Abstract
The prevalence of carbapenemase-producing Enterobacteriaceae (CPE) has been increasing worldwide. blaIMP has been reported to be the predominant carbapenemase-encoding gene within Enterobacteriaceae in Australia. However, there are limited data currently available on CPE from Queensland, Australia. A total of 58 CPE isolates were isolated between July 2009 and March 2014 from Queensland hospitals. The clonality of isolates was determined by Diversilab repetitive sequence-based PCR. The isolates were investigated for the resistance mechanisms carbapenemase, extended-spectrum β-lactamase, and AmpC β-lactamase and for aminoglycoside resistance and plasmid-mediated quinolone resistance genes by PCR. The plasmid types associated with carbapenemase-encoding genes were characterized. The majority of the CPE were Enterobacter cloacae (n = 29). The majority of Queensland CPE isolates were IMP producers and comprised 11 species (n = 48). Nine NDM-producing Enterobacteriaceae were identified. One NDM-producing Klebsiella pneumoniae isolate coproduced OXA-48. One K. pneumoniae isolate was an OXA-181 producer. The incidence of IMP producers increased significantly in 2013. blaIMP-4 was found in all IMP-producing isolates. blaTEM, qnrB, and aacA4 were common among IMP-4 producers. The HI2 (67%) and L/M (21%) replicons were associated with blaIMP-4. All HI2 plasmids were of sequence type 1 (ST1). All but one of the NDM producers possessed blaCTX-M-15. The 16S rRNA methylase genes found among NDM producers were armA, rmtB, rmtC, and rmtF. The substantial increase in the prevalence of CPE in Queensland has been associated mainly with the emergence E. cloacae strains possessing HI2 plasmids carrying blaIMP-4 over the past 2 years. The importation of NDM producers and/or OXA-48-like producers in patients also contributed to the increased emergence of CPE.
INTRODUCTION
The most substantial threat to antibiotic susceptibility in Gram-negative bacteria is the emergence of carbapenemase-producing Enterobacteriaceae (CPE). Some carbapenemases tend to dominate in certain countries of the world. For example, Klebsiella pneumoniae carbapenemase (KPC) is the carbapenemase found most frequently in Enterobacteriaceae in the United States (1), while NDM is frequently found in the Indian subcontinent (2) and SPM is found in Brazil (3). Despite the fact that there have been reports of KPC (4), NDM (5, 6), OXA-48 (7) and even a unique carbapenemase, AIM-1 (8), in Australia, none have become dominant there. In Australia, IMP-producing Enterobacteriaceae, particularly Enterobacter cloacae, have been reported to be the predominant form of CPE (9–11). Antibiotic-resistant E. cloacae not only is a common nosocomial pathogen but also has been reported to cause bloodstream infections (10, 12). The mortality rate for infections by IMP-producing E. cloacae causing bacteremia can be as high as 40% (12).
Carbapenemase-encoding genes are usually located on large plasmids that play an important part in their extensive spread worldwide. Plasmids cause the dissemination of antimicrobial resistance, including carbapenemase-encoding genes, through horizontal transfer among bacterial species of the Enterobacteriaceae family. Plasmids of different replicon types or incompatibility types (Inc), such as IncA/C, FII, L/M, and HI2, have been commonly associated with the carriage and transmission of carbapenemase-encoding genes (13).
In Australia, hospital-based health care is regulated by the state government. A state-owned central microbiology laboratory located in Brisbane, the capital city of Queensland, provides reference service to the majority of public hospitals in Queensland. Our aim was to characterize the mechanisms responsible for carbapenem resistance in CPE isolates in the state of Queensland, Australia. These isolates were referred by hospitals and pathology laboratories to a state reference laboratory between June 2009 and March 2014.
(Part of this study was presented as a poster presentation at the Antimicrobials 2014 Conference of the Australian Society for Antimicrobials [February 2014] [46].)
MATERIALS AND METHODS
Bacterial isolates and susceptibility testing.
We investigated Enterobacteriaceae isolates with meropenem resistance determined by broth microdilution Vitek2 (bioMérieux) methods, using EUCAST breakpoints of >4 μg/ml (14). A total of 165 Enterobacteriaceae isolates with reduced susceptibility to meropenem, as determined by the Vitek2 system using Vitek card AST-N246 (bioMérieux), were referred to our laboratory at the University of Queensland Centre for Clinical Research from seven major hospitals in Queensland between June 2009 and March 2014. Of note, the total numbers of nonrepetitive Enterobacteriaceae isolates tested and of Enterobacteriaceae isolates phenotypically determined to be resistant to meropenem by disk susceptibility testing from the same period were 196,282 and 153 isolates, respectively. The isolates were received for confirmation of the presence of carbapenemase-encoding genes. The specimen types and the geographical locations of the hospitals were recorded. In addition, wherever possible, the patient's travel history was recorded. Only one isolate per species from each patient was included, unless multiple species were isolated. Isolate species identification was performed biochemically by using Vitek2 GN-ID panels and confirmed with protein profiling using Vitek matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (bioMérieux).
Antimicrobial susceptibility testing, MIC determinations, and phenotypic tests for carbapenemase.
All isolates were tested for antibiotic susceptibility to ceftazidime, ceftazidime-clavulanic acid, cefotaxime, cefotaxime-clavulanic acid, cefoxitin, cefepime, ertapenem, meropenem, imipenem, ciprofloxacin, gentamicin, and amikacin by using the Vitek2 system (bioMérieux, VIC, Australia), and results were interpreted according to EUCAST breakpoints (14). MICs of ertapenem, meropenem, and imipenem were determined by using the Etest (bioMérieux, VIC, Australia). All isolates were phenotypically tested for the production of carbapenemase by using the modified Hodge test (MHT) and the Carba NP test (15). The phenotypic detection of metallo-β-lactamase (MBL) was performed by using EDTA as an inhibitor, and the detection of KPC was performed by using boronic acid as an inhibitor, as previously described (6, 16).
Genotypic characterization of antibiotic resistance mechanisms.
PCR for blaNDM, blaIMP, blaVIM, and blaKPC was performed on all suspect isolates by using previously described methods, and sequencing was performed to confirm the carbapenemase genes and the variants of these carbapenemase genes (6, 16, 17). PCR and sequencing to capture the entire integron harboring blaIMP were performed to identify the genetic environment of blaIMP and other antibiotic resistance genes located in the class 1 integron. The class 1 integron containing blaIMP-4 underwent PCR and sequencing using primers HS317 and the HS320, as previously described (18). PCR and sequencing of blaOXA-48-like and blaNDM were performed by using previously described primers (19, 20). All isolates were tested by PCR for the presence of blaTEM, blaCTX-M, blaCMY, blaSHV, aacA4, and 16S rRNA methylase genes (armA and rmtA to rmtF) (21, 22). The variants of β-lactamase genes were determined by sequencing of the PCR products. Plasmid-mediated quinolone resistance (PMQR) was determined by PCR for qnrA, qnrB, and qnrS (23). In addition, PCR to detect the ISCR element was performed as previously described (24).
Clonal analysis of Enterobacter spp., K. pneumoniae, and Escherichia spp.
The clonal relationships of isolates of the same species were characterized by a semiautomated method, repetitive sequence-based PCR (rep-PCR) typing. The clonal relatedness of Enterobacter cloacae and Enterobacter asburiae, K. pneumoniae, and Escherichia coli isolates was determined by Diversilab rep-PCR using appropriate kits according to the manufacturer's instructions (bioMérieux, VIC, Australia) (25). Phylogenetic groups of E. coli were determined as previously described (26). Multilocus sequence typing (MLST) was performed for E. cloacae, K. pneumoniae, and E. coli according to methods described on relevant websites (http://pubmlst.org/ecloacae/, http://bigsdb.web.pasteur.fr/klebsiella/klebsiella.html, and http://mlst.warwick.ac.uk/mlst/, respectively).
Genetic features of blaIMP-carrying plasmids and blaNDM-carrying plasmids.
All isolates were characterized for common replicon types associated with carbapenemase-encoding genes, such as types A/C, L/M, HI2, FII, FIA, FIB, X, I1, and N, by using previously described methods (27). A specific set of primers was designed to target the HI2 replicon in this study, HI2F (5′-CTGGTGGGCATAACTCACCT-3′) and HI2R (5′-TCACCAGGGCTT TCTCTGTT-3′), which gave 942-bp amplicons. pMLST was performed to determine the sequence type (ST) of HI2 plasmids as described on the PubMLST website (http://pubmlst.org/plasmid/primers/incHI2.shtml). Plasmid transfer by liquid mating experiments was performed on 12 representative isolates of IMP producers that were selected based on bacterial species and plasmid types. An E. coli K-12 nalidixic acid-resistant non-lactose fermenter was used as the recipient in the conjugation experiments, as previously described (28). E. coli K-12 isolates acquiring plasmids carrying carbapenemase genes by conjugation were selected on MacConkey agar supplemented with 150 μg/ml ampicillin and 0.1 μg/ml meropenem. The recipients containing IMP plasmids were selected based on the colony morphology of non-lactose fermentation of E. coli K-12. Plasmid transfer by electroporation was performed for NDM and/or OXA-48 producers. Plasmids extracted by alkaline lysis were electroporated into E. coli TOP10 cells (Invitrogen, VIC, Australia). The E. coli TOP10 isolates acquiring plasmids carrying carbapenemase genes were selected on Luria-Bertani agar supplemented with 150 μg/ml ampicillin and 0.1 μg/ml meropenem. The plasmid replicon types were determined for the E. coli recipients acquiring plasmids carrying carbapenemase-encoding genes by PCR-based plasmid replicon typing (29). Other resistance genes carried on the carbapenemase plasmids were also determined.
This work was approved by the Royal Brisbane and Women’s Hospital Human Research Ethics Committee (approval number HREC/13/QRBW/391 [Epidemiology, clinical significance, treatment, and outcome of infections by carbapenem-resistant Enterobacteriaceae and Acinetobacter spp. in Queensland]).
RESULTS
Bacterial isolates and antibiotic resistance mechanisms.
A total of 58 carbapenemase-producing Enterobacteriaceae isolates were included in this study. Two previously reported NDM producers from this region were not included in this study (6, 19). The non-CPE producers (n = 105) had reduced susceptibility to ertapenem but remained susceptible to meropenem and imipenem. The non-CPE producers were negative by MHT and Carba NP testing. Although the confirmed CPE isolates were mainly from southeast Queensland, the region which has the majority of the population in Queensland, a total of 6 CPE isolates were from hospitals further north, including hospitals in Townsville and Rockhampton, which are 600 to 1,300 km away from southeast Queensland.
The majority of cases of CPE infection were caused by carbapenemase-producing E. cloacae (n = 32). Two patients possessed pairs of isolates of different species, K. pneumoniae and E. cloacae as well as Raoultella planticola and Citrobacter freundii. One patient harboring E. cloacae and E. coli was previously described (30). These previously reported pairs of IMP-4-producing E. cloacae and E. coli isolates were included in the molecular characterization and the dendrogram for clonal analysis for comparison with other CPE.
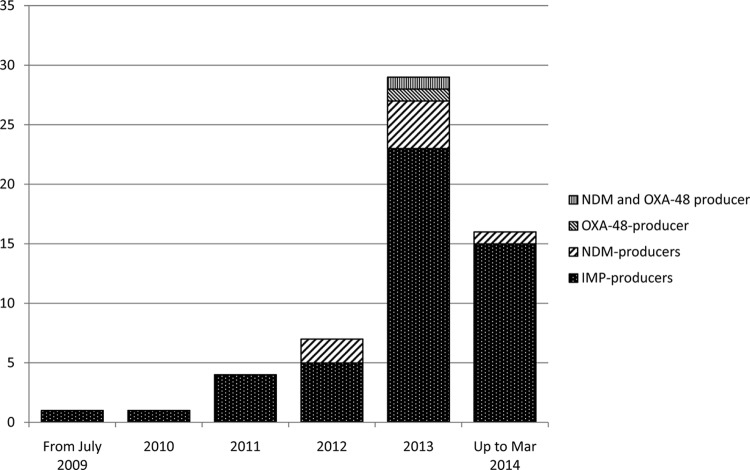
The majority of CPE were isolated in 2013 (n = 27). Seven CPE were isolated in 2012, and only <5 CPE were isolated per annum prior to 2012 (Fig. 1). The majority of isolates were obtained from urine (53%). The other isolates were obtained from blood (14%), respiratory samples (12%), wounds (8%), and rectal swabs (13%). Isolates were initially determined to be meropenem resistant by the Vitek2 system. However, the isolates were not always resistant to meropenem by disk susceptibility testing using EUCAST breakpoints (<22-mm diameter for ertapenem and <16-mm diameter for meropenem disks for carbapenem-resistant isolates) (14). All isolates showed production of carbapenemase by MHT using ertapenem, meropenem, and imipenem disks (10 μg). All isolates showed MBL phenotypes by using EDTA. No KPC phenotype was found. The MIC90s of ertapenem and meropenem for CPE were both >32 μg/ml, and that of imipenem was 24 μg/ml. The MIC50s of ertapenem, meropenem, and imipenem were 16, 4, and 3 μg/ml, respectively.
FIG 1.
Incidence of carbapenemase-producing Enterobacteriaceae in Queensland, Australia, from July 2009 to March 2014. The y axis indicates number of CPE cases.
blaIMP and blaNDM were detected in 48 and 7 isolates, respectively. No isolate harbored blaKPC. The IMP-producing Enterobacteriaceae comprised E. cloacae (n = 29), E. asburiae (n = 2), Enterobacter aerogenes (n = 1), K. pneumoniae (n = 4), E. coli (n = 4), Escherichia hermannii (n = 2), Serratia marcescens (n = 1), C. freundii (n = 2), Citrobacter koseri (n = 1), Proteus mirabilis (n = 1), and R. planticola (n = 1) (Table 1). One NDM-producing K. pneumoniae isolate coproduced OXA-48. One K. pneumoniae isolate produced OXA-181 (Table 2). The NDM producers were probably acquired by patients during overseas travel. In three NDM cases, the patients had a history of travel to India (Table 2). Of the remaining patients, the travel history of one patient could not be retrieved, one patient had a history of travel to Myanmar, and one patient was a migrant from Pakistan. The patient with an NDM isolate coproducing OXA-48 had been transferred from Romania.
TABLE 1.
Characteristics of IMP-4-producing Enterobacteriaceae from Queensland
| Species | No. of isolates | blaIMP variant | No. (%) of isolates carrying gene |
No. (%) of isolates with replicon type |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| blaTEM-1 | blaSHV | blaCTX-M | blaCMY-2-like | qnrB | qnrA | qnrS | aac(6′)-Ib | HI2 | L/M | |||
| E. cloacae | 29 | blaIMP-4 | 27 | 7a | 2c | 1 | 27 | 8 | 2 | 26 | 22 | 7 |
| E. asburiae | 2 | blaIMP-4 | 2 | 2 | 2 | 2 | ||||||
| E. aerogenes | 1 | blaIMP-4 | 1 | 1 | 1 | 1 | ||||||
| E. coli | 4 | blaIMP-4 | 2 | 1 | 1 | 3 | 2 | |||||
| E. hermannii | 2 | blaIMP-4 | 2 | 2a | 3 | 2 | 2 | |||||
| K. pneumoniae | 4 | blaIMP-4 | 4 | 4b | 4 | 1 | 4 | 1 | 2 | |||
| C. freundii | 1 | blaIMP-4 | 1 | 1 | 1c | 1d | 1 | 1 | 1 | 1 | ||
| C. koseri | 1 | blaIMP-4 | 1 | 1 | 1 | 1 | 1 | |||||
| S. marcescens | 1 | blaIMP-4 | 1 | 1 | 1 | |||||||
| P. mirabilis | 1 | blaIMP-4 | 1 | 1a | 1 | 1 | 1 | |||||
| R. planticola | 1 | blaIMP-4 | 1 | 1 | 1 | |||||||
| Total | 48 | 43 (90) | 17 (35) | 3 (6.4) | 2 (4.3) | 41 (85) | 11 (23) | 3 (6.25) | 43 (90) | 32 (67) | 10 (21) | |
blaSHV-12.
blaSHV-1 (n = 2), blaSHV-28 (n = 1), and blaSHV-12 (n = 1).
blaCTX-M-15.
blaCMY-48.
TABLE 2.
Characteristics of NDM- and OXA-48-like-producing Enterobacteriaceae
| Isolate | Yr of isolation | Patient travel history | Specimen type | Sequence type | Gene(s) detected |
Replicon type(s) detected | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| blaNDM variant | blaOXA-48-like variant | blaCTX-M varianta | blaCMY variant | blaSHV variant | blaTEM variant | 16S rRNA methylaseb | aac(6′)-Ib | PMQR | ||||||
| E. coli CR7 | 2012 | India | Urine | 410 | blaNDM-1 | blaCTX-M-15 | blaCMY-42 | armA | aac(6′)-Ib | qnrA | A/C, FII, I1, X, FIA, FIB | |||
| E. coli CR15 | 2013 | Myanmar | Wound | 101 | blaNDM-4 | blaCTX-M-15 | blaCMY-42 | blaTEM-1 | rmtB | aac(6′)-Ib | I1, FIA, FIB, FII, X | |||
| E. coli CR53 | 2014 | Pakistan | Blood | 4450 | blaNDM-4 | blaCMY-42 | blaTEM-1 | rmtB | FII, I1 | |||||
| E. cloacae CR16 | 2012 | India | Urine | 265 | blaNDM-1 | blaCTX-M-15 | blaSHV-12 | armA, rmtC | aac(6′)-Ib | HI2, HI2A, FII, FIB | ||||
| E. cloacae CR37 | 2013 | Not available | Blood | 127 | blaNDM-7 | blaCTX-M-15 | blaTEM-1 | aac(6′)-Ib | qnrB | FII, X | ||||
| K. pneumoniae CR36 | 2013 | India | Urine | 147 | blaNDM-1 | blaCTX-M-15 | blaSHV-11 | blaTEM-1 | rmtF | aac(6′)Ib | qnrB | ColV | ||
| K. pneumoniae CR38 | 2013 | Romania | Urine | 15 | blaNDM-1 | blaOXA-48 | blaCTX-M-15 | blaSHV-28 | blaTEM-1 | rmtC | L/M, FII | |||
| K. pneumoniae CR39 | 2013 | Thailand | Rectal | 231 | blaOXA-181 | blaCTX-M-15 | blaCMY-4 | blaSHV-1 | blaTEM-1 | armA, rmtF | aac-(6′)Ib | qnrS1 | A/C, FII, FIA, FIB | |
All blaCTX-M-15 genes were on separate plasmids carrying blaNDM.
All 16S rRNA methylase genes were on the plasmids carrying blaNDM, except for E. coli CR37.
The blaIMP variants in all isolates were blaIMP-4. The IMP producers were distinct from the NDM producers in the composition of other resistance genes present. These isolates usually carried blaTEM (90%) and the aminoglycoside resistance gene aacA4 (90%). The PMQR gene qnrB was commonly found in IMP producers (85%), while qnrA and qnrS were found in only 23% and 6.3% of IMP producers, respectively (Table 1). blaSHV, blaCTX-M, and blaCMY were less frequently found (Table 1). Other than blaNDM-1, the blaNDM-4 and blaNDM-7 variants were detected (Table 2). PMQR genes (qnrA and qnrB) were less common among NDM producers. The OXA-181-producing K. pneumoniae isolate possessed qnrS. All except one of the NDM producers coproduced CTX-M-15. In addition, blaTEM, blaCMY, and blaSHV were found in 71%, 42%, and 42% of NDM producers, respectively. blaCMY-4, blaCMY-42, blaSHV-1, blaSHV-11, blaSHV-12, and blaSHV-28 were identified among NDM and OXA-48-like producers (Table 2). The NDM producers were usually resistant to amikacin (6 out of 7 isolates), which coincided with the possession of the 16S rRNA methylase genes armA, rmtB, rmtC, and/or rmtF (Table 2). In contrast, none of IMP producers possessed a 16S rRNA methylase gene. All of these IMP producers were susceptible to amikacin.
Clonal analysis of Enterobacter spp., K. pneumoniae, and E. coli.
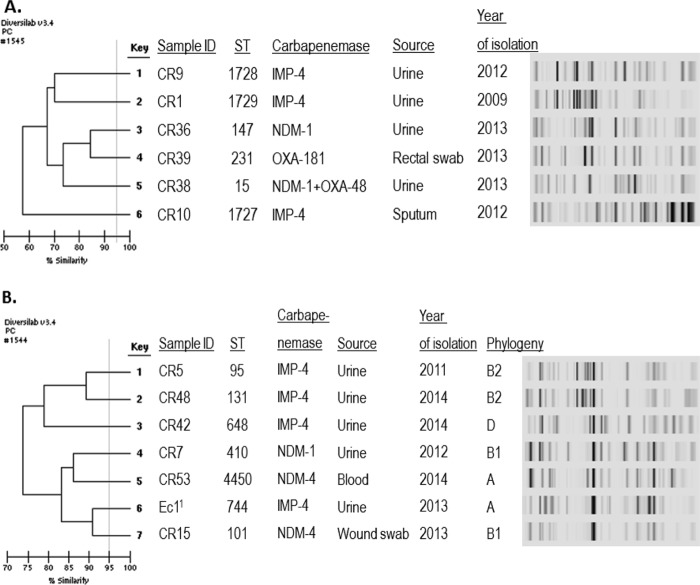
There were no predominant clones observed among Enterobacter spp., using 95% similarity as the cutoff. Several small clusters comprising 2 to 4 isolates with identical rep-PCR patterns were observed (clusters A to G) (Fig. 2). In this study, isolates were considered nonepidemiologically related when they were isolated from different hospitals or isolated >1 year apart. Several E. cloacae isolates were clonal although not epidemiologically related. Examples of these isolates were isolates in clusters B (ST45), C (ST133), and E (ST65). Isolates within these clusters were isolated from different hospitals or in different years. Several identical strains that were closely temporally related potentially indicated person-to-person transmission, such as in cluster A and cluster D. Several new E. cloacae STs were detected in our study isolates, i.e., ST263 and ST265. Interestingly, the two NDM-producing E. cloacae isolates had 94% similarity to each other (ST127 and ST265) and had 90% similarity to IMP-4-producing E. cloacae ST263 isolate CR22 (Fig. 2). Of note, these two NDM producers had only one allele difference. Furthermore, the NDM-producing E. cloacae isolate (CR16) was identical or clonally related to a previously reported NDM-producing E. cloacae isolate from Tasmania (data not shown) (19).
FIG 2.
Dendrogram of carbapenemase-producing Enterobacter spp. Note that all isolates are E. cloacae, except for isolates CR12 and CR13. 1, E. asburiae; 2, Ecl1 was described previously (30).
No clonality was observed among IMP- and NDM-producing E. coli and K. pneumoniae isolates (Fig. 3). Three novel STs were detected among IMP-4-producing K. pneumoniae isolates in our study, ST1727, ST1728, and ST1729 (Fig. 3A). ST101 and ST410 NDM-producing E. coli isolates that were found in this study (Table 2) were previously reported (31, 32). Also, we found NDM-producing E. coli ST4450, which had never been reported to produce NDM previously. Interestingly, the OXA-181-producing K. pneumoniae isolate was of ST231, which was previously reported to produce NDM (33). It is noteworthy that one IMP-4-producing E. coli isolate belonged to the pandemic sequence type ST131 (34). Interestingly, of the four IMP-4-producing E. coli isolates, three belonged to typical virulent extraintestinal pathogenic E. coli phylogenetic groups B2 (n = 2) and D (n = 1) (Fig. 3B). In contrast, all NDM-producing E. coli isolates belonged to commensal groups A and B1.
FIG 3.
Dendrograms of carbapenemase-producing K. pneumoniae (A) and E. coli (B). 1, E. coli Ec1 was described previously (30).
Genetic features of blaIMP-4-carrying plasmids and blaNDM-carrying plasmids.
The majority of clinical IMP-4-producing isolates had HI2 replicon plasmids (67%). Remarkably, the STs of the HI2 plasmids carrying blaIMP-4 were all ST1 (Fig. 2). L/M plasmids were less common (21%). The ISCR1 element was present in all IMP-4 producers. Both HI2 and L/M plasmids were transferred by conjugation. However, the plasmid transformation experiment showed that only L/M plasmids but not HI2 plasmids were transferred (data not shown). In this study, we proved that the blaTEM-1, qnrB2, and aacA4 genes were located in the blaIMP-carrying plasmids. Twelve E. coli K-12 transconjugants acquiring plasmids carrying blaIMP-4 caused an increase of meropenem MICs. The MICs of meropenem for these E. coli K-12 transconjugants ranged from 1 to 4 μg/ml, with an MIC50 of 2 μg/ml. Of note, the meropenem MIC for the E. coli K-12 recipient was 0.012 μg/ml. All the blaIMP-4 and aacA4 genes were located in the class 1 integron. ISCR1 was located downstream of the class 1 integron.
Of the NDM producers, blaNDM-carrying plasmids were transferred successfully into E. coli TOP10 cells. The blaCTX-M-15 genes were carried by plasmids separate from those carrying blaNDM. However, except for one E. coli isolate where rmtB was in a separate plasmid, the 16S rRNA methylase-encoding genes were located on the blaNDM-carrying plasmids.
DISCUSSION
Having lagged behind other countries, Australia is now seeing an increasing incidence of CPE, with the majority being IMP producers. The original IMP producer was initially described in blaIMP-1-producing Serratia marcescens in Japan in 1991 (35). blaIMP-4 was first reported in Acinetobacter baumannii in the 1990s in Hong Kong (36). In Australia, blaIMP-4 was first detected in Melbourne and Sydney in 2002 and has remained the most commonly reported variant of blaIMP (9–11). Here, we describe 11 different species of IMP-4-producing Enterobacteriaceae, including an uncommon species, Raoultella planticola, which has recently been reported to produce IMP-8 in catheter-related cases in Taiwan (37). We also report the first occurrence of IMP-4-producing P. mirabilis. The presence of an endemic carbapenemase-encoding gene, blaIMP-4, in a pandemic strain of E. coli ST131, which is known for its association with CTX-M-15 production and hypervirulence, raises concerns about the potential clonal dissemination of IMP-producing E. coli ST131. An example of spread by clonal expansion is KPC-producing E. coli ST131. KPC producers were initially prevalent among K. pneumoniae isolates. However, since the acquisition of KPC plasmids by E. coli ST131, further extensive spread with a predominance of KPC-producing E. coli ST131 has occurred (38, 39).
Previously, blaIMP-4 was detected in a class 1 integron containing intI1, blaIMP-4, qacG, aacA4, and catB3 (3 to 4 kb in size) (9). The size of the integron in our study was 1.2 kb and consisted of blaIMP-4 and aacA4. A previous study in Australia showed that blaIMP-4 was carried on A/C and L/M plasmids (9), with only one isolate from China carrying an HI2 plasmid (9). Here, we reveal the high incidence of HI2 plasmids carrying blaIMP-4 (67%) in Queensland (Table 1). HI2 plasmids carrying blaIMP-8 were reported in Taiwan (40). It is interesting that these Taiwanese HI2 plasmids carrying blaIMP-4 also carried blaTEM-1, qnrB2, and aacA4. The two plasmid replicon types HI2 and L/M have now shown a strong association with blaIMP. Due to the high incidence of HI2 and L/M plasmids carrying carbapenemase genes in Enterobacteriaceae in Australia, surveillance for Enterobacteriaceae possessing these plasmids in addition to commonly reported carbapenemases is recommended.
It is noteworthy that the variant IMP-4 has also been the predominant MBL reported elsewhere in Australia (9–11). The mechanism of spread of the IMP-4 producers within Queensland is unknown. In other hospitals in Australia, environmental contamination in burn and intensive care units was often identified as the source of the outbreak (10, 41). Our study isolates were referred from a wide geographic coastal area of Queensland from Townsville to Brisbane (1,300 km apart). The facts that the E. cloacae isolates were diverse and that 11 different species were identified in this study showed the capability of blaIMP-4-carrying plasmids to spread horizontally among members of the Enterobacteriaceae. The presence of HI2 plasmid with ST1 harboring blaIMP-4 across different strains of E. cloacae and 7 other species of Enterobacteriaceae in our region has contributed to the increasing number of cases caused by IMP-4 producers. The diverse range of IMP-4-producing Enterobacteriaceae has also been reported in Sydney and Melbourne (9, 11, 42). Initially, Serratia marcescens was the predominant IMP-4 producer in Australia (11); however, E. cloacae has now emerged as the predominant species (10). Our study also describes several novel STs of E. cloacae and the first description of an E. cloacae ST isolated from Australia. The MLST scheme for E. cloacae was relatively recently developed (43). Interestingly, NDM-producing E. cloacae ST265 and ST127 had 4 alleles identical to those of the ST66 and ST114, which were considered high-risk international clones (44). In addition, we reveal three novel STs of K. pneumoniae. There are very scarce data on STs of K. pneumoniae in Australia apart from STs of imported carbapenemase-producing K. pneumoniae isolates (6, 7).
The limitation of this study is that the potential reservoirs and sources of infection by IMP producers as well as clinical outcomes were not investigated, and this warrants further study. blaIMP is locally endemic and is now geographically widespread throughout Australia. Infection control and prevention strategies are an important means of reducing transmission and the overall burden of CPE (45). In conclusion, to control the spread of the CPE in Australia, isolates with reduced susceptibility to carbapenem should undergo molecular confirmation.
ACKNOWLEDGMENTS
We thank all microbiology laboratory staff in Queensland who referred the isolates. We thank the curators of the Institut Pasteur MLST system (Paris, France) for importing novel alleles, profiles, and/or isolates (http://bigsdb.web.pasteur.fr) and Toru Akiyama for the new Enterobacter cloacae alleles and ST (http://pubmlst.org/ecloacae/).
This study is partially funded by the Pathology Queensland Study, Education and Research Trust Fund (grant 4177).
REFERENCES
- 1.Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, Brolund A, Giske CG. 2009. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother 53:3365–3370. doi: 10.1128/AAC.00126-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martins AF, Zavascki AP, Gaspareto PB, Barth AL. 2007. Dissemination of Pseudomonas aeruginosa producing SPM-1-like and IMP-1-like metallo-beta-lactamases in hospitals from southern Brazil. Infection 35:457–460. doi: 10.1007/s15010-007-6289-3. [DOI] [PubMed] [Google Scholar]
- 4.Partridge SR, Ginn AN, Wiklendt AM, Ellem J, Wong JS, Ingram P, Guy S, Garner S, Iredell JR. 2015. Emergence of blaKPC carbapenemase genes in Australia. Int J Antimicrob Agents 45:130–136. doi: 10.1016/j.ijantimicag.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 5.Shoma S, Kamruzzaman M, Ginn AN, Iredell JR, Partridge SR. 2014. Characterization of multidrug-resistant Klebsiella pneumoniae from Australia carrying blaNDM-1. Diagn Microbiol Infect Dis 78:93–97. doi: 10.1016/j.diagmicrobio.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Sidjabat H, Nimmo GR, Walsh TR, Binotto E, Htin A, Hayashi Y, Li J, Nation RL, George N, Paterson DL. 2011. Carbapenem resistance in Klebsiella pneumoniae due to the New Delhi metallo-beta-lactamase. Clin Infect Dis 52:481–484. doi: 10.1093/cid/ciq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Espedido BA, Steen JA, Ziochos H, Grimmond SM, Cooper MA, Gosbell IB, van Hal SJ, Jensen SO. 2013. Whole genome sequence analysis of the first Australian OXA-48-producing outbreak-associated Klebsiella pneumoniae isolates: the resistome and in vivo evolution. PLoS One 8:e59920. doi: 10.1371/journal.pone.0059920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yong D, Toleman MA, Bell J, Ritchie B, Pratt R, Ryley H, Walsh TR. 2012. Genetic and biochemical characterization of an acquired subgroup B3 metallo-beta-lactamase gene, blaAIM-1, and its unique genetic context in Pseudomonas aeruginosa from Australia. Antimicrob Agents Chemother 56:6154–6159. doi: 10.1128/AAC.05654-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Espedido BA, Partridge SR, Iredell JR. 2008. bla(IMP-4) in different genetic contexts in Enterobacteriaceae isolates from Australia. Antimicrob Agents Chemother 52:2984–2987. doi: 10.1128/AAC.01634-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leung GH, Gray TJ, Cheong EY, Haertsch P, Gottlieb T. 2013. Persistence of related bla-IMP-4 metallo-beta-lactamase producing Enterobacteriaceae from clinical and environmental specimens within a burns unit in Australia—a six-year retrospective study. Antimicrob Resist Infect Control 2:35. doi: 10.1186/2047-2994-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peleg AY, Franklin C, Bell JM, Spelman DW. 2005. Dissemination of the metallo-beta-lactamase gene blaIMP-4 among gram-negative pathogens in a clinical setting in Australia. Clin Infect Dis 41:1549–1556. doi: 10.1086/497831. [DOI] [PubMed] [Google Scholar]
- 12.Hayakawa K, Miyoshi-Akiyama T, Kirikae T, Nagamatsu M, Shimada K, Mezaki K, Sugiki Y, Kuroda E, Kubota S, Takeshita N, Kutsuna S, Tojo M, Ohmagari N. 2014. Molecular and epidemiological characterization of IMP-type metallo-beta-lactamase-producing Enterobacter cloacae in a large tertiary care hospital in Japan. Antimicrob Agents Chemother 58:3441–3450. doi: 10.1128/AAC.02652-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carattoli A, Aschbacher R, March A, Larcher C, Livermore DM, Woodford N. 2010. Complete nucleotide sequence of the IncN plasmid pKOX105 encoding VIM-1, QnrS1 and SHV-12 proteins in Enterobacteriaceae from Bolzano, Italy compared with IncN plasmids encoding KPC enzymes in the USA. J Antimicrob Chemother 65:2070–2075. doi: 10.1093/jac/dkq269. [DOI] [PubMed] [Google Scholar]
- 14.EUCAST. 2013. Breakpoint tables for interpretation of MICs and zone diameters. EUCAST, Vaxjo, Sweden: http://www.eucast.org/clinical_breakpoints/. [Google Scholar]
- 15.Dortet L, Poirel L, Nordmann P. 2012. Rapid identification of carbapenemase types in Enterobacteriaceae and Pseudomonas spp. by using a biochemical test. Antimicrob Agents Chemother 56:6437–6440. doi: 10.1128/AAC.01395-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sidjabat HE, Silveira FP, Potoski BA, Abu-Elmagd KM, Adams-Haduch JM, Paterson DL, Doi Y. 2009. Interspecies spread of Klebsiella pneumoniae carbapenemase gene in a single patient. Clin Infect Dis 49:1736–1738. doi: 10.1086/648077. [DOI] [PubMed] [Google Scholar]
- 17.Poirel L, Walsh TR, Cuvillier V, Nordmann P. 2011. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Sidjabat HE, Townsend KM, Hanson ND, Bell JM, Stokes HW, Gobius KS, Moss SM, Trott DJ. 2006. Identification of bla(CMY-7) and associated plasmid-mediated resistance genes in multidrug-resistant Escherichia coli isolated from dogs at a veterinary teaching hospital in Australia. J Antimicrob Chemother 57:840–848. doi: 10.1093/jac/dkl057. [DOI] [PubMed] [Google Scholar]
- 19.Rogers BA, Sidjabat HE, Silvey A, Anderson TL, Perera S, Li J, Paterson DL. 2013. Treatment options for New Delhi metallo-beta-lactamase-harboring enterobacteriaceae. Microb Drug Resist 19:100–103. doi: 10.1089/mdr.2012.0063. [DOI] [PubMed] [Google Scholar]
- 20.Zarakolu P, Day KM, Sidjabat HE, Kamolvit W, Lanyon CV, Cummings SP, Paterson DL, Akova M, Perry JD. 2015. Evaluation of a new chromogenic medium, chromID OXA-48, for recovery of carbapenemase-producing Enterobacteriaceae from patients at a university hospital in Turkey. Eur J Clin Microbiol Infect Dis 34:519–525. doi: 10.1007/s10096-014-2255-z. [DOI] [PubMed] [Google Scholar]
- 21.Sidjabat HE, Paterson DL, Adams-Haduch JM, Ewan L, Pasculle AW, Muto CA, Tian GB, Doi Y. 2009. Molecular epidemiology of CTX-M-producing Escherichia coli isolates at a tertiary medical center in western Pennsylvania. Antimicrob Agents Chemother 53:4733–4739. doi: 10.1128/AAC.00533-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sidjabat HE, Paterson DL, Qureshi ZA, Adams-Haduch JM, O'Keefe A, Pascual A, Rodriguez-Bano J, Doi Y. 2009. Clinical features and molecular epidemiology of CMY-type beta-lactamase-producing Escherichia coli. Clin Infect Dis 48:739–744. doi: 10.1086/597037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibson JS, Cobbold RN, Heisig P, Sidjabat HE, Kyaw-Tanner MT, Trott DJ. 2010. Identification of Qnr and AAC(6′)-1b-cr plasmid-mediated fluoroquinolone resistance determinants in multidrug-resistant Enterobacter spp. isolated from extraintestinal infections in companion animals. Vet Microbiol 143:329–336. doi: 10.1016/j.vetmic.2009.11.031. [DOI] [PubMed] [Google Scholar]
- 24.Toleman MA, Bennett PM, Walsh TR. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol Mol Biol Rev 70:296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sidjabat HE, Derrington P, Nimmo GR, Paterson DL. 2010. Escherichia coli ST131 producing CTX-M-15 in Australia. J Antimicrob Chemother 65:1301–1303. doi: 10.1093/jac/dkq098. [DOI] [PubMed] [Google Scholar]
- 26.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carattoli A, Miriagou V, Bertini A, Loli A, Colinon C, Villa L, Whichard JM, Rossolini GM. 2006. Replicon typing of plasmids encoding resistance to newer beta-lactams. Emerg Infect Dis 12:1145–1148. doi: 10.3201/eid1207.051555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sidjabat HE, Hanson ND, Smith-Moland E, Bell JM, Gibson JS, Filippich LJ, Trott DJ. 2007. Identification of plasmid-mediated extended-spectrum and AmpC beta-lactamases in Enterobacter spp. isolated from dogs. J Med Microbiol 56:426–434. doi: 10.1099/jmm.0.46888-0. [DOI] [PubMed] [Google Scholar]
- 29.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 30.Sidjabat HE, Heney C, George NM, Nimmo GR, Paterson DL. 2014. Interspecies transfer of blaIMP-4 in a patient with prolonged colonization by IMP-4-producing Enterobacteriaceae. J Clin Microbiol 52:3816–3818. doi: 10.1128/JCM.01491-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mushtaq S, Irfan S, Sarma JB, Doumith M, Pike R, Pitout J, Livermore DM, Woodford N. 2011. Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J Antimicrob Chemother 66:2002–2005. doi: 10.1093/jac/dkr226. [DOI] [PubMed] [Google Scholar]
- 32.Sartor AL, Raza MW, Abbasi SA, Day KM, Perry JD, Paterson DL, Sidjabat HE. 2014. Molecular epidemiology of NDM-1-producing Enterobacteriaceae and Acinetobacter baumannii isolates from Pakistan. Antimicrob Agents Chemother 58:5589–5593. doi: 10.1128/AAC.02425-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giske CG, Froding I, Hasan CM, Turlej-Rogacka A, Toleman M, Livermore D, Woodford N, Walsh TR. 2012. Diverse sequence types of Klebsiella pneumoniae contribute to the dissemination of blaNDM-1 in India, Sweden, and the United Kingdom. Antimicrob Agents Chemother 56:2735–2738. doi: 10.1128/AAC.06142-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogers BA, Sidjabat HE, Paterson DL. 2011. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother 66:1–14. doi: 10.1093/jac/dkq415. [DOI] [PubMed] [Google Scholar]
- 35.Osano E, Arakawa Y, Wacharotayankun R, Ohta M, Horii T, Ito H, Yoshimura F, Kato N. 1994. Molecular characterization of an enterobacterial metallo beta-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob Agents Chemother 38:71–78. doi: 10.1128/AAC.38.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu YW, Afzal-Shah M, Houang ET, Palepou MI, Lyon DJ, Woodford N, Livermore DM. 2001. IMP-4, a novel metallo-beta-lactamase from nosocomial Acinetobacter spp. collected in Hong Kong between 1994 and 1998. Antimicrob Agents Chemother 45:710–714. doi: 10.1128/AAC.45.3.710-714.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tseng SP, Wang JT, Liang CY, Lee PS, Chen YC, Lu PL. 2014. First report of bla(IMP-8) in Raoultella planticola. Antimicrob Agents Chemother 58:593–595. doi: 10.1128/AAC.00231-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Hara JA, Hu F, Ahn C, Nelson J, Rivera JI, Pasculle AW, Doi Y. 2014. Molecular epidemiology of KPC-producing Escherichia coli: occurrence of ST131-fimH30 subclone harboring pKpQIL-like IncFIIk plasmid. Antimicrob Agents Chemother 58:4234–4237. doi: 10.1128/AAC.02182-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naas T, Cuzon G, Gaillot O, Courcol R, Nordmann P. 2011. When carbapenem-hydrolyzing beta-lactamase Kpc meets Escherichia coli ST131 in France. Antimicrob Agents Chemother 55:4933–4934. doi: 10.1128/AAC.00719-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen YT, Lauderdale TL, Liao TL, Shiau YR, Shu HY, Wu KM, Yan JJ, Su IJ, Tsai SF. 2007. Sequencing and comparative genomic analysis of pK29, a 269-kilobase conjugative plasmid encoding CMY-8 and CTX-M-3 beta-lactamases in Klebsiella pneumoniae. Antimicrob Agents Chemother 51:3004–3007. doi: 10.1128/AAC.00167-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kotsanas D, Wijesooriya WR, Korman TM, Gillespie EE, Wright L, Snook K, Williams N, Bell JM, Li HY, Stuart RL. 2013. “Down the drain”: carbapenem-resistant bacteria in intensive care unit patients and handwashing sinks. Med J Aust 198:267–269. doi: 10.5694/mja12.11757. [DOI] [PubMed] [Google Scholar]
- 42.Espedido B, Iredell J, Thomas L, Zelynski A. 2005. Wide dissemination of a carbapenemase plasmid among Gram-negative bacteria: implications of the variable phenotype. J Clin Microbiol 43:4918–4919. doi: 10.1128/JCM.43.9.4918-4919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyoshi-Akiyama T, Hayakawa K, Ohmagari N, Shimojima M, Kirikae T. 2013. Multilocus sequence typing (MLST) for characterization of Enterobacter cloacae. PLoS One 8:e66358. doi: 10.1371/journal.pone.0066358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Izdebski R, Baraniak A, Herda M, Fiett J, Bonten MJ, Carmeli Y, Goossens H, Hryniewicz W, Brun-Buisson C, Gniadkowski M, MOSAR WP2, WP3, and WP5 Study Groups 2015. MLST reveals potentially high-risk international clones of Enterobacter cloacae. J Antimicrob Chemother 70:48–56. doi: 10.1093/jac/dku359. [DOI] [PubMed] [Google Scholar]
- 45.Australian Commission on Safety and Quality in Health Care. 2013. Recommendations for the control of multi-drug resistant Gram-negatives: carbapenem resistant Enterobacteriaceae. Commonwealth of Australia, Sydney, Australia. [Google Scholar]
- 46.Sidjabat HE, Townell N, George N, Caffery M, Douglas J, Nimmo GR, Vaska V, Paterson DL. 2014. Increasing emergence of IMP-4 producing Enterobacteriaceae throughout Queensland hospitals, poster P01.18. Antimicrobials 2014 Conference of the Australian Society for Antimicrobials. [Google Scholar]