Abstract
Pathogenically diverse Chlamydia spp. can have surprisingly similar genomes. Chlamydia trachomatis isolates that cause trachoma, sexually transmitted genital tract infections (chlamydia), and invasive lymphogranuloma venereum (LGV) and the murine strain Chlamydia muridarum share 99% of their gene content. A region of high genomic diversity between Chlamydia spp. termed the plasticity zone (PZ) may encode niche-specific virulence determinants that dictate pathogenic diversity. We hypothesized that PZ genes might mediate the greater virulence and gamma interferon (IFN-γ) resistance of C. muridarum compared to C. trachomatis in the murine genital tract. To test this hypothesis, we isolated and characterized a series of C. muridarum PZ nonsense mutants. Strains with nonsense mutations in chlamydial cytotoxins, guaBA-add, and a phospholipase D homolog developed normally in cell culture. Two of the cytotoxin mutants were less cytotoxic than the wild type, suggesting that the cytotoxins may be functional. However, none of the PZ nonsense mutants exhibited increased IFN-γ sensitivity in cell culture or were profoundly attenuated in a murine genital tract infection model. Our results suggest that C. muridarum PZ genes are transcribed—and some may produce functional proteins—but are dispensable for infection of the murine genital tract.
INTRODUCTION
Acquisition of genetic islands encoding the machinery necessary to invade eukaryotic cells was a key event in the evolution of many bacterial pathogens from free-living ancestors. For those pathogens that subsequently lost the ability to replicate outside host cells, a dearth of foreign microbial genetic material and Muller's ratchet may have tilted the balance of niche adaptation toward genomic decay. Reductive evolution appears to have played an especially pivotal role in niche adaptation of the obligate intracellular bacterial pathogens in the family Chlamydiaceae (1). Understanding why extant Chlamydia spp. retained different combinations of the metabolic pathways encoded by their common ancestors could reveal insights into chlamydial pathogenesis and cognate immune defenses against these intracellular pathogens.
Whole-genome comparisons provided early clues concerning the importance of genomic streamlining in chlamydial evolution but less insight into how loss of specific genes and metabolic pathways is adaptive (2–5). What is clear is that the genomes of pathogenically diverse Chlamydia spp. are small, have limited biosynthetic capacity, and are remarkably similar. For example, the murine pathogen Chlamydia muridarum and pathogenically diverse Chlamydia trachomatis isolates share 99% of their gene content (4). Various hypotheses have been put forth to explain this paradox, which was recently described as “divergence without difference” (6). One idea is that polymorphisms in conserved genes are key chlamydial virulence determinants. Some C. trachomatis trachoma strains have no obvious novel genes that differentiate them from genital strains (7), and some trachoma strains that vary in sensitivity to gamma interferon (IFN-γ) differ by only a few polymorphisms in conserved “core” genes (8). Alternatively, but not exclusively, open reading frames (ORFs) that have accumulated in a variable genomic region termed the plasticity zone (PZ) may be determinants of chlamydial pathogenic diversity (4). Comparative genomic and biochemical studies have implied that a partial tryptophan operon in the PZs of some C. trachomatis strains could reflect niche adaptation to immune-regulated tryptophan availability (9–12). Functionality of chlamydial tryptophan synthase, the product of this operon, in cell culture was recently confirmed using a reverse genetic strategy (13). Whether other PZ ORFs similarly mediate chlamydial niche adaptation or the PZ primarily houses inactive and decaying genes remains unclear.
The PZ is consistently located near the region of replication termination in Chlamydia spp. but varies considerably in size and gene content (4). Some PZ ORFs have no homologs outside Chlamydia, whereas others have been grouped into families based upon the homology of their predicted protein products to characterized bacterial and mammalian proteins. Some Chlamydia spp. encode a protein that resembles eukaryotic membrane attack complex (MAC) proteins and perforin (MACPF) in the PZ (4, 14). Various functions have been ascribed to chlamydial MACPFs, including immune evasion by molecular mimicry, protein secretion, and bacterial entry/exit (14, 15). Chlamydial MACPF is also purported to assist in lipid modification because of its proximity to a cluster of phospholipase D (PLD)-like genes (14, 16). Indirect evidence also suggests that PZ PLDs, like their mammalian homologs, are sensitive to primary alcohols and play roles in lipid acquisition or transfer (17, 18). The C. muridarum PZ contains three putative cytotoxins that have similarity to the large clostridial toxins (LCTs) and yersinial YopT (4). The cytotoxin ORFs of C. trachomatis serovars are truncated, disrupted, or absent altogether (2, 19, 20). A guaBA-add operon found in C. muridarum, but not C. trachomatis, could render the mouse pathogen less dependent on host nucleotides (4). These purine interconversion genes are replaced by trpRBA in C. trachomatis (2, 4). Limitations of available animal models and a historic lack of genetic tools have prevented the testing of hypotheses concerning the functions of PZ ORFs in chlamydial virulence and tropism. However, recent breakthroughs in genetic manipulation of Chlamydia now make it possible to address some of these questions directly (13, 21, 22).
Determining the functions of PZ ORFs could provide insight into the molecular basis of chlamydial host and tissue tropism and direct the design of improved animal models of human chlamydial disease. C. muridarum infection of the murine genital tract (GT) is a leading model for the study of adaptive immunity to genital infection because innate immunity is sufficient to clear murine genital infection with human strains of C. trachomatis (23). We and others have speculated that C. muridarum PZ genes play roles analogous to that of C. trachomatis tryptophan synthase in the circumvention of niche-specific innate immune responses (4, 24). If correct, this also implies that specific effectors were retained by C. muridarum because the cognate targets are relevant barriers during the natural course of infection. Murine modeling of C. trachomatis human urogenital tract infection might be improved by use of C. muridarum PZ-null mutants and mice lacking the corresponding targets of these effectors (25). Alternatively, if PZ ORFs are dispensable for GT infection, it would suggest either that these ORFs do not encode functional proteins or that the appropriate targets of the proteins are not relevant during experimental GT infection.
In this study, we used a combination of transcriptional profiling, reverse genetic analysis, and mouse modeling to assess if C. muridarum PZ ORFs are expressed and necessary for murine GT infection. Our results suggest that although PZ ORFs are transcribed, they are dispensable for infection of the murine GT.
MATERIALS AND METHODS
Cell lines, chlamydial culture, and infection.
C. muridarum and C. trachomatis serovar L2 strain 434/Bu (C. trachomatis) were kind gifts from Harlan Caldwell (Rocky Mountain Laboratories, NIAID, Hamilton, MT). Chlamydia organisms were routinely propagated in McCoy mouse fibroblast cells (American Type Culture Collection CRL-1696), and infectious elementary bodies (EB) were purified by density gradient centrifugation (26). McCoy cells and HeLa human cervical epithelial cells (American Type Culture Collection CCL-2) were maintained in Dulbecco's modified Eagle's medium (DMEM) containing high-glucose HEPES, glutamine, and sodium pyruvate (HyClone) and supplemented with 10% fetal bovine serum albumin (Atlanta Biologicals) and 100 μM nonessential amino acids (Gibco). Infections were performed in tissue culture flasks or plates with appropriate dilutions of chlamydial stock in SPG buffer (0.25 M sucrose, 10 mmol/liter phosphate, 5 mmol/liter l-glutamic acid, pH 7.2). The infected flasks were rocked for 2 h at 37°C, whereas infected plates were centrifuged at room temperature for 1 h, followed by rocking at 37°C for 30 min. During large-scale propagation experiments, SPG was replaced with fresh DMEM containing 0.5 μg/ml cycloheximide. In some experiments, DMEM containing 20 U/ml murine IFN-γ (R&D Systems) was added to McCoy cells 24 h prior to infection and postinfection. Cycloheximide was excluded from the cell culture media in experiments where IFN-γ was added.
Recoverable IFU (rIFU) assays.
McCoy cells in 24-well plates were infected with C. muridarum or PZ mutants at a multiplicity of infection (MOI) of 0.25. The infected cell monolayers were frozen in 500 μl SPG at various intervals postinfection (p.i.). Upon thawing, cells were scraped using a pipette tip and EB were harvested by bead beating the cell suspension in microcentrifuge tubes. Inclusion-forming unit (IFU) assays were performed using serial dilutions of the harvests to infect confluent McCoy cell monolayers in 96-well plates. Cells were fixed with methanol and stained with anti-chlamydial lipopolysaccharide (LPS) monoclonal antibody (MAb) (EVIH1), followed by a secondary Alexa Fluor 488-conjugated anti-mouse IgG MAb (Life Technologies) at 24 h p.i. Chlamydial inclusions were imaged and enumerated using an EVOS FL Auto cell-imaging system (Life Technologies).
RT-PCR and qRT-PCR.
McCoy cells in six-well plates were infected with C. muridarum at an MOI of 1. Total RNA was extracted at 0, 6, 12, 18, 24, and 30 h p.i. using TriSure reagent (Bioline), treated with DNase I (Qiagen) for 15 min, and then purified on GeneJet RNA purification columns (Thermo Scientific). Reverse transcriptase (RT) PCRs were performed using 100 ng of total RNA and gene-specific primers (see Table S1 in the supplemental material) or primer pairs spanning intergenic regions (see Table S3 in the supplemental material) using the MyTaq one-step RT-PCR kit (Bioline). For quantitative RT-PCR (qRT-PCR) experiments, total RNA was converted to cDNA using the Maxima H Minus first-strand cDNA synthesis kit (Thermo Scientific). The qRT-PCRs were performed in an Eppendorf Realplex4 thermocycler using SensiFast SYBR No-Rox master mix (Bioline) with gene-specific primers (see Table S2 in the supplemental material). The amplification cycle included a 2-min step at 95°C, followed by 40 cycles of 95°C for 5 s, 60°C for 10 s, and 72°C for 20 s. Absolute transcript copy numbers were calculated by normalizing to standard curves of C. muridarum genomic DNA amplified in parallel with experimental samples. Control reaction mixtures lacking RT were run in parallel with all RT-PCR and qRT-PCR experiments.
PZ cloning and β-galactosidase assays.
Overlapping 500- to 1,000-bp regions of the C. muridarum PZ were PCR amplified from C. muridarum genomic DNA and were cloned upstream of the lacZ gene in pAC-lacZ between the NruI and EagI sites of the vector (27). The sequences of the PCR primers and the genomic coordinates of the cloned PZ regions are listed in Table S4 in the supplemental material. The resulting plasmids were transformed into Escherichia coli DH5α, and the inserts and cloning junctions were confirmed by DNA sequencing. For β-galactosidase activity assays, triplicate samples from overnight cultures of E. coli strains carrying various PZ and control plasmids were grown in Luria-Bertani broth containing 50 μg/ml chloramphenicol and then diluted 1:100 in fresh broth and grown at 37°C until they reached mid-log phase (optical density at 600 nm [OD600] ≈ 0.6). β-Galactosidase expression analysis was performed as described previously (28, 29) with the following modifications. Cells were diluted in a total volume of 500 μl Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4 · H2O, 10 mM KCl, 1 mM MgSO4 · 7H2O) followed by the addition of 50 μl of chloroform and 50 μl of 0.1% sodium dodecyl sulfate. The solution was vortexed for 5 s, and the reactions were equilibrated by incubation at room temperature for 5 min prior to the addition of 100 μl of ortho-nitrophenyl-β-galactoside (4 mg/ml). The reactions were stopped by adding 250 μl of 1 M Na2CO3 and then centrifuged at 14,000 rpm for 5 min in an Eppendorf microcentrifuge to separate the cell pellet and the supernatant. The initial OD600 of the cells and the OD420 of the supernatant were measured and used to calculate β-galactosidase activity.
Preparation of CEL1 endonuclease extract.
CEL1 extract was prepared from celery as described previously (30). Fresh celery stalks (25 kg) were washed, chopped to remove the leaves, and juiced to yield approximately 10 liters of celery juice. Subsequent steps were carried out at 4°C. Five hundred milliliters of buffer A containing 2 M Tris-HCl (pH 7.7) and 0.1 M phenylmethanesulfonyl fluoride (PMSF) was added to the celery juice, and the solution was centrifuged to remove plant debris. Protein was precipitated from the supernatant by ammonium sulfate fractionation in the 25 to 80% saturation range. The protein pellet was resuspended in 100 ml buffer B (0.1 M Tris-HCl, pH 7.7, 0.5 M KCl, 100 μM PMSF, 0.01% Triton X-100) and dialyzed overnight against a large volume of buffer B. The final CEL1 extract was stored in small aliquots at −80°C.
EMS mutagenesis and library construction.
Confluent McCoy cells in a T-75 flask were infected with C. muridarum at an MOI of 0.5. At 16 h p.i., the culture medium was replaced with 5 ml of DMEM containing 8 μg/ml ethyl methanesulfonate (EMS) (Sigma). The cells were exposed to EMS for 1 h and then washed several times with 1× phosphate-buffered saline (PBS), after which fresh DMEM without mutagen was added. EB were bead harvested from the cells at 28 h p.i. and resuspended in 2 ml of SPG. An IFU assay estimated chlamydial survival after EMS treatment at less than 0.1% that of the untreated control. Half of the harvest was stored at −80°C, and the remainder was used to infect a fresh T-75 flask of cells. Four rounds of EMS mutagenesis were performed in total, and the final harvest yielded 9 × 105 EB. These EB were divided into small aliquots and stored at −80°C. To construct mutant libraries, 96-well plates of McCoy cells were infected with 10 EB per well of the mutagenized stock. Mutant chlamydiae were scaled up by four rounds of passaging in 96-well plates. Plates from the third passage were stored at −80°C to serve as the library. Genomic DNA was extracted from the final plates by adding 75 μl of alkaline lysis buffer to each well (0.2 mM EDTA, 25 mM NaOH, pH 12.0) and incubating at 95°C for 10 min, after which the lysis buffer was neutralized by adding an equal volume of 40 mM Tris-HCl (pH 5.0). The genomic DNA plates were stored at 4°C.
TILLING.
Targeting induced local lesions in genomes (TILLING) was used to identify specific mutants in the library and was performed as described previously (13). Specific primers (see Table S6 in the supplemental material) were used to amplify targeted PZ genes from template genomic DNA plates, which were extracted from the mutant libraries. These PCR products were annealed slowly (80°C; 7 s; 60 cycles, decreasing by 0.3°C every cycle) and then were digested with CEL1 endonuclease. Digestions were performed for 10 min at 45°C using an empirically determined volume of CEL1 extract (range, 0.1 to 1 μl) diluted in 1× CEL1 buffer (0.01 M MgSO4, 0.01 M HEPES, pH 7.5, 0.02 M KCl, 0.005% Triton X-100, 2 μg/ml bovine serum albumin). The digestions were stopped by adding 10 μl EDTA (0.15 M, pH 8.0). The digested products were separated on 2% agarose Tris-borate-EDTA (TBE) gels, stained with Midori Green (Bulldog Bio), and visualized using a ChemiDoc XRS+ device (Bio-Rad Laboratories, Inc., Hercules, CA).
Cytotoxicity assays.
McCoy cells seeded on coverslips in 24-well plates were infected with C. trachomatis, C. muridarum, or cytotoxin mutants at an MOI of 10 or 250 in the presence of 0.1 μg/ml rifampin. The cells were fixed in 3.7% formaldehyde at 3 h p.i. and then permeabilized with 1% Triton X-100. Following staining with Alexa Fluor 488-phalloidin for visualization of actin filaments, the coverslips were mounted on slides using ProLong Gold antifade reagent with DAPI (4′,6-diamidino-2-phenylindole) (Life Technologies). The cells were imaged using a Nikon Eclipse Ni-E fluorescence microscope (Nikon Instruments, Melville, NY).
Lactate dehydrogenase (LDH) assays were performed according to the manufacturer's instructions (OPS Diagnostics). McCoy cells were infected with various strains at a range of MOI (10, 25, 100, and 500) in the presence of rifampin. To determine maximal LDH release, cells in control wells were lysed with 10% Triton X-100 10 min prior to supernatant collection. The supernatants were removed from the wells 3 h p.i. after brief centrifugation of the plates to remove debris. Twenty-five microliters of the supernatants was mixed with 75 μl of the dye-buffer solution. The A490 was measured after 15 min incubation at 37°C.
Animal experiments.
C57BL/6J mice were obtained from Jackson Laboratories (Bar Harbor, ME). All animal experiments were approved by the Institutional Animal Care and Use Committees of the University of Arkansas for Medical Sciences and Indiana University. Female mice between 6 and 10 weeks old were treated with 2.5 mg medroxyprogesterone (Depo-Provera) diluted in 100 μl PBS 10 and 3 days prior to infection. Prior to infectious challenge, the vaginal vault was swabbed to remove excess mucus. Mice were inoculated vaginally with C. muridarum or PZ mutants at a concentration of 5 × 104 IFU in 5 μl SPG. The infections were monitored by enumeration of IFU from cervical swabs collected at various time points. At 73 days postinfection, the mice were euthanized and examined visually for hydrosalpinx.
Sequencing library construction and whole-genome sequencing.
Gradient-purified EB were treated with DNase I (NEB) at 37°C for 1 h, and then 5 mM EDTA was added and the mixtures were incubated at 75°C for 10 min to halt digestion. Total DNA was extracted using the DNeasy blood and tissue kit (Qiagen), and DNA concentrations were determined using the Quant-It high-sensitivity double-stranded DNA (dsDNA) assay (Life Technologies Corp.) The dsDNA fragments were generated by treating 1 μg of genomic DNA with NEBNext dsDNA Fragmentase (NEB). DNA sequencing libraries were prepared using the TruSeq Nano DNA sample preparation kit (Illumina Inc.) according to the manufacturer's protocols. Samples were multiplexed using TruSeq Single Index sequencing primers. Equimolar concentrations of the indexed libraries were combined into a single pool and submitted for Illumina HiSeq sequencing at the Tufts University Genomics Core Facility. Paired-end 100-bp sequencing was performed on the Illumina HiSeq 2500 platform.
Genome assembly and sequence analysis.
For single nucleotide polymorphisms (SNP), nucleotide insertion and deletion (indel) analysis, raw sequence data were mapped to the corresponding reference genome (GenBank accession number AE002160.2) using Bowtie2 (56). The resulting SAM files were converted to a BAM file format and sorted. SNPs/indels were called using a Samtools mpileup function. The remaining ambiguous sequences from the data set and mutation calls with low-quality scores were resolved by PCR and Sanger sequencing.
Statistics.
Data were analyzed using Prism 6.0 software (GraphPad). For comparisons of multiple groups with more than one variable, two-way analysis of variance (ANOVA) with Bonferroni posttest was used. Two-way ANOVA with Bonferroni posttest was also used to analyze the differences in chlamydial burdens between groups of infected mice. Multiple comparisons for data with a single variable were analyzed by one-way ANOVA with Dunnett's posttest. Differences in the development of hydrosalpinx between C. muridarum-infected mice and mutant-infected mice were analyzed by Fisher's exact test. Differences were considered statistically significant when the P value was <0.05.
RESULTS
The C. muridarum plasticity zone is transcriptionally active.
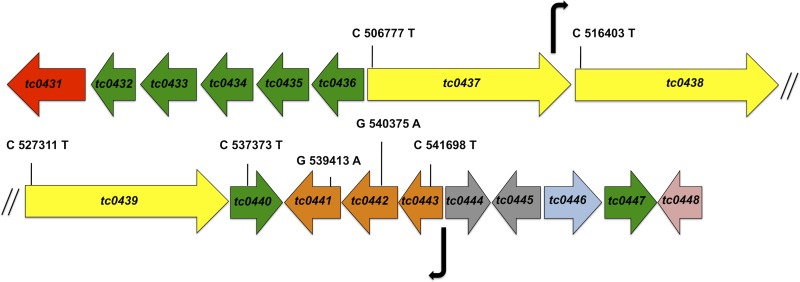
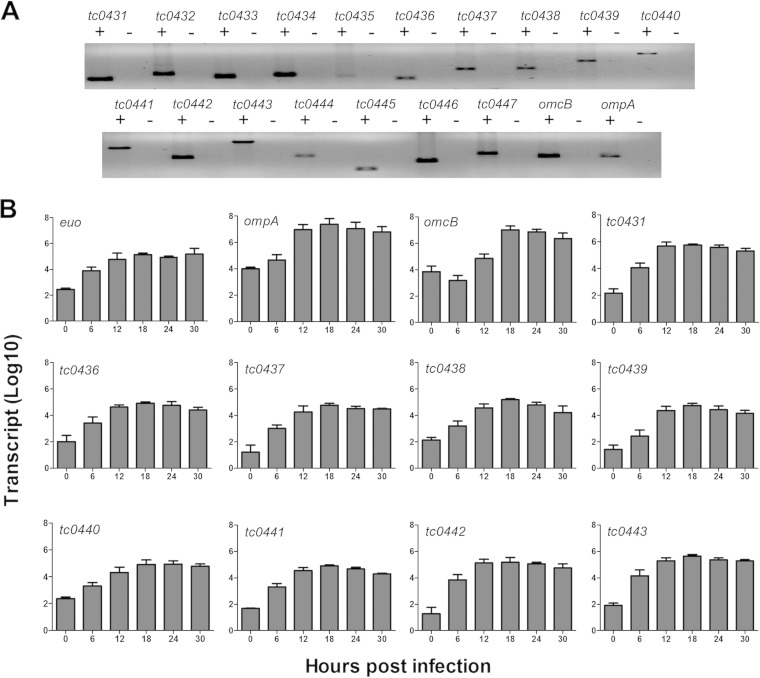
The C. muridarum PZ is defined as the 50-kb locus between tc0431 (MACPF) and tc0448 (dsbB) (Fig. 1). The C. muridarum PZ is poorly characterized, and the only evidence for gene activity in the region comes from a prior transcriptional study of the cytotoxin ORFs (tc0437 to tc0439) (19). To determine if and when other PZ ORFs were expressed, we examined the entire locus for transcriptional activity. RT-PCR analysis of RNA isolated from C. muridarum-infected McCoy cells at 24 h p.i. indicated that all the PZ genes were transcribed (Fig. 2A). Next, we determined the transcriptional and temporal expression profiles of a subset of PZ ORFs (tc0431 and tc0436 to tc0443) using qRT-PCR analysis at various time points p.i. The developmentally regulated genes euo, ompA, and omcB were analyzed as representative chlamydial early, mid-late, and late genes, respectively (19). Increased expression of euo was detected by 6 h p.i., whereas ompA transcripts first increased between 6 and 12 h p.i. and de novo omcB expression was detected only at 12 h p.i. and did not peak until 18 h p.i. (Fig. 2B). The expression trend for all of the PZ genes most closely resembled that of euo, and the absolute peak numbers of PZ ORF transcripts were 1 to 3 log units lower than those of ompA and omcB. Taken together, these results showed that expression of most PZ ORFs initiates in the early to middle developmental cycle and that they are expressed at low levels relative to known highly expressed chlamydial genes.
FIG 1.
Map of the C. muridarum PZ and diagram of PZ ORFs. The ORFs are color coded according to function: red, MACPF; green, PLD; yellow, cytotoxin; orange, purine nucleotide synthesis; gray, conserved hypothetical; blue, peptide ABC transporter; pink, dsbB. The locations of promoters identified by β-galactosidase assays in this study are shown by the bent arrows. The genomic locations and nucleotide changes of the nonsense mutations discussed in this study are indicated in the corresponding ORFs. The diagram is not drawn to scale.
FIG 2.
Transcription of C. muridarum PZ ORFs initiates in the early to middle phase of the developmental cycle. Total RNA was isolated from C. muridarum-infected McCoy cells at 0, 6, 12, 18, 24, and 30 h p.i. (A) RT-PCR at 24 h p.i. indicated that all genes in the PZ are transcribed. Whether amplification was performed with (+) or without (−) reverse transcriptase is indicated. (B) The kinetics of transcription of select PZ ORFs was characterized by qRT-PCR. Transcript levels were normalized to standard curves of dilutions of C. muridarum chromosomes. The data represent the mean transcript levels with standard deviations (SD) from three independent experiments. euo, ompA, and omcB represent early, mid-late, and late genes, respectively.
We next sought to identify the promoters of PZ genes. The abilities of a series of overlapping cloned fragments of the C. muridarum PZ to drive expression of a promoterless lacZ gene were assayed in E. coli to identify promoters. Only two of the constructs exhibited strong promoter activity. One promoter locus (nucleotide positions 542868 to 542368 in GenBank AE002160.2) was located immediately upstream of guaB (see Table S5 in the supplemental material) and was in the correct position and orientation to drive expression of the putative operon guaBA-add (4). RT-PCR analysis of guaB, guaA, and add using primer pairs that amplified across intergenic regions of adjacent genes confirmed that these genes were cotranscribed (see Fig. S1 in the supplemental material). A specific primer pair spanning add and tc0440 also gave rise to an overlapping amplification product from extracted RNA. However, since these two genes are predicted to be located on different strands, the RT-PCR product is likely a result of enzyme run-through. Two putative cytotoxin genes (tc0437 and tc0438) were found to be in a single transcript (see Fig. S1 in the supplemental material), but the presence of an active internal promoter at the 3′ end of tc0437 (nucleotide positions 515330 to 515896) suggested that tc0438 can also be expressed as a separate transcript (see Table S5 in the supplemental material). No promoter likely to drive expression of the operons tc0436-tc0433 and tc0432-tc0431 or the cytotoxin ORFs tc0437 and tc0439 was identified (see Fig. S1 and Table S5 in the supplemental material). Failure to identify other promoters for PZ genes whose expression was confirmed by RT-PCR could indicate that some PZ promoters were not functional in E. coli or were located outside the regions interrogated in the assay.
Members of three PZ gene families are dispensable for C. muridarum survival and proliferation in vitro.
Since little is known about the potential functions of most PZ ORFs, we next tested if they were dispensable using a reverse genetic approach. An EMS-mutagenized C. muridarum library was screened by TILLING for isolates that had nonsense mutations in any of eight C. muridarum-specific PZ ORFs: tc0437, tc0438, tc0439, tc0440, guaA (tc0441), guaB (tc0442), add (tc0443), and MACPF (tc0431) (13). The TILLING library was constructed with a high mutation load to facilitate faster screening. To estimate the number of EMS-induced point mutations in a given gene that would have to be isolated in order to recover at least one nonsense mutation, we assumed a Poisson distribution described as follows: P(x ≥ 1) = 1 − P(x = 0) = 1 − e−a, where a = N × p (N is the number of mutations in gene X that need to be isolated, and p is the probability of obtaining a nonsense mutation in gene X). The majority of mutations induced by EMS are GC→AT transitions. A total of 5 GC transitions in 4 amino acid codons (CAA, CGA, CAG, and TGG) can yield a stop codon. We use tc0442 as an example to illustrate how we arrived at an approximation of screen size (N). The probability p can be calculated as the number of possible nonsense GC transitions in the analyzed region of tc0442 divided by the number of GC base pairs in the same region of tc0442 (p = 19/585 = 0.032). For 95% screen saturation, screen size N was determined as follows: P(x ≥ 1) = 0.95 = 1 − e−0.032N, or n = 94. In other words, if tc0442 is not essential, there is a 95% probability that at least one of 94 isolated mutants is a nonsense mutant. The screen sizes for other PZ genes were calculated by the same method (see Table S7 in the supplemental material).
One or more nonsense mutants in guaBA-add, the C. muridarum-specific PZ PLD tc0440, and the cytotoxins (tc0437 to tc0439) were isolated at the anticipated frequencies, indicating that these ORFs are not essential for chlamydial survival in vitro. Despite attaining 95% saturation in our screen for nonsense mutants of tc0431 (MACPF), all of the mutants that we isolated contained either missense or silent mutations (see Table S7 in the supplemental material). While it is possible that a larger screen of C. muridarum EMS mutants could have revealed a nonsense mutant, the relatively high frequency at which other PZ nonsense mutants were obtained argues against this interpretation. Chlamydia abortus does not encode MACPF (31, 32), and the Chlamydia caviae and human Chlamydia pneumoniae MACPF genes contain frameshifts (5, 32, 33), so our result could indicate that MACPF plays an essential, species-specific role in C. muridarum.
PZ mutants display mild in vitro growth defects.
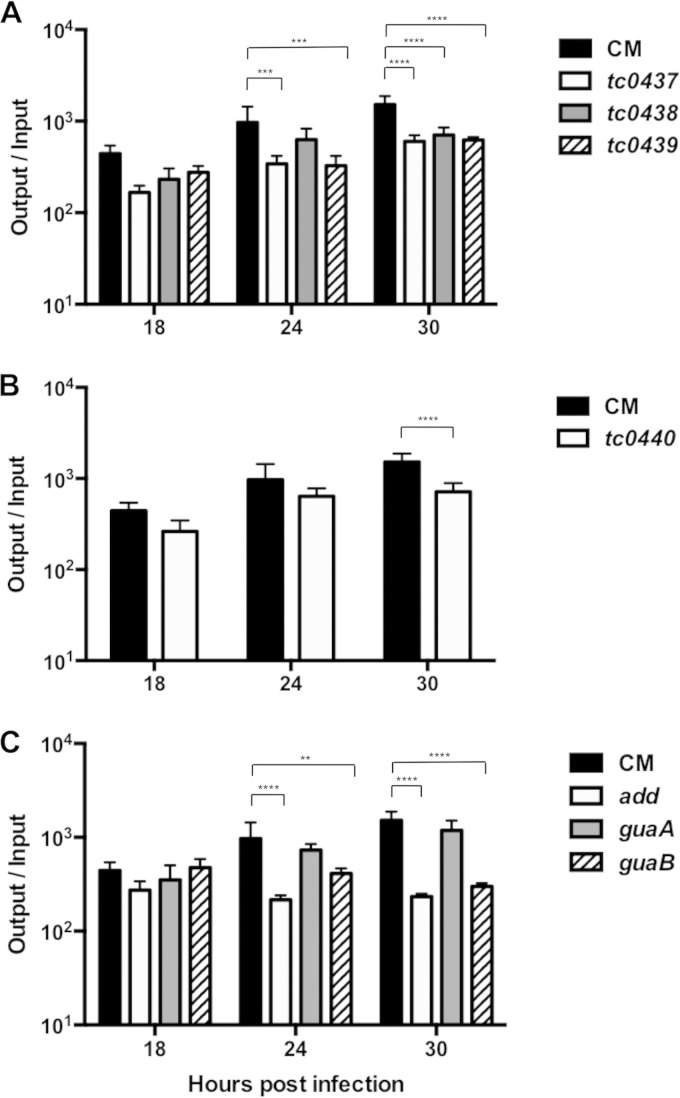
The PZ nonsense mutants were initially characterized using temporal recoverable IFU assays. Since multiple independent mutants of tc0440 and guaA were recovered by TILLING, we selected a representative mutant of each with a nonsense mutation located closest to the predicted 5′ end of these ORFs for further analysis. The growth kinetics of most of the PZ mutants paralleled those of C. muridarum, with the exception of add and guaB, which attained maximal EB production at 18 h p.i. as opposed to 30 h p.i. None of the mutants exhibited dramatic growth defects, although the IFU production of most of the mutants was significantly reduced compared to that of C. muridarum (Fig. 3). This modest reduction in IFU yield could be caused either by mutations in the PZ or by background mutations in the strains. Confocal microscopy failed to reveal obvious differences between the morphology of the mutant and C. muridarum inclusions (data not shown). These results implied that the presence of nonsense mutations in PZ ORFs did not hinder the ability of C. muridarum to form inclusions, complete development, or produce infectious EB.
FIG 3.
C. muridarum PZ mutants exhibit mild growth defects. rIFU analysis was performed for C. muridarum (CM) and PZ mutants at various time intervals following infection of McCoy cells. The data shown are from experiments performed in parallel. The data represent the average (+SD) ratios of input to output IFU from three independent experiments performed in triplicate. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01 by two-way ANOVA with Bonferroni post hoc test.
The cytotoxicity of cytotoxin nonsense mutants is reduced.
Since none of the PZ mutants displayed pronounced growth or morphological defects, we focused on examining phenotypes that have been indirectly attributed to the chlamydial cytotoxins in previous studies. The C. muridarum LCT-like cytotoxins are purported to mediate multiplication-independent cytopathic effects of C. muridarum EB on host cells at high MOI (19). This phenomenon, referred to as “immediate cytotoxicity” (34), resembles the cytoskeletal collapse observed in cultured cells treated with clostridial TcdB (19).
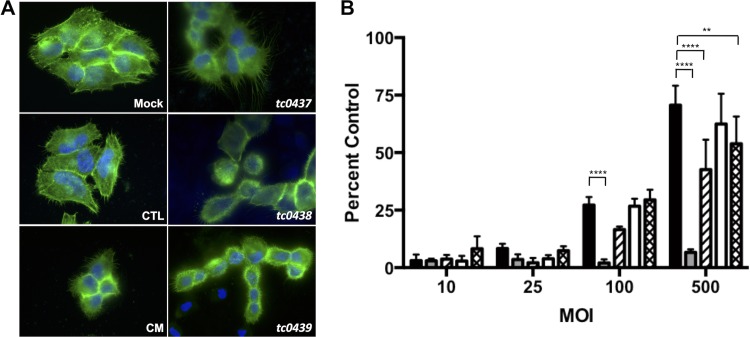
To determine if the C. muridarum cytotoxin gene (tc0437, tc0438, and tc0439) nonsense mutants were less cytotoxic, HeLa cells were infected with EB at an MOI of 250 in the presence of rifampin to block chlamydial multiplication. The cells were fixed and stained with phalloidin at 3 h p.i. and examined by fluorescence microscopy. HeLa cells infected with C. trachomatis serovar L2 strain 434/Bu (C. trachomatis), a cytotoxin-negative strain, did not differ in morphology from uninfected cells. In contrast, cell rounding and alterations in actin filament morphology were observed in infections with C. muridarum and the cytotoxin mutants (Fig. 4 A). This implied that the single cytotoxin mutants retained some cytotoxic activity.
FIG 4.
tc0437 and tc0439 mutants have reduced cytotoxicity. (A) HeLa cells were infected at an MOI of 250 in the presence of rifampin and were fixed 3 h p.i. Cell morphology and actin structure were visualized by staining with Alexa Fluor 488-phalloidin (green) and DAPI (blue). Overlays of fluorescence micrographs of cells that were mock infected or infected with C. muridarum (CM), C. trachomatis (CTL), tc0437, tc0438, or tc0439 are depicted. The images are representative of the results of three independent experiments. (B) LDH in supernatants of HeLa cells infected at various MOI in the presence of rifampin. Shown are C. muridarum (black), C. trachomatis (gray), tc0437 (hatched), tc0438 (white), and tc0439 (crosshatched). Triton X-100-treated cells were used as positive controls for LDH release. The data show the mean percentages (+SD) of the positive control from three independent experiments performed in triplicate. ****, P < 0.0001; **, P < 0.01 by one-way ANOVA with Dunnett's post hoc test.
Microscopy could not distinguish small variations in cytotoxicity, so we measured LDH release at 3 h p.i. from cells infected with C. muridarum, C. trachomatis, and the mutants at various MOI. As expected, C. trachomatis elicited minimal LDH release even at an MOI of 500. In contrast, C. muridarum caused cells to release significantly more LDH at high MOI. Interestingly, the tc0437 and tc0439 mutants elicited lower LDH release than C. muridarum at the highest MOI tested (Fig. 4B). This could indicate that tc0437 and tc0439 wild-type alleles encode active toxins or, alternatively, that background mutations in the mutant strains affected EB cytotoxic activity. Undiminished cytotoxicity of the tc0438 mutant implies that TC0438 may not be functional or has an unrelated function.
IFN-γ resistance is unaltered in C. muridarum PZ mutants.
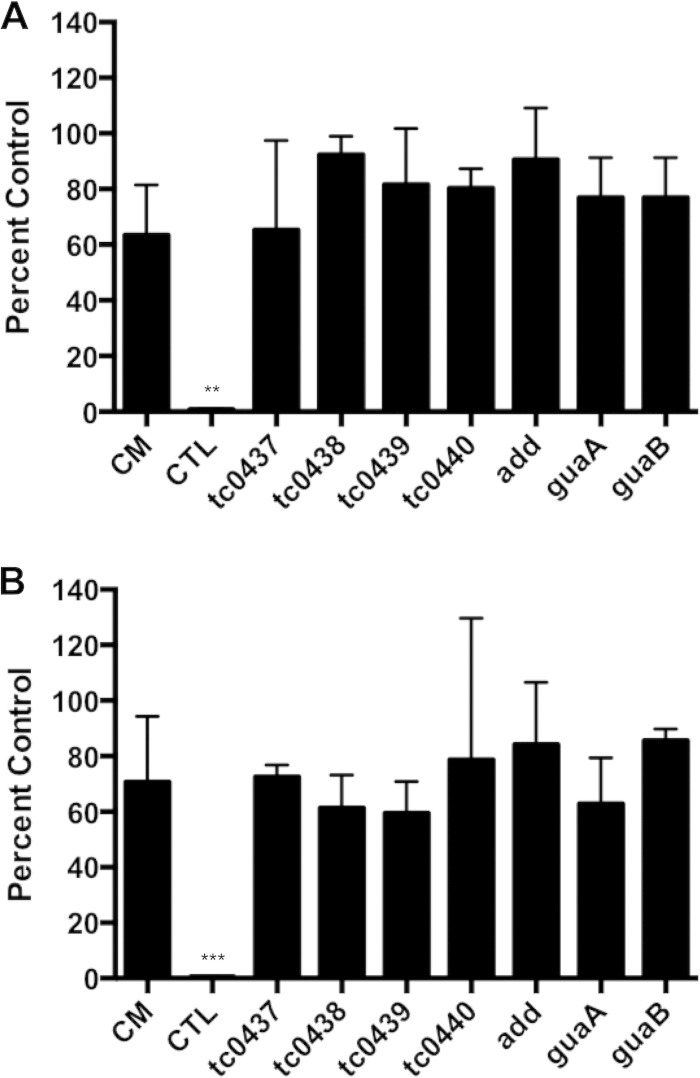
While high-multiplicity infections were important in associating a toxin-like activity with chlamydial EB, these doses may not be relevant in natural chlamydial disease. Roles for the chlamydial cytotoxins in host cell attachment and entry, vesicle trafficking, and modification of host cell GTPases have been reported (35–37). The C. muridarum cytotoxins have also been proposed to mediate evasion of the inhibitory effects of IFN-γ responses in murine cells (12). To test the latter hypothesis, McCoy cells were treated with IFN-γ, followed by infection with C. trachomatis, C. muridarum, or cytotoxin mutants. At 24 h, inclusions were either directly counted (Fig. 5A) or harvested for enumeration of viable EB contained by the inclusions using IFU assays (Fig. 5B). C. trachomatis, but not C. muridarum, was dramatically inhibited by IFN-γ. IFN-γ treatment did not significantly affect the survival or infectivity of the cytotoxin mutants compared to C. muridarum. Parallel experiments with other PZ nonsense mutants indicated that they were also similarly resistant to IFN-γ (Fig. 5). Thus, disruption of individual cytotoxin or other PZ ORFs did not confer IFN-γ sensitivity to C. muridarum.
FIG 5.
PZ mutants are resistant to IFN-γ. The sensitivity to IFN-γ treatment of C. muridarum (CM), C. trachomatis (CTL), and various PZ mutants was assessed by IFU assay (A) and rIFU assay (B). The data are represented as the average (+SD) percentages of untreated (no IFN-γ) infected controls from three independent experiments performed in triplicate. ***, P < 0.001; **, P < 0.01 by one-way ANOVA with Dunnett's post hoc test.
PZ mutants retain virulence in the mouse genital tract infection model.
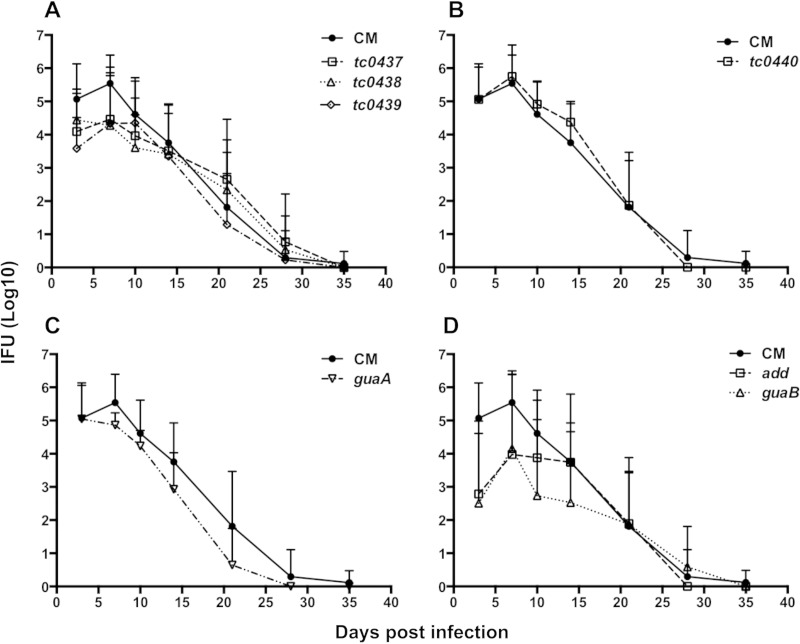
Intravaginal inoculations of mice with human C. trachomatis isolates result in mild infections of short duration with minimal upper genital tract infection and pathology (38). In comparison, C. muridarum is more virulent in this model, and the postinfection sequelae mimic human disease (39). To determine if the C. muridarum PZ genes were niche-specific virulence factors in mice, PZ mutants were analyzed in the murine genital tract infection model. The chlamydial burden in mice infected with the tc0439 cytotoxin mutant was significantly lower at 3 days postinfection than in animals infected with C. muridarum. Infection trajectories in mice inoculated with the tc0437 and tc0438 mutants also trended lower during the first week of infection. However, chlamydial recoveries of all three cytotoxin mutants appeared identical to C. muridarum infection at later time points (Fig. 6A). We also did not observe any significant deviation in bacterial loads of mice infected with the tc0440 mutant compared to mice infected with C. muridarum (Fig. 6B). Interestingly, the guaA nonsense mutant behaved identically to C. muridarum, whereas guaB and add mutants displayed reduced pathogen burdens at early data points (Fig. 6C and D). The guaBA-add operon putatively encodes products that function in the same purine-biosynthetic pathway. The absence of bacterial-shedding phenotypes in the guaA mutant suggested either that the phenotypes of the guaB and add mutants were due to background mutations or that guaBA-add acts noncanonically in C. muridarum. Since none of the PZ mutants were completely attenuated in vivo, we conclude that these genes are dispensable for infection of the murine genital tract.
FIG 6.
Mouse genital tract infections with C. muridarum PZ mutants. Groups of mice were challenged intravaginally with 50,000 IFU of C. muridarum (CM) or various PZ mutants. The infection curve for C. muridarum-infected mice (n = 30) is reproduced in each panel for comparison. (A) Lower genital tract shedding of IFU by mice infected with C. muridarum differed significantly from that by mice infected with the tc0439 mutant (n = 8) at day 3 postinfection (P < 0.01), but not that by mice infected with tc0437 (n = 8) or tc0438 (n = 8). (B and C) IFU shedding from mice infected with CM did not vary significantly from that from mice infected with the t0440 (n = 12) (B) or guaA (n = 16) mutant (C). (D) Shedding by mice infected with the add (n = 8) mutant was significantly less than shedding by mice infected with C. muridarum at 3 days (P < 0.0001) and 7 days (P < 0.01) postinfection. IFU shedding from mice infected with the add (n = 8) mutant was also significantly reduced at 3 days (P < 0.0001), 7 days (P < 0.05), and 10 days (P < 0.001) postinfection. The data are presented as mean numbers of IFU (+SD) for the mice in each group. Statistical differences were analyzed by two-way ANOVA with Bonferroni post hoc test.
Infected mice were also scored at 73 days postinfection for the presence of hydrosalpinx, an immunopathological consequence of chlamydia genital infection. Eighty percent (24/30) of the mice infected with C. muridarum developed hydrosalpinx compared to 63% (10/16) of guaA, 63% (5/8) of guaB, 13% (1/8) of add, 75% (9/12) of pld, 38% (3/8) of tc0437, 50% (4/8) of tc0438, and 38% (3/8) of tc0439 mutant-infected mice. The development of hydrosalpinx was significantly lower in add, tc0437, and tc0439 mutant-infected mice (P < 0.001, P = 0.03, and P = 0.03, respectively). It should be noted, however, that the differences observed in hydrosalpinx cannot necessarily be attributed to specific PZ gene mutations due to other background mutations found in the mutants.
PZ mutants contain multiple background mutations.
To identify a plausible explanation for the differing phenotypes of the purine pathway mutants, we sequenced the genomes of the parent strain and PZ mutants to an average coverage of 2,075× on the Illumina HiSeq platform (Table 1; see Tables S8 to S14 in the supplemental material). Comparison to published C. muridarum genomes indicated that our parent most closely resembled a C. muridarum Weiss isolate that was recently sequenced by K. H. Ramsey et al. (Table 1). On average, the PZ mutants contained 36.4 mutations, 66.67% of which were nonsynonymous substitutions. Most (98.43%) of the mutations were GC→AT transitions, consistent with EMS-induced changes. Several synonymous and nonsynonymous mutations were shared by some PZ mutant strains, which was a consequence of our serial mutagenesis strategy. They included mutations in recA, alaS, tc0237, tc0283, tc0471, tc0490, rpoC, aroA, upp, and tc0877 that were common to the add, tc0437, and tc0438 mutants. The add and tc0437 mutant strains contained identical mutations in tc0035, tc0069, tc0438, tc0575, tc0917, and pfkA-2. The tc0438 mutant shared mutations in murB, tc0330, and tc0575 with the add mutant and a pmpB–pmpC-2 mutation with the tc0437 mutant. Finally, mutations in tc0054, tc0191, pmpD, tc0250, tc0290, tc0312, tc0600, and an intergenic region (GenBank genome position 126188) were present in the tc0439 and guaB mutants. Interestingly, multiple mutant strains had nonsense mutations in tc0412, an ortholog of C. trachomatis ct135 that has been linked to virulence of C. trachomatis in the murine genital tract model (40). None of the tc0412 mutants except add were significantly attenuated in the murine GT, and an identical stop mutation was present in the tc0437 mutant strain, suggesting that attenuation of the add mutant was not linked to this tc0412 allele. No common mutations were present in the guaB and add mutants, which is consistent with the hypothesis that guaB and add might function in a noncanonical pathway independently of guaA.
TABLE 1.
Comparison of the genome of the wild-type C. muridarum isolate used in this study to those of other C. muridarum reference strains
| Gene ID | Description | Position | Base |
||
|---|---|---|---|---|---|
| C. muridarum used in study | C. muridarum Nigga | C. muridarum Weissb | |||
| tc0007 | Exodeoxyribonuclease V, beta chain, putative | 10505 | A | ||
| 10558 | A,C | ||||
| 10564 | A | ||||
| tc0027 | Conserved hypothetical protein | 32915 | G | ||
| tc0052 | Major outer membrane protein, porin; OmpA | 58882 | T | ||
| 58904 | T | ||||
| 59065 | C | C | T | ||
| tc0107 | Lipoprotein | 126406 | G | G | |
| 126416 | A | ||||
| None | Intergenic | 126383 | G | ||
| 126436 | A | ||||
| 126475 | A | ||||
| tc0124 | Transcription repair-coupling factor; TrcF | 151212 | T | T | |
| tc_r05 | rRNA-23S rRNA | 158800 | G | ||
| tc0138 | Phospho-N-acetylmuramoylpentapeptide transferase; MraY | 169451 | TTTT | T | T |
| tc0168 | Ribosomal protein L34; RpmH | 200671 | C | G | C |
| tc0301 | Methionyl tRNA synthetase; MetG | 358414 | T | ||
| tc0338 | ABC transporter, periplasmic substrate binding protein | 401302 | G | ||
| tc0341 | ABC transporter, permease protein | 403623 | T | ||
| 403624 | C | A | A | ||
| 403626 | C | ||||
| 403652 | C,G | T | C | ||
| 403714 | G | ||||
| tc0342 | ABC transporter, permease protein | 404473 | T | ||
| 404884 | A | A | |||
| tc0343 | 1-Deoxy-d-xylulose 5-phosphate reductoisomerase; Dxr | 405819 | G | ||
| tc0359 | Hypothetical protein | 419119 | C | ||
| tc0408 | Hypothetical protein | 468932 | T | G | T |
| tc0412 | Hypothetical protein | 473118 | A | A | |
| tc0437 | Adherence factor | 515204 | A | C | C |
| tc0493 | Phenylacrylic acid decarboxylase; UbiX | 600435 | A | C | C |
| tc0708 | Translation elongation factor; Tuf | 712587 | C | ||
| Hypothetical protein | 846475 | G | G | ||
| tc0727 | Outer membrane protein; OmcB | 866121 | T | G | T |
| tc0832 | DNA polymerase III alpha subunit; DnaE | 967487 | A | A | |
| tc0879 | Hypothetical protein | 1022529 | A | ||
| 1022551 | A | ||||
| tc0893 | groEL-2 | 1035913 | A | ||
C. muridarum Nigg isolate accession number: AE002160.2.
C. muridarum Weiss isolate accession number: ACOW00000000.
DISCUSSION
C. muridarum and C. trachomatis are genetically closely related, and genital infections of their respective hosts, mice and humans, have many similarities (4, 41). While T-cell immunity is critical for the resolution of chlamydial infection in both mice and humans, other aspects of immunity, most notably IFN-γ responses, appear to differ and are incompletely characterized (38). The results of our study suggest that although the PZ is the site of maximal divergence between the genomes of these pathogens, C. muridarum IFN-γ resistance determinants may not localize to the region.
We show that most PZ ORFs are transcribed in the early to middle phase of the developmental cycle and are expressed at relatively low levels. Although we identified two promoter regions that support our RT-PCR findings and that could provide plausible explanations for how some PZ ORFs are transcribed, strong promoters that could drive expression of one of the cytotoxin ORFs (tc0437) and an adjacent operon of PZ PLD (tc0436 to tc0432) that is relatively conserved in Chlamydia spp. were not identified. The putative start sites of tc0436 and tc0437 are located less than 1,000 bp apart in opposite strands of the C. muridarum genome, so perhaps our chosen cloning site disrupted two promoters in the same narrow region on opposing strands, thus compromising their detection. Another possibility is that E. coli sigma factors cannot recognize the chlamydial promoters in this region. For example, the auxiliary C. trachomatis transcription factor GrgA, which is not conserved in E. coli, directs expression from some chlamydial σ66-dependent promoters (42). Chlamydial σ28 also cannot complement an E. coli σ28 mutant, indicating that even conserved E. coli and chlamydial sigma factors are not completely interchangeable (43). We attempted to identify additional promoters using transcriptome sequencing (RNA-seq), but low expression of PZ ORFs relative to other, highly expressed genes yielded insufficient coverage for mapping transcriptional start sites in this region. As costs of deep RNA-seq decrease further and methods for enrichment of chlamydial transcripts from host cell transcripts improve, it might make sense to revisit this approach in the future. Better understanding of transcriptional organization could inform subsequent attempts to inactivate PZ operons.
Two of three cytotoxin mutants displayed reduced cytotoxicity in vitro, which suggests that these alleles are functional. We hope to verify this in future experiments using allelic complementation or by utilizing isogenic cytotoxin mutant strains. The biological role of chlamydial cytotoxins is still largely a matter of speculation. Polymorphisms in the cytotoxins have been associated with differences in the ocular and genital tropisms of C. trachomatis isolates (20). On the basis of their homology to LCTs and YopT, we have previously proposed that the cytotoxins may inactivate interferon-regulated GTPases that inhibit C. trachomatis, but not C. muridarum, in IFN-γ-treated mouse cells (44). Finally, mutants of E. coli efa1, another chlamydial cytotoxin gene homolog, exhibit reduced adherence to epithelial cells and fail to colonize colonic tissue effectively in mice (45). The observed trend toward a lower chlamydial burden during the first week of infection in mice inoculated with the chlamydial cytotoxin mutants could indicate similar roles for C. muridarum cytotoxins in adhesion and colonization, but this hypothesis requires experimental validation. An additional future direction would be to evaluate the IFN-γ resistance and virulence of double and triple cytotoxin mutants.
One of the most surprising findings of our study was that the guaB (IMP dehydrogenase) and add (adenosine deaminase) mutants were moderately attenuated in the GT while a guaA (GMP synthase) mutant was not. Genome sequencing confirmed that the phenotypes of the guaB and add mutants were not caused by a common background mutation. Thus, if unrelated background mutations were not responsible for the very similar GT phenotypes, our results suggest C. muridarum could encode an additional cryptic GMP synthase enzyme or that C. muridarum purine salvage acts noncanonically. The guaBA-add cluster has been retained by several chlamydial species, including those that cause respiratory illness, such as Chlamydia psittaci, Chlamydia felis, and human isolates of C. pneumoniae, but it is unknown if they are functional (33, 46, 47). guaA and guaB are known virulence determinants in other pathogenic organisms, like Borrelia burgdorferi and Francisella tularensis, during intraperitoneal and subcutaneous infection of mice, indicating that one or more purine nucleoside monophosphates (NMP) are limiting in certain mammalian niches (48, 49). Thus, an explanation of our results could be that NMP levels in the murine GT are sufficient to support chlamydial growth, whereas other tissues, like the lung, are starved for purine nucleotides. We hope to differentiate these possibilities by constructing additional guaA, guaB, and add nonsense mutants and comparing their virulence in murine GT and lung infection models.
Although we were unable to identify a role for individual PZ genes in IFN-γ resistance and murine genital infection, functional redundancy in the cytotoxin and PLD superfamily of proteins could have masked the effects of the loss of a single family member. However, our data suggest that cytotoxin functions are not wholly redundant, and true functional redundancy of these alleles is not consistent with the relatively low amino acid identity to one another (40 to 42%). While it would be possible to disrupt all members of the cytotoxin family and the guaBA-add operon sequentially, variable alleles, including tc0412, are a practical impediment to constructing isogenic strains by iterative TILLING (40, 50). Another possible explanation for the lack of phenotype of select PZ mutants is the emergence of suppressor mutations. Although this possibility is difficult to exclude, the fact that all of the PZ mutants were isolated from pools of mutants at the expected frequencies indicates that it is unlikely to have been a significant issue. We are currently investigating the possibility of disrupting the MACPF-PZ PLD (tc0432-tc0431; tc0436 to tc0433), cytotoxin (tc0437-tc0438), and guaBA-add operons using polar transposon insertions by the Targetron method developed by Johnson and Fisher (51).
In summary, our results imply that all the examined PZ ORFs are nonessential for virulence in the mouse GT. However, we cannot rule out their importance in other niches. C. muridarum was originally isolated from the lungs of symptom-free laboratory mice. Disease was observed only upon inoculation of mice with serially passaged lung homogenates, indicating that very low levels of C. muridarum are typically present in pulmonary tissue (52). Later studies demonstrated that C. muridarum is transmitted through the oral-fecal route, which suggests that the gut is a reservoir and that small numbers of C. muridarum organisms enter the lungs during feeding (53). Finally, it is notable that guaBA-add and cytotoxin homologs of other intracellular pathogens modulate infections of the lung and gastrointestinal tract, respectively (45, 48, 54, 55). Thus, examination of C. muridarum PZ mutants in lung or gastrointestinal tract models of mouse infections is warranted. A positive implication of our study is that successful modeling of human disease in the murine GT does not appear to require much of the C. muridarum PZ. Future investigations of polymorphisms outside the PZ will be important in the identification of C. muridarum alleles that confer IFN-γ resistance and dictate virulence in the murine GT model.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants RO1AI099278 and R21AI099307 from the National Institutes of Health (D.E.N.) and in part by grant 1P20GM103625 from the National Institutes of General Medical Sciences (R.P.M.).
We thank Julie Brothwell and Matthew Muramatsu for critical review of the manuscript. Special thanks are also due to Kip Bodi and Albert Tai at the Tufts University Genomics Core Facility for their assistance in whole-genome sequencing.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00106-15.
REFERENCES
- 1.Zomorodipour A, Andersson SGE. 1999. Obligate intracellular parasites: Rickettsia prowazekii and Chlamydia trachomatis. FEBS Lett 452:11–15. doi: 10.1016/S0014-5793(99)00563-3. [DOI] [PubMed] [Google Scholar]
- 2.Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao QX, Koonin EV, Davis RW. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 3.Kalman S, Mitchell W, Marathe R, Lammel C, Fan L, Hyman RW, Olinger L, Grimwood L, Davis RW, Stephens RS. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat Genet 21:385–389. doi: 10.1038/7716. [DOI] [PubMed] [Google Scholar]
- 4.Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, Hickey EK, Peterson J, Utterback T, Berry K, Bass S, Linher K, Weidman J, Khouri H, Craven B, Bowman C, Dodson R, Gwinn M, Nelson W, DeBoy R, Kolonay J, McClarty G, Salzberg SL, Eisen J, Fraser CM. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res 28:1397–1406. doi: 10.1093/nar/28.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Read TD, Myers GSA, Brunham RC, Nelson WC, Paulsen IT, Heidelberg J, Holtzapple E, Khouri H, Federova NB, Carty HA, Umayam LA, Haft DH, Peterson J, Beanan MJ, White O, Salzberg SL, Hsia RC, McClarty G, Rank RG, Bavoil PM, Fraser CM. 2003. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res 31:2134–2147. doi: 10.1093/nar/gkg321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stephens RS, Myers G, Eppinger M, Bavoil PM. 2009. Divergence without difference: phylogenetics and taxonomy of Chlamydia resolved. FEMS Immunol Med Microbiol 55:115–119. doi: 10.1111/j.1574-695X.2008.00516.x. [DOI] [PubMed] [Google Scholar]
- 7.Carlson JH, Porcella SF, McClarty G, Caldwell HD. 2005. Comparative genomic analysis of Chlamydia trachomatis oculotropic and genitotropic strains. Infect Immun 73:6407–6418. doi: 10.1128/IAI.73.10.6407-6418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kari L, Whitmire WM, Carlson JH, Crane DD, Reveneau N, Nelson DE, Mabey DCW, Bailey RL, Holland MJ, McClarty G, Caldwell HD. 2008. Pathogenic diversity among Chlamydia trachomatis ocular strains in nonhuman primates is affected by subtle genomic variations. J Infect Dis 197:449–456. doi: 10.1086/525285. [DOI] [PubMed] [Google Scholar]
- 9.Thomas SM, Garrity LF, Brandt CR, Schobert CS, Feng GS, Taylor MW, Carlin JM, Byrne GI. 1993. IFN-gamma-mediated antimicrobial response. indoleamine 2,3-dioxygenase-deficient mutant host cells no longer inhibit intracellular Chlamydia spp. or Toxoplasma growth. J Immunol 150:5529–5534. [PubMed] [Google Scholar]
- 10.Fehlner-Gardiner C, Roshick C, Carlson JH, Hughes S, Belland RJ, Caldwell HD, McClarty G. 2002. Molecular basis defining human Chlamydia trachomatis tissue tropism: a possible role for tryptophan synthase. J Biol Chem 277:26893–26903. doi: 10.1074/jbc.M203937200. [DOI] [PubMed] [Google Scholar]
- 11.Caldwell HD, Wood H, Crane D, Bailey R, Jones RB, Mabey D, Maclean I, Mohammed Z, Peeling R, Roshick C, Schachter J, Solomon AW, Stamm WE, Suchland RJ, Taylor L, West SK, Quinn TC, Belland RJ, McClarty G. 2003. Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J Clin Invest 111:1757–1769. doi: 10.1172/JCI200317993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson DE, Virok DP, Wood H, Roshick C, Johnson RM, Whitmire WM, Crane DD, Steele-Mortimer O, Kari L, McClarty G, Caldwell HD. 2005. Chlamydial IFN-gamma immune evasion is linked to host infection tropism. Proc Natl Acad Sci U S A 102:10658–10663. doi: 10.1073/pnas.0504198102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kari L, Goheen MM, Randall LB, Taylor LD, Carlson JH, Whitmire WM, Virok D, Rajaram K, Endresz V, McClarty G, Nelson DE, Caldwell HD. 2011. Generation of targeted Chlamydia trachomatis null mutants. Proc Natl Acad Sci U S A 108:7189–7193. doi: 10.1073/pnas.1102229108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ponting CP. 1999. Chlamydial homologues of the MACPF (MAC/peforin) domain. Curr Biol 9:R911–R913. [DOI] [PubMed] [Google Scholar]
- 15.Rosado CJ, Kondos S, Bull TE, Kuiper MJ, Law RHP, Buckle AM, Voskoboinik I, Bird PI, Trapani JA, Whisstock JC, Dunstone MA. 2008. The MACPF/CDC family of pore-forming toxins. Cell Microbiol 10:1765–1774. doi: 10.1111/j.1462-5822.2008.01191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor LD, Nelson DE, Dorward DW, Whitmire WM, Caldwell HD. 2010. Biological characterization of Chlamydia trachomatis plasticity zone MACPF domain family protein CT153. Infect Immun 78:2691–2699. doi: 10.1128/IAI.01455-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson DE, Crane DD, Taylor LD, Dorward DW, Goheen MM, Caldwell HD. 2006. Inhibition of chlamydiae by primary alcohols correlates with the strain-specific complement of plasticity zone phospholipase D genes. Infect Immun 74:73–80. doi: 10.1128/IAI.74.1.73-80.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar Y, Cocchiaro J, Valdivia RH. 2006. The obligate intracellular pathogen Chiamydia trachomatis targets host lipid droplets. Curr Biol 16:1646–1651. doi: 10.1016/j.cub.2006.06.060. [DOI] [PubMed] [Google Scholar]
- 19.Belland RJ, Scidmore MA, Crane DD, Hogan DM, Whitmire W, McClarty G, Caldwell HD. 2001. Chlamydia trachomatis cytotoxicity associated with complete and partial cytotoxin genes. Proc Natl Acad Sci U S A 98:13984–13989. doi: 10.1073/pnas.241377698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carlson JH, Hughes S, Hogan D, Cieplak G, Sturdevant DE, McClarty G, Caldwell HD, Belland RJ. 2004. Polymorphisms in the Chlamydia trachomatis cytotoxin locus associated with ocular and genital isolates. Infect Immun 72:7063–7072. doi: 10.1128/IAI.72.12.7063-7072.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang YB, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Clarke IN. 2011. Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog 7:e1002258. doi: 10.1371/journal.ppat.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kannan RM, Gerard HC, Mishra MK, Mao GZ, Wang SX, Hali M, Whittum-Hudson JA, Hudson AP. 2013. Dendrimer-enabled transformation of Chlamydia trachomatis. Microb Pathog 65:29–35. doi: 10.1016/j.micpath.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 23.Sturdevant JL, Caldwell HD. 2014. Innate immunity is sufficient for the clearance of Chlamydia trachomatis from the female mouse genital tract. Pathog Dis 72:70–73. doi: 10.1111/2049-632X.12164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McClarty G, Caldwell HD, Nelson DE. 2007. Chlamydial interferon gamma immune evasion influences infection tropism. Curr Opin Microbiol 10:47–51. doi: 10.1016/j.mib.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Coers J, Starnbach MN, Howard JC. 2009. Modeling infectious disease in mice: co-adaptation and the role of host-specific IFN gamma responses. PLoS Pathog 5:e1000333. doi: 10.1371/journal.ppat.1000333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caldwell HD, Kromhout J, Schachter J. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun 31:1161–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hefty PS, Stephens RS. 2007. Chlamydial type III secretion system is encoded on ten operons preceded by sigma 70-like promoter elements. J Bacteriol 189:198–206. doi: 10.1128/JB.01034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller JH. 1992. A short course in bacterial genetics: A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- 29.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- 30.Till BJ, Colbert T, Tompa R, Enns LC, Codomo CA, Johnson JE, Reynolds SH, Henikoff JG, Greene EA, Steine MN, Comai L, Henikoff S. 2003. High-throughput TILLING for functional genomics. Methods Mol Biol 236:205–220. doi: 10.1385/1-59259-413-1:205. [DOI] [PubMed] [Google Scholar]
- 31.Thomson NR, Yeats C, Bell K, Holden MTG, Bentley SD, Livingstone M, Cerdeno-Tarraga AM, Harris B, Doggett J, Ormond D, Mungall K, Clarke K, Feltwell T, Hance Z, Sanders M, Quail MA, Price C, Barrell BG, Parkhill J, Longbottom D. 2005. The Chlamlydophila abortus genome sequence reveals an array of variable proteins that contribute to interspecies variation. Genome Res 15:629–640. doi: 10.1101/gr.3684805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voigt A, Schofl G, Saluz HP. 2012. The Chlamydia psittaci genome: a comparative analysis of intracellular pathogens. PLoS One 7:e35097. doi: 10.1371/journal.pone.0035097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell CM, Hovis KM, Bavoil PM, Myers GSA, Carrasco JA, Timms P. 2010. Comparison of koala LPCoLN and human strains of Chlamydia pneumoniae highlights extended genetic diversity in the species. BMC Genomics 11:442. doi: 10.1186/1471-2164-11-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moulder JW, Hatch TP, Byrne GI, Kellogg KR. 1976. Immediate toxicity of high multiplicities of Chlamydia psittaci for mouse fibroblasts (L cells). Infect Immun 14:277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thalmann J, Janik K, May M, Sommer K, Ebeling J, Hofmann F, Genth H, Klos A. 2010. Actin re-organization induced by Chlamydia trachomatis serovar D: evidence for a critical role of the effector protein CT166 targeting Rac. PLoS One 5:e9887. doi: 10.1371/journal.pone.0009887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schramm N, Wyrick PB. 1995. Cytoskeletal requirements in Chlamydia trachomatis infection of host cells. Infect Immun 63:324–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis CH, Wyrick PB. 1997. Differences in the association of Chlamydia trachomatis serovar E and serovar L2 with epithelial cells in vitro may reflect biological differences in vivo. Infect Immun 65:2914–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perry LL, Su H, Feilzer K, Messer R, Hughes S, Whitmire W, Caldwell HD. 1999. Differential sensitivity of distinct Chlamydia trachomatis isolates to IFN-gamma-mediated inhibition. J Immunol 162:3541–3548. [PubMed] [Google Scholar]
- 39.Morrison RP, Feilzer K, Tumas DB. 1995. Gene knockout mice establish a primary protective role for major histocompatibility class II-restricted responses in Chlamydia trachomatis genital tract infection. Infect Immun 63:4661–4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sturdevant GL, Kari L, Gardner DJ, Olivares-Zavaleta N, Randall LB, Whitmire WM, Carlson JH, Goheen MM, Selleck EM, Martens C, Caldwell HD. 2010. Frameshift mutations in a single novel virulence factor alter the in vivo pathogenicity of Chlamydia trachomatis for the female murine genital tract. Infect Immun 78:3660–3668. doi: 10.1128/IAI.00386-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barron AL, White HJ, Rank RG, Soloff BL, Moses EB. 1981. A new animal model for the study of Chlamydia trachomatis genital infections: infection of mice with the agent of mouse pneumonitis. J Infect Dis 143:63–66. doi: 10.1093/infdis/143.1.63. [DOI] [PubMed] [Google Scholar]
- 42.Bao X, Nickels BE, Fan H. 2012. Chlamydia trachomatis protein GrgA activates transcription by contacting the nonconserved region of sigma(66). Proc Natl Acad Sci U S A 109:16870–16875. doi: 10.1073/pnas.1207300109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen L, Li MX, Zhang YX. 2004. Chlamydia trachomatis sigma(28) recognizes the fliC promoter of Escherichia coli and responds to heat shock in chlamydiae. Microbiology 150:205–215. doi: 10.1099/mic.0.26734-0. [DOI] [PubMed] [Google Scholar]
- 44.Nelson DE, Taylor LD, Shannon JG, Whitmire WM, Crane DD, McClarty G, Su H, Kari L, Caldwell HD. 2007. Phenotypic rescue of Chlamydia trachomatis growth in IFN-gamma treated mouse cells by irradiated Chlamydia muridarum. Cell Microbiol 9:2289–2298. doi: 10.1111/j.1462-5822.2007.00959.x. [DOI] [PubMed] [Google Scholar]
- 45.Klapproth JMA, Sasaki M, Sherman M, Babbin B, Donnenberg MS, Fernandes PJ, Scaletsky ICA, Kalman D, Nusrat A, Williams IR. 2005. Citrobacter rodentium lifA/efa1 is essential for colonic colonization and crypt cell hyperplasia in vivo. Infect Immun 73:1441–1451. doi: 10.1128/IAI.73.3.1441-1451.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Lent S, Piet JR, Beeckman D, van der Ende A, Van Nieuwerburgh F, Bavoil P, Myers G, Vanrompay D, Pannekoek Y. 2012. Full genome sequences of all nine Chlamydia psittaci genotype reference strains. J Bacteriol 194:6930–6931. doi: 10.1128/JB.01828-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Azuma Y, Hirakawa H, Yamashita A, Cai Y, Rahman MA, Suzuki H, Mitaku S, Toh H, Goto S, Murakami T, Sugi K, Hayashi H, Fukushi H, Hattori M, Kuhara S, Shirai M. 2006. Genome sequence of the cat pathogen, Chlamydophila felis. DNA Res 13:15–23. doi: 10.1093/dnares/dsi027. [DOI] [PubMed] [Google Scholar]
- 48.Jewett MW, Lawrence KA, Bestor A, Byram R, Gherardini F, Rosa PA. 2009. GuaA and GuaB are essential for Borrelia burgdorferi survival in the tick-mouse infection cycle. J Bacteriol 191:6231–6241. doi: 10.1128/JB.00450-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santiago AE, Cole LE, Franco A, Vogel SN, Levine MM, Barry EM. 2009. Characterization of rationally attenuated Francisella tularensis vaccine strains that harbor deletions in the guaA and guaB genes. Vaccine 27:2426–2436. doi: 10.1016/j.vaccine.2009.02.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramsey KH, Sigar IM, Schripsema JH, Denman CJ, Bowlin AK, Myers GAS, Rank RG. 2009. Strain and virulence diversity in the mouse pathogen Chlamydia muridarum. Infect Immun 77:3284–3293. doi: 10.1128/IAI.00147-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson CM, Fisher DJ. 2013. Site-specific, insertional inactivation of incA in Chlamydia trachomatis using a group II intron. PLoS One 8:e83989. doi: 10.1371/journal.pone.0083989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rank RG. 2006. Chlamydial diseases. The mouse in biomedical research. Elsevier, San Diego, CA. [Google Scholar]
- 53.Cotter TW, Ramsey KH, Miranpuri GS, Poulsen CE, Byrne GI. 1997. Dissemination of Chlamydia trachomatis chronic genital tract infection in gamma interferon gene knockout mice. Infect Immun 65:2145–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stevens MP, Roe AJ, Vlisidou I, van Diemen PM, La Ragione RM, Best A, Woodward MJ, Gally DL, Wallis TS. 2004. Mutation of toxB and a truncated version of the efa-1 gene in Escherichia coli O157:H7 influences the expression and secretion of locus of enterocyte effacement-encoded proteins but not intestinal colonization in calves or sheep. Infect Immun 72:5402–5411. doi: 10.1128/IAI.72.9.5402-5411.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blasi F, Tarsia P, Aliberti S. 2009. Chlamydophila pneumoniae. Clin Microbiol Infect 15:29–35. doi: 10.1111/j.1469-0691.2008.02130.x. [DOI] [PubMed] [Google Scholar]
- 56.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.