNA. The mechanism has been well characterized in animal RNA viruses such as Ebolavirus (1) (EBOV) or Hepatitis C virus (HCV) (2). For plant viruses of the Potyviridae family, polymerase slippage has been proposed as a general process of evolution (3), although a lack of experimental systems has precluded confirmatory data, and most pieces of evidence are indirect (4).
Translation of a large ORF that results in a polyprotein, later processed into mature factors, is the canonical strategy of potyviral protein production. Along with this, in all members of this family, an overlapping ORF named PIPO was identified in the middle of the P3 coding region. The translation of PIPO begins at a specific GA6 motif (5). Interestingly, GA6 and other An motifs (n = ≥6) are misrepresented among members of the Potyviridae family (1.2 A6 motifs in the coding region per viral genome versus the expected 8.1 motifs). This additional ORF of potyviruses produces a P3N-PIPO fusion protein, which was originally identified in Turnip mosaic virus (5) and was shown to be essential for cell-to-cell movement during viral infection (6). Recently, another extra ORF located downstream of a GA6 motif was informatically identified inside the large P1 coding region of sweet-potato-infecting potyviruses (7, 8). This new ORF, named PISPO, harbors the possibility of producing a frameshifted P1N-PISPO gene product, whose existence is still to be determined.
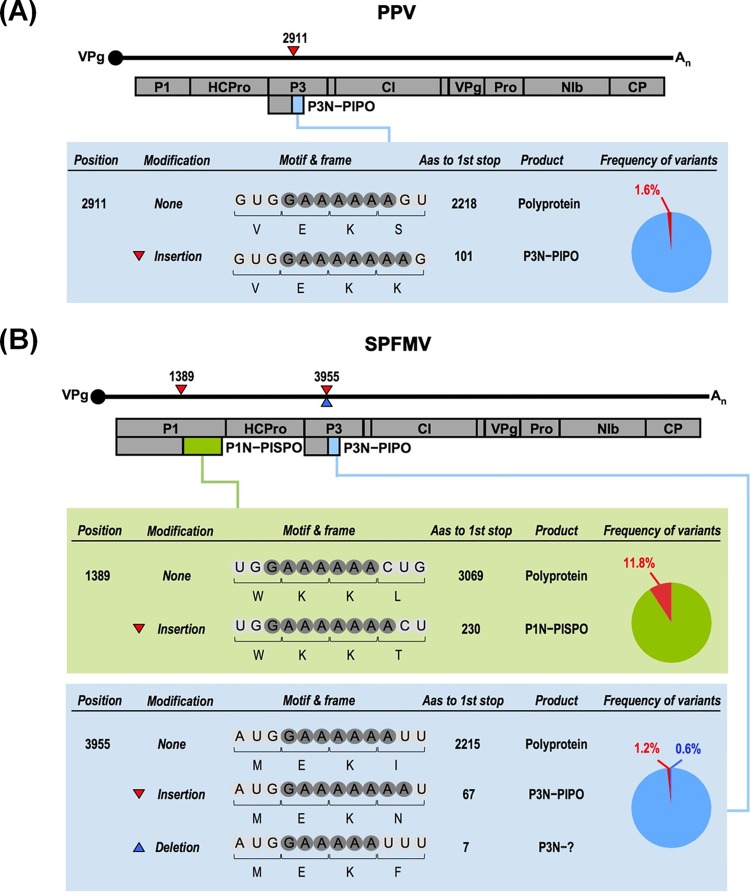
To explore the mechanism by which these additional potyviral proteins can be synthesized, we analyzed available RNA sequencing (RNA-seq) data of two Plum pox virus (PPV) isolates (9). After data filtering (10, 11), sequences were mapped versus the references (12) allowing a maximum of three mismatches per read. The expected indel error was modeled as a Poisson distribution calculating λ from the Illumina indel calling error rate, PCR error rate, and sample indel frequency. This analysis revealed the presence of A residue additions in the PIPO GA6 motif in 1.6% of the reads (Fig. 1A). Interestingly, the presence of an additional A residue in this motif was also detected in libraries of PPV-derived small RNAs (not shown). Besides, published data on another potyvirus, Zucchini yellow mosaic virus, showed a minor variant with an extra A in all samples of a Cucurbita pepo vine studied by deep sequencing of long RNAs (13), and our analysis located this modification in the PIPO GA6 motif as well.
FIG 1.
RNA slippage in viruses of the Potyvirus genus. The genomes of the potyviruses PPV (A) and SPFMV (B) are depicted schematically. Mature gene products are shown as boxes. Additional ORFs corresponding to out-of-frame PIPO and PISPO regions are also depicted. Sites where indels were detected after RNA-seq analysis are shown with red (insertion) or blue (deletion) triangles. Details of the motif, the resulting frame for the modifications, and the length of the expected products, as well as the RNA slippage percentages, are indicated for each modification. Color codes in the pie charts refer to insertions and deletions compared to the genomic sequence.
To assess the scope among potyviruses of the extra A at the PIPO junction, we subjected a sample of sweet potato (Ipomea batatas) infected with the potyvirus Sweet potato feathery mottle virus (SPFMV) to RNA-seq analysis. SPFMV reconstruction (SRR1693230 and SRR1693363) showed that there were 1.8% sequence variants in this PIPO ORF, with the insertion of an A residue as the most prominent modification (Fig. 1B).
Altogether, these data strongly suggest that P3N-PIPO is produced, at least partially, through polymerase slippage. This possibility, previously considered by Chung et al. (5), could not be demonstrated at that time, likely because of the low rate of nucleotide insertion into this site.
Reconstruction of the SPFMV genome confirmed the previously described PISPO ORF imbedded in the P1 coding sequence. But more importantly, the RNA-seq data also revealed the presence of a high proportion of molecules (11.8%) with a single A nucleotide addition in the upstream GA6 motif, which is indicative of polymerase slippage (Fig. 1B). This change would result in the production of the hypothetical P1N-PISPO, and these results not only support the existence of this alternative product but also suggest that this protein might play an important role during sweet potato potyvirus infection.
Considering the evolutionary relatedness of polymerases of the members of the families Picornaviridae and Potyviridae (14), it is reasonable to envision similar behaviors in both viral families and, consistent with that idea, the six-adenine repetition motif is underrepresented in picornaviruses (0.67 motif in the coding region per viral genome versus the expected 1.9 motifs), as was the case in the family Potyviridae. There is no previous report of polymerase slippage in the Picornaviridae family; nonetheless, when an A6 motif was present, as in the case of the enterovirus Human rhinovirus C (SRR363436), there were 2.4% A residue insertions at this location. These data suggest that polymerase slippage can occur in both Picorna-like families but that Potyviridae take more frequent advantage of this mechanism.
A common denominator in RNA slippage is the low fidelity of viral RNA polymerases and their tendency to stutter when encountering repetitive motifs. It is known that polymerases of EBOV, HCV, Vaccinia virus, or T7 bacteriophage, given the appropriate contexts, are prone to slippage (1, 2, 15). Repetitive motifs, however, are not the rule but the exception, probably because of selective negative pressure supported by nonsense-mediated decay (16) or other mechanisms. Nonetheless, in certain situations, slippage of the polymerase would give rise to the production of new protein variants that are used by the virus, opening the door to new ways of adaptation and evolution.
ACKNOWLEDGMENTS
We are grateful to Beatriz García for technical assistance and to the Computational Systems Biology Group of CNB-CSIC for supplying analytical resources.
D.B. is the Royal Society Edward Penley Abraham Research Professor. This work was supported by Spanish Ministerio de Economía y Competitividad grants AGL2013-42537-R to J.J.L.-M. and BIO2013-49053-R and Plant KBBE PCIN-2013-056 to J.A.G.
REFERENCES
- 1.Volchkov VE, Becker S, Volchkova VA, Ternovoj VA, Kotov AN, Netesov SV, Klenk HD. 1995. GP mRNA of Ebola virus is edited by the Ebola virus polymerase and by T7 and vaccinia virus polymerases. Virology 214:421–430. doi: 10.1006/viro.1995.0052. [DOI] [PubMed] [Google Scholar]
- 2.Ratinier M, Boulant S, Combet C, Targett-Adams P, McLauchlan J, Lavergne JP. 2008. Transcriptional slippage prompts recoding in alternate reading frames in the hepatitis C virus (HCV) core sequence from strain HCV-1. J Gen Virol 89:1569–1578. doi: 10.1099/vir.0.83614-0. [DOI] [PubMed] [Google Scholar]
- 3.Hancock JM, Chaleeprom W, Dale J, Gibbs A. 1995. Replication slippage in the evolution of potyviruses. J Gen Virol 76:3229–3232. doi: 10.1099/0022-1317-76-12-3229. [DOI] [PubMed] [Google Scholar]
- 4.Simón-Buela L, Osaba L, García JA, López-Moya JJ. 2000. Preservation of 5′-end integrity of a potyvirus genomic RNA is not dependent on template specificity. Virology 269:377–382. doi: 10.1006/viro.2000.0229. [DOI] [PubMed] [Google Scholar]
- 5.Chung BY, Miller WA, Atkins JF, Firth AE. 2008. An overlapping essential gene in the Potyviridae. Proc Natl Acad Sci U S A 105:5897–5902. doi: 10.1073/pnas.0800468105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei T, Zhang C, Hong J, Xiong R, Kasschau KD, Zhou X, Carrington JC, Wang A. 2010. Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N-PIPO. PLoS Pathog 6:e1000962. doi: 10.1371/journal.ppat.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark CA, Abad JA, Cuellar WJ, Fuentes S, Kreuze JF, Gibson RW, Mukasa SB, Tugume AK, Tairo FD, Valkonen JP. 2012. Sweetpotato viruses: 15 years of progress on understanding and managing complex diseases. Plant Dis 96:168–185. doi: 10.1094/PDIS-07-11-0550. [DOI] [PubMed] [Google Scholar]
- 8.Li F, Xu D, Abad J, Li R. 2012. Phylogenetic relationships of closely related potyviruses infecting sweet potato determined by genomic characterization of Sweet potato virus G and Sweet potato virus 2. Virus Genes 45:118–125. doi: 10.1007/s11262-012-0749-2. [DOI] [PubMed] [Google Scholar]
- 9.Rodamilans B, San León D, Mühlberger L, Candresse T, Neumüller M, Oliveros JC, García JA. 2014. Transcriptomic analysis of Prunus domestica undergoing hypersensitive response to Plum pox virus infection. PLoS One 9:e100477. doi: 10.1371/journal.pone.0100477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.HannonLab. 2014. FASTX toolkit. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: http://hannonlab.cshl.edu/fastx_toolkit/index.html. [Google Scholar]
- 11.Babraham-Bioinformatics. 2014. FASTQC, a quality tool for high throughput sequence data. Babraham Institute, Babraham, United Kingdom: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. [Google Scholar]
- 12.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunham JP, Simmons HE, Holmes EC, Stephenson AG. 2014. Analysis of viral (zucchini yellow mosaic virus) genetic diversity during systemic movement through a Cucurbita pepo vine. Virus Res 191:172–179. doi: 10.1016/j.virusres.2014.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koonin EV, Dolja VV. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol 28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- 15.Shabman RS, Jabado OJ, Mire CE, Stockwell TB, Edwards M, Mahajan M, Geisbert TW, Basler CF. 2014. Deep sequencing identifies noncanonical editing of Ebola and Marburg virus RNAs in infected cells. mBio 5:e02011. doi: 10.1128/mBio.02011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia D, Garcia S, Voinnet O. 2014. Nonsense-mediated decay serves as a general viral restriction mechanism in plants. Cell Host Microbe 16:391–402. doi: 10.1016/j.chom.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]