Abstract
Yersinia enterocolitica is typically considered an extracellular pathogen; however, during the course of an infection, a significant number of bacteria are stably maintained within host cell vacuoles. Little is known about this population and the role it plays during an infection. To address this question and to elucidate the spatially and temporally dynamic gene expression patterns of Y. enterocolitica biovar 1B through the course of an in vitro infection, transcriptome sequencing and differential gene expression analysis of bacteria infecting murine macrophage cells were performed under four distinct conditions. Bacteria were first grown in a nutrient-rich medium at 26°C to establish a baseline of gene expression that is unrelated to infection. The transcriptomes of these bacteria were then compared to bacteria grown in a conditioned cell culture medium at 37°C to identify genes that were differentially expressed in response to the increased temperature and medium but not in response to host cells. Infections were then performed, and the transcriptomes of bacteria found on the extracellular surface and intracellular compartments were analyzed individually. The upregulated genes revealed potential roles for a variety of systems in promoting intracellular virulence, including the Ysa type III secretion system, the Yts2 type II secretion system, and the Tad pilus. It was further determined that mutants of each of these systems had decreased virulence while infecting macrophages. Overall, these results reveal the complete set of genes expressed by Y. enterocolitica in response to infection and provide the groundwork for future virulence studies.
INTRODUCTION
The genus Yersinia includes three species that are pathogenic to humans: Y. pestis, the causative agent of the plague, as well as Y. enterocolitica and Y. pseudotuberculosis, both of which cause gastrointestinal diseases. Of the three organisms, Y. enterocolitica is the species most frequently isolated from humans (1). Y. enterocolitica infections are typically acquired through ingestion of the bacteria in contaminated food or water, especially raw or undercooked pork products (2). After ingestion, the bacteria travel through the gastrointestinal tract to the terminal ileum, where they are able to penetrate the M cells of the Peyer's patches and infect the mesenteric lymph nodes (1–3). This leads to a self-limiting gastroenteritis and mesenteric lymphadenitis in otherwise healthy patients (3). In immunocompromised patients or young children with developing immune systems, the bacteria can spread from the lymph nodes to systemic sites, leading to potentially fatal septicemia (1). Over half of all reported Y. enterocolitica infections occur in children under the age of five, the group that is most predisposed to developing the systemic form of the infection (4).
Pathogenic Y. enterocolitica isolates can be divided into six distinct biovars based on several biochemical and physiological attributes (2, 5). Of the six biovars, the North American isolate, biovar 1B, is the most pathogenic, causing severe illness in humans and high levels of mortality in mice (2, 5, 6). The extreme virulence exhibited by biovar 1B can be partially explained by the presence of the ∼199-kb plasticity zone, a unique island within the biovar 1B genome that encodes a variety of virulence determinants, including an iron uptake system, a type II secretion system (T2SS), a type III secretion system (T3SS), and a type IV pilus (7). It has been demonstrated experimentally that each of these systems contributes to the virulence of the bacteria in mouse models of infection (7–12). In addition to the plasticity zone, there are several other factors that account for the enhanced pathogenicity of biovar 1B over less virulent strains, including additional genomic islands, altered gene expression patterns, and differential regulation of virulence factors (7, 13–15).
Y. enterocolitica, like all pathogenic species of Yersinia, is typically considered an extracellular pathogen, interacting with the host primarily from the surface of the cell rather than from intracellular compartments. This categorization as an extracellular pathogen is likely due to the fact that one of the roles of the Ysc T3SS, the primary Yersinia virulence determinant, is to inhibit phagocytosis (16). There is, however, ample evidence that small populations of bacteria are stably maintained intracellularly in Yersinia-containing vacuoles (YCV) during the infection of macrophages (17). Little is known about the role of these bacteria during infection or the mechanisms they use to defeat the macrophage defenses and replicate within the YCV. Given that there is no defined Yersinia growth medium that accurately mimics conditions in the YCV environment, little work has been done to elucidate the complete set of genes that is expressed under these conditions.
Through the course of an infection, Y. enterocolitica encounters a variety of host environments that it must rapidly and appropriately respond to in order to avoid being cleared by the immune system. To accomplish this, the bacterium must alter the expression of a large number of genes to meet the demands of each host environment it encounters. These changes typically include the induced expression of virulence factors, genes that have been shown to play a role in colonization of the host or the subversion of its immune response. Bacteria must also repress the expression of genes that provide no advantage or constitute a disadvantage in the host environment. Genes that encode proteins like FliC, which is easily detected by the host immune system, fall into this category (18). Among the most important virulence factors expressed by nearly all bacterial pathogens are the secretion systems. These systems are utilized by bacteria to directly interact with the host through the delivery of bacterial proteins into the host cell or into the extracellular space surrounding it. Y. enterocolitica biovar 1B encodes two T2SSs (Yts1 and Yts2) and two T3SSs (Ysc and Ysa). The most critical system is the Ysc T3SS, which is encoded on an ∼70-kb virulence plasmid common to all pathogenic species of Yersinia. The exact roles the other secretion systems play during infection are not well understood.
One way to study the global gene expression patterns of bacterial pathogens exposed to different environments is to use a next-generation RNA sequencing (RNA-Seq)-based approach. RNA-Seq has several advantages over microarray-based techniques, including higher sensitivity, a larger dynamic range, and the ability to discover noncoding and other regulatory RNAs (19, 20). In fact, RNA-Seq is the only technique that allows simultaneous expression measurements of all bacterial transcripts as the bacterium responds to changes in its environment. However, it is exceedingly difficult to sequence the bacterial transcriptome in the context of an infection as the vast majority of transcripts belong to the host (21). To overcome this obstacle, we developed a novel bacterial transcript enrichment strategy that increases the number of bacterial reads, allowing an accurate and unbiased analysis of the entire transcriptome (22, 23). In the present study, we use this technique to analyze the complete transcriptome of Y. enterocolitica biovar 1B through the course of a murine macrophage infection. By focusing on early time points during infection and through comparing gene expression profiles of extracellular versus intracellular bacteria, we identified numerous mechanisms used by the bacteria to respond to macrophages and avoid being eliminated after internalization. Combined with mutational analysis, our results reveal clear roles for the Ysa T3SS, the Yst2 T2SS, and Tad pilus. This is the first study to analyze the complete transcriptome of Y. enterocolitica biovar 1B during an infection and the first in-depth analysis of the many mechanisms the bacterium uses to establish itself within the intracellular environment.
MATERIALS AND METHODS
Bacterial strains, cell lines, and media.
All bacterial strains and plasmids used in the present study can be found in Table S1 in the supplemental material. Y. enterocolitica biovar 1B strain JB580v (24) and derivatives were routinely grown in TYE medium (10 g/liter tryptone, 5 g/liter yeast extract) at 26°C (growth medium) with shaking. For the microscopy experiments, strain GY5718 (25) harboring the plasmid pDW5 (26) was used. P388D1 murine macrophage cells obtained from the American Type Culture Collection (ATCC CCL-46) were grown in RPMI medium supplemented with fetal bovine serum to a final concentration of 10%. All macrophage growth and infections were performed at 37°C in an atmosphere of 5% CO2. Conditioned RPMI was created by filter-sterilizing (using a 0.2-μm-pore-size filter) medium that had been used to culture P388D1 cells for 2 days. Gentamicin (100 μg/ml), ampicillin (100 μg/ml), kanamycin (50 μg/ml), nalidixic acid (20 μg/ml), and chloramphenicol (12.5 μg/ml) were added to the bacterial growth medium where indicated.
Creation of bacterial mutant strains.
Strain GY4478 (JB580 pYV−) (27) was used as the parent strain for making insertion mutants of the Yts2 T2SS and Tad pilus in order to prevent the rapid killing of host cells by the Ysc T3SS. Plasmid pEP185.2 (24) was inserted into genes thought to be critical to the function of the Yts2 T2SS and Tad pilus (28–30), essentially as previously described (31). Briefly, 300- to 500-bp fragments of the genes yts2D (primer pair yts2D-F [5′-TCT AGA CCT ATA GTA GGA TAT GCG GAG AAC-3′] and yts2D-R [5′-TCT AGA CTG GCG TAG TAA CGG AGC-3′]) and tadA (primer pair tadA-F [5′-TCT AGA GCT CGC CAG TAT TGA TAT AGA TC-3′] and tadA-R [5′-TCT AGA CGT CCA ATG CAA TAG GGC-3′]) were PCR amplified and cloned into pCR-BluntII-TOPO (Life Technologies). The insert was then subcloned into the XbaI site of pEP185.2 and electroporated into Escherichia coli strain S17-1λpir (32). The E. coli strain containing the plasmid was then mated to Y. enterocolitica strain GY4478, and insertion mutants were recovered on Luria-Bertani (LB) plates with nalidixic acid and chloramphenicol.
Infection of murine macrophage cells.
P388D1 cells were grown for 2 days in six-well plates, at which point they had formed a nearly confluent monolayer with ∼1.5 × 106 cells/well. Y. enterocolitica was grown overnight and then subcultured in the morning in fresh media at a 1:100 dilution. The bacteria were allowed to grow for ∼3 h, at which point they had entered log-phase growth. Macrophages were infected at a multiplicity of infection (MOI) of 10 (∼1.5 × 107 CFU) with the log-phase bacteria. In parallel, six-well plates containing only conditioned RPMI were infected with an equal amount of bacteria. Infections were initiated by 5 min of centrifugation at 500 × g. Plates were placed back into the CO2 incubator at 37°C, and infections were allowed to proceed for up to 240 min. At 30, 60, 120, and 240 min, triplicate infections were washed twice with 37°C phosphate-buffered saline (PBS) to remove bacteria not associated with the macrophages, followed immediately by the addition of 1 ml of RNAzol (Molecular Research, Inc.). To isolate bacteria that had been internalized by the macrophages, gentamicin was added to triplicate wells after 60 min, and the infections were allowed to proceed for another 60 min (120 min total).
Mutant infections and competition assays.
Infections with Y. enterocolitica insertion mutants were performed as described above with the exception that after the addition of gentamicin, the infections were allowed to proceed for an additional 23 h (24 h total). In parallel, a set of wells had the RPMI medium replaced with fresh medium containing 10 μM cytochalasin D 1 h prior to infection. These wells were infected as before but not treated with gentamicin. After 24 h, triplicate infections were washed twice with 37°C with PBS, followed by lysis in 1 ml of PBS with 1% Triton X-100. In the single infections, this mixture was then serially diluted and plated on LB agar with chloramphenicol, whereas in the competition assays it was plated on nalidixic acid, as well as on plates with both nalidixic acid and chloramphenicol. The plates were incubated at 26°C for 2 days, and then the number of CFU was determined.
Statistical analysis.
For both single infections and competition assays, a Student t test was used to determine the significance of the differences in CFU counts. The results were considered significant if the P value was ≤0.05. Error bars in the single-infection experiments represent standard deviations from the mean.
Microscopy.
P388D1 cells were grown in two-well chamber slides (Nalge Nunc International) and infected with mid-log-phase bacteria at an MOI of ∼10 as in the previous experiment. After 1 h of infection, gentamicin was added to the chamber slides, and incubation continued for 150 min. The medium was then removed and replaced with fresh 37°C RPMI with 1 μg of Hoechst 33342 (Life Technologies)/ml and 100 μg of gentamicin/ml. Incubation was allowed to proceed for another 30 min for a total of 240 min of infection. At this point, the cells were imaged on a fluorescence microscope at ×600 magnification, taking pictures of the bright-field image, as well as the blue and green fluorescent images. As a control, uninfected cells in two-well chamber slides were treated in the same way.
RNA extraction and cDNA synthesis.
Samples (1 ml) in RNAzol from the six-well plates were transferred to 2-ml cryotubes, followed by the addition of 400 μl of molecular-biology-grade H2O (Sigma). Samples were mixed and then frozen at −80°C until extraction could be performed. Tubes were thawed, mixed, and incubated at room temperature for 15 min. They were then centrifuged at 4°C for 15 min at 16,000 × g. A total of 800 μl of the aqueous layer was transferred to a new tube and mixed with 800 μl of 100% ethanol. RNA was extracted from this solution using a Direct-Zol kit (Zymo Research) according to the manufacturer's instructions, followed by quantification using a Qubit (Life Technologies). Total RNA was fragmented using the NEBNext magnesium RNA fragmentation module with a 3-min incubation at 94°C. Fragmented RNA was cleaned using an RNA Clean & Concentrator-5 kit (Zymo). Double-stranded, tagged cDNA was generated from 50 ng of the fragmented RNA according to a previously described method (33). Briefly, oligonucleotides containing a short 22-base tag, followed by 3′ random hexamers, are used to prime reverse transcription by the Moloney murine leukemia virus (MMLV) reverse transcriptase. After completion of the transcript, the MMLV reverse transcriptase adds three Cs to the end of the molecule, and a template-switching reaction occurs (34) using an oligonucleotide with three 3′ ribonucleotide Gs preceded by a 21-base tag as the new template. The single-stranded cDNA is then purified using 1.8× volume of AMPure XP beads (Beckman Coulter). The second strand is generated by PCR using primers that bind to the 5′ and 3′ tags of the cDNA (33). Double-stranded cDNA is then cleaned using the DNA Clean & Concentrator-5 (Zymo).
Enrichment of Y. enterocolitica transcripts and library preparation.
Enrichment of bacterial transcripts from the host-dominated background was performed as previously described with some minor modifications (22, 23). Biotinylated Y. enterocolitica probes were generated from genomic DNA, including the plasmid pYV, using the BioPrime DNA labeling system according to the manufacturer's directions (Life Technologies). A 20-ng portion of double-stranded cDNA from each sample was mixed with 2 μg of biotinylated probe. This mixture was dried using a Vacufuge and resuspended in 10 μl of hybridization buffer (Nimblegen). Samples were denatured at 95°C for 5 min and allowed to rehybridize overnight at 60°C (16 to 20 h). Y. enterocolitica transcripts were captured using the previously described fluidic device and protocol (23) with an elution of the sample in 25 to 40 μl of elution buffer (Pierce). The enriched sample was then cleaned using 1.8× volumes of AMPure XP beads, and the purified product was eluted in H2O. A quantitative PCR was then performed to determine the number of cycles required to generate the final sequencing library, as well as to assess the efficacy of the enrichment (33). The final library was created by PCR amplifying the enriched sample with the full-length sequencing adapters and custom 9-mer barcodes. Each library was cleaned, and size selection was performed using a 0.8× volume of AMPure XP beads, followed by a 0.1× volume, to achieve a final library with an average size of ∼320 bp.
Sequencing.
The Vincent J. Coates Genomic Sequencing Laboratory (University of California at Berkeley) sequenced all of the samples on an Illumina HiSeq2500 using a 100-base single-ended reaction. A 9-base index read was performed to accommodate the custom 9-mer barcodes used in the present study. All sequencing data have been deposited in the NCBI Sequence Read Archive (SRA) under accession number SRP041683.
Bioinformatic analysis of RNA-Seq data.
Raw sequencing reads were filtered to remove low-quality reads and portions of reads derived from sequencing adaptors or primers using a previously described quality filter (35). Sequence alignment was done using TopHat2 (36), which uses the Bowtie2 sequence alignment tool to perform local alignment to the reference genome of Y. enterocolitica 8081 genome (NC_008800.1 [genome] and NC_008791.1 [plasmid]). An estimate of normalized expression values for genes was obtained using Cufflinks (37) to get FPKM (Fragments Per Kilobase of exon per Million fragments mapped) values for the supplied annotations in a GTF (General Transfer Format) format. To perform pairwise comparison .bam analyses, files generated from the alignments were converted to sorted .sam files by Samtools (38). HTseq software (HTSeq 0.6.1) (39) was used to get the counts of mapped reads to the gene feature listed in GFF (General Feature Format) files. The DESeq2 package (40) was used to get differential expression by estimating variance-mean dependence in count data, based on a model using the negative binomial for the desired pairwise analysis. The Clustergram function of MATLAB was used to make the heat maps shown in Fig. 2. This figure shows unsupervised clustering using Euclidean distance matrix.
FIG 2.
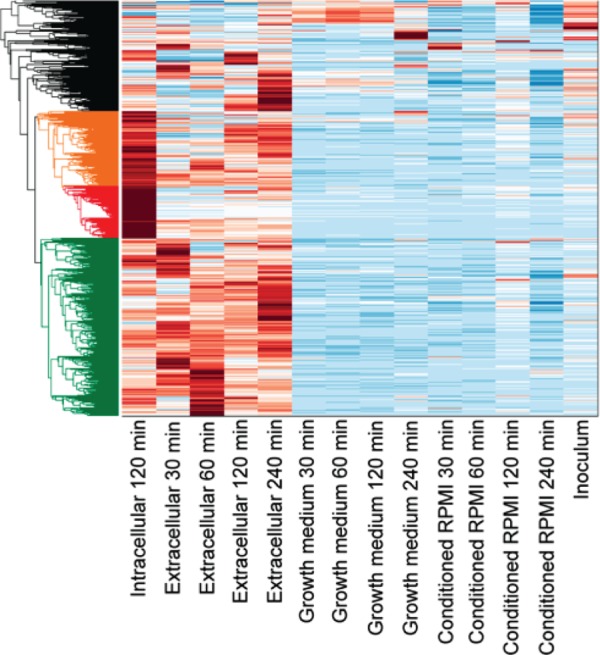
Expression of all Y. enterocolitica genes under each growth condition and over time. FPKM values were determined for each gene under each growth condition and then clustered by the similarity of their expression levels across conditions. The relative levels of expression are indicated by color, with dark blue depicting the lowest levels of expression and dark red indicating the highest. Genes were sorted into four clusters indicated by different colors: those most highly expressed during extracellular infection (green), those most highly expressed by internalized bacteria (red), those highly expressed in both infection conditions (orange), and those that do not fit within the other three clusters (black).
For the differential expression values calculated by the DESeq2 package, we applied robust linear regression on the time series of differential expression data for the infection time course. Genes that showed significant changes in differential expression during the course of infection were selected based on cutoffs on slopes and R2 values calculated from robust linear regression. The Clustergram function in MATLAB was used to cluster these genes to two sets (see Fig. 6). The top set of genes indicated in red are upregulated in early infection and then downregulated in the later phase of infection, and the second set of genes indicated in blue are downregulated in early infection and then upregulated in the later phase of infection.
FIG 6.
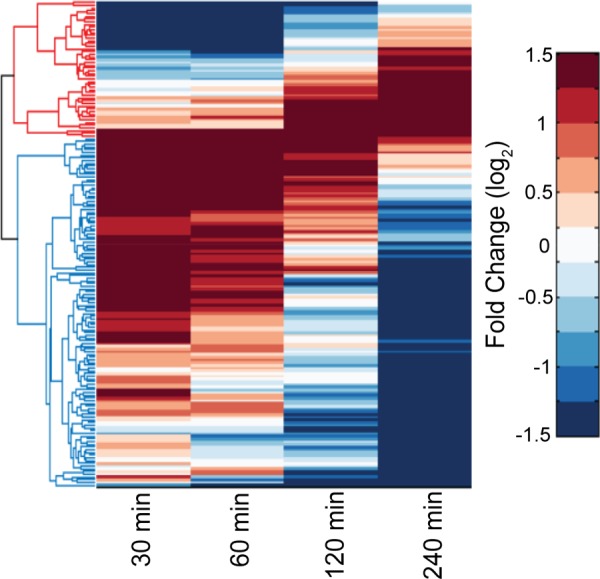
Dynamic expression of genes during the course of infection. The expression of all Y. enterocolitica genes by extracellular bacteria was analyzed at four time points (30, 60, 120, and 240 min) postinfection and compared to bacteria in conditioned RPMI over the same time period. Genes whose expression levels changed from upregulated to downregulated (red cluster in the dendrogram) or vice versa (blue cluster in the dendrogram) through the course of infection are depicted in the heat map. The names and functions of all genes present on the heat map are listed in Table S4 in the supplemental material.
RESULTS
Y. enterocolitica maintains viability after internalization by macrophages.
As Y. enterocolitica changes locations from different growth media, environmental locations, or host niches, the bacteria must rapidly adapt to the new setting by altering the transcription of a large number of genes. We began with bacteria in the exponential growth phase in a nutrient-rich medium at 26°C (growth medium), a common temperature used to grow Y. enterocolitica. This was followed by inoculation into one of four new environments (growth medium, conditioned RPMI, extracellular, and intracellular). The first environment was simply fresh growth medium at 26°C. The second was conditioned tissue culture medium at 37°C. Filter-sterilized RPMI medium that had been used to grow macrophages for 2 days (conditioned RPMI) was used to discover genes whose expression shifted as the bacteria responded to the new medium, the increased temperature, and the soluble host factors present in the medium. This also provided a baseline for the third environment: infection of macrophages (extracellular). We used a murine macrophage line rather than a human line so that we could more directly compare our results to those of other groups using mouse models of yersiniosis. P388D1 cells were used because they have been shown to be more similar to resident and exudate murine macrophages than other comparable cell lines (41). By comparing the transcriptomes of bacteria in conditioned RPMI to those of bacteria in contact with the murine macrophages, we hypothesized that we could identify genes that were differentially expressed in response to the presence of host cells. These genes are likely to be important virulence determinants. The final environment was the intracellular YCV, where the bacteria reside after being taken up by the macrophages (17). This was achieved by treating the infected macrophages with gentamicin to eliminate the extracellular bacteria. Analysis of the gene expression profiles of bacteria in this environment was chosen to elucidate the various mechanisms that Y. enterocolitica biovar 1B uses to survive and replicate within the macrophage.
To verify that the macrophages do not kill Y. enterocolitica during the time course used here, we infected the macrophages with a green fluorescent protein (GFP)-expressing strain and assessed the viability of intracellular bacteria after 4 h of infection. GFP-expressing bacteria observed within the macrophages would have to have been stable for at least 3 h without being degraded, since gentamicin was added after 1 h of infection to prevent extracellular bacteria from continuing to be internalized. Figure 1 depicts a comparison between an uninfected control (Fig. 1A to D) and infection with the GFP-expressing bacteria (Fig. 1E to H) looking at bright-field images, Hoechst staining of the nucleus, GFP-expressing bacteria, and the overlaid image of the three. As expected, live bacteria were found inside the macrophages after 4 h of infection, indicating that they do not kill all of the bacteria that become internalized. Although GFP-expressing bacteria within the macrophages appeared to be viable by live cell imaging, we also performed plate counts comparing the CFU present in our gentamicin-treated samples to an untreated control. After 4 h of infection the gentamicin-treated samples had an average of ∼81% fewer CFU (6.82 × 104 CFU) than the untreated control (5.52 × 106), indicating that ∼19% of the original bacteria were internalized and remained viable. Taken together, these results demonstrate that fairly large populations of bacteria remain alive within the macrophages through the time course of the present study.
FIG 1.
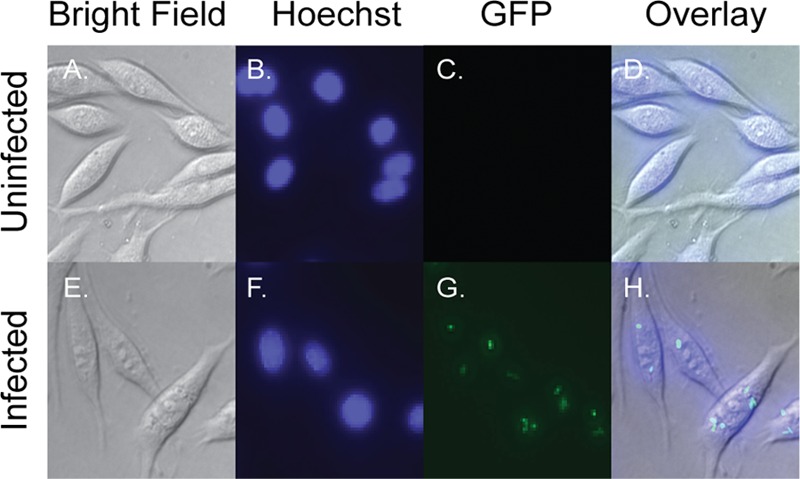
Images of infected and uninfected macrophages. (E to H) Infection of macrophages by Y. enterocolitica. P388D1 cells were infected with a constitutive GFP-expressing strain of Y. enterocolitica and, after 1 h of infection, treated with gentamicin. The infection was allowed to proceed for an additional 3 h, at which point macrophages were stained with the DNA-specific dye Hoechst 33342. (A to D) An uninfected control was treated in the same manner. Live cells are pictured at ×600 magnification.
Y. enterocolitica gene expression dynamics in different environments over time.
To begin to understand the dynamics of the Y. enterocolitica transcriptome, expression levels were determined for every gene in each of the four environments at all of the time points analyzed here. Figure 2 depicts the relative FPKM values for each gene in each environment over the course of 4 h. We found that in the growth medium, as well as in the conditioned RPMI, the majority of genes were consistently expressed at lower levels through the time course. Given the small inoculum (∼1.5 × 107 CFU in 3 ml) and the short time course (4 h), it is unlikely that the bacteria would drastically alter the composition of the mediums during the course of the present study or that the bacteria would reach stationary-phase growth. Therefore, it would not be expected that the bacteria would significantly alter their gene expression patterns in the growth medium or conditioned RPMI over time. In contrast, we see expression patterns that shift dramatically when analyzing the extracellular bacteria through the infection time course. These dynamic gene expression patterns also correspond to higher levels of expression in general than we see in either growth medium or conditioned RPMI. Of particular note is the fact that gene expression patterns in the intracellular environment differ considerably from any of the four time points during extracellular infection. This indicates the intracellular bacteria experience a significantly different set of challenges compared to the bacteria located on the cell surface.
Y. enterocolitica transcriptional changes in response to changing media and temperature.
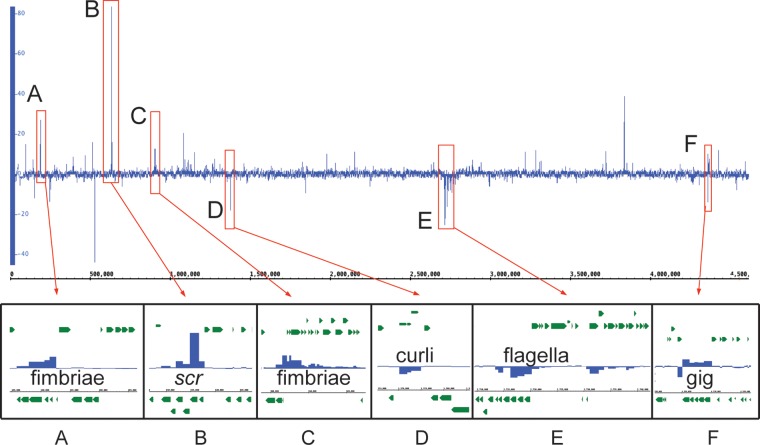
As pathogenic species of Yersinia shift from ambient environmental temperatures to the 37°C temperature of the mammalian host, there is a well-characterized response that relies heavily on genes encoded on the virulence plasmid, particularly the transcriptional activator virF (lcrF in Y. pestis and Y. pseudotuberculosis) (42–44). When analyzing the initial transcriptomic shift from 26°C growth medium to the 37°C conditioned RPMI (Fig. 3A and see Table S2 in the supplemental material) we saw, unsurprisingly, that half of the genes (44 of 87) on the virulence plasmid were upregulated by 4-fold or more. We also observed 176 genes with statistically significant differential expression on the chromosome. A convenient way to analyze the differential expression across an entire bacterial genome at specific times points is to plot the fold changes in the expression of all genes against the chromosomal position. This type of analysis reveals both individual genes and genomic loci with similar changes in expression that may be involved in the same processes. We observed at least six of these loci when comparing the transcriptomes of bacteria in the growth media to those in the conditioned RPMI, which we designated loci A to F (Fig. 4).
FIG 3.
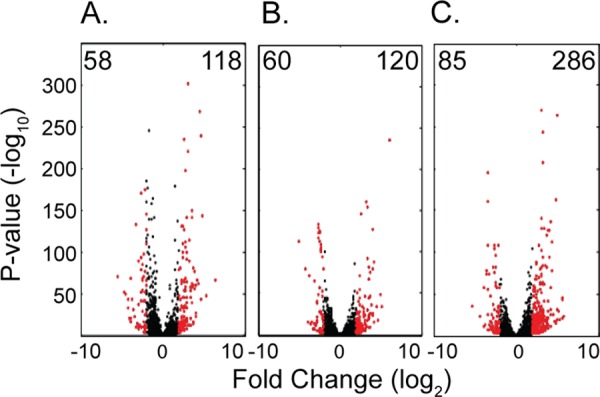
Differential gene expression at 120 min postinfection. Volcano plots were generated by comparing the fold change in expression (log2) of each Y. enterocolitica gene to the corresponding DESeq2 adjusted P value (−log10) of that change. Numbers in the upper left or upper right corners indicate the number of genes upregulated (right) or downregulated (left) by ≥4-fold with a P value of ≤0.01. Genes depicted in black are not differentially expressed by this criterion, whereas genes in red are differentially expressed. (A) Volcano plot depicting differential expression between the conditioned RPMI and the growth medium. (B) Differential expression between extracellular bacteria and the conditioned RPMI. (C) Differential expression between intracellular and extracellular bacteria. All plots compare the differential expression at 120 min postinoculation.
FIG 4.
Differential expression across the genome between conditioned RPMI and growth medium. DESeq2 analysis was performed to discover differentially expressed genes between the conditioned RPMI and the growth medium at 120 min postinoculation. The differential expression of all chromosomal genes is plotted in the top panel. This analysis reveals the presence of at least six chromosomal loci, with high levels of up- or downregulation designated A to F. The lower panels show the operon structures and the differential regulation of the individual genes making up the loci.
Two of the six loci (A and C) contain previously uncharacterized fimbrial operons that are upregulated when comparing growth in conditioned RPMI to the growth medium. These fimbriae and their potential functions are discussed in more detail in the next section. Locus B contains the most highly upregulated gene of this entire study, scrY, which is predicted to encode a sucrose porin. The other genes in locus B contain the transcriptional regulator and the other genes of the scr operon, which encodes the machinery necessary for active transport of sucrose into the bacteria. Oddly, there is no sucrose in the conditioned RPMI medium used in the present study, but there is a significant amount of glucose. Analysis of the homologous porin in Escherichia coli has shown that it can bind glucose, although at about half of the affinity with which it binds sucrose (45). Given the abundance of glucose present in the medium, even low-efficiency uptake may be sufficient to allow the bacteria access to this readily available carbon source. We also observed that genes encoding glucose-specific transporters such as crr, glcA, and malX are not upregulated under this condition, suggesting that in Y. enterocolitica, the ScrY porin may be important for general uptake of sugars rather than specific uptake of sucrose.
The downregulated locus D contains a three-gene operon involved in the synthesis of curli. Curli are common in Enterobacteriaceae and are typically involved in attachment to surfaces and biofilm formation (46). Although curli have not previously been described in Yersinia, it is known that Yersinia do not form biofilms at 37°C. Therefore, this result makes sense if these curli are involved in biofilm formation rather than host-cell binding.
The highly downregulated genes of locus E are all involved in synthesis of the bacterial flagella. It is well established that Y. enterocolitica flagella are downregulated at 37°C, likely as an adaptation to avoid being recognized by Toll-like receptor 5 (TLR5) (18). This expected result confirms the utility of our technique in discovering systems that are differentially expressed upon changing environmental conditions.
A more unexpected finding was seen in locus F with the upregulation of the glg operon, encoding five genes involved in the synthesis and metabolism of glycogen. Although the role of glycogen has not been studied in the context of a Y. enterocolitica infection, it has been demonstrated that E. coli O157:H7 glg operon mutants have a decreased ability to colonize the intestines of mice (47). It has been hypothesized that during colonization of the intestines, E. coli relies on internal stores of carbon during periods of low carbon availability (47). This is likely the case in Y. enterocolitica as well, with bacteria storing available carbon from the RPMI media as glycogen at 37°C in preparation for colonization of the mammalian host.
Y. enterocolitica transcriptional changes in response to extracellular infection.
Bacteria infecting macrophages would be expected to upregulate virulence factors that aid in colonization and evasion of the host immune response. We identified 180 genes with a >4-fold change in expression when comparing the transcriptomes of the extracellular bacteria to those in the conditioned RPMI after 120 min of infection (Fig. 3B), and we observed several loci of genes (A to C, G to I) that encode known and putative virulence factors (Fig. 5 and see Table S3 in the supplemental material). Interestingly, we see that all three of the downregulated loci are the same as upregulated loci in the previous comparison (A to C). The fimbria-encoding operons that were upregulated in response to the conditioned RPMI (A and C) are downregulated by bacteria attached to the macrophages (extracellular). This intriguing observation may indicate that the fimbria proteins are very stable once produced so that after the initial upregulation of the genes there is no need to create more fimbriae. Alternatively, they may be facilitating adherence to other cell types or noncellular host surfaces such as the soluble host factors present in the conditioned RPMI. The genes in locus B encode the sucrose uptake system, which was upregulated in the conditioned RPMI, an observation consistent with the hypothesis that the bacteria are storing carbon while in RPMI in preparation for the infection. Once the bacteria come into contact with the macrophages, they likely rely primarily on the stored glycogen as a carbon source and no longer actively import sugar.
FIG 5.
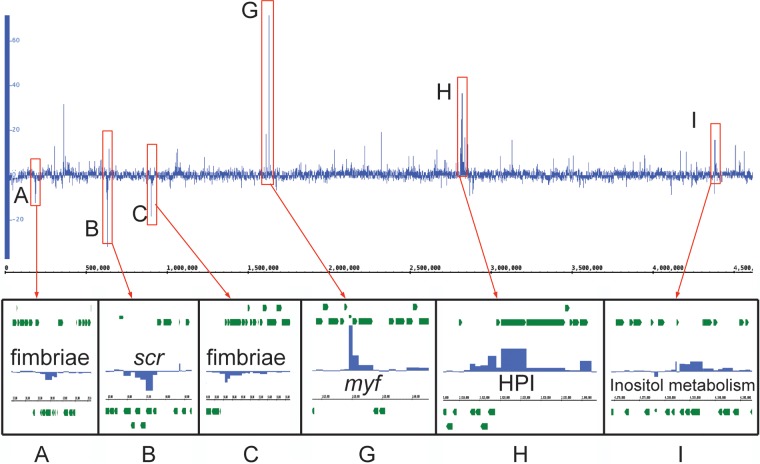
Differential expression across the genome between extracellular bacteria and growth in conditioned RPMI. The expression of all chromosomal genes was analyzed in extracellular bacteria during infection and growth in conditioned RPMI at 120 min postinoculation. Differential gene expression analysis uncovers three previously described loci (A to C) and three new loci (G to I) with interesting changes in expression.
The highly upregulated locus G encodes the pH 6 antigen (Psa [Myf in Y. enterocolitica]), which has been shown in Y. pestis to be important for bacterial attachment to the host cell and the delivery of Yops by the Ysc T3SS into host cells (48). The Myf pilus plays a similar role in Y. enterocolitica, explaining its upregulation in response to host cells (49). The upregulation of the Myf pilus may also explain the downregulation of the locus A and C fimbriae, since the bacteria are unlikely to require all of their pili and fimbriae to be expressed at the same time, preferring the Myf pilus for attachment to macrophages.
The upregulated genes within locus H include most of the genes of the biovar 1B high pathogenicity island (HPI), which encode proteins required for the synthesis, release, and re-uptake of the siderophore yersiniabactin, a crucial virulence factor for overcoming the iron-limited environment found in the host (50, 51). Perhaps the most interesting finding, however, is the highly upregulated locus I, which includes the genes necessary for inositol metabolism. Inositol has multiple roles within host cells, as a building block of several secondary messengers and structural lipids. Work by Sarantis et al. demonstrated that Y. pseudotuberculosis entry into the host cell relied on a phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2]-rich compartment referred to as a “prevacuole” (52). This finding suggests that Y. enterocolitica may make use of inositol derivatives present on the host cell membrane during infection, perhaps as a carbon source or, more interestingly, as a means of disrupting cell signaling or membrane function within the macrophages.
Genes with highly variable expression levels through the course of infection.
Some particularly interesting sets of genes to analyze are those that change their expression levels drastically through the course of an infection. These genes are likely to be the ones most sensitive to the changing environment of the host cell. Using a linear regression on DESeq2 fold changes in expression when comparing the extracellular environment to the conditioned RPMI, we identified 167 genes that change from upregulated to downregulated or vice versa through the course of the infection (Fig. 6). Of the genes with known or predicted functions, the vast majority are involved in either metabolic processes, the uptake of various nutrients, or movement and chemotaxis (see Table S4 in the supplemental material). Among the known virulence factors identified in this analysis, only the putative factors encoded by YE2057 and YE2058 change from upregulated to downregulated. Of the genes that switched from downregulated to upregulated, most were involved in iron uptake. This makes sense if iron becomes a limiting factor as the infection proceeds and more bacteria are present taking up the freely available iron. Given how few of the identified genes are known virulence factors, this suggests that the majority of Y. enterocolitica virulence factors, once upregulated, remain that way for the duration of the infection. Alternatively, virulence factors may have more subtle changes in expression during infection and therefore would not have been found in this analysis. Taken together, these results imply that the nutrients available to bacteria attached to the surface of the macrophages are constantly changing through the course of an infection and that the bacteria differentially express many of their genes to adjust to those changes.
Y. enterocolitica transcriptional changes in response to internalization.
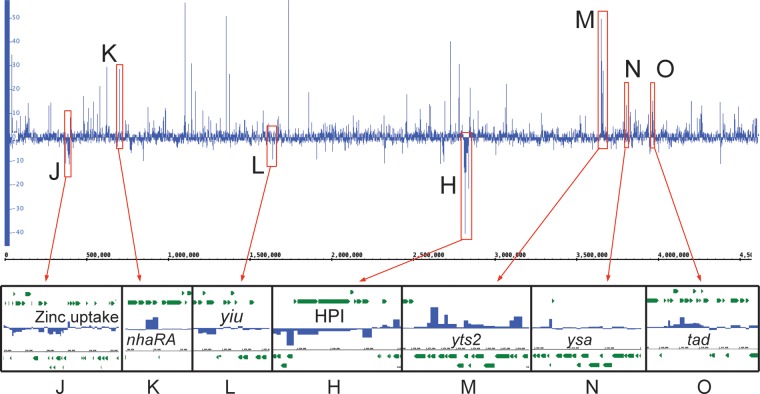
We next performed differential expression analysis comparing the transcriptomes of intracellular bacteria to the extracellular population after 120 min of infection (Fig. 7 and see Table S5 in the supplemental material). There were 371 chromosomal genes with a >4-fold change in expression, more than twice the number we identified when comparing extracellular bacteria to the conditioned RPMI (Fig. 3C). Clustering analysis revealed eight loci of interest, designated loci H and J to O. Strikingly, all three of the downregulated loci (J, L, and H) are involved in metal uptake. The genes in locus J encode a two-component regulatory system that may regulate the expression of the downstream genes involved in a zinc uptake system common to several species of Yersinia (7). The genes within locus L encode the Yiu iron uptake system, which is common to all highly pathogenic species of Yersinia, but have been shown to be of less importance to virulence than the yersiniabactin-encoding Ybt system (locus H) (53, 54). These results suggest that neither zinc nor iron is in limited supply within the macrophages since the bacteria are no longer actively engaged in scavenging the metals from the environment. This result is also consistent with work by Fukuto et al. that showed similar downregulation of iron uptake pathways in Y. pestis bacteria that had been internalized by macrophages (55).
FIG 7.
Differential expression across the genome between intracellular and extracellular bacteria. DESeq2 was used to analyze differential expression across the chromosome between intracellular and extracellular bacteria at 120 min postinfection. Note the numerous differentially expressed genes and loci. Our analysis focused on six new loci designated J to O, as well as the previously discussed locus H.
Locus K contains the highly upregulated (>21-fold) nhaA and nhaR, a pH-dependent Na+/H+ antiporter and its transcriptional regulator. Recent work has shown that nhaA is required for Y. pestis virulence in murine infections (56). The exact mechanism of this requirement is still unknown, although it has been speculated that it functions to prevent Na toxicity (56). Interestingly, the other Y. enterocolitica Na+/H+ antiporter, nhaB, is not upregulated or highly expressed under any condition that we analyzed.
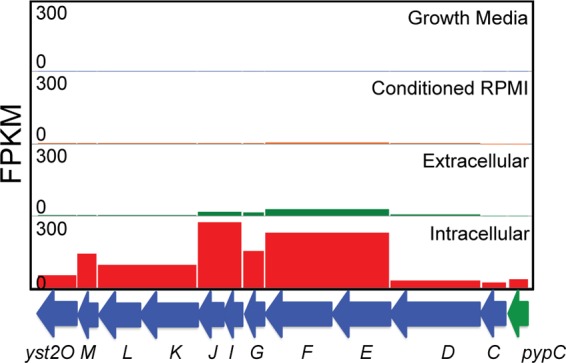
The Yts2 T2SS and Ysa T3SS are found in loci M and N, respectively, and are discussed in detail in subsequent sections due to the novelty of these findings and the importance of secretion systems to bacterial virulence. Another particularly interesting group of genes upregulated in response to internalization is the tad locus (locus O), which encodes a type IVb pilus shown to be important in mediating tight adherence to host cells in bacterial pathogens such as Vibrio cholerae (57). The tad locus is found on the YGI-1 island that is present in Y. pseudotuberculosis and Y. enterocolitica biovar 1B but not in Y. pestis or other biovars of Y. enterocolitica (7). Schilling et al. found that the system was regulated by the upstream transcriptional activator PypB but discovered no condition where they could conclusively demonstrate native expression of the pilus (58). Our results demonstrate that the pilus is likely important during the intracellular phase of infection, perhaps allowing the bacteria to adhere to the inner surface of the YCV and facilitate the translocation of T3SS effectors across the vacuolar membrane. It should be noted that the regulator of the Yts2 T2SS, PypC, also regulates expression of pypB (59), indicating a clear link between the Tad type IVb pilus and the Yts2 system in bacteria that have been internalized by macrophages.
Regulation and expression of the Ysc T3SS and the pYV plasmid.
The most critical virulence determinant of all pathogenic species of Yersinia is the Ysc T3SS, which is encoded on the virulence plasmid pYV, along with the system's transcriptional activator VirF (LcrF in Y. pestis and Y. pseudotuberculosis). It has long been known that any mutation that prevents the proper function of the Ysc system renders the bacterium avirulent (60). Because of its importance to the virulence of all pathogenic Yersinia spp., the Ysc system has been studied as the canonical T3SS. The Ysc T3SS, its effectors, and the role of the virulence plasmid are well understood and have been extensively reviewed (61–65). In the present study, we observed that the most dramatic changes in expression of virulence plasmid genes occurred during the transition from the 26°C growth medium to the 37°C conditioned RPMI. A total of 71 of the 87 genes on the plasmid displayed statistically significant upregulation, with 62% (44 genes) of those genes upregulated by 4-fold or more (see Table S6 in the supplemental material). We observed only one gene with a >4-fold change in expression when comparing the extracellular bacteria to the conditioned RPMI, a putative transposase (YEP0003). This would appear to indicate that expression of the Ysc T3SS relies mainly on the shift to the 37°C temperature and not on contact with the host cell. This finding is consistent with previously published work that demonstrated translation of the transcriptional activator LcrV (VirF) is strongly enhanced by changes in the mRNA conformation at 37°C (43).
Many of the pYV genes that are upregulated at the initial shift to the 37°C conditioned RPMI tend to maintain somewhat similar expression levels throughout the infection, whether the bacteria are attached to the macrophage surface or have been internalized. However, 12 genes showed >4-fold changes in expression when comparing intracellular bacteria to extracellular bacteria, including 4 genes that are upregulated and 8 that are downregulated. Interestingly, two of the most highly downregulated genes, yscO and yscQ, encode proteins that make up critical components of the Ysc T3SS (66), suggesting that the system has decreased importance within the YCV. Table S6 in the supplemental material shows the expression of all of the genes on the virulence plasmid, demonstrating that there is a general trend toward reduced expression of Ysc genes in response to internalization. Despite this general downregulation, the overall expression levels, as indicated by the FPKM values, remain relatively high throughout the course of infection, regardless of the cellular location. The other downregulated gene, yadA, plays a vital role in adherence to host cell surfaces and prevention of complement-mediated inactivation (67, 68), functions that have less importance once internalized. One striking observation about the pYV genes upregulated in response to internalization is that half of them have no known function. These four genes (YEP0007, YEP0011, YEP0049, and YEP0071) may encode previously unidentified virulence factors that play significant roles in pathogenesis only after the bacteria have been internalized.
The Ysa T3SS is expressed by intracellular bacteria.
The Ysa T3SS is encoded by genes within the plasticity zone, and is unique among Y. enterocolitica isolates to biovar 1B (7, 9, 10). Previous work has demonstrated that this secondary T3SS is required for full virulence of the bacterium in a mouse model of infection, but only at very early time points (69). Despite this finding, we recently demonstrated that the Ysa system is expressed throughout the course of murine infections in a contact-dependent manner in every tissue examined (31). Initial studies found the Ysa T3SS to be expressed in vitro only at 26°C in nutrient-rich media containing >150 mmol/liter salt (NaCl or KCl) (69), conditions that would not suggest a role in the infection of a mammalian host. Because of this phenotype of low-temperature induction, Walker et al. examined the expression of the system during the infection of Drosophila S2 cells and found that it was required for growth only when internalized (70).
We determined that the Ysa T3SS is most highly expressed when the bacteria have been internalized by the murine macrophages. Figure 8 shows the normalized expression of each gene within the Ysa locus 2 h postinfection in each of the growth conditions analyzed here. In addition, Table S7 in the supplemental material lists the FPKM values and differential expression results for each of these genes and shows that, on average, the system is upregulated by 3.2-fold when comparing intracellular to extracellular bacteria. The second most strongly upregulated gene was ysaE, which encodes a positive regulator of the Ysa system. We also saw strong upregulation of yspB and yspC. These genes encode the proteins that make up the translocon, which forms a pore in the host cell membrane through which effectors are translocated. The sycB gene, which encodes their chaperone, was also upregulated by >6-fold.
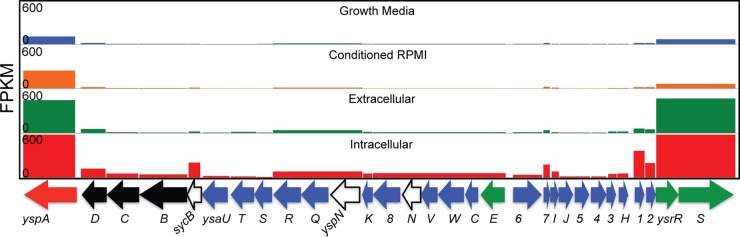
FIG 8.
FPKM values of genes within the Ysa locus under each experimental condition. Pooled FPKM values from three replicates for each gene within the Ysa locus were plotted along the Ysa locus after 120 min of infection under each experimental condition. Genes depicted in blue encode proteins that make up the Ysa T3S apparatus, genes depicted in white encode chaperones, genes depicted in black encode translocon proteins, genes depicted in green encode regulators of the system, and the red gene encodes a secreted effector of the system. Note the general increase in expression across the locus in the intracellular condition.
Except for yspA, all other genes encoding secreted effectors of the Ysa system are located outside the Ysa locus. As shown in Table S7 in the supplemental material, several of these genes also show various degrees of upregulation, with yspM showing the largest change in expression upon internalization (9.3-fold). Combined with the fact that the Ysc T3SS is partially downregulated by intracellular bacteria, these results indicate that the Ysa T3SS is important for the survival of bacteria that have been internalized by the macrophages. This has been observed in other bacteria with multiple T3SS, such as Salmonella enterica serovar Typhimurium, which uses the SPI-1 T3SS to gain entry to host cells and then uses the SPI-2 T3SS to prevent lysosome fusion, thereby allowing the bacteria to replicate within the phagosome (71).
Expression of the Yts1 and Yts2 T2SSs.
The Y. enterocolitica biovar 1B genome encodes two distinct T2SSs with poorly understood roles in virulence. Located directly downstream of the Ysa T3SS locus and within the plasticity zone, the Yts1 T2SS is unique to biovar 1B isolates (7, 11). Iwobi et al. discovered that a Yts1 mutant in strain WA-314 had a decreased ability to colonize the spleens and livers of mice infected orally but not when the animals were infected intravenously (11). This suggests that the system is important for systemic dissemination of the bacteria, at least in strain WA-314. Three secreted effectors of the Yts1 system have been described, encoded by the genes chiY, engY, and YE3650—each of which encodes a protein with the characteristic N-terminal signal sequence for T2S (29). Interestingly, the upstream regulator PclR (PypC-like regulator) appears to play a role in the expression of chiY but not of the genes directly downstream, which encode the structural apparatus of the Yts1 T2SS (29, 30).
Although little is known about the function of the Yts1 system, even less is known about the Yts2 T2SS. This system is common to all species of Yersinia pathogenic to humans; however, expression of the system has never been observed under any in vitro condition, and no secreted effectors have been found (29, 30). Unlike the Yts1 system, the upstream regulator of the Yts2 system, PypC, directly regulates the downstream genes encoding the T2S apparatus (29). Intriguingly, PypC has also been found to positively regulate the genes for the transcriptional regulator pypB, the regulator of the tad locus, and hreP, which encodes a protease required for full virulence of Y. enterocolitica (59, 72).
We found that the Yts1 T2SS showed no significant change in expression during either extracellular or intracellular infection, indicating that it is unlikely to function as an important virulence factor during the infection of macrophages (see Table S8 in the supplemental material). In contrast, we discovered, for the first time, a role for the Yts2 T2SS in the intracellular phase of infection. The system was upregulated by an average of 12.8-fold when comparing intracellular to extracellular bacteria during infection. The gene encoding the positive regulator of the system, pypC, was upregulated 27.7-fold, making it one of the most highly upregulated genes in the intracellular condition (Fig. 9 and see Table S8 in the supplemental material). These results strongly suggest that, like the Ysa T3SS, the Yts2 T2SS is important for the intracellular survival of Y. enterocolitica within macrophages. Given the presence of the system in other pathogenic and nonpathogenic Yersinia species, it seems probable that the system was acquired early in Yersinia evolution and that it has been maintained because it provides a selective advantage in helping to avoid killing by macrophages.
FIG 9.
FPKM values of the genes encoding the Yts2 T2SS. Pooled FPKM values from three biological replicates are shown for each gene encoding the Yts2 T2S apparatus (blue) and the upstream regulator (green), pypC, in each of the experimental conditions. Note the only condition in which all genes are expressed in the intracellular condition.
Analysis of Ysa, Yts2, and Tad mutants.
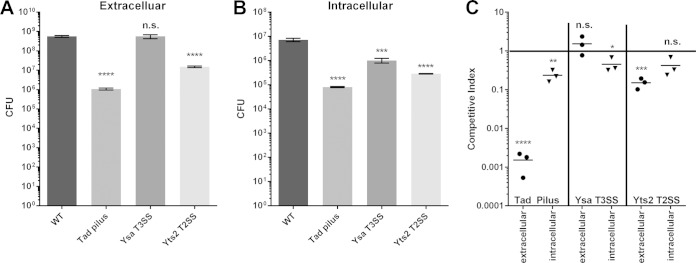
With transcriptomic evidence that that the Ysa T3SS, Yts2 T2SS, and Tad pilus are all highly upregulated in response to internalization, we constructed a series of mutants to determine the importance of these systems during the intracellular phase of infection. The function of the Ysa T3SS was disrupted by creating an insertion in ysaV, a gene encoding a critical component of the secretion apparatus (69). Secretion of effectors from the Yts2 T2SS was blocked by an insertion in yst2D, encoding the outer membrane secretin (29, 30). Assembly of the Tad pilus was prevented through an insertion in tadA, the ATPase responsible for the export of pilus subunits (28). The role of each system was addressed by performing single infections of P388D1 cells in a pYV-cured parent strain. CFU counts of both externally located bacteria and bacteria located within YCV were performed. This was followed by competition assays in which each mutant was mixed 1:1 with the parent strain, and the mixture was used to infect the same P388D1 cells. A pYV-cured parent strain was chosen to avoid rapid killing of the macrophage cells by the Ysc T3SS, preventing long-term bacterial survival in the YCV.
Conditions were set so that the importance of each system could be determined for both extracellular and intracellular conditions. All of the mutants had similar growth rates and grew to the same density as the parent strain in RPMI medium at 37°C (see Fig. S1 in the supplemental material). Figure 10A shows the results of the single-infection experiments in which bacteria were confined to the extracellular environment through the use of cytochalasin D. Interestingly, both the Yts2 T2SS and the Tad pilus, but not the Ysa T3SS, appear to be important for full virulence under this condition, with a 37-fold and a 525-fold reduction in CFU for the Yts2 and Tad mutants, respectively. These mutants also show decreased numbers after internalization (Fig. 10B), but this may be due to the fact that fewer mutants were present to begin with. The Ysa T3SS, however, appears to be important only during the intracellular phase of infection, since we see a relatively modest, but statistically significant, 7-fold drop in the number of CFU present under this condition.
FIG 10.
Single infections and competition assays with tad, ysa, and yts2 mutants. P388D1 murine macrophages were infected for 24 h with tad, ysa, or yts2 mutants either individually (A and B) or in a 1:1 coinfection with the parent strain (C). (A) Cells were treated with cytochalasin D prior to infection to prevent uptake of bacteria into the YCV. (B) After 1 h of infection, cells were treated with gentamicin to eliminate extracellular bacteria. (C) Competition assays were performed with cells treated with cytochalasin D (extracellular) or gentamicin (intracellular). The ratio of mutant to parent was determined by plate counts. Asterisks indicate statistical significance (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001).
To follow up on the single infections, competition assays were set up to determine whether any of the mutations led to distinct competitive defects under either extracellular or intracellular conditions. The competitive index was determined by counting the colonies of each mutant and parent present after 24 h of infection in a 1:1 coinfection with the two strains. Figure 10C shows a pattern similar to what was observed in the single infections, with each mutant, other than the Ysa mutant, demonstrating a competitive defect under both extracellular and intracellular conditions. Intriguingly, the intracellular defect for all three mutants is less severe than the single infections, indicating that the parent strain may be able to partially rescue the mutant. In the case of the secretion systems, the parent's ability to secrete effectors in the presence of the mutant seems to be the obvious explanation. The reason for the difference in the Tad mutant between the single infection and competition assay is less clear.
DISCUSSION
Using RNA-Seq to analyze bacterial gene expression profiles through the course of an infection has proved to be a highly informative method for better understanding the pathways and mechanisms that bacteria use to colonize different host environments (22, 73–75). It has significant advantages over most other transcriptional profiling techniques because it provides an unbiased way to analyze the temporally and spatially dynamic expression of every bacterial gene, in wild-type bacteria, as they inhabit diverse host environments. However, analysis of the bacterial transcriptome during infection is all but impossible without an enrichment step that increases the proportion of bacterial transcripts in the host-dominated sample. In the present study, we utilized our previously described capture-based enrichment strategy (22, 23) to analyze the complete transcriptome of Y. enterocolitica biovar 1B in different media and host environments, including a transcriptomic comparison of bacteria attached to the surface of macrophages to those that had been internalized by the macrophages. This analysis revealed a suite of genes whose expression levels change over the course of an infection, providing insight into the various mechanisms the bacteria use to infect macrophages and remain viable within them after being internalized.
We found that whereas gene expression patterns remain relatively static in the growth medium and conditioned RPMI, they were highly dynamic through the course of infection. Interestingly, the genes with the most significant changes in expression over the course of extracellular infection were involved not necessarily in pathogenesis but in metabolic processes and nutrient uptake. In contrast, most known Yersinia virulence factors were upregulated either in the conditioned RPMI or at the start of macrophage infection and maintained their expression levels through the course of infection. Genes involved in nutrient uptake and utilization had the largest shifts in expression during the course of infection, suggesting that the macrophage cell surface is constantly changing in response to the bacterial infection. In addition, we discovered that inositol metabolism likely plays a role in the bacteria's ability to subvert the host immune response by disrupting cell signaling or membrane function. Further work will be required to test this hypothesis.
Although Yersinia spp. are considered extracellular pathogens, the present study as well as previous work demonstrated that some bacteria remain viable within macrophages for extended periods of time, suggesting that the intracellular bacteria play an active role in the infection (55, 76, 77). The use of an RNA-Seq-based approach to analyze the intracellular bacteria provided an interesting opportunity to see, for the first time, the complete set of genes expressed in response to internalization by macrophages. We hypothesized that the genes upregulated under this condition could be important factors that act to prevent killing by the macrophage and promote intracellular survival. Therefore, some of the most intriguing findings described here were the upregulation of the Ysa T3SS, the Yts2 T2SS, and the coordinately regulated Tad pilus. We also observed downregulation of several metal uptake systems in internalized bacteria, leading to the hypothesis that the intracellular environment may not present the bacteria with the same limited metal availability of the surface.
The function of the Ysa T3SS has long been controversial, due to the low-temperature and high-salt conditions required to induce its expression in vitro. Although the system has been shown to be expressed throughout the infection of mice (31), it has been shown to be important only within the first 24 h after infection (69). It was also recently found to play an important role in the intracellular survival of the bacteria in Drosophila S2 cells (70), although Y. enterocolitica is not known to infect insects. We found that the Ysa system was highly upregulated by bacteria internalized by macrophages but not by bacteria attached to the surfaces of those macrophages or in the conditioned RPMI. We also demonstrate, for the first time in mammalian cells, that mutants of the Ysa T3SS exhibit a decreased ability to survive within murine macrophages. Upon combining the results of previous studies (31, 69) with our present findings, it seems apparent the Ysa system does in fact play a role during the course of an infection, but only when the bacteria have been internalized. This would explain the phenotype of a defect in the initial colonization of mice, since bacteria expressing the system are better able to avoid being killed by the macrophages and are therefore able to more rapidly colonize the terminal ileum and Peyer's patches in the first 24 h after infection. By the time the bacteria have colonized deeper tissues, the initial advantage of macrophage survival has been diminished, although the bacteria still express the Ysa system in these tissues when internalized. Interestingly, we found that only five of the nine Ysp effectors (YspL, YspE, YspK, YspM, and YspP) were significantly upregulated when internalized, suggesting that there may be other cell types or host niches where the other effectors are secreted.
Neither the Yts2 system nor the Tad pilus has ever been shown to be expressed during an infection, although they were both known to be regulated by PypC (29). This coregulation implies that they should be expressed in tandem. We observed that the Yts2 system was among the most highly upregulated loci in the intracellular condition, suggesting its importance in promoting intracellular survival. Although there are currently no known secreted effectors of the Yts2 system, it is possible that the Yts2 T2SS may secrete effectors associated with the Yts1 T2SS. The effectors ChiY and YE3650 are especially interesting given their significant upregulation when internalized. The upregulation of the Tad pilus in response to internalization is also a noteworthy finding given its presence in bacterial pathogens as diverse as Mycobacterium tuberculosis, Pseudomonas aeruginosa, Vibrio cholerae, and Bordetella pertussis (57). The Tad pilus is a type IVb pilus that is typically involved in adhering bacteria to various surfaces, including the surfaces of host cells. Based on our RNA-Seq data, it seems likely that the pilus is acting to adhere the bacteria to the internal surface of the YCV membrane. It has been proposed that there is a direct correlation between the length of the T3S needle and the length of the adhesin on the bacterial surface that acts to stabilize T3S (78). Since the Ysc T3SS relies on YadA to perform this function on the surface of the host cell, our results suggest that the Tad pilus may serve the same function for the Ysa T3SS inside the macrophage. Future studies are necessary to address this hypothesis and determine the length of both the Ysa T3S needle and the Tad pilus.
One of the most remarkable outcomes of the present study was the apparent discordance between the RNA-Seq results and our mutational analysis. Given the high levels of expression of both tad and yts2 genes in the YCV and the low levels of expression on the extracellular surface, we hypothesized that both systems would be important only in promoting intracellular survival. Instead, we observed the opposite phenotype, where, despite low levels of gene expression on the extracellular surface, mutants of both systems appeared to have a greater disadvantage extracellularly than intracellularly. One disadvantage of RNA-Seq is that it provides only a snapshot of gene expression at a specific point in time and, alone, it cannot account for either posttranscriptional or posttranslational regulation. Therefore, it is conceivable that in the YCV the tad and yts2 genes are highly expressed, but they are not translated into proteins or the proteins do not form functional pili or T2SSs due to other downstream events. It is also possible that even though the number of transcripts is low on the cell surface, they are stable enough that numerous functional secretion systems and pili are formed. Taken together, this leads to the conclusion that while RNA-Seq is a powerful technique that may point researchers in the correct direction, such as in the case of the Ysa T3SS, it does not replace the need for traditional molecular microbiology approaches in describing the importance of potential virulence factors.
Due to the relatively limited scope of the present study, it was not possible to describe the expression and differential regulation of every Y. enterocolitica gene that we found. Therefore, we present in Table S9 in the supplemental material the FPKM values of all genes in all four environments at every time point, as well as the relevant DESeq2 comparisons, so that the general research community may benefit from analyses that we were unable to describe here.
Given the numerous virulence factors and other genes we found to be highly expressed by bacteria that had been internalized by the macrophages, it seems apparent that Y. enterocolitica biovar 1B is adapted for an intracellular phase of its pathogenic life cycle. This ability to survive while internalized would seem to be a critical factor in biovar 1B's increased pathogenicity over other Y. enterocolitica strains. It would be highly elucidating to continue this line of research to define the bacterium's transcriptomic response to other host cell types that it encounters as the infection proceeds from the gastrointestinal tract to the lymph nodes and then to deeper tissues such as the spleen and liver. It would also be enlightening to compare these responses to those of less pathogenic strains of Y. enterocolitica to determine to what extent regulatory rather than genetic factors affect the virulence of the organism. Work is under way to evaluate the efficacy of our capture technique to enrich for bacterial transcripts directly from infected host tissues, a sample type in which the bacterial transcripts make up an even smaller percentage of the total. Performing these types of studies in animals will allow researchers to analyze host transcripts as well as the bacterial, providing a truly comprehensive look at host-pathogen interactions in individual tissues through the course of an infection.
Supplementary Material
ACKNOWLEDGMENTS
We used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 Instrumentation Grants S10RR029668 and S10RR027303. Sandia National Laboratories' Laboratory-Directed Research and Development program funded this research. Sandia National Laboratories is a multiprogram laboratory managed and operated by Sandia Corp., a wholly owned subsidiary of Lockheed Martin Corp., for the U.S. Department of Energy's National Nuclear Security Administration under contract DE-AC04-94AL85000.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02922-14.
REFERENCES
- 1.Carniel E, Mollaret HH. 1990. Yersiniosis. Comp Immunol Microbiol Infect Dis 13:51–58. [DOI] [PubMed] [Google Scholar]
- 2.Bottone EJ. 1999. Yersinia enterocolitica: overview and epidemiologic correlates. Microbes Infect 1:323–333. doi: 10.1016/S1286-4579(99)80028-8. [DOI] [PubMed] [Google Scholar]
- 3.Cover TL, Aber RC. 1989. Yersinia enterocolitica. N Engl J Med 321:16–24. [DOI] [PubMed] [Google Scholar]
- 4.Koehler KM, Lasky T, Fein SB, Delong SM, Hawkins MA, Rabatsky-Ehr T, Ray SM, Shiferaw B, Swanson E, Vugia DJ. 2006. Population-based incidence of infection with selected bacterial enteric pathogens in children younger than five years of age, 1996-1998. Pediatr Infect Dis J 25:129–134. doi: 10.1097/01.inf.0000199289.62733.d5. [DOI] [PubMed] [Google Scholar]
- 5.Howard SL, Gaunt MW, Hinds J, Witney AA, Stabler R, Wren BW. 2006. Application of comparative phylogenomics to study the evolution of Yersinia enterocolitica and to identify genetic differences relating to pathogenicity. J Bacteriol 188:3645–3653. doi: 10.1128/JB.188.10.3645-3653.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schiemann DA, Devenish JA, Toma S. 1981. Characteristics of virulence in human isolates of Yersinia enterocolitica. Infect Immun 32:400–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomson NR, Howard S, Wren BW, Holden MT, Crossman L, Challis GL, Churcher C, Mungall K, Brooks K, Chillingworth T, Feltwell T, Abdellah Z, Hauser H, Jagels K, Maddison M, Moule S, Sanders M, Whitehead S, Quail MA, Dougan G, Parkhill J, Prentice MB. 2006. The complete genome sequence and comparative genome analysis of the high pathogenicity Yersinia enterocolitica strain 8081. PLoS Genet 2:e206. doi: 10.1371/journal.pgen.0020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collyn F, Billault A, Mullet C, Simonet M, Marceau M. 2004. YAPI, a new Yersinia pseudotuberculosis pathogenicity island. Infect Immun 72:4784–4790. doi: 10.1128/IAI.72.8.4784-4790.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foultier B, Troisfontaines P, Muller S, Opperdoes FR, Cornelis GR. 2002. Characterization of the ysa pathogenicity locus in the chromosome of Yersinia enterocolitica and phylogeny analysis of type III secretion systems. J Mol Evol 55:37–51. doi: 10.1007/s00239-001-0089-7. [DOI] [PubMed] [Google Scholar]
- 10.Haller JC, Carlson S, Pederson KJ, Pierson DE. 2000. A chromosomally encoded type III secretion pathway in Yersinia enterocolitica is important in virulence. Mol Microbiol 36:1436–1446. [DOI] [PubMed] [Google Scholar]
- 11.Iwobi A, Heesemann J, Garcia E, Igwe E, Noelting C, Rakin A. 2003. Novel virulence-associated type II secretion system unique to high-pathogenicity Yersinia enterocolitica. Infect Immun 71:1872–1879. doi: 10.1128/IAI.71.4.1872-1879.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schubert S, Fischer D, Heesemann J. 1999. Ferric enterochelin transport in Yersinia enterocolitica: molecular and evolutionary aspects. J Bacteriol 181:6387–6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darwin AJ. 2005. Genome-wide screens to identify genes of human pathogenic Yersinia species that are expressed during host infection. Curr Issues Mol Biol 7:135–149. [PubMed] [Google Scholar]
- 14.Dhar MS, Virdi JS. 2014. Strategies used by Yersinia enterocolitica to evade killing by the host: thinking beyond Yops. Microbes Infect 16:87–95. doi: 10.1016/j.micinf.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 15.Thomson NR, Howard S, Wren BW, Prentice MB. 2007. Comparative genome analyses of the pathogenic Yersiniae based on the genome sequence of Yersinia enterocolitica strain 8081. Adv Exp Med Biol 603:2–16. doi: 10.1007/978-0-387-72124-8_1. [DOI] [PubMed] [Google Scholar]
- 16.Black DS, Bliska JB. 2000. The RhoGAP activity of the Yersinia pseudotuberculosis cytotoxin YopE is required for antiphagocytic function and virulence. Mol Microbiol 37:515–527. [DOI] [PubMed] [Google Scholar]
- 17.Pujol C, Bliska JB. 2005. Turning Yersinia pathogenesis outside in: subversion of macrophage function by intracellular yersiniae. Clin Immunol 114:216–226. doi: 10.1016/j.clim.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 18.Minnich SA, Rohde HN. 2007. A rationale for repression and/or loss of motility by pathogenic Yersinia in the mammalian host. Adv Exp Med Biol 603:298–310. doi: 10.1007/978-0-387-72124-8_27. [DOI] [PubMed] [Google Scholar]
- 19.Ozsolak F, Milos PM. 2011. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet 12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Gerstein M, Snyder M. 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westermann AJ, Gorski SA, Vogel J. 2012. Dual RNA-seq of pathogen and host. Nat Rev Microbiol 10:618–630. doi: 10.1038/nrmicro2852. [DOI] [PubMed] [Google Scholar]
- 22.Bent ZW, Brazel DM, Tran-Gyamfi MB, Hamblin RY, VanderNoot VA, Branda SS. 2013. Use of a capture-based pathogen transcript enrichment strategy for RNA-Seq analysis of the Francisella tularensis LVS transcriptome during infection of murine macrophages. PLoS One 8:e77834. doi: 10.1371/journal.pone.0077834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bent ZW, Tran-Gyamfi MB, Langevin SA, Brazel DM, Hamblin RY, Branda SS, Patel KD, Lane TW, VanderNoot VA. 2013. Enriching pathogen transcripts from infected samples: a capture-based approach to enhanced host-pathogen RNA sequencing. Anal Biochem 438:90–96. doi: 10.1016/j.ab.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Kinder SA, Badger JL, Bryant GO, Pepe JC, Miller VL. 1993. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O8 and construction of a transformable R-M+ mutant. Gene 136:271–275. doi: 10.1016/0378-1119(93)90478-L. [DOI] [PubMed] [Google Scholar]
- 25.Bent ZW, Young GM. 2010. Contribution of BlaA and BlaB β-lactamases to antibiotic susceptibility of Yersinia enterocolitica biovar 1B. Antimicrob Agents Chemother 54:4000–4002. doi: 10.1128/AAC.01754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cummings LA, Wilkerson WD, Bergsbaken T, Cookson BT. 2006. In vivo, fliC expression by Salmonella enterica serovar Typhimurium is heterogeneous, regulated by ClpX, and anatomically restricted. Mol Microbiol 61:795–809. doi: 10.1111/j.1365-2958.2006.05271.x. [DOI] [PubMed] [Google Scholar]
- 27.Young BM, Young GM. 2002. YplA is exported by the Ysc, Ysa, and flagellar type III secretion systems of Yersinia enterocolitica. J Bacteriol 184:1324–1334. doi: 10.1128/JB.184.5.1324-1334.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roux N, Spagnolo J, de Bentzmann S. 2012. Neglected but amazingly diverse type IVb pili. Res Microbiol 163:659–673. doi: 10.1016/j.resmic.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 29.Shutinoski B, Schmidt MA, Heusipp G. 2010. Transcriptional regulation of the Yts1 type II secretion system of Yersinia enterocolitica and identification of secretion substrates. Mol Microbiol 75:676–691. doi: 10.1111/j.1365-2958.2009.06998.x. [DOI] [PubMed] [Google Scholar]
- 30.von Tils D, Bladel I, Schmidt MA, Heusipp G. 2012. Type II secretion in Yersinia: a secretion system for pathogenicity and environmental fitness. Front Cell Infect Microbiol 2:160. doi: 10.3389/fcimb.2012.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bent ZW, Branda SS, Young GM. 2013. The Yersinia enterocolitica Ysa type III secretion system is expressed during infections both in vitro and in vivo. Microbiologyopen 2:962–975. doi: 10.1002/mbo3.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol 170:2575–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langevin SA, Bent ZW, Solberg OD, Curtis DJ, Lane PD, Williams KP, Schoeniger JS, Sinha A, Lane TW, Branda SS. 2013. Peregrine: a rapid and unbiased method to produce strand-specific RNA-Seq libraries from small quantities of starting material. RNA Biol 10:502–515. doi: 10.4161/rna.24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matz M, Shagin D, Bogdanova E, Britanova O, Lukyanov S, Diatchenko L, Chenchik A. 1999. Amplification of cDNA ends based on template-switching effect and step-out PCR. Nucleic Acids Res 27:1558–1560. doi: 10.1093/nar/27.6.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vandernoot VA, Langevin SA, Solberg OD, Lane PD, Curtis DJ, Bent ZW, Williams KP, Patel KD, Schoeniger JS, Branda SS, Lane TW. 2012. cDNA normalization by hydroxyapatite chromatography to enrich transcriptome diversity in RNA-seq applications. Biotechniques 53:373–380. doi: 10.2144/000113937. [DOI] [PubMed] [Google Scholar]
- 36.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anders S, Pyl PT, Huber W. 2014. HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059=014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Furth R, van Schadewijk-Nieuwstad M, Elzenga-Claasen I, Cornelisse C, Nibbering P. 1985. Morphological, cytochemical, functional, and proliferative characteristics of four murine macrophage-like cell lines. Cell Immunol 90:339–357. doi: 10.1016/0008-8749(85)90199-6. [DOI] [PubMed] [Google Scholar]
- 42.Hoe NP, Minion FC, Goguen JD. 1992. Temperature sensing in Yersinia pestis: regulation of yopE transcription by lcrF. J Bacteriol 174:4275–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bohme K, Steinmann R, Kortmann J, Seekircher S, Heroven AK, Berger E, Pisano F, Thiermann T, Wolf-Watz H, Narberhaus F, Dersch P. 2012. Concerted actions of a thermo-labile regulator and a unique intergenic RNA thermosensor control Yersinia virulence. PLoS Pathog 8:e1002518. doi: 10.1371/journal.ppat.1002518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cornelis G, Sluiters C, de Rouvroit CL, Michiels T. 1989. Homology between virF, the transcriptional activator of the Yersinia virulence regulon, and AraC, the Escherichia coli arabinose operon regulator. J Bacteriol 171:254–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schulein K, Schmid K, Benzl R. 1991. The sugar-specific outer membrane channel ScrY contains functional characteristics of general diffusion pores and substrate-specific porins. Mol Microbiol 5:2233–2241. doi: 10.1111/j.1365-2958.1991.tb02153.x. [DOI] [PubMed] [Google Scholar]
- 46.Barnhart MM, Chapman MR. 2006. Curli biogenesis and function. Annu Rev Microbiol 60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones SA, Jorgensen M, Chowdhury FZ, Rodgers R, Hartline J, Leatham MP, Struve C, Krogfelt KA, Cohen PS, Conway T. 2008. Glycogen and maltose utilization by Escherichia coli O157:H7 in the mouse intestine. Infect Immun 76:2531–2540. doi: 10.1128/IAI.00096-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Felek S, Tsang TM, Krukonis ES. 2010. Three Yersinia pestis adhesins facilitate Yop delivery to eukaryotic cells and contribute to plague virulence. Infect Immun 78:4134–4150. doi: 10.1128/IAI.00167-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iriarte M, Vanooteghem JC, Delor I, Diaz R, Knutton S, Cornelis GR. 1993. The Myf fibrillae of Yersinia enterocolitica. Mol Microbiol 9:507–520. doi: 10.1111/j.1365-2958.1993.tb01712.x. [DOI] [PubMed] [Google Scholar]
- 50.Carniel E, Guilvout I, Prentice M. 1996. Characterization of a large chromosomal “high-pathogenicity island” in biotype 1B Yersinia enterocolitica. J Bacteriol 178:6743–6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rakin A, Noelting C, Schubert S, Heesemann J. 1999. Common and specific characteristics of the high-pathogenicity island of Yersinia enterocolitica. Infect Immun 67:5265–5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sarantis H, Balkin DM, De Camilli P, Isberg RR, Brumell JH, Grinstein S. 2012. Yersinia entry into host cells requires Rab5-dependent dephosphorylation of PI(4,5)P(2) and membrane scission. Cell Host Microbe 11:117–128. doi: 10.1016/j.chom.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kirillina O, Bobrov AG, Fetherston JD, Perry RD. 2006. Hierarchy of iron uptake systems: Yfu and Yiu are functional in Yersinia pestis. Infect Immun 74:6171–6178. doi: 10.1128/IAI.00874-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pieper R, Huang ST, Parmar PP, Clark DJ, Alami H, Fleischmann RD, Perry RD, Peterson SN. 2010. Proteomic analysis of iron acquisition, metabolic and regulatory responses of Yersinia pestis to iron starvation. BMC Microbiol 10:30. doi: 10.1186/1471-2180-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fukuto HS, Svetlanov A, Palmer LE, Karzai AW, Bliska JB. 2010. Global gene expression profiling of Yersinia pestis replicating inside macrophages reveals the roles of a putative stress-induced operon in regulating type III secretion and intracellular cell division. Infect Immun 78:3700–3715. doi: 10.1128/IAI.00062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Minato Y, Ghosh A, Faulkner WJ, Lind EJ, Schesser Bartra S, Plano GV, Jarrett CO, Hinnebusch BJ, Winogrodzki J, Dibrov P, Hase CC. 2013. Na+/H+ antiport is essential for Yersinia pestis virulence. Infect Immun 81:3163–3172. doi: 10.1128/IAI.00071-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tomich M, Planet PJ, Figurski DH. 2007. The tad locus: postcards from the widespread colonization island. Nat Rev Microbiol 5:363–375. doi: 10.1038/nrmicro1636. [DOI] [PubMed] [Google Scholar]
- 58.Schilling J, Wagner K, Seekircher S, Greune L, Humberg V, Schmidt MA, Heusipp G. 2010. Transcriptional activation of the tad type IVb pilus operon by PypB in Yersinia enterocolitica. J Bacteriol 192:3809–3821. doi: 10.1128/JB.01672-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wagner K, Schilling J, Falker S, Schmidt MA, Heusipp G. 2009. A regulatory network controls expression of the in vivo-expressed HreP protease of Yersinia enterocolitica. J Bacteriol 191:1666–1676. doi: 10.1128/JB.01517-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Portnoy DA, Moseley SL, Falkow S. 1981. Characterization of plasmids and plasmid-associated determinants of Yersinia enterocolitica pathogenesis. Infect Immun 31:775–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matsumoto H, Young GM. 2009. Translocated effectors of Yersinia. Curr Opin Microbiol 12:94–100. doi: 10.1016/j.mib.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mota LJ, Cornelis GR. 2005. The bacterial injection kit: type III secretion systems. Ann Med 37:234–249. doi: 10.1080/07853890510037329. [DOI] [PubMed] [Google Scholar]
- 63.Cornelis GR. 2002. The Yersinia Ysc-Yop ‘type III’ weaponry. Nat Rev Mol Cell Biol 3:742–752. doi: 10.1038/nrm932. [DOI] [PubMed] [Google Scholar]
- 64.Cornelis GR. 2002. The Yersinia Ysc-Yop virulence apparatus. Int J Med Microbiol 291:455–462. [DOI] [PubMed] [Google Scholar]
- 65.Bliska JB, Wang X, Viboud GI, Brodsky IE. 2013. Modulation of innate immune responses by Yersinia type III secretion system translocators and effectors. Cell Microbiol 15:1622–1631. doi: 10.1111/cmi.12164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Diepold A, Wiesand U, Amstutz M, Cornelis GR. 2012. Assembly of the Yersinia injectisome: the missing pieces. Mol Microbiol 85:878–892. doi: 10.1111/j.1365-2958.2012.08146.x. [DOI] [PubMed] [Google Scholar]
- 67.Schindler MK, Schutz MS, Muhlenkamp MC, Rooijakkers SH, Hallstrom T, Zipfel PF, Autenrieth IB. 2012. Yersinia enterocolitica YadA mediates complement evasion by recruitment and inactivation of C3 products. J Immunol 189:4900–4908. doi: 10.4049/jimmunol.1201383. [DOI] [PubMed] [Google Scholar]
- 68.Heise T, Dersch P. 2006. Identification of a domain in Yersinia virulence factor YadA that is crucial for extracellular matrix-specific cell adhesion and uptake. Proc Natl Acad Sci U S A 103:3375–3380. doi: 10.1073/pnas.0507749103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Venecia K, Young GM. 2005. Environmental regulation and virulence attributes of the Ysa type III secretion system of Yersinia enterocolitica biovar 1B. Infect Immun 73:5961–5977. doi: 10.1128/IAI.73.9.5961-5977.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walker KA, Maltez VI, Hall JD, Vitko NP, Miller VL. 2013. A phenotype at last: essential role for the Yersinia enterocolitica Ysa type III secretion system in a Drosophila melanogaster S2 cell model. Infect Immun 81:2478–2487. doi: 10.1128/IAI.01454-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moest TP, Meresse S. 2013. Salmonella T3SSs: successful mission of the secret(ion) agents. Curr Opin Microbiol 16:38–44. doi: 10.1016/j.mib.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 72.Young GM, Miller VL. 1997. Identification of novel chromosomal loci affecting Yersinia enterocolitica pathogenesis. Mol Microbiol 25:319–328. doi: 10.1046/j.1365-2958.1997.4661829.x. [DOI] [PubMed] [Google Scholar]
- 73.Nydam SD, Shah DH, Call DR. 2014. Transcriptome analysis of Vibrio parahaemolyticus in type III secretion system 1 inducing conditions. Front Cell Infect Microbiol 4:1. doi: 10.3389/fcimb.2014.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kroger C, Colgan A, Srikumar S, Handler K, Sivasankaran SK, Hammarlof DL, Canals R, Grissom JE, Conway T, Hokamp K, Hinton JC. 2013. An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe 14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 75.Taveirne ME, Theriot CM, Livny J, DiRita VJ. 2013. The complete Campylobacter jejuni transcriptome during colonization of a natural host determined by RNAseq. PLoS One 8:e73586. doi: 10.1371/journal.pone.0073586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Y, Romanov G, Bliska JB. 2011. Type III secretion system-dependent translocation of ectopically expressed Yop effectors into macrophages by intracellular Yersinia pseudotuberculosis. Infect Immun 79:4322–4331. doi: 10.1128/IAI.05396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Deuretzbacher A, Czymmeck N, Reimer R, Trulzsch K, Gaus K, Hohenberg H, Heesemann J, Aepfelbacher M, Ruckdeschel K. 2009. Beta1 integrin-dependent engulfment of Yersinia enterocolitica by macrophages is coupled to the activation of autophagy and suppressed by type III protein secretion. J Immunol 183:5847–5860. doi: 10.4049/jimmunol.0804242. [DOI] [PubMed] [Google Scholar]
- 78.Mota LJ, Journet L, Sorg I, Agrain C, Cornelis GR. 2005. Bacterial injectisomes: needle length does matter. Science 307:1278. doi: 10.1126/science.1107679. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.