Abstract
We report a novel host-associated virulence plasmid in Rhodococcus equi, pVAPN, carried by bovine isolates of this facultative intracellular pathogenic actinomycete. Surprisingly, pVAPN is a 120-kb invertron-like linear replicon unrelated to the circular virulence plasmids associated with equine (pVAPA) and porcine (pVAPB variant) R. equi isolates. pVAPN is similar to the linear plasmid pNSL1 from Rhodococcus sp. NS1 and harbors six new vap multigene family members (vapN to vapS) in a vap pathogenicity locus presumably acquired via en bloc mobilization from a direct predecessor of equine pVAPA. Loss of pVAPN rendered R. equi avirulent in macrophages and mice. Mating experiments using an in vivo transconjugant selection strategy demonstrated that pVAPN transfer is sufficient to confer virulence to a plasmid-cured R. equi recipient. Phylogenetic analyses assigned the vap multigene family complement from pVAPN, pVAPA, and pVAPB to seven monophyletic clades, each containing plasmid type-specific allelic variants of a precursor vap gene carried by the nearest vap island ancestor. Deletion of vapN, the predicted “bovine-type” allelic counterpart of vapA, essential for virulence in pVAPA, abrogated pVAPN-mediated intramacrophage proliferation and virulence in mice. Our findings support a model in which R. equi virulence is conferred by host-adapted plasmids. Their central role is mediating intracellular proliferation in macrophages, promoted by a key vap determinant present in the common ancestor of the plasmid-specific vap islands, with host tropism as a secondary trait selected during coevolution with specific animal species.
INTRODUCTION
Rhodococcus equi is a Gram-positive aerobic coccobacillus of the Corynebacteriales associated with chronic or subacute pyogenic infections (1, 2). A normal soil inhabitant, the bacterium uses manure as a growth substrate, multiplies in the herbivore's large intestine, and is ubiquitous in the farm environment. Transmission occurs via contaminated dust particles, mostly through airborne exposure (3, 4). R. equi is the causative agent of a major infectious disease of horses that affects young foals worldwide. The infection is characterized by multifocal purulent bronchopneumonia, often accompanied by ulcerative or abcessating lesions in the intestine (1, 5). While best known as an equine pathogen, R. equi also infects other animal species (1, 6–8). In abattoir surveys, R. equi is frequently recovered from porcine submaxillary lymph nodes with granulomatous lesions as well as from apparently healthy pigs (9–11). In cattle, it is typically isolated from caseating abscesses in respiratory lymph nodes resembling bovine tuberculosis (TB) lesions (12). R. equi is also recognized as an opportunistic pathogen in humans, where it causes severe TB-like purulent cavitary pneumonia, bacteremia, and localized extrapulmonary infections (8, 13, 14).
R. equi pathogenesis depends on the capacity of the bacterium to survive and replicate within host macrophages (15–18). In equine isolates, this ability is conferred by a conjugative circular plasmid of 80 kb (19–21) that promotes intravacuolar survival by interfering with phagosome maturation (22). These properties are mediated by the vap pathogenicity island (PAI) (23), a horizontal gene transfer (HGT) locus (24). A hallmark of the vap PAI is the presence of a multigene family encoding homologous virulence-associated proteins (Vaps) (20, 24, 25). One of them, VapA, a 19-kDa secreted protein, is essential for virulence. A single vapA gene deletion causes strong attenuation comparable to that caused by loss of the plasmid, with an inability to both proliferate in macrophages and survive in vivo in mice (26, 27).
Emerging evidence suggests that the virulence plasmid may also play a key role in R. equi host tropism. Early studies showed that VapA-encoding virulence plasmids were typical of equine strains (28, 29), while a second plasmid type, encoding VapB, a VapA variant (24), was common among nonequine (pig and human) isolates (10, 30–33). Recently, the existence of a third type of R. equi virulence plasmid was identified in bovine and human isolates that were initially deemed to be “plasmidless” (because vapA and vapB negative) but that tested positive for a traA plasmid conjugal transfer gene marker (34). Molecular epidemiological analysis of a global collection of R. equi isolates established that the vapA+, vapB+, and novel vapAB-negative plasmid types were each associated with a specific nonhuman host, i.e., equine, porcine, and bovine, respectively (34). Using a unified nomenclature, these plasmids were designated pVAPA, pVAPB, and pVAPN (for “noA-noB”), respectively (1, 24). In contrast to their unique animal species specificity, the three host-adapted virulence plasmid types were commonly detected in human isolates. Besides pointing to a zoonotic origin of infection, this lack of plasmid type selectivity was consistent with humans being an opportunistic, nonadapted host for R. equi (34).
Sequencing of the pVAPA and pVAPB virulence plasmids revealed that they are essentially the same circular replicon (24). The analyzed plasmids, pVAPA1037 and pVAPB1593 (numerical suffix indicates the source strain according to proposed harmonized nomenclature for R. equi virulence plasmids) (24), shared a virtually identical backbone encoding replication/partitioning and conjugal transfer functions. In contrast, the vap PAI was more divergent, differing in both size and vap gene complement, i.e. ≈21 kb and nine vap genes for pVAPA (vapA, -C, -D, -E, -G, and -H and the pseudogenes vapF, -I, and -X) versus ≈15 kb and six vap genes for pVAPB (vapB, -J, -K1, -K2, -L, and -M) (24). In addition to major Vap polypeptide sequence diversification, vap multigene family rearrangements and insertion/deletions affecting adjacent genes accounted for the PAI differences. This suggested that the vap PAIs were evolving at a higher rate than the conserved housekeeping backbone, consistent with diversifying selection and a possible role in host-specific adaptation (24).
Here, we report the genomic analysis and characterization of pVAPN, the bovine-type R. equi virulence plasmid.
MATERIALS AND METHODS
Strains, culture conditions, and reagents.
R. equi PAM1571 is a prototypic traA+ vapAB-negative bovine strain (34) isolated from a heifer's mediastinal lymph node with pyogranulomatous lesions (kindly provided by F. Quigley, Central Veterinary Research Laboratory, Ireland) (12). Its plasmid-cured derivative, PAM1571−, was obtained by subjecting wild-type bacteria to an electroporation pulse of 12.5 kV/cm at 1,000 Ω and 25 μF (GenePulser Xcell; Bio-Rad) followed by six cycles of plating and single-colony subculturing in liquid medium at 37°C (27). These two strains are henceforth designated 1571 and 1571−, respectively. R. equi PAM2012 is another traA+ vapAB-negative bovine strain, isolated in Germany from a case of lymphadenitis in cattle (kindly provided by C. Lämmler, Veterinary Faculty, University of Giessen) (35). R. equi 103S is the reference genome strain, a low-passage clone of equine clinical isolate 103+ used in different laboratories (24). Its isogenic 103SΔvapA and plasmid-cured 103S− derivatives were described previously (27). The presence of the virulence plasmid was routinely checked in all strains by PCR using suitable oligonucleotide primers (see Table S1 in the supplemental material). R. equi was grown in brain heart infusion (BHI; Difco-BD) or Luria-Bertani (LB; Sigma) medium at 30°C unless stated otherwise. The cloning host strain Escherichia coli DH5α was grown at 37°C in LB medium. Media were supplemented with 1.5% (wt/vol) agar and/or antibiotics as appropriate. Fluid cultures were incubated with shaking (200 rpm). Chemicals and primers were purchased from Sigma-Aldrich unless stated otherwise.
DNA techniques.
Total DNA extraction and purification from R. equi, PCR techniques, DNA fragment purification and electrophoresis, recombinant DNA techniques, and plasmid purification and electroporation were performed as previously described (27, 34, 36). For pulsed-field gel electrophoresis (PFGE), plugs were formed by embedding R. equi cells from 1-ml aliquots of stationary (24-h) BHI cultures in melted 1% agarose Tris-EDTA (TE). Plugs were incubated in 1 mg/ml lysozyme, 50 mM NaCl, 10 mM Tris-HCl, pH 8.0, at 37°C for 2 h, washed in 50 mM EDTA, 20 mM Tris-HCl, pH 8.0, and incubated in 1 mg/ml proteinase K, 100 mM EDTA, 0.2 g/ml sucrose, 10 mM Tris-HCl, pH 8.0, at 50°C overnight. Plugs were then loaded into a 1% Pulsed Field Certified agarose gel (Bio-Rad) prepared with 0.5× Tris-borate-EDTA (TBE) buffer. DNA was separated in a Chef-DR II pulsed-field electrophoresis system (Bio-Rad) at a voltage of 5 V/cm2, with a switch time ramping from 20 to 30 s and a 23-h run time at 14°C. Southern blotting was performed by transferring resolved DNA fragments onto a positively charged nylon membrane after treatment of the PFGE gels with 0.25 M HCl for 30 min followed by denaturing solution (1.5 M NaCl, 0.4 M NaOH) for 20 min (twice) and neutralizing solution (1.5 M NaCl, 0.5 M Tris-Cl2 [pH 7.0]) for 20 min (twice). Membranes were hybridized by using a specific vapN-vapQ PCR fragment (see Table S1 in the supplemental material) labeled with digoxigenin (DIG High Prime DNA labeling and detection kit; Roche).
pVAPN sequencing and phylogenetic analyses.
pVAPN1571 was electroeluted from preparative PFGE gels by using a model 422 apparatus (Bio-Rad) and paired-end (2 × 36 bp) sequenced in an Illumina (Solexa) II genome analyzer at the Edinburgh Genomics Facility. To complete the plasmid assembly, host strain 1571 DNA was paired-end (2× 100 bp) sequenced from a 500-bp PCR-free library using an Illumina HiSeq 2000 sequencing system at the Beijing Genomics Institute. pVAPN2012 was entirely sequenced by using the latter approach. Reads were assessed for quality by using FastQC and then trimmed for adaptors by using Scythe and for low-quality reads by using Sickle. De novo assembly was performed by using SPAdes followed by manual verification by PCR mapping and Sanger resequencing of specific regions. The 5′-end telomeric sequence of pVAPN was experimentally confirmed (37) using the suicide vector pSelAct (38) and vector-encoded apramycin resistance for selecting positive clones. The pVAPN sequence was manually curated and annotated in Artemis by using the software and databases listed in Table S2 in the supplemental material. For phylogenetic analyses, orthologs were identified by reciprocal tBlastx analysis with 30% identity over >60% of the protein sequence as a minimum similarity score. Paralogous genes predicted based on the topology of neighbor-joining trees and pseudogenes (except vap pseudogenes) were avoided. Translated products from each ortholog cluster were aligned by Muscle (except where otherwise stated) and back-translated in Mega5. The best evolutionary model for nucleotide substitution was selected according to Akaike information criterion (AIC) in jModelTest. For multilocus sequence alignment (MLSA), gene alignments were concatenated with Seaview. Maximum likelihood (ML) trees were constructed in PhyML. See Table S2 in the supplemental material for bioinformatics and phylogenetic analysis software references/URLs.
Construction of the vapN deletion mutant.
The vapN gene was in-frame deleted from pVAPN1571 by double homologous recombination (36) using 5-fluorocytosine counterselection (38). Briefly, oligonucleotide primer pairs Nmutant_a (EcoRI)/Nmutant_b1 (XmaI) and Nmutant_c1 (XmaI)/Nmutant_d (SpeI) (see Table S1 in the supplemental material) were used to PCR amplify two DNA fragments of 908 and 910 bp carrying the last three 5′-terminal and four 3′-terminal codons of vapN plus adjacent upstream and downstream regions, respectively. The PCR amplicons were joined via the XmaI site introduced by the Nmutant_b1 and Nmutant_c1 primers, the ligation product was inserted into the pSelAct vector (38) using the external SpeI and EcoRI sites introduced by primers Nmutant_a and Nmutant_d, and the resulting plasmid was electroporated into 1571. Allele exchange was checked by PCR mapping using suitable primers (see Table S1 in the supplemental material), and the in-frame deletion was confirmed by DNA sequencing.
Mating experiments.
Transfer of the virulence plasmid between R. equi bacteria was investigated by using a mating protocol essentially as previously described (39). Cultures of donor and recipient R. equi strains grown overnight in BHI were harvested by centrifugation, resuspended in phosphate-buffered saline (PBS) to a cell density of ≈107 CFU, mixed ≈1:1, and spotted in a 5-μl drop onto BHI agar. The recipient 103S− bacteria carried a chromosomal rifampin resistance (Rmpr) marker. 103S−RmpR bacteria were isolated by selection of spontaneously resistant mutants on increasing concentrations of rifampin, from 25 to 100 μg/ml, and stabilization by repeated subculturing in the presence of the highest concentration of the antibiotic. After incubation of the mating mixture at 30°C for 72 h, bacteria were collected in 1 ml of PBS, serially diluted, and plated onto BHI agar without and with supplementation with 100 μg/ml rifampin. At this rifampin concentration, no Rmpr colonies were detected in the donor-only control plates. Transconjugants were identified among Rmpr colonies by simultaneous PCR detection of virulence plasmid-specific markers and of recipient-specific chromosomal gene markers using ad hoc oligonucleotide primers (see Table S1 in the supplemental material).
Macrophage cultures and infection assay.
Low-passage murine J774A.1 macrophages and human monocyte-like THP-1 cells were obtained from the ATCC and cultured at 37°C with 5% CO2 in Dulbecco's minimal essential medium (DMEM) supplemented with 10% decomplemented fetal bovine serum, 2 mM glutamine, and 1 mM pyruvate. THP-1 cells were initially grown in suspension in RPMI 1640 medium with the same supplements. Cells were seeded onto 24-well plates at a density of ≈2 × 105 cells/well and incubated overnight in DMEM and, for THP-1 monocytes, in the presence of 50 ng/ml phorbol 12-myristate 13-acetate (PMA) to allow differentiation into macrophages. Infection assays were performed on ≈80% confluent macrophage monolayers as previously described (40). Intracellular proliferation data were normalized to the initial counts at time zero (t = 0) by using an “intracellular growth coefficient” (IGC) according to the formula IGC = (IBn − IB0)/IB0, where IBn and IB0 are the intracellular bacterial numbers at t = n and t = 0, respectively (40, 41).
Mouse infections.
Experiments were performed at the Animal Facility of the School of Biological Sciences of the University of Edinburgh using in-house-bred 6- to 8-week-old BALB/c mice. Mouse intranasal and intravenous (i.v.) infections and lung competitive virulence assays were performed as previously described (27). The relative proportions of competing bacteria were calculated by analyzing at least 40 random colonies from the plated organ homogenate by PCR using suitable oligonucleotide primers (see Table S1 in the supplemental material). Competitive index (CI) values were calculated by using the formula CI = (test/reference log CFU ratio at t = n)/(input test/reference log CFU ratio in the inoculum) (27). Mouse experiments were approved by the University of Edinburgh Ethical Review Committee and were covered by a project license granted by the United Kingdom Home Office under the Animals (Scientific Procedures) Act of 1986.
Statistics.
Intracellular proliferation and uptake data were compared by using two-way analysis of variance (ANOVA) and one-way ANOVA, respectively, followed by Šidák post hoc multiple-comparison tests. One-sample Student's t tests were used to determine if CI values differed significantly from 1 (i.e., the expected CI value if the ratio of the competing strains remains the same with respect to the ratio at t = 0). Statistical analyses were performed by using Prism 6.0 software (GraphPad, San Diego, CA).
Nucleotide sequence accession numbers.
The pVAPN1571 and pVAPN2012 genome sequences have been deposited in GenBank under accession no. KF439868 and KP851975, respectively.
RESULTS AND DISCUSSION
Identification and sequencing of pVAPN.
Attempts to isolate the novel traA+ vapAB-negative plasmid type (34) from 1571 and other bovine isolates by using the procedure for R. equi circular virulence plasmid extraction (34) were unsuccessful. However, PFGE analysis of undigested genomic DNA revealed a distinct band in the range of ≈100 kb in all bovine strains tested from our collection (n = 22). This band was not detected in R. equi strains carrying a circular virulence plasmid, e.g., 103S harboring pVAPA (Fig. 1). Similarity searches of exploratory low-coverage whole-genome 454 pyrosequencing assemblies from 1571 with the pVAPA reference sequence from R. equi 103S (40) identified contigs harboring vap PAI-homologous genes. Southern blotting using a probe from this novel vap PAI identified the ≈100 kb PFGE band as the putative pVAPN virulence plasmid (Fig. 1). Most (95%) tested traA+ vapAB-negative bovine isolates (34) were positive for pVAPN by PCR using a plasmid-specific vap PAI marker (vapN, the counterpart of the equine vapA and porcine vapB genes [see below]). The ≈100-kb band from strain 1571 was isolated from PFGE gels and shotgun sequenced. The pVAPN1571 genome sequence was completed as described in Materials and Methods.
FIG 1.
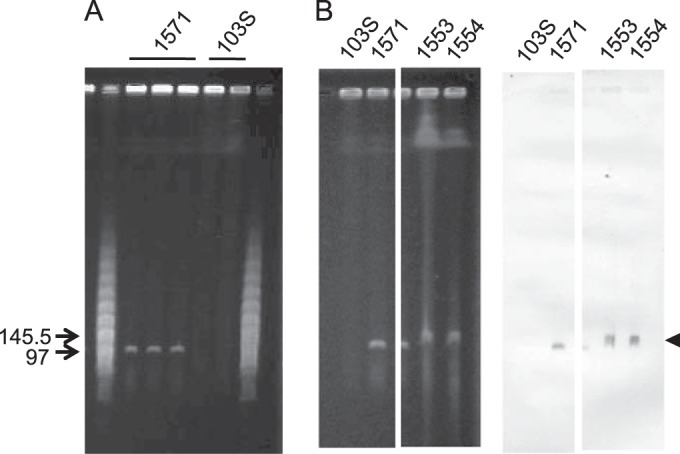
Detection of pVAPN by PFGE. (A) Genomic DNA of bovine isolate 1571 and equine isolate 103S; three and two independent lysates per strain are shown. Relevant positions of the lambda PFGE marker (New England BioLabs) are indicated. pVAPN is observable as a distinct band of ≈100 kb in the bovine isolate. (B) Southern blot analysis of bovine isolates PAM1571, PAM1533, and PAM1554 (strain 103S was used as a negative control). (Left) Relevant sections of the PFGE gel; (right) membrane hybridized with a pVAPN-specific DNA probe (600-bp fragment encompassing the 3′ region of vapN and the 5′ region of vapQ). The arrow indicates the pVAPN band.
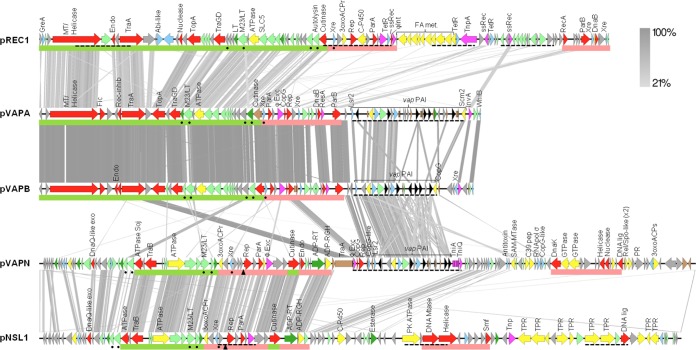
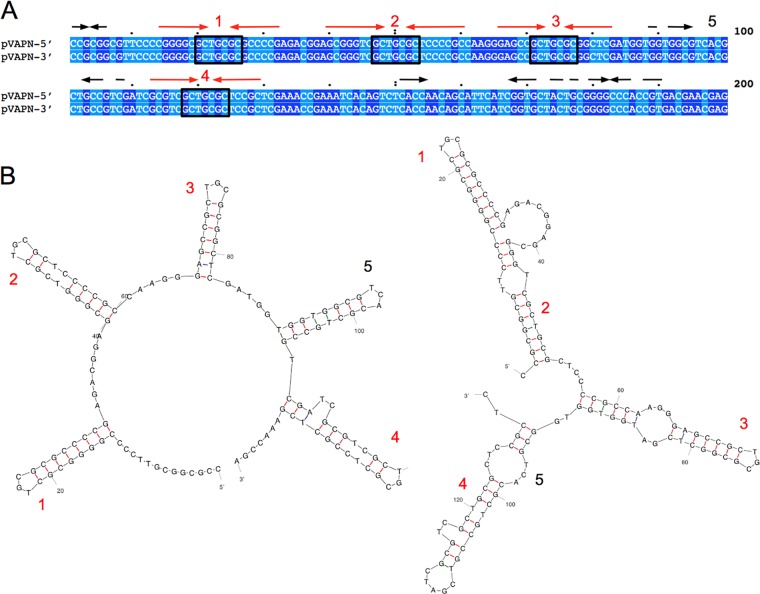
pVAPN1571 is 119,931 bp long and contains 148 open reading frames (ORFs), 10 of which are pseudogenes (Fig. 2). The average G+C content is 66.2%, similar to that of R. equi genomic DNA (68.7%) (40). pVAPN1571 is predicted to be a linear replicon based on its PFGE migration pattern (42), the presence of a traB conjugal translocase determinant (see below), phylogenetic relatedness to other Rhodococcus linear plasmids (Fig. 3A), and the presence of telomeric invertron-like terminal inverted repeats (TIRs) with multiple palindromic secondary structures (Fig. 4) (43–45). The TIR sequences of pVAPN are 569 bp long and 99% identical. The nucleotide sequence of a second example of the pVAPN plasmid, from a bovine isolate from Germany (PAM2012), was virtually identical to that of pVAPN1571 except for the presence of two additional ORFs at the left end (see Fig. S1 in the supplemental material).
FIG 2.
Genome alignments of the linear virulence plasmid pVAPN, circular virulence plasmids pVAPA and pVAPB, and the respective closest homologs from nonpathogenic rhodococcal species (pNSL1 from Rhodococcus sp. NS1 [48] and pREC1 from R. erythropolis [44]). The alignments were built with EasyFig (http://easyfig.sourceforge.net/). The circular plasmids (pVAPA, pVAPB, and pREC1) were linearized starting from the first conserved gene of the housekeeping backbone. Regions with significant similarity between plasmids are connected by gray stripes (tblastx E value threshold of 0.1); grayscale indicates percent similarity. ORFs are color coded as follows: hypothetical proteins (gray), conjugation or DNA replication/recombination/metabolism (red), DNA mobility genes (magenta), transcriptional regulators (blue), secreted proteins (dark green), membrane proteins (pale green), metabolic functions (yellow), vap family genes (black), and pseudogenes (brown). Green and pale red bars below the genes indicate conjugation and replication/partitioning functional modules, respectively; dashed underline indicates HGT regions identified by Alien_hunter (52); and the triangle indicates a putative origin of replication. Abbreviations: 3oxoACPr, 3-oxoacyl-ACP reductase; 3oxoACPs, 3-oxoacyl-ACP synthase; Endo, endonuclease; Exo, exonuclease; Lig, ligase; LT, lytic transglycosylase; MT, methyltransferase; Mtase, methylase; PK, protein kinase; PR, pentapeptide repeat protein; ssRec, site-specific recombinase; TPR, TPR repeat protein; FA met, fatty acid metabolism.
FIG 3.
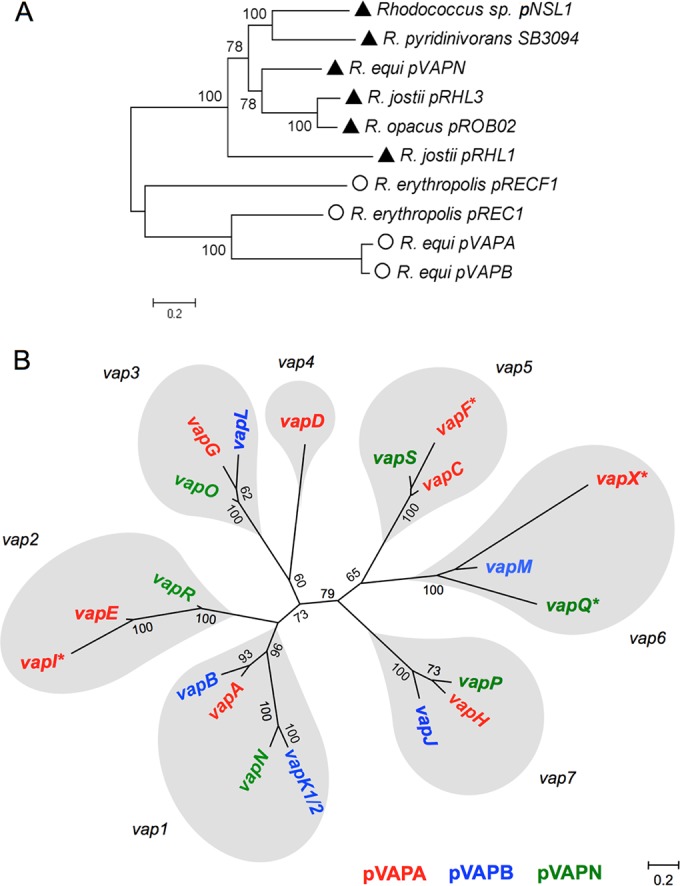
ML trees of Rhodococcus plasmid backbones (A) and the R. equi vap multigene family (B). The Hasegawa-Kishino-Yano with gamma distribution (HKY+G) evolutionary model was used. (A) ML tree based on a concatenated alignment of orthologs from a selection of rhodococcal extrachromosomal replicons (total of 7,802 nucleotides). The genes used are indicated by dots in Fig. 2. Values >50 for 100 bootstrap replicates are indicated. Symbols: triangles, linear plasmids; circles, circular plasmids. (B) vap family members derived from each of the predicted seven precursor vap genes in the MRCA of the extant pVAPA, pVAPB, and pVAPN PAIs are in gray balloons.
FIG 4.
pVAPN telomeric sequences. (A) ClustalΩ alignment of the left- and right-end 200 terminal nucleotides. Identical nucleotides are shaded (dark and light blue, purines and pyrimidines, respectively). Inverted repeats are indicated above the sequence. In red are four conserved palindromic sequences with the central motif GCTNCGC identified in the binding site of telomere-associated proteins involved in Streptomyces linear plasmid replication (72). Several “GCTNCGC” palindromic sequences are normally present in the telomeres of rhodococcal linear plasmids (43–45) (see Fig. S2 in the supplemental material). (B) Secondary structures potentially formed by the palindromic sequences in pVAPN telomeres, as numbered in panel A. Structures were determined with mFold. Free energy (ΔG) values: −33.84 kcal/mol (left), −37.95 kcal/mol (right).
Comparative analysis and functional overview.
Rhodococcus species characteristically possess large plasmids, circular or linear if >100 kb in size (46). These plasmids consist of a vertically evolving backbone, encoding plasmid maintenance and conjugal transfer functions, and a horizontally acquired variable region (VR) providing specific niche-adaptive properties to the host bacterium (24, 37, 44, 45, 47). The housekeeping backbone of pVAPN is unrelated to that of the circular pVAPA/B (equine/porcine-type) R. equi virulence plasmids. pVAPN is instead closely related to the linear plasmid pNSL1 from Rhodococcus sp. NS1 (48) in terms of genetic structure and synteny (Fig. 2). pNSL1 has a size (117,252 bp) similar to that of pVAPN, and its backbone is perfectly colinear with pVAPN. No significant overall similarity with other sequenced linear plasmids from the genus Rhodococcus was detected in pairwise alignments (see Fig. S3 in the supplemental material). However, MLSA analysis of gene orthologs from the housekeeping backbones of a representation of rhodococcal linear and circular plasmids placed pVAPN within a monophyletic clade together with the linear replicons, indicating that they all share a common origin (Fig. 3A).
(i) Conjugation genes.
pVAPN encodes a MOBf (TrwC) family conjugal relaxase (49) homologous to TraA from pVAPA/B (24). Detection of its coding sequence (pVAPN_0650) by PCR using conserved traA target sequences from pVAPA/B allowed the discovery of the traA+ vapAB-negative bovine pVAPN plasmid in the first instance (34). Relaxases play a key role in the conjugation of circular plasmids, nicking the supercoiled double-stranded DNA (dsDNA) and leading the nascent DNA strand into the recipient cell in conjunction with a type IV secretion system (T4SS), which forms the transport channel (50, 51). Indeed, deletion of traA has been shown to prevent the transfer of the equine pVAPA circular virulence plasmid (39). Interestingly, however, the pVAPN traA relaxase gene is corrupted (5′-terminal deletion affecting the first 75 codons, including the gene start and part of the TrwC relaxase domain, and frameshifts in the 3′-terminal region) and probably nonfunctional. This traA pseudogene is located outside the pVAPN conjugation module at the left boundary of the vap PAI. It is immediately contiguous to three ORFs, encoding phage excisionase, Rep, and CopG (regulator of plasmid copy number) homologs, also present in the pVAPA/B backbone (24) but absent in pNSL1. These three ORFs and the adjacent pVAPN vap PAI were identified as HGT acquired based on local DNA compositional bias, as determined by the Alien_hunter program (52) (Fig. 2). This suggests that the traA pseudogene-phage excisionase-rep-copG genes are remnants of a lateral gene exchange, probably the same one that mobilized the vap PAI from the circular virulence plasmid.
Despite traA being a pseudogene and a relaxase-associated T4SS apparatus being absent, pVAPN is transferable by mating (see below). This is probably mediated by pVAPN_0320, encoding a TraB-like plasmid translocase, also present in pNSL1. TraB translocases are evolutionarily related to the septal FtsK/SpoIIE family proteins involved in chromosome segregation (51) and mediate a novel relaxase/T4SS-independent mechanism of conjugation in Streptomyces linear replicons (53). They all share a similar structural arrangement, with an AAA+ motor ATPase domain with characteristic Walker A and B boxes, a transmembrane domain, and a C-terminal DNA-binding winged helix-turn-helix motif (53). In translocase-mediated conjugation, TraB binds to plasmid dsDNA and forms a transmembrane DNA-conducting hexameric channel through which the plasmid is transferred to the recipient bacterium in an ATP-dependent manner. The pVAPN (and pNSL1) conjugation module also comprises (i) an additional AAA+ ATPase with sequence similarity to the conjugative coupling factor TraD (pVAPN_0360); (ii) a homolog of a Soj/ParA family ATPase (pVAPN_0300), involved in chromosomal and plasmid DNA segregation (54) and recruitment of conjugative DNA to the transfer channel (55); (iii) a putative M23 endopeptidase family/lysozyme-like lytic murein transglycosylase/cell wall hydrolase (pVAPN_0390), probably involved in conjugation channel formation (a homolog of which is also present in the circular pVAPA/B virulence plasmids and the related Rhodococcus erythropolis plasmid pREC1) (Fig. 2); and (iv) a number of putative membrane-associated proteins. In addition, pVAPN_0550, at the other side of an interposed plasmid replication/partitioning (rep-parA) module, encodes a putative cutinase. A cutinase gene is also present at the boundary of this replication/partitioning module and the conjugation module in the circular pVAPA/B and pREC1 replicons (Fig. 2). Bacterial cutinase-like proteins, common among mycolic acid-containing actinomycetes (40, 56), have esterase/lipolytic activity (56–58). In phages that infect mycolata, they form part of the LysB lipolytic enzyme complement, thought to aid in the breakdown of the lipid-rich envelope during phage penetration or lytic egress (59). Recently, a cutinase from the R. fascians pFiD188 linear virulence plasmid was shown to be required for efficient conjugation, probably by facilitating the penetration of the DNA translocation complex in the rhodococcal cell envelope (45).
(ii) Replication/partitioning.
The pVAPN self-replication determinant includes a module encoding a Rep protein (pVAPN_0480), which probably directs the bidirectional replication of the plasmid toward the telomeres, and the plasmid-partitioning protein/ATPase ParA (pVAPN_0500). A 26-bp semipalindromic sequence (5′-AAAACCCCCAGGTGGGGGTGGG-TTTT) similar to that determined to be the origin of replication of the pNSL1 plasmid (48) was identified at the same position upstream of the rep gene in pVAPN (Fig. 2). The rep-parA module is detected as HGT genetic material in pNSL1 and is conserved in the circular pVAPA/B and R. erythropolis pREC1 plasmids (also identified as HGT acquired). In pVAPN, it is flanked on the right by a phage excisionase gene (pVAPN_0520), which is conserved in pNSL1 and, interestingly, also in the circular replicons despite these being derived from a different ancestor (Fig. 2). This lends additional support to the hypothesis that the rep-parA determinant forms part of an “exchangeable” gene cassette subjected to HGT between different rhodococcal plasmids (24). This replication/partitioning region appears to serve as an insertion platform for HGT-acquired DNA (24), as suggested by the fact that the VR either is immediately adjacent to (pVAPA/B circular plasmids) or interrupts (pVAPN, pNSL1, and the larger circular plasmid pREC1) it (Fig. 2). Interestingly, in contrast to the circular pVAPA/B plasmids (and pREC1), pVAPN (and pNSL1) does not encode the ParB component of the ParAB replicon segregation system (60). The lack of a parB gene appears to be a hallmark of the ≤400-kb rhodococcal linear extrachromosomal replicons, as exemplified by pREL1 or pBD2 from R. erythropolis (44), pRHL2 and pRHL3 from R. jostii (37), or pFiD188 from R. fascians (45).
(iii) Plasticity region (VR).
The colinearity with pNSL1 is abruptly interrupted at the level of the traA pseudogene, marking the start of the VR. The VR of pVAPN is interrupted by an island of homology with ORFs from the right end of the pNSL1 backbone, suggesting that it was formed by two independent DNA acquisition events (Fig. 2). The left VR section comprises the pVAPA/B-homologous phage excisionase-rep-copG sequence module (see above) plus the vap PAI; the right section encodes rhodococcal/actinobacterial conserved hypothetical proteins and a number of products with various predicted functions (Fig. 2). The complete left VR section with the vap PAI was identified as HGT material (Fig. 2), suggesting that it is a more recent acquisition.
vap PAI.
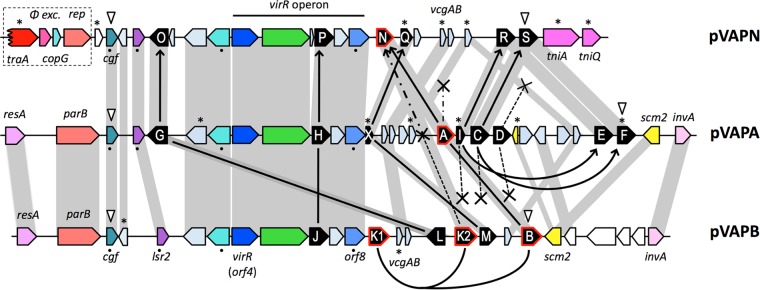
The genetic structure of the vap PAI of pVAPN is similar to that of pVAPA/B (Fig. 5). It is 15.1 kb in length and contains 21 ORFs, including (i) a complement of six vap genes (vapN, vapO, vapP, vapQ pseudogene, vapR, and vapS) encoding polypeptides differing in amino acid sequence identity with pVAPA/B Vaps by 20 to 81% (see Table S3 in the supplemental material); (ii) a vir locus encoding the two key vap PAI transcriptional regulators, VirR (LysR type) and VirS (orphan two-component response regulator) (61, 62), the major facilitator superfamily (MFS) transporter IcgA (63), VapP, and a conserved protein of unknown function; and (iii) several additional non-vap genes (Fig. 5). Four of the latter are conserved as functional genes in the three virulence plasmids, indicating that they are core components of the PAI: pVAPN_0700 (pVAPA/B_0420), encoding a hypothetical protein with similarity to a CopG family transcriptional regulator (here designated cgf), which is probably the first gene of the vap PAI instead of the downstream lsr2 gene considered initially (24); pVAPN_0720 (pVAPA/B_0440), encoding a putative nucleoid-associated protein similar to Lsr2, which in mycobacteria is involved in a number of virulence-related functions (64–66); pVAPN_0760 (pVAPA/B_0470), encoding an S-adenosylmethionine (SAM)-dependent methyltransferase with a potential regulatory role via protein, nucleic acid, or lipid methylation; and pVAPN_870 (pVAPA/B_0570), also known as the vap-coregulated vcgB gene in pVAPA, encoding a hypothetical protein conserved in pathogenic mycobacteria (67) (Fig. 5). At the right end, the putative transposon invertase/resolvase gene invA, found in pVAPA/B and in the VR of the related rhodococcal circular pREC1 plasmid (24), is replaced in pVAPN by tniA-like transposase/integrase and tniQ-like transposase helper protein pseudogenes (Fig. 5).
FIG 5.
Genetic structure of the vap PAIs from pVAPN (15.1 kb), pVAPA (21.5 kb), and pVAPB (15.9 kb). Color codes of genes: vap family (black), DNA conjugation/partitioning (red), DNA mobility/recombination (magenta), transcriptional regulators (blue), other regulators (cyan), membrane proteins (green), metabolic reactions (yellow). Orthologs are in the same color shade and linked by gray bands. ORFs encoding hypothetical proteins are represented in light blue-gray and in white if they are outside the PAI. White arrowheads point to the first and last genes of the consensus PAI. The traA pseudogene/phage excisionase-rep-copG HGT cluster presumably acquired by pVAPN from the pVAPA backbone is boxed. The figure also schematizes the probable evolutionary relationships of the vap multigene family as inferred from phylogenetic analyses (Fig. 3B; see also Fig. S4 and S5 in the supplemental material) and PAI genetic structure; the model minimizes the number of vap gene loss events. Solid lines/arrows connect vap genes belonging to the same monophyletic group (thus likely representing allelic variants of a common vap gene ancestor). Curved lines/arrows indicate vap gene duplications within a PAI. Crosses denote vap genes that were lost, and asterisks indicate pseudogenes. Two alternative evolutionary paths are shown for vapA-vapB-vapK1-vapK2-vapN (see the legend of Fig. S5 in the supplemental material for additional details). The black dots indicate the non-vap genes used for the MLSA shown in Fig. S4B in the supplemental material.
vap PAI evolution.
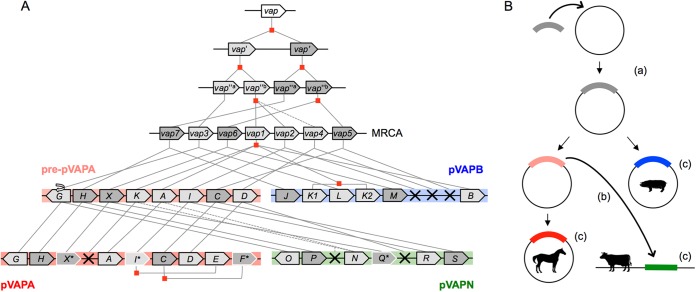
Maximum likelihood (ML) trees grouped the vap multigene family from pVAPN, pVAPA, and pVAPB into several well-supported clades (Fig. 3B; see also Fig. S4A in the supplemental material). vap family members were not clustered by plasmid; instead, vap sequences from different virulence plasmids were grouped under each of the nodes, suggesting that they are allelic variants of a vertically evolving vap precursor gene. Three of the clades contained vap sequences from only one or two of the plasmid types, suggesting a loss of vap alleles. In addition, in two cases, the clades included more than one vap sequence from the same PAI, consistent with instances of vap gene duplication (Fig. 3B; see also Fig. S4A in the supplemental material). To help pinpoint the gene duplication and loss events underlying the evolution of the R. equi vap family, the vap gene tree and a “species” tree of the three PAIs based on their conserved non-vap genes (see Fig. S4B in the supplemental material) were compared by using Notung phylogenetic reconciliation software (68) (see Fig. S5 in the supplemental material). The phylogenetic data were then interpreted in combination with a detailed comparative analysis of the genetic structure of the PAIs (Fig. 5). From these analyses, we inferred that the most recent common ancestor (MRCA) of the three vap PAIs probably comprised seven precursor vap genes. These genes gave rise to the contemporary plasmid type-specific allelic variants as schematized in Fig. 3B (see also Fig. S4A and S5 in the supplemental material for additional details). The seven MRCA vap genes likely originated by successive duplication events from a primordial vap gene (Fig. 6; see also Fig. S5 in the supplemental material), probably acquired by HGT from another organism. Indeed, while being R. equi specific among the actinomycetes, Vap homologs are found in other bacteria from different phyla or even fungi (see Fig. S4A in the supplemental material).
FIG 6.
Hypothetical reconstruction of vap PAI evolution. (A) Model of vap multigene family evolution. Lines indicate the evolutionary path of the vap genes between ancestral PAIs, the most recent common ancestor (MRCA), and extant PAIs. Pre-pVAPA designates the hypothetical direct precursor of the current pVAPA PAI. Gene duplication events are indicated by red squares, gene loss events by crosses, and pseudogenes by asterisks and white borders. (B) Fate of the vap PAI and R. equi virulence plasmid evolution. (a) Acquisition by rhodococcal circular replicon of vap PAI ancestor conferring the ability to colonize macrophages; (b) mobilization of vap PAI from pre-pVAPA plasmid to rhodococcal linear replicon; (c) evolution of species specificity.
The presence in pVAPN, adjacent to the PAI, of an orphan, corrupted copy of traA plus other sequences from the pVAPA/B housekeeping backbone (phage excisionase-rep-copG HGT sequence module) (Fig. 2 and 5) suggests that the vapN PAI was mobilized to the linear replicon from an ancestor of the circular plasmid pVAPA/B and not vice versa. A gene translocation identical to that observed for the allelic variants vapG (pVAPA) and vapO (pVAPN) is unlikely to have occurred twice independently, indicating that the vapN PAI probably originated from a direct precursor of pVAPA after diversification from pVAPB. This interpretation is supported by a phylogenetic analysis performed with the non-vap genes of the PAI (see Fig. S4B in the supplemental material). It also accounts for the presence in the vapN PAI of pVAPA's vapI-vapE and vapC-vapF putative allelic variants vapR and vapS, which are absent in pVAPB (Fig. 5; see also Fig. S5 in the supplemental material). The possibility that the vapA PAI is derived from pVAPN is less plausible because it implies the occurrence, after the mobilization of the PAI, of a second, independent horizontal transfer/recombination event to convey the traA-phage excisionase-rep-copG module from pVAPA/B to pVAPN. The probable evolutionary history of the vap PAI in the three host-adapted R. equi virulence plasmids is schematized in Fig. 6.
pVAPN and its vapN gene are essential for intracellular proliferation in macrophages.
To determine the role of pVAPN in virulence, we obtained an isogenic plasmid-cured derivative of 1571 (1571−) and examined its behavior in in vitro infection assays in mouse J774A.1 and human THP-1 macrophages. Studies with the equine plasmid previously showed that VapA is essential for R. equi virulence (26, 27), in contrast to other pVAPA-encoded Vap products (i.e., VapC, VapD, VapE, VapF [26], VapG [23], or VapH [our unpublished data]), which are dispensable or accessory. Since, according to our data, vapN is pVAPN′s ortholog/allelic variant of vapA, an unmarked, in-frame vapN deletion mutant was also constructed and tested. A plasmidless derivative and a vapA deletion mutant of equine isolate 103S (strains 103S− and 103SΔvapA, respectively) (27) were used as controls.
Figure 7A shows that both 1571− and 1571ΔvapN had lost the ability to proliferate in J774A.1 and THP-1 cells. The effects were essentially identical to those observed for 103S− and 103SΔvapA, respectively (Fig. 7B). These results demonstrate that the bovine plasmid pVAPN is, like the equine plasmid pVAPA, necessary for facilitating R. equi parasitization of host macrophages. These results also show that VapN appears to perform an essential function in pathogenesis, similarly to VapA in the equine plasmid (26). The uptake of 1571− and 1571ΔvapN remained unaffected, as also observed for 103S− and 103SΔvapA (see Fig. S6 in the supplemental material), indicating that the effect of the two plasmids and their cognate VapN and VapA products is specifically related to intracellular survival and/or replication.
FIG 7.
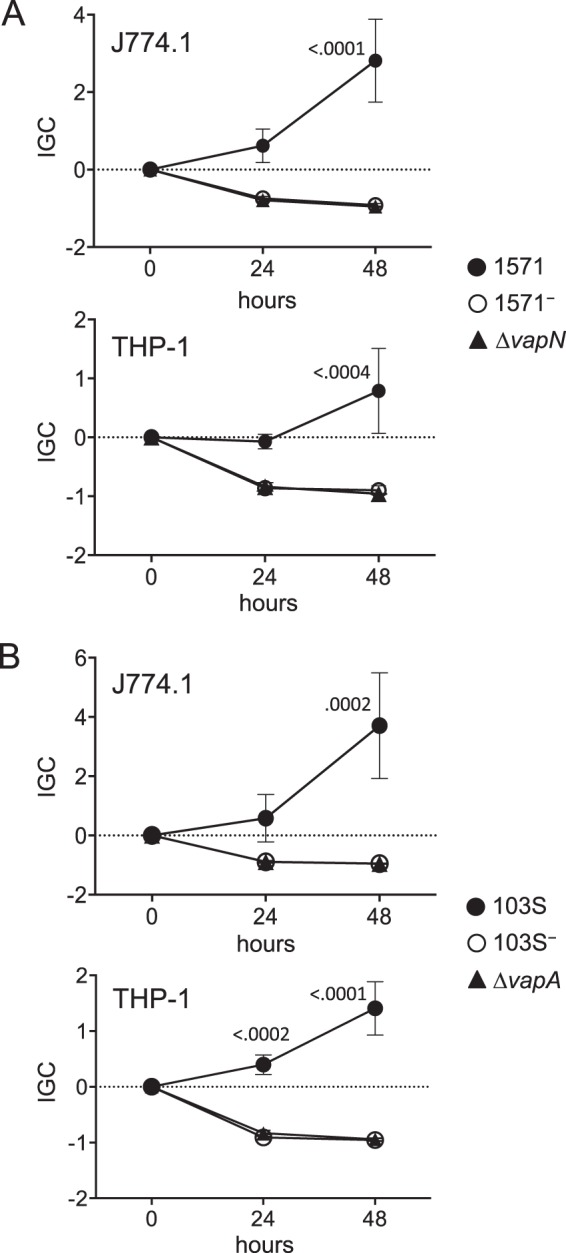
Intracellular proliferation in murine (J774A.1) and human (THP-1) macrophages. Data are expressed as normalized IGC values (see Materials and Methods). Means of data from three duplicate experiments ± standard errors are shown. Statistical significance was analyzed by 2-way ANOVA; P values determined by Šidák post hoc multiple-comparison tests at each time point are shown if ≤0.05. (A) Plasmidless derivative and in-frame ΔvapN mutant of bovine isolate 1571. Two-way ANOVA P values = 0.0007 for J774A.1 and 0.0160 for THP-1. (B) Plasmidless derivative and in-frame ΔvapA mutant of equine isolate 103S. Two-way ANOVA P values = 0.0112 for J774A.1 cells and <0.0001 for THP-1 cells.
Role of pVAPN and vapN in virulence in vivo.
1571 and plasmidless 1571− were also tested in mice by using a competitive lung infection model (27). Immunocompetent BALB/c mice were infected via the intranasal route with a ≈1:1 mix of both bacteria, and R. equi burdens were determined by plate counting over a 4-day period, where R. equi numbers remain stable in the lung (27). The relative proportions of the two strains at each time point were then determined by PCR, and the corresponding CIs were calculated (see Materials and Methods).
The plasmidless 1571− strain was cleared from the lungs at a much higher rate than the 1571 parent strain (Fig. 8A). Except for t = 0, when similar numbers of 1571 and 1571− were recovered, the CI was significantly lower than 1 at all time points (Table 1). By day 3, most (96.3%) of the bacteria were plasmid positive, and at day 4, the 1571− strain was not detected despite total CFU numbers remaining stable in the lungs, indicating that the plasmid-cured bacteria were strongly outcompeted (Fig. 8A). This pattern mirrored the results observed when the same experiment was performed with R. equi 103S and 103S− (27). These data demonstrate that the bovine-type pVAPN plasmid, like equine pVAPA, confers to R. equi the ability to survive in vivo in an animal host.
FIG 8.

Competitive virulence assay in mouse lung. BALB/c mice (n = 4 per time point) were infected intranasally with a ≈1:1 mixture of the test bacteria, and the competing populations were monitored 60 min after infection (t = 0) and then on four consecutive days. Bar height denotes total lung CFU, and the light and dark gray areas within bars indicate the proportions of the competing bacteria. Corresponding CI values are shown in Table 1. (A) Competition between wild-type bovine isolate 1571 and the isogenic plasmidless derivative 1571− at an infection dose of 3.7 × 107 CFU/mouse (2.3 × 107 and 1.4 × 107 CFU, respectively). (B) Competition between the avirulent 1571− strain and an in-frame 1571ΔvapN deletion mutant at an infection dose of 7.8 × 107 CFU/mouse (3.2 × 107 and 4.6 × 107 CFU, respectively).
TABLE 1.
Competitive indexesa
| Competing strains | Mean CI ± SEM (P value) on day: |
||||
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | |
| 1571− and 1571 | 1.54 ± 0.20 (0.0763) | 0.42 ± 0.13 (0.0225) | 0.40 ± 0.12 (0.0407) | 0.06 ± 0.06 (0.0051) | 0.0 (0.0001) |
| ΔvapN and 1571− | 0.88 ± 0.13 (0.4761) | 2.24 ± 0.16 (0.0049) | 3.36 ± 0.41 (0.0107) | 1.47 ± 0.09 (0.0363) | 2.68 ± 0.64 (0.1197) |
Shown are CI values for the mouse infection experiments depicted in Fig. 8. A CI equal to 1 is the theoretical value for two strains with the same competitive ability. Significance of the data was calculated by comparing the experimental CI value at each time point to a theoretical value of 1 (one-sample Student's t test).
We next analyzed the role of VapN in pVAPN-promoted virulence in mice. Since, according to the macrophage data, the lack of VapN was likely to cause strong attenuation in vivo, we compared the competitive ability of 1571ΔvapN against the nonvirulent derivative 1571− (27). This approach takes advantage of the greater sensitivity of competitive tests in assessing small differences in virulence (69, 70). To ascertain the relative importance of VapN and other pVAPN products in R. equi virulence, it is more informative than a comparison with the fully virulent parent strain.
While 1571− was readily outcompeted by the plasmid-positive 1571, sizable numbers of both ΔvapN and 1571− bacteria were recovered at all time points (Fig. 8B). This is similar to the behavior of 103SΔvapA and 103S− under the same experimental conditions (27), demonstrating that the loss of VapN is sufficient to cause a reduction in virulence comparable to that in the absence of its coding pVAPN plasmid. Nevertheless, the CI data showed partial outcompetition of plasmid-cured strain 1571− by the ΔvapN strain, particularly at the two first time points (Table 1). No such differences were previously observed in competition assays between 103SΔvapA and 103S− (27). This suggests that pVAPN products other than VapN are also potentially important for R. equi survival in mice and that there may be some differences in the contributions of VapN and VapA to virulence in their respective plasmid backgrounds.
Collectively, our data support the notion that vapN and vapA are allelic variants of a precursor vap gene essential for R. equi virulence because required for supporting intramacrophage proliferation.
pVAPN transferability by conjugation.
We finally tested whether pVAPN is transferable by mating, as predicted from the sequence data. Experiments were carried out with 1571 as the donor and a rifampin-resistant (Rmpr) plasmidless 103S− (103S−RmpR) as the recipient. The two strains belong to different R. equi chromosomal genogroups (our unpublished data). As a control, conjugation tests with pVAPA from 103S, for which transfer frequencies in the range of 10−2 were previously reported (39), were performed using the same recipient. Transconjugants were determined by screening a total of 900 random Rmpr (100 μg/ml) colonies using suitable recipient- and virulence plasmid-specific PCR markers. Transfer of pVAPA to 103S− was observed at a frequency of 1.25 × 10−2, but transfer of pVAPN could not be detected. We reasoned that the linear plasmid pVAPN could be transferable at a low frequency, unworkable for the PCR-based screening method used. To circumvent this, a transconjugant selection strategy was devised based on the ability of the virulence plasmid to promote R. equi survival in vivo. BALB/c mice were infected i.v. with ≈4 × 108 CFU of a mating mix of 1571 and 103S−RmpR, followed by plating of spleen and liver homogenates onto rifampin-containing plates at days 0, 3, and 5 after infection. Based on previously reported data on i.v. infection in mice (27), this time course was expected to lead to a progressive elimination of plasmid-negative R. equi and a concomitant increase of the plasmid-positive population. The recovered pVAPN-positive/Rmpr bacteria were confirmed to be transconjugants by PCR using suitable strain-specific gene markers and determination of strain-specific DNA sequences (see Materials and Methods; see also Table S1 in the supplemental material).
Figure 9A demonstrates a steady enrichment of pVAPN-positive transconjugants, from 0.5% at day 0 to 34.5% at day 3 and 80.3% at day 5. These data show that pVAPN is transferable between different R. equi strains. The positive selection of 103S− pVAPN transconjugants in mice indicates that the bovine plasmid promotes R. equi virulence irrespective of the strain hosting it. Experiments in J774A.1 cells demonstrated that the acquisition of pVAPN is sufficient to confer to R. equi the capacity for intracellular proliferation in macrophages (Fig. 9B).
FIG 9.
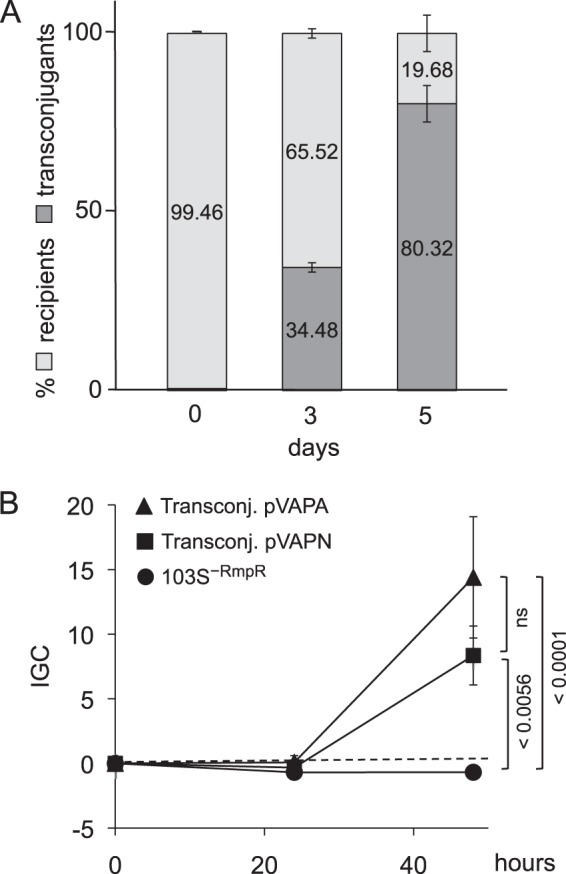
Transfer of pVAPN by mating confers virulence to a plasmid-negative R. equi recipient strain. (A) In vivo selection of pVAPN transconjugants in mice. Note the progressive enrichment of the recipient 103S−RmpR strain upon acquisition of the pVAPN plasmid. Time zero is 60 min after infection. (B) Intracellular proliferation in J774A.1 macrophages. Acquisition of pVAPN (and control pVAPA) promotes intracellular proliferation in the recipient 103S−RmpR strain. Data are expressed as normalized IGC values (see Materials and Methods). Means of data from three duplicate experiments ± standard errors; P values (determined by 2-way ANOVA and Šidák post hoc multiple-comparison tests) are indicated. ns, not significant.
Conclusions.
Our previous work established that the equine-type pVAPA and porcine-type pVAPB virulence plasmids are the same circular replicon in which the HGT-acquired vap PAI evolved divergently, presumably by host-driven selection (24). Our new data show that the bovine pVAPN plasmid originated by horizontal mobilization of the vap PAI to a linear replicon. The pVAPN vap locus, like that of pVAPA/B, is an HGT island and is flanked by DNA mobility genes, consistent with a recent lateral acquisition, probably involving a phage or an integrative element.
Both the circular pVAPA/B (24) and linear pVAPN backbones share common ancestry with other extrachromosomal conjugative replicons found in environmental rhodococci. Rhodococcal plasmids play a key role in facilitating adaptation to different habitats via plasticity regions rich in HGT material. In environmental biodegradative rhodococci, these plasticity regions typically encode catabolic, detoxification, or secondary metabolic determinants, while in pathogenic species (R. equi and R. fascians) they are virulence related (1, 46, 71). In the phytopathogen R. fascians, virulence is conferred by a linear conjugative plasmid (45) without obvious similarity to pVAPN, illustrating that multiple extrachromosomal elements serve as platforms for the expression and dynamic exchange of niche-adaptive traits in the genus Rhodococcus.
Our findings suggest the following hypothetical scenario for the evolution of pathogenicity in R. equi (Fig. 6). First, the acquisition of an ancestral vap PAI by a circular conjugative plasmid endowed a “pre-R. equi” obligate saprotroph with intracellular survival capability in macrophages, promoting its conversion into a facultative parasite by a process known as “co-optive” virulence evolution (40). During coevolution with animal hosts, porcine- and equine-specific tropism evolved as a secondary trait of the PAI in the circular plasmid (Fig. 6B), involving gene duplication and sequence diversification within the vap multigene family (Fig. 6A). Finally, acquisition of the vap PAI by a linear plasmid, presumably from a direct precursor of the equine pVAPA plasmid (Fig. 6B), gave rise to the bovine-adapted pVAPN plasmid, in which another set of specific vap genes evolved (Fig. 6A).
Our analyses with pVAPN confirm the notion that the primary function of the host-adapted R. equi virulence plasmids is to support intracellular proliferation in macrophages. This primordial function is clearly dissociable from host tropism, since epidemiological or experimental evidence indicates that R. equi plasmids promote virulence in accidental (nonadapted) animal hosts, such as humans or mice, regardless of their species-specific type. Our data indicate that a specific vap gene, which was already present in the nearest common ancestor of the contemporary PAIs and evolved into the allelic variants vapA in pVAPA and vapN in pVAPN (and possibly vapB in porcine pVAPB), is critical for intracellular survival in macrophages.
Why bovine host tropism evolved in a linear replicon and not by further host-driven diversification of the PAI in the circular pVAP replicon remains unclear. Since pVAPA and pVAPB share a virtually identical circular backbone, equine- and porcine-specific infectivity most likely resides in their divergent vap PAI. Whether determinants outside the vap PAI in the unrelated pVAPN backbone contribute adaptive features which optimize the interaction of R. equi with the bovine host requires further investigation.
The findings in this study establish R. equi as a novel paradigm of a multihost-adapted pathogen. The pVAPN plasmid reported here, together with the previously characterized equine- and porcine-associated plasmids, provides a unique model system to gain a better understanding of the bacterial mechanisms of intramacrophage survival and host tropism.
Supplementary Material
ACKNOWLEDGMENTS
We thank F. Quigley and C. Lämmler for providing the bovine isolates from which the pVAPN plasmid was sequenced, M. Letek for his early contributions to the R. equi genome program, J. Navas for initial help with mating experiments, P. Iglesias for technical assistance, and U. Fogarty and colleagues at the Irish Equine Centre for encouragement and support. We also thank Edinburgh Genomics (http://genomics.ed.ac.uk/) for pVAP1571 plasmid DNA sequencing and preliminary read assembly.
This work was supported by the Horserace Betting Levy Board (grants vet/prj/712 and 753 to J.A.V.-B.), partially by the Research Stimulus Fund (grant RSF 06 379 to J.A.V.-B. and W.M.), and by core BBSRC funding from the Roslin Institute. A.V.-R. was supported by a European Union Marie Curie postdoctoral fellowship, A.H. and E.A., respectively, by BBSRC-funded Ph.D. and (Zoetis-sponsored) CASE studentships from the Centre for Infectious Diseases of the University of Edinburgh (UoE), and I.M. by a Charles Darwin international scholarship from UoE.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00376-15.
REFERENCES
- 1.Prescott JF. 1991. Rhodococcus equi: an animal and human pathogen. Clin Microbiol Rev 4:20–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vazquez-Boland JA, Giguère S, Hapeshi A, MacArthur I, Anastasi E, Valero-Rello A. 2013. Rhodococcus equi: the many facets of a pathogenic actinomycete. Vet Microbiol 167:9–33. doi: 10.1016/j.vetmic.2013.06.016. [DOI] [PubMed] [Google Scholar]
- 3.Muscatello G, Leadon DP, Klayt M, Ocampo-Sosa A, Lewis DA, Fogarty U, Buckley T, Gilkerson JR, Meijer WG, Vazquez-Boland JA. 2007. Rhodococcus equi infection in foals: the science of ‘rattles’. Equine Vet J 39:470–478. doi: 10.2746/042516407X209217. [DOI] [PubMed] [Google Scholar]
- 4.Cohen ND, Chaffin MK, Kuskie KR, Syndergaard MK, Blodgett GP, Takai S. 2013. Association of perinatal exposure to airborne Rhodococcus equi with risk of pneumonia caused by R. equi in foals. Am J Vet Res 74:102–109. doi: 10.2460/ajvr.74.1.102. [DOI] [PubMed] [Google Scholar]
- 5.Giguère S, Cohen ND, Chaffin MK, Hines SA, Hondalus MK, Prescott JF, Slovis NM. 2011. Rhodococcus equi: clinical manifestations, virulence, and immunity. J Vet Intern Med 25:1221–1230. doi: 10.1111/j.1939-1676.2011.00804.x. [DOI] [PubMed] [Google Scholar]
- 6.Kinne J, Madarame H, Takai S, Jose S, Wernery U. 2011. Disseminated Rhodococcus equi infection in dromedary camels (Camelus dromedarius). Vet Microbiol 149:269–272. doi: 10.1016/j.vetmic.2010.09.037. [DOI] [PubMed] [Google Scholar]
- 7.Takai S, Martens RJ, Julian A, Garcia Ribeiro M, Rodrigues de Farias M, Sasaki Y, Inuzuka K, Kakuda T, Tsubaki S, Prescott JF. 2003. Virulence of Rhodococcus equi isolated from cats and dogs. J Clin Microbiol 41:4468–4470. doi: 10.1128/JCM.41.9.4468-4470.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinstock DM, Brown AE. 2002. Rhodococcus equi: an emerging pathogen. Clin Infect Dis 34:1379–1385. doi: 10.1086/340259. [DOI] [PubMed] [Google Scholar]
- 9.Takai S, Fukunaga N, Ochiai S, Imai Y, Sasaki Y, Tsubaki S, Sekizaki T. 1996. Identification of intermediately virulent Rhodococcus equi isolates from pigs. J Clin Microbiol 34:1034–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makrai L, Takayama S, Denes B, Hajtos I, Sasaki Y, Kakuda T, Tsubaki S, Major A, Fodor L, Varga J, Takai S. 2005. Characterization of virulence plasmids and serotyping of Rhodococcus equi isolates from submaxillary lymph nodes of pigs in Hungary. J Clin Microbiol 43:1246–1250. doi: 10.1128/JCM.43.3.1246-1250.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komijn RE, Wisselink HJ, Rijsman VM, Stockhofe-Zurwieden N, Bakker D, van Zijderveld FG, Eger T, Wagenaar JA, Putirulan FF, Urlings BA. 2007. Granulomatous lesions in lymph nodes of slaughter pigs bacteriologically negative for Mycobacterium avium subsp. avium and positive for Rhodococcus equi. Vet Microbiol 120:352–357. doi: 10.1016/j.vetmic.2006.10.031. [DOI] [PubMed] [Google Scholar]
- 12.Flynn O, Quigley F, Costello E, O'Grady D, Gogarty A, Mc Guirk J, Takai S. 2001. Virulence-associated protein characterisation of Rhodococcus equi isolated from bovine lymph nodes. Vet Microbiol 78:221–228. doi: 10.1016/S0378-1135(00)00297-2. [DOI] [PubMed] [Google Scholar]
- 13.Torres-Tortosa M, Arrizabalaga J, Villanueva JL, Galvez J, Leyes M, Valencia ME, Flores J, Pena JM, Perez-Cecilia E, Quereda C. 2003. Prognosis and clinical evaluation of infection caused by Rhodococcus equi in HIV-infected patients: a multicenter study of 67 cases. Chest 123:1970–1976. doi: 10.1378/chest.123.6.1970. [DOI] [PubMed] [Google Scholar]
- 14.Kedlaya I, Ing MB, Wong SS. 2001. Rhodococcus equi infections in immunocompetent hosts: case report and review. Clin Infect Dis 32:E39–E46. doi: 10.1086/318520. [DOI] [PubMed] [Google Scholar]
- 15.Hondalus MK, Mosser DM. 1994. Survival and replication of Rhodococcus equi in macrophages. Infect Immun 62:4167–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toyooka K, Takai S, Kirikae T. 2005. Rhodococcus equi can survive a phagolysosomal environment in macrophages by suppressing acidification of the phagolysosome. J Med Microbiol 54:1007–1015. doi: 10.1099/jmm.0.46086-0. [DOI] [PubMed] [Google Scholar]
- 17.Zink MC, Yager JA, Prescott JF, Fernando MA. 1987. Electron microscopic investigation of intracellular events after ingestion of Rhodococcus equi by foal alveolar macrophages. Vet Microbiol 14:295–305. doi: 10.1016/0378-1135(87)90117-9. [DOI] [PubMed] [Google Scholar]
- 18.von Bargen K, Haas A. 2009. Molecular and infection biology of the horse pathogen Rhodococcus equi. FEMS Microbiol Rev 33:870–891. doi: 10.1111/j.1574-6976.2009.00181.x. [DOI] [PubMed] [Google Scholar]
- 19.Takai S, Watanabe Y, Ikeda T, Ozawa T, Matsukura S, Tamada Y, Tsubaki S, Sekizaki T. 1993. Virulence-associated plasmids in Rhodococcus equi. J Clin Microbiol 31:1726–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takai S, Hines SA, Sekizaki T, Nicholson VM, Alperin DA, Osaki M, Takamatsu D, Nakamura M, Suzuki K, Ogino N, Kakuda T, Dan H, Prescott JF. 2000. DNA sequence and comparison of virulence plasmids from Rhodococcus equi ATCC 33701 and 103. Infect Immun 68:6840–6847. doi: 10.1128/IAI.68.12.6840-6847.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giguère S, Hondalus MK, Yager JA, Darrah P, Mosser DM, Prescott JF. 1999. Role of the 85-kilobase plasmid and plasmid-encoded virulence-associated protein A in intracellular survival and virulence of Rhodococcus equi. Infect Immun 67:3548–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez-Mora E, Polidori M, Luhrmann A, Schaible UE, Haas A. 2005. Maturation of Rhodococcus equi-containing vacuoles is arrested after completion of the early endosome stage. Traffic 6:635–653. doi: 10.1111/j.1600-0854.2005.00304.x. [DOI] [PubMed] [Google Scholar]
- 23.Coulson GB, Agarwal S, Hondalus MK. 2010. Characterization of the role of the pathogenicity island and vapG in the virulence of the intracellular actinomycete pathogen Rhodococcus equi. Infect Immun 78:3323–3334. doi: 10.1128/IAI.00081-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Letek M, Ocampo-Sosa AA, Sanders M, Fogarty U, Buckley T, Leadon DP, Gonzalez P, Scortti M, Meijer WG, Parkhill J, Bentley S, Vazquez-Boland JA. 2008. Evolution of the Rhodococcus equi vap pathogenicity island seen through comparison of host-associated vapA and vapB virulence plasmids. J Bacteriol 190:5797–5805. doi: 10.1128/JB.00468-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Byrne BA, Prescott JF, Palmer GH, Takai S, Nicholson VM, Alperin DC, Hines SA. 2001. Virulence plasmid of Rhodococcus equi contains inducible gene family encoding secreted proteins. Infect Immun 69:650–656. doi: 10.1128/IAI.69.2.650-656.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain S, Bloom BR, Hondalus MK. 2003. Deletion of vapA encoding virulence associated protein A attenuates the intracellular actinomycete Rhodococcus equi. Mol Microbiol 50:115–128. doi: 10.1046/j.1365-2958.2003.03689.x. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez-Iglesias P, Scortti M, MacArthur I, Hapeshi A, Rodriguez H, Prescott JF, Vazquez-Boland JA. 2014. Mouse lung infection model to assess Rhodococcus equi virulence and vaccine protection. Vet Microbiol 172:256–264. doi: 10.1016/j.vetmic.2014.03.026. [DOI] [PubMed] [Google Scholar]
- 28.Takai S, Sekizaki T, Ozawa T, Sugawara T, Watanabe Y, Tsubaki S. 1991. Association between a large plasmid and 15- to 17-kilodalton antigens in virulent Rhodococcus equi. Infect Immun 59:4056–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sekizaki T, Takai S, Egawa Y, Ikeda T, Ito H, Tsubaki S. 1995. Sequence of the Rhodococcus equi gene encoding the virulence-associated 15-17-kDa antigens. Gene 155:135–136. doi: 10.1016/0378-1119(95)00009-U. [DOI] [PubMed] [Google Scholar]
- 30.Takai S, Anzai T, Fujita Y, Akita O, Shoda M, Tsubaki S, Wada R. 2000. Pathogenicity of Rhodococcus equi expressing a virulence-associated 20 kDa protein (VapB) in foals. Vet Microbiol 76:71–80. doi: 10.1016/S0378-1135(00)00226-1. [DOI] [PubMed] [Google Scholar]
- 31.Oldfield C, Bonella H, Renwick L, Dodson HI, Alderson G, Goodfellow M. 2004. Rapid determination of vapA/vapB genotype in Rhodococcus equi using a differential polymerase chain reaction method. Antonie Van Leeuwenhoek 85:317–326. doi: 10.1023/B:ANTO.0000020383.66622.4d. [DOI] [PubMed] [Google Scholar]
- 32.Makrai L, Takai S, Tamura M, Tsukamoto A, Sekimoto R, Sasaki Y, Kakuda T, Tsubaki S, Varga J, Fodor L, Solymosi N, Major A. 2002. Characterization of virulence plasmid types in Rhodococcus equi isolates from foals, pigs, humans and soil in Hungary. Vet Microbiol 88:377–384. doi: 10.1016/S0378-1135(02)00157-8. [DOI] [PubMed] [Google Scholar]
- 33.Takai S, Tharavichitkul P, Takarn P, Khantawa B, Tamura M, Tsukamoto A, Takayama S, Yamatoda N, Kimura A, Sasaki Y, Kakuda T, Tsubaki S, Maneekarn N, Sirisanthana T, Kirikae T. 2003. Molecular epidemiology of Rhodococcus equi of intermediate virulence isolated from patients with and without acquired immune deficiency syndrome in Chiang Mai, Thailand. J Infect Dis 188:1717–1723. doi: 10.1086/379739. [DOI] [PubMed] [Google Scholar]
- 34.Ocampo-Sosa AA, Lewis DA, Navas J, Quigley F, Callejo R, Scortti M, Leadon DP, Fogarty U, Vazquez-Boland JA. 2007. Molecular epidemiology of Rhodococcus equi based on traA, vapA, and vapB virulence plasmid markers. J Infect Dis 196:763–769. doi: 10.1086/519688. [DOI] [PubMed] [Google Scholar]
- 35.Soedarmanto I, Oliveira R, Lämmler C, Durrling H. 1997. Identification and epidemiological relationship of Rhodococcus equi isolated from cases of lymphadenitis in cattle. Zentralbl Bakteriol 286:457–467. doi: 10.1016/S0934-8840(97)80047-3. [DOI] [PubMed] [Google Scholar]
- 36.Navas J, Gonzalez-Zorn B, Ladron N, Garrido P, Vazquez-Boland JA. 2001. Identification and mutagenesis by allelic exchange of choE, encoding a cholesterol oxidase from the intracellular pathogen Rhodococcus equi. J Bacteriol 183:4796–4805. doi: 10.1128/JB.183.16.4796-4805.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLeod MP, Warren RL, Hsiao WW, Araki N, Myhre M, Fernandes C, Miyazawa D, Wong W, Lillquist AL, Wang D, Dosanjh M, Hara H, Petrescu A, Morin RD, Yang G, Stott JM, Schein JE, Shin H, Smailus D, Siddiqui AS, Marra MA, Jones SJ, Holt R, Brinkman FS, Miyauchi K, Fukuda M, Davies JE, Mohn WW, Eltis LD. 2006. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc Natl Acad Sci U S A 103:15582–15587. doi: 10.1073/pnas.0607048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Geize R, de Jong W, Hessels GI, Grommen AW, Jacobs AA, Dijkhuizen L. 2008. A novel method to generate unmarked gene deletions in the intracellular pathogen Rhodococcus equi using 5-fluorocytosine conditional lethality. Nucleic Acids Res 36:e151. doi: 10.1093/nar/gkn811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tripathi VN, Harding WC, Willingham-Lane JM, Hondalus MK. 2012. Conjugal transfer of a virulence plasmid in the opportunistic intracellular actinomycete Rhodococcus equi. J Bacteriol 194:6790–6801. doi: 10.1128/JB.01210-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Letek M, Gonzalez P, Macarthur I, Rodriguez H, Freeman TC, Valero-Rello A, Blanco M, Buckley T, Cherevach I, Fahey R, Hapeshi A, Holdstock J, Leadon D, Navas J, Ocampo A, Quail MA, Sanders M, Scortti MM, Prescott JF, Fogarty U, Meijer WG, Parkhill J, Bentley SD, Vazquez-Boland JA. 2010. The genome of a pathogenic Rhodococcus: cooptive virulence underpinned by key gene acquisitions. PLoS Genet 6:e1001145. doi: 10.1371/journal.pgen.1001145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deshayes C, Bielecka MK, Cain RJ, Scortti M, de las Heras A, Pietras Z, Luisi BF, Nunez Miguel R, Vazquez-Boland JA. 2012. Allosteric mutants show that PrfA activation is dispensable for vacuole escape but required for efficient spread and Listeria survival in vivo. Mol Microbiol 85:461–477. doi: 10.1111/j.1365-2958.2012.08121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimizu S, Kobayashi H, Masai E, Fukuda M. 2001. Characterization of the 450-kb linear plasmid in a polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl Environ Microbiol 67:2021–2028. doi: 10.1128/AEM.67.5.2021-2028.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang R, Yang Y, Fang P, Jiang C, Xu L, Zhu Y, Shen M, Xia H, Zhao J, Chen T, Qin Z. 2006. Diversity of telomere palindromic sequences and replication genes among Streptomyces linear plasmids. Appl Environ Microbiol 72:5728–5733. doi: 10.1128/AEM.00707-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sekine M, Tanikawa S, Omata S, Saito M, Fujisawa T, Tsukatani N, Tajima T, Sekigawa T, Kosugi H, Matsuo Y, Nishiko R, Imamura K, Ito M, Narita H, Tago S, Fujita N, Harayama S. 2006. Sequence analysis of three plasmids harboured in Rhodococcus erythropolis strain PR4. Environ Microbiol 8:334–346. doi: 10.1111/j.1462-2920.2005.00899.x. [DOI] [PubMed] [Google Scholar]
- 45.Francis I, De Keyser A, De Backer P, Simon-Mateo C, Kalkus J, Pertry I, Ardiles-Diaz W, De Rycke R, Vandeputte OM, El Jaziri M, Holsters M, Vereecke D. 2012. pFiD188, the linear virulence plasmid of Rhodococcus fascians D188. Mol Plant Microbe Interact 25:637–647. doi: 10.1094/MPMI-08-11-0215. [DOI] [PubMed] [Google Scholar]
- 46.Larkin MJ, Kulakov LA, Allen CCR. 2010. Genomes and plasmids in Rhodococcus, p 73–90. In Alvarez HM. (ed), Biology of Rhodococcus. Springer, New York, NY. [Google Scholar]
- 47.Stecker C, Johann A, Herzberg C, Averhoff B, Gottschalk G. 2003. Complete nucleotide sequence and genetic organization of the 210-kilobase linear plasmid of Rhodococcus erythropolis BD2. J Bacteriol 185:5269–5274. doi: 10.1128/JB.185.17.5269-5274.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu Y, Xu M, Shen M, Chen Z, Qin Z. 2010. Cloning, sequencing and identification of replication origin of Rhodococcus linear plasmid pNSL1. Wei Sheng Wu Xue Bao 50:1098–1103. (In Chinese.) [PubMed] [Google Scholar]
- 49.Garcillan-Barcia MP, Francia MV, de la Cruz F. 2009. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Rev 33:657–687. doi: 10.1111/j.1574-6976.2009.00168.x. [DOI] [PubMed] [Google Scholar]
- 50.de la Cruz F, Frost LS, Meyer RJ, Zechner EL. 2010. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol Rev 34:18–40. doi: 10.1111/j.1574-6976.2009.00195.x. [DOI] [PubMed] [Google Scholar]
- 51.Guglielmini J, de la Cruz F, Rocha EP. 2013. Evolution of conjugation and type IV secretion systems. Mol Biol Evol 30:315–331. doi: 10.1093/molbev/mss221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vernikos GS, Parkhill J. 2006. Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics 22:2196–2203. doi: 10.1093/bioinformatics/btl369. [DOI] [PubMed] [Google Scholar]
- 53.Vogelmann J, Ammelburg M, Finger C, Guezguez J, Linke D, Flotenmeyer M, Stierhof YD, Wohlleben W, Muth G. 2011. Conjugal plasmid transfer in Streptomyces resembles bacterial chromosome segregation by FtsK/SpoIIIE. EMBO J 30:2246–2254. doi: 10.1038/emboj.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ebersbach G, Gerdes K. 2005. Plasmid segregation mechanisms. Annu Rev Genet 39:453–479. doi: 10.1146/annurev.genet.38.072902.091252. [DOI] [PubMed] [Google Scholar]
- 55.Atmakuri K, Cascales E, Burton OT, Banta LM, Christie PJ. 2007. Agrobacterium ParA/MinD-like VirC1 spatially coordinates early conjugative DNA transfer reactions. EMBO J 26:2540–2551. doi: 10.1038/sj.emboj.7601696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schue M, Maurin D, Dhouib R, Bakala N′Goma JC, Delorme V, Lambeau G, Carriere F, Canaan S. 2010. Two cutinase-like proteins secreted by Mycobacterium tuberculosis show very different lipolytic activities reflecting their physiological function. FASEB J 24:1893–1903. doi: 10.1096/fj.09-144766. [DOI] [PubMed] [Google Scholar]
- 57.West NP, Chow FM, Randall EJ, Wu J, Chen J, Ribeiro JM, Britton WJ. 2009. Cutinase-like proteins of Mycobacterium tuberculosis: characterization of their variable enzymatic functions and active site identification. FASEB J 23:1694–1704. doi: 10.1096/fj.08-114421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parker SK, Curtin KM, Vasil ML. 2007. Purification and characterization of mycobacterial phospholipase A: an activity associated with mycobacterial cutinase. J Bacteriol 189:4153–4160. doi: 10.1128/JB.01909-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salifu SP, Valero-Rello A, Campbell SA, Inglis NF, Scortti M, Foley S, Vazquez-Boland JA. 2013. Genome and proteome analysis of phage E3 infecting the soil-borne actinomycete Rhodococcus equi. Environ Microbiol Rep 5:170–178. doi: 10.1111/1758-2229.12028. [DOI] [PubMed] [Google Scholar]
- 60.Schumacher MA, Funnell BE. 2005. Structures of ParB bound to DNA reveal mechanism of partition complex formation. Nature 438:516–519. doi: 10.1038/nature04149. [DOI] [PubMed] [Google Scholar]
- 61.Ren J, Prescott JF. 2004. The effect of mutation on Rhodococcus equi virulence plasmid gene expression and mouse virulence. Vet Microbiol 103:219–230. doi: 10.1016/j.vetmic.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 62.Russell DA, Byrne GA, O'Connell EP, Boland CA, Meijer WG. 2004. The LysR-type transcriptional regulator VirR is required for expression of the virulence gene vapA of Rhodococcus equi ATCC 33701. J Bacteriol 186:5576–5584. doi: 10.1128/JB.186.17.5576-5584.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X, Coulson GB, Miranda-Casoluengo AA, Miranda-Casoluengo R, Hondalus MK, Meijer WG. 2014. IcgA is a virulence factor of Rhodococcus equi that modulates intracellular growth. Infect Immun 82:1793–1800. doi: 10.1128/IAI.01670-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen JM, German GJ, Alexander DC, Ren H, Tan T, Liu J. 2006. Roles of Lsr2 in colony morphology and biofilm formation of Mycobacterium smegmatis. J Bacteriol 188:633–641. doi: 10.1128/JB.188.2.633-641.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Colangeli R, Haq A, Arcus VL, Summers E, Magliozzo RS, McBride A, Mitra AK, Radjainia M, Khajo A, Jacobs WR Jr, Salgame P, Alland D. 2009. The multifunctional histone-like protein Lsr2 protects mycobacteria against reactive oxygen intermediates. Proc Natl Acad Sci U S A 106:4414–4418. doi: 10.1073/pnas.0810126106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bartek IL, Woolhiser LK, Baughn AD, Basaraba RJ, Jacobs WR Jr, Lenaerts AJ, Voskuil MI. 2014. Mycobacterium tuberculosis Lsr2 is a global transcriptional regulator required for adaptation to changing oxygen levels and virulence. mBio 5(3):e01106-14. doi: 10.1128/mBio.01106-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miranda-Casoluengo R, Miranda-Casoluengo AA, O'Connell EP, Fahey RJ, Boland CA, Vazquez-Boland JA, Meijer WG. 2011. The vapA co-expressed virulence plasmid gene vcgB (orf10) of the intracellular actinomycete Rhodococcus equi. Microbiology 157:2357–2368. doi: 10.1099/mic.0.049759-0. [DOI] [PubMed] [Google Scholar]
- 68.Durand D, Halldorsson BV, Vernot B. 2006. A hybrid micro-macroevolutionary approach to gene tree reconstruction. J Comput Biol 13:320–335. doi: 10.1089/cmb.2006.13.320. [DOI] [PubMed] [Google Scholar]
- 69.Chiang SL, Mekalanos JJ, Holden DW. 1999. In vivo genetic analysis of bacterial virulence. Annu Rev Microbiol 53:129–154. doi: 10.1146/annurev.micro.53.1.129. [DOI] [PubMed] [Google Scholar]
- 70.Beuzon CR, Holden DW. 2001. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect 3:1345–1352. doi: 10.1016/S1286-4579(01)01496-4. [DOI] [PubMed] [Google Scholar]
- 71.Larkin MJ, Kulakov LA, Allen CC. 2005. Biodegradation and Rhodococcus—masters of catabolic versatility. Curr Opin Biotechnol 16:282–290. doi: 10.1016/j.copbio.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 72.Bao K, Cohen SN. 2003. Recruitment of terminal protein to the ends of Streptomyces linear plasmids and chromosomes by a novel telomere-binding protein essential for linear DNA replication. Genes Dev 17:774–785. doi: 10.1101/gad.1060303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.