Abstract
We report the detection of PER-1 extended-spectrum β-lactamase (ESBL) in a clinical non-O1, non-O139 Vibrio cholerae strain from China. ISCR1-mediated blaPER-1 was embedded in a complex In4 family class 1 integron belonging to the lineage of Tn1696 on a conjugative IncA/C plasmid. A free 8.98-kb circular molecule present with the ISCR1-blaPER-1–truncated 3′-conserved sequence (CS) structure was detected in this isolate. These findings may provide insight into the mobilization of blaPER-1.
TEXT
Extended-spectrum β-lactamases (ESBLs) are uncommon in Vibrio cholerae. However, CTX-M-type, PER-2, and TEM-63 enzymes have been identified in serogroup O1 isolates from Argentina and South Africa (1, 2). PER β-lactamases belong to the ESBL family of enzymes, of which 7 variants have been identified to date. PER-1 was mainly detected in Europe, particularly in Turkey, and Asia (3, 4); PER-2 (86% amino acid sequence identity with PER-1) was reported in South America, mainly in Argentina (5, 6); PER-3, -4, and -5 (all differing by 1 amino acid from PER-1) were reported only in the GenBank database (accession numbers AY740681, EU748544, and EU687473, respectively); PER-6 (92% amino acid sequence identity with PER-2) and PER-7 (differing from PER-1 by 4 amino acids) were identified in Paris, France (7, 8).
In 2005, a non-O1, non-O139 V. cholerae strain, RJ354, was isolated from the blood sample of a hemodialysis patient in the Ruijin Hospital, Shanghai Jiaotong University School of Medicine. In this strain, two novel integron-borne cassettes, dfrA27 and aadA16, were found to be located on a conjugative plasmid in our previous study (9). Initial susceptibility testing via disk diffusion revealed a high level of resistance to ceftazidime in RJ354. The MICs for ceftazidime, cefotaxime, piperacillin-tazobactam, and ceftazidime-clavulanic acid for RJ354, determined by the Etest method (AB bioMérieux, Askim, Sweden), were >256, 256, 2, and 0.25 mg/liter, respectively, indicating the presence of an ESBL. A conjugation experiment using Escherichia coli J53Azr (sodium azide resistant) as the recipient demonstrated that resistance to ceftazidime can be transferred from RJ354 to J53Azr. The susceptibilities of the transconjugant to ceftazidime (MIC, >256 mg/liter), cefotaxime (MIC, 128 mg/liter), piperacillin-tazobactam (MIC, 1 mg/liter), and ceftazidime-clavulanic (MIC, 0.19 mg/liter) were very similar to those of the donor, RJ354.
ESBL genes (blaTEM, blaCTX-M, blaSHV, blaPER, and blaVEB) from strain RJ354 and its transconjugant were screened by PCR (10). Sequencing analysis of blaPER-positive PCR products revealed the presence of PER-1 ESBL in both strains. The plasmid DNA content of isolate RJ354 and its transconjugant was examined using pulsed-field gel electrophoresis (PFGE) with S1 nuclease digestion, as described previously (11). The location of blaPER-1 was determined by blotting the S1 nuclease-linearized PFGE-separated plasmid DNA onto positively charged nylon membranes (Roche Applied Science, Penzberg, Germany), with a digoxigenin-labeled blaPER-1-specific probe. Plasmid incompatibility groups were determined by a PCR-based replicon typing scheme described by Carattoli et al. (12). The results revealed that blaPER-1 was located on a 160-kb broad-host-range IncA/C plasmid within the RJ354 isolate and its transconjugant. In previous studies, blaPER-1 was also found to be located on IncA/C plasmids in Providencia stuartii and Klebsiella pneumoniae strains from Tunisia and South Korea, respectively (10, 13), indicating that the IncA/C plasmids may play a role in the dissemination of blaPER-1.
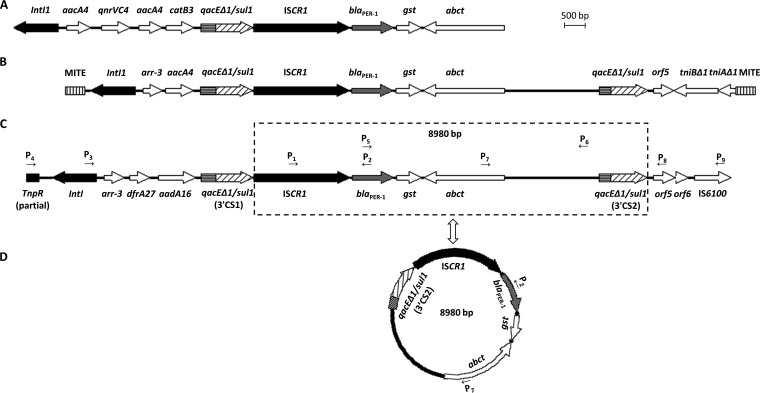
Overlap PCR was performed to determine the genetic context of blaPER-1; primers specific for sequences that typically surround blaPER-1 were used in the PCR experiment (Table 1). PCR amplification and sequencing of the region located upstream of blaPER-1 revealed an ISCR1 element. The spacer between ISCR1 and blaPER-1 was 62 bp long and was identical to the spacer detected in the previously described isolate (14–17). The association of ISCR1 with blaPER-1 was first identified in an Aeromonas punctata strain isolated from China in 2008 (15), 3 years after the isolation of the non-O1, non-O139 V. cholerae strain in the present study. To date, all blaPER-1 genes reported in strains from China were preceded by an ISCR1 element (14–17), while in strains from outside China, the insertion sequence ISPa12 was always located upstream of blaPER-1 (4), suggesting that there may be two divergent paths in the evolution of blaPER-1. Because ISCR1 was identified in the Chinese isolates, PCR primers (P3, P2) located within the 5′-conserved sequence (CS) of class 1 integron and blaPER-1 were used to investigate the sequences further upstream of blaPER-1. As expected, a typical class 1 integron containing an array of arr-3–dfrA27–aadA16 gene cassettes was detected; this array was described previously in the same strain in our laboratory (9). Different Chinese isolates harboring ISCR1-blaPER-1 have distinct arrays of gene cassettes upstream of ISCR1 (Fig. 1) (14, 15), suggesting that different recombination events were responsible for the ISCR1-mediated integration of blaPER-1 into class 1 integrons.
TABLE 1.
Primers used in this study
| Primer | Target gene or region | Primer sequence (5′ to 3′) | PCR product size (kb) |
|---|---|---|---|
| P1 | ISCR1 | GATACTAACTGGCGTGACAAGAG | 0.97 |
| P2 | blaPER-1 | CTCGTCTCCCTGATACGCTTTC | |
| P3 | IntI | CGAACCCAGTGGACATAAGCC | 6.3 |
| P2 | blaPER-1 | CTCGTCTCCCTGATACGCTTTC | |
| P4 | tnpR of Tn1696 | GCCCTTCTTTGACGAACTCCA | 7.8 |
| P2 | blaPER-1 | CTCGTCTCCCTGATACGCTTTC | |
| P5 | blaPER-1 | CGTATCAGGGAGACGAGTTTAGT | 4.6 |
| P6 | Downstream of abct | GGTAGCGAGATTCACAGACAGA | |
| P7 | abct | CCACCACATACACCATCACATCC | 4.0 |
| P8 | orf5 | CATCAGCCGCACAACCTCGTC | |
| P7 | abct | CCACCACATACACCATCACATCC | 5.7 |
| P9 | IS6100 | AGGCGGCTGCTGCGAAATGGTG | |
| P2 | blaPER-1 | CTCGTCTCCCTGATACGCTTTC | 6.4 |
| P7 | abct | CCACCACATACACCATCACATCC |
FIG 1.
Genetic environment of ISCR1-mediated blaPER-1 in Acinetobacter johnsonii XBB1 (GenBank accession no. KF017283) (A), Aeromonas punctate 159 (GenBank accession no. GQ891757) (B), and non-O1, non-O139 Vibrio cholerae of this study (C). Open arrows indicate genetic orientations; thin horizontal arrows indicate primers (P); and the dotted box shows the fragment corresponding to panel D. (D) Small circular molecule detected in this study.
Sequence analysis of the region downstream of blaPER-1 demonstrated the presence of an array of blaPER-1-gst (encoding a glutathione S-transferase)-abct (encoding an ABC-type transporter) identical to the corresponding region described in the Acinetobacter johnsonii XBB1 strain from China in 2014 (14), followed by qacEΔ1/sul1 and orf5, which formed the 3′ end of the novel complex class 1 integron identified in the present study. Notably, at the start of the second copy of 3′ CS (3′ CS2, qacEΔ1/sul1), a 179-bp deletion was observed, in comparison with the full-length copy of 3′ CS1.
In A. johnsonii XBB1, the complex class 1 integron harboring the ISCR1-blaPER-1 structure was flanked by the same miniature inverted-repeat transposable element (MITE) (Fig. 1) (14). However, in the present study, this MITE structure was not detected by PCR. Subsequently, two pairs of primers (P2/P4 and P7/P9) were designed on the basis of the previously reported gene structure of transposon Tn1696 (18), and two unique amplicons of 7.8 kb and 5.7 kb were obtained and sequenced. Analysis of the whole 16,020-bp fragment revealed that the complex class 1 integron was preceded by the tnpR of Tn1696 at the 5′ end and was followed by an IS6100 insertion sequence at the 3′ end (Fig. 1). These findings indicated that ISCR1-mediated blaPER-1 was embedded in a novel complex In4 family class 1 integron belonging to the lineage of Tn1696.
The ISCR1 element is an unusual insertion sequence that demonstrates IS91-like characteristics and may mobilize adjacent DNA sequences via a process called rolling-circle replication (19). Partridge and Hall (20) confirmed that a small circular molecule containing ISCR1-dfrA10-sul1 generated by restriction digestion of the complex class 1 integron In34 can be transposed into a plasmid at a site within the 3′ CS of a cloned class 1 integron. To investigate whether such a circular molecule was present in the RJ354 strain, PCR amplification was performed with the inverse primers (P2, P7) located within blaPER-1 and abct (Fig. 1), and an amplicon of 6.4 kb was obtained, suggesting the presence of a circular molecule. Sequence analysis of the complete 8,980-bp circular molecule was performed to elucidate the structure of ISCR1-blaPER-1-gst-abct-3′ CS2 (with a 179-bp deletion at the beginning of qacEΔ1); the deduced structure was identical to that of the corresponding region in the complex class 1 integron identified in the present study (Fig. 1). Therefore, it is likely that the circular molecule was formed by excision and circularization of the corresponding region of the complex class 1 integron and vice versa. These findings strongly suggest that ISCR1-blaPER-1-truncated 3′ CS may function as a powerful genetic vehicle that can facilitate horizontal dissemination of blaPER-1.
In conclusion, our study describes, for the first time, the identification of a PER-1 ESBL in a non-O1, non-O139 V. cholerae strain. blaPER-1 is carried on a conjugative and broad-host-range IncA/C plasmid; a Tn1696-like transposon is located in this plasmid, and a highly mobile ISCR1 element is embedded in this transposon and is present in the form of a small circular molecule as well.
Nucleotide sequence accession number.
The sequence of the genetic environment of blaPER-1 in non-O1, non-O139 V. cholerae RJ354 has been deposited in GenBank under the accession number KP076293.
ACKNOWLEDGMENTS
This work was supported by the Shanghai Science and Technology Specific Project (grant no. 11ZR1422200) and the National Natural Science Foundation of China (grant no. 81472010).
REFERENCES
- 1.Petroni A, Corso A, Melano R, Cacace ML, Bru AM, Rossi A, Galas M. 2002. Plasmidic extended-spectrum beta-lactamases in Vibrio cholerae O1 El Tor isolates in Argentina. Antimicrob Agents Chemother 46:1462–1468. doi: 10.1128/AAC.46.5.1462-1468.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ismail H, Smith AM, Sooka A, Keddy KH. 2011. Genetic characterization of multidrug-resistant, extended-spectrum-beta-lactamase-producing Vibrio cholerae O1 outbreak strains, Mpumalanga, South Africa, 2008. J Clin Microbiol 49:2976–2979. doi: 10.1128/JCM.00293-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poirel L, Cabanne L, Vahaboglu H, Nordmann P. 2005. Genetic environment and expression of the extended-spectrum beta-lactamase blaPER-1 gene in gram-negative bacteria. Antimicrob Agents Chemother 49:1708–1713. doi: 10.1128/AAC.49.5.1708-1713.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poirel L, Bonnin RA, Nordmann P. 2012. Genetic support and diversity of acquired extended-spectrum beta-lactamases in Gram-negative rods. Infect Genet Evol 12:883–893. doi: 10.1016/j.meegid.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Melano R, Corso A, Petroni A, Centron D, Orman B, Pereyra A, Moreno N, Galas M. 2003. Multiple antibiotic-resistance mechanisms, including a novel combination of extended-spectrum beta-lactamases in a Klebsiella pneumoniae clinical strain isolated in Argentina. J Antimicrob Chemother 52:36–42. doi: 10.1093/jac/dkg281. [DOI] [PubMed] [Google Scholar]
- 6.Quinteros M, Radice M, Gardella N, Rodriguez MM, Costa N, Korbenfeld D, Couto E, Gutkind G, Microbiology Study Group . 2003. Extended-spectrum beta-lactamases in Enterobacteriaceae in Buenos Aires, Argentina, public hospitals. Antimicrob Agents Chemother 47:2864–2867. doi: 10.1128/AAC.47.9.2864-2867.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Girlich D, Poirel L, Nordmann P. 2010. PER-6, an extended-spectrum beta-lactamase from Aeromonas allosaccharophila. Antimicrob Agents Chemother 54:1619–1622. doi: 10.1128/AAC.01585-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnin RA, Potron A, Poirel L, Lecuyer H, Neri R, Nordmann P. 2011. PER-7, an extended-spectrum beta-lactamase with increased activity toward broad-spectrum cephalosporins in Acinetobacter baumannii. Antimicrob Agents Chemother 55:2424–2427. doi: 10.1128/AAC.01795-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun J, Zhou M, Wu Q, Ni Y. 2010. Characterization of two novel gene cassettes, dfrA27 and aadA16, in a non-O1, non-O139 Vibrio cholerae isolate from China. Clin Microbiol Infect 16:1125–1129. doi: 10.1111/j.1469-0691.2009.03060.x. [DOI] [PubMed] [Google Scholar]
- 10.Bae IK, Jang SJ, Kim J, Jeong SH, Cho B, Lee K. 2011. Interspecies dissemination of the bla gene encoding PER-1 extended-spectrum beta-lactamase. Antimicrob Agents Chemother 55:1305–1307. doi: 10.1128/AAC.00994-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barton BM, Harding GP, Zuccarelli AJ. 1995. A general method for detecting and sizing large plasmids. Anal Biochem 226:235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 12.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 13.Mnif B, Ktari S, Chaari A, Medhioub F, Rhimi F, Bouaziz M, Hammami A. 2013. Nosocomial dissemination of Providencia stuartii isolates carrying blaOXA-48, blaPER-1, blaCMY-4 and qnrA6 in a Tunisian hospital. J Antimicrob Chemother 68:329–332. doi: 10.1093/jac/dks386. [DOI] [PubMed] [Google Scholar]
- 14.Zong Z. 2014. The complex genetic context of blaPER-1 flanked by miniature inverted-repeat transposable elements in Acinetobacter johnsonii. PLoS One 9:e90046. doi: 10.1371/journal.pone.0090046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia R, Guo X, Zhang Y, Xu H. 2010. qnrVC-like gene located in a novel complex class 1 integron harboring the ISCR1 element in an Aeromonas punctata strain from an aquatic environment in Shandong Province, China. Antimicrob Agents Chemother 54:3471–3474. doi: 10.1128/AAC.01668-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu M, Wong MH, Chen S. 2013. Molecular characterisation of a multidrug resistance conjugative plasmid from Vibrio parahaemolyticus. Int J Antimicrob Agents 42:575–579. doi: 10.1016/j.ijantimicag.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 17.Wang F, Wu K, Sun J, Wang Q, Chen Q, Yu S, Rui Y. 2012. Novel ISCR1-linked resistance genes found in multidrug-resistant Gram-negative bacteria in southern China. Int J Antimicrob Agents 40:404–408. doi: 10.1016/j.ijantimicag.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 18.Partridge SR, Brown HJ, Stokes HW, Hall RM. 2001. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob Agents Chemother 45:1263–1270. doi: 10.1128/AAC.45.4.1263-1270.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toleman MA, Bennett PM, Walsh TR. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol Mol Biol Rev 70:296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Partridge SR, Hall RM. 2003. In34, a complex In5 family class 1 integron containing orf513 and dfrA10. Antimicrob Agents Chemother 47:342–349. doi: 10.1128/AAC.47.1.342-349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]