Abstract
Purpose.
Photoreceptor degeneration (PRD) is a genetically heterogeneous retinal disorder. Although a number of genes involved in PRD have been identified, their genetic basis remains unknown in a significant number of patients. In this study, we aimed to identify novel disease-causing genes of PRD.
Methods.
Comprehensive ocular examinations were performed in a 2-year-old patient diagnosed with early onset PRD. Retinal capture sequencing was performed to screen causative mutations in known retinal disease-causing loci. Whole-exome sequencing (WES) and a series of variant-filtering strategies were applied for identifying novel disease-causing genes. Retina ATF6 expression was confirmed by immunohistochemistry. RT-PCR was performed to identify ATF6 mRNA in the patient.
Results.
The patient showed typical PRD features, with macular involvement and ellipsoid zone irregularities. Results of retinal capture sequencing were negative. WES data led to identification of biallelic loss-of-function mutations in the ATF6 gene. The first variant generates a premature stop codon (NCBI accession no. NM_007348: c.1126C>T, p.R376*) and the second variant affects a splicing donor site (NM_007348: c.1533+1G>C). Sanger sequencing confirmed the 2 alleles are from 1 parent each. Both of the variants are extremely rare in the population. The splicing variant causes either intron inclusion or exon skipping in the patient, thus severely disrupting ATF6 functional domains. ATF6 is expressed in three neuronal cell layers of mouse retina.
Conclusions.
Our results support ATF6 as a novel disease-causing gene for PRD and suggest that disrupted protein quality control mechanisms may be a novel pathological mechanism underlying human retinal degeneration.
Keywords: ATF6, next-generation sequencing, photoreceptor degeneration, unfolded protein response
This study suggests ATF6 is a novel disease-causing gene for human retinal degeneration.
Protein homeostasis in the cell is achieved through highly coordinated processes including protein synthesis, folding, post-translational modifications, and transport. The endoplasmic reticulum (ER) is the central organelle where protein folding takes place and is subsequently regulated. Accumulation of unfolded proteins in ER triggers intracellular signal transduction pathways, cumulatively known as the unfolded protein response (UPR) pathway, and leads to degradation of misfolded proteins.1 The UPR is a highly conserved signal transduction system which consists of three major branches mediated by the stress sensors IRE1α, PERK, and ATF6, respectively. These branches work either cooperatively or independently and regulate downstream gene expression in order to cope with the fluctuating protein folding load in the ER.1 Despite the general roles played by the UPR pathway components, only three of them have so far been associated with human Mendelian disorders, and these include EIF2AK3,2 WFS13 and SIL1.4 As expected, mutations in these genes lead to syndromic abnormalities in human patients that involve multiple organs or systems including skeleton, brain, liver, eye, and others.2–4 In contrast, human disease correlations remain largely unknown for the remaining UPR pathway components, including 1 of the 3 core mediators, ATF6.
ATF6 encodes a transmembrane protein first identified as an ER stress response element-binding factor.5 As one of the key signaling mediators activated by ER stress, ATF6 localizes on the ER membrane at the normal state. When cells undergo ER stress, ATF6 senses the signals and translocates to Golgi body where it is processed by the proteases S1P and S2P to release its cytoplasmic fragment ATF6f.1 ATF6f then induces upregulation of genes involved in ER-associated degradation to buffer ER stress and protect the cell from apoptosis.1 Surprisingly, although it plays an essential role as an ER stress sensor, ATF6 deficiency in mice shows no gross developmental abnormalities.6,7 This observation raises the possibility that mutations in ATF6 might lead to more subtle non-syndromic defects, possibly due to functional compensation by other UPR branches. Interestingly, a recent study showed that selective activation of ATF6 can help ameliorate the protein folding stress caused by mislocalized rhodopsin (RHO), a photoreceptor-specific protein,8 suggesting that ATF6 might play an important role in retina and photoreceptor survival.
One of the most common genetic disorders representing photoreceptor degeneration (PRD) is retinitis pigmentosa (RP; Mendelian Inheritance in Man [MIM]# 268000; OMIM database).9 Its prevalence is approximately 1 in 3000 to 7000.9 The genetic cause of RP is highly complicated, with at least 74 RP-causing genes identified (Retinal Information Network).10 However, mutations in known RP-causing genes account for only 60% of RP cases, indicating novel loci have yet to be discovered.11 In this study, we identified ATF6 loss-of-function (LOF) mutations in a patient diagnosed with early onset PRD. These findings provide the first link between a UPR pathway gene and a human retinal genetic disorder.
Methods
Clinical Diagnosis
A 2-year-old patient who was noted to have poor visual behavior underwent clinical examination of visual function, external evaluation, retinoscopy, and dilated funduscopy. Visual acuity was tested using Allen pictures. An examination with the patient under anesthesia was performed for full retinal examination with scleral depression. While the patient was under light anesthesia, electrophysiology (electroretinograms [ERGs] and visual evoked potentials) was performed. ERG conditions were mesopic lighting using light-emitting diode goggles for stimulation. A Burian type speculum contact lens was used for recording ERGs. Anesthesia was deepened, and then wide-angle fundus photography (Clarity Medical Systems, Inc., Pleasanton, CA, USA), fluorescein angiography (with 0.077 mg/kg 100 mg/mL sodium fluorescein), and spectral-domain optical coherence tomography (SDOCT) were performed.
DNA Sample Collection
Informed written consent was obtained from all participating individuals, and this study adhered to the tenet of Declaration of Helsinki. Blood samples of the patient and parents were collected and shipped to eyeGENE coordinating center Clinical Laboratory Improvement Amendments laboratory. Genomic DNA was extracted by Gentra Puregene (Qiagen, Valencia, CA, USA). DNA concentration was measured using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA).
DNA Library Preparation and Next-Generation Sequencing (NGS)
One microgram of genomic DNA was sheared into 200- to 500-bp fragments. The fragments were processed and amplified as previously described.11 For each capture reaction, we pooled 50 precapture DNA libraries. The targeted DNA was captured with a customized retinal disease gene panel11 for retinal capture sequencing to screen for variants in known disease-causing genes. If no causative variants were identified, we performed whole-exome sequencing (WES) by capturing the DNA with NimbleGenSeqCap EZ hybridization and wash kit (NimblegenSeqCap EZ human exome library version 2.0) following the manufacturer's instructions. Illumina HiSequation 2000 (Illumina, San Diego, CA, USA) was used to sequence captured libraries.
Bioinformatics Analysis
For each sample, we obtained 100 bp of paired-end sequencing reads. Reads were mapped to human reference genome hg19 by using the Burrows-Wheeler aligner.12 Base quality recalibration, local realignment, and variant calling were performed as in previous reports.11 Because PRD is a rare Mendelian disease, variants with a frequency higher than 1/200 in a series of public databases and internal databases were eliminated. We also obtained variant frequencies from the Exome Aggregation Consortium (ExAC)13 database. After the frequency-based filtering step, we filtered out synonymous variants, identified known retinal disease-causing variants, and annotated the damaging effect of variants by Sorting Intolerant From Tolerant (SIFT),14 Polyphen2,15 likelihood ratio test (LRT),16 MutationTaster17; MutationAssessor18 as described previously.11
Sanger Sequencing
A 500-bp sequence flanking both sides of candidate variants were retrieved from the University of California, Santa Cruz (UCSC) genome browser (hg19 assembly). We masked the repetitive sequences using RepeatMasker.19 Primer 320 was used to design primers for amplifying a 500-bp PCR product to sequence the variant and at least 50-bp region surrounding it. After PCR, a 3730xl unit (ABI) was used to sequence the amplicons.
Immunohistochemistry
Modified Davidson's fixative was used to fix eyes overnight for paraffin embedding. Eye sections of 7 μm were cut (Microtome, Leica). Slides were deparaffinized, and antigen retrieval was performed by boiling sections in 0.01 M citrate buffer (pH 6.0) for 30 minutes, followed by cooling for 30 minutes at room temperature. Slides were washed in phosphate-buffered saline (PBS), incubated for 1 hour at room temperature in hybridization buffer (10% normal goat serum, 0.1% Triton X-100, PBS), and then incubated overnight in primary antibody (anti-mouse ATF6; 1:50 dilution; Novus, Littleton, CO, USA) diluted in hybridization buffer. Slides were then washed in PBS, incubated with secondary antibody (Alexa 488 anti-mouse; 1:500 dilution; Invitrogen, Carlsbad, CA, USA) diluted in hybridization buffer at room temperature for 2 hours, washed in PBS, mounted with anti-fade medium (Prolong; Invitrogen) to reduce bleaching, and cover-slipped. 4′,6-Diamidino-2-phenylindole1:1000 dilution; Life Technology, Carlsbad, CA, USA) was used for nuclear counterstaining. Fluorescent images were produced with an Apotome 2 microscope (Zeiss, Pleasanton, CA, USA) and processed using ZEN software and Photoshop CS4 software (Adobe, San Jose, CA, USA).
RT-PCR of Peripheral Blood RNA
Peripheral blood samples from the patient and control were collected in a PAXgene blood RNA tube (Becton Dickinson, Franklin Lakes, NJ, USA). A PAXgene blood RNA kit (Qiagen) was used to extract RNA according to the manufacturer's instructions. Complimentary DNA (cDNA) was synthesized using SuperScript III reverse transcriptase (Invitrogen). RT-PCRs were performed using Multiplex polymerase (Qiagen) and primer sets exon 8-F (GCTTGTCAGTCTCGCAAGAAG) and exon 13-R (AATTTGAGCCCTGTTCCAGAG). PCR products were run on 2% agarose gel stained with ethidium bromide along with 1-kb-plus ladder (New England Biolabs, Ipswich, MA, USA).
Results
Clinical Findings
The patient we investigated (eyeGENE ID: WPZ+E.93) was a 2-year-old girl of Caucasian ethnicity with a diagnosis of early onset PRD. She was referred for poor vision and congenital nystagmus. The girl was initially falling, but parents did not feel she had difficulty in poor light or at night. There was no family history of any ocular diseases. During the examinations in the clinic, the patient was found to have a distance visual acuity of 20/200 to 20/400 (OD-OS, respectively). She did not demonstrate any ocular misalignment but did have bilateral, symmetrical pendular nystagmus. Refractive error was +7.00 in the right eye and +7.75 in the left eye. Anterior segment and pupillary examination results were normal. Intraocular pressures were normal (9 mm Hg OD and 12 mm Hg OS).
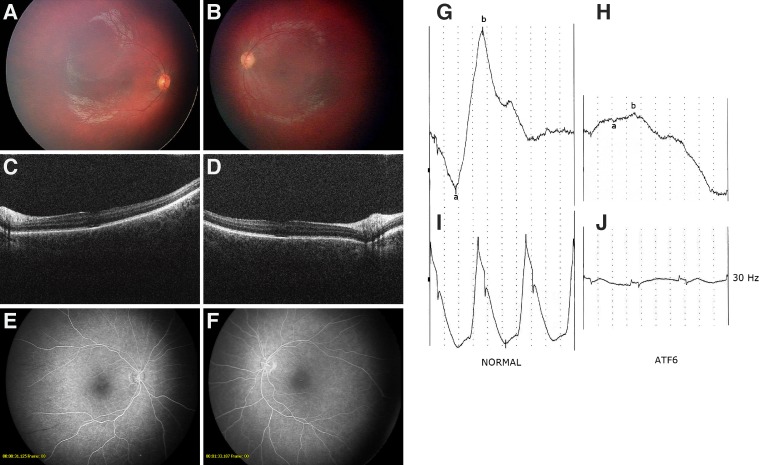
Retinal funduscopic examination showed pigmentary changes in the macular region of both eyes, but peripheral retinae appeared relatively normal and without obvious pigmentation, and mildly narrowed retinal arterioles (Figs. 1A, 1B). ERG results were symmetrical with very poor single-flash ERG responses featuring no significant a- or b-waves (Fig. 1H). The 30-Hz flickers following ERGs were present but at very low amplitude (Fig. 1J). The visual evoked potentials were within normal ranges. The SDOCT showed poor foveal contours with some apparent retention of the inner layers of the retina. There was patchy loss of the ellipsoid zone in both foveae (Figs. 1C, 1D). Otherwise the SDOCTs appeared to be unremarkable. Fluorescein angiography showed window defects in both foveae and possibly an area of avascular retina in the temporal periphery in the right eye (Figs. 1E, 1F). The patient did not show syndromic abnormalities.
Figure 1.
Clinical manifestations in a patient with early onset PRD. Fundus images of maculae and optic nerves of right (A) and left (B) eye showed symmetrically reduced foveal reflex and pigmentary mottling of the fovea areas. SDOCT of maculae and optic nerves of right (C) and left (D) eyes showed symmetrical abnormalities in the ellipsoid zone within the fovea. Also noted are reduced contours of the fovea for age and the appearance of retained layers of the inner foveae. Fluorescein angiography of the maculae and optic nerves of right (E) and left (F) eyes showed window defects in the foveae. Mesopic single-flash (G, H) and 30-Hz flicker ERGs (I, J) recorded in a normal child (left) and the patient with ATF6 mutations (right). Horizontal scale: 10 ms. Vertical scale: 5 μV. Responses were significantly impaired in the patient.
Genetic Findings
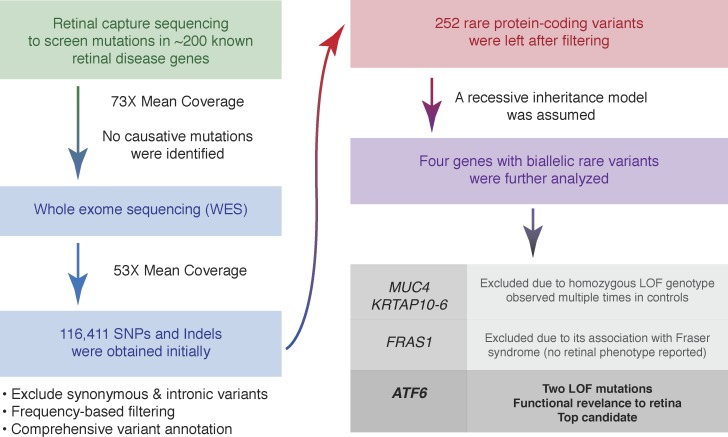
We first applied retinal capture sequencing in this patient to screen for mutations in all known retinal disease-causing genes. High coverage at 99% of designed regions was achieved, but no causative mutation was identified, suggesting that the disease might be caused by mutations in a novel disease gene. To test this hypothesis, we performed WES and obtained high quality data with mean coverage of 53× for exonic regions. More than 116,000 variants were obtained initially. After a series of filtering and annotations (described in Methods), only 252 rare and protein-encoding change variants remained. We focused on genes with biallelic variants, and only four candidate genes were left (MUC4, KRTAP10-6, FRAS1, and ATF6) (Fig. 2). Truncating mutations of MUC4 and KRTAP10-6 genes occurred in homozygous states for 54 and 3,119 times, respectively13; thus, they were not likely to be linked to human Mendelian disease. FRAS1 mutations are known to cause Fraser syndrome, an extracellular matrix disorder with no retinal phenotype reported in patients.21 Hence, ATF6 appeared to be the top candidate in our analysis (Fig. 2).
Figure 2.
Next-generation sequencing strategy for identifying potential disease-causing mutations.
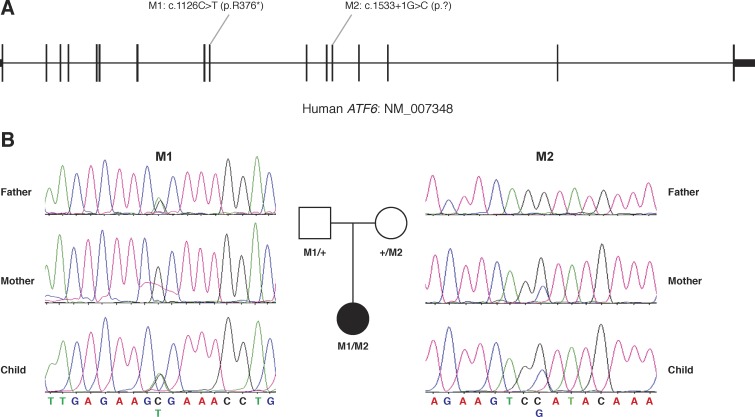
Strikingly, the ATF6 variants are both LOF (Fig. 3A). The first variant is a nonsense mutation which leads to protein truncation (NCBI database accession no. NM_007348: c.1126C>T, p.R376*). It was observed only once in 121,356 control chromosomes.13 The second variant disrupts the splicing donor site in exon 12 (accession no. NM_007348: c.1533+1G>C). It also is very rare because the variant is absent in the control databases. As both of the variants will result in premature stoppage of protein translation, it is likely they will also trigger nonsense-mediated decay of the transcript, therefore resulting in a complete LOF of ATF6. To further test whether the mutant alleles indeed segregated with the phenotype, we performed Sanger sequencing on these two alleles in both parents as well as the patient. As expected, the patient inherited the nonsense variant from one parent (her father) and the splicing mutation from the other (her mother), confirming the fact that these two alleles are in trans and supporting a recessive inheritance model of the disease (Fig. 3B). To further assess how frequently ATF6 mutations are observed in retinal disease patients, we screened an additional 549 patients with retinal diseases, including 215 patients with Leber's congenital amaurosis, 260 with RP, 36 with cone-rod dystrophy, 7 with achromatopsia, and 31 with Stargardt's disease. However, no additional patients with biallelic ATF6 mutations were found, indicating that mutation in ATF6 is a rare cause for human inherited retinal diseases.
Figure 3.
Genetic findings in the early onset PRD patient. (A) Two ATF6 LOF variants were identified in the principal ATF6 transcript. (B) Sanger sequencing confirmed both the variants and the inheritance from one parent each.
ATF6 Is Expressed in Mouse Photoreceptor Perinuclear Region
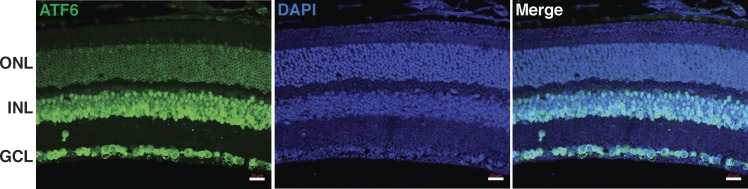
A previous study detected ATF6 expression in the retina by Western blotting.22 To further explore the expression pattern of ATF6 in the retina, we performed immunohistochemistry in adult wild-type (WT) mice. The results showed apparent ATF6 expression in ganglion cell layer, inner nuclear layer, and outer nuclear layer (Fig. 4). Specifically, in the outer nuclear layer, ATF6 was expressed in the perinuclear region of photoreceptors (Fig. 4), agreeing with the subcellular localization of ER, where ATF6 primarily localizes for protein folding surveillance.
Figure 4.
ATF6 expression pattern in the retina. Immunohistochemistry showed ATF6 (green) expresses in all three neuronal cell layers of retina. ATF6 ring-shaped distribution surrounding photoreceptor nuclei indicates perinuclear localization. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar: 20 μm.
Splicing Variant Causes Abnormal ATF6 mRNA Splicing in the Patient
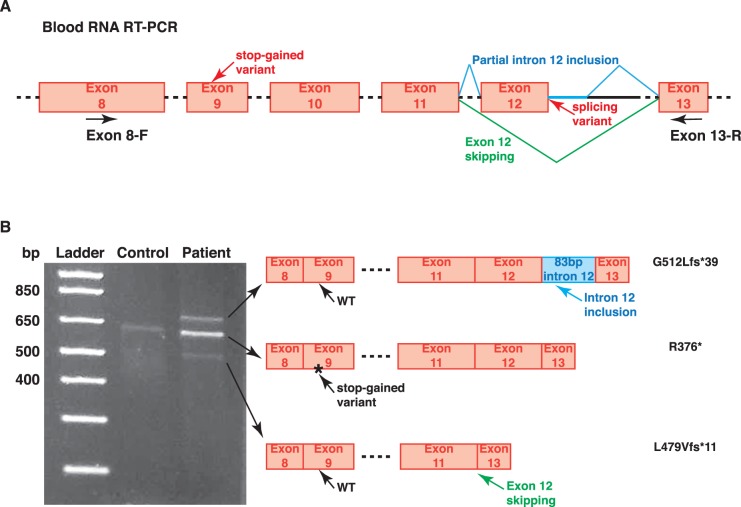
The stop-gained variant on the paternal allele completely abolishes the transmembrane and C-terminal regions that are critical for ATF6 function.23 However, the effect of the maternal allele, which harbors a splicing variant (c.1533+1G>C) was difficult to predict. Thus, we performed RT-PCR of patient blood RNA, using a primer set which amplifies ATF6 exons 8 to 13 (Fig. 5A). In the control RT-PCR, one 600-bp-long band was observed, corresponding to the expected WT RNA product (Fig. 5B). However, in the patient RT-PCR, 2 additional bands were observed, with 1 700-bp long and the other 500-bp long (Fig. 5B). All three bands in the patient RT-PCR were Sanger sequenced to confirm their identities. The uppermost band included an 83-bp-long flanking intronic sequence of ATF6 intron 12. The lowest band completely lacked any exon 12 sequence (Fig. 5B, Supplementary Fig. S1). We also confirmed that the WT-size band in the patient RT-PCR was indeed derived from the paternal stop-gained allele instead of the maternal splicing allele (Fig. 5B, Supplementary Fig. S1). Hence, the splicing variant causes either an 83-bp intron inclusion or exon 12 skipping. No WT RNA product was generated by this maternal allele. Both of the mRNA products resulting from abnormal splicing led to frameshift and premature stop codons (G512Lfs*39 for intron inclusion and L479Vfs*11 for exon skipping), abolishing the ATF6 C-terminal glycosylation sites that are critical for sensing the ER stress signals.24 With the presented functional evidence, we argue that both the stop-gained and the splicing variants in the patient will cause severely impaired function of the ATF6 protein.
Figure 5.
Patient peripheral blood RNA RT-PCR. (A) A primer set amplifying exon 8 to exon 13 was used to cover both the stop-gained and the splicing variant. (B) In the control RT-PCR, only the WT-sized band was observed. In the patient RT-PCR, three bands were observed; the WT-sized band was the stop-gained allele, and other were two caused by either exon skipping or intron inclusion. Protein-coding changes were inferred and labeled after each RT-PCR product. Chromatograms of Sanger sequencing are shown in Supplementary Figure S1.
Discussion
Our study presents the first evidence that links ATF6, the core component of the UPR pathway, to human inherited retinal diseases. This finding is surprising as protein homeostasis and the UPR play essential roles for all cell types. Consistent with our finding of a human patient with early onset PRD without other syndromic abnormalities, Atf6 null mice were shown to be viable and to have grossly normal phenotypes. These observations indicated that, although ATF6 is an important player in regulating the UPR pathway, its function can be largely compensated, probably through other branches of UPR pathways. In contrast, ATF6 plays an essential role in the retina, particularly in photoreceptor cells. Photoreceptor cells are highly specialized cells that continuously undergo intensive protein turnover with up to 10% of the rod outer segment contents renewed daily.25 Thus, it requires a much higher level of protein synthesis, folding, and transport than other tissues,26 and this suggests that the retina can be particularly vulnerable to disruption of the UPR pathway. In addition, the progression of the degeneration remains unknown in our patient. Early cases of PRD can be rapidly progressive compared to that in later onset cases of PRD. However, if compensatory mechanisms are in play in the eye as in other organ systems, then it would be predicted that PRD may be less affected.
Consistent with this idea, a recent study specifically linked ATF6 to correct folding of retinal proteins. Point mutations in RHO that result in mislocalization of RHO in photoreceptor cells is a major cause for autosomal dominant RP in human.27 These mutations can produce misfolded RHO proteins retained in the ER of photoreceptor cells and subsequently lead to cell death when ER stress cannot be resolved.27 Selective activation of ATF6 in cells overexpressing these mutant RHO proteins can prevent their accumulation, while the effect on WT RHO is milder.8 In contrast, activation of PERK, which is another UPR branch mediator, lacked the specific actions on mutant RHO proteins.8 These results suggested that ATF6 could play a retina-specific role in ameliorating ER stress in photoreceptors by preferentially affecting misfolded proteins.
Both of the ATF6 variants are rare in healthy controls, at frequencies of less than 10−6. By screening our cohort of more than 500 patients, only 1 patient with ATF6 biallelic mutations was observed, indicating that mutations in ATF6 account for only a small fraction of patients with retinal diseases. Another way to assess the contribution of ATF6 mutations to human disease is by looking at the allele frequency in healthy controls. Based on ExAC data,13 the frequency of all ATF6 LOF variants (excluding those in the last exon) was only 13 in 120,000 in control individuals. From a population genetics perspective using Hardy-Weinberg equilibrium, the probability that we would observe an individual who loses two copies of the ATF6 gene would be extremely low at <1 in 300,000,000, or 100,000 times less frequent than the incidence of RP. Hence, our finding of a 2-copy ATF6 loss in 1 patient in our cohort strongly supports its association with the retinal disease.
Mutations that cause protein misfolding can lead to a number of retinal degeneration cases.27 In our case, we cannot completely exclude the possibility that a common variant that causes mild protein misfolding might also contribute to the patient phenotype. However, the potential of multifactorial or oligogenic causes underlying human retinal degeneration can only be further confirmed by more precise genotype-phenotype characterizations that involve multiple families with mutations in UPR pathway components.
In summary, by identifying ATF6 LOF mutations in a patient with sporadic early onset PRD, we suggest ATF6 is a novel gene involved in this retinal degenerative disorder. This conclusion is supported by the importance of protein homeostasis specifically in the retina, previous functional studies on the ATF6-Rhodopsin association and compelling human genetic data. Upon revision of our manuscript, ATF6 mutations in achromatopsia patients and photoreceptor dysfunction in Atf6-null mice were reported (Kohl S, et al. IOVS 2015; ARVO E-Abstract 2865). The patient in our study was too young to complete a color vision test, but the foveal abnormalities highly agree with the phenotypes observed in achromatopsia patients with ATF6 mutations (Kohl S, et al. IOVS 2015; ARVO E-Abstract 2865). Given the very early retinal phenotype observed in our patient with ATF6 LOF mutations, it is plausible that ATF6 hypomorphic variants might be involved in milder or age-related retinal disorders. Although a rare cause for retinal disease cohorts, our finding of ATF6 suggests that a defect in the UPR pathway components is a new mechanism underlying human retinal degeneration. As a number of drugs have been developed to target the UPR pathway,28 further studies of the involvement of the UPR pathway in the retina might present excellent opportunities for translational studies.
Acknowledgments
The authors thank the patient and her parents for participating in this study, the eyeGENE Working Group (https://nei.nih.gov/eyegene/staff_eyegene), the Exome Aggregation Consortium, which provided exome variant data, Graeme Mardon for suggestions on this project, and Evan M. Jones for reviewing and editing the manuscript.
Supported by National Eye Institute (NEI) Grants R01EY022356 and R01EY018571; Retinal Research Foundation; Foundation Fighting Blindness Grant BR-GE-0613-0618-BCM (RC); Cullen Foundation endowment to the Molecular and Human Genetics Graduate Program, Baylor College of Medicine (MX); a predoctoral fellowship funded by the Burroughs Wellcome Trust Fund (FW); National Institutes of Health (NIH) T32 funding 2T32EY007102-21A1 (ZG); and Houston Laboratory and Population Sciences Training Program in Gene Environment Interaction. The eyeGENE study was supported by Department of Health and Human Services/NIH/NEI intramural program under eyeGENE Protocol 06-EI-0236 and 10-EI-N164, funded in part under Contract HHS-N-260-2007-00001-C. Next-Generation Sequencing was conducted at the Functional Genomic Core facility, Baylor College of Medicine, and was supported by NIH shared instrument Grant 1S10RR026550 (RC).
Disclosure: M. Xu, None; V. Gelowani, None; A. Eblimit, None; F. Wang, None; M.P. Young, None; B.L. Sawyer, None; L. Zhao, None; G. Jenkins, None; D.J. Creel, None; K. Wang, None; Z. Ge, None; H. Wang, None; Y. Li, None; M.E. Hartnett, None; R. Chen, None
References
- 1. Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol. 2012; 13: 89–102. [DOI] [PubMed] [Google Scholar]
- 2. Delepine M,, Nicolino M,, Barrett T,, Golamaully M,, Lathrop GM,, Julier C. EIF2AK3, encoding translation initiation factor 2-alpha kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nat Genet. 2000; 25: 406–409. [DOI] [PubMed] [Google Scholar]
- 3. Strom TM,, Hortnagel K,, Hofmann S,, et al. Diabetes insipidus, diabetes mellitus, optic atrophy and deafness (DIDMOAD) caused by mutations in a novel gene (wolframin) coding for a predicted transmembrane protein. Hum Mol Genet. 1998; 7: 2021–2028. [DOI] [PubMed] [Google Scholar]
- 4. Anttonen AK,, Mahjneh I,, Hamalainen RH,, et al. The gene disrupted in Marinesco-Sjogren syndrome encodes SIL1, an HSPA5 cochaperone. Nat Genet. 2005; 37: 1309–1311. [DOI] [PubMed] [Google Scholar]
- 5. Yoshida H,, Haze K,, Yanagi H,, Yura T,, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J Biol Chem. 1998; 273: 33741–33749. [DOI] [PubMed] [Google Scholar]
- 6. Yamamoto K,, Sato T,, Matsui T,, et al. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007; 13: 365–376. [DOI] [PubMed] [Google Scholar]
- 7. Usui M,, Yamaguchi S,, Tanji Y,, et al. Atf6alpha-null mice are glucose intolerant due to pancreatic beta-cell failure on a high-fat diet but partially resistant to diet-induced insulin resistance. Metabolism. 2012; 61: 1118–1128. [DOI] [PubMed] [Google Scholar]
- 8. Chiang WC,, Hiramatsu N,, Messah C,, Kroeger H,, Lin JH. Selective activation of ATF6 and PERK endoplasmic reticulum stress signaling pathways prevent mutant rhodopsin accumulation. Invest Ophthalmol Vis Sci. 2012; 53: 7159–7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ferrari S,, Di Iorio E,, Barbaro V,, Ponzin D,, Sorrentino FS,, Parmeggiani F. Retinitis pigmentosa: genes and disease mechanisms. Curr Genomics. 2011; 12: 238–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daiger BR SP,, Greenberg J,, Christoffels A,, Hide W. RetNet. http://www.sph.uth.tmc.edu/RetNet/. Accessed April 23, 2015.
- 11. Wang F,, Wang H,, Tuan HF,, et al. Next generation sequencing-based molecular diagnosis of retinitis pigmentosa: identification of a novel genotype-phenotype correlation and clinical refinements. Hum Genet. 2014; 133: 331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li H,, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Exome Aggregation Consortium (ExAC). Available at: http://exac.broadinstitute.org Accessed February Ruary 15 2015.
- 14. Ng PC,, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003; 31: 3812–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Adzhubei IA,, Schmidt S,, Peshkin L,, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010; 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chun S,, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009; 19: 1553–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schwarz JM,, Rodelsperger C,, Schuelke M,, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010; 7: 575–576. [DOI] [PubMed] [Google Scholar]
- 18. Reva B,, Antipin Y,, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011; 39: e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smit A,, Hubley, R,, Green P. RepeatMasker Open-3.0. 1996-2010. Available at: http://www.repeatmasker.org Accessed December 30, 2014.
- 20. Untergasser A,, Cutcutache I,, Koressaar T,, et al. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012; 40: e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vrontou S,, Petrou P,, Meyer BI,, et al. Fras1 deficiency results in cryptophthalmos, renal agenesis and blebbed phenotype in mice. Nat Genet. 2003; 34: 209–214. [DOI] [PubMed] [Google Scholar]
- 22. Choudhury S,, Nashine S,, Bhootada Y,, et al. Modulation of the rate of retinal degeneration in T17M RHO mice by reprogramming the unfolded protein response. Adv Exp Med Biol. 2014; 801: 455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Haze K,, Yoshida H,, Yanagi H,, Yura T,, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999; 10: 3787–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hong M,, Luo S,, Baumeister P,, et al. Underglycosylation of ATF6 as a novel sensing mechanism for activation of the unfolded protein response. J Biol Chem. 2004; 279: 11354–11363. [DOI] [PubMed] [Google Scholar]
- 25. Young RW. The renewal of rod and cone outer segments in the rhesus monkey. J Cell Biol. 1971; 49: 303–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tzekov R,, Stein L,, Kaushal S. Protein misfolding and retinal degeneration. Cold Spring Harb Perspect Biol. 2011; 3: a007492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin JH,, Lavail MM. Misfolded proteins and retinal dystrophies. Adv Exp Med Biol. 2010; 664: 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hetz C,, Chevet E,, Harding HP. Targeting the unfolded protein response in disease. Nat Rev Drug Discov. 2013; 12: 703–719. [DOI] [PubMed] [Google Scholar]