Abstract
Staphylococcus aureus is a major cause of nosocomial and community-acquired infections. The success of S. aureus as a pathogen is due in part to its many virulence determinants and resistance to antimicrobials. In particular, methicillin-resistant S. aureus has emerged as a major cause of infections and led to increased use of the antibiotics vancomycin and daptomycin, which has increased the isolation of vancomycin-intermediate S. aureus and daptomycin-nonsusceptible S. aureus strains. The most common mechanism by which S. aureus acquires intermediate resistance to antibiotics is by adapting its physiology and metabolism to permit growth in the presence of these antibiotics, a process known as adaptive resistance. To better understand the physiological and metabolic changes associated with adaptive resistance, six daptomycin-susceptible and -nonsusceptible isogenic strain pairs were examined for changes in growth, competitive fitness, and metabolic alterations. Interestingly, daptomycin nonsusceptibility coincides with a slightly delayed transition to the postexponential growth phase and alterations in metabolism. Specifically, daptomycin-nonsusceptible strains have decreased tricarboxylic acid cycle activity, which correlates with increased synthesis of pyrimidines and purines and increased carbon flow to pathways associated with wall teichoic acid and peptidoglycan biosynthesis. Importantly, these data provided an opportunity to alter the daptomycin nonsusceptibility phenotype by manipulating bacterial metabolism, a first step in developing compounds that target metabolic pathways that can be used in combination with daptomycin to reduce treatment failures.
INTRODUCTION
Daptomycin is a calcium-dependent cyclic lipopeptide antibiotic that has become a common therapeutic option in the treatment of serious Staphylococcus aureus infections (1–5). As daptomycin use has increased, patient treatment failures have increased because of the emergence of S. aureus strains that can grow in the presence of low concentrations of daptomycin (3, 6–14). This increase in daptomycin-nonsusceptible (DapNS) S. aureus isolates has enhanced interest in discovering the changes that occur during the transition to a nonsusceptibility phenotype. Insight into this transition can be gained by understanding the mechanism by which daptomycin acts on bacteria. Primarily, daptomycin disrupts membrane function by redirecting cell division and cell wall synthetic proteins, which causes leakage of ions and membrane depolarization (15, 16) and induction of the cell wall stress stimulon via VraSR (17–19). Because of daptomycin's mode of action, it was reasonable to predict that DapNS S. aureus would have alterations in cell wall-related and membrane genes. This prediction was borne out when comparisons of daptomycin-susceptible (DapS) and DapNS isogenic strain sets identified single nucleotide polymorphisms in the mprF gene, the yycFG (also known as, walKR) operon, and genes encoding subunits of RNA polymerase (i.e., rpoB and rpoC) (10, 13, 20–27). Similarly, phenotypic studies have identified several mechanisms that contribute to daptomycin nonsusceptibility, specifically, a thickened cell wall (26, 28, 29), enhanced cell surface charge (i.e., increased activity of MprF and d-alanylation of wall teichoic acids [WTAs]) (23, 30–33), and/or altered cell membrane composition, fluidity, and permeabilization (21, 29, 34).
In addition to genetic and phenotypic analyses of DapNS strains, global transcriptional and proteomic analyses have identified differences in virulence determinants, lipid metabolism, and teichoic acid and cell wall biosynthesis (18, 22, 25–27, 35, 36). Additionally, several metabolic pathways, including intermediary, fermentative, amino acid, vitamin, and nucleoside metabolism, are altered during the transition to a daptomycin nonsusceptibility phenotype (18, 25, 27, 35, 36). While transcriptional and proteomic analyses provide valuable data, they offer an incomplete view of the metabolic changes due to posttranscriptional and posttranslational regulation and feedback and feedforward changes caused by substrate and cofactor availability. In order to understand the metabolic changes that permit growth in the presence of daptomycin, we assessed the physiology and metabolism of six isogenic DapS and DapNS S. aureus clinical strain pairs by using nuclear magnetic resonance (NMR)-based metabolic profiling and growth, enzyme, and metabolite analyses. This approach allowed us to gain new insight into the metabolic changes that accompany the transition to a daptomycin nonsusceptibility phenotype, but it also permits the integration of our NMR metabolomic data with the transcriptional and proteomic studies. Lastly, these data provided insight into methods to reverse the daptomycin nonsusceptibility phenotype.
MATERIALS AND METHODS
Bacterial strains.
Six different isogenic strain sets isolated from patients before daptomycin therapy and after treatment failure were used in this study (Table 1). The strains represent two methicillin-susceptible S. aureus (MSSA) and four methicillin-resistant S. aureus (MRSA) pairs with or without mprF and/or yyc operon mutations. Antibiotic susceptibility or nonsusceptibility to daptomycin was verified with Etest strips (bioMérieux) and the broth microdilution method (Table 2). Briefly, daptomycin susceptibility was determined with Etest strips on Mueller-Hinton (Becton Dickinson and Company) agar plates inoculated with 100 μl of a 1:100-diluted 0.5 McFarland suspension of bacteria in 0.85% saline. Additionally, the MIC of daptomycin was determined in triplicate by the broth microdilution method in Mueller-Hinton broth supplemented with Ca2+. The MIC was defined as the lowest antibiotic concentration that inhibited visible growth after 24 h of incubation at 37°C. The potential influences of daptomycin and vancomycin cross-resistance (26, 28, 29, 37, 38) have been addressed by determining the vancomycin MICs (Table 2). Only strain pair CB1663/CB1664 had a significant change in vancomycin susceptibility, with strain CB1664 having an intermediate vancomycin susceptibility phenotype (vancomycin-intermediate S. aureus [VISA]). In addition to determining MICs, the thicknesses of cell wall sacculi were determined by atomic force microscopy (AFM) (Table 2).
TABLE 1.
Isogenic DapS and DapNS S. aureus strain pairs used in this study
| DapS/DapNS strain paira | Methicillin susceptibility | SNPs of DapNS strain |
Source and/or reference | ||
|---|---|---|---|---|---|
| mprF | yycG | rpoB/rpoC | |||
| 616/703 | MSSA | S295Lc | NDd | ND | 21 |
| Q2819/Q2818 | MRSA | S337Lc | Nonee | None | 22 |
| BOY755/BOY300 | MSSA | S295Lc | ND | ND | 30 |
| CB5011/CB5012 | MRSA | L826Fb | None | ND | Cubist Pharmaceuticals, 25 |
| CB5062/CB5063 | MRSA | None | None | None | Cubist Pharmaceuticals, 34 |
| CB1663/CB1664 | MRSA | L826Fb | R86H | None | Cubist Pharmaceuticals, 34 |
All clinical strain pairs were bloodstream isolates.
Mutation in putative mprF synthase domain.
Mutation in putative mprF translocase domain.
ND, not determined.
None, no mutation determined.
TABLE 2.
Daptomycin and vancomycin MICs and thickness of cell wall sacculi of DapS and DapNS strain pairs
| DapS/DapNS strain pair | MIC (μg/ml) |
Cell wall sacculus thickness (nm) | ||
|---|---|---|---|---|
| Daptomycin |
Vancomycin (broth microdilution) | |||
| Etest | Broth microdilution | |||
| 616/703a | 0.094/1.5 | 0.5/2 | 0.5–1/0.5–1 | 16.3 ± 2.0/21.8 ± 2.2 |
| Q2819/Q2818a | 0.094/1–1.5 | 0.5/2 | 0.5–1/1 | 19.5 ± 2.1/19.7 ± 2.3c |
| BOY755/BOY300b | 0.094/1–1.5 | 0.5/2 | 1/1 | 19.9 ± 3.8/21.6 ± 0.9 |
| CB5011/CB5012 | 0.094/5 | 0.5/2 | 0.5/1 | 21.3 ± 2.3/24.6 ± 3.1 |
| CB5062/CB5063 | 0.094/2 | 0.5/4 | 0.5/1 | 20.4 ± 3.1/21.9 ± 1.5 |
| CB1663/CB1664 | 0.125/3 | 0.5/4 | 0.5–1/2–4 | 21.2 ± 2.1/31.4 ± 4.1 |
Patients were treated with vancomycin prior to daptomycin treatment.
No vancomycin treatment of the patient because of allergies.
Not significant. All other strain pairs have P values of < 0.05.
The availability of isogenic DapS and DapNS strain pairs is very limited. In five of the six strain pairs, the DapNS strain had a mutation in mprF (Table 1). This raises the possibility that mprF mutations could be responsible for any common metabolic changes observed in the DapNS strains relative to the susceptible strains. That being said, heterogeneity in the bacterial responses to fitness challenges and the tricarboxylic acid (TCA) cycle inhibitor fluorocitrate suggests that the common metabolic changes in the DapNS strains are due to multiple genetic and/or epigenetic changes.
Bacterial growth conditions.
S. aureus strains were grown in filter-sterilized tryptic soy broth without dextrose (TSB; Becton Dickinson and Company) supplemented with 0.25% glucose (Sigma-Aldrich) or 0.25% [13C6]glucose (Cambridge Isotope Laboratories) or on TSB plates containing 1.5% agar. Bacteria from overnight cultures were diluted 1:100 in TSB and incubated for 1.5 to 2 h. These precultures were centrifuged for 5 min at 5,000 rpm, and the exponentially growing cells were inoculated into prewarmed TSB to an optical density at 600 nm (OD600) of 0.07. All bacterial cultures were incubated at 37°C and aerated at 225 rpm with a flask-to-medium ratio of 10:1. The growth rate (μ) of the S. aureus strains was calculated by the formula (ln OD2 − ln OD1)/(t2 − t1), where OD1 and OD2 are the ODs calculated from the exponential growth phase at times t1 and t2, respectively. The generation time of each strain was determined with the formula ln 2/μ.
For experiments performed with daptomycin (Cubist Pharmaceuticals), the culture medium was supplemented with CaCl2 to a final Ca2+ concentration of 50 μg/ml, as recommended by the manufacturer. In these cultures, daptomycin (0.5 μg/ml for DapS strains and 1.5 μg/ml for DapNS strains) was added to produce a growth reduction of approximately 50% after 2.5 h of incubation.
Determination of competitive fitness.
The relative fitness of the DapS parental strains and the isogenic DapNS isolates was determined in paired competition experiments (39). Precultures were prepared as described above. The precultures were diluted 1:1,000, and equal volumes of DapS and DapNS cultures were used to create a mixed culture that was incubated for 20 to 22 h. The CFU counts of DapS and DapNS strains were determined at the beginning and end of the incubation by spotting 10-μl aliquots of serial dilutions onto nonselective TSB agar plates and onto Ca2+-supplemented plates containing 1 μg of daptomycin/ml. As previously described (40), the number of generations of the competing strains was calculated by the formula (log B − log A)/log2, where A and B are the numbers of CFU per milliliter at the beginning and end of the culture period, respectively. The relative fitness of each strain was determined from the ratio of the number of generations from the DapNS strain to the DapS strain.
Measurement of acetate and ammonium in culture supernatants.
Aliquots (1.5 ml) of bacterial cultures were centrifuged for 2 min at 13,200 rpm, and supernatants were removed and stored at −20°C until use. Acetate and ammonia concentrations were determined with kits purchased from R-Biopharm and used according to the manufacturer's directions. The metabolite concentrations were measured in duplicate in three independent experiments.
Aconitase activity assay.
Bacteria were harvested during the postexponential growth phase (6 h) by centrifugation, suspended in ACN buffer (100 μM fluorocitrate, 90 mM Tris/HCl, pH 8.0), and lysed with lysing matrix B tubes and a FastPrep instrument (MP Biomedicals). The lysate was centrifuged for 5 min at 13,200 rpm at 4°C, and the aconitase activity in the cell-free lysate was measured by the method of Kennedy et al. (41). One unit of aconitase activity is defined as the amount of enzyme necessary to give a ΔA240 min−1 of 0.0033. Protein concentrations were determined by the method of Bradford (42).
NMR sample preparation.
Samples for intracellular metabolite analysis were prepared from independent cultures in the exponential (2.5 h) and postexponential (6 h) growth phases. For two-dimensional (2D) 1H-13C heteronuclear single quantum coherence (HSQC) analysis, bacteria were cultivated in TSB medium containing 0.25% [13C6]glucose. Bacteria were harvested by vacuum filtration with 0.45-μm-pore-size Microfil V filters (Millipore) that were prewashed with extraction buffer (20 mM phosphate buffer; uncorrected pH 7.2). The bacteria containing filters were placed into precooled 50-ml conical tubes and immersed in liquid nitrogen. The bacteria were suspended from the filter with ice-cold extraction buffer and washed to remove residual medium components. Cells were adjusted to 20 OD600 units and lysed with precooled lysing matrix B tubes and a FastPrep instrument (MP Biomedicals). The resulting lysates were centrifuged at −9°C to remove cell debris and glass beads. A second extraction step was performed with the same extraction buffer, and the samples were pooled. A final volume of 1.5 ml of cell-free lysate was frozen in dry ice, lyophilized, and kept at −80°C until use for NMR analysis. At the time of use, the samples were dissolved in 600 μl of D2O containing 50 μM 3-(trimethylsilyl)propionic-2,2,3,3-d4 acid sodium salt (TMSP) for one-dimensional (1D) 1H spectra or 500 μM TMSP for 2D 1H-13C HSQC spectra and transferred to NMR tubes.
NMR data collection and analysis.
The 1D and 2D NMR spectra were collected as described previously (43, 44). NMR spectra were collected on a Bruker 500-MHz Avance DRX spectrometer equipped with a 5-mm triple-resonance cryoprobe (1H, 13C, 15N) with a z axis gradient, a BACS-120 sample changer, an automatic tuning and matching accessory, and Bruker Icon NMR software.
1D 1H NMR spectra were processed with our MVAPACK software suite (http://bionmr.unl.edu/mvapack.php) (45). Each spectrum was Fourier transformed and then automatically phased (46), binned (47), normalized by standard normal variate normalization, denoised with removal of solvent and buffer peaks, and unit variance scaled (for principal-component analysis [PCA]) or Pareto scaled (for orthogonal partial least-squares discriminant analysis [OPLS-DA]). PCA, linear discriminant analysis (LDA), and OPLS-DA models were calculated with MVAPACK. Hotelling 95% confidence ellipsoids, PCA score dendrograms, and corresponding Mahalanobis P values were generated with our PCA/PLS-DA utilities (http://bionmr.unl.edu/pca-utils.php) (48, 49) to determine the statistical significance of group separation in the PCA score plots. An observed P value of ≤0.05 was used to identify statistically significant group separation. The OPLS-DA models were validated by analysis of variance of the cross-validated residuals (CV-ANOVA) (50) and 7-fold Monte Carlo single cross-validation (51).
The 2D 1H-13C HSQC spectra were processed with NMRPipe (52) and analyzed with NMRViewJ (53). Peak intensities were normalized and referenced to TMSP. The relative concentration of each metabolite was defined on the basis of the average intensity of all of the NMR peaks assigned to the metabolite. Peak annotation was accomplished by comparing the observed 1H and 13C chemical shifts against chemical shifts for known metabolites deposited in three metabolomic databases, the Platform for RIKEN Metabolomics (http://prime.psc.riken.jp/) (54), the Human Metabolome Database (http://www.hmdb.ca/) (55), and the BiomagResBank (http://www.bmrb.wisc.edu/) (56). Error tolerances of 0.08 and 0.25 ppm for 1H and 13C chemical shifts, respectively, were used to make a peak assignment.
Peptidoglycan preparation.
Peptidoglycan purification was based on the method of Jonge and colleagues (57). Briefly, bacterial cultures (10 ml) from the postexponential growth phase (6 h) were harvested by centrifugation for 10 min at 4,000 rpm (4°C) and suspended in 5 ml of ice-cold 50 mM Tris-HCl (pH 7.0). The cell suspension was transferred dropwise into 15 ml of 5% boiling sodium dodecyl sulfate (SDS), boiled for 30 min, and concentrated by centrifugation for 10 min at 4,000 rpm. The pellet was washed twice with 1 M NaCl and three times with hot deionized water (60°C) to remove the SDS. Cells were broken with lysing matrix B tubes and a FastPrep instrument (PreCellys 24; Peqlab). Broken cell walls and glass beads were separated by pipetting the suspension through a cell sieve (40-μm mesh size). The cell walls were concentrated by centrifugation at 10,000 rpm for 30 min at room temperature; suspended in 750 μl of 100 mM Tris-HCl (pH 8.0) containing 20 mM MgSO4, 10 μg/ml DNase A, and 50 μg/ml RNase I; and then incubated at 37°C for 2 h. After incubation, 100 μg/ml proteinase K and 10 mM CaCl2 were added and the mixture was incubated overnight at 37°C. After protein digestion, the peptidoglycan was collected by centrifugation as described above and washed two times with deionized water, once with 8 M LiCl, once with 10 mM EDTA, and twice with deionized water. For AFM imaging, 10 μl of the diluted peptidoglycan suspension was placed on freshly cleaved mica and dried under a soft stream of water-free nitrogen gas (grade 5.0 or higher).
AFM imaging.
For AFM imaging, a Bioscope Catalyst Atomic Force Microscope (Bruker Nano Surface, Karlsruhe, Germany) mounted on an inverted light microscope (Leica DMI 4000B; Leica Microsystems GmbH) operating in phase-contrast mode was used. The ScanAsyst mode was used to visualize peptidoglycan. All scans were carried out in air (20°C with 43% relative humidity) with a tip velocity of 20 μm/s and a resolution of 1,024 by 1,024 pixels. ScanAsyst Air Cantilevers (Bruker AXS S.A.S.) featuring tip curvatures of <7 nm were used for imaging, and the spring constant was calibrated by the thermal tuning method described by Hutter and Bechhoefer (58). Image processing was done with Gywiddion open-source image processing software (59). The heights of 50 individual cell walls were measured and used for statistical analysis with SigmaPlot 11 (Systat Software GmbH) and one-way ANOVA (Table 2).
RESULTS
DapNS strains have a delayed transition into postexponential growth.
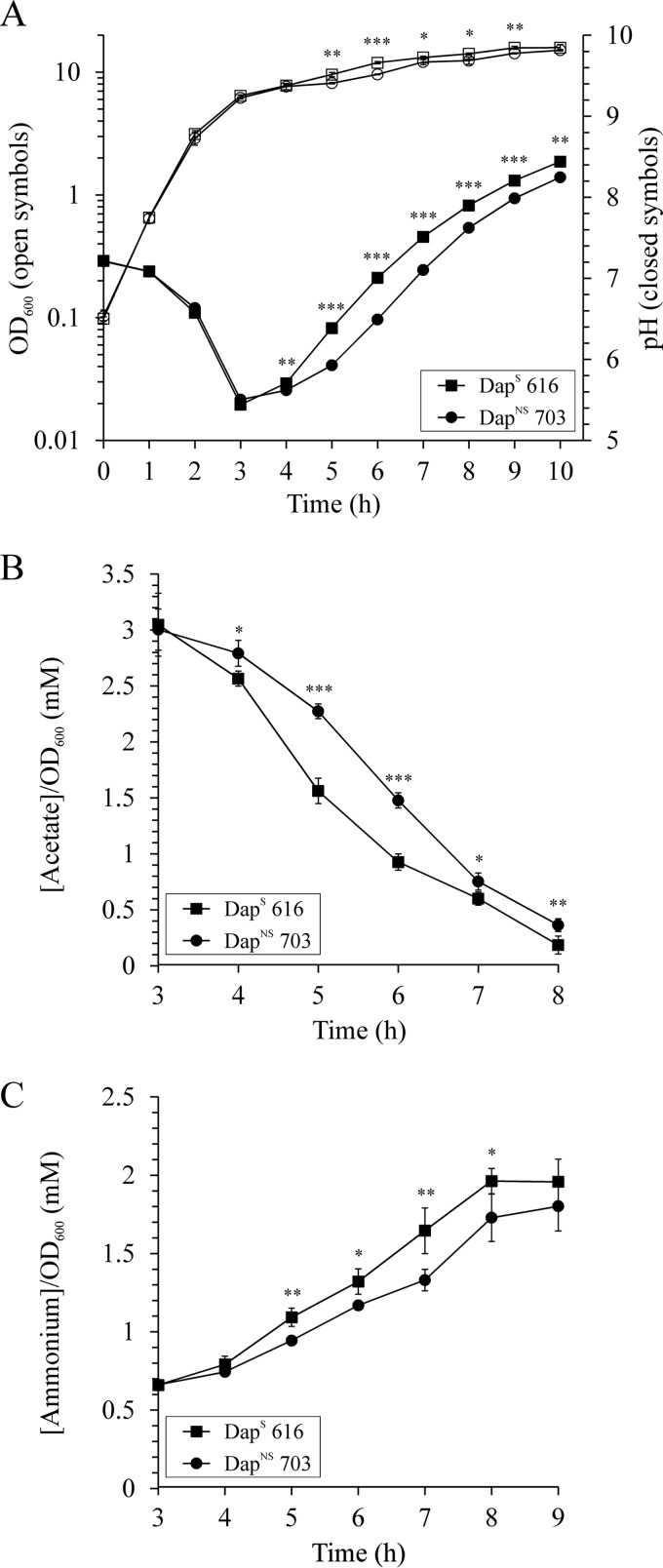
The transition from antibiotic susceptibility to nonsusceptibility can coincide with growth alterations in S. aureus (60). To determine if the transition from a DapS to a DapNS state affected bacterial growth, we assessed the growth of six DapNS strains and their isogenic parental DapS strains. Five strain pairs revealed similar exponential growth phase profiles with generation times of 24 to 27 min and comparable acidification of the culture medium with pH minima after 3 to 4 h of growth (e.g., DapS 616 and DapNS 703, Fig. 1A and Table 3; see Fig. S1 in the supplemental material). In contrast to the growth similarities of most DapS and DapNS strain pairs, the generation time of DapNS strain CB1664 was significantly higher than that of isogenic DapS strain CB1663 (37 ± 1 and 26 ± 0 min, respectively); this resulted in a late pH minimum for strain CB1664 after 6 h of growth (see Fig. S1E in the supplemental material). Interestingly, strain CB1664 is the only strain that also has an intermediate vancomycin resistance phenotype. Although the growth profiles of most of the strain pairs were similar, a slight difference in growth was observed at the transition between the exponential and postexponential growth phases. These slight differences often reflect a metabolic transition similar to a diauxic shift, which occurs when bacteria are switching from the catabolism of one carbon source to that of another.
FIG 1.
Growth and physiological characteristics of DapS (strain 616) and isogenic derivative DapNS (strain 703) bacteria. (A) Growth and pH profile of strain pair 616/703 grown aerobically in TSB. (B and C) Acetate depletion (B) and net ammonium accumulation (C) in the culture supernatants of strain pair 616/703. The results presented are the averages and standard deviations of three independent experiments. Symbols are defined in the insets. The statistical significance of differences between strains 616 and 703, determined by Student's t test, is indicated (*, P <0.1; **, P <0.05; ***, P <0.005).
TABLE 3.
Summary growth characteristics and extracellular metabolites of DapS and DapNS strain pairsa
| Strain | Generation time (min) | OD600b | pHb | Acetateb concn (mM) | Ammoniumb concn (mM) |
|---|---|---|---|---|---|
| 616 | 24 ± 0 | 11.9 ± 0.3 (0.0021) | 7.0 ± 0.0 (0.0002) | 11.1 ± 0.7 (0.0055) | 15.8 ± 0.6 (0.0025) |
| 703 | 25 ± 1 | 9.6 ± 0.1 | 6.5 ± 0.0 | 14.2 ± 0.6 | 11.2 ± 0.3 |
| Q2819 | 25 ± 0 | 11.0 ± 0.4 (0.0096) | 9.6 ± 0.9 (0.0068) | 12.6 ± 0.6 (0.0150) | |
| Q2818 | 26 ± 1 | 9.7 ± 0.3 | 6.5 ± 0.0 | 13.7 ± 0.4 | 10.3 ± 0.2 |
| BOY755 | 25 ± 1 | 10.6 ± 0.3 (0.0095) | 6.7 ± 0.0 (0.0006) | 11.8 ± 0.7 (0.0120) | 12.1 ± 0.4 (0.0204) |
| BOY300 | 26 ± 1 | 9.4 ± 0.3 | 6.4 ± 0.0 | 15.2 ± 0.1 | 9.6 ± 0.9 |
| CB5011 | 25 ± 0 | 10.2 ± 0.8 (0.1120) | 6.9 ± 0.1 (0.0347) | 10.8 ± 1.4 (0.0575) | NDc |
| CB5012 | 26 ± 0 | 8.9 ± 0.2 | 6.5 ± 0.1 | 13.6 ± 0.8 | |
| CB5062 | 26 ± 0 | 9.5 ± 0.6 (0.3721) | 6.1 ± 0.1 (0.2799) | ND | ND |
| CB5063 | 27 ± 0 | 9.1 ± 0.5 | 6.2 ± 0.0 | ||
| CB1663 | 26 ± 0 | 9.7 ± 1.2 (0.3400) | 6.5 ± 0.3 (0.0404) | ND | ND |
| CB1664 | 37 ± 1 | 8.8 ± 0.1 | 5.8 ± 0.1 |
Average values of three independent experiments ± the standard deviations are shown.
Values were determined after 6 h of growth in TSB under aerobic conditions. The statistical significance (P value) of the difference of the DapS value from that of the corresponding DapNS strain, determined by the Student t test, is shown in parentheses.
ND, not determined.
During rapid aerobic growth in glucose-containing medium, S. aureus incompletely oxidizes glucose to acetyl coenzyme A (acetyl-CoA), which causes acetate to accumulate in the culture medium, resulting in a decreased pH (61–63). Induction of the TCA cycle facilitates the utilization of acetate, a process that requires anaplerotic reactions to offset the withdrawal of biosynthetic intermediates. Commonly, amino acids are used to provide the anaplerotic carbons, a process that requires deamination of the amino acids, which causes ammonia to accumulate in the culture medium (62). The combined acid extraction from the culture medium and the accumulation of ammonia causes the medium to alkalinize. The exponential growth phase acidification of the culture medium was similar for five of the six strain pairs examined in this study; however, the postexponential growth phase alkalization was delayed in four of the six DapNS strains (Fig. 1A and Table 3; see Fig. S1 in the supplemental material). This delay in alkalization of the culture medium is likely a reflection of that diauxic-shift-like change observed at the exponential-to-postexponential growth phase transition in the growth profiles. Taken together, most DapNS strains had slightly delayed transitions into the postexponential growth phase.
Competitive fitness.
The similar generation times observed for most DapNS and DapS strain pairs led us to examine if a biological fitness cost was associated with daptomycin nonsusceptibility in S. aureus. For this analysis, mixed-culture competition assays of DapNS and DapS strain pairs were performed in the absence of selective pressure. The relative fitness of most of the isogenic pairs ranged from 0.761 ± 0.004 to 0.98 ± 0.02; however, the CB1663/CB1664 strain pair had a more pronounced relative fitness difference of 0.63 ± 0.02 (Table. 4). This difference is likely a reflection of the longer generation time of VISA DapNS strain CB1664 than that of DapS strain CB1663 (Tables 2 and 3; see Fig. S1E in the supplemental material). In other words, the biological cost of daptomycin nonsusceptibility ranged from no or little loss of fitness to a considerable loss of fitness. This may be of importance with respect to the persistence of DapNS strains in the clinical setting.
TABLE 4.
Relative fitness of DapNS S. aureus strains
| DapS/DapNS strain pair | Mean relative fitness ± SEMa | P valueb |
|---|---|---|
| 616/703 | 0.953 ± 0.037 | 0.4708 |
| Q2819/Q2818 | 0.761 ± 0.004 | <0.0001 |
| BOY755/BOY300 | 0.914 ± 0.039 | 0.0684 |
| CB5011/CB5012 | 0.801 ± 0.030 | 0.0010 |
| CB5062/CB5063 | 0.982 ± 0.021 | 0.2312 |
| CB1663/CB1664 | 0.625 ± 0.019 | <0.0001 |
Values were derived from at least four independent experiments.
The statistical significance (P value) of the difference between the formed generations of the DapS strain and those of the corresponding DapNS strain was determined by Student's t test.
Delayed transition into postexponential growth of DapNS strains correlates with decreased TCA cycle activity.
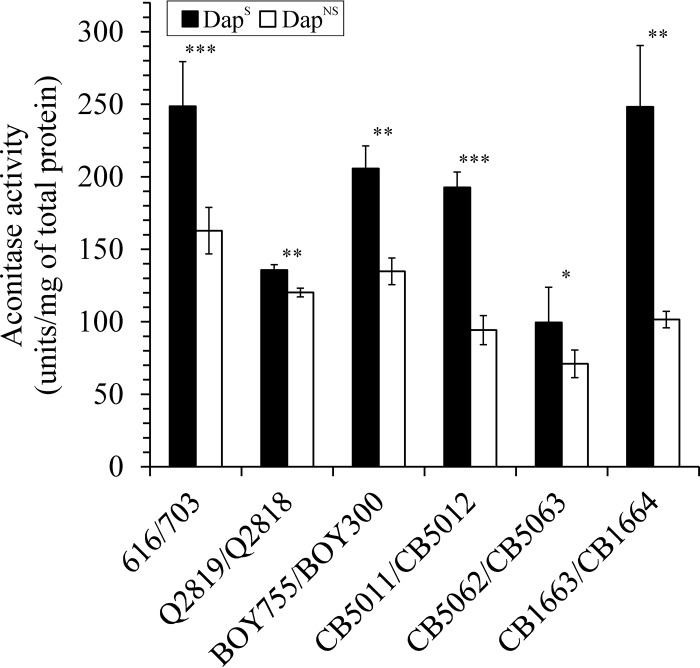
The diauxic-shift-like change in the growth of the DapNS strains and the altered pH profiles in the postexponential growth phase suggested that catabolism of incompletely oxidized carbon sources (i.e., acetate) was altered during the transition to a DapNS state. To assess if the catabolism of carbon sources was altered, the concentrations of acetate and ammonium in the culture medium were determined in the postexponential growth phase. As expected, the postexponential growth phase catabolism of acetate and generation of ammonia was significantly lower in DapNS strains than in the isogenic DapS strains (Fig. 1B and C and Table 3). As stated above, the catabolism of acetate requires a functioning TCA cycle; hence, the delayed alkalization of the medium and acetate catabolism suggested that nonsusceptibility to daptomycin corresponds to decreased carbon flow through the TCA cycle. To test this suggestion, the activity of the TCA cycle enzyme aconitase was assessed in DapS and DapNS S. aureus strains after 6 h of incubation. Consistent with the physiological differences at the transition to the postexponential growth phase, the aconitase activity was significantly lower in all six DapNS strains than in the parental DapS strains (Fig. 2). Taken together, the subtle physiological differences between the DapNS and DapS strains suggest that the delayed transition to postexponential growth corresponded to altered central metabolism, specifically, the TCA cycle.
FIG 2.
DapNS strains have less aconitase activity than DapS parental strains. Aconitase activities were determined in triplicate in at least three independent cultures after 6 h (postexponential phase) of growth. The results presented are averages, and error bars represent the standard deviations. The statistical significance of differences between a DapS strain and the corresponding DapNS strain, determined by Student's t test, is indicated (*, P < 0.1; **, P < 0.05; ***, P < 0.005).
DapS and DapNS strains have significantly different metabolic states.
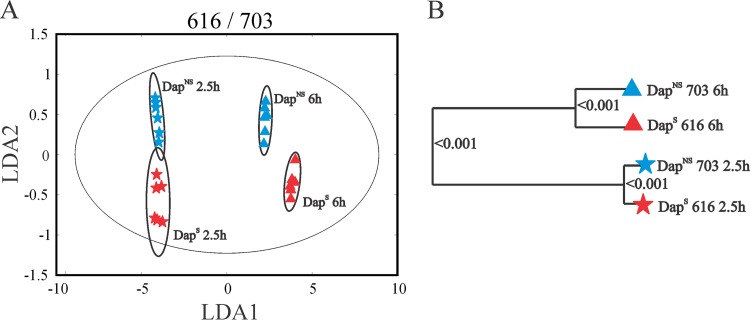
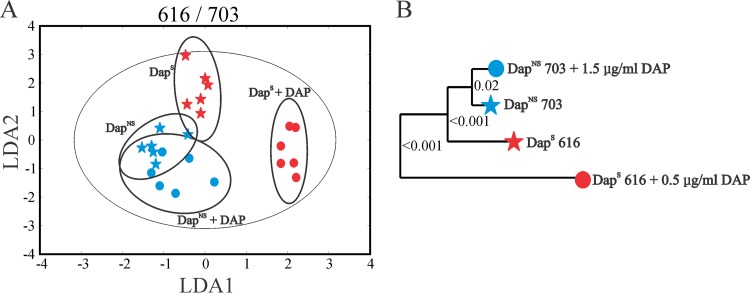
The physiologic and enzymatic data strongly indicated that the transition of S. aureus to a daptomycin nonsusceptibility phenotype coincided with metabolic changes. To examine these metabolic changes in more detail, the metabolomes of three strain pairs (i.e., 616/703, Q2819/Q2818, and BOY755/BOY300) were assessed by NMR metabolomics during the exponential (2.5 h) and postexponential (6 h) growth phases. The 1D 1H NMR metabolic profiles were first analyzed by PCA, and then LDA was used to convert the 3D PCA score plot into a 2D plot to simplify data visualization. LDA is used to identify a rotational orientation that provides an optimal view of a 3D data set, which is then projected onto a 2D plane (64, 65). The LDA plot of the 616/703 strain pair (Fig. 3A) revealed four distinct clusters formed by the strain and growth phase, demonstrating that the transition to a DapNS state significantly altered the metabolome of strain 703 relative to that of strain 616. The corresponding metabolomic tree diagram generated from the PCA scores and associated P values for each cluster were determined to assess the significance and the distance between groups (Fig. 3B). As expected, the largest separation was observed between the exponential and postexponential growth phases, as this growth phase transition coincides with major metabolic changes, specifically, derepression of the TCA cycle and oxidative phosphorylation. The separation between DapS and DapNS was apparent during both the exponential and postexponential growth phases; however, the metabolic differences were more pronounced after 6 h of cultivation. This metabolic difference is consistent with the postexponential-phase physiological observations (Fig. 1 and Table 3). Similar clustering differences in the PCA score plots were observed for strain pairs Q2819/Q2818 and BOY755/BOY300 (see Fig. S2 in the supplemental material).
FIG 3.
Metabolic profiles of DapS strain 616 and isogenic derivative DapNS strain 703. Clustering of metabolomes from the exponential (2.5 h) and postexponential (6 h) growth phases examined by PCA of 1D 1H-NMR spectra and then presented by LDA demonstrates significantly different metabolic profiles in the 2D LDA plot (A) and the dendrogram plot (B), both of which are generated from 3D PCA scores. The ellipses correspond to the 95% confidence limits of a normal distribution for each cluster. Each dendrogram node is labeled with a P value; a lower P value indicates a larger separation between the clades.
OPLS-DA was used to identify the spectral features (metabolites) that significantly contribute to the observed class separations. OPLS-DA models were calculated by using one predictive and two orthogonal components. The quality of OPLS-DA models was evaluated on the basis of cross-validation by a Monte Carlo leave-n-out procedure (66) and CV-ANOVA (see Table S1 in the supplemental material). As an example, the R2 (degree of fit), Q2 (predictive ability), and CV-ANOVA P value for the comparisons between the 616 and 703 strains after 2.5 or 6 h of growth were 0.9949, 0.8093, and 4.9 × 10−3 and 0.9972, 0.9280, and 1.2 × 10−3, respectively. Importantly, the validated OPLS-DA model further supports the trends seen in the 3D PCA score plots (Fig. 3; see Fig. S2 in the supplemental material). In addition, OPLS-DA identified the relative contributions of each NMR bin (i.e., 1H NMR chemical shift and associated metabolites) to the group separation based on an associated S plot (see Fig. S3 in the supplemental material). Each NMR bin with a high correlation {p(corr)[1] > 0.8 or ≤ 0.8} and covariance (p[1] > 1.0 or ≤ 0.1) was considered to be a major contributor to the class separation in the OPLS-DA score plot. A comparison of these major contributors to group separation revealed that 45% of the NMR bins were common to at least two of the three strain pairs during postexponential growth. Similarly, 30% of the NMR bins were common to at least two of the three strain pairs during exponential growth. This analysis suggests a common difference between the metabolomes of the three DapS and DapNS strain pairs (see Table S2 in the supplemental material). Overall, these results further support the notion that the transition of S. aureus to a DapNS state coincides with an altered metabolome relative to that of DapS parental strains.
The transition to a daptomycin nonsusceptibility phenotype alters central, amino acid, pyrimidine, and purine metabolism.
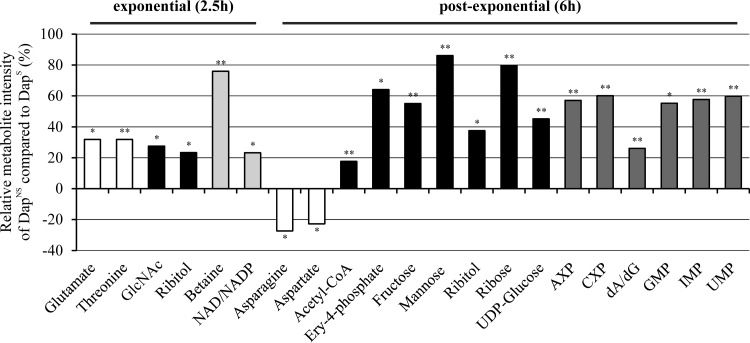
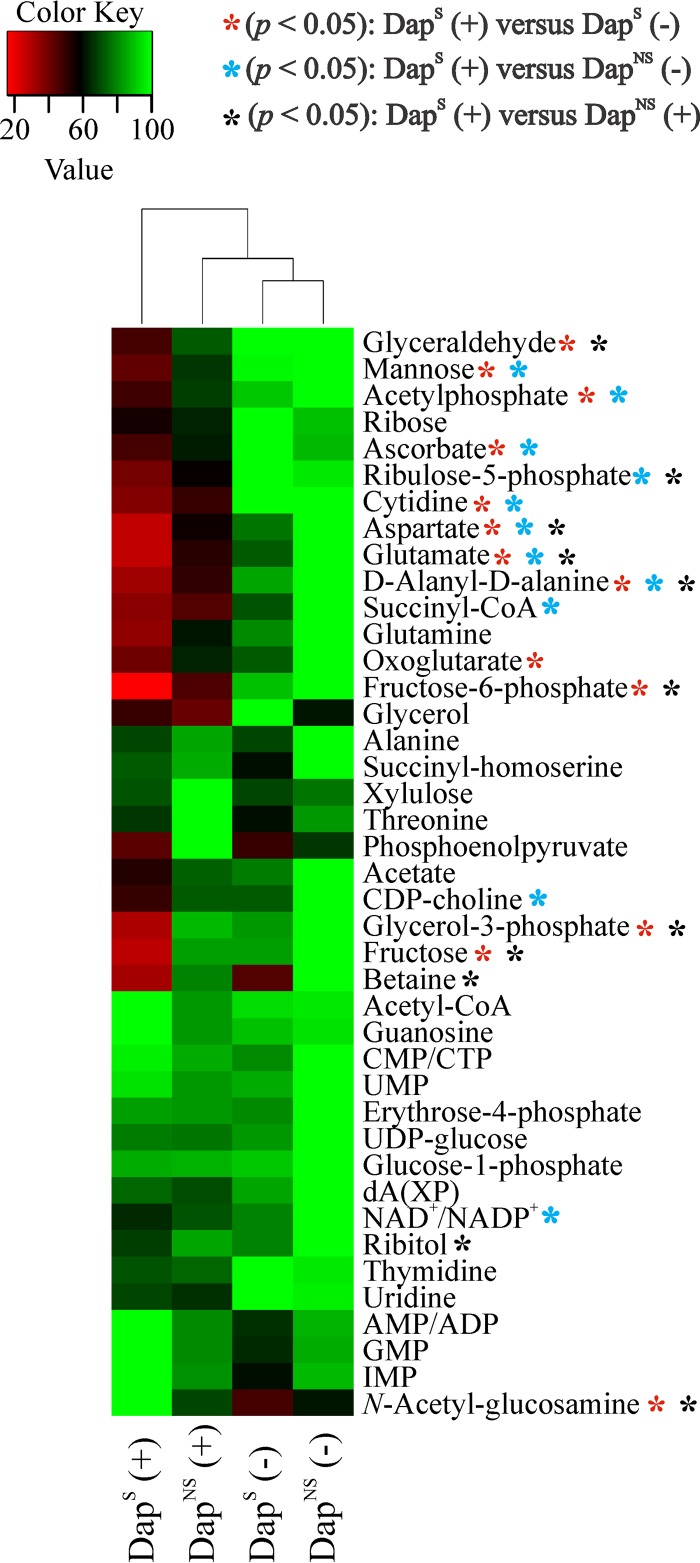
Because of the large number and significant overlap of peaks in a 1D 1H NMR spectrum and the chemical shift degeneracy of metabolites, the assignment of metabolites based solely on 1D 1H NMR can be challenging. To facilitate the identification of altered metabolites between strain pairs, 2D 1H-13C HSQC NMR was used to identify metabolic changes between strains 616 and 703. The reduced complexity of the 2D 1H-13C HQSC spectra and the two correlated and distinct chemical shifts allowed for improved metabolite assignments. In addition to aiding in metabolite identification, the 13C-labeled glucose provides information about the flow of carbon through the metabolome. By this approach, 41 and 34 metabolites were identified in the exponential and postexponential growth phases (see Table S3 in the supplemental material), respectively, accounting for about 60% of the peaks in the 2D 1H-13C HSQC spectra. Similar to the physiological data and PCA and OPLS-DA models (Table 3 and Fig. 3; see Table S2 in the supplemental material), the majority of the metabolic changes between DapS and DapNS strains were found in the postexponential growth phase (Fig. 4).
FIG 4.
Significantly altered metabolites of DapNS strain 703 compared to those of parental DapS strain 616. Metabolite concentration changes were identified by 2D 1H-13C HSQC NMR analyses in triplicate of bacteria grown aerobically in TSB medium containing [13C]glucose. Metabolites are highlighted according to their chemical nature as amino acids (white), amino sugar/sugars/sugar alcohol (black), pyrimidines/purines (dark gray), or others (bright gray). Positive values represent higher concentrations, and negative values represent lower concentrations, of the metabolites in the DapNS strain than those of the metabolites in the DapS strain. Abbreviations: GlcNAc, N-acetyl-d-glucosamine; Ery-4-phosphate, erythrose-4-phophate; AXP, AMP, ADP, or ATP; CXP, CMP, CDP, or CTP; dA/dG, deoxyadenosine/deoxyguanosine. Each bar is shown with the statistical significance determined by Student's t test at the 90% (*, P < 0.1) or 95% (**, P < 0.05) confidence level.
All exponential growth phase metabolites whose concentrations were significantly altered were more abundant in DapNS strain 703. Importantly, N-acetyl-d-glucosamine was increased in DapNS strains. UDP-activated N-acetylglucosamine is synthesized from the glycolysis/gluconeogenesis intermediate fructose-6-phosphate and serves as a precursor for the synthesis of the exopolysaccharides polysaccharide intercellular adhesion (PIA) and capsule, which are associated with biofilm formation (67) and virulence (68). In addition, N-acetylglucosamine is a component of peptidoglycan and WTA and a biosynthetic intermediate used in the synthesis of N-acetylmuramic acid and N-acetylmannosamine, major components of peptidoglycan and WTA (69), respectively. The concentration of ribitol, a precursor of CDP-ribitol that is used in WTA biosynthesis (70), was also significantly increased in DapNS strains during both the exponential and postexponential growth phases. In contrast, the concentration of the WTA component mannose was significantly altered only in the postexponential growth phase. Other metabolites with increased concentrations in DapNS strains were erythrose-4-phosphate, ribose, and fructose, which relate to the pentose phosphate pathway that is necessary for the synthesis of the WTA precursor ribitol. Overall, these data suggest an increased carbon flow of glucose into pathways associated with the synthesis of cell wall components in DapNS.
An increase in carbon flow to pathways that provide biosynthetic intermediates necessary for cell wall precursors implies that carbon is directed away from pathways not involved in cell wall precursor synthesis. Consistent with this implication, the reduced activity of the TCA cycle (Fig. 2) suggests that carbon cannot enter into the TCA cycle in DapNS strains at a rate equivalent to that at which it does in susceptible strains. This is reflected in higher concentrations of acetyl-CoA in DapNS strains and in the lower concentrations of the TCA cycle-derived amino acids glutamate, aspartate, and asparagine in DapNS strain 703 than in susceptible strain 616 in the postexponential phase (Fig. 4; see Table S2 in the supplemental material).
In addition to the changes in central and amino acid metabolism, there were alterations in pyrimidine and purine metabolism, with higher concentrations of several nucleotides in DapNS strains than in DapS strains. Taken together, the metabolic perturbations associated with daptomycin nonsusceptibility in S. aureus involve (i) decreased TCA cycle activity, (ii) increased synthesis of pyrimidines and purines, and (iii) increased carbon flow in pathways associated with WTA and peptidoglycan biosynthesis.
DapNS strain metabolism is minimally altered during growth in the presence of daptomycin.
For most S. aureus strains, the transition to the daptomycin nonsusceptibility phenotype caused little or no loss of fitness in the absence of daptomycin in the cultivation medium (Table 4). To determine if the transition to the daptomycin nonsusceptibility phenotype alters metabolism in DapS and DapNS strains, the strain pairs 616/703, Q2819/Q2818, and BOY755/BOY300 were treated with a concentration of daptomycin sufficient to produce an ∼50% reduction in growth and the metabolomes were harvested and analyzed by 1D 1H NMR. Daptomycin concentrations of 0.5 and 1.5 μg/ml for DapS and DapNS strains, respectively, were empirically determined to reduce bacterial growth to comparable levels (45% ± 15%) after 2.5 h of incubation. As expected, challenging S. aureus cultures with daptomycin resulted in metabolic alterations that caused group separation in the LDA plots generated from the 1D 1H NMR spectra (Fig. 5A). Importantly, the corresponding dendrogram illustrates that the metabolomes of DapNS strain 703, when challenged with daptomycin, clustered closer to the untreated metabolomes than did treated parental strain 616 (Fig. 5B). Similar patterns were also observed in strain pairs Q2819/Q2818 and BOY755/BOY300 (see Fig. S3 in the supplemental material). These results were also verified by OPLS-DA (see Table S1 in the supplemental material).
FIG 5.
Exposure to daptomycin alters the metabolome of DapS strain 616 to a greater extent than that of DapNS strain 703. S. aureus cultures were challenged with daptomycin at concentrations leading to a growth reduction of approximately 50% at the time of harvest after 2.5 h. Differences in the intracellular metabolomes were examined by PCA of the 1D 1H-NMR spectra and are presented as a 2D LDA plot (A) and a dendrogram plot (B), both of which were generated from 3D PCA scores. The ellipses correspond to the 95% confidence limits of a normal distribution for each cluster. Each dendrogram node is labeled with a P value; a lower P value indicates greater separation between the clades.
For all three strain pairs analyzed, the groups of daptomycin-treated and untreated DapNS cells were closer to each other than the groups of treated and untreated DapS cells. This was apparent from the clustering patterns of the groups and the P values reported for each clade separation in the dendrogram, in which the P values were used to quantify the distance between separated clades. A pairwise P value was also calculated for each pair of analyzed groups in order to measure the relative distance (see Table S4 in the supplemental material). As an example, the groups of bacteria of strain 616 with or without daptomycin treatment were separated with a P value of 1.06 × 10−6, while the higher P value of 1.70 × 10−2 for DapNS strain 703 represented a shorter distance; hence, fewer metabolic alterations were associated with daptomycin challenge in strain 703 than in strain 616. The P values for challenged and unchallenged cultures of strain Q2819 (2.79 × 10−6) versus strain Q2818 (2.23 × 10−3) and strain BOY755 (2.85 × 10−10) versus strain BOY300 (1.03 × 10−2) revealed patterns similar to that of strain pair 616/703. Additionally, the metabolomes of daptomycin-treated DapNS cells were more similar to those of the untreated parental DapS cells (P values of 3.98 × 10−4, 7.48 × 10−6, and 1.86 × 10−4 for strain pairs 616/703, Q2819/Q2818, and BOY755/BOY300, respectively) than the metabolomes of daptomycin-treated DapS cultures (P values of 1.06 × 10−6, 2.79 × 10−6, and 2.85 × 10−10 for strains 616, Q2819, and BOY755, respectively). These data suggest that the metabolism of DapNS strains is preadapted to permit growth in the presence of daptomycin.
The 2D 1H-13C HSQC analysis of DapS (616) and DapNS (703) with 13C6-labeled glucose revealed a broad impact of daptomycin on the metabolomes with decreased concentrations for the majority of metabolites compared to untreated bacteria (Fig. 6). Challenging bacteria of both DapS and DapNS strains with daptomycin also resulted in reduced concentrations of the TCA cycle intermediate 2-oxoglutarate and TCA cycle-derived amino acids (i.e., Asp, Glu). While the concentrations of Asp and Glu were decreased in both DapS and DapNS strains, significantly lower concentrations were detected in DapS strain 616 than in strain 703. Similar patterns of significantly lower metabolite concentrations in strain 616 than in strain 703 after treatment with daptomycin were observed for metabolites associated with glycolysis (i.e., fructose, fructose-6-phosphate, glyceraldehyde, and acetylphosphate), the pentose phosphate pathway (i.e., ribulose-5-phosphate), and the peptidoglycan precursor d-alanyl–d-alanine and for WTA precursors (i.e., ribitol, glycerol-3-phosphate, and mannose). In contrast, the concentration of N-acetylglucosamine was higher in DapS cells exposed to daptomycin than in unchallenged DapS strain 616 or challenged DapNS strain 703. Notably, the higher concentration of N-acetylglucosamine in DapNS strains than in DapS strains observed under unchallenged conditions (Fig. 4) was not significantly altered during a daptomycin challenge (Fig. 6; see Table S3 in the supplemental material), suggesting that the metabolism of DapNS strains is better prepared for growth in the presence of daptomycin.
FIG 6.
Metabolic alterations of S. aureus due to a daptomycin challenge. The metabolomes of DapS strain 616 and DapNS strain 703 with (+) or without (−) a daptomycin challenge were analyzed by 2D 1H-13C-HSQC after 2.5 h of aerobic growth in TSB containing 13C-labeled glucose in three independent experiments. The relative concentrations of the metabolites identified in three independent replicates are shown on a color scale heat map, and hierarchical clustering is depicted by dendrograms. The statistical significance of differences between the metabolite concentrations of treated and untreated DapS (red) or DapNS (blue) bacteria or treated DapS and treated DapNS (black) bacteria, determined by Student's t test, is indicated (*, P < 0.1; **, P < 0.05).
Metabolic intervention to increase the daptomycin sensitivity of DapNS strains.
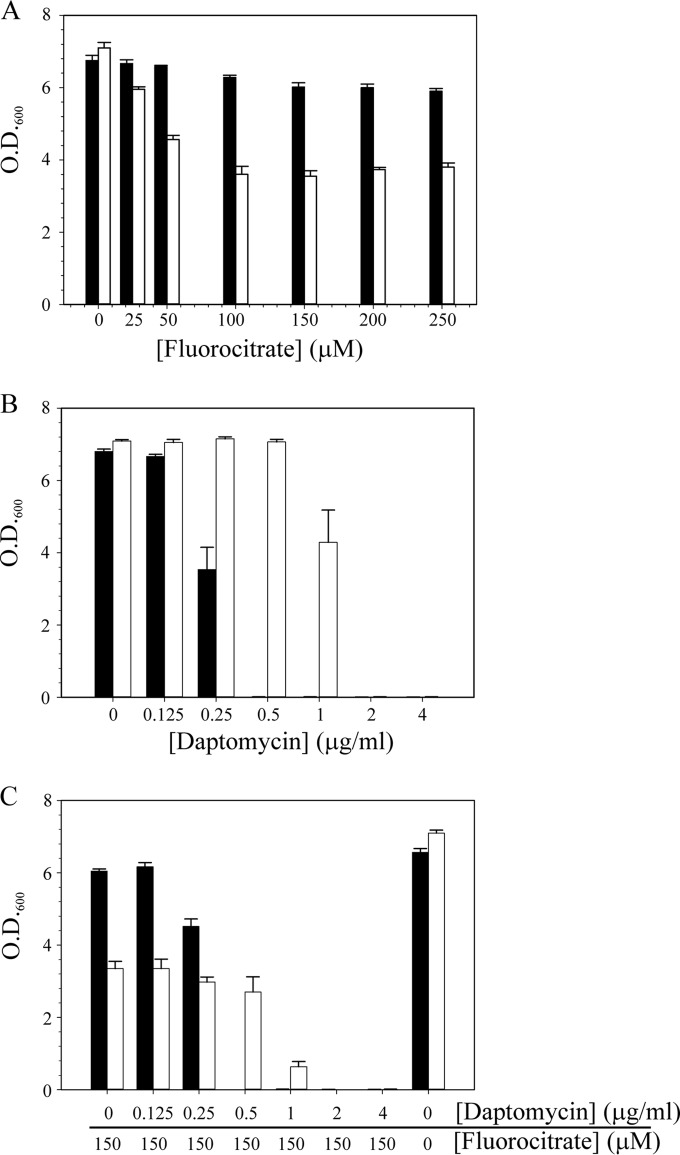
Transition to the daptomycin nonsusceptibility phenotype alters metabolism (Fig. 3 and 4). When bacteria alter carbon flow through a metabolic pathway, they can become more or less sensitive to inhibitors of those enzymes or pathways. If the level of an enzyme is decreased, the bacteria might become more resistant to an inhibitor because of a decrease in the number of targets for that inhibitor. Conversely, bacteria can become more sensitive to an inhibitor because the inhibitor concentration overwhelms the reduced enzyme level. To determine if inhibiting an enzyme or pathway would alter the daptomycin susceptibility of DapNS strains, the aconitase-specific inhibitor fluorocitrate was added to the culture medium in the presence or absence of daptomycin and growth was assessed after 12 h of incubation (Fig. 7). The addition of fluorocitrate to the culture medium significantly decreased the growth yield of S. aureus DapNS strain 703 relative to that of DapS strain 603 (Fig. 7A). In fact, the DapS strain was largely unaffected by the addition of fluorocitrate. As expected, in the absence of fluorocitrate, the DapNS strain was more resistant to daptomycin than the susceptible strain was (Fig. 7B). In contrast, when fluorocitrate and daptomycin were combined, the DapNS strain became significantly (P ≤ 0.05) more sensitive to daptomycin (Fig. 7C). Analysis of the remaining strain pairs found that the sensitivity of the DapNS strains to fluorocitrate varied from none (strains BOY755, CB5012, and Q2818) to a sensitivity resembling that of strain 703 (strains CB1664 and, to a lesser extent, CB5063). Taken together, these data demonstrate that the daptomycin nonsusceptibility phenotype can be altered by manipulating metabolism. In addition, the heterogeneity of responses to fluorocitrate suggests that the TCA cycle changes are not the primary metabolic adaptation that confers the daptomycin nonsusceptibility phenotype on S. aureus. These data also demonstrate that susceptibility to fluorocitrate cannot be attributed to mutations in mprF because strains with the S295L mutation (i.e., 703 and BOY300) differ profoundly in fluorocitrate susceptibility.
FIG 7.
Fluorocitrate increases the daptomycin susceptibility of DapNS strain 703. Fluorocitrate (A), daptomycin (B), or both fluorocitrate and daptomycin (C) were added to the cultivation medium, Mueller-Hinton broth, at the concentrations indicated, and the OD600 was determined after 12 h of aerobic growth. The data represent the mean and SEM of at least three independent experiments. Strain 616 is represented by black bars, and strain 703 is represented by white bars.
DISCUSSION
The transition from antibiotic susceptibility to nonsusceptibility often coincides with a reduced growth rate and/or reduced fitness. As examples, the acquisition of new genetic determinants such as the staphylococcal chromosomal cassette (SCCmec) that confers resistance to β-lactam antibiotics caused severe growth reductions in the first MRSA strains (71). Accordingly, these strains were restricted to clinical settings where high selective antibiotic pressure predominates. Evolution has reduced the size of SCCmec elements, and the fitness of these MRSA strains has increased, allowing these strains to persist in the community (72, 73). Antibiotics such as mupirocin (74) or rifampin (40, 75) drive resistance by selecting for bacteria that have adapted their normal physiological processes to permit growth in the presence of these antibiotics. Interestingly, the transition to a low-level DapNS state of the MSSA and MRSA strain pairs revealed similar generation times of DapNS and DapS strains and a minimal fitness burden. These data suggest that once DapNS strains have evolved, they have the capability to persist in the environment. This is in contrast to that of S. aureus strains with intermediate vancomycin susceptibility, where resistance usually coincides with increased generation times (strain pair CB1663/CB1664 in Table 3) (60, 76). Despite the cross-resistance between vancomycin and daptomycin, these differences indicate considerable variation in the adaptation process and further underscore the need for a deeper understanding of daptomycin nonsusceptibility and the subsequent development of new strategies to counteract it.
Several studies have attempted to unravel the changes associated with the daptomycin nonsusceptibility phenotype on a global scale by transcriptional and proteomic profiling (18, 22, 25, 26, 35, 36). Although the transcriptomes of different S. aureus strains showed limited similarities, these studies all noted altered transcription of genes involved in metabolism. These studies focused on the gene and protein levels; however, metabolism is also influenced by posttranscriptional and posttranslational regulation and substrate and cofactor availability. That being said, these studies described transcriptional and proteomic alterations that were quite moderate, similar to our observation regarding the slightly delayed transition of DapNS strains to the postexponential growth phase (Fig. 1; see Fig. S1 in the supplemental material). This transition from rapidly dividing bacteria with an abundant supply of nutrients to postexponential growth and catabolism of secondary metabolites coincides with a fundamental metabolic switch from substrate level phosphorylation and secretion of incompletely oxidized metabolites to activation of TCA cycle function and oxidative phosphorylation in order to catabolize nonpreferred carbon sources (61–63). The delay in the transition to TCA cycle-based metabolism was evident from the decreased aconitase activities and delayed acetate catabolism in the DapNS strains (Fig. 1B and 2).
NMR metabolomics confirmed that the transition of S. aureus to a daptomycin nonsusceptibility phenotype is associated with altered physiology and metabolism (Fig. 3; see Fig. S2 in the supplemental material). Specifically, the data suggest that daptomycin nonsusceptibility coincides with a redirection of carbon flow from the TCA cycle into the pentose phosphate pathway, providing intermediates for the biosynthesis of WTAs, peptidoglycan, and nucleosides/nucleotides (Fig. 4 and 6; see Table S2 in the supplemental material). This suggested that the function of the TCA cycle is impaired in DapNS strains, which is supported by the observations of Fischer et al. (35) showing that levels of subunits of the TCA cycle enzyme succinate dehydrogenase are lower in DapNS strain 701 than in isogenic DapS strain 616. In addition, they observed that the activities of the glycolytic enzymes enolase and glyceraldehyde-3-phosphate dehydrogenase were higher in the isogenic strain 701 (DapNS) pair than in strain 616 (DapS) (35). Another factor potentially contributing to decreased TCA cycle activity (Fig. 1 and 2) is the reported mutation in DapNS strains in the upstream region of the gene for acetyl-CoA synthetase (E.C.6.2.1.1), which is involved in the conversion of acetate to acetyl-CoA (20). Importantly, the TCA cycle changes in some DapNS strains can be exploited to increase the efficacy of daptomycin in these strains (Fig. 7).
In S. aureus, WTA consists of peptidoglycan-anchored N-acetyl-d-glucosamine, N-acetyl-d-mannosamine, glycerol-3-phosphate, and polyribitol-phosphate (69). The extent to which a thickened cell wall contributes to daptomycin nonsusceptibility is still a matter of debate (23); however, we note that precursors of WTA are increased in DapNS strains (Fig. 4). These precursors include intermediates and derivatives of the pentose phosphate pathway such as erythrose-4-phosphate, ribose, ribitol, and mannose. In addition, UDP-N-acetyl-d-mannosamine is most likely synthesized from UDP-N-acetyl-d-glucosamine and both cell wall precursors originate from the glycolytic intermediate fructose-6-phosphate. UDP-N-acetyl-d-glucosamine is also a key intermediate required for the biosynthesis of the major structural polysaccharides peptidoglycan PIA and capsule. Together, these data suggest that intracellular concentrations of WTA precursors and intermediates are increased in DapNS strains, which coincides with decreased carbon flow through the TCA cycle. The increase in WTA precursors is consistent with previous studies that demonstrated increased transcription and production of WTA in DapNS strains (33, 35). In addition, we observed an increase in the concentration of d-alanyl–d-alanine, which is important in the proposed charge repulsion model of daptomycin nonsusceptibility that involves increased d-alanylation of WTA (29, 30).
Alterations in the purine and pyrimidine metabolism associated with the daptomycin nonsusceptibility phenotype have been reported previously (27, 35). Specifically, in vitro-derived DapNS strains revealed a mutation in the ribose phosphate pyrophosphokinase gene, prs, which is involved in the biosynthesis of purines and pyrimidines (36). Transcriptional profiling of DapNS strains also indicated that the transcription of purine and pyrimidine biosynthesis genes was reduced (36). Similarly, a strain with an rpoB mutation conferring reduced susceptibility to daptomycin exhibited lower transcription of genes in purine and pyrimidine biosynthetic pathways (27). In contrast to the studies suggesting that purine and pyrimidine biosynthesis was decreased in DapNS strains, one study found that the purine biosynthesis protein PurH was more abundant in DapNS strain 701 than in DapS strain 616 (35). These seemingly conflicting studies highlight the value of a metabolomic approach, specifically, examination of the products of genes and enzymes. Consistent with the latter study (35) nucleoside and nucleotide concentrations were significantly increased in DapNS strains (Fig. 4).
The biosynthesis of purine bases (guanosine, adenosine) and derivatives requires the synthesis of IMP, which is derived from histidine, glutamine, and 5-phospho-d-ribosyl-1-pyrophosphate (PRPP). Histidine and PRPP both require the activity of the PPP for IMP synthesis, and the concentration of several associated metabolites (i.e., erythrose-4-phosphate, ribose, ribulose-5-phosphate) has been significantly affected in DapNS strain 703 with or without challenging the bacteria with daptomycin. Together with orotate, PRPP is involved in the synthesis of UMP, a central intermediate of pyrimidine metabolism (i.e., thymidine, cytosine, uracil). Orotate synthesis itself requires carbamoylphosphate, an intermediate of amino acid degradation that can be further metabolized to urea in the urea cycle. Some of the intermediates related to pyrimidine synthesis; specifically, dihydroorotate and the urea cycle metabolites arginine and citrulline, were identified as major contributors to group separation between DapS and DapNS strains by 1D 1H-NMR OPLS-DA analyses (see Table S2 in the supplemental material).
Lastly, the concentration of betaine was significantly higher in DapS strains than in DapNS strains. In bacteria, glycine betaine confers osmoprotection and facilitates growth at high salt concentrations (77–79). Our observation is consistent with transcriptional data of DapNS strains showing higher transcription of genes involved in the uptake and/or synthesis of glycine betaine (36); in other words, DapNS strains are trying to overcome this deficiency. While this observation may be important, the advantage that this bacterial adaptation confers has not been investigated in more detail.
Conclusions.
Vancomycin continues to be a drug of choice for treating MRSA infections (80); hence, the emergence of vancomycin-nonsusceptible strains is of great concern. While vancomycin-resistant S. aureus (VRSA) isolates are rare, the prevalence of VISA and heterogeneous intermediate-level vancomycin resistance (hVISA) is increasing (81). When vancomycin is not a viable patient treatment option (e.g., when a patient is allergic to the drug or the infection is due to VRSA or VISA), then daptomycin is a therapeutic option (80). As a consequence of increased therapeutic use of daptomycin, the in vivo development of daptomycin nonsusceptibility has also increased (82). This in vivo development of daptomycin nonsusceptibility has generated a limited number of isogenic DapS and DapNS strain pairs that could be used to gain insight into the metabolic changes associated with the adaptive resistance transition. As expected (26, 28, 29), the transition to the daptomycin nonsusceptibility phenotype correlated with an increased thickness of the peptidoglycan in most of the nonsusceptible strains (Table 2), the exception being strain Q2818, which is consistent with previous observations (22). Peptidoglycan contains the amino sugars N-acetylglucosamine and N-acetylmuramic acid; hence, an increase in peptidoglycan thickness would require increased availability of amino sugars. The concentration of N-acetylglucosamine increased in DapNS strains, and the concentrations of other potential amino sugar precursor metabolites (e.g., UDP-glucose) were also increased (Fig. 4 and 6). In other words, the transition to a daptomycin nonsusceptibility phenotype coincides with increased carbon flow into amino sugar biosynthesis. The alteration in carbon flow appears to be at the expense of basal TCA cycle activity during the exponential growth phase and maximal TCA cycle activity during the postexponential growth phase (Fig. 2). Redirecting carbon flow away from the TCA cycle can have deleterious fitness costs, specifically, a decreased ability to place progeny into the next generation (61). DapNS strains have largely avoided this fitness cost (Table 4) by maintaining sufficient TCA cycle activity to balance the need for increased biosynthesis of cell wall components and bacterial growth. This required balance may also explain why DapNS strains do not arise more frequently.
Supplementary Material
ACKNOWLEDGMENTS
G.A.S. was supported by funds provided through the Hatch Act to the University of Nebraska Institute of Agriculture and Natural Resources and by funds provided through the National Institutes of Health (AI087668) to G.A.S. and R.P. R.P. was supported by funds provided through the NIH (P20 RR-17675 and P30 GM103335) and from the American Heart Association (0860033Z). Portions of this research were performed in facilities renovated with support from the NIH (RR015468-01). H.P., M.B., and M.H. were supported by Deutsche Forschungsgemeinschaft grants SFB 1027 B2 and BI 1350/1-2. S.B.-V. and R.S.D. were supported by funds provided by the NIH (AI11176). A.S.B. is supported in part by NIH-NIAID grant 5RO1-AI039108-18. We thank Cubist Pharmaceuticals for supplying strains and daptomycin.
R.G. contributed the overall experimental design, execution, and interpretation. S.L. contributed the NMR metabolomic experimental design, execution, and interpretation.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00160-15.
REFERENCES
- 1.Sakoulas G, Eliopoulos GM, Alder J, Eliopoulos CT. 2003. Efficacy of daptomycin in experimental endocarditis due to methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 47:1714–1718. doi: 10.1128/AAC.47.5.1714-1718.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wootton M, MacGowan AP, Walsh TR. 2006. Comparative bactericidal activities of daptomycin and vancomycin against glycopeptide-intermediate Staphylococcus aureus (GISA) and heterogeneous GISA isolates. Antimicrob Agents Chemother 50:4195–4197. doi: 10.1128/AAC.00678-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fowler VG Jr, Boucher HW, Corey GR, Abrutyn E, Karchmer AW, Rupp ME, Levine DP, Chambers HF, Tally FP, Vigliani GA, Cabell CH, Link AS, DeMeyer I, Filler SG, Zervos M, Cook P, Parsonnet J, Bernstein JM, Price CS, Forrest GN, Fätkenheuer G, Gareca M, Rehm SJ, Brodt HR, Tice A, Cosgrove SE. 2006. Daptomycin versus standard therapy for bacteremia and endocarditis caused by Staphylococcus aureus. N Engl J Med 355:653–665. doi: 10.1056/NEJMoa053783. [DOI] [PubMed] [Google Scholar]
- 4.Falagas ME, Giannopoulou KP, Ntziora F, Vardakas KZ. 2007. Daptomycin for endocarditis and/or bacteraemia: a systematic review of the experimental and clinical evidence. J Antimicrob Chemother 60:7–19. doi: 10.1093/jac/dkm137. [DOI] [PubMed] [Google Scholar]
- 5.Steenbergen JN, Alder J, Thorne GM, Tally FP. 2005. Daptomycin: a lipopeptide antibiotic for the treatment of serious Gram-positive infections. J Antimicrob Chemother 55:283–288. doi: 10.1093/jac/dkh546. [DOI] [PubMed] [Google Scholar]
- 6.Hayden MK, Rezai K, Hayes RA, Lolans K, Quinn JP, Weinstein RA. 2005. Development of daptomycin resistance in vivo in methicillin-resistant Staphylococcus aureus. J Clin Microbiol 43:5285–5287. doi: 10.1128/JCM.43.10.5285-5287.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mangili A, Bica I, Snydman DR, Hamer DH. 2005. Daptomycin-resistant, methicillin-resistant Staphylococcus aureus bacteremia. Clin Infect Dis 40:1058–1060. doi: 10.1086/428616. [DOI] [PubMed] [Google Scholar]
- 8.Marty FM, Yeh WW, Wennersten CB, Venkataraman L, Albano E, Alyea EP, Gold HS, Baden LR, Pillai SK. 2006. Emergence of a clinical daptomycin-resistant Staphylococcus aureus isolate during treatment of methicillin-resistant Staphylococcus aureus bacteremia and osteomyelitis. J Clin Microbiol 44:595–597. doi: 10.1128/JCM.44.2.595-597.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skiest DJ. 2006. Treatment failure resulting from resistance of Staphylococcus aureus to daptomycin. J Clin Microbiol 44:655–656. doi: 10.1128/JCM.44.2.655-656.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Julian K, Kosowska-Shick K, Whitener C, Roos M, Labischinski H, Rubio A, Parent L, Ednie L, Koeth L, Bogdanovich T, Appelbaum PC. 2007. Characterization of a daptomycin-nonsusceptible vancomycin-intermediate Staphylococcus aureus strain in a patient with endocarditis. Antimicrob Agents Chemother 51:3445–3448. doi: 10.1128/AAC.00559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vikram HR, Havill NL, Koeth LM, Boyce JM. 2005. Clinical progression of methicillin-resistant Staphylococcus aureus vertebral osteomyelitis associated with reduced susceptibility to daptomycin. J Clin Microbiol 43:5384–5387. doi: 10.1128/JCM.43.10.5384-5387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirby A, Mohandas K, Broughton C, Neal TJ, Smith GW, Pai P, Nistal de Paz C. 2009. In vivo development of heterogeneous glycopeptide-intermediate Staphylococcus aureus (hGISA), GISA and daptomycin resistance in a patient with meticillin-resistant S. aureus endocarditis. J Med Microbiol 58:376–380. doi: 10.1099/jmm.0.006486-0. [DOI] [PubMed] [Google Scholar]
- 13.Murthy MH, Olson ME, Wickert RW, Fey PD, Jalali Z. 2008. Daptomycin non-susceptible meticillin-resistant Staphylococcus aureus U S A 300 isolate. J Med Microbiol 57:1036–1038. doi: 10.1099/jmm.0.2008/000588-0. [DOI] [PubMed] [Google Scholar]
- 14.Sakoulas G, Brown J, Lamp KC, Friedrich LV, Lindfield KC. 2009. Clinical outcomes of patients receiving daptomycin for the treatment of Staphylococcus aureus infections and assessment of clinical factors for daptomycin failure: a retrospective cohort study utilizing the Cubicin Outcomes Registry and Experience. Clin Ther 31:1936–1945. doi: 10.1016/j.clinthera.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 15.Silverman JA, Perlmutter NG, Shapiro HM. 2003. Correlation of daptomycin bactericidal activity and membrane depolarization in Staphylococcus aureus. Antimicrob Agents Chemother 47:2538–2544. doi: 10.1128/AAC.47.8.2538-2544.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pogliano J, Pogliano N, Silverman JA. 2012. Daptomycin-mediated reorganization of membrane architecture causes mislocalization of essential cell division proteins. J Bacteriol 194:4494–4504. doi: 10.1128/JB.00011-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuroda M, Kuroda H, Oshima T, Takeuchi F, Mori H, Hiramatsu K. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol Microbiol 49:807–821. doi: 10.1046/j.1365-2958.2003.03599.x. [DOI] [PubMed] [Google Scholar]
- 18.Muthaiyan A, Silverman JA, Jayaswal RK, Wilkinson BJ. 2008. Transcriptional profiling reveals that daptomycin induces the Staphylococcus aureus cell wall stress stimulon and genes responsive to membrane depolarization. Antimicrob Agents Chemother 52:980–990. doi: 10.1128/AAC.01121-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dengler V, Meier PS, Heusser R, Berger-Bächi B, McCallum N. 2011. Induction kinetics of the Staphylococcus aureus cell wall stress stimulon in response to different cell wall active antibiotics. BMC Microbiol 11:16. doi: 10.1186/1471-2180-11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman L, Alder JD, Silverman JA. 2006. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob Agents Chemother 50:2137–2145. doi: 10.1128/AAC.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones T, Yeaman MR, Sakoulas G, Yang SJ, Proctor RA, Sahl HG, Schrenzel J, Xiong YQ, Bayer AS. 2008. Failures in clinical treatment of Staphylococcus aureus infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob Agents Chemother 52:269–278. doi: 10.1128/AAC.00719-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyle-Vavra S, Jones M, Gourley BL, Holmes M, Ruf R, Balsam AR, Boulware DR, Kline S, Jawahir S, Devries A, Peterson SN, Daum RS. 2011. Comparative genome sequencing of an isogenic pair of USA800 clinical methicillin-resistant Staphylococcus aureus isolates obtained before and after daptomycin treatment failure. Antimicrob Agents Chemother 55:2018–2025. doi: 10.1128/AAC.01593-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang SJ, Nast CC, Mishra NN, Yeaman MR, Fey PD, Bayer AS. 2010. Cell wall thickening is not a universal accompaniment of the daptomycin nonsusceptibility phenotype in Staphylococcus aureus: evidence for multiple resistance mechanisms. Antimicrob Agents Chemother 54:3079–3085. doi: 10.1128/AAC.00122-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pillai SK, Gold HS, Sakoulas G, Wennersten C, Moellering RC Jr, Eliopoulos GM. 2007. Daptomycin nonsusceptibility in Staphylococcus aureus with reduced vancomycin susceptibility is independent of alterations in MprF. Antimicrob Agents Chemother 51:2223–2225. doi: 10.1128/AAC.00202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mehta S, Cuirolo AX, Plata KB, Riosa S, Silverman JA, Rubio A, Rosato RR, Rosato AE. 2012. VraSR two-component regulatory system contributes to mprF-mediated decreased susceptibility to daptomycin in in vivo-selected clinical strains of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 56:92–102. doi: 10.1128/AAC.00432-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Camargo IL, Neoh HM, Cui L, Hiramatsu K. 2008. Serial daptomycin selection generates daptomycin-nonsusceptible Staphylococcus aureus strains with a heterogeneous vancomycin-intermediate phenotype. Antimicrob Agents Chemother 52:4289–4299. doi: 10.1128/AAC.00417-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui L, Isii T, Fukuda M, Ochiai T, Neoh HM, Camargo IL, Watanabe Y, Shoji M, Hishinuma T, Hiramatsu K. 2010. An RpoB mutation confers dual heteroresistance to daptomycin and vancomycin in Staphylococcus aureus. Antimicrob Agents Chemother 54:5222–5233. doi: 10.1128/AAC.00437-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui L, Tominaga E, Neoh HM, Hiramatsu K. 2006. Correlation between reduced daptomycin susceptibility and vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother 50:1079–1082. doi: 10.1128/AAC.50.3.1079-1082.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mishra NN, Yang SJ, Sawa A, Rubio A, Nast CC, Yeaman MR, Bayer AS. 2009. Analysis of cell membrane characteristics of in vitro-selected daptomycin-resistant strains of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 53:2312–2318. doi: 10.1128/AAC.01682-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang SJ, Kreiswirth BN, Sakoulas G, Yeaman MR, Xiong YQ, Sawa A, Bayer AS. 2009. Enhanced expression of dltABCD is associated with the development of daptomycin nonsusceptibility in a clinical endocarditis isolate of Staphylococcus aureus. J Infect Dis 200:1916–1920. doi: 10.1086/648473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang SJ, Xiong YQ, Dunman PM, Schrenzel J, Francois P, Peschel A, Bayer AS. 2009. Regulation of mprF in daptomycin-nonsusceptible Staphylococcus aureus strains. Antimicrob Agents Chemother 53:2636–2637. doi: 10.1128/AAC.01415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rubio A, Conrad M, Haselbeck RJ, Kedar GC, Brown-Driver V, Finn J, Silverman JA. 2011. Regulation of mprF by antisense RNA restores daptomycin susceptibility to daptomycin-resistant isolates of Staphylococcus aureus. Antimicrob Agents Chemother 55:364–367. doi: 10.1128/AAC.00429-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bertsche U, Weidenmaier C, Kuehner D, Yang SJ, Baur S, Wanner S, François P, Schrenzel J, Yeaman MR, Bayer AS. 2011. Correlation of daptomycin resistance in a clinical Staphylococcus aureus strain with increased cell wall teichoic acid production and d-alanylation. Antimicrob Agents Chemother 55:3922–3928. doi: 10.1128/AAC.01226-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mishra NN, McKinnell J, Yeaman MR, Rubio A, Nast CC, Chen L, Kreiswirth BN, Bayer AS. 2011. In vitro cross-resistance to daptomycin and host defense cationic antimicrobial peptides in clinical methicillin-resistant Staphylococcus aureus isolates. Antimicrob Agents Chemother 55:4012–4018. doi: 10.1128/AAC.00223-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fischer A, Yang SJ, Bayer AS, Vaezzadeh AR, Herzig S, Stenz L, Girard M, Sakoulas G, Scherl A, Yeaman MR, Proctor RA, Schrenzel J, François P. 2011. Daptomycin resistance mechanisms in clinically derived Staphylococcus aureus strains assessed by a combined transcriptomics and proteomics approach. J Antimicrob Chemother 66:1696–1711. doi: 10.1093/jac/dkr195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song Y, Rubio A, Jayaswal RK, Silverman JA, Wilkinson BJ. 2013. Additional routes to Staphylococcus aureus daptomycin resistance as revealed by comparative genome sequencing, transcriptional profiling, and phenotypic studies. PLoS One 8:e58469. doi: 10.1371/journal.pone.0058469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel JB, Jevitt LA, Hageman J, McDonald LC, Tenover FC. 2006. An association between reduced susceptibility to daptomycin and reduced susceptibility to vancomycin in Staphylococcus aureus. Clin Infect Dis 42:1652–1653. doi: 10.1086/504084. [DOI] [PubMed] [Google Scholar]
- 38.Sakoulas G, Alder J, Thauvin-Eliopoulos C, Moellering RC Jr, Eliopoulos GM. 2006. Induction of daptomycin heterogeneous susceptibility in Staphylococcus aureus by exposure to vancomycin. Antimicrob Agents Chemother 50:1581–1585. doi: 10.1128/AAC.50.4.1581-1585.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lenski RE. 1988. Experimental studies of pleiotropy and epistasis in Escherichia coli 0.1. Variation in competitive fitness among mutants resistant to virus-T4. Evolution 42:425–432. [DOI] [PubMed] [Google Scholar]
- 40.Wichelhaus TA, Böddinghaus B, Besier S, Schäfer V, Brade V, Ludwig A. 2002. Biological cost of rifampin resistance from the perspective of Staphylococcus aureus. Antimicrob Agents Chemother 46:3381–3385. doi: 10.1128/AAC.46.11.3381-3385.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kennedy MC, Emptage MH, Dreyer JL, Beinert H. 1983. The role of iron in the activation-inactivation of aconitase. J Biol Chem 258:11098–11105. [PubMed] [Google Scholar]
- 42.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 43.Halouska S, Zhang B, Gaupp R, Lei S, Snell E, Fenton RJ, Barletta RG, Somerville GA, Powers R. 2013. Revisiting protocols for the NMR analysis of bacterial metabolomes. J Integr OMICS 3:120–137. doi: 10.5584/jiomics.v3i2.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang B, Halouska S, Schiaffo CE, Sadykov MR, Somerville GA, Powers R. 2011. NMR analysis of a stress response metabolic signaling network. J Proteome Res 10:3743–3754. doi: 10.1021/pr200360w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Worley B, Powers R. 2014. MVAPACK: a complete data handling package for NMR metabolomics. ACS Chem Biol 9:1138–1144. doi: 10.1021/cb4008937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Worley B, Powers R. 2014. Simultaneous phase and scatter correction for NMR datasets. Chemometr Intell Lab Syst 131:1–6. doi: 10.1016/j.chemolab.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Meyer T, Sinnaeve D, Van Gasse B, Tsiporkova E, Rietzschel ER, De Buyzere ML, Gillebert TC, Bekaert S, Martins JC, Van Criekinge W. 2008. NMR-based characterization of metabolic alterations in hypertension using an adaptive, intelligent binning algorithm. Anal Chem 80:3783–3790. doi: 10.1021/ac7025964. [DOI] [PubMed] [Google Scholar]
- 48.Worley B, Halouska S, Powers R. 2013. Utilities for quantifying separation in PCA/PLS-DA score plots. Anal Biochem 433:102–104. doi: 10.1016/j.ab.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Werth MT, Halouska S, Shortridge MD, Zhang B, Powers R. 2010. Analysis of metabolomic PCA data using tree diagrams. Anal Biochem 399:58–63. doi: 10.1016/j.ab.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eriksson L, Trygg J, Wold S. 2008. CV-ANOVA for significance testing of PLS and OPLS® models. J Chemom 22:594–600. doi: 10.1002/cem.1187. [DOI] [Google Scholar]
- 51.Shao J. 1993. Linear model selection by cross-validation. J Am Stat Assoc 88:486–494. doi: 10.1080/01621459.1993.10476299. [DOI] [Google Scholar]
- 52.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293. [DOI] [PubMed] [Google Scholar]
- 53.Johnson BA. 2004. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol 278:313–352. [DOI] [PubMed] [Google Scholar]
- 54.Akiyama K, Chikayama E, Yuasa H, Shimada Y, Tohge T, Shinozaki K, Hirai MY, Sakurai T, Kikuchi J, Saito K. 2008. PRIMe: a web site that assembles tools for metabolomics and transcriptomics. In Silico Biol 8:339–345. [PubMed] [Google Scholar]
- 55.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, Mandal R, Sinelnikov I, Xia J, Jia L, Cruz JA, Lim E, Sobsey CA, Shrivastava S, Huang P, Liu P, Fang L, Peng J, Fradette R, Cheng D, Tzur D, Clements M, Lewis A, De Souza A, Zuniga A, Dawe M, Xiong Y, Clive D, Greiner R, Nazyrova A, Shaykhutdinov R, Li L, Vogel HJ, Forsythe I. 2009. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res 37:D603–D610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Kent Wenger R, Yao H, Markley JL. 2008. BioMagResBank. Nucleic Acids Res 36:D402–D408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Jonge BL, Chang YS, Gage D, Tomasz A. 1992. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A. J Biol Chem 267:11248–11254. [PubMed] [Google Scholar]
- 58.Hutter JL, Bechhoefer J. 1993. Calibration of atomic-force microscope tips. Rev Sci Instrum 64:1868–1873. doi: 10.1063/1.1143970. [DOI] [Google Scholar]
- 59.Nečas D, Klapetek P. 2012. Gwyddion: an open-source software for SPM data analysis. Cent Eur J Physics 10:181–188. doi: 10.2478/s11534-011-0096-2. [DOI] [Google Scholar]
- 60.Nelson JL, Rice KC, Slater SR, Fox PM, Archer GL, Bayles KW, Fey PD, Kreiswirth BN, Somerville GA. 2007. Vancomycin-intermediate Staphylococcus aureus strains have impaired acetate catabolism: implications for polysaccharide intercellular adhesin synthesis and autolysis. Antimicrob Agents Chemother 51:616–622. doi: 10.1128/AAC.01057-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Somerville GA, Chaussee MS, Morgan CI, Fitzgerald JR, Dorward DW, Reitzer LJ, Musser JM. 2002. Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect Immun 70:6373–6382. doi: 10.1128/IAI.70.11.6373-6382.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Somerville GA, Saïd-Salim B, Wickman JM, Raffel SJ, Kreiswirth BN, Musser JM. 2003. Correlation of acetate catabolism and growth yield in Staphylococcus aureus: implications for host-pathogen interactions. Infect Immun 71:4724–4732. doi: 10.1128/IAI.71.8.4724-4732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gaupp R, Schlag S, Liebeke M, Lalk M, Götz F. 2010. Advantage of up-regulation of succinate dehydrogenase in Staphylococcus aureus biofilm. J Bacteriol 192:2385–2394. doi: 10.1128/JB.01472-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Duda RO, Hart PE, Stork DG. 2001. Pattern classification, 2nd ed Wiley, New York, NY. [Google Scholar]
- 65.Yu H, Yang J. 2001. A direct LDA algorithm for high-dimensional data—with application to face recognition. Pattern Recognit 34:2067–2070. doi: 10.1016/S0031-3203(00)00162-X. [DOI] [Google Scholar]
- 66.Xu Q-S, Liang Y-Z, Du Y-P. 2004. Monte Carlo cross-validation for selecting a model and estimating the prediction error in multivariate calibration. J Chemom 18:112–120. doi: 10.1002/cem.858. [DOI] [Google Scholar]
- 67.Cramton SE, Gerke C, Schnell NF, Nichols WW, Götz F. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun 67:5427–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O'Riordan K, Lee JC. 2004. Staphylococcus aureus capsular polysaccharides. Clin Microbiol Rev 17:218–234. doi: 10.1128/CMR.17.1.218-234.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xia G, Kohler T, Peschel A. 2010. The wall teichoic acid and lipoteichoic acid polymers of Staphylococcus aureus. Int J Med Microbiol 300:148–154. doi: 10.1016/j.ijmm.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 70.Pereira MP, Brown ED. 2004. Bifunctional catalysis by CDP-ribitol synthase: convergent recruitment of reductase and cytidylyltransferase activities in Haemophilus influenzae and Staphylococcus aureus. Biochemistry 43:11802–11812. doi: 10.1021/bi048866v. [DOI] [PubMed] [Google Scholar]
- 71.Ender M, McCallum N, Adhikari R, Berger-Bächi B. 2004. Fitness cost of SCCmec and methicillin resistance levels in Staphylococcus aureus. Antimicrob Agents Chemother 48:2295–2297. doi: 10.1128/AAC.48.6.2295-2297.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hiramatsu K, Cui L, Kuroda M, Ito T. 2001. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol 9:486–493. doi: 10.1016/S0966-842X(01)02175-8. [DOI] [PubMed] [Google Scholar]
- 73.Lee SM, Ender M, Adhikari R, Smith JM, Berger-Bächi B, Cook GM. 2007. Fitness cost of staphylococcal cassette chromosome mec in methicillin-resistant Staphylococcus aureus by way of continuous culture. Antimicrob Agents Chemother 51:1497–1499. doi: 10.1128/AAC.01239-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hurdle JG, O'Neill AJ, Ingham E, Fishwick C, Chopra I. 2004. Analysis of mupirocin resistance and fitness in Staphylococcus aureus by molecular genetic and structural modeling techniques. Antimicrob Agents Chemother 48:4366–4376. doi: 10.1128/AAC.48.11.4366-4376.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O'Neill AJ, Huovinen T, Fishwick CW, Chopra I. 2006. Molecular genetic and structural modeling studies of Staphylococcus aureus RNA polymerase and the fitness of rifampin resistance genotypes in relation to clinical prevalence. Antimicrob Agents Chemother 50:298–309. doi: 10.1128/AAC.50.1.298-309.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sieradzki K, Leski T, Dick J, Borio L, Tomasz A. 2003. Evolution of a vancomycin-intermediate Staphylococcus aureus strain in vivo: multiple changes in the antibiotic resistance phenotypes of a single lineage of methicillin-resistant S. aureus under the impact of antibiotics administered for chemotherapy. J Clin Microbiol 41:1687–1693. doi: 10.1128/JCM.41.4.1687-1693.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Graham JE, Wilkinson BJ. 1992. Staphylococcus aureus osmoregulation: roles for choline, glycine betaine, proline, and taurine. J Bacteriol 174:2711–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Miller KJ, Zelt SC, Bae J-H. 1991. Glycine betaine and proline are the principal compatible solutes of Staphylococcus aureus. Curr Microbiol 23:131–137. doi: 10.1007/BF02091971. [DOI] [Google Scholar]
- 79.Kunin CM, Rudy J. 1991. Effect of NaCl-induced osmotic stress on intracellular concentrations of glycine betaine and potassium in Escherichia coli, Enterococcus faecalis, and staphylococci. J Lab Clin Med 118:217–224. [PubMed] [Google Scholar]
- 80.Liu C, Bayer A, Cosgrove SE, Daum RS, Fridkin SK, Gorwitz RJ, Kaplan SL, Karchmer AW, Levine DP, Murray BE, Rybak MJ, Talan DA , Chambers HF. 2011. Clinical practice guidelines by the Infectious Diseases Society of America for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children: executive summary. Clin Infect Dis 52:285–292. doi: 10.1093/cid/cir034. [DOI] [PubMed] [Google Scholar]
- 81.Richter SS, Diekema DJ, Heilmann KP, Dohrn CL, Crispell EK, Riahi F, McDanel JS, Satola SW, Doern GV. 2014. Activity of vancomycin, ceftaroline, and mupirocin against Staphylococcus aureus from a 2011 national surveillance study in the United States. Antimicrob Agents Chemother 58:740–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bayer AS, Schneider T, Sahl HG. 2013. Mechanisms of daptomycin resistance in Staphylococcus aureus: role of the cell membrane and cell wall. Ann N Y Acad Sci 1277:139–158. doi: 10.1111/j.1749-6632.2012.06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.