Abstract
Chlamydia pneumoniae is a Gram-negative bacterium that causes acute or chronic respiratory infections. As obligate intracellular pathogens, chlamydiae efficiently manipulate host cell processes to ensure their intracellular development. Here we focused on the interaction of chlamydiae with the host cell transcription factor activator protein 1 (AP-1) and its consequence on chlamydial development. During Chlamydia pneumoniae infection, the expression and activity of AP-1 family proteins c-Jun, c-Fos, and ATF-2 were regulated in a time- and dose-dependent manner. We observed that the c-Jun protein and its phosphorylation level significantly increased during C. pneumoniae development. Small interfering RNA knockdown of the c-Jun protein in HEp-2 cells reduced the chlamydial load, resulting in smaller inclusions and significantly lower chlamydial recovery. Furthermore, inhibition of the c-Jun-containing AP-1 complexes using tanshinone IIA changed the replicative infection phenotype into a persistent one. Tanshinone IIA-dependent persistence was characterized by smaller, aberrant inclusions, a strong decrease in the chlamydial load, and significantly reduced chlamydial recovery, as well as by the reversibility of the reduced recovery after the removal of tanshinone IIA. Interestingly, not only was tanshinone IIA treatment accompanied by a significant decrease of ATP levels, but fluorescence live cell imaging analysis by two-photon microscopy revealed that tanshinone IIA treatment also resulted in a decreased fluorescence lifetime of protein-bound NAD(P)H inside the chlamydial inclusion, indicating that chlamydial reticulate bodies have decreased metabolic activity. In all, these data demonstrate that the AP-1 transcription factor is involved in C. pneumoniae development, with tanshinone IIA treatment resulting in persistence.
INTRODUCTION
Chlamydia pneumoniae is an obligate intracellular bacterium that causes acute and chronic infections of the upper and lower respiratory tract. The rate of seropositivity in most adult populations ranges from 60 to 90%, indicating the high worldwide prevalence of C. pneumoniae (1). During infection, chlamydiae exhibit a biphasic replication cycle with two distinct developmental forms, the infectious form, called the elementary body (EB), and the reticulate body (RB), which is the metabolically active and dividing form. Differentiation of EBs into RBs followed by replication occurs in a specialized vesicular compartment known as the inclusion (2). After replication the RBs start to redifferentiate into infectious EBs, which leave the cell, resulting in disease dissemination. In contrast, chlamydiae can also enter a persistent state, resulting in reduced infectivity and metabolic activity (3, 4). Some chlamydial proteins have been suggested to be indicators of chlamydial persistence, such as the major outer membrane protein (MOMP) and chlamydial heat shock protein 60 (cHsp60) (5, 6). Furthermore, persistence is characterized by an incomplete developmental cycle in line with the formation of a smaller and aberrant inclusion harboring only a few but large and aberrant RBs (7–9). Redifferentiation into infectious EBs does not occur, making direct cultivation of persistent chlamydiae impossible, although they remain viable (8, 10, 11). Persistence is reversible, as removal of the stimuli, e.g., iron deprivation, gamma interferon (IFN-γ) treatment, or application of certain antibiotics (e.g., penicillin), leads to reactivation and the return to replicative development (5, 12). In general, the molecular switch determining persistent or replicative infection is poorly understood.
Chlamydiae are discussed to be energy parasites, relying on the host cell for the ATP and metabolites required for development (13). Therefore, chlamydiae manipulate the energy balance of the host cell. Moreover, chlamydiae are known to modulate various host cell pathways, such as delaying apoptosis and preventing lysosomal fusion, to ensure their survival (14–17). A key player in these host cell processes is the dimeric transcription factor activator protein 1 (AP-1), making it an ideal target for chlamydiae to manipulate (18, 19). AP-1, composed of proteins from the Jun, Fos, and activating transcription factor (ATF) families, induces changes in the expression of host genes harboring AP-1 binding sites, influencing, e.g., inflammation, cell stress, and survival (19–21). A role for AP-1 upon infection with microbes or viruses has already been described (22, 23). Indeed, AP-1 was regulated during C. pneumoniae infection in smooth vascular muscle cells, while for the sexually transmittable organism Chlamydia trachomatis, AP-1 has been demonstrated to be crucial for bacterial growth and development (24, 25).
In this study, we aimed to investigate the role of AP-1 in the development of either a replicative or a persistent C. pneumoniae infection. We could demonstrate that proteins of the AP-1 family are modulated during C. pneumoniae infection and that small interfering RNA (siRNA) knockdown of c-Jun results in reduced chlamydial growth. Furthermore, application of the AP-1 inhibitor tanshinone IIA, which prevents the c-Jun/c-Fos AP-1 complex from binding to DNA, strongly alters the C. pneumoniae infection phenotype from a replicative to a persistent one. Moreover, we demonstrate that tanshinone IIA significantly reduces ATP levels within the first 12 h after infection, and two-photon microscopy revealed that tanshinone IIA treatment significantly decreases the amount of protein-bound NAD(P)H [τ2-NAD(P)H] in the infected cells. In all, these data demonstrate that AP-1 activity plays a role in C. pneumoniae development and that tanshinone IIA treatment contributes to persistence development.
MATERIALS AND METHODS
Organisms and cell culture.
Chlamydia pneumoniae (CWL strain 029 [ATCC VR-1310]) was propagated in HEp-2 cells as described previously (26). Cells were cultured over serial passages in high-glucose Dulbecco's modified Eagle's medium (DMEM; Lonza) supplemented with 10% fetal calf serum (FCS; Sigma), l-glutamine (2 mM; Biochrom), HEPES (33 mM; in-house facility Paul Ehrlich Institute [PEI]), and gentamicin (10 μg/ml; Sigma). Cells were grown in T75 flasks at 37°C with 5% CO2.
Infections.
Cells were infected with Chlamydia pneumoniae at 1, 10, 25, or 30 inclusion-forming units (IFUs) per cell in culture medium supplemented with 1 μg/ml cycloheximide (Sigma). After centrifugation for 1 h at 37°C, the cells were incubated for 4 h, 24 h, 48 h, or 72 h at 37°C with 5% CO2. For infection in chamber slides, the cells were not centrifuged. Cells cultured in medium alone were used as a control for each time point.
Chlamydial recovery.
The amount of infectious C. pneumoniae elementary bodies after treatment with either c-Jun-specific siRNA (described above) or tanshinone IIA or after no treatment was determined by titration experiments, as described before (7). After infection with C. pneumoniae for 65 to 72 h, the cells were harvested and vortexed with glass beads to release the chlamydiae, followed by several centrifugation steps. The supernatant was then used for reinfection of fresh cells. After another 48 h of incubation, cells were methanol fixed and stained as described above, before quantification using a Zeiss Observer microscope with a 40× oil immersion objective (Axiovision software; Carl Zeiss). The number of infectious progeny was determined by calculating the number of IFUs relative to the number of untreated infected cells.
Inhibitor treatment.
Tanshinone IIA (Sigma) was resuspended in dimethyl sulfoxide and used in parallel with C. pneumoniae infection at a final concentration of 25 μM.
Immunofluorescence analysis.
Cells were seeded in chamber slides at 24 h prior to infection and fixed with methanol at 48 h after infection. For immunofluorescence staining, an Imagen chlamydia kit (Oxoid) containing a fluorescein isothiocyanate (FITC)-labeled monoclonal antibody against chlamydia lipopolysaccharide (LPS) and a red stain for cytoplasm was used. Cells were counterstained with DAPI (4′,6-diamidino-2-phenylindole; Invitrogen) to visualize the nuclei and chlamydial DNA. Chamber slides were analyzed using a Zeiss Observer microscope with a 40× oil immersion objective (Axiovision software; Carl Zeiss) and a Zeiss LSM7 Live microscope with a 63× oil immersion objective (ZEN 2009 software; Carl Zeiss). Quantification of infection was assessed by randomly counting at least 300 cells. The inclusion size was measured using ImageJ software. Briefly, a threshold algorithm was run over the FITC-DAPI-stained images following measurement of the area of the inclusions.
Electron microscopy.
For transmission electron microscopy, differentially infected and/or treated HEp-2 cells were prepared as described previously (27). Briefly, infected HEp-2 cells with and without tanshinone IIA treatment were incubated for 48 h. Afterwards, the medium was removed and the cells were fixed using 5% glutaraldehyde in phosphate-buffered saline (pH 7.4) for 1 h at 4°C. Samples were then treated with 1% OsO4 for 2 h, dehydrated in a graded ethanol series, and embedded in araldite (Fluka). A contrasting agent was applied to the ultrathin sections, and the developmental stage and morphology of the inclusions were analyzed with a Philips EM 400 microscope.
Fluorescence live cell imaging (FLIM) of NAD(P)H by two-photon microscopy.
Two-photon microscopy studies were performed as previously described (28). Briefly, for two-photon microscopy studies, HEp-2 cells were grown on cover glasses in 50-mm culture dishes and infected with C. pneumoniae for 48 h. The cover glasses were examined in a MiniCeM (JenLab) chamber for microscopy fitted to a heated stage, which enabled live cell imaging. The two-photon microscope (DermaInspect; Jenlab) was equipped with a Chroma 640DCSPXR dichroic mirror (AHF Analysentechnik AG) and a 40× (numerical aperture, 1.3) Plan-Apochromat oil immersion objective (Zeiss). Fluorescence lifetimes were analyzed using SPCImage software (version 2.9.5.2996; Becker & Hickl). For image analysis, a region of interest (ROI) inside the chlamydial inclusion was selected. The lifetimes of pixels (5 by 5) in the ROI had been averaged before. The lifetime decay curves were fit to a double-decay model, in which the fast-decaying component corresponds to free NAD(P)H [τ1-NAD(P)H] and the slowly decaying component corresponds to protein-bound NAD(P)H [τ2-NAD(P)H]. The instrument response function (IRF) was measured from the second harmonic generation signal of a beta-barium-borate crystal, and it was used in the lifetime fit model. The mean values for τ2-NAD(P)H for all pixels inside the ROIs were calculated.
ATP assay.
For ATP measurement, a PhosphoWorks luminometric ATP assay kit (Biomol) was used following the manufacturer's instructions. Cells were seeded 24 h before infection and treatment in Nunclon plates with a white surface (Nunc). Cells were infected with 10 IFUs and treated with or without tanshinone IIA, as described above. As a control, uninfected and untreated cells were used. Directly after infection and treatment (0 h postinfection [hpi]) and at 6 and 12 hpi, ATP amounts were measured with a Tecan Infinite M200PRO microplate reader (Tecan).
siRNA transfection.
For knockdown (KD) experiments, cells were treated with a c-Jun-specific siRNA smart pool (Qiagen), a transfection reagent (mock-transfected control; Stemfect RNA transfection kit; Stemgent), or nontarget siRNA (Allstars control siRNA; AllStars negative control; Qiagen) or were left untreated (medium control) according to the manufacturer's instructions. HEp-2 cells were seeded into 6-well plates 24 h before transfection. At 24 h posttransfection (hpt), cells were harvested using trypsin-EDTA (in-house facility PEI) and counted with a cell counter (CASY cell counter; Roche). Knockdown efficiency was assessed by quantitative real-time PCR (qRT-PCR) and Western blot analysis. Furthermore, cells were seeded for infection experiments with 10 IFUs either in 24-well plates (for qRT-PCR and Western blot analysis) or in chamber slides (for immunofluorescence analysis) for the desired time points (24 hpi, 48 hpi, and 72 hpi). For data analysis, normalization was performed against the results of the control.
Western blot analysis.
For determination of protein amounts, pellets of uninfected or infected HEp-2 cells with and without tanshinone IIA treatment were lysed with Laemmli buffer and denatured at 95°C for 10 min. Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes (GE Healthcare). Membranes were blocked in Tris-buffered saline with Tween 20 (TBST; in-house facility PEI)–5% fat-free skimmed milk, followed by incubation with primary antibodies. Primary antibodies were rabbit monoclonal c-Jun antibody (catalog number 60A8; Cell Signaling), rabbit monoclonal phospho-c-Jun antibody (catalog number D47G9; Cell Signaling), rabbit monoclonal c-Fos antibody (catalog number 9F6; Cell Signaling), rabbit monoclonal phospho-c-Fos antibody (catalog number D82C12; Cell Signaling), rabbit monoclonal ATF-2 antibody (catalog number 20F1; Cell Signaling), and rabbit monoclonal phospho-ATF-2 antibody (catalog number 11G2; Cell Signaling), each of which was diluted 1:1,000 in TBST–5% bovine serum albumin (BSA); mouse monoclonal MOMP antibody (hybridoma supernatant; generated and kindly provided by G. Zhong and partly described elsewhere [29]) diluted 1:25 in TBST–5% BSA; and mouse monoclonal chlamydial Hsp60 antibody (hybridoma supernatant; generated and kindly provided by G. Zhong) diluted 1:500 in TBST–5% BSA. Equal loading and blotting efficiencies were verified by the use of mouse monoclonal β-actin antibody (catalog number AC-15; Sigma). For protein detection, goat anti-rabbit IgG or goat anti-mouse IgG horseradish peroxidase-conjugated secondary antibodies (catalog numbers sc-2004 and sc-2005; Santa Cruz Biotechnology) were used, followed by incubation with chemiluminescence protein detection reagents (GE Healthcare). Protein amounts were quantified using ImageJ software.
RNA isolation, reverse transcription-PCR, and qRT-PCR.
Eukaryotic and bacterial RNA was isolated using an RNeasy Plus RNA minikit (Qiagen) according to the manufacturer's instructions. cDNA was generated by reverse transcription-PCR using an ImProm-II reverse transcription system (Promega) following the manufacturer's instructions. cDNA was then used for measuring gene expression using a LightCycler 480 apparatus (Roche); SYBR green (Mesa Blue quantitative PCR Master Mix Plus for SYBR assay; NoRox kit); primers specific for c-Jun (5′-ATCGACATGGAGTCCCAG-3′, 5′-CGATTCTCTCCAGCTTCC-3′), c-Fos (5′-AACCTGTCAAGAGCATCAGC-3′, 5′-CCCAGTCTGCTGCATAGAAG-3′), MOMP (5′-GCCACAGCATTGTCTACTACTGAT-3′, 5′-AATCTGAACTGACCAGATACGTGA-3′), cHsp60 (5′-AGGACGTCACGTAGTTATAGATAA-3′, 5′-AGTTTTGCTGGCGACTTCT-3′), or Chlamydia pneumoniae 16S RNA (5′-TCGCCTGGGAATAAGAGAGA-3′, 5′-AATGCTGACTTGGGGTTGAG-3′); and the 2−ΔΔCT threshold cycle (CT) method for calculation of relative gene expression (30). GAPDH (glyceraldehyde-3-phosphate dehydrogenase; 5′-GAGTCAACGGATTTGGTCGT-3′, 5′-TTGATTTTGGAGGGATCTCG-3′) was used as a housekeeping gene. The level of gene expression relative to that of uninfected cells was determined at each time point.
Statistical analysis.
Numerical data are presented as the mean ± standard deviation (SD) and were calculated by a paired Student's t test using Microsoft Excel and GraphPad Prism (version 4) software, with P values being considered significant when they were less than 0.05.
RESULTS
AP-1 proteins are regulated by C. pneumoniae infection.
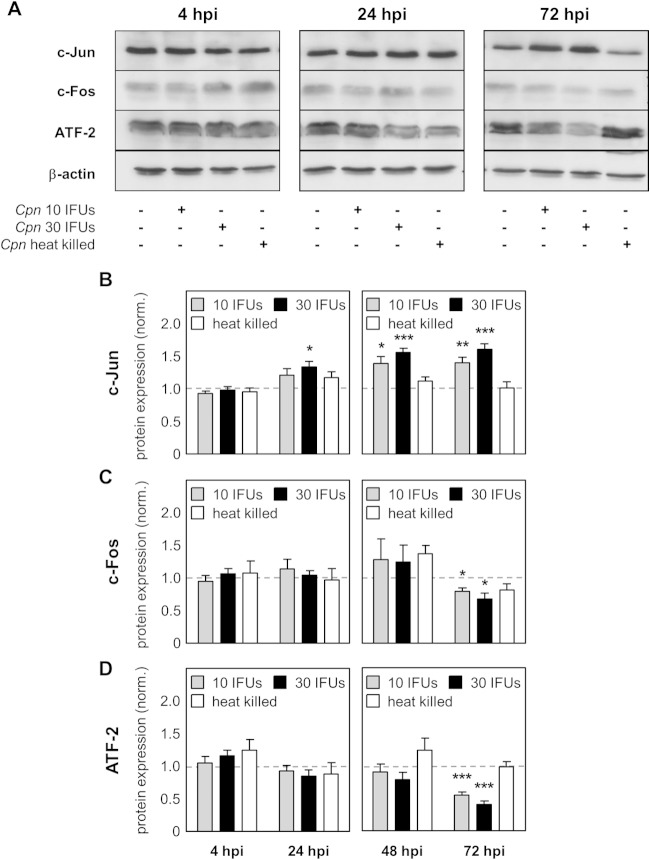
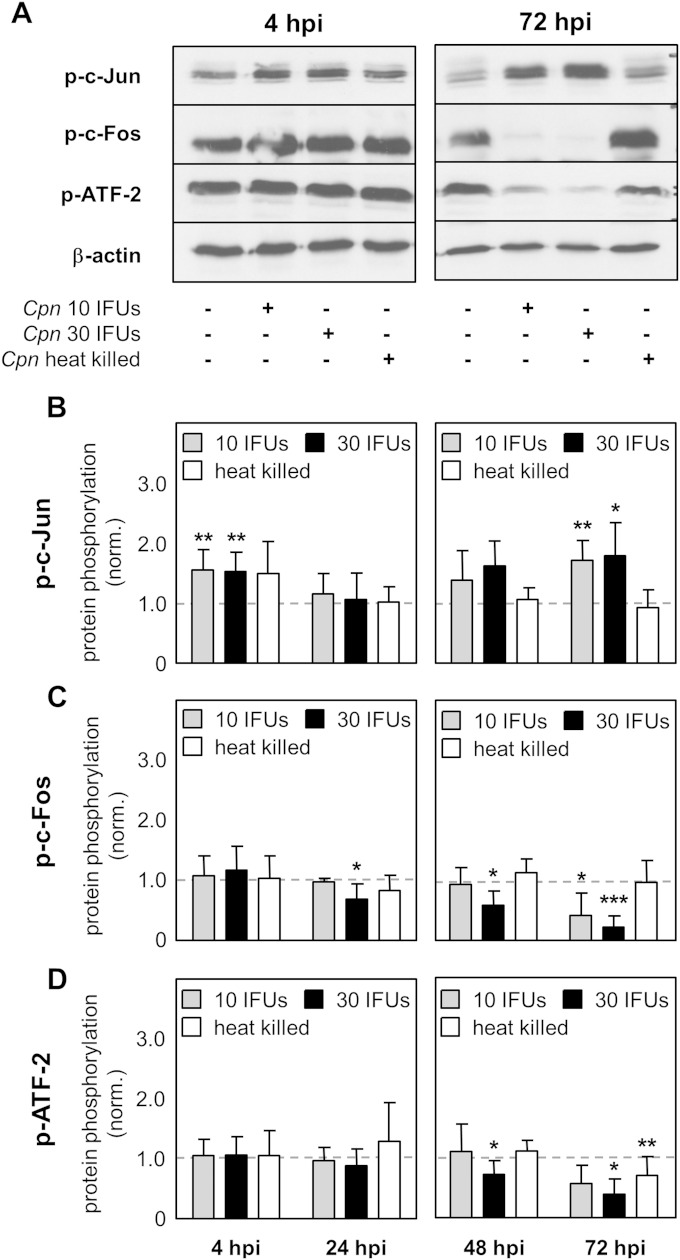
To investigate if AP-1 proteins are regulated during C. pneumoniae infection, the level of expression of the c-Fos, ATF-2, and c-Jun proteins over time was determined using Western blot analysis. Upon infection of HEp-2 cells with either a low dose (10 IFUs) or a high dose (30 IFUs) of C. pneumoniae or after stimulation with heat-killed C. pneumoniae, it was observed that the indicated proteins were differentially regulated. Over time, a decrease in c-Fos and ATF-2 protein expression and an increase in c-Jun protein expression were detectable (Fig. 1A). In more detail, using densitometry analysis, we found no differences in c-Fos and ATF-2 protein expression at 4 hpi, 24 hpi, and 48 hpi between infected and control cells, whereas protein expression was significantly downregulated at 72 hpi (Fig. 1C and D). Focusing on c-Jun, we also did not observe a difference in protein expression at 4 hpi. In contrast, at 24 h after infection with a low dose, the level of the c-Jun protein increased and the increase was significant with a high dose of infection. At later time points, 48 and 72 h after infection, c-Jun protein expression remained significantly upregulated (Fig. 1B). Of note, stimulation with heat-killed C. pneumoniae did not alter AP-1 protein expression. Moreover, phosphorylation of the AP-1 components was in complete accordance with the regulation of c-Jun, c-Fos, or ATF-2 protein expression (Fig. 2A). While the levels of c-Fos and ATF-2 phosphorylation significantly decreased over time, the quantity of phosphorylated c-Jun was upregulated at 4, 48, and 72 hpi (Fig. 2B to D). In all, we found that C. pneumoniae regulates AP-1 protein expression and activity levels in a time- and dose-dependent manner.
FIG 1.
Differential regulation of AP-1 proteins after C. pneumoniae infection. Lysates from uninfected HEp-2 cells (control), HEp-2 cells infected with 10 IFUs (low dose) or 30 IFUs (high dose) of C. pneumoniae (Cpn), or HEp-2 cells infected with heat-killed C. pneumoniae were prepared at various time points after infection (4 hpi, 24 hpi, 48 hpi, and 72 hpi). (A) Western blot of the indicated proteins at 4 hpi, 24 hpi, and 72 hpi. (B to D) Using densitometry, expression of the c-Jun (B), c-Fos (C), and ATF-2 (D) proteins was quantified during C. pneumoniae infection of HEp-2 cells. Data were normalized (norm.) against those for the uninfected control (dashed gray lines). Data, presented as the mean ± SD, and immunoblots are representative of those from at least four independent experiments (n = 4 to 6). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
FIG 2.
Phosphorylation of AP-1 proteins after C. pneumoniae infection. Lysates from uninfected HEp-2 cells (control), HEp-2 cells infected with 10 IFUs (low dose) or 30 IFUs (high dose) of C. pneumoniae (Cpn), or HEp-2 cells infected with heat-killed C. pneumoniae were prepared at various time points after infection (4 hpi, 24 hpi, 48 hpi, and 72 hpi). (A) The phosphorylation of the c-Jun, c-Fos, and ATF-2 proteins and the expression of β-actin were analyzed by Western blot analysis at the indicated time points (4 hpi and 72 hpi). (B to D) Using densitometry, phosphorylation of the c-Jun (B), c-Fos (C), and ATF-2 (D) proteins was quantified during C. pneumoniae infection of HEp-2 cells. Data were normalized against those for the uninfected control (dashed gray lines). Data are presented as the mean ± SD. Data and immunoblots are representative of those from at least four independent experiments (n = 4 to 6). p-c-Jun, p-c-Fos, and p-ATF-2, phosphorylated c-Jun, c-Fos, and ATF-2, respectively. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Knockdown of the c-Jun protein influences C. pneumoniae infection.
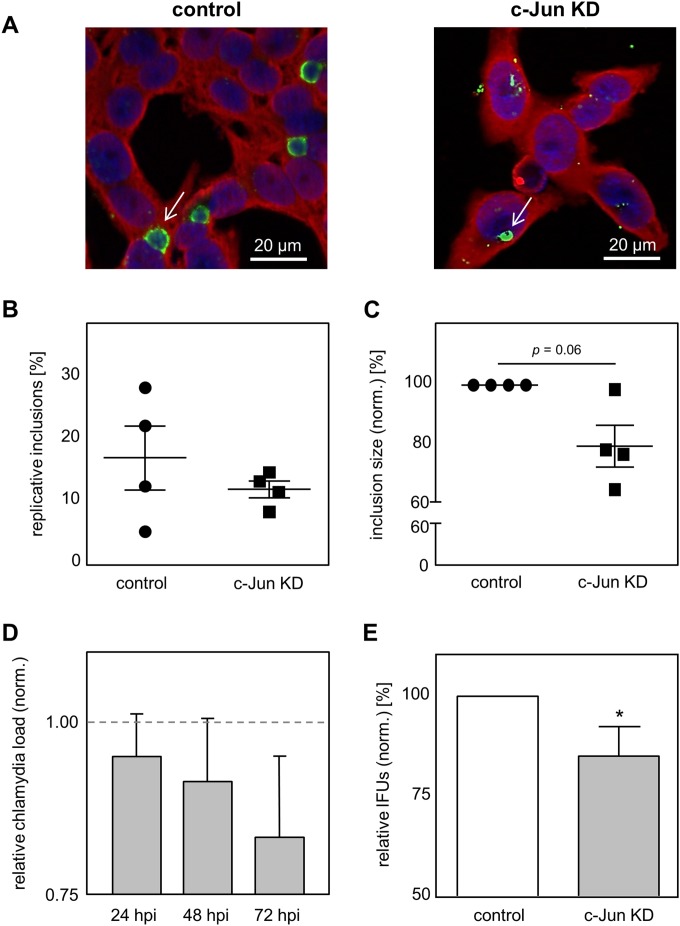
Infection with C. pneumoniae was shown to result in the significant upregulation of c-Jun protein expression. To investigate whether increased expression of c-Jun is beneficial for intracellular growth and the progeny of C. pneumoniae, we performed c-Jun-specific siRNA knockdown experiments in HEp-2 cells. By qRT-PCR and Western blot analysis, we could confirm c-Jun protein knockdown efficiency. We demonstrated that c-Jun mRNA expression is significantly reduced at 24 h posttransfection (hpt). In addition, the amount of c-Jun mRNA remained significantly reduced during C. pneumoniae infection over time (24, 48, and 72 hpi) (data not shown). In agreement with those findings, quantification of c-Jun protein levels using Western blot analysis showed a strong decrease in the amount of the c-Jun protein at 24 hpt, as well as at 24 h, 48 h, and 72 h after C. pneumoniae infection, compared to the amount produced by control cells (see Fig. S1A and B in the supplemental material). The quantification of protein expression by densitometry revealed that the levels of the c-Jun protein were significantly decreased in the c-Jun-knockdown cells (data not shown). To assess the consequence of the decreased c-Jun protein level on C. pneumoniae infection, we analyzed the infection using chlamydia-specific staining and immunofluorescence microscopy (Fig. 3A). We observed that more pathological inclusions, characterized by a large and round morphology, were present in the control cells than in the cells treated with c-Jun-specific siRNA (Fig. 3B). Moreover, after quantification of the inclusion size, we found that in the c-Jun-specific siRNA-treated cells, the size of the inclusions was reduced by 20% ± 13.7% compared to that of the inclusions in the control cells (Fig. 3C). This decrease in inclusion size was accompanied by a reduction in the level of MOMP protein expression (74.6% ± 24.6%) in comparison to that for the control (100%) determined by Western blot analysis and densitometry (see Fig. S1C and D in the supplemental material). To further characterize the effect of c-Jun knockdown on C. pneumoniae infection, we analyzed the chlamydial load in either control or knockdown cells via qRT-PCR. In concordance with the smaller inclusions, we detected a reduction in the amount of C. pneumoniae 16S RNA in the c-Jun-knockdown cells (Fig. 3D). Additionally, we assessed the impact of the c-Jun knockdown on the ability of the chlamydiae to recover. We observed significantly fewer inclusions after infection with C. pneumoniae extracted from c-Jun-knockdown cells (85.1% ± 7.3%) than after infection with C. pneumoniae obtained from control cells (100%) (Fig. 3E). Taken together, we can demonstrate that a diminished level of c-Jun protein during infection impairs C. pneumoniae development.
FIG 3.
Knockdown of the c-Jun protein impairs C. pneumoniae infection. (A) Infected cells with and without c-Jun knockdown (c-Jun KD) were methanol fixed at 48 hpi, immunostained by use of an Imagen chlamydia kit (green, chlamydial LPS; red, cytoplasm), and counterstained with DAPI (blue). White arrows, C. pneumoniae inclusions. (B and C) From the immunofluorescence images, the amount of pathological inclusions (B) and the size (C) of control and c-Jun-KD cells were quantified during infection. (D) Quantification of the relative C. pneumoniae load in c-Jun KD cells using qRT-PCR. (E) Chlamydial recovery after infection with C. pneumoniae cells obtained from infected control or c-Jun-specific siRNA-treated cells. The data in panels C to E were normalized against those for the control (dashed gray line in panel D). Data represent the mean ± SD. Data, immunoblots, and immunofluorescence images are representative of those from at least three independent experiments (n = 3 to 7). *, P < 0.05.
Blocking AP-1-mediated transcription results in C. pneumoniae growth restriction.
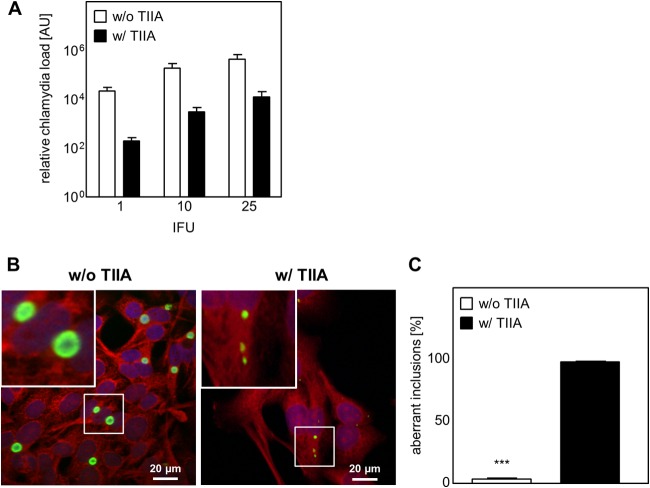
We demonstrated that c-Jun knockdown has a negative effect on C. pneumoniae development. As c-Jun is part of the AP-1 complex, we next wanted to check if the role of c-Jun in AP-1-mediated transcription is essential for chlamydial development. Therefore, we used tanshinone IIA, which prevents AP-1 DNA binding, as a specific inhibitor of c-Jun/c-Fos AP-1 complexes (31). We observed that the application of tanshinone IIA during a time span of up to 12 h after C. pneumoniae infection already had a strong effect on chlamydial recovery and chlamydial inclusion morphology (data not shown). In our experiments, we applied tanshinone IIA in parallel with C. pneumoniae infection. To assess the effect of tanshinone IIA treatment on chlamydial development, we first determined MOMP and cHsp60 protein expression by Western blot analysis (see Fig. S2A in the supplemental material). Tanshinone IIA treatment strongly altered MOMP and, consequently, cHsp60 protein expression, resulting in a significant reduction of both proteins compared to those in untreated cells (100%) at 48 hpi (MOMP, 17.2% ± 7.3%; cHsp60, 41.7% ± 13.9%) and the amount of MOMP as well at 72 hpi (MOMP, 19.5% ± 7.9%; cHsp60, 67.7% ± 19.0%) (see Fig. S2B in the supplemental material). Even though MOMP and cHsp60 protein expression decreased, the expression of the cHsp60 protein was always significantly higher than that of the MOMP protein. Subsequently, we determined the MOMP/cHsp60 ratio, based on the protein expression data, as an indicator of persistence. In tanshinone IIA-treated cells, we found the MOMP/cHsp60 ratio to be significantly decreased in contrast to that in the infected but untreated cells (see Fig. S2C in the supplemental material). Additionally, we assessed MOMP and cHsp60 mRNA expression after tanshinone IIA treatment (see Fig. S2D in the supplemental material). We could show that the mRNA level of MOMP significantly decreased over time starting at 24 hpi, while cHsp60 mRNA was regulated inversely, with its level already increasing significantly after 4 hpi. In line with the level of protein expression, a decrease in the level of MOMP mRNA was again accompanied by a higher level of cHsp60 mRNA expression. In addition, we could show that after tanshinone IIA treatment the load of C. pneumoniae inside the HEp-2 cells strongly decreased (1 IFU, 157-fold; 10 IFUs, 62-fold; 25 IFUs, 46-fold) (Fig. 4A). Furthermore, analyzing the infection by immunofluorescence microscopy, we could observe that tanshinone IIA-treated cells showed an infection phenotype different from that of untreated cells (Fig. 4B). The inclusions were smaller and had an aberrant shape (Fig. 4B, right) compared to the shape of the control cells (which were not treated with tanshinone IIA), which exhibited a typical replicative infection phenotype. After tanshinone IIA treatment, the majority (98.4% ± 0.6%) of inclusions exhibited an aberrant shape compared to the shape of the untreated cells, where only a few (4.1% ± 0.6%) inclusions had such an aberrant morphology (Fig. 4C).
FIG 4.
Tanshinone IIA treatment decreases the chlamydial load and limits inclusion development. HEp-2 cells were infected with C. pneumoniae (1, 10, or 25 IFUs) and in parallel treated with (w/) or without (w/o) tanshinone IIA (TIIA; 25 μM). After 48 h the cells were harvested, followed by mRNA isolation. (A) Quantification of relative chlamydial load using qRT-PCR. AU, arbitrary units. (B) Infected cells (10 IFUs) with and without tanshinone IIA treatment were fixed with methanol after 48 h, immunostained by use of an Imagen chlamydia kit (green, chlamydial LPS; red, cytoplasm), and counterstained with DAPI (blue). (Insets) Magnified images of the boxed areas. (C) Aberrant inclusions were quantified in C. pneumoniae-infected cells with and without tanshinone IIA treatment. Data, presented as the mean ± SD, and immunofluorescence images are representative of those from at least three independent experiments (n = 3 to 4). ***, P < 0.001.
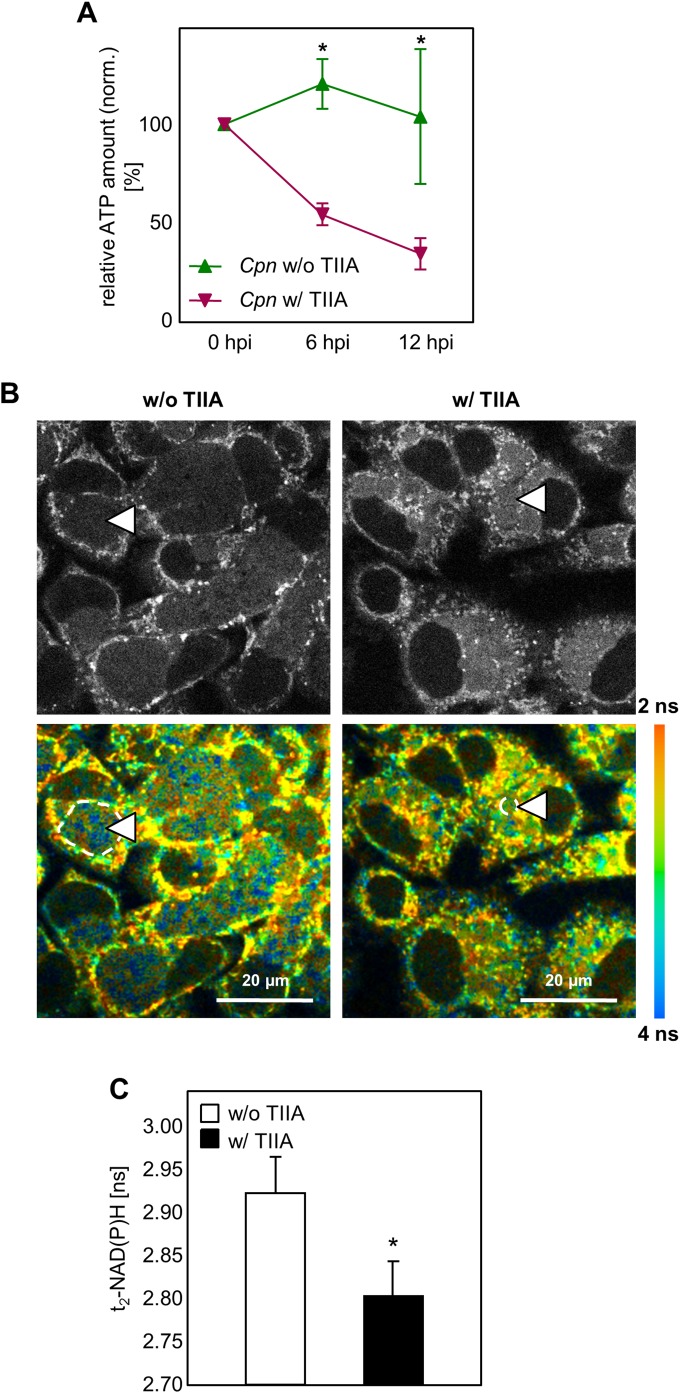
In addition to quantitative and morphological analysis of C. pneumoniae infection upon tanshinone IIA treatment, we assessed the ATP level as an indicator of changes in energy metabolism. We found that at 6 and 12 h after infection, the ATP level increased, while in the presence of tanshinone IIA, the ATP level was significantly reduced (Fig. 5A). For later time points of infection, we used the NAD(P)H measurement as an indicator of the metabolic activity of the chlamydial RBs (32). Therefore, we assessed the fluorescence lifetime of protein-bound NAD(P)H [τ2-NAD(P)H] in the C. pneumoniae inclusions using two-photon microscopy and fluorescence live cell imaging (FLIM). We found that, using this method, it was clearly visible that tanshinone IIA treatment resulted in a decrease in the amount of τ2-NAD(P)H compared to that in the untreated controls (Fig. 5B). Quantifying τ2-NAD(P)H from multiple infected cells and their corresponding ROIs, we found significantly lower τ2-NAD(P)H levels (Fig. 5C). These data clearly demonstrate that AP-1 transcription activity, especially that of the c-Jun-containing AP-1 complex, is of major importance for C. pneumoniae development.
FIG 5.
The ATP level and protein-bound NAD(P)H fluorescence lifetime decreased after tanshinone IIA treatment. (A) Quantification of the relative amount of ATP. HEp-2 cells were infected with 10 IFUs of C. pneumoniae (Cpn) and treated with or without tanshinone IIA (TIIA; 25 μM). Relative ATP levels were measured directly (0 hpi) and at 6 hpi and 12 hpi. (B and C) HEp-2 cells were infected with C. pneumoniae, and tanshinone IIA was applied in parallel, followed by incubation for 48 h. (B) Gray-scale images of the NAD(P)H fluorescence intensity (top) and color-coded images of the protein-bound NAD(P)H fluorescence lifetime (τ2-NAD(P)H) (bottom) in C. pneumoniae-infected cells not treated (left) and treated (right) with tanshinone IIA. Dashed lines and arrowheads indicate representative chlamydial inclusions which were selected for quantitative analysis. ns, nanoseconds. (C) Quantitative analysis of τ2-NAD(P)H inside the chlamydial inclusion. Chlamydial inclusions (n = 30) from three different samples were analyzed on two independent experimental days. Data, presented as the mean ± SEM, are representative of those from three independent experiments (n = 3). *, P < 0.05.
Tanshinone IIA treatment induces a persistence-like C. pneumoniae phenotype.
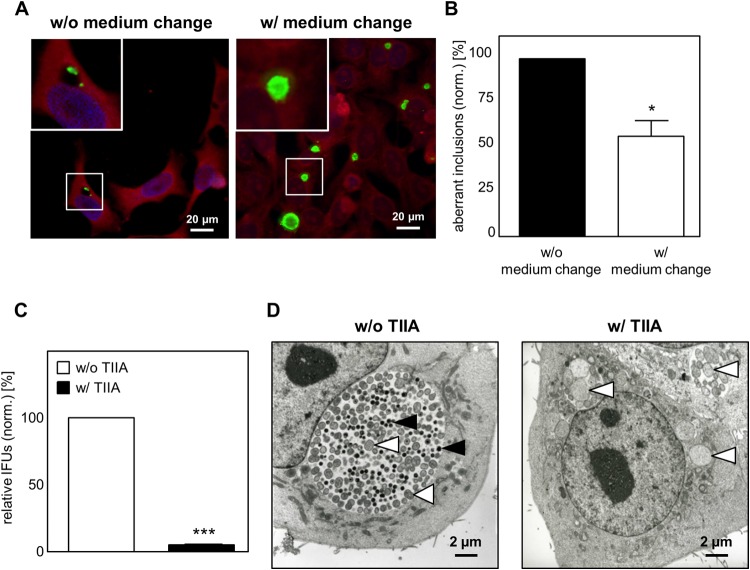
To assess if tanshinone IIA treatment causes a persistent C. pneumoniae infection, we first examined whether this phenotype is reversible. After C. pneumoniae infection and application of tanshinone IIA for 48 h, we removed the inhibitor by changing the medium. After an additional 48 h of culturing, the phenotype of infection and the inclusion morphology were analyzed using immunofluorescence microscopy. Without a medium change, all inclusions (100% ± 0.0%) remained small with an aberrant shape (Fig. 6A). In contrast, at 48 h after the medium change, almost half of all inclusions (43.2% ± 15.2%) increased in size, indicating that they returned to their replicative developmental cycle (Fig. 6B). In addition, recovery of infectious C. pneumoniae after tanshinone IIA removal resulted in the formation of typical developing inclusions, while in the presence of the inhibitor, recovery was hardly possible (4.9% ± 0.1%) (Fig. 6C). To clearly discriminate between the elementary and reticulate bodies present in the inclusion, we used electron microscopy. The untreated cells showed a typical replicative infection characterized by large inclusions containing a mixture of EBs and RBs (Fig. 6D, left). In contrast, tanshinone IIA treatment during C. pneumoniae infection resulted in small inclusions harboring only a few but large and aberrant RBs, showing the typical persistence phenotype (Fig. 6D, right). These data demonstrate that inhibition of the c-Jun-containing AP-1 complex by tanshinone IIA turns a replicative C. pneumoniae infection into a persistent infection.
FIG 6.
Inhibition of AP-1 (c-Jun/c-Fos) transcription activity results in persistent C. pneumoniae infection. HEp-2 cells were infected with C. pneumoniae (10 IFUs), and tanshinone IIA (TIIA; 25 μM) was applied in parallel, followed by incubation for 48 h. Cells were cultured with or without tanshinone IIA for an additional 48 h. (A) Cells were fixed with methanol, immunostained by use of an Imagen chlamydia kit (green, Chlamydia LPS; red, cytoplasm), and counterstained with DAPI (blue). (Insets) Magnified images of the boxed areas. (B) Quantification of aberrant inclusions during infection without and with a medium change. (C) Chlamydial recovery 48 h after infection with and without tanshinone IIA treatment. (D) Electron microscope micrographs of infected HEp-2 cells without and with tanshinone IIA treatment. Black arrowheads, elementary bodies; white arrowheads, reticular bodies. Data represent the mean ± SD from at least three independent experiments. Immunofluorescence and electron microscope images are representative of those from three independent experiments (n = 3). *, P < 0.05; ***, P < 0.001.
DISCUSSION
Chlamydia pneumoniae is a Gram-negative bacterium with an obligate intracellular life cycle, forcing it to manipulate host cell pathways. The ubiquitous transcription factor AP-1 is a central player in regulating various host cell pathways and an ideal target for chlamydiae to modulate. Indeed, we show that AP-1 is involved in the development of C. pneumoniae. Proteins of the AP-1 family were phosphorylated and subsequently regulated in a time- and dose-dependent manner upon infection. In addition, knockdown of c-Jun with specific siRNA altered the infection, causing the formation of smaller inclusions; reduced the chlamydial load; and significantly decreased chlamydial recovery. Finally, application of a specific c-Jun/c-Fos AP-1 complex inhibitor, tanshinone IIA, induced the persistence of C. pneumoniae infection and a corresponding decrease in the ATP level and the τ2-NAD(P)H fluorescence lifetime.
The role of AP-1 during inflammation has already been highlighted for other pathogens (33–35). The hepatitis C virus was found to modify AP-1, supporting viral development by promoting cell cycle progression (36). In addition, Bacillus anthracis produces the edema toxin, which was shown to induce AP-1 and to restrict tumor necrosis factor alpha production (37). In concordance with those findings, we demonstrated that proteins of the AP-1 family are regulated upon chlamydia infection. We found that C. pneumoniae regulates AP-1 proteins mainly at late time points during development, a finding in line with that from a recent study focusing on Chlamydia trachomatis (25). It was demonstrated that the phosphorylation of c-Jun and c-Fos with subsequent AP-1-dependent transcription is induced at a late developmental stage, suggesting that AP-1 is involved in replication or during the redifferentiation process.
Since expression of the c-Jun protein was modulated earlier than that of c-Fos and ATF-2 and protein expression was increased, our data suggest that c-Jun plays a more crucial role in chlamydial development. To strengthen this idea, we applied an approach with siRNA knockdown of c-Jun to assess the requirement of this AP-1 component for chlamydial development. Knockdown of the c-Jun protein altered C. pneumoniae infection, resulting in (i) 20% smaller inclusions, (ii) downregulation of MOMP, (iii) reduction of the chlamydial load, and (iv) a significant decrease in chlamydial recovery. Our findings indicate that growth conditions are suboptimal in the absence of the c-Jun protein and are in line with those described in the report of Olive et al., who found that c-Jun knockdown results in a significant reduction in the level of IFU production (25). In our study, the knockdown effect on infection was possibly limited, since the siRNA approach resulted in only a partial reduction in the level of the c-Jun protein. Therefore, we can speculate that the remaining c-Jun proteins can still dimerize with c-Fos, resulting in AP-1-related transcription.
Subsequently, we wanted to evaluate the function of c-Jun as part of c-Jun/c-Fos AP-1, as we observed increased AP-1 DNA binding following C. pneumoniae infection (data not shown). We used tanshinone IIA, an isolate of the root of Salvia miltiorrhiza Bunge, which was shown to specifically inhibit the c-Jun/c-Fos AP-1 complex, preventing DNA binding and the subsequent transcriptional activity (31, 38). Blocking AP-1 DNA binding, using tanshinone IIA, resulted in a drastic change in infectivity and chlamydial development. We could show that the application of tanshinone IIA not only led to growth restriction, as recently shown for Chlamydia trachomatis, but also changed a replicative infection into a persistent one (31). We first determined the levels of MOMP and cHsp60 protein expression and subsequently calculated the MOMP/cHsp60 ratio (as described by Beatty et al. [10]) as an indicator for persistence. We found that after tanshinone IIA treatment the MOMP/cHsp60 ratio decreased significantly, and such a decrease is usually associated with the persistence of C. trachomatis. Studies concerning C. trachomatis persistence have often found the expression of MOMP to be decreased, while cHsp60 expression has been found to remain stable or increase during persistence (2, 7, 10, 39). It was shown that, besides the chlamydial species, an inducer of persistence, such as IFN-γ or penicillin, is of essential importance for the transcriptional persistence profile (40, 41). The results presented in previous publications are in agreement with the finding that the MOMP/cHsp60 ratio cannot be used as a universal persistence marker (5, 6, 42). Tanshinone IIA, as an inducer of persistence, might be responsible for this unusual C. pneumoniae persistence profile, as has been found for C. trachomatis.
A possible explanation for the persistent phenotype induced by tanshinone IIA is the fact that AP-1 is involved in processes coordinating cell proliferation, which requires a functional host cell metabolism. Moreover, intracellular chlamydiae also have a high energy demand during their replicative cycle. As inhibition of AP-1 leads to the suppression of cell proliferation, our data suggest that interference with AP-1-regulated host cell metabolism affects chlamydial development (43, 44). In line with that suggestion, it is known that AP-1 regulates genes responsible for cellular energy balance, such as glut-1 (45). As chlamydiae are hypothesized to be energy parasites, they are highly dependent on the host cell ATP/glucose supply (46). Indeed, upregulation of glut-1 mRNA expression following chlamydial infection was already described to take place in a later phase of infection (47). In the late stage of development, metabolically active RBs in need of ATP for replication and redifferentiation are present (48). In accordance with that finding, we demonstrated that ATP is upregulated early (6 and 12 hpi) after C. pneumoniae infection. In contrast, in the presence of tanshinone IIA, ATP levels were significantly decreased. The lack of ATP in the phase where differentiation from EBs to RBs occurs might cause the induction of persistence, as the requirements for proper chlamydial development are not provided anymore (49). Analysis of a later developmental stage (48 hpi) revealed a decrease in the lifetime of NAD(P)H fluorescence, serving as an indicator of the reduced metabolic activity of the chlamydial RBs. Therefore, we hypothesize that tanshinone IIA treatment results in limitations in the glucose supply and the subsequent ATP supply, which is responsible for incomplete development and leads to persistence (47, 50).
In summary, we demonstrated that AP-1 is involved in Chlamydia pneumoniae development. Knockdown of c-Jun altered the infection, resulting in a decreased inclusion size, chlamydial load, and chlamydial recovery, emphasizing the importance of c-Jun for chlamydial development. Furthermore, inhibition of AP-1 (c-Jun/c-Fos)-related transcription using tanshinone IIA dramatically changed the infection morphology as well as the bacterial load and chlamydial recovery, inducing persistence. These data demonstrate a role for the c-Jun-containing AP-1 complex and its transcriptional activity in Chlamydia pneumoniae development.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kristin Wischnat, Bianca Walber, Julia Heger, Henner Zirpel, Laura Roßmann, and Katharina Schmitt for technical support.
This project was financially supported by the German Research Foundation (DFG ZA-533/7-1 and DFG Ru1436/2-1).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.03083-14.
REFERENCES
- 1.Koh WP, Taylor MB, Hughes K, Chew SK, Fong CW, Phoon MC. 2002. Seroprevalence of IgG antibodies against Chlamydia pneumoniae in Chinese, Malays and Asian Indians in Singapore. Int J Epidemiol 31:1001–1007. doi: 10.1093/ije/31.5.1001. [DOI] [PubMed] [Google Scholar]
- 2.Scidmore-Carlson MA, Shaw EI, Dooley CA, Fischer ER, Hackstadt T. 1999. Identification and characterization of a Chlamydia trachomatis early operon encoding four novel inclusion membrane proteins. Mol Microbiol 33:753–765. doi: 10.1046/j.1365-2958.1999.01523.x. [DOI] [PubMed] [Google Scholar]
- 3.Harper A, Pogson CI, Jones ML, Pearce JH. 2000. Chlamydial development is adversely affected by minor changes in amino acid supply, blood plasma amino acid levels, and glucose deprivation. Infect Immun 68:1457–1464. doi: 10.1128/IAI.68.3.1457-1464.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson DE, Virok DP, Wood H, Roshick C, Johnson RM, Whitmire WM, Crane DD, Steele-Mortimer O, Kari L, McClarty G, Caldwell HD. 2005. Chlamydial IFN-gamma immune evasion is linked to host infection tropism. Proc Natl Acad Sci U S A 102:10658–10663. doi: 10.1073/pnas.0504198102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hogan RJ, Mathews SA, Mukhopadhyay S, Summersgill JT, Timms P. 2004. Chlamydial persistence: beyond the biphasic paradigm. Infect Immun 72:1843–1855. doi: 10.1128/IAI.72.4.1843-1855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villegas E, Camacho A, Sorlózano A, Rojas J. 2008. Emerging strategies in the diagnosis, prevention and treatment of Chlamydophila pneumoniae. Expert Opin Ther Patents 18:1–15. doi: 10.1517/13543776.18.1.1. [DOI] [Google Scholar]
- 7.Beatty WL, Byrne GI, Morrison RP. 1993. Morphologic and antigenic characterization of interferon gamma-mediated persistent Chlamydia trachomatis infection in vitro. Proc Natl Acad Sci U S A 90:3998–4002. doi: 10.1073/pnas.90.9.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borel N, Pospischil A, Hudson AP, Rupp J, Schoborg RV. 2014. The role of viable but non-infectious developmental forms in chlamydial biology. Front Cell Infect Microbiol 4:97. doi: 10.3389/fcimb.2014.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beatty WL, Morrison RP, Byrne GI. 1994. Persistent chlamydiae: from cell culture to a paradigm for chlamydial pathogenesis. Microbiol Rev 58:686–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beatty WL, Morrison RP, Byrne GI. 1994. Immunoelectron-microscopic quantitation of differential levels of chlamydial proteins in a cell culture model of persistent Chlamydia trachomatis infection. Infect Immun 62:4059–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beatty WL, Morrison RP, Byrne GI. 1995. Reactivation of persistent Chlamydia trachomatis infection in cell culture. Infect Immun 63:199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wyrick PB. 2010. Chlamydia trachomatis persistence in vitro: an overview. J Infect Dis 201(Suppl):S88–S95. doi: 10.1086/652394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moulder JW. 1991. Interaction of chlamydiae and host cells in vitro. Microbiol Rev 55:143–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Zandbergen G, Gieffers J, Kothe H, Rupp J, Bollinger A, Aga E, Klinger M, Brade H, Dalhoff K, Maass M, Solbach W, Laskay T. 2004. Chlamydia pneumoniae multiply in neutrophil granulocytes and delay their spontaneous apoptosis. J Immunol 172:1768–1776. doi: 10.4049/jimmunol.172.3.1768. [DOI] [PubMed] [Google Scholar]
- 15.Scidmore MA, Fischer ER, Hackstadt T. 2003. Restricted fusion of Chlamydia trachomatis vesicles with endocytic compartments during the initial stages of infection. Infect Immun 71:973–984. doi: 10.1128/IAI.71.2.973-984.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Small PLC, Ramakrishnan L, Falkow S. 1994. Remodeling schemes of intracellular pathogens. Science 263:637–639. [DOI] [PubMed] [Google Scholar]
- 17.Ham H, Sreelatha A, Orth K. 2011. Manipulation of host membranes by bacterial effectors. Nat Rev Microbiol 9:635–646. doi: 10.1038/nrmicro2602. [DOI] [PubMed] [Google Scholar]
- 18.Eferl R, Wagner EF. 2003. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer 3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 19.Schonthaler HB, Guinea-Viniegra J, Wagner EF. 2011. Targeting inflammation by modulating the Jun/AP-1 pathway. Ann Rheum Dis 70(Suppl 1):i109–i112. doi: 10.1136/ard.2010.140533. [DOI] [PubMed] [Google Scholar]
- 20.Shaulian E, Karin M. 2001. AP-1 in cell proliferation and survival. Oncogene 20:2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- 21.Shaulian E, Karin M. 2002. AP-1 as a regulator of cell life and death. Nat Cell Biol 4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 22.Seo JH, Lim JW, Kim H, Kim KH. 2003. Helicobacter pylori in a Korean isolate activates mitogen-activated protein kinases, AP-1, and NF-κB and induces chemokine expression in gastric epithelial AGS cells. Lab Invest 84:49–62. doi: 10.1038/labinvest.3700010. [DOI] [PubMed] [Google Scholar]
- 23.Xie J, Pan H, Yoo S, Gao S-J. 2005. Kaposi's sarcoma-associated herpesvirus induction of AP-1 and interleukin 6 during primary infection mediated by multiple mitogen-activated protein kinase pathways. J Virol 79:15027–15037. doi: 10.1128/JVI.79.24.15027-15037.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller SA, Selzman CH, Shames BD, Barton HA, Johnson SM, Harken AH. 2000. Chlamydia pneumoniae activates nuclear factor kappaB and activator protein 1 in human vascular smooth muscle and induces cellular proliferation. J Surg Res 90:76–81. doi: 10.1006/jsre.2000.5847. [DOI] [PubMed] [Google Scholar]
- 25.Olive AJ, Haff MG, Emanuele MJ, Sack LM, Barker JR, Elledge SJ, Starnbach MN. 2014. Chlamydia trachomatis-induced alterations in the host cell proteome are required for intracellular growth. Cell Host Microbe 15:113–124. doi: 10.1016/j.chom.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gieffers J, van Zandbergen G, Rupp J, Sayk F, Kruger S, Ehlers S, Solbach W, Maass M. 2004. Phagocytes transmit Chlamydia pneumoniae from the lungs to the vasculature. Eur Respir J 23:506–510. doi: 10.1183/09031936.04.00093304. [DOI] [PubMed] [Google Scholar]
- 27.Van Zandbergen G, Klinger M, Mueller A, Dannenberg S, Gebert A, Solbach W, Laskay T. 2004. Cutting edge: neutrophil granulocyte serves as a vector for Leishmania entry into macrophages. J Immunol 173:6521–6525. doi: 10.4049/jimmunol.173.11.6521. [DOI] [PubMed] [Google Scholar]
- 28.Szaszák M, Shima K, Käding N, Hannus M, Solbach W, Rupp J. 2013. Host metabolism promotes growth of Chlamydia pneumoniae in a low oxygen environment. Int J Med Microbiol 303:239–246. doi: 10.1016/j.ijmm.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 29.Wolf K, Fischer E, Mead D, Zhong G, Peeling R, Whitmire B, Caldwell HD. 2001. Chlamydia pneumoniae major outer membrane protein is a surface-exposed antigen that elicits antibodies primarily directed against conformation-dependent determinants. Infect Immun 69:3082–3091. doi: 10.1128/IAI.69.5.3082-3091.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Park S, Song J-S, Lee D-K, Yang C-H. 1999. Suppression of AP-1 activity by tanshinone and cancer cell growth inhibition. Bull Korean Chem Soc 20:925–928. [Google Scholar]
- 32.Szaszák M, Steven P, Shima K, Orzekowsky-Schröder R, Hüttmann G, König IR, Solbach W, Rupp J. 2011. Fluorescence lifetime imaging unravels C. trachomatis metabolism and its crosstalk with the host cell. PLoS Pathog 7:e1002108. doi: 10.1371/journal.ppat.1002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghosh S, Bhattacharyya S, Sirkar M, Sa GS, Das T, Majumdar D, Roy S, Majumdar S. 2002. Leishmania donovani suppresses activated protein 1 and NF-kappaB activation in host macrophages via ceramide generation: involvement of extracellular signal-regulated kinase. Infect Immun 70:6828–6838. doi: 10.1128/IAI.70.12.6828-6838.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zachos G, Clements B, Conner J. 1999. Herpes simplex virus type 1 infection stimulates p38/c-Jun N-terminal mitogen-activated protein kinase pathways and activates transcription factor AP-1. J Biol Chem 274:5097–5103. doi: 10.1074/jbc.274.8.5097. [DOI] [PubMed] [Google Scholar]
- 35.Vallejo JG, Knuefermann P, Mann DL, Sivasubramanian N. 2000. Group B streptococcus induces TNF-gene expression and activation of the transcription factors NF-B and activator protein-1 in human cord blood monocytes. J Immunol 165:419–425. doi: 10.4049/jimmunol.165.1.419. [DOI] [PubMed] [Google Scholar]
- 36.Koike K. 2007. Pathogenesis of HCV-associated HCC: dual-pass carcinogenesis through activation of oxidative stress and intracellular signaling. Hepatol Res 37(Suppl 2):S115–S120. doi: 10.1111/j.1872-034X.2007.00173.x. [DOI] [PubMed] [Google Scholar]
- 37.Comer JE, Galindo CL, Zhang F, Wenglikowski AM, Bush KL, Garner HR, Peterson JW, Chopra AK. 2006. Murine macrophage transcriptional and functional responses to Bacillus anthracis edema toxin. Microb Pathog 41:96–110. doi: 10.1016/j.micpath.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 38.Lee C-Y, Sher H-F, Chen H-W, Liu C-C, Chen C-H, Lin C-S, Yang P-C, Tsay H-S, Chen JJW. 2008. Anticancer effects of tanshinone I in human non-small cell lung cancer. Mol Cancer Ther 7:3527–3538. doi: 10.1158/1535-7163.MCT-07-2288. [DOI] [PubMed] [Google Scholar]
- 39.Cevenini R, Donati M, La Placa M. 1988. Effects of penicillin on the synthesis of membrane proteins of Chlamydia trachomatis LGV2 serotype. FEMS Microbiol Lett 56:41–45. doi: 10.1111/j.1574-6968.1988.tb03147.x. [DOI] [Google Scholar]
- 40.Jones ML, Gaston JS, Pearce JH. 2001. Induction of abnormal Chlamydia trachomatis by exposure to interferon-gamma or amino acid deprivation and comparative antigenic analysis. Microb Pathog 30:299–309. doi: 10.1006/mpat.2001.0433. [DOI] [PubMed] [Google Scholar]
- 41.Mathews S, George C, Flegg C, Stenzel D, Timms P. 2001. Differential expression of ompA, ompB, pyk, nlpD and Cpn0585 genes between normal and interferon-gamma treated cultures of Chlamydia pneumoniae. Microb Pathog 30:337–345. doi: 10.1006/mpat.2000.0435. [DOI] [PubMed] [Google Scholar]
- 42.Wyrick PB. 2010. Chlamydia trachomatis persistence in vitro: an overview. J Infect Dis 201(Suppl 2):S88–S95. doi: 10.1086/652394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H, Gao X, Zhang B. 2005. Tanshinone: an inhibitor of proliferation of vascular smooth muscle cells. J Ethnopharmacol 99:93–98. doi: 10.1016/j.jep.2005.01.057. [DOI] [PubMed] [Google Scholar]
- 44.Li X, Du J-R, Yu Y, Bai B, Zheng X-Y. 2010. Tanshinone IIA inhibits smooth muscle proliferation and intimal hyperplasia in the rat carotid balloon-injured model through inhibition of MAPK signaling pathway. J Ethnopharmacol 129:273–279. doi: 10.1016/j.jep.2010.03.021. [DOI] [PubMed] [Google Scholar]
- 45.Santalucía T, Christmann M, Yacoub MH, Brand NJ. 2003. Hypertrophic agonists induce the binding of c-Fos to an AP-1 site in cardiac myocytes: implications for the expression of GLUT1. Cardiovasc Res 59:639–648. doi: 10.1016/S0008-6363(03)00472-3. [DOI] [PubMed] [Google Scholar]
- 46.Moulder JW. 1970. Glucose metabolism of L cells before and after infection with Chlamydia psittaci. J Bacteriol 104:1189–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ojcius DM. 1998. Enhancement of ATP levels and glucose metabolism during an infection by Chlamydia. NMR studies of living cells. J Biol Chem 273:7052–7058. [DOI] [PubMed] [Google Scholar]
- 48.Omsland A, Sager J, Nair V, Sturdevant DE, Hackstadt T. 2012. Developmental stage-specific metabolic and transcriptional activity of Chlamydia trachomatis in an axenic medium. Proc Natl Acad Sci U S A 109:19781–19785. doi: 10.1073/pnas.1212831109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolf K, Fischer E. 2000. Ultrastructural analysis of developmental events in Chlamydia pneumoniae-infected cells. Infect Immun 68:2379–2385. doi: 10.1128/IAI.68.4.2379-2385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee SK, Kim BS, Yang WS, Kim SB, Park SK, Park JS. 2001. High glucose induces MCP-1 expression partly via tyrosine kinase-AP-1 pathway in peritoneal mesothelial cells. Kidney Int 60:55–64. doi: 10.1046/j.1523-1755.2001.00770.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.