Abstract
MicroRNA-124 contributes to neurogenesis through regulating its targets, but its expression both in the brain of Huntington’s disease mouse models and patients is decreased. However, the effects of microRNA-124 on the progression of Huntington’s disease have not been reported. Results from this study showed that microRNA-124 increased the latency to fall for each R6/2 Huntington’s disease transgenic mouse in the rotarod test. 5-Bromo-2’-deoxyuridine (BrdU) staining of the striatum shows an increase in neurogenesis. In addition, brain-derived neurotrophic factor and peroxisome proliferator-activated receptor gamma coactivator 1-alpha protein levels in the striatum were increased and SRY-related HMG box transcription factor 9 protein level was decreased. These findings suggest that microRNA-124 slows down the progression of Huntington’s disease possibly through its important role in neuronal differentiation and survival.
Keywords: nerve regeneration, microRNA-124, neurogenesis, neuronal survival, Huntington's disease, SRY-related HMG box transcription factor 9, brain-derived neurotrophic factor; peroxisome, proliferator-activated receptor gamma coactivator 1-alpha, mutant huntingtin
Introduction
Huntington’s disease (HD) is a fatal, incurable neurodegenerative disease caused by a cytosine-adenine-guanine (CAG) expansion in the gene encoding protein huntingtin (HTT). HD presents with cognitive deficit and motor control impairment, and ultimately results in death which is caused by the neuronal dysfunction from progressive loss of cortical and striatal neurons (Johnson et al., 2008). Increasing evidence indicates that the mutant huntingtin (mHTT) distorts the normal transcriptional program of the susceptible neurons, indicating that the transcriptional dysregulation might be a crucial pathogenic mechanism in HD (Cha, 2000; Luthi-Carter and Cha, 2003).
MicroRNA is a class of small non-coding RNA molecules, which function in transcriptional and post-transcriptional regulation of target gene expression by degrading their target mRNAs or repressing their translation (Chen and Rajewsky, 2007; Bartel, 2009). MicroRNAs are expressed in the central nervous system and temporally and/or spatially regulate their target genes during the development (Kapsimali et al., 2007; Landgraf et al., 2007; Bak et al., 2008), and further lead to alterations of the translation of target genes. Recently, it has been reported that several cell survival-associated microRNA expression levels are aberrant compared with the normal mice (Hoss et al., 2014). MicroRNA-128a is dysregulated through affecting HTT and Huntington interaction protein1 in HD monkeys (Kocerha et al., 2014). It has been shown that microRNA is associated with neuronal differentiation in mice, and microRNA-124, a brain specific microRNA, is thought to be one of the crucial regulators for neuronal differentiation in neurodegeneration (Roshan et al., 2009). In addition, microRNA-124 also controls neuronal survival by regulating pro-survival and pro-apoptotic signals (Gascon and Gao, 2012). MicroRNA-124 can regulate the proliferation of neural progenitor cells (Liu et al., 2011). The role of microRNA-124 in HD is shown by its expression being repressed in the brains of human patients and mouse models, which leads to the dysregulation of RE1-Silencing Transcription Factor (REST) (Johnson and Buckley, 2009). REST nuclear translocation leads to the transcriptional repression of brain-derived neurotrophic factor (BDNF) (Abuhatzira et al., 2007), resulting in the neuronal damage (Lipsky and Marini, 2007). During the neuronal differentiation, SRY-related HMG box transcription factor 9 (SOX9) is a physiological target of micrRNA-124 and microRNA-124-mediated repression of SOX9 is also essential for neuron formation from neural stem cells (Cheng et al., 2009). It is also well known that peroxisome proliferator-activated receptor gamma coactivator-1 alpha (PGC-1α is an important factor to promote cell survival and improve neurological defects in HD through regulating mitochondrial functions and mHTT (mutant huntingtin) aggregation (Chaturvedi et al., 2009; Sen et al., 2011; Jones et al., 2012; Tsunemi et al., 2012).
In this study, we attempted to determine whether the microRNA-124 can slow down the progression of HD in transgenic mice and promote neuronal differentiation in the striatum of the mouse brain. We also tested whether microRNA-124 could modulate SOX-9, PGC-1α, and BDNF expressions in the brain, which are imbalanced in HD. To confirm the effects of microRNA-124 on neurogenesis in vivo, 5-bromo-2′-deoxyuridine (BrdU) staining of striatum tissue was performed.
Materials and Methods
MicroRNA-124
MicroRNA-124 was synthesized by Bioneer Company (Bioneer, Daejeon, Korea). Its sequence is as follows: Guide, UUA AGG CAC GCG GUG AAU GCC A; passenger, GCA UUC ACC GCG UGC CUU AAU U.
HD mouse model and microRNA-124 & BrdU injection schedule
Transgenic HD mice of the R6/2 line (B6CBA-Tg (HD exon1) 62Gpb/3J, 111 CAGs) and their widetype littermates (Jackson Laboratories, Barr Harbor, ME, USA) were used (Mangiarini et al., 1996). These mice were obtained by crossing ovarian transplant of hemizygote females with B6CBAF1/J males. The mice were housed in groups with ad libitum access to food and water with 12-hour light/dark cycles. R6/2: six males, six females; littermate: two males, two females. Weight: 15–20 g, age: 4–12 weeks. The genotype was assessed using PCR assay. Temperature: 94°C for 4 minutes; 94°C for 30 seconds, 58°C for 30 seconds, 72°C for 60 seconds: 28 cycles; 72°C for 5 minutes; 4°C forever. The following primer sequences were used: Forward, 5′-CCG CTC AGG TTC TGC TTT TA-3′; reverse, 5′-TGG AAG GAC TTG AGG GAC TC-3′.
In R6/2 mice, disease phenotype appeared at 8 weeks of age. MicroRNA-124 injection into bilateral striata of R6/2 mice was started at 8 weeks old. 20 nmol of microRNA-124 was used for each mouse. The microRNA-124 was injected only once. 150 mg/kg of BrdU (Thermo Fisher Scientific, Boston, MA, USA) was intraperitoneally injected for 5 days after injecting miRNA. Four mice were used for miRNA-124 injection and four mice for vehicle injection (N/C miRNA).
All animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) of Seoul National University Hospital accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International.
Immunohistochemistry
Immunocytochemistry of brain section was performed as described previously (Im et al., 2013). R6/2 mice were anesthetized and transcardially perfused with 10 mL of cold saline and 10 mL of 4% paraformaldehyde in 0.1 M PBS at 12 weeks of age. Brains were removed from the skull, cryoprotected in 30% sucrose solution at 4°C, and sectioned at 20 μm. Free-floating sections were washed and blocked with normal goat serum, then stained with the anti-BrdU antibody (1:100; Abcam, Cambridge, MA, USA). On the following day, the sections were washed in PBS with three times and incubated with FITC conjugated anti-rabbit IgG (1:500; Jackson ImmunoResearch Laboratories, PA) for 2 hours.
MicroRNA injection
For in vivo R6/2 microRNA-124 (n = 4, two males and two females), R6/2 N/C microRNA (n = 4, two males and two females) or Cy3-labeled microRNA-124 injection, mice at 8 weeks of age were anesthetized by an intraperitoneal injection of 1% ketamine (30 mg/kg) and xylazine hydrochloride (4 mg/kg), and then positioned in a stereotaxic apparatus. MicroRNA-124, N/C microRNA or Cy3-labeled microRNA-124 (2 μL) was injected into the bilateral striata using a 30-guage Hamilton syringe at the following coordinates: anterior-posterior +0.38 mm, medial-lateral ±2.0 mm, and dorsal-ventral –3.5 mm from the bregma.
Rotarod performance and weight measurement
As described previously (Im et al., 2013), the rotarod test was performed by using a rotarod apparatus (Jungdo Instruments, Seoul, Korea). Mice were placed on the rod with an accelerating rotation speed from 4 to 40 r/min over a period of 3 minutes with 15 minutes of rest in between the trials. Rotarod evaluation measurement was performed every week from 4 to 12 weeks. Three trials were performed and the mean latencies to falls were recorded. Rotarod evaluation and weight measurements were performed every week from 4 to 12 weeks (Figure 1).
Figure 1.
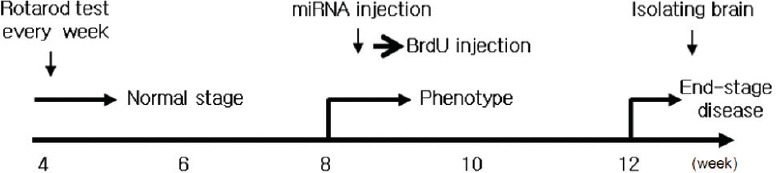
Experimental schedule for microRNA-124 and 5-bromo-2′-deoxyuridine (BrdU) injection, behavior test, and weight measurement.
Protein extracts and western blot analysis
The whole brains of of R6/2 mice were isolated, immediately frozen on liquid nitrogen, and stored at −70°C until protein extraction. Protein extracts were prepared by using RIPA buffer (Thermo Fisher Scientific) with freshly added protease inhibitor and phosphatase inhibitor (Roche, Nutley, NJ, USA). Total protein concentration was quantified by a colorimetric detection assay (BCA Protein Assay, Thermo Scientific Pierce, Rockford, IL, USA). Aliquots of 30 μg protein were separated on 12% sodium dodecyl sulphate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membrane. The blots were probed with primary antibodies anti-mouse PGC1α (1:1,000; Santa Cruz Biotechnology, Santa Cruz, CA, USA), SOX9 (1:1,000; Abcam), and BDNF (1:1,000; Santa Cruz Biotechnology) at 4°C for 24 hours, followed by horseradish peroxidase conjugated secondary anti-mouse or rabbit antibody (1:5,000; GE Healthcare, UK), and the blots were developed by ECL solution (Thermo Fisher Scientific). Western blots were scanned and intensity values were obtained using ImageJ software (NIH, MD, USA).
Statistical analysis
All data were statistically analyzed using SPSS 17.0 software (SPSS, Chicago, IL, USA) and are expressed as the mean ± SEM. Student’s t-test was used. The differences were considered significant when P < 0.05.
Results
Injection of microRNA-124 improved behavioral phenotypes of R6/2 mice model
To determine the effects of microRNA-124 on behavioral deficits of R6/2 mice, microRNA-124 was injected on 8-week-old mice and phenotypes were tested. Results show that although significant difference of weight loss was not detected between the microRNA-124 injected R6/2 mice group and the N/C microRNA-injected R6/2 mice group in the rotarod test, microRNA-124 injections significantly increased latency to fall compared to N/C microRNA-injected R6/2 mice group at 10 and 11 weeks of age (P < 0.05) (Figure 2A, B). Therefore, microRNA-124 improved behavioral phenotypes of R6/2 mice.
Figure 2.
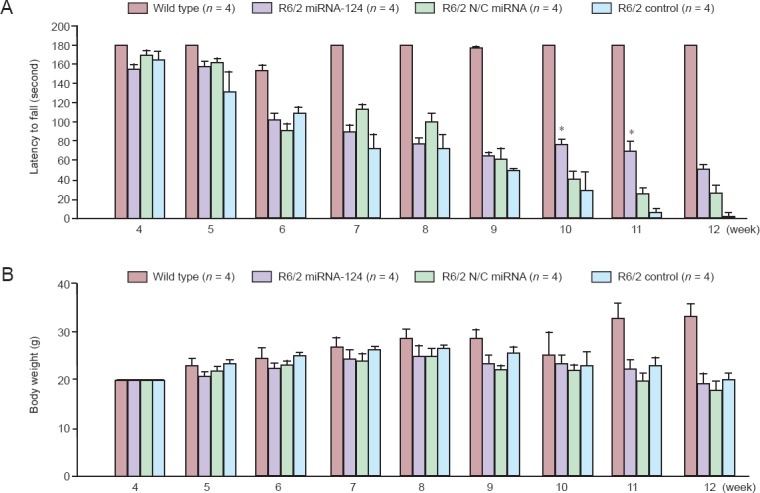
Injection of microRNA-124 (miRNA-124) slows down behavioral phenotypes of R6/2 Huntington’s disease transgenic mice.
(A) Rotarod test showed better motor performance at 10 and 11 weeks of age in miRNA-124 injected R6/2 mice compared to R6/2 N/C miRNA (*P < 0.05; Student’s t-test). The data are expressed as the mean ± SEM from four rats per group. (B) Measurement of body weight showed no difference among 10, 11, and 12 weeks of age in miRNA-124 injected R6/2 mice compared to control. R6/2 miRNA-124: miRNA-124 injected mice; R6/2 N/C miRNA: scramble miRNA injected mice; R6/2 control; no injection.
MicroRNA-124 increased the neurogenesis in the striatum and cortex of R6/2 mouse brain
Since the BrdU was injected from 8 weeks, brain tissues were obtained at 12 weeks and the BrdU signals were captured (FITC). With the microRNA-124 injection, more BrdU-positive cells in both the striatum and cortex were noted than those of the control (R6/2 mice only) group (Figure 3A). The quantitative graphs also showed the increase in number of BrdU-positive cells in the striatum and cortex (Figure 3B, C). To further show that microRNA-124 was successfully injected into the striatum, microRNA-124 labeled with Cy3 dye was injected into the R6/2 mouse striatum for 2 weeks; red Cy3-postive signal showed that microRNA-124 was successfully injected into the mouse brain (Figure 3D). Taken together, microRNA-124 increased the neurogenesis both in the striatum and cortex.
Figure 3.
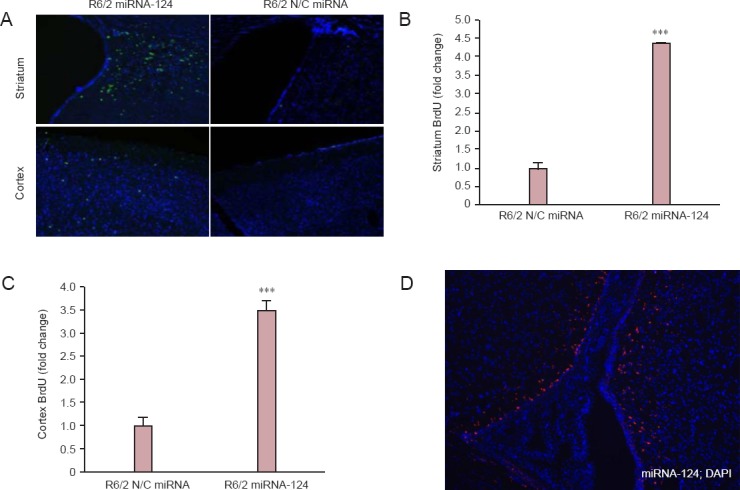
MicroRNA-124 (miRNA-124) increased the neurogenesis of neuronal cells in the striatum and cortex of R6/2 Huntington’s disease transgenic mice.
(A) The striatum and cortex were sectioned and stained with BrdU-FITC (BrdU, green and DAPI, blue). (B) In the striatum, the number of BrdU-positive cells was significantly higher in miRNA-124 injected group compared with control-injected group (***P < 0.001; Student’s t-test; n = 3). (C) In the cortex, the number of BrdU-positive cells was significantly higher in miRNA-124 injected group compared with control-injected group (***P < 0.001; Student’s t-test; n = 3). The data are expressed as the mean ± SEM. (D) MiRNA-124 labeled with Cy3 dye was injected into the R6/2 mouse striatum (red color means cells which have Cy3-labeld miRNA-124).
MicroRNA-124 altered related protein expression in the striatum of R6/2 mouse brain
The 12-week-old R6/2 mice injected with N/C microRNA or microRNA-124 for 4 weeks were sacrificed, and proteins were obtained from the striatum of the mouse brain. The direct target of microRNA-124, SOX9 protein was decreased. BDNF proteins were up-regulated in microRNA-124-injected R6/2 mice compared to the N/C control-injected R6/2 mice. Expression of PGC-1α was promoted compared to the control-injected R6/2 mice. Overall, expression levels of neuroprotective PGC-1α and BDNF were activated and SOX9, the repressor of cell differentiation, was down-regulated by injection of microRNA-124 (Figure 4).
Figure 4.
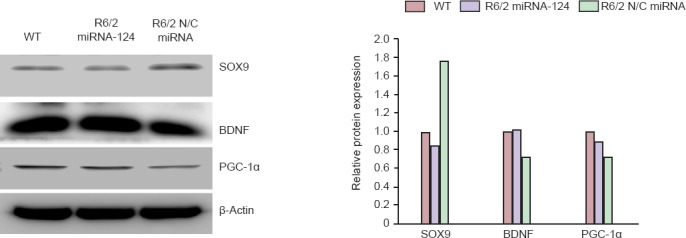
MicroRNA-124 (miRNA-124) altered related protein expression in the striatum of R6/2 Huntington’s disease transgenic mice.
The 12-week-old R6/2 mice injected with N/C miRNA or miRNA-124 for 4 weeks were sacrificed, and proteins were obtained from the striatum of the mouse brain. Western blot analysis confirmed that BDNF protein was up-regulated in miRNA-124-injected R6/2 mice compared to the control-injected R6/2 mice. Expression of PGC-1α was increased, while expression of SOX9 was decreased compared to the control-injected R6/2 mice. SOX9: SRY-related HMG box transcription factor 9; BDNF: brain-derived neurotrophic factor; PGC-1α: peroxisome proliferator-activated receptor gamma coactivator-1 alpha.
Discussion
In this study, we investigated the role of microRNA-124 in R6/2 mice, by implanting the microRNA-124 to the striatum of R6/2 mouse brain, to further understand the neuroprotective function of microRNA-124 in HD. MicroRNA-124 improved latency to fall of R6/2 mice and increased the cell proliferation at the subventricular zone, indicating the increase of neurogenesis. MicroRNA-124 also up-regulated PGC1 and BDNF expressions and down-regulated SOX9 expression in the striatum of R6/2 mouse brain, which implies the improvement of neuronal survival and differentiation in the striatum.
Although many studies have shown that microRNA-124 is down-regulated in the brain of HD patients and mouse models (Das et al., 2013), few studies investigated whether the microRNA-124 can affect the progression of HD in vivo. In this study, microRNA-124 increased the latency to fall of R6/2 mice at 10 and 11 weeks, which first indicates that microRNA-124 may slow down the progression of HD.
Many in vitro studies have shown that the forced expression of microRNA-124 in some non-neuronal cells can promote neuronal differentiation (Lim et al., 2005; Visvanathan et al., 2007; Yu et al., 2008). A recent study shows that microRNA-124 is a determinant of neuronal fate in the subvenricular zone, in which in vivo inhibition of microRNA-124 blocked the neurogenesis at the subventricular zone (Akerblom et al., 2012). The persistence of neurogenesis at the subventricular zone is the premise for the striatum to get mature or well-differentiated into neurons (Parent et al., 2002). Therefore, microRNA-124 may potentially benefit for neurogenesis, which is implicated in this study. The down-regulation of SOX9 protein expression in the striatum and increasing BrdU-positive signals in the striatum suggest a promotion of neurogenesis by microRNA-124. Notably, the REST proteins from the whole cell lysate were up-regulated in microRNA-124-injected R6/2 mice, but it is unknown if the increased REST proteins were from the nucleus or cytoplasm. Therefore, it could not be determined that whether the REST has repressing activation. However, the BDNF, the target of REST, was increased by microRNA-124 injection which implies that the increase of REST did not translocate to the nucleus to repress the transcription.
In HD, PGC-1α was found to be a therapeutic target and regulation for mitochondrial function in cell survival (McGill and Beal, 2006). Lower brain BDNF levels are detected in both HD patients and mouse models, and the loss of BDNF in the brain leads to more neuronal loss (Zuccato et al., 2001, 2011; Ciammola et al., 2007; Strand et al., 2007; Zuccato and Cattaneo, 2007), indicating an essential role of BDNF in the neuronal survival. In this study, microRNA-124 markedly increased the PGC-1α and BDNF protein levels, suggesting its protective role in neuronal survival.
In summary, microRNA-124 has shown positive effects on slowing down the progression of HD, promoting neurogenesis in the striatum, and improving neuronal survival. Collectively, microRNA-124 is a promising therapeutic strategy for HD.
Footnotes
Funding: This study was supported by a grant (A121911 and HI14C2348) of the Korean Health Technology R&D Project, Ministry of Health & Welfare, and National Research Foundation of Korea (NRF) (2011-0012728 and 2014R1A2A1A11051520).
Conflicts of Interest: None declared.
Copyedited by Yang SH, Jin J, Li CH, Song LP, Zhao M
References
- Abuhatzira L, Makedonski K, Kaufman Y, Razin A, Shemer R. MeCP2 deficiency in the brain decreases BDNF levels by REST/CoREST-mediated repression and increases TRKB production. Epigenetics. 2007;2:214–222. doi: 10.4161/epi.2.4.5212. [DOI] [PubMed] [Google Scholar]
- Akerblom M, Sachdeva R, Barde I, Verp S, Gentner B, Trono D, Jakobsson J. MicroRNA-124 is a subventricular zone neuronal fate determinant. J Neurosci. 2012;32:8879–8889. doi: 10.1523/JNEUROSCI.0558-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak M, Silahtaroglu A, Moller M, Christensen M, Rath MF, Skryabin B, Tommerup N, Kauppinen S. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14:432–444. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha JHJ. Transcriptional dysregulation in Huntington's disease. Trends Neurosci. 2000;23:387–392. doi: 10.1016/s0166-2236(00)01609-x. [DOI] [PubMed] [Google Scholar]
- Chaturvedi RK, Adhihetty P, Shukla S, Hennessy T, Calingasan N, Yang L, Starkov A, Kiaei M, Cannella M, Sassone J, Ciammola A, Squitieri F, Beal MF. Impaired PGC-1alpha function in muscle in Huntington's disease. Hum Mol Genet. 2009;18:3048–3065. doi: 10.1093/hmg/ddp243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciammola A, Sassone J, Cannella M, Calza S, Poletti B, Frati L, Squitieri F, Silani V. Low brain-derived neurotrophic factor (BDNF) levels in serum of Huntington's disease patients. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:574–577. doi: 10.1002/ajmg.b.30501. [DOI] [PubMed] [Google Scholar]
- Das E, Jana NR, Bhattacharyya NP. MicroRNA-124 targets CCNA2 and regulates cell cycle in STHdh(Q111)/Hdh(Q111) cells. Biochem Biophys Res Commun. 2013;437:217–224. doi: 10.1016/j.bbrc.2013.06.041. [DOI] [PubMed] [Google Scholar]
- Gascon E, Gao FB. Cause or effect: misregulation of microRNA pathways in neurodegeneration. Front Neurosci. 2012;6:48. doi: 10.3389/fnins.2012.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoss AG, Kartha VK, Dong X, Latourelle JC, Dumitriu A, Hadzi TC, Macdonald ME, Gusella JF, Akbarian S, Chen JF, Weng Z, Myers RH. MicroRNAs located in the Hox gene clusters are implicated in huntington's disease pathogenesis. PLoS Genet. 2014;10:e1004188. doi: 10.1371/journal.pgen.1004188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im W, Ban J, Lim J, Lee M, Lee ST, Chu K, Kim M. Extracts of adipose derived stem cells slows progression in the R6/2 Model of Huntington's disease. PloS One. 2013;8:e59438. doi: 10.1371/journal.pone.0059438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R, Buckley NJ. Gene dysregulation in Huntington's disease: REST, microRNAs and beyond. Neuromol Med. 2009;11:183–199. doi: 10.1007/s12017-009-8063-4. [DOI] [PubMed] [Google Scholar]
- Johnson R, Zuccato C, Belyaev ND, Guest DJ, Cattaneo E, Buckley NJ. A microRNA-based gene dysregulation pathway in Huntington's disease. Neurobiol Dis. 2008;29:438–445. doi: 10.1016/j.nbd.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Jones AW, Yao Z, Vicencio JM, Karkucinska-Wieckowska A, Szabadkai G. PGC-1 family coactivators and cell fate: roles in cancer, neurodegeneration, cardiovascular disease and retrograde mitochondria-nucleus signalling. Mitochondrion. 2012;12:86–99. doi: 10.1016/j.mito.2011.09.009. [DOI] [PubMed] [Google Scholar]
- Kapsimali M, Kloosterman WP, de Bruijn E, Rosa F, Plasterk RHA, Wilson SW. MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system. Genome Biol. 2007;8:R173. doi: 10.1186/gb-2007-8-8-r173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocerha J, Xu Y, Prucha MS, Zhao D, Chan AW. microRNA-128a dysregulation in transgenic Huntington's disease monkeys. Mol Brain. 2014;7:46. doi: 10.1186/1756-6606-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foà R, Schliwka J, Fuchs U, Novosel A, Müller RU, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lipsky RH, Marini AM. Brain-derived neurotrophic factor in neuronal survival and behavior-related plasticity. Ann N Y Acad Sci. 2007;1122:130–143. doi: 10.1196/annals.1403.009. [DOI] [PubMed] [Google Scholar]
- Liu XS, Chopp M, Zhang RL, Tao T, Wang XL, Kassis H, Hozeska-Solgot A, Zhang L, Chen C, Zhang ZG. MicroRNA profiling in subventricular zone after stroke: MiR-124a regulates proliferation of neural progenitor cells through Notch signaling pathway. PloS One. 2011;6:e23461. doi: 10.1371/journal.pone.0023461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthi-Carter R, Cha JH. Mechanisms of transcriptional dysregulation in Huntington's disease. Clin Neurosci Res. 2003;3:165–177. [Google Scholar]
- Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, Lawton M, Trottier Y, Lehrach H, Davies SW, Bates GP. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- McGill JK, Beal MF. PGC-1alpha, a new therapeutic target in Huntington's disease? Cell. 2006;127:465–468. doi: 10.1016/j.cell.2006.10.023. [DOI] [PubMed] [Google Scholar]
- Parent JM, Vexler ZS, Gong C, Derugin N, Ferriero DM. Rat forebrain neurogenesis and striatal neuron replacement after focal stroke. Ann Neurol. 2002;52:802–813. doi: 10.1002/ana.10393. [DOI] [PubMed] [Google Scholar]
- Roshan R, Ghosh T, Scaria V, Pillai B. MicroRNAs: novel therapeutic targets in neurodegenerative diseases. Drug Discov Today. 2009;14:1123–1129. doi: 10.1016/j.drudis.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Sen N, Satija YK, Das S. PGC-1alpha, a key modulator of p53, promotes cell survival upon metabolic stress. Mol Cell. 2011;44:621–634. doi: 10.1016/j.molcel.2011.08.044. [DOI] [PubMed] [Google Scholar]
- Strand AD, Baquet ZC, Aragaki AK, Holmans P, Yang L, Cleren C, Beal MF, Jones L, Kooperberg C, Olson JM, Jones KR. Expression profiling of Huntington's disease models suggests that brain-derived neurotrophic factor depletion plays a major role in striatal degeneration. J Neurosci. 2007;27:11758–11768. doi: 10.1523/JNEUROSCI.2461-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunemi T, Ashe TD, Morrison BE, Soriano KR, Au J, Roque RA, Lazarowski ER, Damian VA, Masliah E, La Spada AR. PGC-1alpha rescues Huntington's disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci Transl Med. 2012;4:142ra197. doi: 10.1126/scitranslmed.3003799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Gene Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JY, Chung KH, Deo M, Thompson RC, Turner DL. MicroRNA miR-124 regulates neurite outgrowth during neuronal differentiation. Exp Cell Res. 2008;314:2618–2633. doi: 10.1016/j.yexcr.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C, Cattaneo E. Role of brain-derived neurotrophic factor in Huntington's disease. Prog Neurobiol. 2007;81:294–330. doi: 10.1016/j.pneurobio.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L, MacDonald ME, Friedlander RM, Silani V, Hayden MR, Timmusk T, Sipione S, Cattaneo E. Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Marullo M, Vitali B, Tarditi A, Mariotti C, Valenza M, Lahiri N, Wild EJ, Sassone J, Ciammola A, Bachoud-Levi AC, Tabrizi SJ, Di Donato S, Cattaneo E. Brain-derived neurotrophic factor in patients with Huntington's disease. PLoS One. 2011;6:e22966. doi: 10.1371/journal.pone.0022966. [DOI] [PMC free article] [PubMed] [Google Scholar]


