Abstract
Purpose.
We minimized the influence of image quality variability, as measured by signal strength (SS), on optical coherence tomography (OCT) thickness measurements using the histogram matching (HM) method.
Methods.
We scanned 12 eyes from 12 healthy subjects with the Cirrus HD-OCT device to obtain a series of OCT images with a wide range of SS (maximal range, 1–10) at the same visit. For each eye, the histogram of an image with the highest SS (best image quality) was set as the reference. We applied HM to the images with lower SS by shaping the input histogram into the reference histogram. Retinal nerve fiber layer (RNFL) thickness was automatically measured before and after HM processing (defined as original and HM measurements), and compared to the device output (device measurements). Nonlinear mixed effects models were used to analyze the relationship between RNFL thickness and SS. In addition, the lowest tolerable SSs, which gave the RNFL thickness within the variability margin of manufacturer recommended SS range (6–10), were determined for device, original, and HM measurements.
Results.
The HM measurements showed less variability across a wide range of image quality than the original and device measurements (slope = 1.17 vs. 4.89 and 1.72 μm/SS, respectively). The lowest tolerable SS was successfully reduced to 4.5 after HM processing.
Conclusions.
The HM method successfully extended the acceptable SS range on OCT images. This would qualify more OCT images with low SS for clinical assessment, broadening the OCT application to a wider range of subjects.
Keywords: OCT, Image processing, retinal nerve fiber layer, histogram matching
A novel histogram matching method was developed for optical coherence tomography images to reduce the measurement variability in retinal nerve fiber layer thickness related to signal strength (SS, an image quality index) variation, and successfully extended the acceptable SS range.
Retinal nerve fiber layer (RNFL) thickness measurements obtained by optical coherence tomography (OCT) have been widely acknowledged as one of the essential clinical parameters for glaucoma assessment.1–4 However, many studies have shown that signal quality of OCT images is associated strongly with RNFL thickness measurements as well as the reliability of segmentation.5–7 This limits the range of acceptable OCT signal quality and usable OCT image data, thereby limiting the applicability of OCT measurements, especially to older or diseased subjects, whose images tend to show lower best-achievable signal quality than young and healthy subjects.8,9
Histogram matching (HM) is an image processing technique to calibrate the differences in intensity contrast when capturing with different cameras, image acquisition equipment, settings, and different light sources.10–12 By shaping an input image histogram to a reference histogram, HM is able to compensate the differences in intensity and image contrast, and even enhances the image quality. The HM technique has been used widely as a preprocessing step in cellular imaging and many medical imaging modalities, such as positron emission tomography (PET), single photon emission computed tomography (SPECT), and magnetic resonance imaging (MRI), to correct the difference in background intensity, improve registration, and reduce analysis variability.13–16
We hypothesized that applying HM on OCT image data enhances image quality of images with lower signal quality that would reduce RNFL thickness measurement variability related to image quality variations. The purpose of this study was to develop a method for minimizing the influence of signal quality–related OCT RNFL thickness measurement variability using a novel OCT image processing method based on HM technique.
Methods
Subjects and Image Acquisition
This was an observational cross-sectional study. Healthy volunteers were recruited at the University of Pittsburgh Medical Center Eye Center. The University of Pittsburgh Review Board and ethics committee approval was obtained for the study, and informed consent was obtained from all subjects. This study followed the tenets of the Declaration of Helsinki and was conducted in compliance with the Health Insurance Portability and Accountability Act.
The circumpapillary region from all eyes was scanned using Cirrus HD-OCT (software version 6.5; Carl Zeiss Meditec, Dublin, CA, USA) with Optic Disc Cube 200 × 200 scan pattern to acquire the three-dimensional (3D) cube data. The scanning protocol collected 200 × 200 sampling points from a 6 × 6 mm2 area centered on the optic disc with 1024 data points along the 2.0 mm axial scan depth. A series of OCT scans with various signal strengths (SSs) were acquired from each eye at the same visit by intentionally defocusing and changing the refraction settings. The SS is a proprietary metric of OCT image quality provided by the device manufacturer. The SS can range from 0 (no signal) to 10 (very strong signal) with an arbitrary unit. It appears on the standard output of the Cirrus device and has been used widely for evaluating OCT image quality in a clinical setting.7,9,17 Due to the limitations of practical scanning, such as the eye condition of subjects, images with full SS range (SS = 1–10) are not always available. Multiple OCT scans (at least 10 scans) were acquired from each eye to achieve a wide range of SS (at least ranging from SS = 3–9). Images with apparent eye movement during scanning were discarded. Eye movement was defined subjectively as image artifacts on OCT en face (or OCT fundus) images showing a horizontal frame shift larger than a diameter of a major retinal blood vessel (approximately 125 μm or 4 pixels)18 or a major distortion of the optic disc (disruption of the natural oval shape). Raw OCT image data were exported to a standalone computer for further processing and analysis.
Histogram Matching Processing
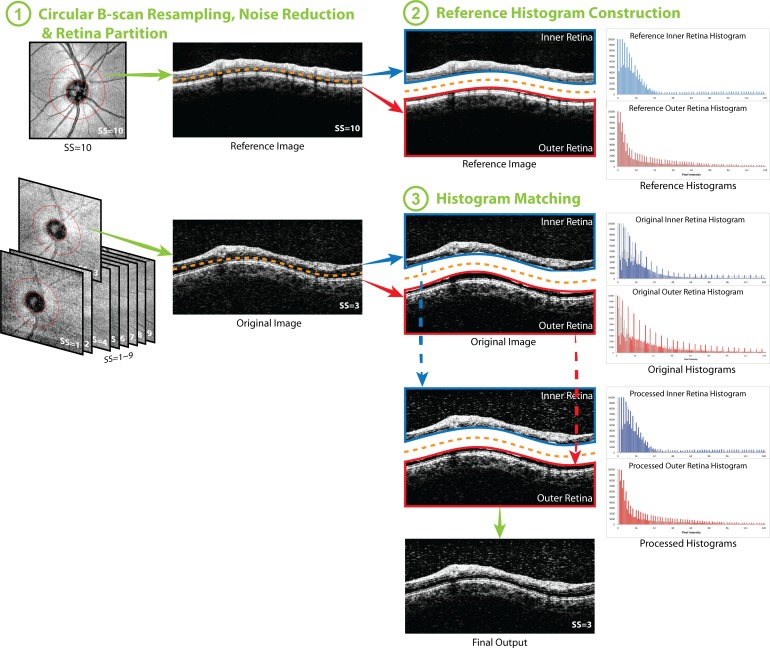
The overall flow of HM processing is presented in Figure 1. The processing is divided into three parts: circular B-scan resampling and speckle noise reduction, followed by either reference histogram construction or HM processing, depending on the SS. The details of each step are described below:
Figure 1.
Flow chart of the histogram matching (HM) method. (1) Circular B-scan was resampled along the 3.4-mm diameter circle (the red circle on the en face image, left column) centered to the optic nerve head for each image. After speckle noise reduction, the circular scans were partitioned into two halves: inner and outer retina. (2) Histograms of inner and outer retina from the image with highest signal strength were set as the reference histograms. (3) The HM was applied to inner and outer retina respectively, and finally combined together to generate the histogram matched image (final output). The ranges of vertical and horizontal axes were adjusted in the figure for better visualization of the histogram and the effect of the processing. After HM, the processed histograms almost overlapped with the reference histograms, which are presented as the gray shadow in the histograms in (3).
Circular B-Scan Resampling.
For each 3D cube image data, the geometric center of the optic nerve head was determined automatically based on the manually delineated disc margin. We sampled 512 equally spaced A-scans along the 3.4-mm diameter circle to generate a virtual circular B-scan (Fig. 1.1).
Speckle Noise Reduction.
A custom speckle noise reduction method was applied to the resampled circular B-scan to reduce the speckle noise. The details of the noise reduction method have been described previously.19 In brief, the custom speckle noise reduction method was a localized high amplitude signal removing method based on a selective smoothing method, where only spiky OCT signal is suppressed to the level of its heavily smoothed counterpart. In this way, high frequency components with relatively high amplitude, considered as speckle noise, were removed, while high frequency components with low amplitude, recognized as retinal tissue signals, were preserved so that more details of the tissue structures remained in the OCT signals after noise reduction.19
Reference Histogram Construction (Applied Only for an Image With the Highest SS for Each Eye).
After circular B-scan resampling and speckle noise reduction, the pixel intensity histogram of the resampled image with the highest SS (best available image quality) of each image series was set as the reference histogram (Fig. 1.2). Images with lower SS were processed with the HM method (see below) so that their histograms had matching shapes with the reference histogram.
To take the clinical reality into account, where for some elder or diseased eye, images with good SS (SS > 6) cannot be achieved, reference histograms were constructed in two ways (Figs. 2, 3).
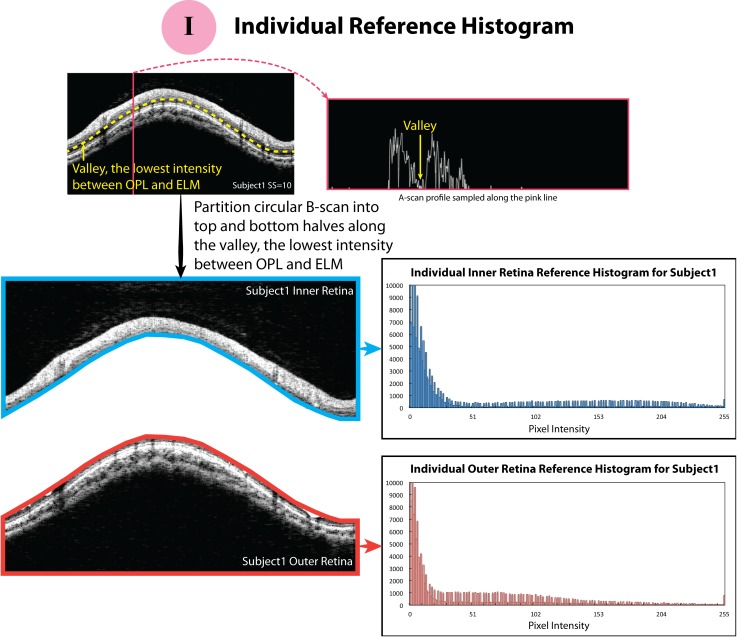
Figure 2.
The schematic figures of how the individual reference histograms were constructed. The blue and red borders surrounding the circular B-scan indicate the region where the reference histograms were calculated.
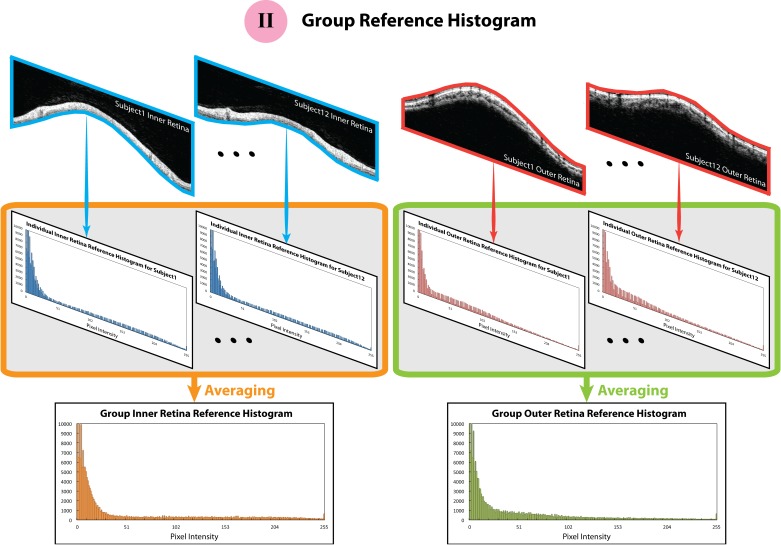
Figure 3.
The schematic figures of how the group reference histograms were constructed. The blue and red borders surrounding the circular B-scan indicate the region where the reference histograms were calculated.
Individual Reference Histogram.
With our observations that histogram statistics differed among retinal layers, we partitioned the circular B-scan image into top and bottom halves along the “valley,” where the OCT signal amplitude was the lowest between the outer plexiform layer (OPL) and external limiting membrane (ELM; the yellow dash curve in Fig. 2, or as the yellow arrow indicates in the A-scan profile in Fig. 2). The valley was automatically detected by our own segmentation software, which is based on the algorithm described previously.2 The top half contained the vitreous body, RNFL, ganglion cell layer, inner plexiform layer, inner nuclear layer, and OPL, and, therefore, also was called the inner retina. The bottom half included the outer nuclear layer, inner and outer segments of the photoreceptors, RPE, and the region below the RPE, and, therefore, also was noted as the outer retina (Fig. 2). The total pixel numbers of the inner and outer retina were matched to half of the entire circular B-scan by padding (with the lowest signal of a given image) or cropping signals from the region in the vitreous or below the RPE so that the proportion of actual retinal signal was consistent across all the subjects. For each image series, the histograms of inner and outer retina of a resampled image with the highest SS were set as the reference histograms for inner and outer retina, respectively. In other words, each image series of a given case had its own inner and outer retina reference histograms.
Group Reference Histogram.
To reflect the clinical reality, where images with good SS simply cannot be obtained on some elder or diseased eyes, mean histogram patterns of the recruited 12 subjects were generated and used as the group reference histograms for all the images (Fig. 3). Group reference histograms were generated for inner and outer retina separately.
Histogram Matching (Applied to the Rest of the Images).
The HM has been known as an image processing technique, where a series of histogram equalization and inverse equalization steps are used to match the statistical information, or histogram shape, of two images.10,11 For each image series on each individual subject, all images except the one with the highest SS (best image quality) were processed with HM (Fig. 1.3). Images, which were the subjects of HM processing, were preprocessed and partitioned in the same way as the reference histogram.
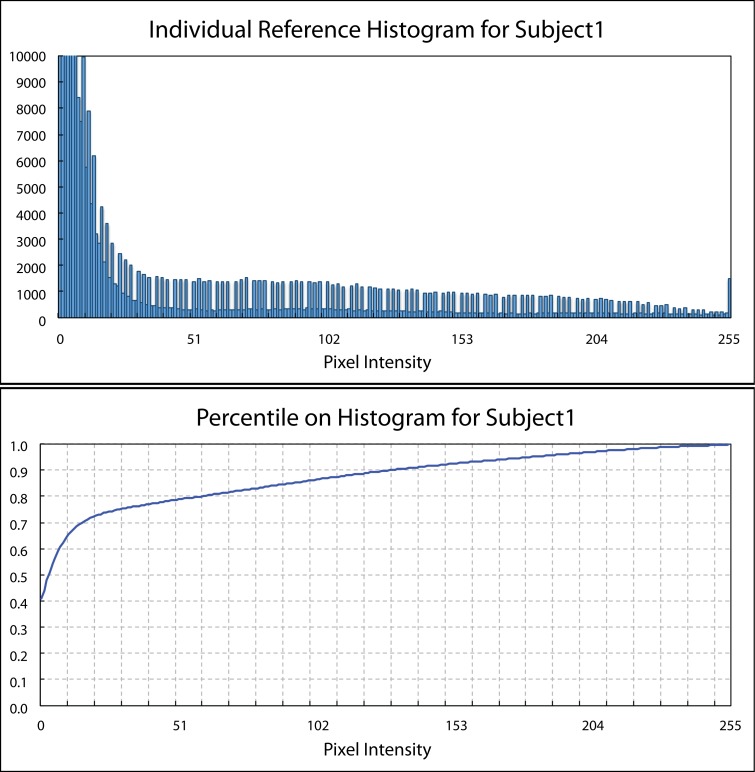
First, the percentile information at each intensity on a given histogram was calculated by Equation 1 (Fig. 4).20
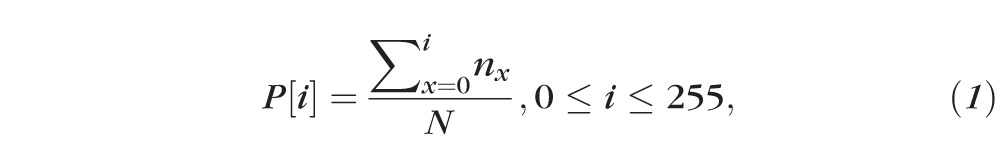 |
where P[i] indicates the percentile information at intensity i , while nx shows the number of pixel having intensity x, and N is the total pixel number within the region. For Cirrus data, the full intensity dynamic range is from 0 to 255.
Figure 4.
An illustration of the reference histogram and corresponding percentile information on the histogram.
Based on the percentile information, a mapping matrix used to convert the shape of input histogram to the shape of reference histogram then was generated by matching or minimizing the distance in percentile at each intensity on the histograms between reference and input histograms, as shown in Equation 2.
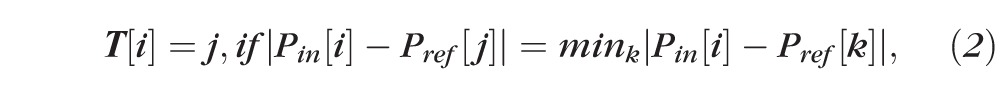 |
where T[i] is the resulted mapping matrix, Pin[i] and Pref[j] are the percentile information on the input and reference histograms at intensities i and j, respectively. For each intensity i in the input histogram, we found a corresponding intensity j in the reference histogram, so that the percentile Pin[i] and Pref[j] had minimal difference. All the sample points in the input image data with intensity i then were mapped to intensity j to generate the output image data.
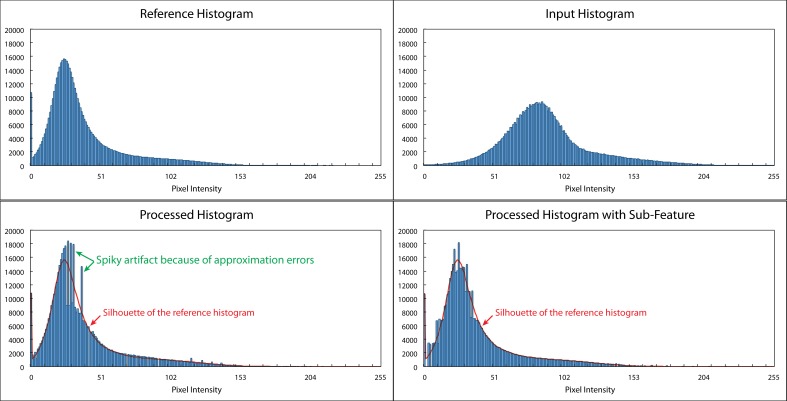
Conventional HM procedure views the sampling points with the same intensity as a group and, thus, cannot distinguish pixels with the same intensity but having different characteristics, and should not be classified into the same group. This generates the approximation errors due to quantization and rounding-off, which can be observed as the spiky shape of the outcome histogram as Figure 5 shows. To solve the approximation errors and be able to separate pixels with the same intensity, we introduced a subfeature besides the intensity to each sampling points.12,21,22 The subfeature virtually made the histogram bin finer than the minimal intensity unit and allowed us to have more flexibility to model the histogram shape.
Figure 5.
Histogram matching with and without a subfeature. Top left: The reference histogram. Top right: The input histogram. Bottom left: The histogram using conventional HM. Bottom right: The histogram after HM with subfeature. Conventional HM has to move the pixels with the same intensity as a group and, therefore, results in the quantization and rounding-off errors, which can be observed as the spiky artifacts on the processed histogram as indicated by the green arrows in the bottom left figure. The errors can be reduced with the subfeature, which enables us to separate pixels with the same intensity but in different retinal layers, as shown in the bottom right figure. The red curve in the bottom left and bottom right figures presents the shape of the reference histogram.
To select a suitable subfeature, it was necessary to consider what information can help distinguish two pixels with the same intensity but different characteristics. Any feature that is able to differentiate such pixels can be used as a subfeature, such as the location of the pixel in the retina, the mean neighbor intensity (which can be calculated from various neighboring regions, such as 3 × 3, 5 × 5, or 1 × 3 neighboring pixels within the same A-scan), or the intensity variance among neighboring pixels.21 The more distinguishable the subfeature is, the more flexibility we have, and, thus, the finer HM we can achieve. However, simply applying the most distinct subfeature, for example the axial position of each pixel, did not improve the results because the axial location of a specific retinal layer varies within a frame. An ideal subfeature is expected to reflect the underlying differences between pixels with the same intensity. The most important pixel characteristic here was whether the target pixel belonged to the same tissue structure or not, which can be assessed by looking at the neighboring pixel information. To add this information, we used the mean and variance of intensity within a pixel's 3 × 3 neighborhood as the subfeature. Our preliminary results showed that using the variance as the subfeature did not reduce the quantization and rounding-off errors. Variance of neighboring pixels represents the homogeneity of the neighborhood. However, as the variance does not vary from layer to layer, it does not provide additional information. On the other hand, using the mean intensity successfully reduced the quantization and rounding-off errors from our preliminary results. By using mean intensity as the subfeature, the contextual information (surrounding tissue information) can be included in the HM process to improve the outcome. As a consequence, we chose to use the mean intensity of a pixel's 3 × 3 neighborhood as the subfeature.
To integrate the subfeature (the mean intensity of a pixel's 3 × 3 neighbors) to the original pixel intensity, each sampling point had a new value, Inew, as presented in Equation 3:
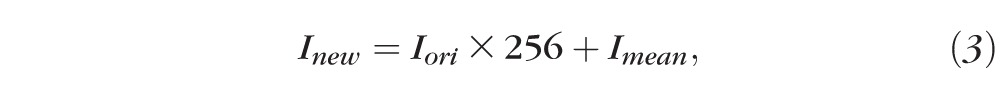 |
where 256 is the full intensity dynamic range for Cirrus data, while Iori and Imean stand for the original intensity of the sampling point and the mean intensity of its 3 × 3 neighbors. We upsampled the original pixel intensity, Iori, by multiplying by the full intensity dynamic range and put the subfeature, Imean (also with a dynamic range of 256), in the gap to increase the intensity resolution.
The HM and mapping matrices were performed and generated based on the new intensity and corresponded histogram. The effects of the individual and group reference histograms were tested separately.
RNFL Thickness Measurements
To test if the HM processing reduces the RNFL thickness measurement variability related to SS variation, the circumpapillary RNFL thicknesses were measured using our custom segmentation software before and after HM (original and HM measurements). The thickness measurements reported from the native segmentation analysis of Cirrus (device measurement) also were collected for comparison. The relationships between device, original, and HM measurements and SS were investigated further.
Statistical Analysis
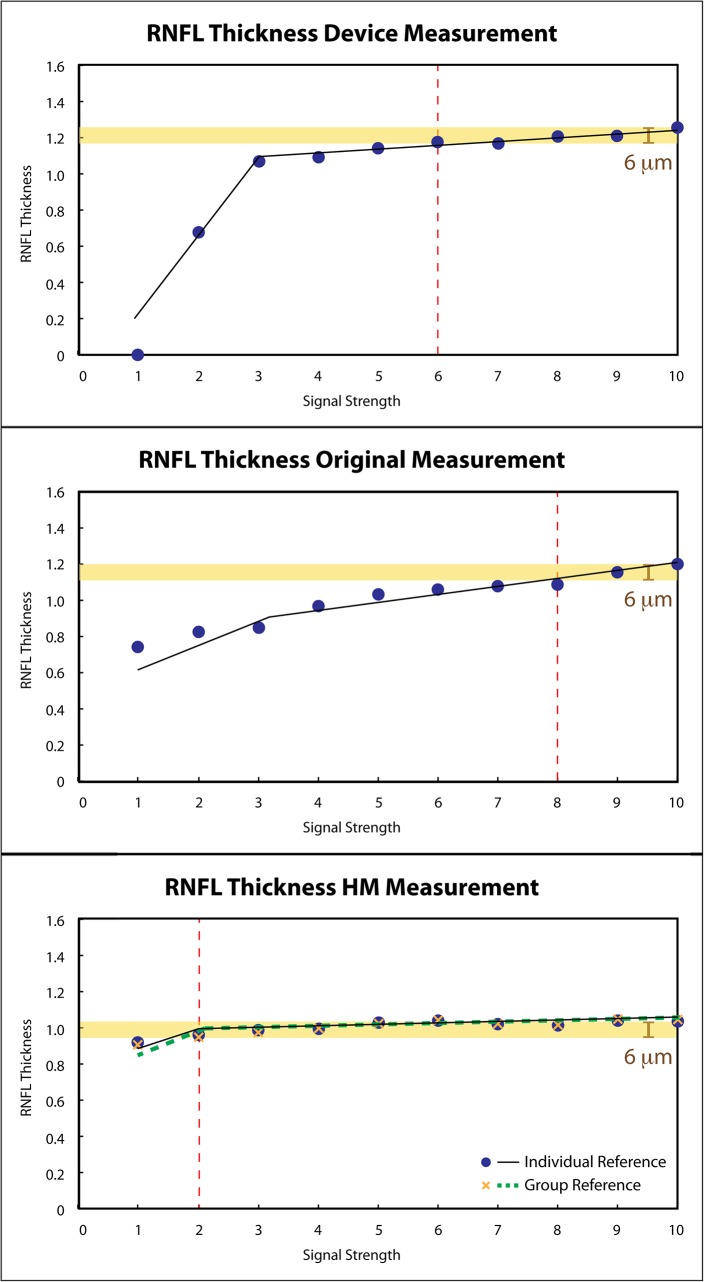
Nonlinear mixed effects models were constructed to analyze the relationship between RNFL thickness and SS for device, original, and HM measurements. To minimize the systematic differences in RNFL thickness measurements between our custom segmentation algorithm and the segmentation algorithm used in the commercial device, the mean RNFL thicknesses of various SSs for each eye was calculated for each method, and then the RNFL thickness at each SS was normalized by dividing by the mean RNFL thickness value. The maximum RNFL thickness difference, defined as the thickest RNFL minus the thinnest RNFL (both from device measurements) within the manufacturer recommended acceptable SS range (SS from 6–10) (yellow band in Fig. 6), was calculated on each case, and set as a comparison reference. The lowest tolerable SS that achieved the same RNFL thickness difference on original, HM, as well as on device measurements was detected on each case based on quadratic regression model to compare the acceptable SS range and test if the acceptable SS range could be extended with HM processing. Paired t-tests were used to compare the lowest SS achieving the same RNFL thickness differences among device, original, and HM measurements. In addition, Wilcoxon tests were used to compare the total measurement variability (the largest RNFL thickness measurement differences with SS range from 1–10) among device, original, and HM measurements. A value of P < 0.05 was considered as statistically significant.
Figure 6.
Relationships between thickness measurements and SS. Circumpapillary RNFL thicknesses from one subject were plotted against SS with two segments of the linear relationships on the device (top), original (middle), and HM (bottom) measurements. The original measurements show larger absolute measurement difference in the higher SS segment. While with HM, the same maximum absolute difference (yellow band) within the manufacturer recommended acceptable SS range on the device (top, SS 6–10) was achieved at a lower SS (SS = 2). Red vertical dash lines in the original and HM plots indicate the lowest SS that has the same RNFL measurement variability as the standard SS range in device measurements. The RNFL thicknesses were normalized to mean thickness value for each method.
Results
A total of 12 right eyes from 12 healthy volunteers (4 males and 8 females) was recruited at the University of Pittsburgh Medical Center Eye Center. The average age was 31.7 ± 11.1 years.
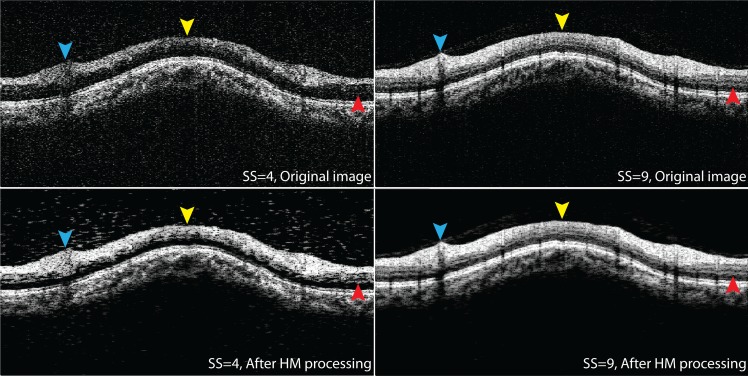
Notable image quality improvements in terms of retinal layer visibility and signal strength were subjectively observed, as shown in Figure 7. The top left figure in Figure 7 shows an original cross-section with low SS (SS = 4). The bottom left figure shows the same cross-section after HM processing with individual reference histogram. In the original low SS cross-sectional image, weak signals were observed at superior (blue arrowhead), nasal (yellow arrowhead), and temporal (red arrowhead) quadrants, and the retinal layers were hard to differentiate. The signals at the same region were significantly enhanced after HM processing and became comparable to the cross-sectional image with the highest available SS (SS = 9) from the same eye same subject (top right). A cross-section of the highest available SS (SS = 9) after HM processing also is shown on the bottom right, indicating that HM processing maintained the signal quality for images with qualified signal strength and does not generate additional artifacts or noise.
Figure 7.
HM processing enhances image quality. Top row: Original cross-sectional images from the same eye and same subject with low and the highest available SS (SS = 4 [left] and SS = 9 [right]). Bottom row: Cross-sectional images after HM processing for low (left) and high (right) SS. The HM processing significantly enhanced the image quality in terms of signal strength and retinal layer visibility (arrowheads).
For the relationships between RNFL thickness and SS, two segments of linear relationship were fitted on device, original, and HM measurements (Fig. 6). Table 1 summarizes the slopes and intercepts of both segments, and the breaking points of all the measurements. The slope in the first segment (β1) presents the linear relationship in the lower SS range (range from 1 to the breaking point), while the slope in the second segment (β2) presents the linear relationship in the higher SS range (range from the breaking point to 10). The original measurements had statistically significantly larger slope in the second segment (β2) than the device measurements (4.89 vs. 1.72 μm/SS), indicating that original measurements were more sensitive to SS in the higher SS segment. However, with HM processing, the slope in the higher SS segment was statistically significantly smaller than both original measurements regardless of using individual or group reference histograms (1.17 and 1.06 μm/SS, Table 1).
Table 1.
Summarization of the Relationships Between Measurements and SS
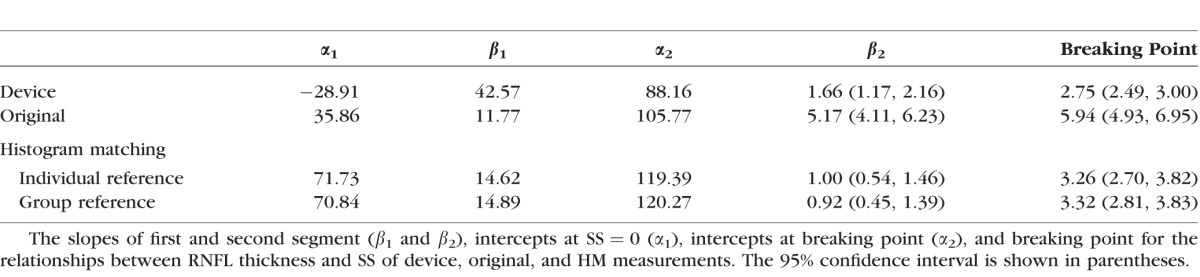
For the breaking point, the original measurements had statistically significantly higher breaking point than the device measurements (5.94 vs. 2.75 SS), while it became similar after applying HM (3.26 with individual and 3.32 with group reference), suggesting that HM stabilized the measurement variability in wider SS range.
The lowest SS that maintained the maximum RNFL thickness differences was detected for each subject (Table 2). For device measurement, the lowest SS that achieved the same maximum difference was 5.1, while original measurement showed statistically significantly higher lowest SS, which was 8.5 (P < 0.0001, paired t-test). The lowest SS was statistically significantly reduced from the device measurements to 4.5 after HM processing for individual and group reference histograms (P < 0.03, paired t-test), indicating that HM processing successfully extended the acceptable SS range. The trend also can be observed in Figure 6.
Table 2.
Summarization of the Minimum Acceptable SS Range and the Measurement Variability
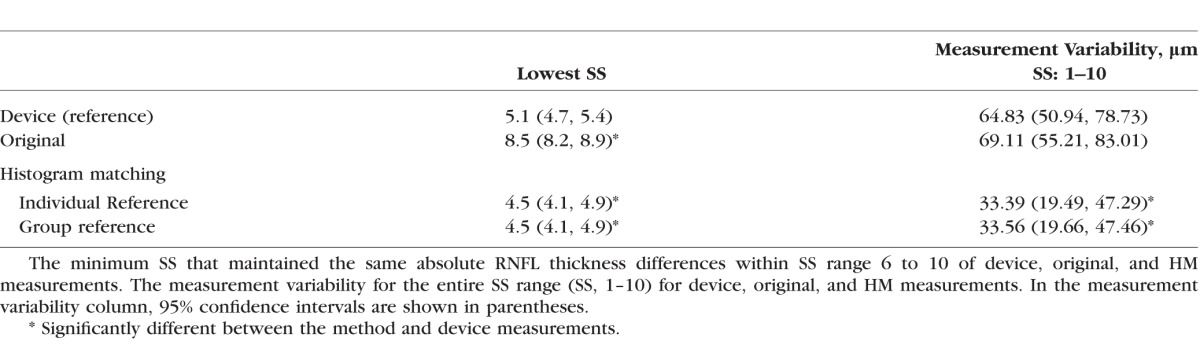
No statistically significant differences in total measurement variability (SS range, 1–10) were found between device and original measurements (64.8 vs. 69.1 μm, P = 0.86, Wilcoxon tests; Table 2). However, HM measurements showed statistically significantly smaller total measurement variability than the device measurements on individual and group references (64.8 vs. 33.4 and 33.6 μm, both P < 0.038, respectively, Wilcoxon tests).
Discussion
A novel HM-based OCT image enhancement method was developed in this study. The proposed method successfully enhanced OCT images with lower image quality and reduced the RNFL thickness measurement variability related to image quality variation. The HM extended the acceptable signal quality range and further reduced the thickness measurement variability, which would broaden the application of OCT to elder or diseased subjects who tend to have lower best-achievable image quality.
Compared to other histogram manipulation methods, such as contrast and brightness adjustment, histogram matching is more effective in reducing the RNFL thickness measurement variability. Contrast/brightness adjustment is applying linear/nonlinear transformation to pixel values, sometimes with thresholds. While it is possible to optimize such adjustments mathematically, it is difficult to standardize the output contrast/brightness systematically, especially on images with specific intensity levels that need to be clarified, like OCT images. Within the OCT image intensity level, the range of meaningful signals that represent retinal tissues is relatively small. As contrast/brightness adjustment is applied more or less uniformly across the entire intensity level (even with nonlinear transformation), it often generates unwanted side effects (e.g., boosted noise level in the vitreous cavity). Histogram matching takes a reference histogram, which has a well-balanced histogram shape optimized for the best tissue visualization on OCT images. This type of signal modification cannot be achieved with contrast/brightness adjustment.
Separating the retina into two parts and applying individual HM on each part improved the image enhancement performance. We observed that the OCT signal from the outer retina generally was stronger than the signal from the inner retina along all the SS variation. In other words, the signal from the inner retina degraded more than the signal from the outer retina as SS decreased. Therefore, if simply applying HM to the entire image, the signal from the inner retina remained weak even after enhancement. This affected the segmentation performance as the inner retina is supposed to have relatively high reflectivity. When only the inner retinal section was used as a reference to adjust the entire image, unwanted clipping artifacts (over saturation) were observed in the outer retinal section. Only when histogram matching was applied separately to the inner and outer retina sections was the signal level normalized between the inner and outer retinal sections, so that the segmentation performance was more stable and reliable, leading to the reduced measurement variability.
No statistically significant differences were detected in the slope of the higher SS segment and the breaking point when using individual and group reference histograms. This indicates that the specific reference histogram for each subject can be replaced by a group reference histogram and reach similar results or that we did not have enough power to detect a difference.
A potential limitation of this method is our assumption that the disease does not change the histogram characteristics of the scan. If the histogram characteristics are different from the group reference histogram, then the outcomes may not reflect the actual disease status. In that case, an individual reference histogram or a separated group reference histogram with similar histogram characteristics (pathology) is required. Further investigation for the validation of such references with a larger number of subjects is warranted.
Another limitation of this study is that the group reference histogram was constructed from all of the recruited eyes. Further investigation with a larger number of subjects is required.
With HM processing, similar RNFL thickness measurements were generated across a wider SS range, suggesting less measurement variability expected even with lower SS image than the current manufacturer's recommendation. In addition, the measurement variability was also reduced across the recommended SS range (SS, 6–10). The reduced measurement variability may help reduce the false-positive reading due to acceptable, but low SS, and may further improve the ability to detect smaller changes over time.
In this study, we used our custom segmentation algorithm to test the proposed HM method rather than the native segmentation software of the device. This inevitable limitation is because the device software does not accept modified OCT image files for processing. Our custom segmentation algorithm tends to report thicker RNFL thickness value compare to the native segmentation software of the device. This variation was minimized by normalizing RNFL thickness to the mean value of each method when performing the statistical analyses. Strictly speaking, the observed improvement is limited to the custom algorithm. However, overall trend of correlation between segmented thickness measurements and image quality is observed regardless of the difference in algorithm approach. Therefore, it is reasonable to speculate that the HM method may expand the acceptable SS range without affecting the OCT measurement variability.
In conclusion, the proposed HM method successfully extended the acceptable SS range on OCT images, while maintaining the similar measurement variability within the manufacturer recommended SS range. With the potential to achieve wider acceptable SS range, HM would qualify more OCT images with relatively low SS for clinical assessment and further broaden the OCT application to a wider range of subjects.
Acknowledgments
Presented in part at the annual meeting of the Association for Research in Vision and Ophthalmology, Orlando, Florida, United States, May 2014.
Supported in part by National Institutes of Health (NIH; Bethesda MD, USA) Contracts NIH R01-EY013178, P30-EY008098; Eye and Ear Foundation (Pittsburgh, PA, USA); and Research to Prevent Blindness (New York, NY, USA).
Disclosure: C.-L. Chen, None; H. Ishikawa, None; G. Wollstein, None; R.A. Bilonick, None; I.A. Sigal, None; L. Kagemann, None; J.S. Schuman, P
References
- 1. Koozekanani D,, Boyer K,, Roberts C. Retinal thickness measurements from optical coherence tomography using a Markov boundary model. IEEE Trans Med Imaging. 2001; 20: 900–916. [DOI] [PubMed] [Google Scholar]
- 2. Ishikawa H,, Stein DM,, Wollstein G,, et al. Macular segmentation with optical coherence tomography. Invest Ophthalmol Vis Sci. 2005; 46: 2012–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gabriele ML,, Wollstein G,, Ishikawa H,, et al. Optical coherence tomography: history, current status, and laboratory work. Invest Ophthalmol Vis Sci. 2011; 52: 2425–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. DeBuc DC. A review of algorithms for segmentation of retinal image data using optical coherence tomography. Image Segmentation. 2011; 15–54.
- 5. Cheung CY,, Leung CK,, Lin D,, et al. Relationship between retinal nerve fiber layer measurement and signal strength in optical coherence tomography. Ophthalmology. 2008; 115: 1347–1351. [DOI] [PubMed] [Google Scholar]
- 6. Stein DM,, Wollstein G,, Ishikawa H,, et al. Effect of corneal drying on optical coherence tomography. Ophthalmology. 2006; 113: 985–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Folio LS,, Wollstein G,, Ishikawa H,, et al. Variation in optical coherence tomography signal quality as an indicator of retinal nerve fiber layer segmentation error. Br J Ophthalmol. 2012; 96: 514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ray R,, Stinnett SS,, Jaffe GJ. Evaluation of image artifact produced by optical coherence tomography of retinal pathology. Am J Ophthalmol. 2005; 139: 18–29. [DOI] [PubMed] [Google Scholar]
- 9. Na JH,, Sung KR,, Lee Y. Factors associated with the signal strengths obtained by spectral domain optical coherence tomography. Korean J Ophthalmol. 2012; 26: 169–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. González RC,, Woods RE. Digital Image Processing. Upper Saddle River, NJ: Prentice Hall; 2002. [Google Scholar]
- 11. Sintorn IM,, Bischof L,, Jackway P,, et al. Gradient based intensity normalization. J Microsc. 2010; 240: 249–258. [DOI] [PubMed] [Google Scholar]
- 12. Stanciu SG,, Stanciu GA,, Coltuc D. Automated compensation of light attenuation in confocal microscopy by exact histogram specification. Microsc Res Tech. 2010; 73: 165–175. [DOI] [PubMed] [Google Scholar]
- 13. Valous NA,, Lahrmann B,, Zhou W,, et al. Multistage histopathological image segmentation of Iba1-stained murine microglias in a focal ischemia model: methodological workflow and expert validation. J Neurosci Methods. 2013; 213: 250–262. [DOI] [PubMed] [Google Scholar]
- 14. Salas-Gonzalez D,, Górriz JM,, Ramírez J,, et al. Improving the convergence rate in affine registration of PET and SPECT brain images using histogram equalization. Comput Math Methods Med. 2013; 2013: 760903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shinohara RT,, Sweeney EM,, Goldsmith J,, et al. Statistical normalization techniques for magnetic resonance imaging. Neuroimage Clin. 2014; 6: 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mehta SB,, Chaudhury S,, Bhattacharyya A,, et al. A soft-segmentation visualization scheme for magnetic resonance images. Magn Reson Imaging. 2005; 23: 817–828. [DOI] [PubMed] [Google Scholar]
- 17. Russell DJ,, Fallah S,, Leor CJ,, et al. A comprehensive model for correcting RNFL readings of varying signal strengths in cirrus optical coherence tomography. Invest Ophthalmol Vis Sci. 2014; 55: 7297–7302. [DOI] [PubMed] [Google Scholar]
- 18. Jonas JB,, Nguyen XN,, Naumann GO. Parapapillary retinal vessel diameter in normal and glaucoma eyes. I. Morphometric data. Invest Ophthalmol Vis Sci. 1989; 30: 1599–1603. [PubMed] [Google Scholar]
- 19. Chen CL,, Ishikawa H,, Wollstein G,, et al. Individual A-scan signal normalization between two spectral domain optical coherence tomography devices. Invest Ophthalmol Vis Sci. 2013; 54: 3463–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang YJ. Improving the accuracy of direct histogram specification. J Electron Imaging. 1992; 28: 213–214. [Google Scholar]
- 21. Coltuc D,, Bolon P,, Chassery JM. Exact histogram specification. IEEE Trans Image Process. 2006; 15: 1143–1152. [DOI] [PubMed] [Google Scholar]
- 22. Bevilacqua A,, Azzari P. A high performance exact histogram specification algorithm. 14th Int Conf Image Anal Proc. 2007; 2007: 623–628. [Google Scholar]