Abstract
Fungi can shield surface pathogen-associated molecular patterns (PAMPs) for evading host immune attack. The most common and opportunistic human pathogen, Candida albicans, can shield β-(1 3)-glucan on the cell wall, one of the major PAMPs, to avoid host phagocyte Dectin-1 recognition. The way to interfere in the shielding process for more effective antifungal defense is not well established. In this study, we found that deletion of the C. albicans GPI7 gene, which was responsible for adding ethanolaminephosphate to the second mannose in glycosylphosphatidylinositol (GPI) biosynthesis, could block the attachment of most GPI-anchored cell wall proteins (GPI-CWPs) to the cell wall and subsequently unmask the concealed β-(1,3)-glucan. Neutrophils could kill the uncloaked gpi7 mutant more efficiently with an augmented respiratory burst. The gpi7 mutant also stimulated Dectin-1-dependent immune responses of macrophages, including activation of nuclear factor-κB (NF-κB) and mitogen-activated protein kinase (MAPK) pathways and secretion of specific cytokines, such as tumor necrosis factor alpha (TNF-α), interleukin 6 (IL-6), and IL-12p40. Furthermore, the gpi7 null mutant could induce an enhanced inflammatory response through promoting significant recruitment of neutrophils and monocytes and could stimulate stronger Th1 and Th17 cell responses to fungal infections in vivo. These in vivo phenotypes also were Dectin-1 dependent. Thus, we assume that GPI-CWPs are involved in the immune mechanism of C. albicans escaping from host recognition by Dectin-1. Our studies also indicate that the blockage of GPI anchor synthesis is a strategy to inhibit C. albicans evading host recognition.
INTRODUCTION
Candida albicans colonizes the skin, genital mucosa, and intestinal mucosa of healthy individuals. In immunocompromised individuals, C. albicans can disseminate into the bloodstream, causing life-threatening systemic candidiasis (1–4). Although hosts developed immune defenses aimed at pathogen clearance and blocking it from invading into deeper tissues, C. albicans also has evolved numerous efficient strategies to evade host immune attacks (5). How to disturb the immune-evading process of the fungus to prevent invasive infections remains poorly understood.
The polysaccharides on the cell wall of fungi, such as β-glucan and mannans, serve as pathogen-associated molecular patterns (PAMPs) that can be recognized by host-expressed pattern recognition receptors (PRRs), such as Toll-like receptors (TLRs), nucleotide-oligomerization domain (Nod)-like receptors (NLRs), and the recently emerging family of spleen tyrosine kinase (Syk)-coupled C-type lectin receptors (CLRs) (5). This PRR engagement by PAMPs triggers innate immune cell responses and renders antigen-presenting cells competent to prime T cells, thereby initiating adaptive immunity (6, 7).
Dectin-1, a Syk-coupled CLR expressed on myeloid cells, recognizes β-(1,3)-glucan carbohydrates on various fungi (8–12). The Y238X polymorphism in human Dectin-1 is associated with human mucosal C. albicans infection (13). However, live C. albicans, including yeast and filament forms, binds to Dectin-1 in a more restricted pattern in vitro, except in the region between the parent yeast cell and the mature bud (10). During disseminated infection, β-glucan of C. albicans was absolutely masked in earlier stages, while large percentages were exposed in later stages in a morphotype-independent fashion, with no difference in β-glucan exposure between yeast and hyphal forms (14). The shielding of β-(1,3)-glucan favors fungus escaping from recognition by Dectin-1 for survival and persistence (1). Therefore, the possibility of unmasking β-(1,3)-glucan could provide a therapeutic opportunity for fungal infection.
Cell wall proteins (CWPs) that are covalently linked to the skeletal polysaccharides also constitute the fungal cell wall. Most covalently linked CWPs of C. albicans are glycosylphosphatidylinositol (GPI)-anchored cell wall proteins (GPI-CWPs). They link to cell wall β-(1,6)-glucan through a GPI remnant, and the GPI anchor is responsible for targeting all of these proteins to the cell wall (15–17). Extensive studies previously have indicated that GPI-CWPs can contribute to cell wall integrity, promote biofilm formation, mediate adherence to host cells and abiotic medical devices, and promote invasion of epithelial layers and acquisition of iron (16, 18–23). McLellan et al. reported that a new small molecule, christened gepinacin, could inhibit GPI-CWPs in C. albicans and elicit enhanced immune responses (24). We hypothesized here that abolishing GPI-CWPs, the outermost cellular structure, could “uncloak” the cells for β-(1,3)-glucan exposure.
In the present study, we demonstrated that abolishing GPI-CWPs of C. albicans through blocking GPI anchor synthesis could expose its surface β-(1,3)-glucan. Host phagocyte Dectin-1 could recognize the uncloaked C. albicans cells and mediate effective immune responses. Our studies suggested blocking GPI anchor synthesis would be an ideal strategy to disturb C. albicans evading host Dectin-1 recognition.
MATERIALS AND METHODS
Mice.
C57BL/6 mice were obtained from Shanghai Laboratory Animal Center of the Chinese Academy of Sciences. Dectin-1-deficient mice on a C57BL/6 background were kindly provided by Gordon D. Brown (the Dectin-1-deficient mice were generated on a mixed 129/Sv × C57BL/6 genetic background in Gordon Brown's laboratory and backcrossed for nine generations on the C57BL/6 background) (25). All of the animal experiments were performed in compliance with institutional guidelines and according to the protocol approved by the Institutional Animal Use and Care Committee of Tongji University.
Antibodies.
Antibodies against Syk, phospho-Syk, phospho-ERK, p38, phospho-p38, JNK, phospho-JNK, phospho-IκBα, p65, and PCNA were purchased from Cell Signaling Technologies. Antibodies against ERK and IκBα were purchased from Santa Cruz Biotechnology. Antibodies against β-actin and hemagglutinin (HA) tag were purchased from Abmart. Alexa-488-labeled and Cy3-labeled goat anti-mouse antibodies were purchased from Life Technologies. Antibody against β-(1,3)-glucan was purchased from Biosupplies Inc. The following antibodies, along with the appropriate isotype controls, were used in flow cytometry: fluorescein isothiocyanate-conjugated 7/4 (Serotec), phycoerythrin-Cy7-conjugated anti-CD11b (clone M1/70; BioLegend), phycoerythrin-conjugated anti-Gr-1 (anti-Ly6G/C; clone RB6-8C5; BioLegend), and allophycocyanin-conjugated anti-F4/80 (clone BM8; BioLegend).
Strain growth conditions and construction.
All strains were maintained on SDA agar (1% peptone, 4% dextrose, and 1.8% agar) plates and grown in YPD broth (1% yeast extract, 2% peptone, and 2% dextrose) at 30°C. For hyphal growth, 1 × 106 C. albicans cells were cultured in RPMI 1640 medium (10.4 g RPMI 1640 [Gibco BRL], 2.0 g NaHCO3, and 34.5 g morpholinepropanesulfonic acid [Sigma], pH 7.0, in 1 liter double-distilled water sterilized by filtration) at 37°C for 4 h.
The entire coding sequence of GPI7 was deleted from strain SN152 by two-step homologous recombination using a fusion PCR-based strategy as described previously (26, 27). The GPI7-reconstituted strain (gpi7Δ/Δ::GPI7) was constructed by the SAT1 flipping method (28). Strains expressing HA tag-fused Als1p (gpi7Δ/Δ als1Δ/ALS1-HA and als1Δ/ALS1-HA) were generated in strains gpi7Δ/Δ and SN152, respectively. One allele of ALS1 was deleted according to the fusion PCR-based strategy, and a 102-bp fragment containing three repeated sequences of HA epitope tag was inserted into the ALS1 gene allele by the SAT1 flipping method.
All of the strains and the primers used for strain construction are listed in Tables S1 and S2 in the supplemental material.
Isolation and analysis of CWPs and cytosol proteins.
HF-pyridine specifically cleaves phosphodiester bonds, through which GPI-CWPs are linked to β-1,6-glucan chains (29). The GPI-CWPs were extracted from C. albicans cells by HF-pyridine (Sigma-Aldrich) as described previously (30). The cellular proteins were extracted by cell lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [PMSF], and protease inhibitors, pH 7.4) (31). Equal amount of CWPs and cytosol proteins then were used in immunoblot analysis.
GPI-CWPs are highly mannosylated in C. albicans, so we used peroxidase-linked concanavalin A (ConA; Sigma-Aldrich), which could bind to surface mannose residues, to reveal them. The CWPs and cytosol proteins were analyzed by immunoblotting with 1 μg/ml peroxidase-labeled ConA for mannosylated protein staining (32). The bands were analyzed using ImageJ analysis software.
C. albicans yeast cells expressing HA tag-fused Als1p were stained with anti-HA tag antibody followed by Alexa-488-labeled goat anti-mouse antibody, and micrograph pictures were acquired and analyzed by the LAS AF Lite program (version 2.1.1, build 4443; Leica Microsystems) (n = 100 for yeast cells).
β-Glucan exposure assays.
Exponentially growing C. albicans yeast cells were washed in PBS (for hyphal-form assays, 1 × 106 C. albicans cells were cultured in RPMI 1640 medium at 37°C for 4 h on a microscope slide in a 6-well plate) and blocked in PBS plus 2% bovine serum albumin for 1 h at room temperature. After blocking, the yeast and hyphal cells were stained with anti-β-(1,3)-glucan primary antibody overnight at 4°C on a rotator and Cy3-labeled goat anti-mouse antibody for 1 h at 30°C. The cells then were washed three times with PBS. The microscope slides which were adhered to the stained yeast or hyphae cells described above were scanned with a confocal laser scanning microscope (TCS SP5; Leica). Micrograph pictures were acquired and analyzed by the LAS AF Lite program (version 2.1.1, build 4443; Leica Microsystems) (n = 100 for yeast cells and n = 20 for hyphae).
We also quantified the exposure of β-glucan on C. albicans yeast by flow cytometry. The anti-β-(1,3)-glucan primary antibody-staining yeast cells were incubated with Alexa-488-labeled goat anti-mouse antibody for 1 h at 30°C. After being washed with PBS, the stained cells described above were fixed with 1% formaldehyde in PBS and then analyzed by flow cytometry (BD FACSVerse).
Thioglycolate-elicited peritoneal macrophage and neutrophil preparation.
C57BL/6 mice were injected intraperitoneally with 2 ml of 3% (wt/vol) thioglycolate (Merck). Peritoneal cells were collected by washing with PBS containing 0.5 mM EDTA 14 h later and 72 h later for neutrophil and macrophage isolation, respectively. The cells were cultured in RPMI 1640 plus 10% (vol/vol) heat-inactivated fetal bovine serum.
Macrophage-yeast interaction.
C. albicans yeast cells were harvested and exposed to four doses of 100,000 μJ/cm2 in a CL-1000 UV cross-linker (UVP), with agitation between each dose to treat cells evenly (33). The thioglycolate-elicited macrophages were stimulated with the UV-inactivated C. albicans (multiplicity of infection [MOI] of 5) for the indicated time.
For nuclear extracts, 1 × 107 murine macrophage cells were incubated in 6-cm plates and stimulated with the UV-inactivated C. albicans for 30 min and 1 h, respectively, whereas, for total cell lysates, 3 × 106 cells were incubated in 12-well plates and stimulated with the UV-inactivated C. albicans for 15, 30, and 45 min, respectively. For cytokine production assay, 1 × 107 cells were incubated in 6-well plates and stimulated with the UV-inactivated C. albicans for 6 h, and then cell supernatants were collected for the cytokine production assay.
Western blotting.
Thioglycolate-elicited peritoneal macrophages were lysed in the lysis buffer (250 mM NaCl, 50 mM HEPES, 1 mM EDTA, 1% Nonidet P-40, 1 mM Na3VO4, 1 mM NaF, and protease inhibitors, pH 7.4) for total cell lysates. For nuclear extracts, thioglycolate-elicited peritoneal macrophages were lysed in the lysis buffer (10 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol [DTT], 0.05% Nonidet P-40, and protease inhibitors, pH 7.9). The nuclear pellet were harvested by centrifugation (4°C, 17,000 rpm) and then washed with the lysis buffer. The nuclear pellets then were resuspended in the extraction buffer (5 mM HEPES, 300 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, pH 7.9) and incubated with vortexing at 4°C for 40 min. Equal amounts of protein obtained from nuclear extracts or total cell lysates were subjected to SDS-PAGE, blotted with the indicated primary antibodies and secondary antibodies, and then developed with the chemiluminescence method according to the manufacturer's instructions (Millipore) using the ECL detection system (GE Healthcare).
Cytokine production assay.
Concentrations of tumor necrosis factor alpha (TNF-α), interleukin-6 (IL-6), IL-12p40, IL-10, gamma interferon (IFN-γ), and IL-17 in murine kidney homogenates or thioglycolate-elicited peritoneal macrophages culture supernatant were determined with commercially available Ready-Set-Go cytokine kits (eBioscience) in triplicate according to the manufacturer's instructions.
Neutrophil killing assay.
Thioglycolate-elicited peritoneal neutrophils (0.6 × 106) were cultured in RPMI 1640 plus 10% (vol/vol) heat-inactivated fetal bovine serum. The cells were mixed with unopsonized live C. albicans (1 × 104) in the wells of a 24-well plate and were kept for 60 min at 4°C to allow the cells to settle before being transferred to 37°C for another 60 min. Control plates were kept in parallel at 4°C during the incubation. The cells then were mixed, with scraping, and were plated on YPD agar for counting viable C. albicans cells after incubation for 48 h at 30°C.
Respiratory burst assay.
For analysis of hydrogen peroxide (H2O2) generation, neutrophils were loaded with dihydrorhodamine 123 at a final concentration of 1 mM. After incubation with live C. albicans at 37°C for 1 h, the conversion of dihydrorhodamine 123 was assessed by a multimode microplate reader (SpectraMax; Molecular Devices, USA) and was expressed as mean fluorescent intensity. Cells loaded with dihydrorhodamine 123 but not treated with C. albicans were used to assess background H2O2 production.
Murine systemic candidiasis model.
C57BL/6 female mice (6 to 8 weeks) were anesthetized intraperitoneally (1% pentobarbital sodium, 50 mg/kg) before infection. A total of 2 × 105 live C. albicans cells was administered intravenously in a final volume of 100 μl in PBS. Mice were monitored daily and were killed by CO2 asphyxiation after 7 days of infection. The kidneys were removed and disrupted in 0.5 ml PBS. Supernatants of kidney homogenates were harvested and stored at −80°C for measurement of cytokine production.
Murine peritoneal infection model.
Female C57BL/6 or Dectin-1-deficient mice (8 to 10 weeks) were injected intraperitoneally with UV-inactivated 2 × 105 C. albicans cells and were killed by CO2 asphyxiation after 4 h. The inflammatory infiltrate was collected by lavage with ice-cold PBS containing 5 mM EDTA, and red blood cells were lysed. Cells were counted and blocked with PBS containing 5% heat-inactivated FBS and 1 mM sodium azide at 4°C. The cells then were incubated with primary antibodies for 30 min at 4°C. After being fixed with 1% formaldehyde in PBS, cells were analyzed by flow cytometry (BD FACSVerse) to determine the leukocyte composition as described before (8). Contents of IL-6, monocyte chemoattractant protein-1 (MCP-1), and granulocyte-macrophage colony-stimulating factor (GM-CSF) in peritoneal lavage fluid were determined with commercially available cytokine kits (eBioscience) in triplicate according to the manufacturer's instructions.
Splenocyte recall response assay.
Female C57BL/6 and Dectin-1-deficient mice were injected intravenously with 3 × 104 exponentially growing live C. albicans SN152, gpi7Δ/Δ::GPI7 and gpi7Δ/Δ cells, respectively, and killed by CO2 asphyxiation 7 days later. Total splenocytes were harvested in RPMI 1640 plus 10% (vol/vol) heat-inactivated fetal bovine serum and 1% penicillin-streptomycin (Gibco). The splenocytes were cultured in a 96-well U-bottomed plate with 2 × 106 cells/well and restimulated with 1 × 105 UV-inactivated C. albicans SC5314 for 48 h. Cytokines in the supernatant were detected according to the method described above.
Statistical analysis.
At least three biological replicates were performed for all experiments unless otherwise indicated. One-way analysis of variance (ANOVA) with Bonferroni's posttest was used when multiple groups were analyzed. The two-tailed Student's t test was used for analysis of two groups, with paired analysis when appropriate. For analysis of nonparametrically distributed data, the Mann-Whitney test or Kruskal-Wallis test was used. Statistical significance was set at a P value of 0.05 or 0.01.
RESULTS
GPI7 gene deletion in C. albicans could abolish most GPI-CWPs.
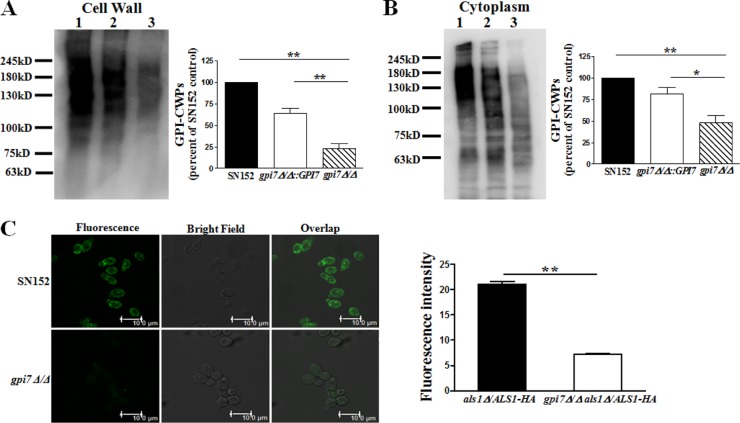
The GPI7 gene is responsible for adding ethanolaminephosphate to the second mannose in GPI biosynthesis (17, 34). We generated a mutant strain of C. albicans (gpi7Δ/Δ) and confirmed deletion of the GPI7 ORF (open reading frame) by PCR (see Fig. S1 in the supplemental material). The highly mannosylated GPI-CWPs were extracted from C. albicans cells by HF-pyridine and were revealed by peroxidase-linked ConA, which could bind to mannose residues. Immunoblotting of the parent strain GPI-CWP extract, using peroxidase-linked ConA, revealed marked protein bands (Fig. 1A). The gpi7 mutants exhibited a marked reduction (75%) of ConA-stained material, suggesting fewer GPI-anchored proteins linked to the cell wall (Fig. 1A). In addition, we also found that the level of GPI-anchored proteins in the cytoplasm of gpi7 mutants was significantly reduced (50%) (Fig. 1B). It is reasonable that reintegration of the GPI7 gene was sufficient to reproduce the expected GPI-CWPs and cytoplasmic GPI-anchored proteins (Fig. 1A and B).
FIG 1.
Deletion of GPI7 gene in C. albicans could block GPI-anchored protein biosynthesis and anchorage on cell wall. (A) Representative immunoblotting with peroxidase-labeled ConA staining of HF-pyridine-released C. albicans GPI-CWPs (lane 1, SN152; lane 2, gpi7Δ/Δ::GPI7; lane 3, gpi7Δ/Δ) and the ratio of gpi7Δ/Δ and gpi7Δ/Δ::GPI7 to SN152. (B) Representative immunoblotting with peroxidase-labeled ConA staining of C. albicans cytosol proteins (lane 1, SN152; lane 2, gpi7Δ/Δ::GPI7; lane 3, gpi7Δ/Δ) and the ratio of gpi7Δ/Δ and gpi7Δ/Δ::GPI7 to SN152. The mannosylated protein level of C. albicans SN152 was set to 100% as a control (A and B). (C) Representative observation of HA tag-fused Als1p, a well-characterized GPI-anchor protein, on the cell surface of the indicated strains (SN152 and gpi7Δ/Δ) by confocal laser scanning microscopy and fluorescence intensity analysis. The scale bar represents 10 μm. Data shown in panels A and B were quantified by ImageJ. Data shown in panel C were quantified by the Leica LAS AF Lite program. Data represent means (± standard errors of the means [SEM]) from three independent experiments. *, P < 0.05; **, P < 0.01 (one-way ANOVA with Bonferroni's posttest [A and B] or by Student's t test [C]).
Als1p, a well-characterized GPI-anchored protein, is known to be able to mediate C. albicans adhesion to the host cell (16, 22). We stained the C. albicans strains expressing HA tag-fused Als1p with anti-HA tag antibody and found a markedly reduced Als1p attachment on the cell wall (65%) in the gpi7 mutant compared with that of the parent strain by confocal microscopy (Fig. 1C).
Results described above indicated that GPI7 gene deletion in C. albicans could significantly reduce GPI-anchored protein synthesis in the cytoplasm and abolish GPI-CWPs on the cell wall.
GPI7 gene deletion in C. albicans exposed cell wall β-(1,3)-glucan.
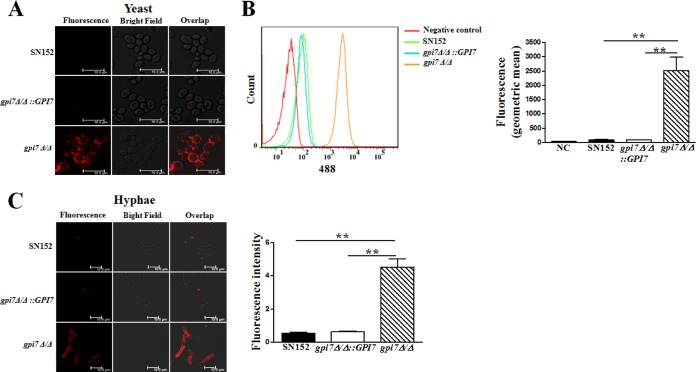
β-(1,3)-Glucan, a well-characterized PAMP of C. albicans, is buried underneath the outer layer of the cell wall with highly glycosylated mannoproteins (1, 35). We speculated that abolishing GPI-CWPs could disrupt the intricate architecture of the cell wall and increase β-glucan exposure. To examine this possibility, we detected exposure of β-(1,3)-glucan by anti-β-(1,3)-glucan primary antibody. We found a remarkable exposure of β-(1,3)-glucan on the yeast cell surface of the gpi7 mutant compared to that of the parent strain (Fig. 2A). The gpi7 mutant yeast cells had a 30-fold greater reactivity with the anti-β-glucan antibody than the parent strain by flow cytometry (Fig. 2B). We also examined the effect of GPI7 gene deletion on cells grown in RPMI 1640, a culture medium associated with strong hyphal growth and β-glucan masking. Our results suggested that abolishing GPI-CWPs also could cause a dramatic increase of β-(1,3)-glucan exposure in the hyphal form and had an 8-fold greater reactivity with the anti-β-glucan antibody than the parent strain (Fig. 2C). Concurrently, the C. albicans cells were labeled with propidium iodide to assess their viability by FACS (fluorescence-activated cell sorting). The gpi7 mutant did not show increased cell death, suggesting that increased β-glucan exposure was not due to the loss of viability (data not shown).
FIG 2.
Cell wall β-(1,3)-glucan was highly exposed in the C. albicans gpi7 mutant in both yeast and hyphal forms. Yeast-form growing cells (A and B) and hyphal growing cells (C) were stained with anti-β-(1,3)-glucan primary antibody to visualize β-(1,3)-glucan. The fluorescence intensity was quantified by flow cytometry (B, right) and the Leica LAS AF Lite program (C, right). The scale bar represents 10 μm. Data represent means (± SD) from triplicates of one representative experiment of three. **, P < 0.01 (one-way ANOVA with Bonferroni's posttest).
Neutrophils killed C. albicans gpi7 mutant more efficiently with an augmented respiratory burst.
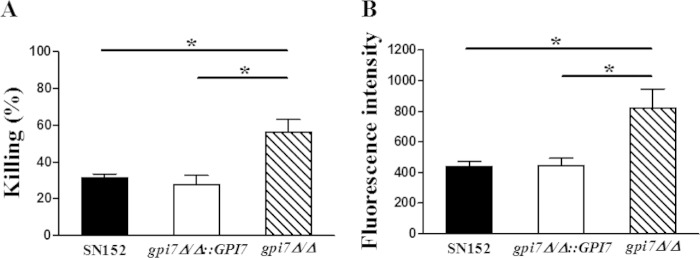
Neutrophils are the most abundant leukocyte in humans and are essential to the innate immune response against invading fungus (36). The polymorphonuclear leukocyte (PMN) Dectin-1 plays a key role in the recognition and killing of fungal pathogens by the innate immune system (37). As increased β-(1,3)-glucan exposure was seen on the cell surface, we explored whether the neutrophil response to the C. albicans gpi7 mutant would be increased as well. We found that thioglycolate-elicited peritoneal neutrophils could kill the gpi7 mutant more efficiently, and the killing ability was weakened when the GPI7 gene was reverted (Fig. 3A). Neutrophils also produced significantly more reactive oxygen species (ROS) when challenged by the C. albicans gpi7 mutant strain than by the parent and revertant strains (Fig. 3B).
FIG 3.
Neutrophils killed the uncloaked gpi7 mutant more efficiently with an augmented respiratory burst. Thioglycolate-elicited peritoneal neutrophils (0.6 × 106 cells) were infected with viable C. albicans (1 × 104 cells) for 60 min. (A) C. albicans then was plated on YPD agar for 48 h to measure viable CFU. (B) The respiratory burst was measured by assessing the conversion of dihydrorhodamine 123 to rhodamine. Data represent means (± SD) from triplicates of one representative experiment of three. *, P < 0.05 (one-way ANOVA with Bonferroni's posttest).
C. albicans gpi7 mutant could stimulate Dectin-1-mediated antifungal innate immune responses.
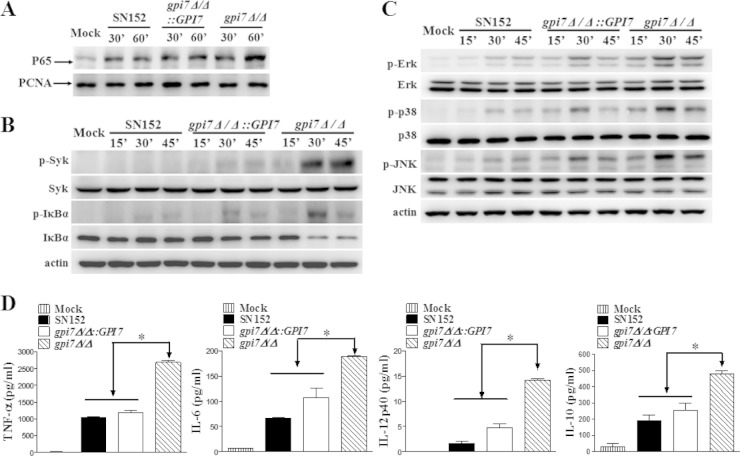
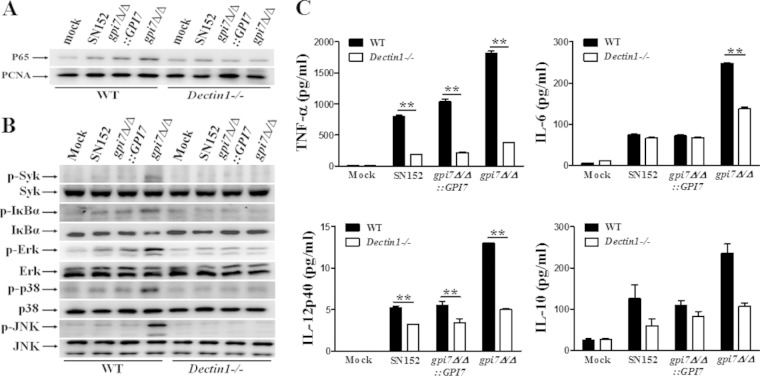
Macrophages are essential to initiate antifungal innate immune responses and prime adaptive immune responses (5, 6). We tested whether the β-glucan exposure induced by abolishing GPI-CWPs could stimulate an enhanced macrophage response. We found that the gpi7 mutant stimulation induced more nuclear translocation of NF-κB (p65), Syk phosphorylation, and IκBα phosphorylation and degradation in thioglycolate-elicited peritoneal macrophages, suggesting NF-κB signaling activation (Fig. 4A and B). Consistent with this, gpi7 mutant stimulation induced more ERK phosphorylation, JNK phosphorylation, and p38 phosphorylation in macrophages than the parent strain, indicating mitogen-activated protein kinase (MAPK) signaling activation (Fig. 4C). In addition, the enhanced activation of NF-κB and MAPK signaling were downregulated when the GPI7 gene was reverted. NF-κB and MAPK activation could promote the expression of various inflammatory cytokines. We also found that the gpi7 mutant stimulation induced robust production of inflammatory cytokines, including TNF-α, IL-6, IL-12p40, and IL-10, in macrophages, while the production of these cytokines was significantly decreased when parent and revertant strains were stimulated (Fig. 4D).
FIG 4.
C. albicans gpi7 mutant could be recognized by host macrophages. gpi7Δ/Δ could induce NF-κB and MAPK activation and inflammatory responses in macrophages. Thioglycolate-elicited peritoneal macrophages were stimulated by UV-inactivated C. albicans yeast SN152, gpi7Δ/Δ::GPI7, and gpi7Δ/Δ (MOI of 5) for the indicated times. The nuclear extracts (A) and cell lysates (B and C) were analyzed by immunoblotting with the indicated antibodies. (D) Enzyme-linked immunosorbent assay (ELISA) results for TNF-α, IL-6, IL-12p40, and IL-10 in supernatants of thioglycolate-elicited peritoneal macrophages, which were stimulated with the indicated C. albicans strains (MOI of 5) for 6 h. Mock, unstimulated macrophages. Data represent means (± SD) from triplicates of one representative experiment of three. *, P < 0.05 (one-way ANOVA with Bonferroni's posttest).
To confirm that the enhanced macrophage immune response induced by the gpi7 mutant is through β-(1,3)-glucan exposure and is host Dectin-1 dependent, we stimulated thioglycolate-elicited peritoneal macrophages from both wild-type (Clec7a+/+) and Dectin-1-deficient (Clec7a−/−) mice with the gpi7 mutant yeast cells and found that the activation of NF-κB and MAPK signaling was defective in Dectin-1-deficient macrophage cells (Fig. 5A and B). Consequently, the gpi7 mutant yeasts could not significantly increase the production of inflammatory cytokines in Dectin-1-deficient macrophage cells (Fig. 5C). We also found that laminarin, a Dectin-1 receptor inhibitor, could downregulate the NF-κB and MAPK activation, as well as inflammatory cytokine production, in macrophages challenged by the gpi7 mutant (see Fig. S2 in the supplemental material).
FIG 5.
Inflammatory responses in macrophage stimulated by C. albicans gpi7Δ/Δ were Dectin-1 dependent. Thioglycolate-elicited peritoneal macrophages from wild-type or Dectin-1-deficient mice were stimulated with UV-inactivated C. albicans yeast SN152, gpi7Δ/Δ::GPI7, or gpi7Δ/Δ (MOI of 5) for 60 min (for nuclear extracts analysis) or 30 min (for total cell lysate analysis). The nuclear extracts (A) and total cell lysates (B) were analyzed by immunoblotting with the indicated antibodies. (C) ELISA results for TNF-α, IL-6, IL-12p40, and IL-10 in supernatants of thioglycolate-elicited peritoneal macrophages from wild-type mice or Dectin-1-deficient mice, which were stimulated with the indicated C. albicans yeast strains (MOI of 5) for 6 h. Mock, unstimulated macrophages. Data represent means (± SD) from triplicates of one representative experiment of three. **, P < 0.01 (two-way ANOVA with Bonferroni's posttest).
C. albicans gpi7 mutant could induce stronger inflammatory response in vivo.
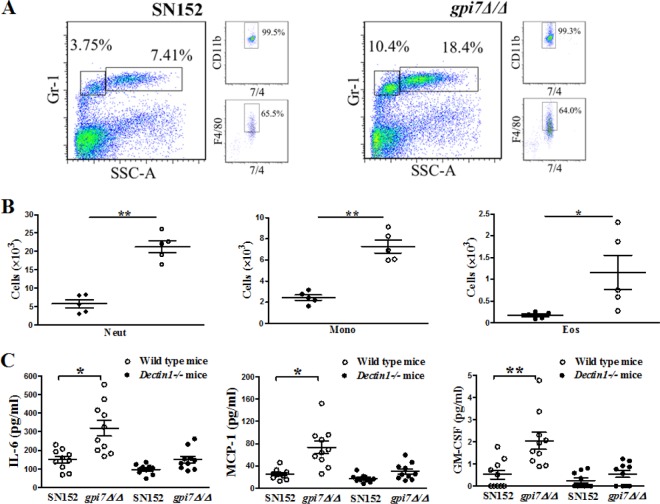
We further explored how the enhanced leukocyte recognition induced by exposure of β-glucan affected inflammation response in vivo using a peritoneal infection model. After 4 h of intraperitoneal infection with 2 × 105 UV-inactivated C. albicans cells, we found that gpi7 mutant-infected mice recruited more cells than the parent strain-infected mice, including Gr-1hi7/4hi neutrophils, Gr-1+7/4hi F4/80+ inflammatory monocytes (Fig. 6A and B), and Gr-1int7/4loF4/80+ side-scatter-high eosinophils (Fig. 6B) in the peritoneum. The increased inflammatory cell recruitment also was associated with increased production of specific cytokines and growth factors, including IL-6, MCP-1, and GM-CSF (Fig. 6C). However, IL-6, MCP-1, and GM-CSF production did not show a significant difference in the peritoneum when Dectin-1-deficient mice were infected with the gpi7 mutant strain or parent strain SN152, which suggested that the in vivo phenotypes were Dectin-1 dependent (Fig. 6C). In addition, we also detected TNF-α, IL-12p40, and IL-10 production in the peritoneal infection model, which was detected by the in vitro response described above. TNF-α production did not show statistical significance when challenged by C. albicans gpi7Δ/Δ compared to that with the parent strain (data not shown). However, IL12p40 and IL-10 were not detectable in the experiments.
FIG 6.
Host increased inflammatory responses in vivo against C. albicans gpi7Δ/Δ. C57BL/6 or Dectin-1-deficient mice were intraperitoneally infected with 2 × 105 UV-inactivated C. albicans organisms of SN152, gpi7Δ/Δ::GPI7, or gpi7Δ/Δ for 4 h. (A) Flow cytometry for Gr-1hi7/4hi neutrophils and Gr-1+7/4hi F4/80+ inflammatory monocytes in the peritoneum. (B) Scatter plots of myeloid cell subsets in the peritoneum. Each symbol represents an individual mouse. Neut, neutrophil; Mono, monocyte; Eos, eosinophil. (C) ELISAs for cytokines, chemokines, and growth factors in lavage fluid from the peritoneal cavities. MCP-1, chemokine CCL2; GM-CSF, granulocyte-monocyte colony-stimulating factor. Data are representative of three independent experiments. P < 0.05 (*) and P < 0.01 (**) (Mann-Whitney nonparametric t test [B] and Kruskal-Wallis nonparametric one-way ANOVA with Dunn's posttest [C]).
C. albicans gpi7 mutant could stimulate stronger Th cell responses.
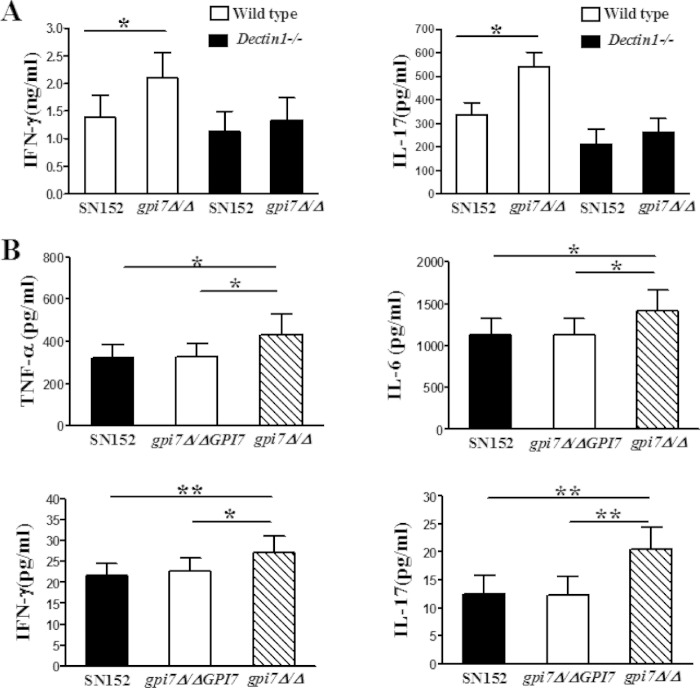
Engagement of PRRs in innate immune cells could render them competent to prime T cells and thereby drive the adaptive immune response (7). Thus, we further investigated the contribution of β-(1,3)-glucan exposure to adaptive immunity. We collected splenocytes from live C. albicans-infected mice. The splenocytes then were restimulated with UV-inactivated C. albicans SC5314 for 48 h, and Th1 and Th17 responses were monitored by measuring IFN-γ and IL-17 in cell supernatant, respectively. The splenocytes from uninfected mice, including wild-type and Dectin-1-deficient mice, have a Th cell response after C. albicans restimulation that is similar to that from parent strain-infected mice (data not shown). The levels of IFN-γ and IL-17 production of splenocytes from gpi7Δ/Δ-infected mice were significantly higher than those from parent strain- or revertant strain-infected mice when restimulated by C. albicans 5314 (Fig. 7A), indicating that the gpi7 mutant induced a stronger recall Th cell response in the host. In Dectin-1-deficient mice, the stronger Th cell recall response induced by C. albicans gpi7Δ/Δ was weakened, suggesting that the phenotype was Dectin-1 dependent (Fig. 7A). We also detected significantly enhanced TNF-α, IL-6, IFN-γ, and IL-17 levels in the kidneys of gpi7 mutant-infected mice compared to those of the parent and revertant strain-infected mice 7 days after intravenous infection (Fig. 7B). Accordingly, the kidney fungal burdens of mice inoculated with the gpi7 mutant were significantly lower than those of mice infected with either the parent or revertant strain (see Fig. S3 in the supplemental material). Infiltrating cells in kidneys of mice infected with gpi7Δ/Δ also were significantly fewer than those of mice infected with the parent or revertant strain (see Fig. S3).
FIG 7.
C. albicans gpi7 mutant could stimulate enhanced host Th cell responses. (A) Splenocyte recall assay. C57BL/6 and Dectin-1-deficient mice were infected with 3 × 104 viable C. albicans organisms (SN152, gpi7Δ/Δ::GPI7, or gpi7Δ/Δ) for 7 days. Splenocytes were restimulated with UV-killed C. albicans SC5314 for 48 h, and IFN-γ and IL-17 accumulation in the supernatants was measured by ELISA (n = 10). Data represent means (± SD) from triplicates of one representative experiment of three. (B) ELISAs for cytokines in kidneys from C57BL/6 mice intravenously infected with 2 × 105 viable C. albicans for 7 days (n = 12). Data are representative of three independent experiments. *, P < 0.05; **, P < 0.01 (Kruskal-Wallis nonparametric one-way ANOVA with Dunn's posttest).
DISCUSSION
The C. albicans cell wall is a complex dynamic structure based on a core structure of β-(1,3)-glucan that is covalently linked to β-(1,6)-glucan, chitin, and an outer layer of the matrix composed of mannoproteins (mainly GPI-anchored proteins) (38). Previous studies have indicated that GPI-anchored proteins are critical for the virulence and pathogenicity of C. albicans, and the carbohydrates serve as PAMPs to mediate immune recognition by the host (1, 39–41). Fungi could shield the surface PAMPs to evade host immune attacks. Histoplasma capsulatum can mask the cell wall β-(1,3)-glucan by the outer layer α-(1,3)-glucan. Previous studies have found increased Dectin-1-dependent host immune recognition after abolishing cell wall α-(1,3)-glucan in Histoplasma capsulatum (42, 43). As the main PAMP, β-(1,3)-glucan also is concealed in C. albicans (10). The antifungal drug caspofungin and a new small molecule, gepinacin, which target the cell wall, can cause the exposure of β-(1,3)-glucan in C. albicans and elicit a stronger host immune response (24, 33). In the present study, we found that GPI-CWPs also were involved in C. albicans escaping from host Dectin-1 recognition and further demonstrated that blocking GPI anchor synthesis to abolish GPI-CWPs was a potential strategy to disturb C. albicans evading host immune attack.
Recognition of β-(1,3)-glucan by Dectin-1 has been shown to be crucial in host antifungal defense (8, 11, 44). However, C. albicans shields β-glucan from being recognized by Dectin-1 on innate immune cells (1, 14, 32). The outer-layer mannoproteins of C. albicans are mainly GPI-anchored proteins. The GPI anchor, which is responsible for transportation and attachment of all of the GPI-anchored proteins to the cell wall, is synthesized and added to the proteins in the endoplasmic reticulum (15). C. albicans GPI7 is responsible for adding ethanolaminephosphate to the second mannose in GPI anchor biosynthesis (17). Richard and Plaine reported that the GPI anchor modification by GPI7 mainly influenced the linkage of GPI-anchored protein to the cell wall, while the plasma membrane-targeted GPI-anchored proteins were not affected (16). Therefore, we chose the gpi7 mutant as a tool to investigate the role of GPI-CWPs in the immune-escaping characteristics of C. albicans. We further confirmed that the GPI7 gene was the key to normal GPI-anchored protein synthesis. GPI7 gene deletion in C. albicans could block most GPI-anchored protein biosynthesis and its attachment to the cell surface (Fig. 1). We further demonstrated that abolishing the GPI-anchored proteins on the cell wall through deletion of the C. albicans GPI7 gene could unmask the concealed β-(1,3)-glucan in either yeast or hyphal form (Fig. 2). The exposure of β-(1,3)-glucan may be due to the fact that cell surface-glycosylated proteins of the gpi7 mutant do not adequately conceal the inner layer. Thus, we hypothesized that the unmasking of β-(1,3)-glucan on C. albicans cell surfaces could increase host Dectin-1-mediated immune recognition.
Neutrophils contribute to the initial steps of fungal killing, which plays a special role in neutropenic and immunosuppressed populations (36, 45). CARD9 signaling and ROS formation by the NADPH oxidase system were involved in neutrophil killing of C. albicans (46). CR3 and the Dectin-1 receptor have been reported to be able to recognize cell wall β-glucan of C. albicans and mediate killing (46). Our results showed that neutrophils killed the gpi7 mutant more efficiently and produced significantly more ROS when challenged by the gpi7 mutant (Fig. 3). This suggested that blocking GPI anchor synthesis to abolish GPI-CWPs of C. albicans increased the uptake and killing functions of neutrophils, which might depend on the recognition of exposed β-(1,3)-glucan by the Dection-1 receptor, as well as ROS formation.
After encountering fungal pathogens, host macrophages secrete cytokines and chemokines and engulf the pathogen in phagosomes (38). Here, we showed that gpi7 mutant with the loss of GPI-CWPs elicited increased recognition by macrophages (Fig. 4). The enhanced macrophage cellular responses consisted of activation of NF-κB and MAPK signaling and production of specific cytokines (TNF-α, IL-6, IL-12p40, and IL-10). IL-6 and IL-23 (consisting of IL-12p40 and p19) are key cytokines leading to induction of Th17 cell differentiation (47). TNF-α and IL-6 are involved in innate immune responses against Candida infection through promoting neutrophil production and activation (48, 49). The enhanced innate immune responses were Dectin-1 dependent, suggesting that they were attributed to β-glucan exposure caused by GPI anchor synthesis inhibition (Fig. 5; also see Fig. S2 in the supplemental material). However, Plaine et al. reported that laminarin failed to inhibit the enhanced proinflammatory response (TNF-α) to the C. albicans gpi7 mutant in murine cells, indicating that the Dectin-1 receptor was not involved in the process (50). We consider that the controversial results may be attributed to different experimental protocols. Plaine et al. challenged resident peritoneal macrophages by gpi7 mutant in the presence of laminarin for 24 h and measured proinflammatory cytokines, while we stimulated thioglycolate-elicited peritoneal macrophages by gpi7 mutant for 6 h. In the present study, we also confirmed that the enhanced innate immune response stimulated by gpi7 mutant was Dectin-1 dependent using Dectin-1-deficient mice besides laminarin blocking. Richard and Plaine (16) investigated the interaction between live gpi7 mutant and mouse macrophage-like cell line J774 and found that gpi7 mutant could upregulate Erk1/2 phosphorylation, which was consistent with our findings. In our experiments, we also found that gpi7 mutant stimulation induced an enhanced activation of NF-κB and MAPK signaling (phosphorylation of both p38 and JNK) in thioglycolate-elicited peritoneal macrophages. We further demonstrated that gpi7 mutant stimulation could induce more production of specific cytokines in murine macrophages. Nonetheless, immune sensing of C. albicans requires the cooperative recognition of mannans and glucan by PRRs (38), so we cannot exclude the participation of other PRRs in this process. Further studies are needed to evaluate cross talk between Dectin-1 and other PRRs in our experiment. It has been reported that glycogen also can induce PMNs and mineral oil can induce macrophages (51, 52). It may be worthwhile to compare innate immune cells induced by different methods in a further study to make sure that the results are comparable.
We further confirmed this finding in vivo. Both Th1 and Th17 cells have been proposed to mediate protection against C. albicans infection (53–55). IFN-γ and IL-17 are the key cytokines produced by Th1 and Th17 cells, respectively. IFN-γ and IL-17 elevation in kidneys and the enhanced splenocyte recall responses of gpi7 mutant-infected mice indicated that the C. albicans gpi7 mutant could stimulate stronger host Th1 and Th17 responses (Fig. 7A). TNF-α and IL-6 have been considered pivotal cytokines in anticandidal defense through promoting neutrophil production and activation (48, 49). The elevation of proinflammatory cytokines in kidneys of gpi7 mutant-infected mice (Fig. 7B) might result in the recruitment of innate immune cells, such as neutrophils, and priming of adaptive immune response to clear the infected fungi. We determined this process in a peritoneal infection model, demonstrating the marked elevation of cytokines and chemokines, including IL-6, GM-CSF, and MCP-1 in the peritoneal cavity, which resulted in subsequent recruitment of neutrophils and monocytes (Fig. 6). In addition to promoting neutrophil production and activation, IL-6 also plays a critical role in the induction of Th17 cell differentiation (56). GM-CSF has been reported to be important for enhancement of neutrophil maturation and potentiating neutrophil functions (57–59). MCP-1 is an important mediator for monocyte recruitment (60). These in vivo phenotypes were weakened in Dectin-1-deficient mice, suggesting the host-enhanced inflammatory responses and Th cell responses induced by C. albicans with the loss of GPI-CWPs were Dectin-1 dependent (Fig. 6C and 7A).
In this study, our data showed that abolishing GPI-CWPs could uncloak C. albicans and expose it to the host immune system. The antifungal effects, including enhanced recognition by innate immune cells and Th1 and Th17 responses, were host Dectin-1 dependent and attributed to β-(1,3) glucan exposure. The present study provided a potential antifungal therapeutic strategy in modulating the host immune response to C. albicans. In addition, abolishing GPI-CWPs in C. albicans by blocking GPI anchor synthesis also destroyed the cell wall integrity and increased the susceptibility of C. albicans to cell membrane-targeting drugs, such as azoles, and cell wall-targeting drugs, such as echinocandin (data not shown). Therefore, the key genes encoding GPI anchor synthesis may provide drug targets for broad-spectrum treatment of antifungal infections.
Supplementary Material
ACKNOWLEDGMENTS
We thank Gordon D. Brown (University of Aberdeen) for providing Dectin1−/− mice, Suzanne M. Noble (University of California-San Francisco) for C. albicans SN152 and plasmids pSN40 and pSN52, and Dominique Sanglard (Centre Hospitalier Universitaire Vaudois) for C. albicans SC5314. We also thank Jürgen Wendland (Carlsberg Laboratory in Denmark) for providing plasmid pFA-ARG4 and Joachim Morschhauser (University of Wurzburg) for providing plasmid pSFS2A.
This study was supported by the National Key Basic Research Program of China (no. 2013CB531602), the National Science Foundation of China (81471924, 81202563, and 81330083), the National Science and Technology Major Project for the Creation of New Drugs (no. 2012ZX09103101-003 and 2013ZX09J13108-03B), the Shanghai Science and Technology Support Program (no. 14401902200 and 14431902200), and the Shanghai Manufacture-Education-Research-Medical Cooperative Project (no. 12DZ1930505).
We declare that no competing interests exist.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00097-15.
REFERENCES
- 1.Gow NA, van de Veerdonk FL, Brown AJ, Netea MG. 2012. Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat Rev Microbiol 10:112–122. doi: 10.1038/nrmicro2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miceli M, Díaz J, Lee S. 2011. Emerging opportunistic yeast infections. Lancet Infect Dis 11:142–151. doi: 10.1016/S1473-3099(10)70218-8. [DOI] [PubMed] [Google Scholar]
- 3.Hsu FC, Lin PC, Chi CY, Ho MW, Ho CM, Wang JH. 2009. Prognostic factors for patients with culture-positive Candida infection undergoing abdominal surgery. J Microbiol Immunol Infect 42:378–384. [PubMed] [Google Scholar]
- 4.Horn DL, Neofytos D, Anaissie EJ, Fishman JA, Steinbach WJ, Olyaei AJ, Marr KA, Pfaller MA, Chang CH, Webster KM. 2009. Epidemiology and outcomes of candidemia in 2019 patients: data from the prospective antifungal therapy alliance registry. Clin Infect Dis 48:1695–1703. doi: 10.1086/599039. [DOI] [PubMed] [Google Scholar]
- 5.Romani L. 2011. Immunity to fungal infections. Nat Rev Immunol 11:275–288. doi: 10.1038/nri2939. [DOI] [PubMed] [Google Scholar]
- 6.Brown GD. 2011. Innate antifungal immunity: the key role of phagocytes. Annu Rev Immunol 29:1–21. doi: 10.1146/annurev-immunol-030409-101229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wüthrich M, Deepe GS Jr, Klein B. 2012. Adaptive immunity to fungi. Annu Rev Immunol 30:115–148. doi: 10.1146/annurev-immunol-020711-074958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor PR, Tsoni SV, Willment JA, Dennehy KM, Rosas M, Findon H, Haynes K, Steele C, Botto M, Gordon S, Brown GD. 2007. Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat Immunol 8:31–38. doi: 10.1038/ni1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heinsbroek SE, Brown GD, Gordon S. 2005. Dectin-1 escape by fungal dimorphism. Trends Immunol 26:352–354. doi: 10.1016/j.it.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 10.Gantner BN, Simmons RM, Underhill DM. 2005. Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. EMBO J 24:1277–1286. doi: 10.1038/sj.emboj.7600594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodridge HS, Reyes CN, Becker CA, Katsumoto TR, Ma J, Wolf AJ, Bose N, Chan AS, Magee AS, Danielson ME, Weiss A, Vasilakos JP, Underhill DM. 2011. Activation of the innate immune receptor Dectin-1 upon formation of a “phagocytic synapse.” Nature 472:471–475. doi: 10.1038/nature10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tam JM, Mansour MK, Khan NS, Seward M, Puranam S, Tanne A, Sokolovska A, Becker CE, Acharya M, Baird MA, Choi AM, Davidson MW, Segal BH, Lacy-Hulbert A, Stuart LM, Xavier RJ, Vyas JM. 2014. Dectin-1-dependent LC3 recruitment to phagosomes enhances fungicidal activity in macrophages. J Infect Dis 210:1844–1854. doi: 10.1093/infdis/jiu290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferwerda B, Ferwerda G, Plantinga TS, Willment JA, van Spriel AB, Venselaar H, Elbers CC, Johnson MD, Cambi A, Huysamen C, Jacobs L, Jansen T, Verheijen K, Masthoff L, Morré SA, Vriend G, Williams DL, Perfect JR, Joosten LA, Wijmenga C, van der Meer JW, Adema GJ, Kullberg BJ, Brown GD, Netea MG. 2009. Human dectin-1 deficiency and mucocutaneous fungal infections. N Engl J Med 361:1760–1767. doi: 10.1056/NEJMoa0901053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wheeler RT, Kombe D, Agarwala SD, Fink GR. 2008. Dynamic, morphotype-specific Candida albicans beta-glucan exposure during infection and drug treatment. PLoS Pathog 4:e1000227. doi: 10.1371/journal.ppat.1000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaffin WL. 2008. Candida albicans cell wall proteins. Microbiol Mol Biol Rev 72:495–544. doi: 10.1128/MMBR.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richard ML, Plaine A. 2007. Comprehensive analysis of glycosylphosphatidylinositol-anchored proteins in Candida albicans. Eukaryot Cell 6:119–133. doi: 10.1128/EC.00297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richard M, Ibata-Ombetta S, Dromer F, Bordon-Pallier F, Jouault T, Gaillardin C. 2002. Complete glycosylphosphatidylinositol anchors are required in Candida albicans for full morphogenesis, virulence and resistance to macrophages. Mol Microbiol 44:841–853. doi: 10.1046/j.1365-2958.2002.02926.x. [DOI] [PubMed] [Google Scholar]
- 18.Hoyer LL. 2001. The ALS gene family of Candida albicans. Trends Microbiol 9:176–180. doi: 10.1016/S0966-842X(01)01984-9. [DOI] [PubMed] [Google Scholar]
- 19.Fonzi WA. 1999. PHR1 and PHR2 of Candida albicans encodes putativeglycosidases required for proper cross-linking of beta-1,3- and beta-1,6-glucans. J Bacteriol 181:7070–7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staab JF, Bradway SD, Fidel PL, Sundstrom P. 1999. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 283:1535–1538. doi: 10.1126/science.283.5407.1535. [DOI] [PubMed] [Google Scholar]
- 21.Martinez-Lopez R, Park H, Myers CL, Gil C, Filler SG. 2006. Candida albicans Ecm33p is important for normal cell wall architecture and interactions with host cells. Eukaryot Cell 5:140–147. doi: 10.1128/EC.5.1.140-147.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu Y, Ibrahim AS, Sheppard DC, Chen YC, French SW, Cutler JE, Filler SG, Edwards JE Jr. 2002. Candida albicans Als1p: an adhesin that is a downstream effector of the EFG1 filamentation pathway. Mol Microbiol 44:61–72. doi: 10.1046/j.1365-2958.2002.02873.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhao X, Daniels KJ, Oh SH, Green CB, Yeater KM, Soll DR, Hoyer LL. 2006. Candida albicans Als3p is required for wild-type biofilm formation on silicone elastomer surfaces. Microbiology 152:2287–2299. doi: 10.1099/mic.0.28959-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLellan CA, Whitesell L, King OD, Lancaster AK, Mazitschek R, Lindquist S. 2012. Inhibiting GPI anchor biosynthesis in fungi stresses the endoplasmic reticulum and enhances immunogenicity. ACS Chem Biol 7:1520–1528. doi: 10.1021/cb300235m. [DOI] [PubMed] [Google Scholar]
- 25.Vautier S, Drummond RA, Redelinghuys P, Murray GI, MacCallum DM, Brown GD. 2012. Dectin-1 is not required for controlling Candida albicans colonization of the gastrointestinal tract. Infect Immun 80:4216–4222. doi: 10.1128/IAI.00559-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan L, Côte P, Li XX, Jiang YY, Whiteway M. 2012. PalI domain proteins of Saccharomyces cerevisiae and Candida albicans. Microbiol Res 167:422–432. doi: 10.1016/j.micres.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 27.Noble SM, Johnson AD. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell 4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reuss O, Vik A, Kolter R, Morschhäuser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 29.Klis FM, de Koster CG, Brul S. 2011. A mass spectrometric view of the fungal wall proteome. Future Microbiol 6:941–951. doi: 10.2217/fmb.11.72. [DOI] [PubMed] [Google Scholar]
- 30.Castillo L, Calvo E, Martínez AI, Ruiz-Herrera J, Valentín E, Lopez JA, Sentandreu R. 2008. A study of the Candida albicans cell wall proteome. Proteomics 8:3871–3781. doi: 10.1002/pmic.200800110. [DOI] [PubMed] [Google Scholar]
- 31.Hamada K, Terashima H, Arisawa M, Kitada K. 1998. Amino acid sequence requirement for efficient incorporation of glycosylphosphatidylinositol-associated proteins into the cell wall of Saccharomyces cerevisiae. J Biol Chem 273:26946–26953. doi: 10.1074/jbc.273.41.26946. [DOI] [PubMed] [Google Scholar]
- 32.Kapteyn JC, Hoyer LL, Hecht JE, Müller WH, Andel A, Verkleij AJ, Makarow M, Van Den Ende H, Klis FM. 2000. The cell wall architecture of Candida albicans wild-type cells and cell wall-defective mutants. Mol Microbiol 35:601–611. doi: 10.1046/j.1365-2958.2000.01729.x. [DOI] [PubMed] [Google Scholar]
- 33.Wheeler RT, Fink GR. 2006. A drug-sensitive genetic network masks fungi from the immune system. PLoS Pathog 2:e35. doi: 10.1371/journal.ppat.0020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imhof I, Flury I, Vionnet C, Roubaty C, Egger D, Conzelmann A. 2004. Glycosylphosphatidylinositol (GPI) proteins of Saccharomyces cerevisiae contain ethanolamine phosphate groups on the alpha1,4-linked mannose of the GPI anchor. J Biol Chem 279:19614–19627. doi: 10.1074/jbc.M401873200. [DOI] [PubMed] [Google Scholar]
- 35.Davis SE, Hopke A, Minkin SC Jr, Montedonico AE, Wheeler RT, Reynolds TB. 2014. Masking of β(1-3)-glucan in the cell wall of Candida albicans from detection by innate immune cells depends on phosphatidylserine. Infect Immun 82:4405–4413. doi: 10.1128/IAI.01612-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Low CY, Rotstein C. 2011. Emerging fungal infections in immunocompromised patients. F1000 Med Rep 3:14. doi: 10.3410/M3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kennedy AD, Willment JA, Dorward DW, Williams DL, Brown GD, DeLeo FR. 2007. Dectin-1 promotes fungicidal activity of human neutrophils. Eur J Immunol 37:467–478. doi: 10.1002/eji.200636653. [DOI] [PubMed] [Google Scholar]
- 38.Netea MG, Brown GD, Kullberg BJ, Gow NA. 2008. An integrated model of the recognition of Candida albicans by the innate immune system. Nat Rev Microbiol 6:67–78. doi: 10.1038/nrmicro1815. [DOI] [PubMed] [Google Scholar]
- 39.Poulain D, Jouault T. 2004. Candida albicans cell wall glycans, host receptors and responses: elements for a decisive crosstalk. Curr Opin Microbiol 7:342–349. doi: 10.1016/j.mib.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 40.Galán-Díez M, Arana DM, Serrano-Gómez D, Kremer L, Casasnovas JM, Ortega M, Cuesta-Domínguez A, Corbí AL, Pla J, Fernández-Ruiz E. 2010. Candida albicans beta-glucan exposure is controlled by the fungal CEK1-mediated mitogen-activated protein kinase pathway that modulates immune responses triggered through dectin-1. Infect Immun 78:1426–1436. doi: 10.1128/IAI.00989-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klis FM, Sosinska GJ, de Groot PW, Brul S. 2009. Covalently linked cell wall proteins of Candida albicans and their role in fitness and virulence. FEMS Yeast Res 9:1013–1028. doi: 10.1111/j.1567-1364.2009.00541.x. [DOI] [PubMed] [Google Scholar]
- 42.Rappleye CA, Eissenberg LG, Goldman WE. 2007. Histoplasma capsulatum alpha-(1,3)-glucan blocks innate immune recognition by the beta-glucan receptor. Proc Natl Acad Sci U S A 104:1366–1370. doi: 10.1073/pnas.0609848104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edwards JA, Alore EA, Rappleye CA. 2011. The yeast-phase virulence requirement for α-glucan synthase differs among Histoplasma capsulatum chemotypes. Eukaryot Cell 10:87–97. doi: 10.1128/EC.00214-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goodridge HS, Wolf AJ, Underhill DM. 2009. Beta-glucan recognition by the innate immune system. Immunol Rev 230:38–50. doi: 10.1111/j.1600-065X.2009.00793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arendrup MC. 2010. Epidemiology of invasive candidiasis. Curr Opin Crit Care 16:445–452. doi: 10.1097/MCC.0b013e32833e84d2. [DOI] [PubMed] [Google Scholar]
- 46.Gazendam RP, van Hamme JL, Tool AT, van Houdt M, Verkuijlen PJ, Herbst M, Liese JG, van de Veerdonk FL, Roos D, van den Berg TK, Kuijpers TW. 2014. Two independent killing mechanisms of Candida albicans by human neutrophils: evidence from innate immunity defects. Blood 124:590–597. doi: 10.1182/blood-2014-01-551473. [DOI] [PubMed] [Google Scholar]
- 47.LeibundGut-Landmann S, Gross O, Robinson MJ, Osorio F, Slack EC, Tsoni SV, Schweighoffer E, Tybulewicz V, Brown GD, Ruland J, Reis e Sousa C. 2007. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat Immunol 8:630–638. doi: 10.1038/ni1460. [DOI] [PubMed] [Google Scholar]
- 48.Basu S, Quilici C, Zhang HH, Grail D, Dunn AR. 2008. Mice lacking both G-CSF and IL-6 are more susceptible to Candida albicans infection: critical role of neutrophils in defense against Candida albicans. Growth Factors 26:23–34. doi: 10.1080/08977190801987513. [DOI] [PubMed] [Google Scholar]
- 49.Netea MG, van Tits LJ, Curfs JH, Amiot F, Meis JF, van der Meer JW, Kullberg BJ. 1999. Increased susceptibility of TNF-α lymphotoxin-α double knockout mice to systemic candidiasis through impaired recruitment of neutrophils and phagocytosis of Candida albicans. J Immunol 163:1498–1505. [PubMed] [Google Scholar]
- 50.Plaine A, Yáñez A, Murciano C, Gaillardin C, Gil ML, Richard ML, Gozalbo D. 2008. Enhanced proinflammatory response to the Candida albicans gpi7 null mutant by murine cells. Microbes Infect 10:382–389. doi: 10.1016/j.micinf.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 51.Marques da Silva C, Miranda Rodrigues L, Passos da Silva Gomes A, Mantuano Barradas M, Sarmento Vieira F, Persechini PM, Coutinho-Silva R. 2008. Modulation of P2X7 receptor expression in macrophages from mineral oil-injected mice. Immunobiology 213:481–492. doi: 10.1016/j.imbio.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 52.Cockrell A, Laroux FS, Jourd'heuil D, Kawachi S, Gray L, Van der Heyde H, Grisham MB. 1999. Role of inducible nitric oxide synthase in leukocyte extravasation in vivo. Biochem Biophys Res Commun 257:684–686. doi: 10.1006/bbrc.1999.0484. [DOI] [PubMed] [Google Scholar]
- 53.Odds FC. 1988. Candida and candidosis. Baillere-Tindall, London, United Kingdom. [Google Scholar]
- 54.Huang W, Na L, Fidel PL, Schwarzenberger P. 2004. Requirement of interleukin-17A for systemic anti-Candida albicans host defense in mice. J Infect Dis 190:624–631. doi: 10.1086/422329. [DOI] [PubMed] [Google Scholar]
- 55.Conti HR, Shen F, Nayyar N, Stocum E, Sun JN, Lindemann MJ, Ho AW, Hai JH, Yu JJ, Jung JW, Filler SG, Masso-Welch P, Edgerton M, Gaffen SL. 2009. Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J Exp Med 206:299–311. doi: 10.1084/jem.20081463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saijo S, Ikeda S, Yamabe K, Kakuta S, Ishigame H, Akitsu A, Fujikado N, Kusaka T, Kubo S, Chung SH, Komatsu R, Miura N, Adachi Y, Ohno N, Shibuya K, Yamamoto N, Kawakami K, Yamasaki S, Saito T, Akira S, Iwakura Y. 2010. Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity 32:681–691. doi: 10.1016/j.immuni.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 57.Lord BI, Molineux G, Pojda Z, Souza LM, Mermod JJ, Dexter TM. 1991. Myeloid cell kinetics in mice treated with recombinant interleukin-3, granulocyte colony-stimulating factor (CSF), or granulocyte-macrophage CSF in vivo. Blood 77:2154–2159. [PubMed] [Google Scholar]
- 58.Weisbart RH, Kwan L, Golde DW, Gasson JC. 1987. Human GM-CSF primes neutrophils for enhanced oxidative metabolism in response to the major physiological chemoattractants. Blood 69:18–21. [PubMed] [Google Scholar]
- 59.Metcalf D, Begley CG, Williamson DJ, Nice EC, De Lamarter J, Mermod JJ, Thatcher D, Schmidt A. 1987. Hemopoietic responses in mice injected with purified recombinant murine GM-CSF. Exp Hematol 15:1–9. [PubMed] [Google Scholar]
- 60.Wenzel U, Schneider A, Valente AJ, Abboud HE, Thaiss F, Helmchen UM, Stahl RA. 1997. Monocyte chemoattractant protein-1 mediates monocyte/ macrophage influx in anti-thymocyte antibody-induced glomerulonephritis. Kidney Int 51:770–776. doi: 10.1038/ki.1997.108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.