ABSTRACT
It has been proposed that viral cell-to-cell transmission plays a role in establishing and maintaining chronic infections. Thus, understanding the mechanisms and kinetics of cell-to-cell spread is fundamental to elucidating the dynamics of infection and may provide insight into factors that determine chronicity. Because hepatitis C virus (HCV) spreads from cell to cell and has a chronicity rate of up to 80% in exposed individuals, we examined the dynamics of HCV cell-to-cell spread in vitro and quantified the effect of inhibiting individual host factors. Using a multidisciplinary approach, we performed HCV spread assays and assessed the appropriateness of different stochastic models for describing HCV focus expansion. To evaluate the effect of blocking specific host cell factors on HCV cell-to-cell transmission, assays were performed in the presence of blocking antibodies and/or small-molecule inhibitors targeting different cellular HCV entry factors. In all experiments, HCV-positive cells were identified by immunohistochemical staining and the number of HCV-positive cells per focus was assessed to determine focus size. We found that HCV focus expansion can best be explained by mathematical models assuming focus size-dependent growth. Consistent with previous reports suggesting that some factors impact HCV cell-to-cell spread to different extents, modeling results estimate a hierarchy of efficacies for blocking HCV cell-to-cell spread when targeting different host factors (e.g., CLDN1 > NPC1L1 > TfR1). This approach can be adapted to describe focus expansion dynamics under a variety of experimental conditions as a means to quantify cell-to-cell transmission and assess the impact of cellular factors, viral factors, and antivirals.
IMPORTANCE The ability of viruses to efficiently spread by direct cell-to-cell transmission is thought to play an important role in the establishment and maintenance of viral persistence. As such, elucidating the dynamics of cell-to-cell spread and quantifying the effect of blocking the factors involved has important implications for the design of potent antiviral strategies and controlling viral escape. Mathematical modeling has been widely used to understand HCV infection dynamics and treatment response; however, these models typically assume only cell-free virus infection mechanisms. Here, we used stochastic models describing focus expansion as a means to understand and quantify the dynamics of HCV cell-to-cell spread in vitro and determined the degree to which cell-to-cell spread is reduced when individual HCV entry factors are blocked. The results demonstrate the ability of this approach to recapitulate and quantify cell-to-cell transmission, as well as the impact of specific factors and potential antivirals.
INTRODUCTION
Viral entry into permissive cells is the first step in establishing infection and is thus a common and often effective target for antiviral therapy. However, after replication and assembly of viral particles in an infected cell, many viruses, including hepatitis C virus (HCV), can spread to infect additional cells by two routes of transmission: cell-free and cell-to-cell spread (1, 2). Although viral dissemination is facilitated by the release of “cell-free” virus from infected cells, which can then travel throughout the body to enter distant cells, direct cell-to-cell transmission allows the virus to spread to neighboring cells while remaining shielded from host neutralizing antibodies and other extracellular viral clearance mechanisms (3, 4). It is the latter advantage that implicates cell-to-cell spread in the establishment and maintenance of persistent infections and therapy failure (3, 5, 6).
Different mechanisms of direct viral cell-to-cell transmission have been described (reviewed in references 1 and 2). In cell culture, cell-to-cell spread of HIV-1 has been found to be much more efficient than cell-free virus uptake, with estimates ranging between 10-fold- and 18,000-fold-higher efficiency in mediating viral spread (7–11). However, while HIV-1 cell-to-cell transmission is more efficient, it appears to be mediated by the same factors that are involved in cell-free virus entry, as it has been found to be equally sensitive to inhibition by the same fusion inhibitors and receptor-blocking antibodies that inhibit cell-free virus entry (8). Consistent with cell-to-cell spread being efficient, cell-to-cell transmission has been implicated in mediating HCV persistence (12–14) and possibly playing a role in viral escape during therapy by allowing the spread of viral variants resistant to direct-acting antivirals (15). For HCV infection, cell-to-cell transmission has been studied in vitro using HCV focus spread assays in which cell-free virus uptake is blocked by neutralizing antibodies against the HCV E2 glycoprotein (6, 13, 16, 17). Based on these in vitro studies, it appears that the factors that mediate HCV cell-free viral spread do not completely overlap those involved in cell-to-cell spread, and thus some inhibitors block cell-free entry but not cell-to-cell spread (6, 16). However, the dynamics of cell-to-cell spread has not been determined and the involvement of different HCV entry factors on the kinetics of cell-to-cell spread has not been quantified.
Mathematical modeling has been widely used to understand viral infection dynamics and treatment response; however, these models typically assume only cell-free virus infection mechanisms (18–23). In vitro mathematical models were developed to understand intracellular HCV RNA kinetics during infection and treatment (24–26) and cell-free HCV entry (27). Models accounting for the dynamics of cell-to-cell viral spread have been developed for HIV-1 (3, 28–31), as well as for oncolytic virotherapy (32). However, there are no mathematical models specifically addressing the kinetics of HCV cell-to-cell spread in vitro.
In this study, we performed experiments to monitor the kinetics of HCV cell-to-cell spread and the effect of blocking different HCV entry factors on those kinetics. To quantify and provide insight into the dynamics of cell-to-cell spread, we developed mathematical models that describe focus growth assuming different modes of expansion and that account for stochastic variability in the initiation and the growth rates of individual foci. Fitting our models to the observed experimental data, we determined that models assuming size-dependent focus growth best recapitulate the data. To quantify the effect of blocking different HCV entry factors, such as the tetraspanin CD81, claudin-1 (CLDN1), the Niemann-Pick C1-like 1 receptor (NPC1L1), and transferrin receptor 1 (TfR1), on focus expansion, we performed experiments in the presence of blocking antibodies and small-molecule inhibitors. Consistent with previous reports suggesting differential requirements for individual cellular factors in HCV cell-to-cell spread, blocking CLDN1 and NPC1L1 potently reduced focus expansion, while blocking TfR1 and CD81 resulted in more intermediate effects. The results demonstrate the ability of this approach to recapitulate and quantify cell-to-cell transmission, as well as the impact of specific factors and potential antivirals.
MATERIALS AND METHODS
Cells.
Huh7 cells (33) (also known as Huh7/scr cells [34, 35] and Huh7-1 cells [36]) were obtained from Francis Chisari (The Scripps Research Institute, La Jolla, CA). Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Mediatech) supplemented with 10% fetal bovine serum (FBS) (HyClone, Logan, UT), 100 units/ml penicillin, 100 mg/ml streptomycin, and 2 mM l-glutamine (Gibco Invitrogen, Carlsbad, CA).
Virus.
Plasmid DNA containing the full-length HCV JFH-1 genome (pUC-JFH1) was provided by Takaji Wakita (National Institute of Infectious Diseases, Tokyo, Japan) (37–39). Methods for HCV RNA in vitro transcription, HCV RNA electroporation, as well as propagation and titration of HCV cell culture (HCVcc) stocks have been described in detail elsewhere (40).
Reagents and antibodies.
Dimethyl sulfoxide (DMSO) stocks of ezetimibe (Sequoia Research Products) and the TfR1 inhibitor ferristatin (NSC306711; National Cancer Institute) were diluted into culture medium to the indicated concentrations. Rabbit anti-human CLDN1 polyclonal antibody was obtained from Abcam (Cambridge, MA). Rabbit anti-human NPC1L1 polyclonal antibody was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). TfR1 monoclonal antibody (clone M-A712) was obtained from BD Pharmingen (San Jose, CA). Mouse anti-human CD81 monoclonal antibody (clone 1D6) was purchased from AbD Serotec (Raleigh, NC). Inhibition of all these HCV entry factors by antibodies/inhibitors has been previously reported by us or others to block HCV cell-free entry without affecting HCV RNA replication (6, 16, 41, 42). The human anti-HCV E2 monoclonal antibody (MAb) AR3A (or C1), obtained from Dennis Burton and Mansun Law (The Scripps Research Institute, La Jolla, CA), was described previously (33, 43, 44). Horseradish peroxidase (HRP)-conjugated anti-human, anti-mouse, and anti-rabbit secondary antibodies were purchased from Pierce (Rockford, IL). Negative-control IgG antibodies were obtained from Santa Cruz Biotechnology.
Cell-to-cell spread “focus size” assay.
Confluent monolayers of Huh7 cells were infected with 50 focus-forming units (FFU) of HCVcc. After 17 h of incubation, the viral inoculum was removed, cells were rinsed with phosphate-buffered saline (PBS), and medium containing 10 μg/ml anti-E2 was added to neutralize cell-free virus infection as previously determined (6, 13, 16). Additional indicated treatments were performed in parallel with anti-E2 treatment for the duration of the experiment to assess the effect of blocking different host factors on HCV cell-to-cell spread. For antibody inhibition experiments, all blocking antibodies were used at 25 μg/ml in a total volume of 100 μl, a concentration that ensured that the maximal effect of each antibody was achieved. Triplicate wells were fixed and stained to detect HCV-positive foci. The number of HCV-positive cells per focus was counted as a readout of HCV cell-to-cell spread. To determine the degree of cell division during the assay period, cell counts from equivalently infected and treated triplicate wells were performed at the time of infection and 72 h postinoculation at the time of fixing.
Immunostaining of HCV positive foci.
Immunostaining of HCV-positive cells was described previously (40). Briefly, endogenous peroxidases were blocked by incubating fixed cells in 1× PBS containing 0.3% (vol/vol) hydrogen peroxide (Fisher, Fairlawn, NJ). Following three washes with 1 × PBS, cells were permeabilized and blocked for 1 h with 1× PBS containing 0.5% (vol/vol) Triton X-100 (Fisher), 3% (wt/vol) bovine serum albumin (BSA) (Sigma), and 10% (vol/vol) FBS. The primary human HCV E2 MAb AR3A was diluted to 2.3 μg/ml in 1× PBS containing 0.5% (vol/vol) Triton X-100 and 3% (wt/vol) BSA and incubated with cells for 1 h at room temperature. Bound anti-E2 was subsequently detected with a 1:1,000 dilution of an HRP-conjugated anti-human antibody (Pierce) followed by a 30-min incubation with a 3-amino-9-ethyl-carbazole (AEC) detection substrate (BD Biosciences). Cells were washed with distilled water (dH2O), and foci were quantified and photographed using a Nikon TE2000U microscope (Nikon Instruments).
Mathematical models of HCV focus expansion.
We assume that each focus of HCV-infected cells is founded by a single infected cell that became infected at time t0. The size of each focus, measured in number of cells [N(t)], changes over time according to a stochastic model representing the propagation of infection to other cells: the probability of a cluster of size k existing at time t to expand to a cluster of size k + 1, with k = 1,2,…, in a small interval of time h given that the first cell became infected at time t0, is denoted by
| 1 |
Here, ρk represents the growth rate of the cluster from size k to k + 1. The first cell of each focus is assumed to have become infected in a time window of 0 to 17 h postinoculation before anti-E2 and individual antibody/inhibitor treatments were applied; i.e., t0 is between 0 and 17 h. The initiation of infection could have occurred at any point during this time interval. Thus, the time of infection, t0, is assumed to follow a probability distribution truncated on the time interval from 0 to 17 h. The actual distribution for t0 with density function f(t) (see equation 8) is inferred from our experimental data (45) as described below. Based on the lack of any observed cell death, we also assume that no cell death occurs during the 72-h assay.
The transition probabilities described in equation 1 lead to the following equations for the conditional state probabilities pk(t|s) = P[N(t) = k|s]:
| 2 |
where pk, k = 1,2, … is shorthand for the probability that a focus contains k cells at time t given that the focus was founded at time s, pk(t|s). We assume that each focus is founded by one infected cell at time s, which is equivalent to the initial conditions p1(s|s) = 1 and pk(s|s) = 0, k > 1. For all t<s, pk(t|s) = 0, k = 1,2,…., as no cell is infected before time s. The system of equation 2 can be solved once ρk is specified (see below) to determine the probability generating function for the conditional state probabilities pk(t|s) (46). The probability that we would observe a cluster of size k at a time t after anti-E2 treatment (t > 17 h), given that the first cell of the focus became infected at time s in the time interval from 0 to 17 h postinoculation, can then be calculated by integrating over all possible times at which a focus could have been initiated, i.e.,
| 3 |
With this, the probability of observing, for example, a focus of size 12 or larger is then given by
As stated earlier, each focus is assumed to be established by a single cell that became infected by an HCV particle administered to the culture at time zero. Because there is an eclipse phase of ∼ 24 h (42) between the time a cell becomes infected and the time progeny virus are secreted into the medium and we removed the original viral inoculum when neutralizing anti-E2 was added to the culture at 17 h postinoculation, we assume that all virus transmission in a specific focus is due to cell-to-cell transmission of progeny virus from the founding cell. We then made the following different assumptions about the growth rate, ρk, with each corresponding to different assumed focus growth dynamics.
(i) Poisson process.
In the simplest scenario, we assume a constant growth rate ρ independent of the size of the focus, hence, ρk = ρ, k = 1,2, …. In this case, foci grow according to a Poisson process, where the time between infection events is exponentially distributed with parameter ρ (46). In this scenario, the probability of observing a cluster of size k at time t > 17 h (equation 2) is given by
| 4 |
This formulation represents focus growth dynamics that is not dependent on the limitations that would be inherent in a growth process solely depending on cell-to-cell transmission, especially for larger focus sizes.
(ii) Birth process.
A second possible assumption would be a birth process where each infected cell in a focus can give rise to another infected cell with a constant rate ρ, i.e., ρk = kρ (46). In this case, the probability of observing a cluster of size k at time t is given by
| 5 |
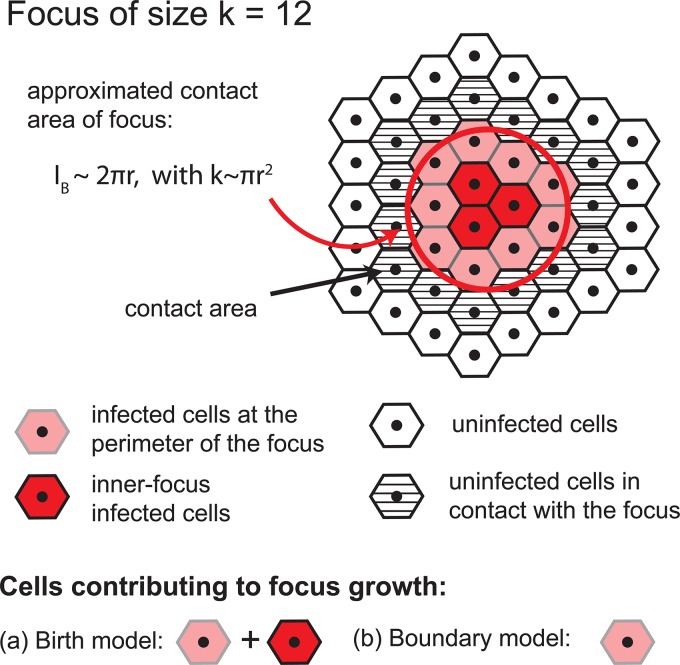
Under this assumption, at later times even when infected cells are completely surrounded by other infected cells (termed inner focus-infected cells), these cells would still contribute to the expansion of the focus (Fig. 1).
FIG 1.
Depiction of cells that contribute to focus expansion in the birth and boundary models. Shown is a sample 12-cell focus. In the birth model, all HCV-infected cells of a focus contribute to focus expansion (i.e., red and light red cells); in the boundary model, only cells at the perimeter of a focus can mediate focus growth (i.e., only light red cells). Of note, the boundary model assumes a compact focus shape, i.e., infected cells are approximately distributed in a circle.
(iii) Boundary process.
A third possible assumption is a boundary expansion process, where only infected cells at the perimeter of the focus contribute to focus expansion (Fig. 1). Thus, the growth rate of a focus of size k, ρk, depends on the number of infected cells at the edge of the focus that are in direct contact with uninfected hepatocytes, IB. This size of the focus boundary and, thus, the growth rate vary with increasing cluster size. Considering compact focus expansion, cluster growth can be assumed to occur radially. A focus comprising k infected cells would cover an area (A) of πr2, where r is the radius of the cluster, and hence, k ∼ πr2. Analogously, we can approximate the boundary cells by IB ∼ 2πr. As only cells at the edge of the cluster would contribute to focus growth, the growth rate per infected cell of the total cluster is given by IB ρ0/k, where ρ0 defines the basic infection rate per cell. The growth rate of a focus of size k is then defined by
| 6 |
with . Defining , i.e., , we can calculate the probability of observing a focus of size k at time t given that the focus was established at time t0 by
| 7 |
with ak,j defined by
The detailed derivations and mathematical proofs for the state probabilities in equation 7 are given in the Appendix. Using equation 7 in equation 3, we can derive the probability of observing a focus of size k at the end of the experiment, i.e., t = 72 h.
Distribution for the time to infection of the first cell in a focus, t0.
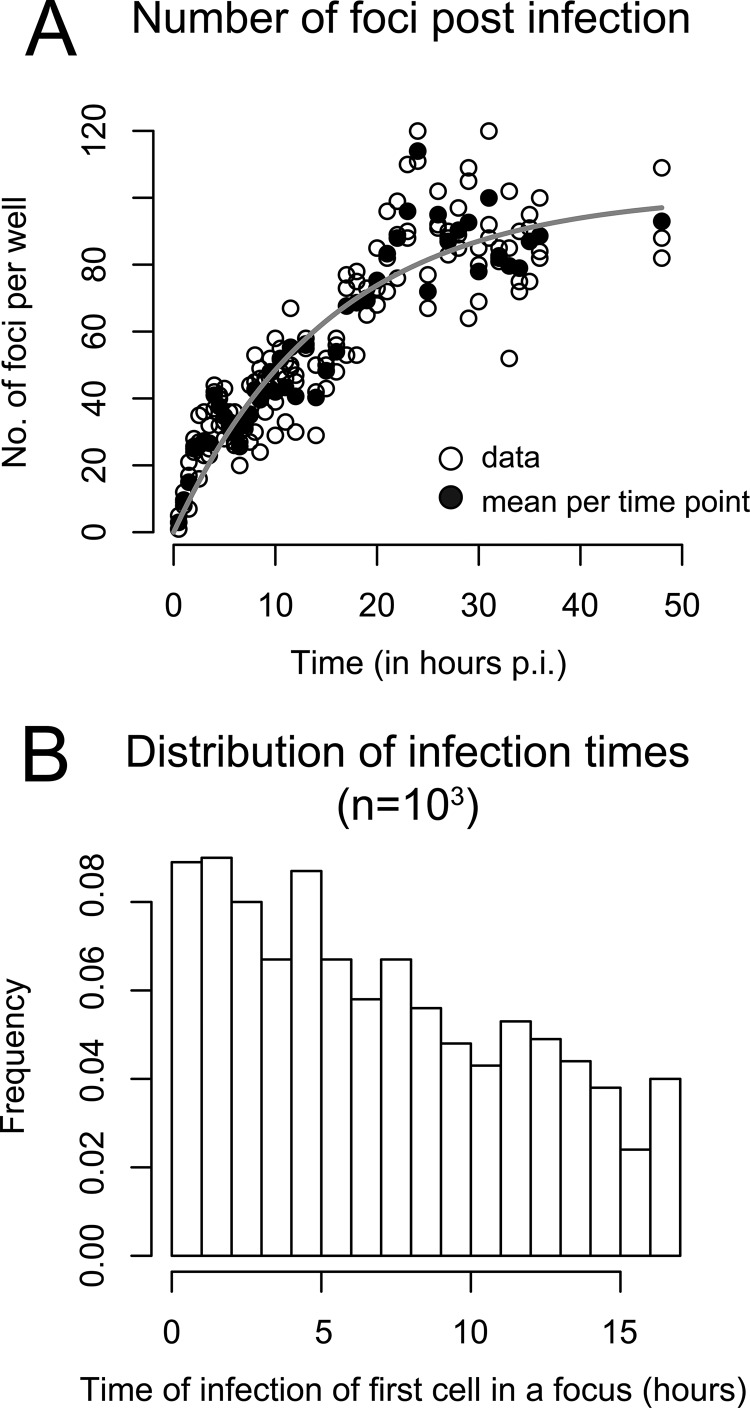
We previously demonstrated that the rate of HCVcc infection initiation after viral inoculation is not constant but rather decreases over time (see Fig. 1 in reference 45). If the infection rate was constant, then the number of foci would increase linearly over time. However, as shown in reference 45, the number of foci increases more slowly than linearly. To match the data, we assume that the infection rate λ0 decreases exponentially over time with a loss rate α; hence, λ(t) = λ0e-αt. This is equivalent to assuming that virions lose infectivity at rate α. Fitting λ(t) to the data, we estimated λ0 and α to determine the distribution of the time to infection of the first cell of a focus. The data and the best fit of λ(t) using our derived estimates of α = 0.065 h−1 and λ0 = 6.59 foci per hour are shown in Fig. 2A. A possible distribution of t0 at which the first cell of a focus becomes infected after inoculation of the culture with HCVcc and before antibody treatment, i.e., t0 ∈ [0, 17 h], is shown in Fig. 2B.
FIG 2.
Time of infection of the first cell in a focus. (A) Analysis of the experimental data from reference 45 indicates that the number of foci per well depends on the time postinoculation (p.i.). Open circles indicate numbers of foci counted in triplicate wells. Filled circles are means for each time point. The gray curve shows the fit of a time-dependent infection rate, λ(t), to the data, assuming an exponential loss of infectivity over time, λ(t) = λ0e−αt. (B) Distribution of the time of first cell infection in a focus in the period from 0 to 17 h. The plot shows the histogram for 1,000 random draws.
The corresponding density function for the time to infection of the founding cell of a focus, f(t), truncated on the time interval from 0 to 17 h is then defined by
| 8 |
The definition of f(t) as given in equation 8 is then used in equation 3 when the focus growth rate ρ is estimated. Assuming that the infection rate decays in a multiphasic manner, as observed in reference 45, which leads to a biphasic infection rate on the interval from 0 to 17 h, does not lead to a significant change in our results (data not shown).
Data fitting.
We fitted equation 3 with the different scenarios for the focus expansion rate, ρk, given by equations 4 to 7, to the data using a maximum likelihood approach. The log likelihood of observing n different foci in the culture at 72 h, with each focus comprising ki, i = 1,…,n infected cells, is then denoted by
| 9 |
Equation 9 was maximized to estimate ρ using the fitting routine optim in the R language of statistical computing (47).
RESULTS
HCV focus expansion is best described by mathematical models assuming focus size-dependent growth rates.
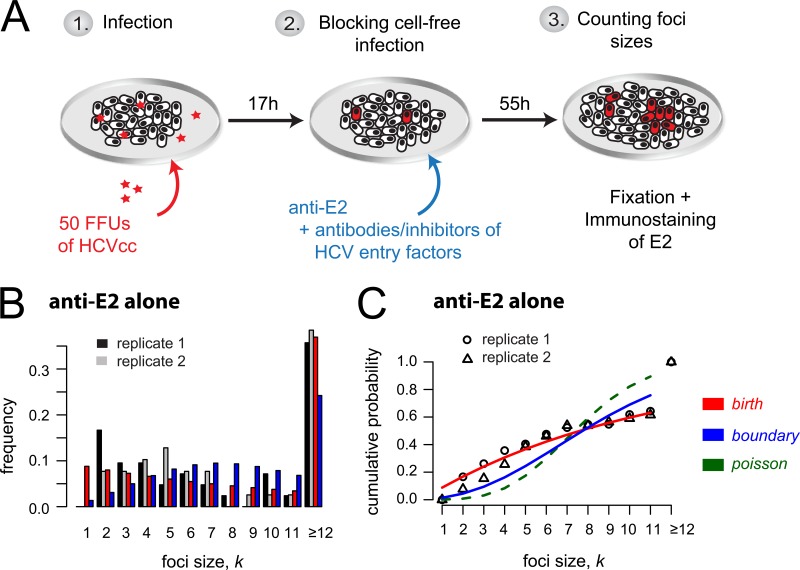
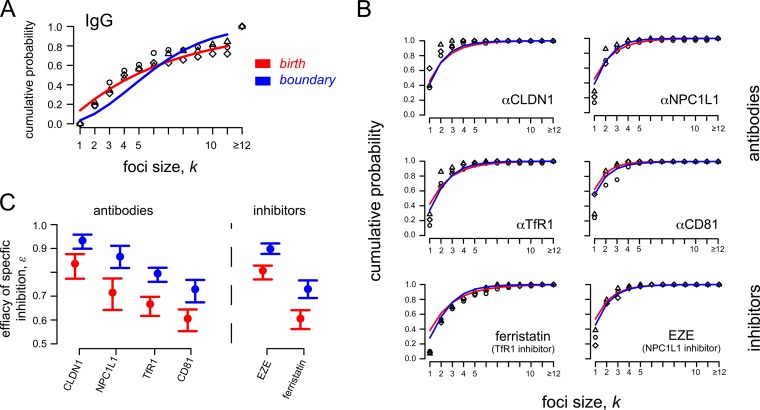
In order to monitor the kinetics of HCV cell-to-cell spread, we performed focus formation assays in which a neutralizing antibody to the HCV E2 glycoprotein is used to block cell-free virus infection such that focus expansion is dependent on cell-to-cell viral transmission (6, 13, 16, 17). Specifically, as depicted in Fig. 3A, confluent Huh7 cell cultures were inoculated with 50 FFU of HCVcc for 17 h to allow time for ∼80% viral entry into ∼40 individual single cells (45). Cells were then washed, and cell-free virus spread was blocked by the addition of anti-E2 at a concentration that neutralizes extracellular virus infection of cells (6, 13, 16). At 72 h postinoculation, cell monolayers were fixed and stained for HCV-positive cells. The extent of cell-to-cell spread was assessed by counting the number of HCV-positive cells per focus. Across experiments, under these conditions (i.e., anti-E2-only treatment), we observed an average of 41 ± 2 (standard deviation [SD]) individual foci per well, with ∼35% of the foci consisting of 12 or more infected cells (Fig. 3B).
FIG 3.
HCV cell-to-cell spread assays with observed and predicted focus size distribution. (A) Sketch of the experimental protocol. (B) Distribution of focus sizes at 72 h postinoculation, when cell-free virus is neutralized by anti-E2 treatment. Foci comprising 12 or more cells were combined. Observed data from two replicate wells (black and gray) compared to model predictions (red, birth model; blue, boundary model) are shown. Results are representative of over 5 experiments in which the anti-E2-alone condition was included. (C) Agreement of model fits (lines) and data (symbols) can best be determined when looking at the cumulative probability of observing a focus that comprises up to k infected cells. The birth model (red line) provides the best fit to the data.
Despite the fact that the assay was performed in confluent monolayers, limited cell division still occurred under these experimental conditions, and thus, net focus expansion could be affected not only by cell-to-cell virus transmission but also by proliferation of infected cells. Hence, in parallel equivalently treated wells, we counted the number of cells present at the time of infection and at 72 h postinoculation, when the assay ended, to estimate the number of cell divisions that had occurred. Though the precise cell count varied between experiments, we observed an average ∼5.3-fold (± 0.7-fold) increase in the number of cells during the 72-h experiment in untreated cultures and in those treated with anti-E2 alone, indicating that approximately 2 to 2.5 cell divisions had occurred. As such, cell division alone could in theory result in foci containing up to 4 to 6 cells; however, because HCV-infected cells divide more slowly than noninfected cells (48), we assume that cell proliferation could account for foci containing up to 4 cells.
To elucidate the dynamics of focus expansion observed and more accurately quantify the kinetics of spread, we fitted three mathematical models to the observed data that describe focus expansion as a stochastic process according to different assumptions (see Materials and Methods). The different models explicitly account for the fact that individual foci are initiated at different time points during the 17 h before anti-E2 is added to the culture (Fig. 2). The Poisson model, which assumes that foci grow at a constant rate independent of focus size, ρk = ρ, did not fit the observed data well (Fig. 3C). In contrast, the birth and boundary models, which are based on the assumption that the rate of focus expansion depends on the current size of the focus, led to better fits of the experimental data (Fig. 3B and C). Ruling out the Poisson model is consistent with the anti-E2 treatment inhibiting cell-free virus infection, confirming that the focus expansion observed was mediated solely by cell-to-cell transmission and potentially proliferation of infected cells.
With a model assuming that individual foci grow according to a birth process where each infected cell of a focus can contribute to focus expansion, ρk = kρ, we estimate a focus expansion rate of ρ = 0.038 h−1 [0.034, 0.041] (Table 1). This model provides a good explanation of the experimental data; however, it overestimates the frequency of foci of size k = 1 (Fig. 3B and C). As a modification of the birth model, the boundary model assumes that focus expansion is mainly driven by infected cells at the boundary of the focus. While both models fit the data reasonably well, the boundary model overestimates intermediate focus sizes (Fig. 3B) and provides a slightly impaired fit to the data compared to the birth model as measured by the Akaike information criterion (AIC), where a lower AIC value indicates a better fit of the model (49) (corrected AIC for the birth model, 348.9; that for the boundary model, 369.7). The boundary model leads to estimates of the focus expansion rate that are roughly twice as high as those for the birth model, ρ = 0.067 h−1 [0.062, 0.073] (Table 1). Because the birth and the boundary models represent the two extreme scenarios of size-dependent focus growth, we proceeded under the assumption that the actual growth rate lies between the values estimated by these two models.
TABLE 1.
Estimated HCV focus expansion rates using different mathematical modelsa
| Treatment | Parameter | Value with model |
||
|---|---|---|---|---|
| Poisson | Birth | Boundary | ||
| Anti-E2 | ρ (10−2 per hour) | 10.83 (9.79, 12.13) | 3.76 (3.49, 4.02) | 6.70 (6.22, 7.27) |
| LL | −224.5 | −173.4 | −183.8 | |
| AIC | 453.2 | 348.9 | 369.7 | |
| Anti-E2 + IgG | ρ (10−2 per hour) | 7.91 (7.10, 8.66) | 3.07 (2.84, 3.27) | 5.24 (4.85, 5.69) |
| LL | −276.6 | −222.4 | −233.2 | |
| AIC | 557.4 | 446.8 | 468.5 | |
Focus expansion rates during anti-E2 treatment alone and with additional IgG control treatment were estimated assuming constant focus expansion, ρk = ρ (Poisson), or size-dependent focus expansion, ρk = kρ (birth) or ρk = ρ (boundary) after 17 h p.i. Triplicates of each condition were combined in the fitting procedure. Numbers in parentheses are the 10% and 90% percentiles over 200 bootstrap replicates of the data. The maximized value of the log likelihood function (equation 8) (LL) determines the quality of the fit. The higher the LL, the better the model explanation of the data. In addition, the corrected Akaike information criterion (AIC) for model comparison is shown, indicating a consistent improper fit of the Poisson model. Analysis of one representative experiment of 5 is shown.
Quantification of focus expansion when blocking specific host factors with antibodies.
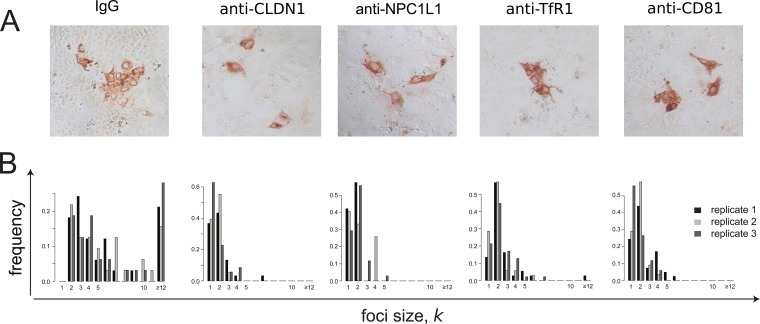
It has been previously reported that blocking host factors, such as NPC1L1 (6), CLDN1 (4, 13), TfR1 (16), and possibly CD81 (4, 13, 14, 50–52), inhibits HCV cell-to-cell spread. In order to quantify the effect of blocking these host cell factors on HCV cell-to-cell spread, we performed the same focus cell-to-cell spread assay described above, but cultures were additionally cotreated with antibodies that block individual HCV entry factors, in particular, NPC1L1, CLDN1, TfR1, and CD81. As a negative control, parallel cultures were cotreated with nonspecific IgG. At 72 h postinoculation, cell monolayers were fixed and stained for HCV E2 (Fig. 4A), so that the number of cells per focus could be counted (Fig. 4B). Again, in parallel infected and equivalently treated wells, we counted the number of cells present at the time of infection and at 72 h postinoculation, when the assay ended, to estimate the number of cell divisions that occurred under each treatment condition. In contrast to the average ∼5.3-fold ± 0.7-fold increase in cell number observed in the untreated and anti-E2-alone-treated cultures, cell numbers increased only ∼2-fold ± 0.4-fold during the 72-h experiment in cultures treated with any of the other antibodies or small-molecule inhibitors (Table 2). Thus, in this case, foci comprising 2 cells could be due to cell division alone. Under all of these conditions, we observed on average 37 ± 7 individual foci per well. However, foci in cultures treated with anti-NPC1L1 or anti-CLDN1 had an average focus size of only 1 or 2 cells with a maximum of 5 or 7 cells, respectively, consistent with high levels of inhibition of cell-to-cell spread.
FIG 4.
Blocking different cellular HCV entry factors exhibits differential effects on HCV focus expansion. HCV cell-to-cell spread assays were performed in the presence of neutralizing anti-E2 plus IgG, anti-CLDN1, anti-NPC1L1, anti-TfR1, or anti-CD81. (A) Representative images of foci formed 72 h postinoculation. HCV-infected cells were stained for E2 (red). (B) Distribution of focus sizes at 72 h postinoculation. For each condition, 3 replicate wells were analyzed. Results of one representative experiment of three are shown.
TABLE 2.
Proliferation of cells during the in vitro focus assaya
| Treatment | Fold increase (mean ± SD) | Average fold increase |
|---|---|---|
| None | 5.4 ± 1.0 | Untreated, 5.3 ± 0.7 |
| Anti-E2 | 5.1 | |
| IgG | 2.4 ± 1.0 | Specifically treated, 2.0 ± 0.4 |
| Anti-CLDN1 | 1.4 ± 0.4 | |
| Anti-NPC1L1 | 1.7 ± 0.4 | |
| Anti-TfR1 | 1.9 ± 0.2 | |
| Anti-CD81 | 1.9 ± 0.1 | |
| EZE | 1.9 ± 0.7 | |
| Ferristatin | 2.7 ± 0.2 |
During each experiment, cell number was counted in equivalently treated cell cultures at the time of infection and at 72 h p.i. The averages for triplicate wells from one representative experiment of three are shown.
Since the appropriateness of the Poisson model was ruled out above because it generated poor fits to the observed data, we quantified the effect of the various antibodies on HCV focus expansion rates using the birth and the boundary models. Both models indicate that cotreatment of cultures with control IgG led to an ∼20% nonspecific reduction of focus expansion rates (Table 1), which is at least in part due to the reduced cell proliferation observed during treatment with the IgG (Table 2). To control for this reduced cell proliferation and estimate the effect of blocking the targeted host factors, in these assays we defined the focus expansion rate by ρk = (1 − ε)ρkIgG, k = 1,…,n, where ρkIgG defines the baseline expansion rate calculated from the IgG control (Table 1). The parameter ε represents the degree to which focus expansion was reduced in the presence of the different antibody treatments, with ε = 0 indicating no specific effect and ε = 1 defining 100% inhibition of focus growth. Model fits are shown in Fig. 5A and B. Both models indicate a similar hierarchy of efficacies for reducing HCV focus expansion when these different host factors are targeted (Table 3 and Fig. 5C). Blocking CLDN1 with antibodies showed the largest reduction of focus growth of ∼ 93% for boundary (∼84% for birth). Antibody blocking of NPC1L1 exhibited a slightly less potent reduction of ∼87% (∼72%). Antibodies against TfR1 resulted in even less inhibition of ∼80% (∼67%), and antibodies against CD81 resulted in the least inhibition, ∼77% (∼64%). As the amount of cell proliferation is comparable among the different treatment regimens (Table 2), the estimated efficacy ε mainly represents the effect of the treatments on cell-to-cell transmission rates.
FIG 5.
Observed and predicted focus size distributions under different treatment conditions. HCV cell-to-cell spread assays were performed in the presence of neutralizing anti-E2 plus antibodies (anti-CLDN1, anti-NPC1L1, anti-TfR1, and anti-CD81) or small-molecule inhibitors (ezetimibe [EZE] and ferristatin) against specific HCV entry factors. The birth (red) and boundary (blue) models were used to analyze the data. (A) Data and model predictions of foci in control cultures treated with anti-E2 plus IgG. (B) Data and model predictions of focus sizes in cultures treated with antibodies or small-molecule inhibitors blocking specific HCV entry factors. (C) Both the birth and boundary models indicate the same hierarchy in the estimated focus growth reduction when the different HCV entry factors are blocked. Circles and bars indicate the estimated effectiveness of blocking specific host factors (ε) and the 95% confidence interval, respectively (see Table 3).
TABLE 3.
Effect of blocking host factors on HCV focus expansiona
| Factor | Birth model |
Boundary model |
||||
|---|---|---|---|---|---|---|
| ε (%) | LL | AIC | ε (%) | LL | AIC | |
| CLDN1 | 83.6 (77.3, 87.6) | −119.2 | 240.3 | 93.3 (89.9, 95.8) | −117.7 | 237.4 |
| NPC1L1 | 71.5 (64.2, 77.4) | −96.7 | 195.5 | 86.6 (81.8, 91.1) | −94.5 | 191.1 |
| TfR1 | 66.7 (61.7, 69.7) | −188.4 | 378.9 | 79.5 (76.0, 81.9) | −178.2 | 358.5 |
| CD81 | 60.6 (55.3, 64.4) | −176.7 | 355.5 | 72.9 (67.4, 76.8) | −169.3 | 340.7 |
| EZE | 80.7 (77.0, 82.8) | −271.3 | 339.6 | 89.8 (87.7, 92.1) | −256.2 | 329.3 |
| Ferristatin | 60.6 (56.2, 64.1) | −168.8 | 544.6 | 73.0 (69.2, 76.6) | −163.6 | 514.3 |
The estimated effectiveness (ε) of host antibodies and small inhibitors in blocking focus expansion using the birth and boundary models is shown. Triplicates of each treatment were combined in the fitting procedure. Numbers in parentheses are the 10% and 90% percentiles over 200 bootstrap replicates of the data. LL and AIC parameters are as described in Table 1. Analysis of one representative experiment of 3 is shown.
Quantification of focus expansion when specific host factors are blocked with small-molecule inhibitors.
To confirm the above antibody results, we performed the same focus assay but used an alternative blocking strategy utilizing the small-molecule inhibitors ezetimibe and ferristatin, which specifically inhibit NPC1L1 and TfR1, respectively. In this case, estimated focus expansion rates with anti-E2 treatment alone, ρkanti-E2, were considered the baseline expansion rates, and the focus expansion rate was defined as ρk = (1 − ε)ρkanti-E2. Importantly, similar to what was observed with anti-NPC1L1, ezetimibe blocking of NPC1L1 led to a similar reduction in focus growth (∼90% [∼81%]). Likewise, the inhibition of focus growth under ferristatin treatment was comparable to the efficacy of anti-TfR1, with an ∼73% (∼61%) reduction in focus growth (Fig. 5B and Table 3). Of note, although the effects measured were similar to that observed in response to antibody blocking of these receptors, the anti-E2-only control cultures exhibited more cell proliferation during the assay, and thus, the model estimates of focus growth inhibition by these small-molecule inhibitors are influenced to some extent by inhibition of cell proliferation (Table 2).
Modeling predicts that the growth of larger foci is best explained by the birth model, while the dynamics of smaller foci could be better described by the boundary model.
We observed that some HCV foci appeared to grow in a more irregular (i.e., noncircular) pattern (Fig. 6). In these cases, the boundary model, which assumes a circular focus shape, would underestimate the number of infected cells that contribute to focus growth. In order to better define the differences in how these two models explain the observed focus size distributions, we performed a detailed comparison of the birth and the boundary models using the current 72-h assay data. Initial analysis suggested that the birth model provides a better fit to data when larger focus sizes are observed (e.g., for anti-E2-alone treatment, AICbirth = 348.9 and AICboundary = 369.7 [Table 1]), while the boundary model performs better for scenarios with smaller average focus sizes. We tested this observation by fitting the data to a model with a growth rate defined by ρk = kαρ, where the exponent 0 ≤ α ≤ 1 defines the dependence of ρ on focus size. For α = 1, the model would be equivalent to the birth model, while for α = 0.5, the relationship is equivalent to the one assumed by the boundary model. Fitting such a model to the observed data and estimating ρ and α, we estimated α = 1 for anti-E2 alone and anti-E2 + IgG with a confidence interval of 0.92 to 1.0 based on 10% and 90% percentiles over 200 bootstrap replicates of the data. With CLDN1 and NPC1L1 inhibitors, which led to smallest focus sizes, we estimated α ≈ 0.5 for CLDN1 (α = 0.43) and NPC1L1 (α = 0.45), but with large confidence intervals ranging from 0.0 to 1.0. These results corroborate our initial observations, i.e., that scenarios leading to larger focus sizes during the 72-h experiment are best explained by a birth model, while the dynamics of smaller focus sizes seems to be better described by the boundary model.
FIG 6.
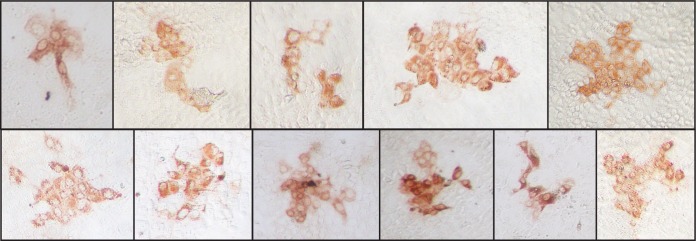
Irregular shape of HCV foci. HCV cell-to-cell spread assays were performed in the present of neutralizing anti-E2. At 72 h postinoculation, cells were fixed and stained for E2 (red). Photographs were taken at a magnification of ×20 using a Nikon TE2000U microscope. Representative noncircular images are shown.
DISCUSSION
Understanding the mechanisms and dynamics by which viruses spread within a host has important implications for pathogenesis, viral escape, and the design of potent antiviral therapies (6, 12, 15, 53). Hence, in this study, we analyzed data from in vitro HCV cell-to-cell spread assays using simple stochastic models describing focus expansion as a means to understand and quantify the dynamics of HCV cell-to-cell spread and the degree to which spread is reduced when individual HCV entry factors are blocked. Specifically, we tested three different mathematical models and found that focus expansion in the presence of neutralizing anti-E2 is best explained by models based on size-dependent growth rates (Fig. 3). Cell-free infection should lead to focus growth rates that are independent of focus size. Because the Poisson model, which assumes size-independent focus growth rates, does not explain the observed focus size distribution, our analysis supports the empirical determination that 10 μg/ml of AR3A anti-E2 is effective at blocking cell-free HCV infection and thus confirms that the focus expansion observed in our assay is due to cell-to-cell spread of HCV and/or proliferation of infected cells.
Since our data indicate that cell proliferation in anti-E2 treated cultures can increase cell numbers 5.3-fold ± 0.7-fold (Table 2), proliferation of infected cells can in theory account for foci up to size k = 4 to 6. However, most foci formed under anti-E2 treatment alone are larger than k = 4, suggesting that cell-to-cell transmission is the main mechanism of the focus expansion observed under these assay conditions. The birth and the boundary models tested represent two extremes of size-dependent focus growth, with the birth model assuming that every cell in a focus contributes to expansion while the boundary model only allows cells at the edge of a focus to contribute. Despite this difference, both models fit the current data reasonably well. This might be because most cells in the small foci formed during the short (72-h) experiment are in contact with uninfected cells for the majority of the assay period.
Interestingly, the boundary model provided a better fit to experimental data where there were smaller average focus sizes, while scenarios with the growth of larger foci were better described by the birth model. Two possible explanations for this observation can be suggested. (i) The boundary model assumes compact focus growth, with the surface of a focus of size k approximated by a circle. Because some HCV foci appear to grow in a more irregular (i.e., noncircular) pattern (Fig. 6), this might underestimate the number of cells that contribute to focus expansion. While this would not affect the analysis of relatively small foci (e.g., k < 4 cells), where all cells are by default adjacent to uninfected cells, as observed for CLDN1 and NPC1L1, it might play a role as foci become larger. (ii) Although most of the cell proliferation detected likely occurred early in the assay before cells became increasingly compressed, the relative contribution of cell proliferation on focus expansion might increase proportionally to focus size, which would more easily violate the assumption that foci expand only by cell-to-cell transmission along the boundaries of a focus. Additionally, our data indicate that cell proliferation is significantly larger in untreated cultures (Table 1), suggesting that cell proliferation may perhaps have played a more significant role on the larger foci formed in these untreated cultures. Further experimental and modeling efforts are needed to estimate the effect of cell-to-cell spread in conjunction with cell proliferation, not only accounting for focus size but also controlling for observed focus shapes (Fig. 6) and possibly decreased proliferation rates of infected cells (54).
The actual influence of cell proliferation on the observed focus sizes is difficult to quantify in these assays. For cultures cotreated with anti-E2 plus antibodies against NPC1L1 or CLDN1, we observed average focus sizes of only 1 or 2 infected cells. In addition, we found that incubation with any nonhuman antibody reduced cell proliferation slightly, such that only a roughly 2-fold increase in cell numbers occurred during the 72 h experiment (Table 2). This seems to indicate that inhibition of CLDN1 or NPC1L1 blocks the majority of cell-to-cell spread, with the observed focus sizes being solely due to proliferation of infected cells. Performing the same HCV cell-to-cell spread assays in a nongrowing Huh7 cell culture model (40, 55, 56) could be useful to eliminate the influence of cell proliferation on focus size.
One factor not considered in our analysis is the possibility that two or more individual foci may merge at some point during the experiment, which might happen if closely situated foci grow large enough, as previously analyzed (32). However, because the average numbers of foci per well are comparable between the different conditions tested (anti-E2 and IgG, ∼36 foci per well; all other scenarios, ∼38 foci per well; Wilcoxon test, P = 0.5), we can assume that this did not affect our comparisons of the efficacies of different antibody treatments on cell-to-cell spread. For longer experiments monitoring more extensive focus expansion, even less virus per square millimeter of cell monolayer could be inoculated to allow more room for individual foci to grow. Additionally, live cell imaging and/or counting the number of foci at subsequent time points over the course of the experiment would help to determine if merging of individual foci occurs (e.g., focus number would decrease over time).
Despite the difficulties and limitations of mathematical models in capturing the dynamics of individual focus growth, estimates of focus growth rates obtained by the birth and the boundary models describing size-dependent focus growth represent the potential upper and lower limits of the actual growth rates, respectively. Importantly, when various inhibitors targeting different host cell factors are assessed, both the birth and the boundary models indicate the same hierarchy of cell-to-cell inhibition, with blocking of CLDN1 reducing focus expansion the most, followed by NPC1L1, TfR1, and finally CD81 (Fig. 5C). As foci up to a size of k = 2 cells could be solely due to cell proliferation (Table 2), antibodies against CLDN1 and NPC1L1 may in fact block all cell-to-cell transmission of infection, given the observed focus sizes in these scenarios. Blocking TfR1 either with an antibody or a small-molecule inhibitor does not exhibit as great a reduction of focus expansion, consistent with TfR1 being less essential for cell-to-cell transmission of HCV. Likewise, our anti-CD81 data suggest that CD81 is not absolutely required for HCV cell-to-cell spread; however, further analysis with additional antibodies or other types of CD81-targeted inhibitors is needed to resolve the ongoing debate (6, 13, 14, 16) regarding the involvement of CD81 in HCV cell-to-cell transmission.
In summary, in the current study we developed stochastic models to explain the dynamics of HCV spread in an in vitro focus expansion assay and to quantify the effect that blocking specific host factors has on cell-to-cell spread. This mathematical approach confirms that our anti-E2 treatment blocks cell-free infection and indicates a hierarchy of effectiveness of the tested entry factor-targeted inhibitors in slowing focus expansion/cell-to-cell viral spread. This type of approach could be used to quantify in vitro cell-to-cell transmission of other viruses and to determine the effectiveness of antivirals on blocking viral cell-to-cell spread.
APPENDIX
Detailed derivation and calculation of the state probabilities for the boundary model.
Here, we prove the state probability of observing a focus of size k at time t for the boundary model as defined in equation 7. For simplicity, we assume that the first cell of the focus was infected at time t0 = 0. Given ρ1 = ρ, the probability of observing a focus of size k = 1 at time t, p1(t), is given by
| A1 |
As given in equation 7, the general probability of observing a focus of size k at time t, pk(t), k ≥ 2, is then defined by
| A2 |
To prove equation A2, we can use the following relationship between pk(t) and pk + 1(t). To observe a focus of size k + 1 at time t, the focus needs to have expanded to a size k by time s < t, one additional cell must become infected at time s with rate ρk, and there must be no further expansion of the focus until time t. Thus, the probability of observing a focus of size k + 1 at time t, pk + 1(t), is defined by
| A3 |
The proof that equation A2 holds true for all k ≤ N follows by induction. With equation A3 and equation A1, it is easy to show that equation A2 holds for k = 2. For k + 1, we then obtain
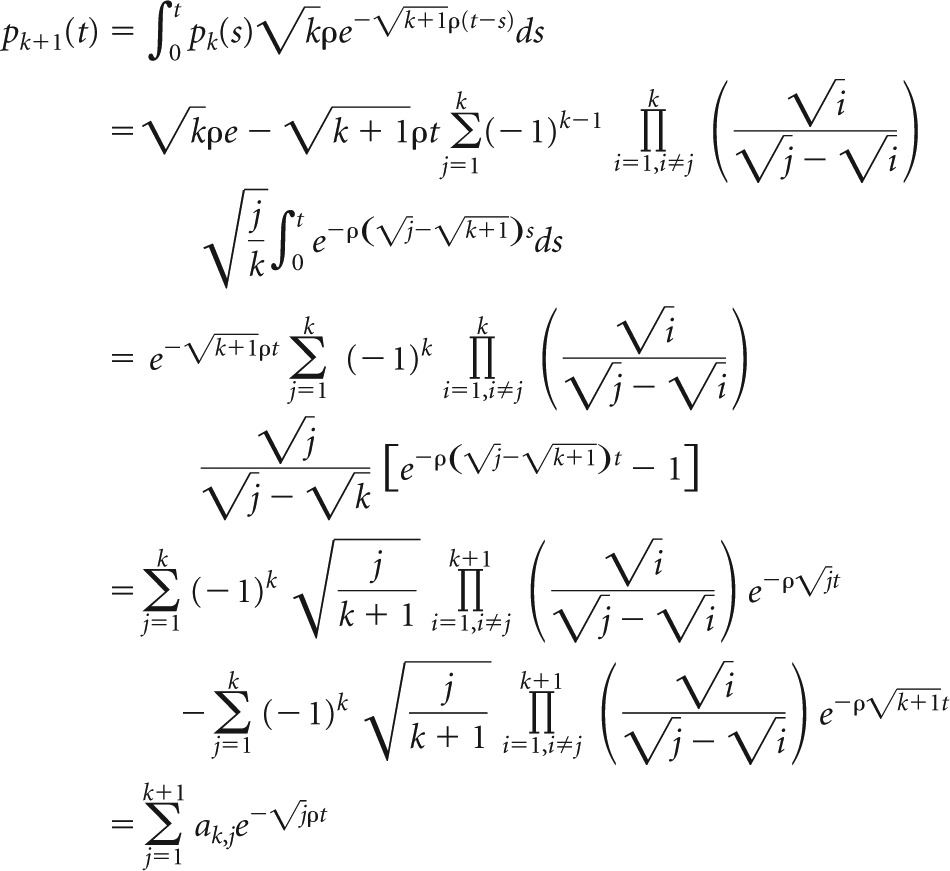 |
Thereby, the last equation follows, as
With this, equation 7 has been shown.
ACKNOWLEDGMENTS
We thank Gitanjali Subramanya and Beverly Chrabot for excellent technical support.
F.G. was supported by the Center for Modeling and Simulation in the Biosciences (BIOMS). Portions of this work were performed under the auspices of the U.S. Department of Energy under contract DE-AC52-06NA25396 and supported by NIH grant R01-AI07881.
REFERENCES
- 1.Sattentau Q. 2008. Avoiding the void: cell-to-cell spread of human viruses. Nat Rev Microbiol 6:815–826. doi: 10.1038/nrmicro1972. [DOI] [PubMed] [Google Scholar]
- 2.Mothes W, Sherer NM, Jin J, Zhong P. 2010. Virus cell-to-cell transmission. J Virol 84:8360–8368. doi: 10.1128/JVI.00443-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sigal A, Kim JT, Balazs AB, Dekel E, Mayo A, Milo R, Baltimore D. 2011. Cell-to-cell spread of HIV permits ongoing replication despite antiretroviral therapy. Nature 477:95–98. doi: 10.1038/nature10347. [DOI] [PubMed] [Google Scholar]
- 4.Brimacombe CL, Grove J, Meredith LW, Hu K, Syder AJ, Flores MV, Timpe JM, Krieger SE, Baumert TF, Tellinghuisen TL, Wong-Staal F, Balfe P, McKeating JA. 2011. Neutralizing antibody-resistant hepatitis C virus cell-to-cell transmission. J Virol 85:596–605. doi: 10.1128/JVI.01592-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abela IA, Berlinger L, Schanz M, Reynell L, Gunthard HF, Rusert P, Trkola A. 2012. Cell-cell transmission enables HIV-1 to evade inhibition by potent CD4bs directed antibodies. PLoS Pathog 8:e1002634. doi: 10.1371/journal.ppat.1002634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barretto N, Sainz B Jr, Hussain S, Uprichard SL. 2014. Determining the involvement and therapeutic implications of host cellular factors in hepatitis C virus cell-to-cell spread. J Virol 88:5050–5061. doi: 10.1128/JVI.03241-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen P, Hubner W, Spinelli MA, Chen BK. 2007. Predominant mode of human immunodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. J Virol 81:12582–12595. doi: 10.1128/JVI.00381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin N, Welsch S, Jolly C, Briggs JA, Vaux D, Sattentau QJ. 2010. Virological synapse-mediated spread of human immunodeficiency virus type 1 between T cells is sensitive to entry inhibition. J Virol 84:3516–3527. doi: 10.1128/JVI.02651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sato H, Orenstein J, Dimitrov D, Martin M. 1992. Cell-to-cell spread of HIV-1 occurs within minutes and may not involve the participation of virus particles. Virology 186:712–724. doi: 10.1016/0042-6822(92)90038-Q. [DOI] [PubMed] [Google Scholar]
- 10.Sourisseau M, Sol-Foulon N, Porrot F, Blanchet F, Schwartz O. 2007. Inefficient human immunodeficiency virus replication in mobile lymphocytes. J Virol 81:1000–1012. doi: 10.1128/JVI.01629-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolodkin-Gal D, Hulot SL, Korioth-Schmitz B, Gombos RB, Zheng Y, Owuor J, Lifton MA, Ayeni C, Najarian RM, Yeh WW, Asmal M, Zamir G, Letvin NL. 2013. Efficiency of cell-free and cell-associated virus in mucosal transmission of human immunodeficiency virus type 1 and simian immunodeficiency virus. J Virol 87:13589–13597. doi: 10.1128/JVI.03108-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeisel MB, Lupberger J, Fofana I, Baumert TF. 2013. Host-targeting agents for prevention and treatment of chronic hepatitis C—perspectives and challenges. J Hepatol 58:375–384. doi: 10.1016/j.jhep.2012.09.022. [DOI] [PubMed] [Google Scholar]
- 13.Timpe JM, Stamataki Z, Jennings A, Hu K, Farquhar MJ, Harris HJ, Schwarz A, Desombere I, Roels GL, Balfe P, McKeating JA. 2008. Hepatitis C virus cell-cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology 47:17–24. doi: 10.1002/hep.21959. [DOI] [PubMed] [Google Scholar]
- 14.Witteveldt J, Evans MJ, Bitzegeio J, Koutsoudakis G, Owsianka AM, Angus AG, Keck ZY, Foung SK, Pietschmann T, Rice CM, Patel AH. 2009. CD81 is dispensable for hepatitis C virus cell-to-cell transmission in hepatoma cells. J Gen Virol 90:48–58. doi: 10.1099/vir.0.006700-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao F, Fofana I, Heydmann L, Barth H, Soulier E, Habersetzer F, Doffoel M, Bukh J, Patel AH, Zeisel MB, Baumert TF. 2014. Hepatitis C virus cell-cell transmission and resistance to direct-acting antiviral agents. PLoS Pathog 10:e1004128. doi: 10.1371/journal.ppat.1004128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin DN, Uprichard SL. 2013. Identification of transferrin receptor 1 as a hepatitis C virus entry factor. Proc Natl Acad Sci U S A 110:10777–10782. doi: 10.1073/pnas.1301764110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barretto N, Uprichard SL. 2015. Hepatitis C virus cell-to-cell spread assays. Bio Protoc 4:e1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perelson AS. 2002. Modelling viral and immune system dynamics. Nat Rev Immunol 2:28–36. doi: 10.1038/nri700. [DOI] [PubMed] [Google Scholar]
- 19.Canini L, Perelson AS. 2014. Viral kinetic modeling: state of the art. J Pharmacokinet Pharmacodyn 41:431–443. doi: 10.1007/s10928-014-9363-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guedj J, Dahari H, Rong L, Sansone ND, Nettles RE, Cotler SJ, Layden TJ, Uprichard SL, Perelson AS. 2013. Modeling shows that the NS5A inhibitor daclatasvir has two modes of action and yields a shorter estimate of the hepatitis C virus half-life. Proc Natl Acad Sci U S A 110:3991–3996. doi: 10.1073/pnas.1203110110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nowak MA, May RM. 2000. Virus dynamics: mathematical principles of immunology and virology. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 22.Dahari H, Shteingart S, Gafanovich I, Cotler SJ, D'Amato M, Pohl RT, Weiss G, Ashkenazi YJ, Tichler T, Goldin E, Lurie Y. 2015. Sustained virological response with intravenous silibinin: individualized IFN-free therapy via real-time modelling of HCV kinetics. Liver Int 35:289–294. doi: 10.1111/liv.12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neumann AU, Lam NP, Dahari H, Gretch DR, Wiley TE, Layden TJ, Perelson AS. 1998. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 282:103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 24.Dahari H, Ribeiro RM, Rice CM, Perelson AS. 2007. Mathematical modeling of subgenomic hepatitis C virus replication in Huh-7 cells. J Virol 81:750–760. doi: 10.1128/JVI.01304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dahari H, Sainz B Jr, Perelson AS, Uprichard SL. 2009. Modeling subgenomic hepatitis C virus RNA kinetics during treatment with alpha interferon. J Virol 83:6383–6390. doi: 10.1128/JVI.02612-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Binder M, Sulaimanov N, Clausznitzer D, Schulze M, Huber CM, Lenz SM, Schloder JP, Trippler M, Bartenschlager R, Lohmann V, Kaderali L. 2013. Replication vesicles are load- and choke-points in the hepatitis C virus lifecycle. PLoS Pathog 9:e1003561. doi: 10.1371/journal.ppat.1003561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Padmanabhan P, Dixit NM. 2012. Viral kinetics suggests a reconciliation of the disparate observations of the modulation of claudin-1 expression on cells exposed to hepatitis C virus. PLoS One 7:e36107. doi: 10.1371/journal.pone.0036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Culshaw RV, Ruan S, Webb G. 2003. A mathematical model of cell-to-cell spread of HIV-1 that includes a time delay. J Math Biol 46:425–444. doi: 10.1007/s00285-002-0191-5. [DOI] [PubMed] [Google Scholar]
- 29.Komarova NL, Levy DN, Wodarz D. 2013. Synaptic transmission and the susceptibility of HIV infection to anti-viral drugs. Sci Rep 3:2103. doi: 10.1038/srep02103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dixit NM, Perelson AS. 2004. Multiplicity of human immunodeficiency virus infections in lymphoid tissue. J Virol 78:8942–8945. doi: 10.1128/JVI.78.16.8942-8945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wodarz D, Levy DN. 2011. Effect of different modes of viral spread on the dynamics of multiply infected cells in human immunodeficiency virus infection. J R Soc Interface 8:289–300. doi: 10.1098/rsif.2010.0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey K, Kirk A, Naik S, Nace R, Steele MB, Suksanpaisan L, Li X, Federspiel MJ, Peng KW, Kirk D, Russell SJ. 2013. Mathematical model for radial expansion and conflation of intratumoral infectious centers predicts curative oncolytic virotherapy parameters. PLoS One 8:e73759. doi: 10.1371/journal.pone.0073759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. 2005. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A 102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong J, Gastaminza P, Chung J, Stamataki Z, Isogawa M, Cheng G, McKeating JA, Chisari FV. 2006. Persistent hepatitis C virus infection in vitro: coevolution of virus and host. J Virol 80:11082–11093. doi: 10.1128/JVI.01307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gastaminza P, Kapadia SB, Chisari FV. 2006. Differential biophysical properties of infectious intracellular and secreted hepatitis C virus particles. J Virol 80:11074–11081. doi: 10.1128/JVI.01150-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sainz B Jr, Barretto N, Uprichard SL. 2009. Hepatitis C virus infection in phenotypically distinct Huh7 cell lines. PLoS One 4:e6561. doi: 10.1371/journal.pone.0006561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kato T, Date T, Miyamoto M, Furusaka A, Tokushige K, Mizokami M, Wakita T. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808–1817. doi: 10.1053/j.gastro.2003.09.023. [DOI] [PubMed] [Google Scholar]
- 38.Kato T, Furusaka A, Miyamoto M, Date T, Yasui K, Hiramoto J, Nagayama K, Tanaka T, Wakita T. 2001. Sequence analysis of hepatitis C virus isolated from a fulminant hepatitis patient. J Med Virol 64:334–339. doi: 10.1002/jmv.1055. [DOI] [PubMed] [Google Scholar]
- 39.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu X, Uprichard SL. 2010. Cell-based hepatitis C virus infection fluorescence resonance energy transfer (FRET) assay for antiviral compound screening. >Curr Protoc Microbiol Chapter 17:Unit 17.15. doi: 10.1002/9780471729259.mc1705s18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Si Y, Liu S, Liu X, Jacobs JL, Cheng M, Niu Y, Jin Q, Wang T, Yang W. 2012. A human claudin-1-derived peptide inhibits hepatitis C virus entry. Hepatology 56:507–515. doi: 10.1002/hep.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sainz B Jr, Barretto N, Martin DN, Hiraga N, Imamura M, Hussain S, Marsh KA, Yu X, Chayama K, Alrefai WA, Uprichard SL. 2012. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat Med 18:281–285. doi: 10.1038/nm.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 44.Law M, Maruyama T, Lewis J, Giang E, Tarr AW, Stamataki Z, Gastaminza P, Chisari FV, Jones IM, Fox RI, Ball JK, McKeating JA, Kneteman NM, Burton DR. 2008. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat Med 14:25–27. doi: 10.1038/nm1698. [DOI] [PubMed] [Google Scholar]
- 45.Sabahi A, Marsh KA, Dahari H, Corcoran P, Lamora JM, Yu X, Garry RF, Uprichard SL. 2010. The rate of hepatitis C virus infection initiation in vitro is directly related to particle density. Virology 407:110–119. doi: 10.1016/j.virol.2010.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karlin S, Taylor HM. 1975. A first course in stochastic processes, 2nd ed Academic Press, New York, NY. [Google Scholar]
- 47.Team RDC. 2006. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 48.Munakata T, Nakamura M, Liang Y, Li K, Lemon SM. 2005. Down-regulation of the retinoblastoma tumor suppressor by the hepatitis C virus NS5B RNA-dependent RNA polymerase. Proc Natl Acad Sci U S A 102:18159–18164. doi: 10.1073/pnas.0505605102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akaike H. 1974. A new look at the statistical model identification. IEEE Trans Automat Contr 19:716–723. doi: 10.1109/TAC.1974.1100705. [DOI] [Google Scholar]
- 50.Jones CT, Catanese MT, Law LM, Khetani SR, Syder AJ, Ploss A, Oh TS, Schoggins JW, MacDonald MR, Bhatia SN, Rice CM. 2010. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat Biotechnol 28:167–171. doi: 10.1038/nbt.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fofana I, Xiao F, Thumann C, Turek M, Zona L, Tawar RG, Grunert F, Thompson J, Zeisel MB, Baumert TF. 2013. A novel monoclonal anti-CD81 antibody produced by genetic immunization efficiently inhibits hepatitis C virus cell-cell transmission. PLoS One 8:e64221. doi: 10.1371/journal.pone.0064221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Catanese MT, Loureiro J, Jones CT, Dorner M, von Hahn T, Rice CM. 2013. Different requirements for scavenger receptor class B type I in hepatitis C virus cell-free versus cell-to-cell transmission. J Virol 87:8282–8293. doi: 10.1128/JVI.01102-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhong P, Agosto LM, Ilinskaya A, Dorjbal B, Truong R, Derse D, Uchil PD, Heidecker G, Mothes W. 2013. Cell-to-cell transmission can overcome multiple donor and target cell barriers imposed on cell-free HIV. PLoS One 8:e53138. doi: 10.1371/journal.pone.0053138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kannan RP, Hensley LL, Evers LE, Lemon SM, McGivern DR. 2011. Hepatitis C virus infection causes cell cycle arrest at the level of initiation of mitosis. J Virol 85:7989–8001. doi: 10.1128/JVI.00280-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu X, Sainz B Jr, Uprichard SL. 2009. Development of a cell-based hepatitis C virus infection fluorescent resonance energy transfer assay for high-throughput antiviral compound screening. Antimicrob Agents Chemother 53:4311–4319. doi: 10.1128/AAC.00495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sainz B Jr, Chisari FV. 2006. Production of infectious hepatitis C virus by well-differentiated, growth-arrested human hepatoma-derived cells. J Virol 80:10253–10257. doi: 10.1128/JVI.01059-06. [DOI] [PMC free article] [PubMed] [Google Scholar]