Abstract
The type VI secretion system (T6SS) as a virulence factor-releasing system contributes to virulence development of various pathogens and is often activated upon contact with target cells. Citrobacter freundii strain CF74 has a complete T6SS genomic island (GI) that contains clpV, hcp-2, and vgr T6SS genes. We constructed clpV, hcp-2, vgr, and T6SS GI deletion mutants in CF74 and analyzed their effects on the transcriptome overall and, specifically, on the flagellar system at the levels of transcription and translation. Deletion of the T6SS GI affected the transcription of 84 genes, with 15 and 69 genes exhibiting higher and lower levels of transcription, respectively. Members of the cell motility class of downregulated genes of the CF74ΔT6SS mutant were mainly flagellar genes, including effector proteins, chaperones, and regulators. Moreover, the production and secretion of FliC were also decreased in clpV, hcp-2, vgr, or T6SS GI deletion mutants in CF74 and were restored upon complementation. In swimming motility assays, the mutant strains were found to be less motile than the wild type, and motility was restored by complementation. The mutant strains were defective in adhesion to HEp-2 cells and were restored partially upon complementation. Further, the CF74ΔT6SS, CF74ΔclpV, and CF74Δhcp-2 mutants induced lower cytotoxicity to HEp-2 cells than the wild type. These results suggested that the T6SS GI in CF74 regulates the flagellar system, enhances motility, is involved in adherence to host cells, and induces cytotoxicity to host cells. Thus, the T6SS plays a wide-ranging role in C. freundii.
INTRODUCTION
The type VI secretion system (T6SS) is present in a number of bacterial species, including important pathogens, Pseudomonas aeruginosa (1), Vibrio cholerae (2), enteroaggregative Escherichia coli (3), Burkholderia thailandensis (4), Serratia marcescens (5), Burkholderia mallei (6), and Salmonella enterica (7). The T6SS consists of a core structure formed by IAHP (intracellular multiplication protein F [IcmF]-associated homologous protein), the inner membrane proteins IcmF and DotU/IcmH, the lipoprotein SciN, and the ATPase ClpV. IcmF is predicted to be located in the inner membrane and consists of a cytosolic domain and a large periplasmic domain. The cytosolic part harbors a conserved Walker A motif (phosphate-binding loop), implying that IcmF functions as an ATPase during type VI protein secretion. icmF mutants have been shown to prevent Hcp secretion (2, 8–10). ClpV is a member of the AAA+ (ATPases associated with various cellular activities) protein family (8, 11). It forms oligomeric complexes to energize the system for the secretion of effector proteins, which include the secreted VgrG (valine glycine repeat) and Hcp (hemolysin-coregulated protein) proteins (1, 12–17).
It has been reported that the T6SS contributes to the virulence development of various pathogens and is often activated upon contact with target cells for the secretion of effector proteins (1–7, 14–17). The expression and assembly of the T6SS are tightly controlled at both the transcriptional and posttranscriptional levels (12, 13, 18). Furthermore, the IcmF protein of the T6SS has been shown to be involved in flagellar regulation and affects motility and biofilm formation (15). The flagellum is composed of a basal body, a hook, and a filament. FliC, a component of the filament, is transported from the cytoplasm by a number of transport systems in different bacterial species, including the SPI1 type III secretion system (T3SS), the Dot/Icm type IV secretion system (T4SS), and the locus of enterocyte effacement (LEE)-encoded T3SS in Salmonella enterica serovar Typhimurium, Legionella pneumophila, and enteropathogenic E. coli, respectively (19, 20).
Citrobacter freundii is considered a commensal of the intestinal tract in humans and other animals (21). However, C. freundii can also cause diarrhea and other infections in humans (22–25). Relatively little is known about the virulence of C. freundii. Recently, we identified a C. freundii strain, CF74, that showed an aggregative adherence pattern and cytotoxicity to HEp-2 cells. The strain was found to contain a complete T6SS located on a genomic island (GI), and the T6SS gene cluster consists of 16 genes, including key T6SS genes (clpV, hcp-2, and vgr) (26). In this study, we constructed clpV, hcp-2, vgr, and T6SS GI deletion mutants in CF74 and demonstrate that the T6SS affects the transcription of over 84 genes, and especially the flagellar system, at the levels of transcription and translation and is involved in the secretion of FliC. The T6SS was also found to contribute to adhesion and cytotoxicity to host cells.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
C. freundii strain CF74 was isolated from a fecal sample from a goat, as reported previously (26). All strains were grown aerobically at 37°C in Luria-Bertani (LB) medium. Antibiotics were added at the following concentrations: 100 mg/ml for ampicillin and streptomycin and 30 mg/ml for chloramphenicol where appropriate. All strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| Strains | ||
| CF74 | C. freundii isolate from fecal samples from a goat | Laboratory stock |
| CF74ΔT6SS | CF74 mutant with T6SS core component gene cluster deletion | This study |
| CF74ΔclpV | CF74 mutant with clpV gene deletion | This study |
| CF74Δhcp-2 | CF74 mutant with hcp-2 gene deletion | This study |
| CF74Δvgr | CF74 mutant with vgr gene deletion | This study |
| E. coli SM10λpir | supE recA::rp4-2-Tc::Mu Kmr λpir | Laboratory stock |
| E. coli DH5αλpir | RP4-2-tet Mu-Km::Tn7 integrant leu-63::IS10 recA1 creC510 hsdR17 endA1 zbf-5 uidA(ΔmluI)::pir+ thi | Laboratory stock |
| CF74ΔclpV(pBAD24-clpV) | ΔclpV(pBAD24-clpV) | This study |
| CF74Δhcp-2(pBAD24-hcp-2) | Δhcp-2(pBAD24-hcp-2) | This study |
| CF74Δvgr(pBAD24-vgr) | Δvgr(pBAD24-vgr) | This study |
| Plasmids | ||
| pBAD24 | pMB1 Ampr; pBAD promoter | Laboratory stock |
| pBAD24-clpV | The clpV coding region was cloned into the EcoRI and HindIII sites of pBAD24 | This study |
| pBAD24-hcp-2 | The hcp-2 coding region was cloned into the KpnI and HindIII sites of pBAD24 | This study |
| pBAD24-vgr | The vgr coding region was cloned into the KpnI and HindIII sites of pBAD24 | This study |
| pWM91 | Ampr f1+ ori lacZa of pBluescript II SK(+); oriRR6Kγ oriTRP4 sacB Sucs | Laboratory stock |
| pWM91ΔT6SS | pWM91-T6SSup::T6SSdown; Ampr | This study |
| pWM91ΔclpV | pWM91-clpVup::clpvdown; Ampr | This study |
| pWM91Δhcp-2 | pWM91-hcp-2up::hcpdown; Ampr | This study |
| pWM91Δvgr | pWM91-vgrup::vgrdown; Ampr | This study |
Construction of isogenic mutants and plasmids.
All in-frame deletion mutants were generated in C. freundii strain CF74 via double crossover using the suicide plasmid pWM91 as described previously (27, 28). The upstream and downstream regions of the gene of interest were amplified using the primers shown in Table S1 in the supplemental material. Using fusion PCR of these two fragments, we generated a fragment that was cloned into pWM91, a plasmid containing the counterselectable sacB gene. The recombinant plasmids were then purified and introduced into E. coli SM10 λpir and conjugally transferred into CF74, and deletion of the genes of interest was selected in LB agar with 10% sucrose and without NaCl. Chromosomal deletion mutants were identified by colony PCR and quantitative reverse transcription (qRT)-PCR.
CF74 deletion mutants were complemented by pBAD24 harboring the genes of interest, allowing arabinose-controlled gene expression. For transformation of plasmids into CF74, electrocompetent cells were prepared as described previously (29).
RNA extraction and qRT-PCR.
To prepare cells for RNA extraction, fresh LB medium was inoculated from an overnight culture of CF74, deletion mutants, or their complementation mutants (1:100) and incubated at 37°C with shaking at 220 rpm. The strains were collected at an optical density at 600 nm (OD600) of 1.0. Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. RNA samples were further purified using the RNeasy minikit (Qiagen, Valencia, CA), followed by treatment with DNase I (Qiagen, Valencia, CA) to eliminate genomic DNA contamination. The RNA size, integrity, and total amount were measured using a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA).
To measure gene transcription in different strains, qRT-PCR was performed using specific primers (see Table S1 in the supplemental material) based on the targeted genes. Total RNA (1.0 mg) was reverse transcribed to generate cDNA as the template for qRT-PCR. qRT-PCR was carried out using SYBR Premix Ex Taq II (Perfect Real Time; TaKaRa) using a Rotor-Gene Q thermal cycler (Qiagen, Valencia, CA). Data were analyzed with Rotor-Gene Q series software version 1.7 (Qiagen, Valencia, CA). The data were normalized to the endogenous reference gene gapA and analyzed by the cycle threshold method (2−ΔΔCT) (30). Three independent technical replicates were carried out for each target.
RNA-Seq and transcriptional data analysis.
rRNA was depleted using the MicrobEnrich kit and the Microbexpress bacterial mRNA enrichment kit (both from Ambion, Austin, TX) according to the manufacturer's specifications. Sequencing libraries were prepared using the mRNA-Seq 8-Sample Prep kit (Illumina, San Diego, CA) according to the manufacturer's protocol. The RNA-Seq libraries were sequenced using IlluminaGA IIx and the paired-end sequencing module.
All raw FastQ files were trimmed and sorted by SolexQA (31). To obtain estimates of transcription levels, Burrows-Wheeler alignment (BWA) (32) was used to map the trimmed sequencing reads against the genome sequence of C. freundii CF74. Cufflinks (v1.1.0) (33) was then used to estimate gene transcription levels. Reads per kilobase of exon model per million mapped reads (RPKM) (34) was used as a normalized metric to present the gene expression levels.
Swimming motility assays.
Motility was evaluated using a medium based on Difco-Tryptone (BD Diagnostic Systems) broth containing 1% (wt/vol) NaCl (Sigma-Aldrich) with Difco Bacto agar at 0.3% (wt/vol) to 1% (wt/vol). For plasmid-based complementation experiments, 0.2% arabinose and 100 μg/ml ampicillin were added to the plates, and all plates were poured the night before use and allowed to air dry on the bench. The plates were stab inoculated and incubated at 30°C for 16 h, and halo diameter measurements were recorded. All strains were tested in triplicate, and each experiment was carried out on three separate occasions (35, 36).
Preparation and analysis of whole-cell extracts and culture supernatant proteins.
Bacterial culture supernatants were collected at an OD600 of 1.0 and filtered through 0.22-μm low-protein-binding filters (Millipore). A 10% (vol/vol) final concentration of trichloroacetic acid (TCA) (Sigma-Aldrich) was used to precipitate the proteins. The supernatants were incubated overnight at 4°C, and the aggregated proteins were precipitated by centrifugation at 4°C, washed with cold acetone, air dried, and stored at −80°C until use.
Whole-cell lysates (WCL) were generated by sonicating the cell pellets. Briefly, the cell pellets were suspended in lysis buffer containing 7 M urea, 2 M thiourea, 4% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 65 mM dithiothreitol (DTT), and 40 mM Tris with 1/100 (vol/vol) protease inhibitor cocktail (Roche), and the suspension was sonicated.
Western blotting.
WCL and culture supernatant proteins were subsequently analyzed by SDS-10% PAGE, and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore) for immunoblotting. The immobilized proteins were incubated with primary antibodies, namely, rabbit anti-FliC (made by Abzome Co., Ltd., China; 1:5,000) and mouse anti-GapA (Thermo Fisher Scientific, Waltham, MA; 1:5,000), followed by incubation with horseradish peroxidase (HRP)-conjugated goat anti-rabbit (MBL; 1:10,000) and goat anti-mouse IgG (MBL; 1:3,000), respectively, and then detected on ECL chemiluminescence film (Kodak, China) using a Protec automatic film processor (Optimax).
In vitro adhesion and cytotoxicity assays.
Comparison of in vitro adhesion to host cells was performed using the human epidermoid carcinoma cell line HEp-2 (CCC0068; Beijing Union Medical College cell resource center), as previously described (26). Infections were performed at a multiplicity of infection (MOI) of 100:1. After 3 h of incubation at 37°C with 5% CO2, the infected monolayers were rinsed with phosphate-buffered saline (PBS), and then the cells were lysed in PBS containing 0.25% Triton X-100. The lysates were plated onto agar plates, and the number of adhesive bacteria was determined by counting the CFU after overnight incubation at 37°C. Each sample determination was performed in triplicate, and experiments were repeated two times.
The lactate dehydrogenase (LDH) released by the HEp-2 cells was determined using the Cytotox96 kit (Promega) according to the manufacturer's instructions. The relative amount of cytotoxicity was expressed as follows: (experimental release − spontaneous release)/(maximum release − spontaneous release) × 100, where the spontaneous release was the amount of LDH activity in the supernatant of uninfected cells and the maximum release was that when cells were lysed with the lysis buffer provided by the manufacturer. All experiments were performed two times in duplicate (26).
Statistical analysis.
SPSS software version 13.0 (SPSS Inc., Chicago, IL, USA) was used to conduct all statistical comparisons. A nonparametric test (Mann-Whitney U test) was employed to compare the different groups. A two-tailed P value of 0.05 or less was considered to be statistically significant.
RESULTS
Transcriptome comparison of C. freundii strain CF74 and T6SS deletion mutant CF74ΔT6SS.
A CF74 T6SS deletion mutant (CF74ΔT6SS) was created by allelic exchange, and the deletion from gene CFCDC_0705 to gene CFCDC_0726 was confirmed by PCR sequencing. To analyze differential expression of transcripts between C. freundii strain CF74 and the deletion mutant CF74ΔT6SS, an RNA-Seq was performed. Over 20 million reads each were obtained for CF74 and CF74ΔT6SS. Approximately 99% of the reads were mapped to the reference genome, CF74 (Table 2). A total of 84 genes were found to be differentially transcribed between CF74 and CF74ΔT6SS; 15 and 69 genes exhibited higher and lower levels of transcription in CF74ΔT6SS, respectively (see Tables S2 and S3 in the supplemental material).
TABLE 2.
Summary of RNA-Seq coverage data
| Statistic | Value |
|
|---|---|---|
| CF74 | CF74ΔT6SS | |
| No. of reads that aligned | 22,113,030 | 20,751,152 |
| No. of reads that did not align | 236,198 | 171,780 |
| Total no. of reads in the sample | 22,349,228 | 20,922,932 |
| % alignments | 98.94 | 99.18 |
| No. of reads that aligned to the 16S rRNA gene | 82,443 | 3,312 |
| No. of reads that aligned to the 23S rRNA gene | 144,083 | 5,112 |
| % of all reads that aligned to 16S and 23S rRNA genes | 1.01 | 0.04 |
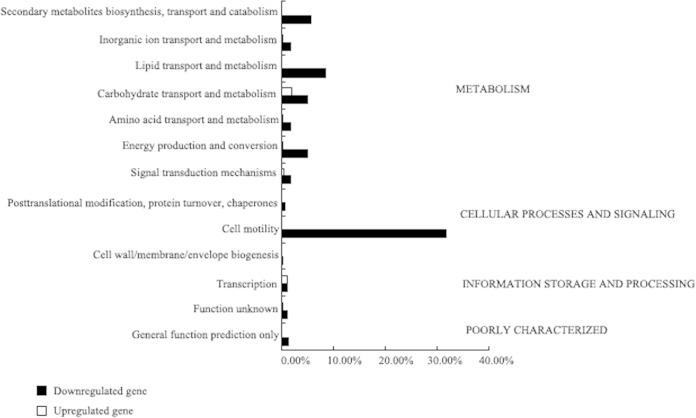
The genes differentially transcribed were classified into functional categories based on clusters of orthologous groups (COGs) (http://www.ncbi.nlm.nih.gov/COG), and the percentage of significantly upregulated and downregulated genes in CF74ΔT6SS in each COG category was determined. In CF74ΔT6SS, 12 COGs contained more repressed than activated genes. Many of these differentially regulated genes are involved in metabolism, including the utilization of glycerol, ethanolamine, and maltose (see Tables S2 and S3 in the supplemental material). The COG with the largest proportion of T6SS-regulated genes was the cell motility class, in which approximately 31.82% of the genes were downregulated significantly, with no genes in the class upregulated significantly (Fig. 1). The downregulated cell motility genes of CF74ΔT6SS were mainly flagellar genes, including fliC, flgM, flgK, fliD, flgL, motA, and fliT (see Fig. S1 in the supplemental material).
FIG 1.
T6SS-regulated genes in CF74. Shown is COG analysis of T6SS-regulated genes in CF74. Major COG categories are indicated on the right, while subcategories are listed on the left. The x axis represents the percentage of genes in the corresponding class. Genes that are activated or repressed in CF74ΔT6SS are indicated by white or black bars, respectively.
The T6SS affects the flagellar system at the transcriptional level.
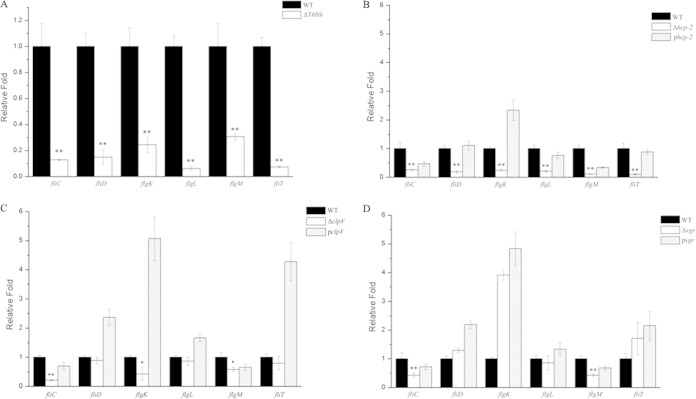
To confirm the RNA-Seq results, we selected six flagellar genes (fliC, flgM, flgK, fliD, flgL, and fliT) for qRT-PCR analysis of the wild-type CF74 and the mutant CF74ΔT6SS under the same culture conditions as for the RNA-Seq experiments. The relative expression levels of flagellar genes were normalized to that of a housekeeping gene, gapA. The results corresponded well to the RNA-Seq data (Table 3 and Fig. 2A). These results indicated that the loss of the T6SS resulted in changes in the expression of flagellar genes at the level of transcription.
TABLE 3.
Levels of expression of flagellar genes affected by T6SS
| Gene no. | Gene name | Function | Orientationa | Expression ratio |
|
|---|---|---|---|---|---|
| RNA-Seqb | RT-PCRc | ||||
| CFCDC_3602 | fliC | Bacterial flagellin C-terminal helical region | − | 2.940 | 2.966 |
| CFCDC_3603 | fliD | Flagellar capping protein | + | 2.471 | 2.745 |
| CFCDC_3605 | fliT | Flagellar biosynthesis protein | + | 2.878 | 3.772 |
| CFCDC_2668 | flgM | Anti-sigma28 factor | − | 2.127 | 1.709 |
| CFCDC_2679 | flgK | Flagellar hook-associated protein | + | 2.730 | 2.042 |
| CFCDC_2680 | flgL | Flagellar hook-associated protein | + | 1.911 | 4.004 |
Transcription direction (+, Watson strand; −, Crick strand).
Log2 of expression ratios (ratio of RPKM in wild-type and T6SS GI deletion mutant samples) obtained from RNA-Seq.
Log2 of expression ratios (ratio of RPKM in wild-type and T6SS GI deletion mutant samples) obtained from the RT-PCR.
FIG 2.
Relative expression levels of six flagellar genes (fliC, flgM, flgK, fliD, flgL, and fliT) at the level of transcription in different mutants in comparison to the wild type. (A) Effect of deletion of the T6SS GI (ΔT6SS) in comparison to the wild type (WT). (B to D) Effects of deletion of the individual T6SS genes clpV, hcp, and vgr. The mutants are marked as ΔclpV (CF74ΔclpV), Δhcp-2 (CF74Δhcp-2), and Δvgr (CF74Δvgr) and the complemented mutants as pclpV [CF74ΔclpV(pBAD24-clpV)], phcp-2 [CF74Δhcp-2(pBAD24-hcp-2)], and pvgr [CF74Δvgr(pBAD24-vgr)]. The y axis represents the relative fold change among different strains, and the x axis represents different flagellar genes. The statistical significance between the wild type and mutants was determined by Mann-Whitney U test. *, P < 0.05; **, P < 0.01. The error bars indicate 1 standard deviation.
The T6SS has three key genes, clpV, vgr, and hcp-2. ClpV is an ATPase that forms oligomeric complexes to energize the system for the secretion of effector proteins, while Vgr and Hcp-2 are effector proteins (1, 12–14, 17). To understand which of these proteins affected the expression of the flagellar genes at the level of transcription, we constructed three mutants (CF74ΔclpV, CF74Δhcp-2, and CF74Δvgr) and complemented strains [CF74ΔclpV(pBAD24-clpV), CF74Δhcp-2(pBAD24-hcp-2), and CF74Δvgr(pBAD24-vgr)], in which the expression of the six flagellar genes were measured by qRT-PCR analysis. It was shown (Fig. 2B to D) that fliC and flgM were downregulated in all three mutant strains and were restored upon complementation (P < 0.05). flgK was downregulated in all three mutants but significantly only in CF74Δhcp-2 (P < 0.01). fliD, flgL, and fliT were downregulated only in CF74Δhcp-2 and complemented back in CF74Δhcp-2(pBAD24-hcp-2) (P < 0.01). Overall, the six flagellar genes were significantly downregulated only in CF74Δhcp-2, similar to the outcome of the deletion of the entire T6SS in CF74ΔT6SS (P < 0.01) (Fig. 2A and B). Therefore, it seems that hcp-2 is the key gene of the T6SS that affects the expression of flagellar genes at the level of transcription.
The T6SS is involved in the secretion of FliC.
We examined the effect of the T6SS on FliC secretion by comparing the wild type, the mutants (CF74ΔT6SS, CF74ΔclpV, CF74Δvgr, and CF74Δhcp-2), and the complemented strains [CF74ΔclpV(pBAD24-clpV), CF74Δhcp-2(pBAD24-hcp-2), and CF74Δvgr(pBAD24-vgr)]. The strains were grown to an OD600 of 1.0 in LB medium, and the proteins from both culture supernatants and WCL were analyzed by Western blotting with a rabbit anti-flagellin antiserum that recognized the FliC protein. As shown in Fig. 3, in the culture supernatant and WCL, the bands of FliC in CF74ΔT6SS were obviously weaker than in the wild type. By densitometric quantification of relative band intensities normalized to GapA, we found that the FliC bands in the culture supernatants of CF74ΔclpV, CF74Δvgr, and CF74Δhcp-2 were decreased significantly (ranging from 30% to 66%) relative to the wild type and restored partially upon complementation. In the WCL, the FliC bands of CF74Δvgr and CF74Δhcp-2 were weaker than those of the wild type, but the FliC band of CF74ΔclpV was not clearly weaker. The absence of the cytosolic GapA protein in the culture supernatants indicates that the appearance of FliC in the culture supernatants was not a consequence of bacterial cell lysis. These results showed that the T6SS and its component proteins participated in the production and export of FliC.
FIG 3.
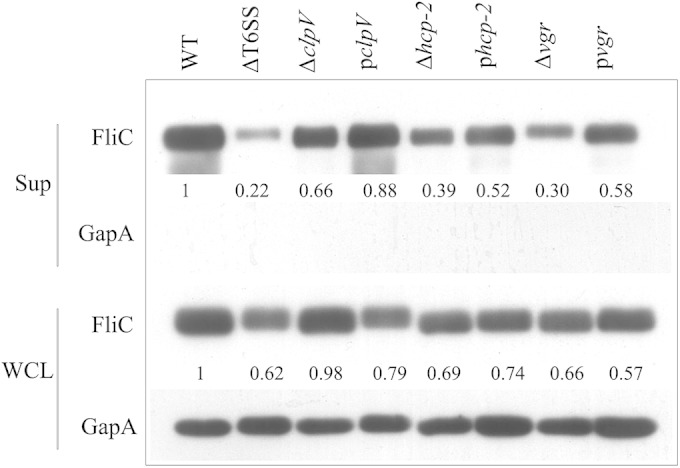
Effect of the T6SS on secretion of FliC. Shown is immunoblot analysis of proteins in the culture supernatant (Sup) and whole-cell lysates (WCL) prepared from mutants of C. freundii CF74 grown in LB medium. The WT, CF74ΔT6SS, CF74ΔclpV, CF74Δvgr, and CF74Δhcp-2 strains were uninduced; CF74ΔclpV(pBAD24-clpV), CF74Δhcp-2(pBAD24-hcp-2), and CF74Δvgr(pBAD24-vgr) were induced with 0.2% arabinose for 30 min. The position of a reactive band corresponding to FliC was detected with anti-FliC antibodies, and GapA was detected with anti-GapA antibodies. GapA was used as a loading control. The numbers represent densitometric quantifications of relative band intensities normalized to GapA and the wild type. pclpV, CF74ΔclpV(pBAD24-clpV); phcp-2, CF74Δhcp-2(pBAD24-hcp-2); pvgr, CF74Δvgr(pBAD24-vgr).
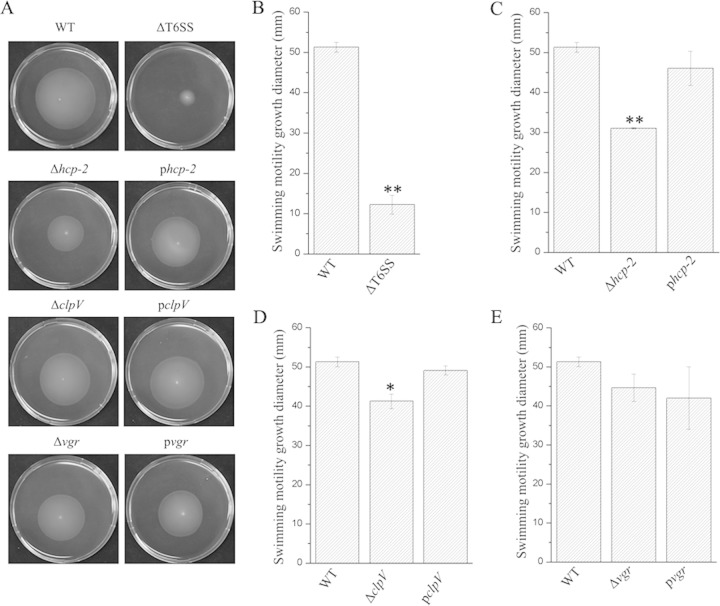
The T6SS and its component proteins affect motility.
To determine whether the T6SS affects motility, we used swimming motility assays to compare the wild-type strain, the mutants (CF74ΔT6SS, CF74ΔclpV, CF74Δvgr, and CF74Δhcp-2), and the complemented mutants [CF74ΔclpV(pBAD24-clpV), CF74Δhcp-2(pBAD24-hcp-2), and CF74Δvgr(pBAD24-vgr)]. The mutant strains were less motile than the wild type, and the motility was restored upon complementation (Fig. 4A). For CF74ΔT6SS, the diameter of the swimming motility halo was 12.99 mm, which was significantly decreased in comparison to a halo of 54.59 mm for the wild type (P < 0.01) (Fig. 4B). The halo of CF74Δhcp-2 was reduced more than those of the CF74Δvgr and CF74ΔclpV mutants (P < 0.05). Although the halo of CF74Δvgr was decreased compared with the wild type, it was not statistically significant (Fig. 4C to E). These data suggest that the T6SS, and in particular its effector Hcp, affected motility.
FIG 4.
Effect of the T6SS on motility. (A) Representative images of swimming motility of C. freundii CF74, CF74ΔT6SS, CF74ΔclpV, CF74Δvgr, and CF74Δhcp-2 and their complemented mutants [CF74ΔclpV(pBAD24-clpV), CF74Δhcp-2(pBAD24-hcp-2), and CF74Δvgr(pBAD24-vgr)]. Cells were inoculated with a toothpick from an overnight LB agar plate onto a swim plate (tryptone broth plus 0.3% agar) and photographed after 16 h of incubation at 30°C. The complemented mutants were induced with swim plates with 0.2% arabinose in the medium. (B to E) Quantification of the inhibition of the swimming motility halo by C. freundii CF74 mutants in swim plates. Statistical significance between the wild-type strain and mutants (CF74ΔT6SS, CF74ΔclpV, CF74Δvgr, and CF74Δhcp-2) was determined by a Mann-Whitney U test. *, P < 0.05; **, P < 0.01. pclpV, CF74ΔclpV(pBAD24-clpV); phcp-2, CF74Δhcp-2(pBAD24-hcp-2); pvgr, CF74Δvgr(pBAD24-vgr). The error bars indicate 1 standard deviation.
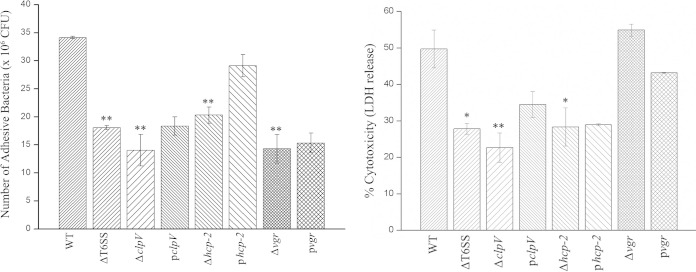
T6SS and its component proteins affect adhesion and cytotoxicity in HEp-2 cells.
The capacities to adhere to host cells were compared among the wild type, the deletion mutants, and the complemented strains under the same conditions. The deletion mutants of the entire T6SS and its component genes were defective in adhesion to HEp-2 cells (P < 0.01), which was restored partially upon complementation (Fig. 5, left). These results indicated that the T6SS was involved in the adherence of CF74 to host cells.
FIG 5.
Effects of the T6SS on adhesion and cytotoxicity to HEp-2 cells. (Left) Effects of T6SS on CF74 adherence to HEp-2 cells (MOI, 100). The performances of mutant strains were compared statistically to that of the wild-type strain, CF74. **, P < 0.01. (Right) The LDH released from HEp-2 cells was measured after exposure to C. freundii CF74, mutants, and their complemented mutants at 10 h using the optical density reading (A490/A630) (vertical axis). Significant differences between the wild type and mutants are indicated with asterisks: **, P < 0.01; *, P < 0.05. pclpV, CF74ΔclpV(pBAD24-clpV); phcp-2, CF74Δhcp-2(pBAD24-hcp-2); pvgr, CF74Δvgr(pBAD24-vgr). The error bars indicate 1 standard deviation.
CF74 showed high cytotoxicity to cultured HEp-2 cells (26). We hypothesized that the T6SS may play a role in cytotoxicity. Using an LDH release assay, we observed that, unlike CF74Δvgr, CF74ΔT6SS, CF74ΔclpV, and CF74Δhcp-2 induced lower LDH release by the HEp-2 cells than the wild-type, CF74 (P < 0.05) (Fig. 5, right), which was restored partially upon complementation, indicating that the T6SS contributes to the apoptotic death of HEp-2 cells in vitro.
DISCUSSION
C. freundii CF74 has a complete T6SS system located on a GI. We analyzed the effect of the T6SS GI on global gene expression at the level of transcription and showed that over 84 genes are affected by the loss of the T6SS in CF74. The expression of 82.14% of these genes was repressed in CF74ΔT6SS, indicating that in CF74, the T6SS or proteins secreted by the T6SS preferentially mediate positive regulation of gene expression. Further work is required to determine why so many genes are regulated by the T6SS or other genes on the GI. One or more of the T6SS genes or other genes on the GI must have affected the expression of such a large number of genes. There are several open reading frames (ORFs) of unknown function in the region deleted in CF74ΔT6SS. There is also an rhsA gene and 3 rhs-like elements. RhsA, a nuclease, has been shown to affect the expression of genes involved in transcription, RNA processing, nucleotide biosynthesis and metabolism, and amino acid biosynthesis in E. coli (37). rhsA is also embedded in the T6SS gene cluster in Dickeya dadantii and is likely to be exported through the T6SS (38). Therefore, it is possible that RhsA secreted through the T6SS affects gene expression.
Genes significantly downregulated in CF74ΔT6SS include those involved in glycerol and ethanolamine metabolism, while the significantly upregulated genes include those involved in maltose metabolism. It is interesting that the T6SS GI upregulates the ethanolamine utilization genes. Ethanolamine is abundant in the intestines of animals and can serve as both a carbon and a nitrogen source. Ethanolamine utilization has been associated with virulence (39, 40). Ethanolamine utilization genes have been shown to be upregulated in the intestines of mice in experimental Listeria monocytogenes infection (41) and similarly in the intestines of chickens in experimental S. enterica serovar Typhimurium infection (42). Ethanolamine can also induce virulence gene expression in E. coli O157:H7 (43). Therefore, it is likely that the upregulation of the ethanolamine utilization genes by the T6SS in C. freundii CF74 gives it a competitive advantage in the intestinal or environmental niche, where there is a rich source of ethanolamine. Note that the effect observed with CF74ΔT6SS may have been exerted by non-T6SS genes or regulatory elements on the GI rather than directly by the T6SS, as discussed above, which requires further investigation.
The cell motility COG has the largest proportion of T6SS-regulated genes, with approximately 31.82% of its genes being downregulated significantly in the T6SS deletion mutant, the majority of which are flagellar genes, including those encoding effector proteins, chaperones, and regulators. Mainly fliC, flgM, flgK, fliD, flgL, motA, and fliT were regulated by the T6SS in CF74. No flagellar genes were upregulated in the CF74ΔT6SS mutant.
The deletion of vgr, hcp-2, and clpV, key genes of the T6SS, resulted in changes in the expression of flagellar genes at the level of transcription. The expression of flagellar genes was downregulated significantly in CF74Δhcp-2, but not to the same extent as in CF74ΔT6SS. Other T6SS genes are also possibly important, such as icmF, which was not examined in this study. IcmF is an ATPase involved in T6SS protein secretion, and IcmF mutants prevent Hcp secretion (2, 8–10). It has been reported that the ΔicmF strain decreased the expression of flhC, flhD, flgM, and fliA flagellar genes at the level of transcription in avian-pathogenic E. coli (APEC) (15). Our results suggested that deletion of T6SS or its key genes was detrimental to the expression of the flagellar regulon at the level of transcription, as was found in E. coli (15).
FliC, a component of the flagellum filament, is transported from the cytoplasm by specific transport systems, such as the T3SS and T4SS (19, 20), and is polymerized with the help of the cap protein FliD, producing long helical flagellar filaments (44). In the present study, we found that FliC was reduced, not only at the level of transcription, but also at the level of translation in the T6SS mutant, indicating that the reduction of FliC was due to not only altered expression of fliC, but also the inability to transport FliC, possibly via the type VI secretion system. Thus, it is likely that the T6SS is involved in secretion of FliC in C. freundii. Further studies are needed to clarify how the T6SS contributes to the production and secretion of FliC.
Swimming motility is dependent on flagella. In the present study, motility tests showed that CF74ΔT6SS was less motile, which corroborates a previous report that deletion of the T6SS component, icmF, in V. cholerae and APEC produced reduced motility (15, 45). Among the 3 T6SS genes tested—vgr, hcp-2, and clpV—hcp-2 seems to have the greatest effect, while clpV has a less significant effect on motility. Interestingly, both ClpV and IcmF are ATPases, with the latter reported to affect the production and secretion of the FliC protein in both E. coli and V. cholerae (11, 45). It is likely that IcmF has a similar role in C. freundii, which may complement the activity of clpV in the clpV deletion mutant.
Reduced motility in T6SS deletion mutants is attributed to reduced production and secretion of the FliC protein. We found that the loss of Hcp-2 resulted in changes in FliC at the level of transcription and translation and affected motility. Thus, Hcp-2 is likely to be one of the important T6SS proteins that play a role in FliC protein production and export, and the mechanisms remain to be determined.
Our previous study showed that CF74 has strong adhesion and high cytotoxicity to HEp-2 cells (26). In this study, we found that the T6SS played a role in the adhesion and cytotoxicity of CF74 to host cells. All 3 T6SS genes tested, vgr, clpV, and hcp2, adversely affected the adhesion of C. freundii to HEp-2 cells, and two of the three genes tested, clpV and hcp-2, contributed to the cytotoxicity of C. freundii to HEp-2 cells, while vgr seems to have no effect on cytotoxicity. Our results are consistent with findings in other bacterial pathogens of the role of T6SS in adherence and cytotoxicity to host cells. It has been reported that the T6SS in APEC is involved in adherence to host cells (46), the Bordetella bronchiseptica T6SS mediates cytotoxicity in murine macrophages (47), and the Campylobacter jejuni T6SS confers cytotoxicity to red blood cells (48). Since FliC contributes to cytotoxicity (49), it is possible that the T6SS of CF74 contributes to the apoptotic death of host cells in vitro indirectly by affecting the production and secretion of FliC.
In conclusion, we show that deletion of the T6SS of C. freundii CF74 and its key proteins (ClpV, Hcp-2, and Vgr) affected the production and secretion of the flagellin protein FliC and affected motility. Deletion of the T6SS also affected the adhesion and cytotoxicity of CF74 to HEp-2 cells. Further, the T6SS GI has a much greater effect on the global expression of genes, with more than 84 genes differentially affected, which remains to be further investigated.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (no. 81301401) and the Project of the State Key Laboratory for Infectious Disease Prevention and Control (2011SKLID209).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.03071-14.
REFERENCES
- 1.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordonez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312:1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A 103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dudley EG, Thomson NR, Parkhill J, Morin NP, Nataro JP. 2006. Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol Microbiol 61:1267–1282. doi: 10.1111/j.1365-2958.2006.05281.x. [DOI] [PubMed] [Google Scholar]
- 4.Schwarz S, West TE, Boyer F, Chiang WC, Carl MA, Hood RD, Rohmer L, Tolker-Nielsen T, Skerrett SJ, Mougous JD. 2010. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog 6:e1001068. doi: 10.1371/journal.ppat.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murdoch SL, Trunk K, English G, Fritsch MJ, Pourkarimi E, Coulthurst SJ. 2011. The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. J Bacteriol 193:6057–6069. doi: 10.1128/JB.05671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schell MA, Ulrich RL, Ribot WJ, Brueggemann EE, Hines HB, Chen D, Lipscomb L, Kim HS, Mrazek J, Nierman WC, Deshazer D. 2007. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol Microbiol 64:1466–1485. doi: 10.1111/j.1365-2958.2007.05734.x. [DOI] [PubMed] [Google Scholar]
- 7.Parsons DA, Heffron F. 2005. sciS, an icmF homolog in Salmonella enterica serovar Typhimurium, limits intracellular replication and decreases virulence. Infect Immun 73:4338–4345. doi: 10.1128/IAI.73.7.4338-4345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A. 2009. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J 28:315–325. doi: 10.1038/emboj.2008.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu HY, Chung PC, Shih HW, Wen SR, Lai EM. 2008. Secretome analysis uncovers an Hcp-family protein secreted via a type VI secretion system in Agrobacterium tumefaciens. J Bacteriol 190:2841–2850. doi: 10.1128/JB.01775-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng J, Leung KY. 2007. Dissection of a type VI secretion system in Edwardsiella tarda. Mol Microbiol 66:1192–1206. doi: 10.1111/j.1365-2958.2007.05993.x. [DOI] [PubMed] [Google Scholar]
- 11.Schlieker C, Zentgraf H, Dersch P, Mogk A. 2005. ClpV, a unique Hsp100/Clp member of pathogenic proteobacteria. Biol Chem 386:1115–1127. doi: 10.1515/BC.2005.128. [DOI] [PubMed] [Google Scholar]
- 12.Bingle LE, Bailey CM, Pallen MJ. 2008. Type VI secretion: a beginner's guide. Curr Opin Microbiol 11:3–8. doi: 10.1016/j.mib.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Bonemann G, Pietrosiuk A, Mogk A. 2010. Tubules and donuts: a type VI secretion story. Mol Microbiol 76:815–821. doi: 10.1111/j.1365-2958.2010.07171.x. [DOI] [PubMed] [Google Scholar]
- 14.Cascales E. 2008. The type VI secretion toolkit. EMBO Rep 9:735–741. doi: 10.1038/embor.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Pace F, Boldrin de Paiva J, Nakazato G, Lancellotti M, Sircili MP, Guedes Stehling E, Dias da Silveira W, Sperandio V. 2011. Characterization of IcmF of the type VI secretion system in an avian pathogenic Escherichia coli (APEC) strain. Microbiology 157:2954–2962. doi: 10.1099/mic.0.050005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Solomon JM, Rupper A, Cardelli JA, Isberg RR. 2000. Intracellular growth of Legionella pneumophila in Dictyostelium discoideum, a system for genetic analysis of host-pathogen interactions. Infect Immun 68:2939–2947. doi: 10.1128/IAI.68.5.2939-2947.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y, Tao J, Yu H, Ni J, Zeng L, Teng Q, Kim KS, Zhao GP, Guo X, Yao Y. 2012. Hcp family proteins secreted via the type VI secretion system coordinately regulate Escherichia coli K1 interaction with human brain microvascular endothelial cells. Infect Immun 80:1243–1251. doi: 10.1128/IAI.05994-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filloux A, Hachani A, Bleves S. 2008. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 154:1570–1583. doi: 10.1099/mic.0.2008/016840-0. [DOI] [PubMed] [Google Scholar]
- 19.Badea L, Beatson SA, Kaparakis M, Ferrero RL, Hartland EL. 2009. Secretion of flagellin by the LEE-encoded type III secretion system of enteropathogenic Escherichia coli. BMC Microbiol 9:30. doi: 10.1186/1471-2180-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miao EA, Andersen-Nissen E, Warren SE, Aderem A. 2007. TLR5 and Ipaf: dual sensors of bacterial flagellin in the innate immune system. Semin Immunopathol 29:275–288. doi: 10.1007/s00281-007-0078-z. [DOI] [PubMed] [Google Scholar]
- 21.Guerrant RL, Dickens MD, Wenzel RP, Kapikian AZ. 1976. Toxigenic bacterial diarrhea: nursery outbreak involving multiple bacterial strains. J Pediatr 89:885–891. doi: 10.1016/S0022-3476(76)80591-4. [DOI] [PubMed] [Google Scholar]
- 22.Guarino A, Capano G, Malamisura B, Alessio M, Guandalini S, Rubino A. 1987. Production of Escherichia coli STa-like heat-stable enterotoxin by Citrobacter freundii isolated from humans. J Clin Microbiol 25:110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karasawa T, Ito H, Tsukamoto T, Yamasaki S, Kurazono H, Faruque SM, Nair GB, Nishibuchi M, Takeda Y. 2002. Cloning and characterization of genes encoding homologues of the B subunit of cholera toxin and the Escherichia coli heat-labile enterotoxin from clinical isolates of Citrobacter freundii and E. coli. Infect Immun 70:7153–7155. doi: 10.1128/IAI.70.12.7153-7155.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parida SN, Verma IC, Deb M, Bhujwala RA. 1980. An outbreak of diarrhea due to Citrobacter freundii in a neonatal special care nursery. Indian J Pediatr 47:81–84. doi: 10.1007/BF02900180. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt H, Montag M, Bockemuhl J, Heesemann J, Karch H. 1993. Shiga-like toxin II-related cytotoxins in Citrobacter freundii strains from humans and beef samples. Infect Immun 61:534–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bai L, Xia S, Lan R, Liu L, Ye C, Wang Y, Jin D, Cui Z, Jing H, Xiong Y, Bai X, Sun H, Zhang J, Wang L, Xu J. 2012. Isolation and characterization of cytotoxic, aggregative Citrobacter freundii. PLoS One 7:e33054. doi: 10.1371/journal.pone.0033054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komeili A, Vali H, Beveridge TJ, Newman DK. 2004. Magnetosome vesicles are present before magnetite formation, and MamA is required for their activation. Proc Natl Acad Sci U S A 101:3839–3844. doi: 10.1073/pnas.0400391101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Philippe N, Alcaraz JP, Coursange E, Geiselmann J, Schneider D. 2004. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51:246–255. doi: 10.1016/j.plasmid.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 29.Hamashima H, Iwasaki M, Arai T. 1995. A simple and rapid method for transformation of Vibrio species by electroporation. Methods Mol Biol 47:155–160. [DOI] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Cox MP, Peterson DA, Biggs PJ. 2010. SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 11:485. doi: 10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. 2011. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol 12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 35.Cong Y, Wang J, Chen Z, Xiong K, Xu Q, Hu F. 2011. Characterization of swarming motility in Citrobacter freundii. FEMS Microbiol Lett 317:160–171. doi: 10.1111/j.1574-6968.2011.02225.x. [DOI] [PubMed] [Google Scholar]
- 36.Rashid MH, Kornberg A. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 97:4885–4890. doi: 10.1073/pnas.060030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aggarwal K, Lee KH. 2011. Overexpression of cloned RhsA sequences perturbs the cellular translational machinery in Escherichia coli. J Bacteriol 193:4869–4880. doi: 10.1128/JB.05061-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koskiniemi S, Lamoureux JG, Nikolakakis KC, t' Kint de Roodenbeke C, Kaplan MD, Low DA, Hayes CS. 2013. Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci U S A 110:7032–7037. doi: 10.1073/pnas.1300627110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garsin DA. 2010. Ethanolamine utilization in bacterial pathogens: roles and regulation. Nat Rev Microbiol 8:290–295. doi: 10.1038/nrmicro2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Staib L, Fuchs TM. 2014. From food to cell: nutrient exploitation strategies of enteropathogens. Microbiology 160:1020–1039. doi: 10.1099/mic.0.078105-0. [DOI] [PubMed] [Google Scholar]
- 41.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, Barthelemy M, Vergassola M, Nahori MA, Soubigou G, Regnault B, Coppee JY, Lecuit M, Johansson J, Cossart P. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 42.Harvey PC, Watson M, Hulme S, Jones MA, Lovell M, Berchieri A Jr, Young J, Bumstead N, Barrow P. 2011. Salmonella enterica serovar Typhimurium colonizing the lumen of the chicken intestine grows slowly and upregulates a unique set of virulence and metabolism genes. Infect Immun 79:4105–4121. doi: 10.1128/IAI.01390-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luzader DH, Clark DE, Gonyar LA, Kendall MM. 2013. EutR is a direct regulator of genes that contribute to metabolism and virulence in enterohemorrhagic Escherichia coli O157:H7. J Bacteriol 195:4947–4953. doi: 10.1128/JB.00937-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ikeda T, Oosawa K, Hotani H. 1996. Self-assembly of the filament capping protein, FliD, of bacterial flagella into an annular structure. J Mol Biol 259:679–686. doi: 10.1006/jmbi.1996.0349. [DOI] [PubMed] [Google Scholar]
- 45.Das S, Chakrabortty A, Banerjee R, Chaudhuri K. 2002. Involvement of in vivo induced icmF gene of Vibrio cholerae in motility, adherence to epithelial cells, and conjugation frequency. Biochem Biophys Res Commun 295:922–928. doi: 10.1016/S0006-291X(02)00782-9. [DOI] [PubMed] [Google Scholar]
- 46.Ma J, Bao Y, Sun M, Dong W, Pan Z, Zhang W, Lu C, Yao H. 2014. Two functional type VI secretion systems in avian pathogenic Escherichia coli are involved in different pathogenic pathways. Infect Immun 82:3867–3879. doi: 10.1128/IAI.01769-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weyrich LS, Rolin OY, Muse SJ, Park J, Spidale N, Kennett MJ, Hester SE, Chen C, Dudley EG, Harvill ET. 2012. A Type VI secretion system encoding locus is required for Bordetella bronchiseptica immunomodulation and persistence in vivo. PLoS One 7:e45892. doi: 10.1371/journal.pone.0045892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bleumink-Pluym NM, van Alphen LB, Bouwman LI, Wosten MM, van Putten JP. 2013. Identification of a functional type VI secretion system in Campylobacter jejuni conferring capsule polysaccharide sensitive cytotoxicity. PLoS Pathog 9:e1003393. doi: 10.1371/journal.ppat.1003393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olsen JE, Hoegh-Andersen KH, Casadesus J, Rosenkranzt J, Chadfield MS, Thomsen LE. 2013. The role of flagella and chemotaxis genes in host pathogen interaction of the host adapted Salmonella enterica serovar Dublin compared to the broad host range serovar S. Typhimurium. BMC Microbiol 13:67. doi: 10.1186/1471-2180-13-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.