Abstract
Inflammation is a major cause of respiratory impairment during Pneumocystis pneumonia. Studies support a significant role for cell wall β-glucans in stimulating inflammatory responses. Fungal β-glucans are comprised of d-glucose homopolymers containing β-1,3-linked glucose backbones with β-1,6-linked glucose side chains. Prior studies in Pneumocystis carinii have characterized β-1,3 glucan components of the organism. However, recent investigations in other organisms support important roles for β-1,6 glucans, predominantly in mediating host cellular activation. Accordingly, we sought to characterize β-1,6 glucans in the cell wall of Pneumocystis and to establish their activity in lung cell inflammation. Immune staining revealed specific β-1,6 localization in P. carinii cyst walls. Homology-based cloning facilitated characterization of a functional P. carinii kre6 (Pckre6) β-1,6 glucan synthase in Pneumocystis that, when expressed in kre6-deficient Saccharomyces cerevisiae, restored cell wall stability. Recently synthesized β-1,6 glucan synthase inhibitors decreased the ability of isolated P. carinii preparations to generate β-1,6 carbohydrate. In addition, isolated β-1,6 glucan fractions from Pneumocystis elicited vigorous tumor necrosis factor alpha (TNF-α) responses from macrophages. These inflammatory responses were significantly dampened by inhibition of host cell plasma membrane microdomain function. Together, these studies indicate that β-1,6 glucans are present in the P. carinii cell wall and contribute to lung cell inflammatory activation during infection.
INTRODUCTION
Pneumocystis organisms are opportunistic fungi that produce significant morbidity and mortality in immunocompromised hosts, with infection-related fatalities ranging between 10% and 45% (1). Pneumocystis jirovecii is the species known to infect humans, while Pneumocystis carinii represents the parallel species studied widely in rodents (2). Pneumocystis pneumonia remains a significant cause of mortality during AIDS, despite highly active antiretroviral therapy (3–5). Severe Pneumocystis pneumonia is characterized by intense lung inflammation involving CD8+ cells and neutrophils, impairing gas exchange (6–9).
The P. carinii cell walls contain abundant β-glucan molecules (10). Fungal cell wall β-glucans are homopolymers of d-glucose consisting of β-1,3 core chains with variable numbers of β-1,6 glucose side chains. The variable inflammatory activities of different glucan preparations have been postulated to be related to the relative amounts and configurations of these two major structures (β-1,3 versus β-1,6) (11). Almost all of the initial studies in fungi have largely focused on unfractionated glucans (10). In fact, all prior studies in P. carinii previously utilized only unfractionated β-1,3/β-1,6 glucans. Interestingly, recent investigations in Saccharomyces cerevisiae indicate major roles for β-1,6 glucans in strongly mediating cellular activation and inflammation (11). Our investigations of unfractionated P. carinii β-glucans indicate that innate recognition of these cell wall components is an important factor driving lung inflammation during Pneumocystis pneumonia (10, 12–16).
The roles of β-1,6 glucan components in mediating lung inflammation have remained unknown. In fact, due to larger amounts of β-1,3 glucans in many fungi, the β-1,3 components have been presumed, perhaps incorrectly, to be responsible for most β-glucan effects (11, 17). However, recent studies using S. cerevisiae-derived cell wall components have shown that β-1,6 (rather than β-1,3) glucans trigger cell activation (11). The extent to which β-1,6 glucans participate in lung inflammatory responses during Pneumocystis challenge remains unknown. Our studies further revealed that lung cell membrane microdomains on lung cells containing lactosylceramide participate in the response to Pneumocystis β-glucans (10, 12–16). It also has not yet been determined whether β-1,6 glucans further interact with lactosylceramide in host membrane microdomains.
These studies were undertaken to investigate whether P. carinii organisms indeed contain β-1,6 glucans and whether P. carinii expresses the related enzymatic machinery necessary to synthesize β-1,6 glucan carbohydrates. Once it was established that P. carinii does express β-1,6 glucans on its surface, we next tested novel β-1,6 glucan synthase inhibitors against P. carinii preparations. We further isolated a β-1,6 glucan-enriched P. carinii cell wall fraction and evaluated its activity in activating inflammatory activity in macrophages. Here, we provide the first evidence of the presence of β-1,6 glucans in P. carinii and demonstrate activity of this cell wall component in triggering host cell inflammatory responses.
MATERIALS AND METHODS
Reagents and strains.
Unless otherwise specified, all reagents were from Sigma-Aldrich (St. Louis, MO). Standard yeast molecular techniques were implemented to generate the strains and plasmids. P. carinii organisms were obtained from the American Type Culture Collection (ATCC) (Manassas, VA) (18). All animal studies were reviewed and approved by the Mayo Clinic Institutional Animal Care and Usage Committee. P. carinii was propagated for 8 to 10 weeks in immunosuppressed, corticosteroid-treated rats, as we reported previously (14, 19). Whole populations of P. carinii containing both trophic forms and cysts were purified from chronically infected rat lungs by homogenization and filtration through 10-μm filters and further fractionated into enriched populations of trophic forms (99.5% pure) and cysts (>40-fold enriched) by differential filtration through 3-μm filters (20). To exclude the presence of other infectious organisms in the P. carinii isolates, the preparations were stained (Diff Quick Modified Wright-Giemsa stain; Dade Diagnostics, Aguada, PR) and examined to exclude concurrent infection with bacteria or fungi. Isolates with significant contamination by other microorganisms were discarded (10). Small test quantities of the β-1,6 glucan synthase inhibitors D75-4590 and compound 16b were kindly provided by Akihiro Kitamura and Junichi Kuroyanagi, Daiichi Sankyo Co., Ltd., Tokyo, Japan.
Reference β-1,6 glucan isolate.
As a reference source of β-1,6 glucan, pustulan (EMD Millipore, Billerica, MA) was used in this study. Pustulan is derived from the lichen Umbilicaria pustulata. Hydrolysis and fractionation techniques used routinely to determine the polysaccharide content of the sugar of interest have demonstrated that not even trace amounts of di-, tri-, or tetrasaccharides are observed in pustulan, indicating that the molecule is linear in nature and contains exclusive β-1,6 glucan linkages (21). Other researchers have routinely used pustulan to study β-1,6 glucan–C-type lectin receptor (CLR) interactions and yeast β-1,6 glucan cell wall biology (22–26).
Generation of a polyclonal antibody recognizing β-1,6 glucan.
An anti-β-1,6 glucan antibody was generated in rabbits (Bethyl Laboratories) by immunizing them with purified β-1,6 glucan (pustulan) conjugated to keyhole limpet hemocyanin (KLH) (Solulink, Inc., San Diego, CA) (27). The pustulan-KLH conjugate (2 mg) was used to immunize rabbits (Bethyl Laboratories, Inc., Montgomery, TX) to generate antiserum to β-1,6 glucan. IgG was purified from the antiserum with protein A-Sepharose. The polyclonal antibody was tested against β-1,3 glucan, as well as mannan and related sugars, and showed no cross-reactivity.
Immune localization of β-1,6 and β-1,3 glucans in P. carinii cyst walls.
To initially evaluate the surface expression of P. carinii β-1,6 and β-1,3 glucans, freshly isolated P. carinii organisms were fixed for 90 min in 4% formaldehyde without detergent permeabilization, specifically to detect surface-expressed carbohydrate antigens (28, 29). The fixed cells were washed three times with 0.1 M KH2PO4, pH 6.5. The cells were then resuspended in 1.2 M sorbitol, 0.12 M KH2PO4, 33 mM citric acid, pH 5.9. The cells were then washed three times in sorbitol buffer and applied to poly-l-lysine-coated slides for 15 min. To initially detect β-1,6 glucan epitopes, we used a rabbit polyclonal antibody that was the kind gift of Frans Klis, University of Amsterdam (30). In subsequent experiments, we used our own polyclonal anti-β-1,6 glucan antibody generated in rabbits (Bethyl Laboratories). To contrast the localization of β-1,3 glucans, we next employed a mouse monoclonal antibody from Biosupplies Australia (Victoria, Australia) (30, 31). The slides were incubated with a 1:1,000 dilution (of a 1.0-mg/ml stock) of the primary anti-β-1,6 glucan antibody or with a 1:2,500 dilution (of a 1.0-mg/ml stock) of the primary anti-β-1,3 glucan antibody for 2 h at room temperature. The slides were then washed three times in 3% bovine serum albumin (BSA)-1× phosphate-buffered saline (PBS) for 5 min. An anti-rabbit IgG-fluorescein isothiocyanate (FITC) antibody produced in goat (1:32 dilution; Sigma, St. Louis, MO) and conjugated with FITC or a donkey anti-mouse antibody (1:32 dilution; Sigma-Aldrich, St. Louis, MO) conjugated with Texas Red was added to the slides and incubated for an additional 1 h. The slides were again washed as described above and visualized and photographed under oil immersion using an Olympus IX70 microscope with phase contrast and filters for FITC and Texas Red detection.
To further determine the ultrastructural localization of β-1,6 glucan carbohydrates in Pneumocystis organisms, immune electron microscopy was utilized. To prepare these samples, P. carinii cysts and trophic forms were fixed in buffer containing 4% formaldehyde and 2% glutaraldehyde in phosphate-buffered saline. Electron microscopy was performed using the β-1,6 glucan antibody at the Mayo electron microscopy core facility (Rochester, MN) using methods we reported previously (32).
Cloning and characterization of a Pckre6 β-1,6 glucan synthase in P. carinii.
A partial, though incomplete, DNA sequence of a gene homologous to a yeast kre6 β-1,6 glucan-synthetic gene was obtained by analysis of the Pneumocystis Genome Project Database (http://pgp.cchmc.org/). Primers for P. carinii kre6 (Pckre6) were synthesized, and the rapid amplification of cDNA ends (RACE) technique (GeneRacer kit; Life Technologies, Carlsbad, CA) was used to generate full-length cDNA sequences. We originally cloned the Pckre6 gene a number of years ago, but it was not until recently that we were able to fully characterize the gene and report its activity in generation of the P. carinii cell wall. The predicted amino acid sequence of PcKre6 was compared with the amino acid sequences of S. cerevisiae ScKre6 and Schizosaccharomyces pombe SpKre6, respectively, by employing the ClustalW2 tool (http://www.ebi.ac.uk/Tools/msa/clustalw2/) to derive sequence alignments.
Southern hybridization of Pckre6.
P. carinii genomic DNA was derived from heavily infected rats with P. carinii pneumonia using TRIzol reagent (Life Technologies). Genomic DNA was also extracted from the lungs of uninfected rats. Both rat DNA and P. carinii DNA were digested with HindIII, XbaI, and XhoI incubated at 37°C overnight. The digested products were separated by agarose gel electrophoresis and transferred to nitrocellulose. To generate radiolabeled probes, amplicons from PCR were labeled with Amersham [α-32P]dATP (GE Healthcare, Piscataway, NJ) according to the RadPrime DNA Labeling System protocol (Life Technologies). Quick Spin columns (Roche, Indianapolis, IN) were used to remove unincorporated [α-32P]dATP. The radiolabeled probes and the nitrocellulose membranes were used for Southern hybridization. The nitrocellulose membranes were air dried and exposed to X-ray films at −70°C for 48 h.
Relative expression of Pckre6 in isolated life cycle forms of the organism.
Our previous studies demonstrated that certain Pneumocystis cell wall-regulatory genes are differentially expressed under conditions of varying pH (33, 34). Accordingly, the pH-regulated expression of P. carinii was evaluated by Northern blotting. P. carinii organisms were maintained for 2 h under various pH conditions, and total RNA was isolated. Equal amounts of RNA (5.0 μg) were separated through a 1.0% agarose gel in the presence of 2.2 M formaldehyde, transferred to nitrocellulose, and hybridized with a 90-bp probe labeled with [α-32P]dATP (Amersham, Inc., Piscataway, NJ). After hybridization, the membranes were washed and visualized by autoradiography.
While the life cycle of Pneumocystis remains incompletely understood, it involves both smaller trophic forms, which strongly adhere to alveolar epithelial cells, and the larger cystic forms that are believed to be the transmissible form of the organism. Therefore, to gain initial insights into potential activity of Pckre6 during the life cycle of the organism, we next evaluated the relative expression of Pckre6 in isolated life cycle forms of the organism. To accomplish this, P. carinii organisms were separated into cystic and trophic forms by differential filtration. P. carinii-infected rats were sacrificed after 8 weeks of immunosuppression. The P. carinii organisms were purified by homogenization and filtration through 10-μm filters that retained lung cells but allowed passage of P. carinii. The P. carinii filtrates were centrifuged (1,500 × g for 30 min), and the pellets were resuspended in Hanks' balanced salt solution. Next, differential filtration was used to separate P. carinii cysts and trophic forms. In brief, P. carinii cysts were retained by a 3-μm Nuclepore filter and resuspended after exhaustive washing. P. carinii trophic forms passed through the filter and were recovered by centrifugation. This separation procedure yielded trophic form populations containing 99.5% trophic forms, with the remainder representing nonviable host cell fragments. The trophic preparations were essentially devoid of cysts. The cyst preparations were ∼90% enriched for cysts. Any residual trophic form that might have carried over into the cyst fractions would act only to reduce any observed magnitude of differential expression in Pckre6 between the two populations. RNA from the separated P. carinii populations was extracted from the separated forms using TRIzol reagent (Life Technologies), and an equal amount of RNA (5.0 μg) was separated by electrophoresis through 1.2% agarose in the presence of 2.2 M formaldehyde. The separated RNA was transferred to nitrocellulose and hybridized with a 90-bp probe as described above, using the ExpressHyb protocol (Clontech, Inc., Mountain View, CA). Densitometry of the hybridization was performed on a GS-800 calibrated densitometer (Bio-Rad) with Quantity One 1-D analysis software, and the relative density abundance of the Pckre6 mRNA was compared to that of P. carinii actin in the cyst and trophic form blots.
Functional activity of Pckre6 in mediating fungal cell wall integrity.
Strains of S. cerevisiae with deletions of kre6 exhibit defects in cell wall integrity and, hence, have impaired growth in the presence of calcofluor white M2R (35). We therefore studied the growth of the BY4741 kre6Δ S. cerevisiae strain transformed with either a control vector (pYES2.1/V5-His/lacZ) or pYES2.1 containing the in-frame full-length Pckre6 cDNA (pYES2.1/Pckre6/V5/6XHis). Cells were grown overnight in synthetic complete medium containing 2% galactose and supplemented with appropriate amino acids but lacking uracil to select and maintain the plasmids. Colony morphology studies were conducted by plating wild-type S. cerevisiae with a control vector or the S. cerevisiae kre6Δ (kre6-deficient) or Pckre6-complemented S. cerevisiae kre6Δ strain on synthetic complete medium without uracil and containing calcofluor white M2R (100 μg/ml). Colonies were observed after growing the strains for 3 days at 30°C (36).
Isolation of a β-1,6 glucan-enriched cell wall component from P. carinii.
A β-glucan-rich cell wall fraction from P. carinii containing mixed β-1,3/β-1,6 glucans was prepared as we previously reported (10, 37). P. carinii organisms were isolated from the lungs of heavily infected animals by homogenization and filtration through 10-μm filters. The organisms (∼30 g [wet weight]) were autoclaved (120°C; 20 min), and the glucans were isolated by NaOH digestion and characterized as previously detailed (10). The alkali-insoluble cell wall fraction was neutralized with 10 mM phosphate buffer, pH 6.8, before digestion with Zymolyase 100T (predominantly containing a β-1,3 glucanase; Seikagaku Corp., Tokyo, Japan) at 37°C for 18 h with rotation to liberate soluble β-1,6 glucans (38). The digestion was stopped after 24 h by incubation for 15 min at 75°C, and undigested glucans were collected by centrifugation. The soluble β-1,6 glucan-enriched fraction contained in the supernatant was dialyzed against sterile water in Spectra/Por (Spectrum Laboratories, Inc., Rancho Dominguez, CA) tubing with a molecular mass cutoff of 6 to 8 kDa for 24 h and then lyophilized. Extensive measures were employed to ensure that the fractions were free of endotoxin that might have been carried over from the lungs of rats with P. carinii pneumonia. Prior to use in studies, the Pneumocystis β-1,6 glucan fractions were treated with polymyxin B-agarose (Sigma) and then separated from the agarose by centrifugation. The final preparation was assayed for endotoxin with the Limulus amebocyte lysate assay (Associates of Cape Cod, East Falmouth, MA) and found to consistently contain <0.125 units of endotoxin.
Competitive dot blot assay for P. carinii β-1,6 glucans.
To verify the β-1,6 glucan contents of the carbohydrates contained in the P. carinii cell wall isolates, competitive dot blot assays were performed (39). Serial dilutions of P. carinii cell wall preparations (CWP) isolated as described above were spotted (2 μl) onto Hybond-C nitrocellulose (Amersham) and air dried. The membranes were blocked with 3% BSA in 1× Tris-buffered saline (TBS) with 0.1% (vol/vol) Tween 20 (TBST). The β-1,6 glucan antibody used in the dot blot assay, as well as in the inhibition enzyme immunoassay (IEIA) described below, was generated in rabbits (Bethyl Laboratories) as detailed above. The anti-β-1,6 glucan antibody working solution (1:5,000 in TBST) was mixed with various soluble competing carbohydrates, including diluent alone (no competing carbohydrate) or pustulan (soluble β-1,6 glucan), laminarin (soluble β-1,3 glucans), or yeast mannan (soluble mannoproteins), and rocked at 25°C for 1 h prior to addition of the mixture to the spotted membrane (39). Each competing carbohydrate was present at a concentration of 5 μg/ml. The membranes were subsequently probed for 1 h at 25°C, washed, and then incubated with goat anti-rabbit-horseradish peroxidase (HRP) (1:10,000 in TBST) for 1 h at 25°C. The blots were developed with ECL detection reagent (Amersham).
β-1,6 Glucanase digestion of P. carinii cell wall isolates.
The P. carinii cell wall isolates were further treated with a specific Trichoderma harzianum β-1,6 glucanase that was expressed in S. cerevisiae and purified from the transformed yeast (40). To accomplish this, T. harzianum (ATCC) was cultured on potato dextrose agar (PDA) at 30°C for 48 h, and the mycelia were collected. The cell walls were disrupted with 0.5-mm glass beads in the provided buffer from the RNAeasy kit, and the RNA was isolated using an RNeasy kit (Qiagen, Madison, WI). Next, cDNA was synthesized from 100 ng of RNA using a Superscript III First Strand cDNA synthesis kit and oligo(dT) primers (Life Technologies) according to the kit recommendations. The full-length coding sequence of the bgn16.2 gene was amplified using oligonucleotides 5′-ATGAAGTACTCCATCGTTGCTC-3′ and 5′-CCTGTATCCAGAGCAGACATC-3′ (0.3 μM), designed using MacVector software (Cary, NC). The PCR conditions were as follows: 94°C for 2 min for 1 cycle (hot start), followed by 94°C for 30 s, 62°C for 30 s, and 72°C for 2 min for 35 cycles, with a final extension at 72°C for 10 min, using Platinum Taq DNA polymerase (Life Technologies). The product was cloned into pYES2.1/V5-His-TOPO TA (Life Technologies), and the plasmid sequences were verified and compared to the published sequence (GenBank accession no. X79196.1) and subsequently subcloned into S. cerevisiae strain BY4741 (ATCC). The expressed β-1,6 glucanase enzyme was purified from approximately 1 liter of transformed yeast cultured in 2% galactose minimal medium after the cell walls had been disrupted using a French press cell disruptor before passing over a Y-PER-His affinity column (Pierce Chemical, Rockford, IL) according to the manufacturer's instructions. The purified protein fractions were dialyzed overnight in Slide-A-Lyzer dialysis cassettes with a molecular mass cutoff of 10 kDa (Pierce) against a bath of 50 mM potassium acetate plus 10% glycerol, pH 5.5. The various P. carinii cell wall isolates were digested with the purified β-1,6 glucanase at 45°C overnight in the same buffer and then submitted to dot blot assay with the β-1,6 glucan antibody as detailed above.
Determination of β-1,6 glucan synthase activity in P. carinii.
Pneumocystis organisms were isolated from the lungs of heavily infected rats (approximately 1 g), and cell wall fractions were isolated as previously described (20, 41). Purified P. carinii organisms were sonicated on ice in 50 mM Tris-HCl, 150 mM NaCl buffer with 1 mM EDTA three times for 30 s each sonication, with 1 min on ice between sonications. The sonications were performed at 35% maximal power. The preparations were then centrifuged at 2,000 × g for 5 min at 4°C to remove cellular debris. The supernatant was then centrifuged at 100,000 × g for 45 min at 4°C. The resulting pellet was washed gently with cold 50 mM Tris-HCl buffer containing 1 mM EDTA and 1 mM mercaptoethanol (pH 7.5; buffer A) and finally resuspended in buffer A containing 33% glycerol. The protein content of the cell wall isolate was determined by a bicinchoninic acid (BCA) assay (Pierce). The 40-μl assay mixture contained 30 to 60 μg isolated cell wall protein, 2.5 mM UDP–d-glucose (Sigma), 150 μM GTP (Sigma), 1.3 mM EDTA-Na, 0.75% (wt/vol) BSA, 4.1% (vol/vol) glycerol, and 100 mM 2-(N-morpholino)ethanesulfonic acid (MES)–Na, pH 6.5 (Sigma). The assay mixture was incubated for 4 h at 30°C, and the assay was stopped by incubation at 75°C for 15 min. Detection of β-1,6 glucose products was performed using a competitive enzyme immunoassay (41, 42). The wells of a 96-well Nunc Maxi-Sorb plate (eBioscience, San Diego, CA) were coated by adding 200 μl at 2 μg/ml of purified β-1,6 glucan (pustulan; EMD Millipore, Billerica, MA) in 0.5 M sodium bicarbonate-sodium carbonate buffer, pH 9.6, and stored overnight at 4°C. Newly synthesized β-1,6 glucans from the P. carinii preparations were boiled extensively prior to testing to completely dissolve the material. The plates were washed 4 times with 1× PBS containing 0.05% (vol/vol) Tween 20 (PBT) and then blocked with 200 μl 1% BSA (Pierce) in PBT (PBTB) for 1 h at room temperature. Pustulan (β-1,6 glucan) standards (12 dilutions; 1,000 to 9.7 ng/ml in PBTB) or P. carinii β-1,6 glucan assay preps were mixed with an equal volume of anti-β-1,6 glucan antibody diluted 1:100,000 in PBTB, which were preincubated for 20 min at 37°C with rotation. After washing the wells 4 times with PBT, 100 μl of the glucan samples or standards-plus-antibody mixture was applied to the β-1,6 glucan (pustulan)-coated wells and incubated for 1 h at room temperature. The wells were washed before adding 100 μl of the secondary goat anti-rabbit HRP-conjugated antibody (Southern Biotechnology, Birmingham, AL) diluted 1:20,000 in PBTB and incubated for 1 h at room temperature. The wells were washed 6 times before adding 100 μl of 1× TMB substrate (eBioscience). After 5 min, the reaction was terminated by adding 50 μl 2N H2SO4. The absorbance at 450 nm was measured in a VersaMax plate reader (Molecular Devices, Sunnyvale, CA), and standards were plotted using the 4-parameter curve-fitting setting of the SoftMax Pro software. Additional assays were performed in the presence of newly described β-1,6 glucan synthase inhibitors (0.06 μg/ml each) that inhibit β-1,6 glucan synthesis in yeast and Candida albicans (41–45).
Activity of the P. carinii β-1,6 glucan-enriched cell wall fraction in stimulating host cell inflammatory responses.
We further sought to determine whether the P. carinii β-1,6 glucan-enriched cell wall fraction possessed activity to stimulate host cell inflammatory responses. To accomplish this, we coated 10-μm latex beads with the P. carinii β-1,6 glucan-enriched cell wall fraction dissolved in 0.1 M NaHCO3 buffer according to the manufacturer's protocol (Polysciences, Inc., Warrington, PA). These 10-μm beads were used to mimic the size of intact P. carinii cysts. Bead coating was assessed by the orcinol-sulfuric acid method (46). Either P. carinii β-1,6 glucan-coated or uncoated beads were incubated at the indicated concentrations with RAW 264.7 mouse macrophages (ATCC) overnight, and tumor necrosis factor alpha (TNF-α) release into the media was determined by enzyme-linked immunosorbent assay (ELISA) (eBiosciences, Inc., San Diego, CA). In addition, since our prior work had indicated that whole P. carinii organisms and unfractionated P. carinii β-1,6 glucans mediate cell inflammatory activation through glycosphingolipids present in host cell membrane microdomains (13), we further examined TNF-α release in the presence of the glucosylceramide synthesis inhibitor d-threo-1-phenyl-2-decanoylamino-3-morpholino-1-propanol·HCl (PDMP) at the indicated concentrations.
Statistical analysis.
All the data shown are expressed as the means and standard errors of the mean (SEM) derived from multiple experimental runs. Statistical differences between various data groups were first determined using analysis of variance (ANOVA) and Student's t test, as indicated. The corresponding nonparametric statistics were applied when the data were not distributed in a Gaussian fashion. Statistical testing was performed using GraphPad Prism version 5.0b software, and statistical differences were considered to be significant at a P value of <0.05.
Nucleotide sequence accession number.
The complete sequence of Pckre6 was analyzed and deposited in GenBank under accession number AY371293.1.
RESULTS
The P. carinii cell wall contains both β-1,3 and β-1,6 glucans, as determined by fluorescence and immune electron microscopy.
Isolated P. carinii cyst preparations contain significant quantities of both β-1,3 and β-1,6 carbohydrates. We first assessed the presence of these carbohydrates in freshly isolated P. carinii cyst forms with immunofluorescence (Fig. 1). Similar to S. cerevisiae, S. pombe, and Histoplasma capsulatum, P. carinii cysts appear to have relatively homogeneous deposition of β-1,3 glucan in the cell wall outside the plasma membrane (Fig. 1B) (31, 47, 48). Interestingly, β-1,6 staining was also noted widely in the P. carinii cyst life form, but rather than being present as a continuous layer of the polysaccharide in the cell wall, the sugar appeared to be distributed discontinuously in the P. carinii cell wall, with greater amounts in some regions than others, as demonstrated by a punctate staining pattern (Fig. 1C). Furthermore, as the merged image reveals (Fig. 1D), β-1,6 glucans overlap some of the β-1,3 glucan, but not continuously throughout the cyst wall. Immune electron microscopy of P. carinii cyst forms further verified the presence of β-1,6 glucan on the outside of the plasma membrane in the P. carinii cyst wall (Fig. 1E). These results are consistent with studies in yeast that document that β-1,6 glucan represents a less abundant component of the overall cell wall by mass (49).
FIG 1.
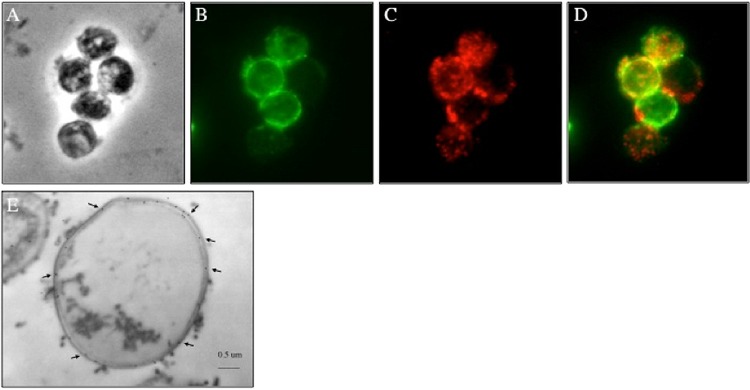
β-1,3 and β-1,6 glucans are present in the outer cell wall layer of P. carinii. (A) Phase-contrast microscopy image of P. carinii cyst forms. (B and C) P. carinii cysts were fixed, and antibodies specific for β-1,3 glucan (B) and β-1,6 glucan (C) were used to localize the respective carbohydrates. (D) Merged β-1,3 glucan and β-1,6 glucan localization. (E) Representative transmission immune electron micrograph (n = 20) utilizing 18-nm gold particles showing labeling of β-1,6 glucans in the P. carinii cyst wall (arrows).
P. carinii contains a kre6 β-1,6 glucan synthase gene.
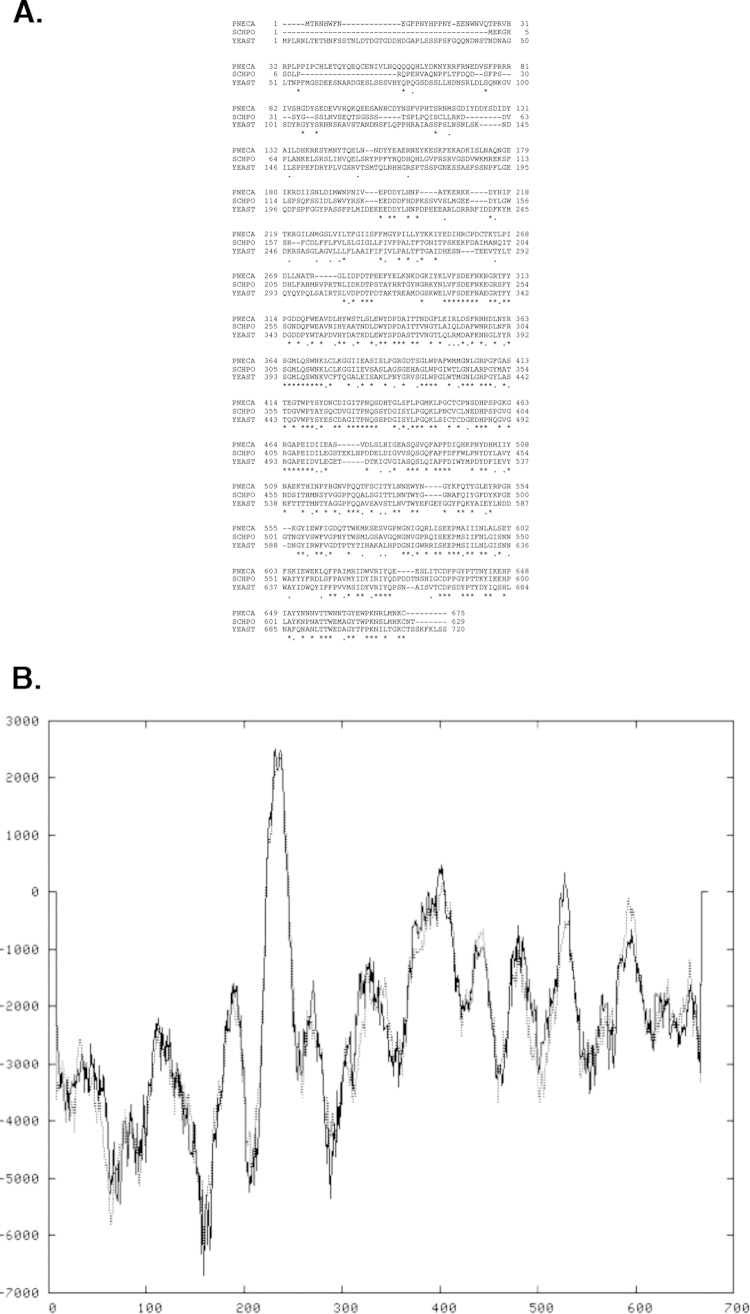
Interrogation of the P. carinii genome revealed a partial sequence for a kre6-like gene. Using the RACE technique described above to uncover full-length P. carinii reading frames (33, 50), we isolated the remaining portion of the Pckre6 gene. The predicted PcKre6 protein has a theoretical molecular mass of ∼79 kDa and a pI of 5.93. Alignment of protein sequences demonstrated that the amino acid sequence of the predicted protein has significant homology to proteins from S. pombe (64% homology by BLASTX) and S. cerevisiae (62% homology by BLASTX) (Fig. 2A). The specific region of amino acids 299 to 626 of the PcKre6 predicted protein is a conserved domain related to members of glycosyl hydrolase family 16. This family includes S. cerevisiae Kre6 (51). Unlike S. pombe and S. cerevisiae Kre6 proteins, which contain three predicted transmembrane motifs, PcKre6, in a fashion similar to that of the pathogen Cryptococcus neoformans β-1,6 glucan synthase, is predicted to have only one transmembrane-spanning region, as analyzed by TMpred software (Fig. 2B) (52).
FIG 2.
(A) Alignment of the predicted PcKre6 amino acid sequence with sequences from other relevant fungal species. PNECA, P. carinii; SCHPO, S. pombe; YEAST, S. cerevisiae. Multiple-sequence alignments of the fungal Kre6 β-1,6 glucan synthases performed with ClustalW2 (MacVector 8.1.2) demonstrating significant amino acid homology. The asterisks denote identically conserved amino acid residues. The periods indicate similar charged amino acids, and the dashes indicate gaps in the protein alignment to allow matching of conserved amino acids. (B) Transmembrane hydrophobicity plot of PcKre6 protein. A value of 1,000 or above represents a strongly preferred model for transmembrane domain architecture (52).
After identifying the complete Pckre6 open reading frame, a cognate 90-bp probe was generated and hybridized to a restriction-digested panel of P. carinii genomic DNA to verify the presence of the gene in P. carinii organisms (Fig. 3). Strong hybridization of the Pckre6 probe was noted in HindIII-, XbaI-, and XhoI-digested P. carinii genomic DNA, but no specific hybridization was demonstrated in similarly digested rat lung genomic DNA. These data suggest that the Pckre6 gene we amplified in our RACE procedure is contained within and specific to the P. carinii genome and does not represent a contaminating rat host DNA fragment.
FIG 3.
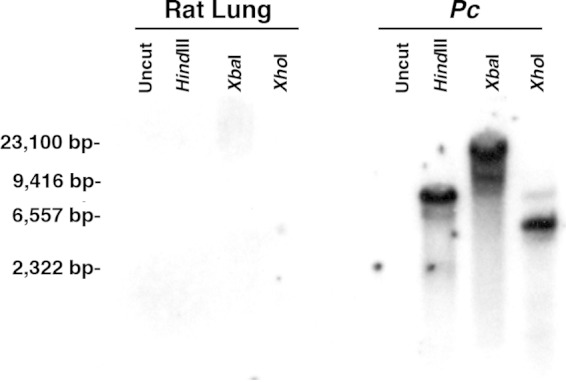
The Pckre6 homolog is represented in the P. carinii genome. The full-length Pckre6 cDNA homolog was radiolabeled and hybridized to P. carinii genomic DNA digested with HindIII, XbaI, and XhoI, as indicated. Pckre6 was present in one location on each Southern analysis but was not present in similarly restriction endonuclease-digested host lung DNA.
Pckre6 is expressed at physiological and higher pH and in the cyst form of the organism.
Our laboratory has shown previously that P. carinii life forms express cell wall-remodeling transcripts differently based on the environmental conditions that they are exposed to (33, 34, 53). Therefore, we addressed whether Pckre6 expression was also regulated by environmental pH, examining a pH range of 4 to 8. Similar to the other three Pneumocystis cell wall genes tested previously, expression of the Pckre6 transcript was detected by Northern blotting at pH ∼7.0, a pH relevant to the host lung, where the organism lives (Fig. 4, left). However, somewhat greater expression was also observed at a higher pH of 8.0.
FIG 4.
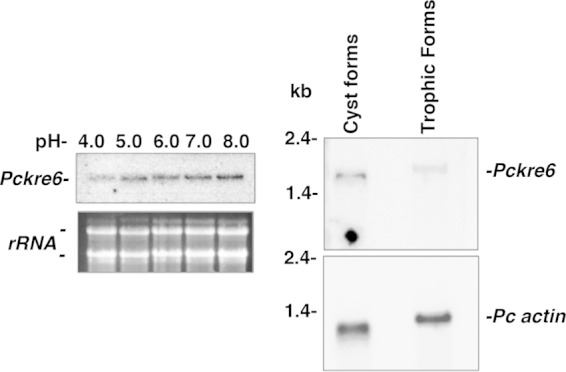
Pneumocystis Pckre6 is differently expressed in response to pH. Total P. carinii life forms were isolated and resuspended in Ham's F-12 medium with 10% fetal calf serum at pH 4.0 to 8.0 at 37°C over 2 h. Steady-state mRNA levels were detected by Northern hybridization with the 90-bp Pckre6 amplicon. (Left) Hybridization of Pckre6 to the nylon membrane. Below is a photograph of the two Pneumocystis major ribosomal subunits, demonstrating equal RNA loading. (Right) P. carinii Pckre6 is expressed at slightly higher levels in cystic life forms of P. carinii versus trophic life forms of the organism. (Top) Hybridization of the Pckre6 probe. (Bottom) Repeat hybridization of the membrane with P. carinii actin to confirm equal loading.
Finally, based on our previous observations that Pcgsc-1, the major β-1,3 glucan synthase gene of P. carinii organisms, is predominantly expressed in the cyst life form of the organism (20), we further investigated whether Pckre6 gene expression was also regulated over the P. carinii life cycle. Cyst and trophic forms were separated, and Pckre6 mRNA expression was evaluated by Northern analysis (Fig. 4, right). Pckre6 expression was slightly higher in the P. carinii cyst form than in the trophic forms. Repeated hybridizations with the same blots utilizing specific actin verified that the abundance of Pckre6 mRNA was not a consequence of RNA loading. Using densitometry, we observed that Pckre6 represented ∼34% of the density of P. carinii actin for the cystic forms compared to ∼16% for the trophic forms. Hence, there appeared to be roughly 2-fold greater Pckre6 expression in the cysts than in the trophic forms. We also observed slightly different relative migration rates of the Pckre6 transcripts in the trophic forms than in the cyst form isolates. We have also previously observed slightly different migration rates of trophic and cyst RNA products for other genes, with no evidence that they represent splice variants (33, 34). We believe that this may be related to the fact that the RNA partially retains secondary structure in the trophic form-derived compared to the cyst form-derived RNA, causing relative migration at different rates with the same molecular mass (54).
Pckre6 can rescue cell wall sensitivity to calcofluor in S. cerevisiae cells lacking kre6.
Studies were next performed to determine whether Pckre6 cDNA encoded a fungal Kre6-like protein. Due to the inability to culture and genetically manipulate P. carinii in vitro, traditional methods to characterize gene function in the organism are not possible at this time. To circumvent this, we used the heterologous expression and complementation of genetic knockouts in tractable fungi, such as Candida glabrata, S. cerevisiae, or S. pombe, to assess gene function in P. carinii (33, 50, 55, 56). Using this approach, the biological activity of Pckre6 was determined by assessing the gene's ability to complement budding S. cerevisiae kre6Δ cells in the presence of calcofluor white M2R, a compound that binds strongly to cell wall cellulose and chitins (57). In kre6Δ yeast, this reagent can inhibit organism growth by binding exposed chitin residues due to improper or deficient β-1,6 glucan cell wall deposition (35). The wild-type parent strain and kre6Δ mutant strains containing the control empty vector, along with kre6Δ yeast cells containing Pckre6 cDNA, were cultured overnight under inducing conditions and were subsequently plated on medium containing 2% galactose minimal medium alone (yeast plating control) or medium containing 100 μg/ml calcofluor white M2R. All the plates also lacked uracil for the maintenance of the respective plasmids. Under these conditions, the presence of Pckre6 cDNA efficiently complemented the yeast kre6Δ strain to levels similar to the growth of wild-type yeast, further indicating that PcKre6 functions as a β-1,6 glucan synthase protein in the fungus (Fig. 5).
FIG 5.
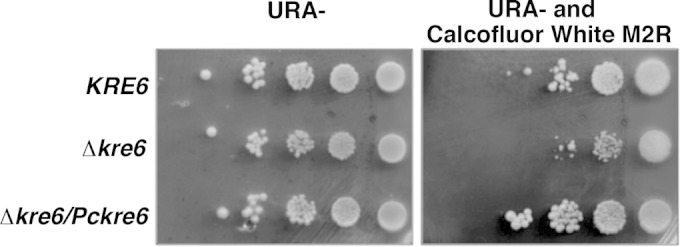
Pckre6 can restore S. cerevisiae kre6Δ sensitivity to calcofluor white M2R. Wild-type yeast, kre6 mutant yeast, and kre6 mutant yeast transformed with Pckre6 were cultured in minimal medium to an A600 of 1 and serially 10-fold diluted. Ten microliters of each sample was spotted either onto minimal medium without uracil (URA−) and without calcofluor white M2R or onto minimal medium without uracil but containing calcofluor white M2R (100 μg/ml) and incubated at 30°C for 3 days.
P. carinii cell wall preparations contain β-1,6 glucans and can incorporate UDP-glucose into β-1,6 polysaccharide.
To determine the presence of β-1,6 glucans in P. carinii, we isolated a β-1,6 glucan-enriched cell wall fraction and implemented a competitive dot blot assay (39) to verify its β-1,6 glucan content, using a polyclonal β-1,6 glucan antibody we generated. Similar to what others have shown with β-1,6 glucans derived from C. neoformans (39), isolated P. carinii β-1,6 preparations immobilized on the membrane bound the β-1,6 glucan antibody. Furthermore, the soluble β-1,6 glucan pustulan was the single polysaccharide that could significantly compete with the P. carinii β-1,6 glucan immobilized on the membrane (Fig. 6A). To further confirm the presence of β-1,6 glucans in P. carinii, we digested our P. carinii β-1,6 glucan samples with recombinant T. harzianum β-1,6 glucanase enzyme. The substrate specificity of the enzyme for β-1,6 glucan linkages (pustulan) was confirmed, with no glucanase activity noted against β-1,3 glucans, such as laminarin and pachyman, or other fungal sugar substrates (40). Similar to the control β-1,6 glucan pustulan, the P. carinii β-1,6 glucan preparations were digested with the T. harzianum β-1,6 glucanase and tested again in the dot blot assay. Digestion of the P. carinii cell wall preparation with β-1,6 glucanase resulted in significant reduction in binding of the β-1,6 glucan antibody (Fig. 6B).
FIG 6.
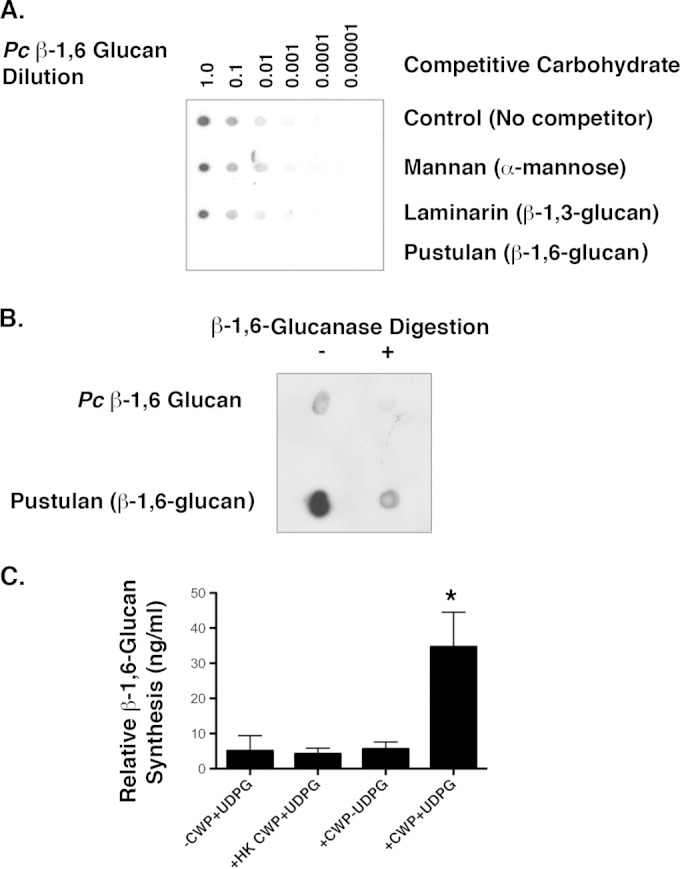
P. carinii contains a β-1,6 glucan cell wall fraction. (A) β-1,6 Glucan dot blot assay comparing the abilities of various carbohydrates to compete with a P. carinii β-1,6 glucan-enriched cell wall fraction immobilized onto nitrocellulose for detection with a specific anti-β-1,6 glucan antibody. Cell wall material was serially diluted onto the membrane. The primary anti-β-1,6 glucan antibody was incubated with the competitive polysaccharides before being applied to the nitrocellulose membrane. The blots were incubated for 1 h, washed, and incubated with secondary goat anti-rabbit-HRP. Only the known β-1,6 glucan pustulan successfully competed with the P. carinii cell wall isolate to eliminate detection by the antibody. (B) The P. carinii β-1,6 glucan cell wall isolate and pustulan were digested with β-1,6 glucanase overnight. The P. carinii β-1,6 glucan was largely digested by the β-1,6 glucanase. Pustulan was also largely (though not completely) digested by the β-1,6 glucanase. (C) Finally, P. carinii CWP were demonstrated to generate β-1,6 glucan polysaccharide in the presence of UDP-glucose (UDPG), as measured by IEIA. Heat-killed P. carinii cell wall preparations (HK CWP) did not exhibit significant β-1,6 glucan synthesis activity (P < 0.05 by one-way ANOVA across all groups). *, P < 0.05 by unpaired Student's t test comparing UDPG-driven β-1,6 glucan synthesis from viable CWP in the presence and absence of UDPG. The data are expressed as means and standard errors of the mean (SEM) derived from multiple experimental runs.
Finally, to establish whether P. carinii CWP could actively generate β-1,6 glucan, we incubated P. carinii membrane preparations with UDP-glucose, the substrate common to other fungal Kre6-like proteins, along with appropriate reaction buffer. We subsequently assayed for the generation of insoluble β-1,6 glucan via IEIA. Over a 4-h incubation period, P. carinii cell wall preparations incorporated significant UDP-glucose into insoluble β-1,6 glucan, as detected by IEIA (Fig. 6C) (P = 0.0274) comparing UDP-glucose incorporation to reaction buffer alone with either no P. carinii cell wall preparation or heat-killed CWP, reaction buffer without UDP-glucose, and P. carinii CWP plus UDP-glucose (Fig. 6C).
Inhibition of P. carinii β-1,6 glucan synthase activity with specific β-1,6 glucan synthase inhibitors.
Recent work by others has identified specific inhibitors that reduce β-1,6 glucan synthase activity (43–45). Such agents represent a potential antifungal strategy capable of inhibiting β-1,6 glucan synthase activity in vitro, as well as in vivo (43–45). Accordingly, we next sought to investigate whether the production of β-1,6 glucans by P. carinii could be suppressed with these recently identified β-1,6 glucan synthase inhibitors. Two recently described β-1,6 glucan synthase inhibitors were able to suppress β-1,6 glucan production by isolated P. carinii membrane preparations, with the greatest inhibition noted with compound 16b, followed by D75-4590 (Fig. 7). These results further support the idea that P. carinii organisms assemble β-1,6 glucan through a Kre6-like synthase.
FIG 7.
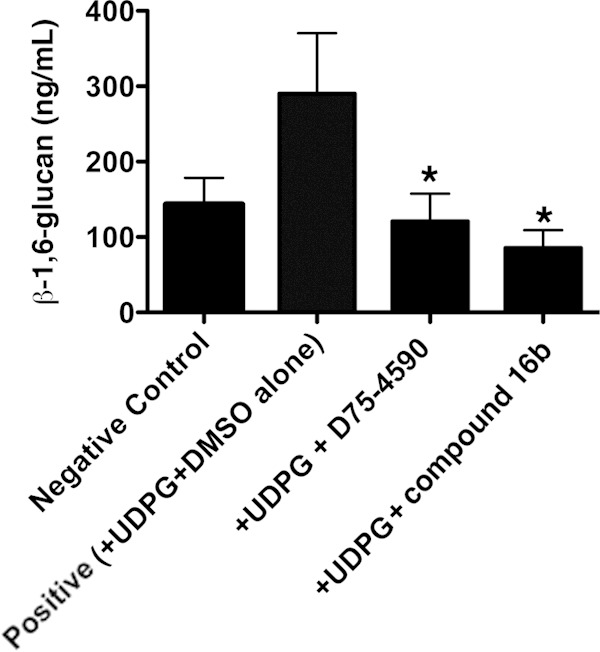
P. carinii cell wall preparations incorporate β-1,6 glucans, which can be inhibited by the β-1,6 glucan synthase inhibitors D75-4590 and compound 16b. To assess whether P. carinii actively incorporates UDP-glucose into insoluble β-1,6-containing carbohydrates, P. carinii organisms were purified and cell wall preparations were generated and reacted with UDPG as described in Materials and Methods. The resulting derived β-1,6 glucans were assayed by IEIA. The assays were conducted in the presence of either DMSO diluent alone (positive control) or D75-4590 or compound 16b (0.06 μg/ml each in DMSO diluent), which are agents that specifically inhibit β-1,6 glucan synthase-type proteins. The negative control was P. carinii cell wall preparations alone, without UDPG substrate, which represents the basal β-1,6 glucans already present in these preparations. The positive control was P. carinii cell wall preparations incubated in reaction mixtures containing both UDPG and DMSO diluent. Both D75-4590 and compound 16b significantly inhibited the generation of new P. carinii β-1,6 glucan compared to positive-control reaction mixtures. Shown are data derived from three experimental runs, each performed in duplicate. *, P < 0.05, contrasting β-1,6 glucan generated in the presence of UPDG and DMSO diluent alone but with no inhibitor (positive control) and reactions performed with UPDG with inhibitor diluted in DMSO. The data are expressed as means and SEM derived from multiple experimental runs.
Isolated P. carinii β-1,6 glucans induce vigorous TNF-α responses that are mediated by host cell glycosphingolipids.
Serum-free cultured RAW264.7 mouse macrophages were challenged for 18 h with P. carinii β-1,6 glucan-coated polystyrene beads. These beads are approximately 10 μm in size, similar to the size of P. carinii cyst forms (58). From these studies, we observed that surface-bound P. carinii β-1,6 glucan elicits significant TNF-α release from cultured macrophages. Furthermore, similar to our prior studies performed with whole, unfractionated P. carinii β-1,3/β-1,6 glucans, treatment of the macrophages for 24 h with the glycosphingolipid synthesis inhibitor PDMP significantly reduced TNF-α release in a dose-responsive manner (Fig. 8). Results from the study strongly suggest that P. carinii β-1,6 glucans participate in inflammatory responses in the host and that reducing glycosphingolipid synthesis in the host may represent a potential anti-inflammatory strategy during P. carinii pneumonia.
FIG 8.
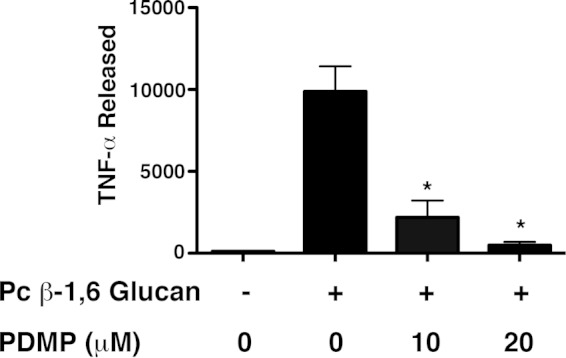
P. carinii β-1,6 glucan immobilized on 10-μm beads stimulates macrophage TNF-α protein production, which is suppressed by inhibition of glycosphingolipid synthesis. RAW 264.7 mouse macrophages were stimulated with bead-immobilized P. carinii β-1,6 glucan overnight, and macrophage TNF-α protein production was measured by ELISA in the presence or absence of the glycosphingolipid inhibitor PDMP. PDMP significantly suppressed generation of the chemokine in a dose-dependent manner (*, P < 0.05 compared with the control). The data are expressed as means and SEM derived from multiple experimental runs.
DISCUSSION
Prior studies indicated that the P. carinii cyst wall is comprised of β-1,3 glucans, major surface glycoprotein-glycoprotein A (MSG-gpA), melanin, and potentially also chitin components (20, 59–64). We have previously shown that β-1,3 glucans comprise a large portion of the P. carinii cyst wall. In particular, β-1,3 glucans represent a major carbohydrate present on P. carinii, as determined by immune electron and fluorescence microscopy (58, 59). Furthermore, P. carinii β-glucan has been shown to be a significant inflammatory stimulant for lung dendritic, epithelial, and alveolar macrophage cellular responses (13, 14, 65). Finally, a gene responsible for P. carinii β-1,3 glucan synthesis, termed Pcgsc-1, was isolated and shown by immunoprecipitation assays to possess β-1,3 glucan synthase activity, providing direct evidence of β-1,3 glucan machinery in the organism (20).
More recently, another glucan component of the fungal and yeast cell walls, termed β-1,6 glucan, has been shown both to possess relevant cell wall structural activity and to promote host inflammatory responses and organism virulence (11, 39). β-1,6 Glucan linkages are thought to be involved in anchoring glycophosphatidylinositol (GPI)-linked membrane-associated proteins to β-1,3 glucans (66, 67). Significantly, the P. carinii surface MSG-gpA mannoprotein complex has been shown to be such a GPI-anchored protein (68). In yeast, β-1,6 glucans can make up as much as 20% of the total β-glucan present in the cell wall (69). Although β-1,6 glucans make up a relatively small percentage of total β-glucan by mass, their immunological properties impacting host innate immune response are extremely pronounced (11). Indeed, when isolated yeast β-1,6 glucans stimulate human neutrophils, their effectiveness in eliciting free radicals, phagocytic ability, and transcription of heat shock protein mRNA are substantially greater than the responses resulting from β-1,3 glucans alone (11). Furthermore, when C. albicans β-1,6 glucans are presented to human neutrophils, these polysaccharides can induce neutrophil migration through nonreceptor sarcoma (Src) tyrosine kinase protein signal transduction pathways (70). Not only are β-1,6 glucans important in inflammatory signaling, they also have been shown to be important in animal models of virulence. For example, in C. neoformans, knockout of the β-1,6 glucan synthase genetic machinery renders these cryptococcal strains avirulent in mouse models of systemic infection (39).
With this background in mind, we sought to investigate the presence of β-1,6 glucans in P. carinii. Our findings are the first description of this carbohydrate component of the P. carinii cyst wall. Immune electron and fluorescence microscopy with a specific β-1,6 glucan-targeted antibody documented the presence of the carbohydrate predominantly in cystic forms of the fungus. However, unlike β-1,3 glucan, which is homogeneously distributed throughout the cyst wall, the β-1,6 glucan linkages appear to be localized in a more discrete, heterogeneous, punctate pattern on the organism's surface, with some cyst areas having heavier deposition than others. Immune electron microscopic analysis also revealed that the β-1,6 glucans are localized in discrete regions of the cyst wall. These observations are consistent with previous investigations of this polysaccharide in S. cerevisiae indicating that the β-1,6 glucan linkages represent a smaller percentage of total β-glucan in the fungal cell wall (69). We estimate that β-1,6 glucans represent approximately 10% of the total Pneumocystis cell wall carbohydrate in our preparations. At this time, we have not yet succeeded in isolating P. carinii β-1,3 glucans entirely free of β-1,6 glucans. Hence, we cannot determine their relative proinflammatory activity on an equal mass basis. However, studies with S. cerevisiae glucans indicate that β-1,6 glucans exhibit substantial proinflammatory activities compared to the β-1,3 glucan components of the organism's cell wall (65).
Molecular cloning of the Pckre6 gene predicted a mature protein with homology to the β-1,6 glucan synthase proteins of other fungi and possessing at least one transmembrane domain, as determined by Kyte-Doolittle plots, a widely applied analytical tool for determining the hydrophobic characteristics of a protein. Similarly to Pcgsc-1, Pckre6 gene expression seems to be predominantly restricted to the cyst form of the organism (20). Other fungi, including other members of the Ascomycota, utilize cell wall assembly processes continuously throughout the cell cycle (71–73). Northern blot analysis by our laboratory indicated that Pckre6 gene expression is highest in the P. carinii cyst life form. We also demonstrated that under conditions of normal lung physiological pH, high levels of expression of Pckre6 are noted. We have shown previously that other cell wall-regulating genes, namely, Pccbk1 and Pckre6, are also most highly expressed at lung physiological pH, further suggesting coordination of P. carinii cell wall-modifying genes in the host lung (33, 34).
We further demonstrated that P. carinii organisms are capable of generating β-1,6 glucans, as determined using a competitive enzyme immunoassay. Freshly isolated P. carinii cytoplasmic membrane preparations exhibited the ability to incorporate UDP-glucose into soluble β-1,6 carbohydrate, as measured by competitive immunoassay.
Of considerable importance is the concept that mammalian hosts lack a kre6 homolog equivalent and therefore do not generate β-1,6 glucans. Accordingly, pharmacological inhibition of β-1,6 glucan synthesis may represent a highly attractive target for the treatment of fungal infections. Kitamura and colleagues recently described a novel cell-based assay to screen large numbers of yeast cell wall component inhibitors, including those that suppress β-1,6 glucan synthesis (44). Several pyridobenzimidazole derivatives with activities against S. cerevisiae kre6 activity were uncovered (44, 45). These β-1,6 glucan inhibitors were further demonstrated to possess activity against Candida spp., C. neoformans, and Aspergillus fumigatus (43–45).
Of note, in the current investigation, we report that two such inhibitors also suppress the generation of β-1,6 carbohydrate by P. carinii cell wall preparations. Recent whole-genome-sequencing projects of both P. jirovecii (human pathogen) and Pneumocystis murina (mouse pathogen) have been published (74, 75). Using information derived from Pckre6 sequences, we searched the P. jirovecii and P. murina genomes and demonstrated the presence of a single conserved Pneumocystis kre6 gene in each Pneumocystis species. Therefore, this new class of cell wall inhibitors represents potential new antifungal medications, which may be used either alone or in conjunction with other antifungal agents. Indeed, against C. albicans, the β-1,6 glucan inhibitor D11-2040 has been shown to have synergistic activity when administered along with other antifungal agents (45).
Exposure of β-1,6 glucans on the surfaces of fungi and yeast also represents an important mechanism of host recognition and inflammatory response (11, 70). In whole-blood cultures, β-1,6 glucans were found to be strong inducers of the cytokines interleukin 1β (IL-1β), IL-6, IL-8, and TNF-α (26). Furthermore, when human monocytes were exposed to β-1,6 glucans from C. albicans, they differentiated into dendritic cells with altered biological activity, similar to monocytes exposed to whole, intact C. albicans organisms, further indicating that the polysaccharide can have a profound impact on host innate immune responses (76). Our studies with P. carinii-derived β-1,6 glucan preparations further support the proinflammatory activities of these cell wall components. Stimulation of cultured macrophages with P. carinii β-1,6 glucans resulted in a robust TNF-α response. In addition, the vigorous TNF-α response was dampened by inhibition of glycosphingolipid synthesis (PDMP), in a fashion similar to our observations using whole, unfractionated β-glucans, suggesting the importance of host cell membrane glycosphingolipids in mediating responses to the fungus (37). Vigorous host cell responses to β-1,6 glucans from rat-derived P. carinii by murine RAW macrophages also support cross-reactivity of the fungal surface carbohydrates across species. Whether the contents of β-1,6 glucans vary in Pneumocystis organisms derived from rats, mice, and humans remains unknown.
In summary, we observed the presence of β-1,6 glucans in P. carinii and the ability of P. carinii organisms to generate the polysaccharide in vitro. A newly discovered class of anti-β-1,6 glucan agents inhibited this synthetic ability. In addition, we performed an initial isolation and characterization of the PcKre6-like β-1,6 glucan synthase gene from P. carinii and showed that its transcription can be environmentally regulated, with the highest expression in P. carinii cyst forms. Finally, initial studies with purified P. carinii β-1,6 glucans support the idea that these components are highly stimulatory to the host. Due to the importance of β-1,6 glucans in cell wall integrity and the ability to target their generation with β-1,6 glucan inhibitors, PcKre6 deserves further attention as an attractive antifungal therapeutic target.
ACKNOWLEDGMENTS
We greatly appreciate the efforts of Jon Charlesworth in the Mayo Clinic electron microscopy core in assisting with the generation of the immune electron microscopy. We thank Akihiro Kitamura, Junichi Kuroyanagi, and colleagues at Daiichi Sankyo Company, Tokyo, Japan, for providing the test β-1,6 glucan-inhibitory compounds. Finally, we also appreciate the assistance of Seher Iqbal in the early development of this body of research.
None of us has any conflict of interest with the content of the manuscript.
These studies were funded by the Mayo Foundation, the Walter and Leonore Annenberg Foundation, and NIH grant R01-HL62150 to A.H.L.
REFERENCES
- 1.Thomas CF Jr, Limper AH. 2007. Current insights into the biology and pathogenesis of Pneumocystis pneumonia. Nat Rev Microbiol 5:298–308. doi: 10.1038/nrmicro1621. [DOI] [PubMed] [Google Scholar]
- 2.Stringer JR, Beard CB, Miller RF, Wakefield AE. 2002. A new name (Pneumocystis jiroveci) for Pneumocystis from humans. Emerg Infect Dis 8:891–896. doi: 10.3201/eid0809.020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang L, Cattamanchi A, Davis JL, den Boon S, Kovacs J, Meshnick S, Miller RF, Walzer PD, Worodria W, Masur H, International HIV-Associated Opportunistic Pneumonias (IHOP) Study, Lung HIV Study . 2011. HIV-associated Pneumocystis pneumonia. Proc Am Thorac Soc 8:294–300. doi: 10.1513/pats.201009-062WR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller RF, Huang L, Walzer PD. 2013. Pneumocystis pneumonia associated with human immunodeficiency virus. Clin Chest Med 34:229–241. doi: 10.1016/j.ccm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Huang L, Morris A, Limper AH, Beck JM, ATS Pneumocystis Workshop Participants . 2006. An official ATS workshop summary: recent advances and future directions in pneumocystis pneumonia (PCP). Proc Am Thorac Soc 3:655–664. doi: 10.1513/pats.200602-015MS. [DOI] [PubMed] [Google Scholar]
- 6.Bhagwat SP, Wright TW, Gigliotti F. 2010. Anti-CD3 antibody decreases inflammation and improves outcome in a murine model of Pneumocystis pneumonia. J Immunol 184:497–502. doi: 10.4049/jimmunol.0901864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Limper AH, Offord KP, Smith TF, Martin WJ II. 1989. Pneumocystis carinii pneumonia. Differences in lung parasite number and inflammation in patients with and without AIDS. Am Rev Respir Dis 140:1204–1209. [DOI] [PubMed] [Google Scholar]
- 8.Bang D, Emborg J, Elkjaer J, Lundgren JD, Benfield TL. 2001. Independent risk of mechanical ventilation for AIDS-related Pneumocystis carinii pneumonia associated with bronchoalveolar lavage neutrophilia. Respir Med 95:661–665. doi: 10.1053/rmed.2001.1119. [DOI] [PubMed] [Google Scholar]
- 9.Benfield TL, Vestbo J, Junge J, Nielsen TL, Jensen AB, Lundgren JD. 1995. Prognostic value of interleukin-8 in AIDS-associated Pneumocystis carinii pneumonia. Am J Respir Crit Care Med 151:1058–1062. [DOI] [PubMed] [Google Scholar]
- 10.Vassallo R, Standing JE, Limper AH. 2000. Isolated Pneumocystis carinii cell wall glucan provokes lower respiratory tract inflammatory responses. J Immunol 164:3755–3763. doi: 10.4049/jimmunol.164.7.3755. [DOI] [PubMed] [Google Scholar]
- 11.Rubin-Bejerano I, Abeijon C, Magnelli P, Grisafi P, Fink GR. 2007. Phagocytosis by human neutrophils is stimulated by a unique fungal cell wall component. Cell Host Microbe 2:55–67. doi: 10.1016/j.chom.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans SE, Hahn PY, McCann F, Kottom TJ, Pavlovic ZV, Limper AH. 2005. Pneumocystis cell wall beta-glucans stimulate alveolar epithelial cell chemokine generation through nuclear factor-kappaB-dependent mechanisms. Am J Respir Cell Mol Biol 32:490–497. doi: 10.1165/rcmb.2004-0300OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans SE, Kottom TJ, Pagano RE, Limper AH. 2012. Primary alveolar epithelial cell surface membrane microdomain function is required for Pneumocystis beta-glucan-induced inflammatory responses. Innate Immun 18:709–716. doi: 10.1177/1753425912436763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn PY, Evans SE, Kottom TJ, Standing JE, Pagano RE, Limper AH. 2003. Pneumocystis carinii cell wall beta-glucan induces release of macrophage inflammatory protein-2 from alveolar epithelial cells via a lactosylceramide-mediated mechanism. J Biol Chem 278:2043–2050. doi: 10.1074/jbc.M209715200. [DOI] [PubMed] [Google Scholar]
- 15.Limper AH, Lebron F, Evans SE, Hahn RY. 2003. Pneumocystis carinii: cell wall beta-glucan-mediated pulmonary inflammation. J Eukaryot Microbiol 50(Suppl):646. doi: 10.1111/j.1550-7408.2003.tb00667.x. [DOI] [PubMed] [Google Scholar]
- 16.McCann F, Carmona E, Puri V, Pagano RE, Limper AH. 2005. Macrophage internalization of fungal beta-glucans is not necessary for initiation of related inflammatory responses. Infect Immun 73:6340–6349. doi: 10.1128/IAI.73.10.6340-6349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wheeler RT, Kombe D, Agarwala SD, Fink GR. 2008. Dynamic, morphotype-specific Candida albicans beta-glucan exposure during infection and drug treatment. PLoS Pathog 4:e1000227. doi: 10.1371/journal.ppat.1000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krajicek BJ, Kottom TJ, Villegas L, Limper AH. 2010. Characterization of the PcCdc42 small G protein from Pneumocystis carinii, which interacts with the PcSte20 life cycle regulatory kinase. Am J Physiol Lung Cell Mol Physiol 298:L252–L260. doi: 10.1152/ajplung.00191.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kottom TJ, Thomas CF Jr, Mubarak KK, Leof EB, Limper AH. 2000. Pneumocystis carinii uses a functional cdc13 B-type cyclin complex during its life cycle. Am J Respir Cell Mol Biol 22:722–731. doi: 10.1165/ajrcmb.22.6.3838. [DOI] [PubMed] [Google Scholar]
- 20.Kottom TJ, Limper AH. 2000. Cell wall assembly by Pneumocystis carinii. Evidence for a unique gsc-1 subunit mediating beta -1,3-glucan deposition. J Biol Chem 275:40628–40634. doi: 10.1074/jbc.M002103200. [DOI] [PubMed] [Google Scholar]
- 21.Lindberg B, McPherson J. 1954. Studies on the chemistry of lichens. Acta Chem Scand 8:985–988. doi: 10.3891/acta.chem.scand.08-0985. [DOI] [Google Scholar]
- 22.Brown GD, Gordon S. 2001. Immune recognition. A new receptor for beta-glucans. Nature 413:36–37. doi: 10.1038/35092620. [DOI] [PubMed] [Google Scholar]
- 23.Luksa J, Podoliankaite M, Vepstaite I, Strazdaite-Zieliene Z, Urbonavicius J, Serviene E. 2015. Yeast beta-1,6-glucan is a primary target for the Saccharomyces cerevisiae K2 toxin. Eukaryot Cell 14:406–414. doi: 10.1128/EC.00287-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polonelli L, Beninati C, Teti G, Felici F, Ciociola T, Giovati L, Sperinde M, Lo Passo C, Pernice I, Domina M, Arigo M, Papasergi S, Mancuso G, Conti S, Magliani W. 2014. Yeast killer toxin-like candidacidal Ab6 antibodies elicited through the manipulation of the idiotypic cascade. PLoS One 9:e105727. doi: 10.1371/journal.pone.0105727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palma AS, Feizi T, Zhang Y, Stoll MS, Lawson AM, Diaz-Rodriguez E, Campanero-Rhodes MA, Costa J, Gordon S, Brown GD, Chai W. 2006. Ligands for the beta-glucan receptor, Dectin-1, assigned using “designer” microarrays of oligosaccharide probes (neoglycolipids) generated from glucan polysaccharides. J Biol Chem 281:5771–5779. doi: 10.1074/jbc.M511461200. [DOI] [PubMed] [Google Scholar]
- 26.Noss I, Doekes G, Thorne PS, Heederik DJ, Wouters IM. 2013. Comparison of the potency of a variety of beta-glucans to induce cytokine production in human whole blood. Innate Immun 19:10–19. doi: 10.1177/1753425912447129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maneesri J, Azuma M, Sakai Y, Igarashi K, Matsumoto T, Fukuda H, Kondo A, Ooshima H. 2005. Deletion of MCD 4 involved in glycosylphosphatidylinositol (GPI) anchor synthesis leads to an increase in beta-1,6-glucan level and a decrease in GPI-anchored protein and mannan levels in the cell wall of Saccharomyces cerevisiae. J Biosci Bioeng 99:354–360. doi: 10.1263/jbb.99.354. [DOI] [PubMed] [Google Scholar]
- 28.Roman J, LaChance RM, Broekelmann TJ, Kennedy CJ, Wayner EA, Carter WG, McDonald JA. 1989. The fibronectin receptor is organized by extracellular matrix fibronectin: implications for oncogenic transformation and for cell recognition of fibronectin matrices. J Cell Biol 108:2529–2543. doi: 10.1083/jcb.108.6.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Icenhour CR, Kottom TJ, Limper AH. 2006. Pneumocystis melanins confer enhanced organism viability. Eukaryot Cell 5:916–923. doi: 10.1128/EC.00176-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montijn RC, van Rinsum J, van Schagen FA, Klis FM. 1994. Glucomannoproteins in the cell wall of Saccharomyces cerevisiae contain a novel type of carbohydrate side chain. J Biol Chem 269:19338–19342. [PubMed] [Google Scholar]
- 31.Rappleye CA, Eissenberg LG, Goldman WE. 2007. Histoplasma capsulatum alpha-(1,3)-glucan blocks innate immune recognition by the beta-glucan receptor. Proc Natl Acad Sci U S A 104:1366–1370. doi: 10.1073/pnas.0609848104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Riordan DM, Standing JE, Kwon KY, Chang D, Crouch EC, Limper AH. 1995. Surfactant protein D interacts with Pneumocystis carinii and mediates organism adherence to alveolar macrophages. J Clin Invest 95:2699–2710. doi: 10.1172/JCI117972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kottom TJ, Limper AH. 2004. Pneumocystis carinii cell wall biosynthesis kinase gene CBK1 is an environmentally responsive gene that complements cell wall defects of cbk-deficient yeast. Infect Immun 72:4628–4636. doi: 10.1128/IAI.72.8.4628-4636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kottom TJ, Thomas CF Jr, Limper AH. 2001. Characterization of Pneumocystis carinii PHR1, a pH-regulated gene important for cell wall integrity. J Bacteriol 183:6740–6745. doi: 10.1128/JB.183.23.6740-6745.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roemer T, Bussey H. 1991. Yeast beta-glucan synthesis: KRE6 encodes a predicted type II membrane protein required for glucan synthesis in vivo and for glucan synthase activity in vitro. Proc Natl Acad Sci U S A 88:11295–11299. doi: 10.1073/pnas.88.24.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamata K, Kurita T, Bhuiyan MS, Sato K, Noda Y, Yoda K. 2007. KEG1/YFR042w encodes a novel Kre6-binding endoplasmic reticulum membrane protein responsible for beta-1,6-glucan synthesis in Saccharomyces cerevisiae. J Biol Chem 282:34315–34324. doi: 10.1074/jbc.M706486200. [DOI] [PubMed] [Google Scholar]
- 37.Carmona EM, Kottom TJ, Hebrink DM, Moua T, Singh RD, Pagano RE, Limper AH. 2012. Glycosphingolipids mediate pneumocystis cell wall beta-glucan activation of the IL-23/IL-17 axis in human dendritic cells. Am J Respir Cell Mol Biol 47:50–59. doi: 10.1165/rcmb.2011-0159OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aimanianda V, Clavaud C, Simenel C, Fontaine T, Delepierre M, Latge JP. 2009. Cell wall beta-(1,6)-glucan of Saccharomyces cerevisiae: structural characterization and in situ synthesis. J Biol Chem 284:13401–13412. doi: 10.1074/jbc.M807667200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilbert NM, Donlin MJ, Gerik KJ, Specht CA, Djordjevic JT, Wilson CF, Sorrell TC, Lodge JK. 2010. KRE genes are required for beta-1,6-glucan synthesis, maintenance of capsule architecture and cell wall protein anchoring in Cryptococcus neoformans. Mol Microbiol 76:517–534. doi: 10.1111/j.1365-2958.2010.07119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de la Cruz J, Pintor-Toro JA, Benitez T, Llobell A. 1995. Purification and characterization of an endo-beta-1,6-glucanase from Trichoderma harzianum that is related to its mycoparasitism. J Bacteriol 177:1864–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vink E, Rodriguez-Suarez RJ, Gerard-Vincent M, Ribas JC, de Nobel H, van den Ende H, Duran A, Klis FM, Bussey H. 2004. An in vitro assay for (1→6)-beta-d-glucan synthesis in Saccharomyces cerevisiae. Yeast 21:1121–1131. doi: 10.1002/yea.1156. [DOI] [PubMed] [Google Scholar]
- 42.Douwes J, Doekes G, Montijn R, Heederik D, Brunekreef B. 1996. Measurement of beta(1→3)-glucans in occupational and home environments with an inhibition enzyme immunoassay. Appl Environ Microbiol 62:3176–3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kitamura A, Higuchi S, Hata M, Kawakami K, Yoshida K, Namba K, Nakajima R. 2009. Effect of beta-1,6-glucan inhibitors on the invasion process of Candida albicans: potential mechanism of their in vivo efficacy. Antimicrob Agents Chemother 53:3963–3971. doi: 10.1128/AAC.00435-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kitamura A, Someya K, Hata M, Nakajima R, Takemura M. 2009. Discovery of a small-molecule inhibitor of {beta}-1,6-glucan synthesis. Antimicrob Agents Chemother 53:670–677. doi: 10.1128/AAC.00844-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kitamura A, Someya K, Okumura R, Hata M, Takeshita H, Nakajima R. 2010. In vitro antifungal activities of D11-2040, a beta-1,6-glucan inhibitor, with or without currently available antifungal drugs. Biol Pharm Bull 33:192–197. doi: 10.1248/bpb.33.192. [DOI] [PubMed] [Google Scholar]
- 46.Bruckner J. 1955. Estimation of monosaccharides by the orcinol-sulphuric acid reaction. Biochem J 60:200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iwamoto MA, Fairclough SR, Rudge SA, Engebrecht J. 2005. Saccharomyces cerevisiae Sps1p regulates trafficking of enzymes required for spore wall synthesis. Eukaryot Cell 4:536–544. doi: 10.1128/EC.4.3.536-544.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Humbel BM, Konomi M, Takagi T, Kamasawa N, Ishijima SA, Osumi M. 2001. In situ localization of beta-glucans in the cell wall of Schizosaccharomyces pombe. Yeast 18:433–444. doi: 10.1002/yea.694. [DOI] [PubMed] [Google Scholar]
- 49.Utsugi T, Minemura M, Hirata A, Abe M, Watanabe D, Ohya Y. 2002. Movement of yeast 1,3-beta-glucan synthase is essential for uniform cell wall synthesis. Genes Cells 7:1–9. doi: 10.1046/j.1356-9597.2001.00495.x. [DOI] [PubMed] [Google Scholar]
- 50.Kottom TJ, Limper AH. 2013. The Pneumocystis Ace2 transcription factor regulates cell wall-remodeling genes and organism virulence. J Biol Chem 288:23893–23902. doi: 10.1074/jbc.M113.471243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shahinian S, Bussey H. 2000. β-1,6-Glucan synthesis in Saccharomyces cerevisiae. Mol Microbiol 35:477–489. doi: 10.1046/j.1365-2958.2000.01713.x. [DOI] [PubMed] [Google Scholar]
- 52.Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng 10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 53.Kottom TJ, Han J, Zhang Z, Limper AH. 2011. Pneumocystis carinii expresses an active Rtt109 histone acetyltransferase. Am J Respir Cell Mol Biol 44:768–776. doi: 10.1165/rcmb.2009-0443OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buratti E, Baralle FE. 2004. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol Cell Biol 24:10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cockell MM, Lo Presti L, Cerutti L, Cano Del Rosario E, Hauser PM, Simanis V. 2009. Functional differentiation of tbf1 orthologues in fission and budding yeasts. Eukaryot Cell 8:207–216. doi: 10.1128/EC.00174-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hauser PM, Lo Presti L, Cockell M, Cerutti L, Simanis V. 2006. Analysis of Pneumocystis carinii gene function by complementation in yeast mutants. J Eukaryot Microbiol 53(Suppl 1):S149–S150. doi: 10.1111/j.1550-7408.2006.00210.x. [DOI] [PubMed] [Google Scholar]
- 57.Hoch HC, Galvani CD, Szarowski DH, Turner JN. 2005. Two new fluorescent dyes applicable for visualization of fungal cell walls. Mycologia 97:580–588. doi: 10.3852/mycologia.97.3.580. [DOI] [PubMed] [Google Scholar]
- 58.Parada D, Caleiras E, Farias RM, Escorihuela-Garcia S, Garcia Tamayo J. 1998. An ultrastructural evaluation of Pneumocystis carinii. Invest Clin 39:293–306. [PubMed] [Google Scholar]
- 59.Cushion MT, Linke MJ, Ashbaugh A, Sesterhenn T, Collins MS, Lynch K, Brubaker R, Walzer PD. 2010. Echinocandin treatment of pneumocystis pneumonia in rodent models depletes cysts leaving trophic burdens that cannot transmit the infection. PLoS One 5:e8524. doi: 10.1371/journal.pone.0008524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haidaris CG, Wright TW, Gigliotti F, Haidaris PJ. 1991. Molecular cloning and characterization of ferret Pneumocystis carinii gp120. J Protozool 38:5S–6S. [PubMed] [Google Scholar]
- 61.Haidaris PJ, Wright TW, Gigliotti F, Haidaris CG. 1992. Expression and characterization of a cDNA clone encoding an immunodominant surface glycoprotein of Pneumocystis carinii. J Infect Dis 166:1113–1123. doi: 10.1093/infdis/166.5.1113. [DOI] [PubMed] [Google Scholar]
- 62.Icenhour CR, Kottom TJ, Limper AH. 2003. Evidence for a melanin cell wall component in Pneumocystis carinii. Infect Immun 71:5360–5363. doi: 10.1128/IAI.71.9.5360-5363.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walker AN, Garner RE, Horst MN. 1990. Immunocytochemical detection of chitin in Pneumocystis carinii. Infect Immun 58:412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Villegas LR, Kottom TJ, Limper AH. 2012. Chitinases in Pneumocystis carinii pneumonia. Med Microbiol Immunol 201:337–348. doi: 10.1007/s00430-012-0239-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carmona EM, Vassallo R, Vuk-Pavlovic Z, Standing JE, Kottom TJ, Limper AH. 2006. Pneumocystis cell wall beta-glucans induce dendritic cell costimulatory molecule expression and inflammatory activation through a Fas-Fas ligand mechanism. J Immunol 177:459–467. doi: 10.4049/jimmunol.177.1.459. [DOI] [PubMed] [Google Scholar]
- 66.Klis FM, de Groot P, Hellingwerf K. 2001. Molecular organization of the cell wall of Candida albicans. Med Mycol 39(Suppl 1):S1–S8. [PubMed] [Google Scholar]
- 67.Klis FM, Mol P, Hellingwerf K, Brul S. 2002. Dynamics of cell wall structure in Saccharomyces cerevisiae. FEMS Microbiol Rev 26:239–256. doi: 10.1111/j.1574-6976.2002.tb00613.x. [DOI] [PubMed] [Google Scholar]
- 68.Guadiz G, Haidaris CG, Maine GN, Simpson-Haidaris PJ. 1998. The carboxyl terminus of Pneumocystis carinii glycoprotein A encodes a functional glycosylphosphatidylinositol signal sequence. J Biol Chem 273:26202–26209. doi: 10.1074/jbc.273.40.26202. [DOI] [PubMed] [Google Scholar]
- 69.Magnelli P, Cipollo JF, Abeijon C. 2002. A refined method for the determination of Saccharomyces cerevisiae cell wall composition and beta-1,6-glucan fine structure. Anal Biochem 301:136–150. doi: 10.1006/abio.2001.5473. [DOI] [PubMed] [Google Scholar]
- 70.Sato T, Iwabuchi K, Nagaoka I, Adachi Y, Ohno N, Tamura H, Seyama K, Fukuchi Y, Nakayama H, Yoshizaki F, Takamori K, Ogawa H. 2006. Induction of human neutrophil chemotaxis by Candida albicans-derived beta-1,6-long glycoside side-chain-branched beta-glucan. J Leukoc Biol 80:204–211. doi: 10.1189/jlb.0106069. [DOI] [PubMed] [Google Scholar]
- 71.Castro C, Ribas JC, Valdivieso MH, Varona R, del Rey F, Duran A. 1995. Papulacandin B resistance in budding and fission yeasts: isolation and characterization of a gene involved in (1,3)beta-d-glucan synthesis in Saccharomyces cerevisiae. J Bacteriol 177:5732–5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Douglas CM, Foor F, Marrinan JA, Morin N, Nielsen JB, Dahl AM, Mazur P, Baginsky W, Li W, el-Sherbeini M. 1994. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-beta-d-glucan synthase. Proc Natl Acad Sci U S A 91:12907–12911. doi: 10.1073/pnas.91.26.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kelly R, Register E, Hsu MJ, Kurtz M, Nielsen J. 1996. Isolation of a gene involved in 1,3-beta-glucan synthesis in Aspergillus nidulans and purification of the corresponding protein. J Bacteriol 178:4381–4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cisse OH, Pagni M, Hauser PM. 2012. De novo assembly of the Pneumocystis jirovecii genome from a single bronchoalveolar lavage fluid specimen from a patient. mBio 4:e00428–00412. doi: 10.1128/mBio.00428-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ma L, Huang DW, Cuomo CA, Sykes S, Fantoni G, Das B, Sherman BT, Yang J, Huber C, Xia Y, Davey E, Kutty G, Bishop L, Sassi M, Lempicki RA, Kovacs JA. 2013. Sequencing and characterization of the complete mitochondrial genomes of three Pneumocystis species provide new insights into divergence between human and rodent Pneumocystis. FASEB J 27:1962–1972. doi: 10.1096/fj.12-224444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nisini R, Torosantucci A, Romagnoli G, Chiani P, Donati S, Gagliardi MC, Teloni R, Sargentini V, Mariotti S, Iorio E, Cassone A. 2007. β-Glucan of Candida albicans cell wall causes the subversion of human monocyte differentiation into dendritic cells. J Leukoc Biol 82:1136–1142. doi: 10.1189/jlb.0307160. [DOI] [PubMed] [Google Scholar]