Abstract
Background
Potentially pathogenic microorganisms can be detected by quantitative real-time polymerase chain reaction (qPCR) in sputum from patients with COPD, although how this technique relates to culture and clinical measures of disease is unclear. We used cross-sectional and longitudinal data to test the hypotheses that qPCR is a more sensitive measure of bacterial presence and is associated with neutrophilic airway inflammation and adverse clinical outcomes.
Methods
Sputum was collected from 174 stable COPD subjects longitudinally over 12 months. Microbial sampling using culture and qPCR was performed. Spirometry and sputum measures of airway inflammation were assessed.
Findings
Sputum was qPCR-positive (>106 copies/mL) in 77/152 samples (Haemophilus influenzae [n=52], Moraxella catarrhalis [n=24], Streptococcus pneumoniae [n=19], and Staphylococcus aureus [n=7]). Sputum was culture-positive in 50/174 samples, with 49 out of 50 culture-positive samples having pathogen-specific qPCR bacterial loads >106 copies/mL. Samples that had qPCR copy numbers >106/mL, whether culture-positive or not, had increased sputum neutrophil counts. H. influenzae qPCR copy numbers correlated with sputum neutrophil counts (r=0.37, P<0.001), were repeatable within subjects, and were >106/mL three or more times in 19 patients, eight of whom were repeatedly sputum culture-positive. Persistence, whether defined by culture, qPCR, or both, was associated with a higher sputum neutrophil count, lower forced expiratory volume in 1 second (FEV1), and worsened quality of life.
Interpretation
qPCR identifies a significant number of patients with potentially bacteria-associated neutrophilic airway inflammation and disease that are not identified by traditional culture-based methods.
Keywords: H. influenzae, qPCR, sputum
Introduction
The role of bacteria in the pathogenesis of airway inflammation and symptoms in stable COPD remains unclear. Potentially pathogenic microorganisms (PPMs) have been identified at stable state in bronchoalveolar lavage fluid1,2 and sputum.3 The most commonly identified species are Haemophilus influenzae and Moraxella catarrhalis.1,2 Although often assumed to reflect colonization, previous work has shown that a single positive sputum culture for a PPM is associated with increased neutrophilic airway inflammation,4,5 worsened health status,6 increased exacerbations,7 and an increased risk of bacteria-associated exacerbations.8 Moreover, antibiotic therapy in patients with stable COPD is associated with a reduction in sputum neutrophil counts in those with elevated bacterial loads.9 These findings suggest that PPMs contribute directly to neutrophilic airway inflammation in patients with stable COPD.
More recently, quantitative real-time polymerase chain reaction (qPCR) has been used to detect respiratory PPMs, with the potential advantage of allowing rapid quantification of bacteria. How this measure compares with culture-dependent methods and whether it relates to airway inflammation and clinically important outcomes are largely unknown. We tested the hypothesis that the presence of PPMs in sputum measured by qPCR in subjects with stable COPD is a more sensitive measure of bacteria-associated neutrophilic airway inflammation by assessing the cross-sectional and longitudinal relationship between bacterial load, sputum neutrophil count, and clinical outcomes.
Methods
Data was collected from a prospective COPD cohort study performed as previously described.8,10 In summary, subjects were aged 40 or over and had a physician diagnosis of COPD and a postbronchodilator forced expiratory volume in 1 second (FEV1)/forced vital capacity (FVC) of less than 0.7. All subjects had experienced at least one exacerbation requiring emergency health care utilization in the 12 months prior to recruitment. Subjects were seen for four visits when stable, at 3-monthly intervals, for the study duration. A “stable” visit was defined as one where subjects were free from an exacerbation episode (identified according to health care utilization and daily diary cards) for at least 6 weeks prior to the visit. Subjects with a clinical diagnosis of asthma or on prophylactic antibiotic therapy were excluded. All subjects gave written informed consent, and the study was approved by the Regional Ethics Committee of Leicestershire, Northamptonshire, and Rutland (reference number: 07/H0406/157).
Measurements
At each visit, spirometry was performed according to guidelines.11 Health-related quality of life and symptom assessments were measured using a visual analog scale (VAS)12 for the domains of dyspnea, cough, sputum production, and sputum purulence; the Chronic Respiratory Disease Questionnaire (CRQ),13 and the St George’s Respiratory Questionnaire (SGRQ).14 Venous blood was obtained for measurement of a full blood count and C-reactive protein (CRP). Spontaneous or induced sputum was collected and processed for differential cell counts and detection of bacteria. Identification of PPMs in sputum culture (H. influenzae, M. catarrhalis, Streptococcus pneumoniae, Staphylococcus aureus, and Pseudomonas aeruginosa) was performed according to the Health Protection Agency (HPA) standard operating procedures.15 Sputum culture that did not show a dominant pathogenic respiratory pathogen was classified as having nonsignificant growth (NSG). Total aerobic counts were obtained by first plating homogenized sputum, serially diluted in Dulbecco’s phosphate-buffered saline, on chocolate and blood agar plates, and then, counting the number of colonies (colony-forming units [CFUs]) as previously described.16 qPCR was used to estimate bacterial load based on the abundance of 16S ribosomal subunit, variable region V4, and V5 encoding genes, and genome copy numbers of H. influenzae, M. catarrhalis, S. pneumoniae, and S. aureus, as described previously.8 In brief, total bacterial DNA was extracted from 500 μL of homogenized sputum using the QIAmp DNA Mini Kit (QIAGEN Ltd, Venlo, the Netherlands). DNA standards for qPCR were prepared from pure DNA cultures of Escherichia coli, H. influenzae, M. catarrhalis, S. pneumoniae, and S. aureus. The SYBR® green assay (Applied Biosystems®; Life Technologies Corp, Carlsbad, CA, USA) was used to quantify qPCR total bacterial load (16S) and qPCR pathogen-specific H. influenzae and S. aureus bacterial load. The TaqMan® assay (Applied Biosystems; Life Technologies Corp) was used for quantification of qPCR pathogen-specific M. catarrhalis and S. pneumonia bacterial loads. The lower detection limit of a CFU was ≥1×105/mL of sputum, and a threshold of ≥1×106 genome copies/mL was regarded as clinically significant. Neutrophilic airway inflammation was defined as a sputum neutrophil count >60%.17
Statistical analysis
Statistical analysis was performed using PASW Statistics for Windows, Version 18.0 (SPSS Inc. Chicago, IL, USA) and GraphPad Prism version 6 (GraphPad Software Inc., La Jolla, CA, USA). Normality tests were performed using the Kolmogorov–Smirnov test. Parametric and nonparametric data were presented as mean (standard deviation [SD]) and median (interquartile range). Log-transformed data were expressed as geometric mean (95% confidence interval [CI]). Repeatability was assessed using intraclass correlation coefficients (ICCs) and a one-way random effects average measures model. Association was measured using Pearson and Spearman rank correlation coefficients. One-way analysis of variance, Kruskal–Wallis, and chi-square tests were used to compare clinical characteristics across groups. P-values of <0.05 were considered to be statistically significant.
Results
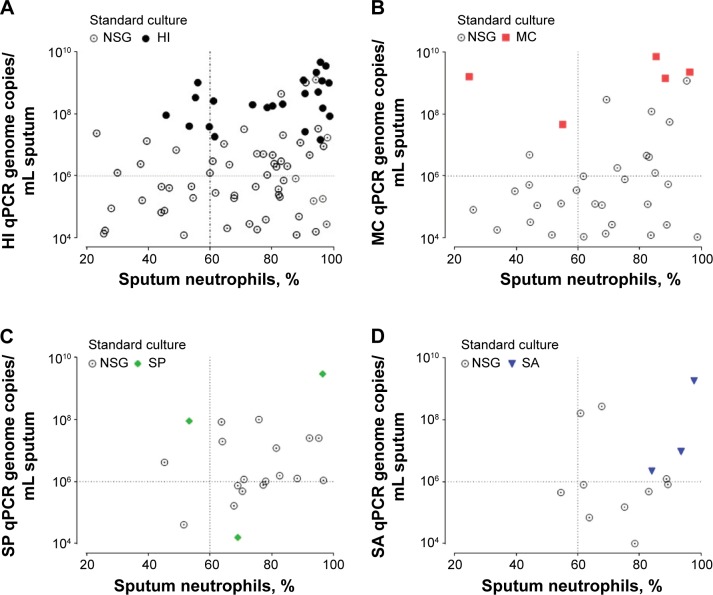
At study entry, 174 subjects provided an adequate sputum sample for any quantification. The most common pathogen detected was H. influenzae, occurring in 52/152 (34%) of qPCR samples and 29/174 (17%) of culture samples. M. catarrhalis, S. pneumoniae, and S. aureus occurred in 24 (16%), 19 (13%), and seven (5%) of the qPCR pathogen-specific samples and in six (8%), four (2%), and three (2%) of all cultures, respectively. P. aeruginosa was detected in eight (5%) culture samples and was not quantified by qPCR. There was 98% concordance between the corresponding PPM on routine culture and the qPCR pathogen-specific pathogen; all but one culture (S. pneumoniae culture) (Figure 1) had a qPCR pathogen-specific bacterial load >106 copies/mL. In PPM-positive culture samples compared with NSG culture, there was a 4.6-fold increase in CFUs (95% CI of fold difference, 2.0 to 10.5; P<0.001) and a 4.1-fold increase in total qPCR 16S bacterial load (95% CI of fold difference, 1.9 to 8.8; P<0.001). Subjects that were positive for PPM at culture or qPCR had more severe airflow limitation, increased symptoms, and increased inflammatory indices (Table 1).
Figure 1.
Scatter correlation plot of sputum neutrophils (x-axis) with pathogen-specific bacterial load (y-axis) for samples above the limit of detection (qPCR LLD >1×104 copies/mL of sputum).
Notes: Data were categorized into respective sputum culture pathogens (NSG, HI, MC, SP, and SA). (A) HI. (B) MC. (C) SP. (D) SA pathogen-specific qPCR. The horizontal dashed line on the y-axis related to pathogen-specific qPCR bacterial load of 106. The vertical dashed line on the x-axis related to sputum neutrophil count at 60% (neutrophilic airway inflammation).
Abbreviations: NSG, nonsignificant growth; HI, Haemophilus influenzae; LLD, lower limit of detection; MC, Moraxella catarrhalis; qPCR, quantitative real-time polymerase chain reaction; SP, Streptococcus pneumoniae; SA, Staphylococcus aureus.
Table 1.
Baseline clinical and bacterial characteristics of subjects with and without PPM, on routine culture and by qPCR
| Routine culture
|
qPCR PPM detection
|
|||
|---|---|---|---|---|
| −PPM n=124 |
+PPM n=50 |
−PPM n=44 |
+PPM n=77 |
|
| Males, n (%) | 88 (71) | 35 (70) | 27 (61) | 59 (77) |
| Age, years | 68 (9) | 69 (8) | 70 (8) | 67 (9) |
| Pack-years smoked, mean (range) | 52 (10–159) | 54 (10–153) | 53 (10–159) | 54 (10–156) |
| FEV1 (L)# | 1.38 (0.54) | 1.18 (0.53)¥ | 1.29 (0.48) | 1.29 (0.55) |
| FEV1 (% predicted)# | 52 (20) | 46 (18)¥ | 52 (18) | 48 (19) |
| FEV1/FVC ratio (%)# | 52 (12) | 46 (13)¥ | 51 (12) | 47 (13)ᴥ |
| Exacerbations frequency/yr | 3.5 (2.7) | 4.1 (2.7) | 3.5 (2.7) | 3.6 (2.6) |
| Inhaled corticosteroid dose, μg | 1,279 (748) | 1,675 (749)¥ | 1,416 (651) | 1,493 (768) |
| SGRQ, units | 52.3 (17.5) | 54.6 (16.9) | 50.5 (13.7) | 53.0 (19.4) |
| CRQ, units | 4.1 (1.1) | 4.0 (1.2) | 4.8 (1.3) | 4.0 (1.2) |
| VAS cough, mm | 36 (26) | 46 (27)¥ | 37 (27) | 41 (26) |
| VAS dyspnea, mm | 49 (26) | 51 (26) | 53 (27) | 48 (26) |
| VAS sputum production, mm | 35 (26) | 49 (26)¥ | 31 (26) | 44 (26)ᴥ |
| VAS sputum purulence, mm | 31 (25) | 39 (27) | 33 (26) | 28 (27) |
| Peripheral leukocyte count, ×109 cells/mL* | 8.2 (7.8–8.5) | 8.9 (8.2–9.7)¥ | 8.7 (8.1–9.5) | 8.4 (7.9–8.9) |
| Peripheral neutrophil count, ×109 cells/mL* | 5.2 (4.9–5.5) | 5.8 (5.3–6.5)¥ | 5.7 (5.1–6.3) | 7.3 (4.9–5.8) |
| Peripheral eosinophil count, ×109 cells/mL* | 0.21 (0.18–0.24) | 0.21 (0.18–0.25) | 0.22 (0.18–0.28) | 0.22 (0.19–0.25) |
| C-reactive protein, mg/L† | 3 (3–11) | 6 (3–18) | 3 (3–12) | 6 (3–14) |
| Total cell count, ×106 cells/g sputum* | 2.5 (2.0–3.1) | 6.0 (4.2–8.5)¥ | 2.7 (1.9–4.1) | 4.1 (3.1–5.5)ᴥ |
| Sputum neutrophil differential, % | 64 (22) | 81 (19)¥ | 65 (18) | 75 (22)ᴥ |
| Sputum eosinophil differential, %* | 1.4 (1.1–1.8) | 0.9 (0.6–1.2)¥ | 1.6 (1.0–2.6) | 1.2 (0.8–1.6) |
| Proportion with neutrophilic inflammation | 62% | 86%¥ | 63% | 79%ᴥ |
| Log colony forming units/mL of sputum˫ | 6.9 (6.7–7.1) | 7.6 (7.2–7.9)¥ | 6.8 (6.5–7.1) | 7.3 (7.0–7.6)ᴥ |
| Total 16S log genome copies/mL of sputumⱶ,** | 8.3 (8.1–8.4) | 8.9 (7.2–9.2)¥ | 8.1 (7.8–8.3) | 8.8 (8.6–9.0)ᴥ |
| Proportion HI qPCR codetection | 47% | 70%¥ | 0% | 82%ᴥ |
| Proportion MC qPCR codetection | 28% | 54%¥ | 0% | 51%ᴥ |
| Proportion SP qPCR codetection | 18% | 39%¥ | 0% | 28%ᴥ |
| Proportion SA qPCR codetection | 9% | 10% | 0% | 14%ᴥ |
| HI log genome copies/mL qPCRⱶ,** | 4.3 (3.9–4.6) | 6.5 (5.7–7.3)¥ | 2.8 (2.7–2.8) | 6.4 (5.9–6.9)ᴥ |
| MC log genome copies/mL qPCRⱶ,** | 3.5 (3.3–3.8) | 5.4 (4.6–6.3)¥ | 2.7 (2.7–2.7) | 5.0 (4.4–5.5)ᴥ |
| SP log genome copies/mL qPCRⱶ,** | 3.5 (3.2–3.7) | 4.0 (3.4–4.5) | 2.7 (2.7–2.7) | 4.2 (3.8–4.7)ᴥ |
| SA log genome copies/mL qPCRⱶ,** | 3.0 (2.8–3.2) | 3.1 (2.7–3.6) | 2.7 (2.7–2.7) | 3.3 (2.9–3.6)ᴥ |
Notes: Data were presented as mean (standard deviation) unless stated.
Postbronchodilator.
Geometric mean (95% confidence interval).
Median (interquartile range).
Mean (95% confidence interval).
Based on 16S qPCR log genome copies/mL.
P<0.05 between −PPM and +PPM culture groups (Student’s t-test, Mann–Whitney, or chi-square, as appropriate).
P<0.05 between −PPM and +PPM qPCR groups, >106 copies/mL (Student’s t-test, Mann–Whitney, or chi-square, as appropriate). Neutrophilic airway inflammation was defined as greater than 60% sputum neutrophils.
Abbreviations: CRQ, Chronic Respiratory Disease Questionnaire; FEV1, forced expiratory volume in 1 second; FVC, forced vital capacity; HI, Haemophilus influenzae; MC, Moraxella catarrhalis; PPM, potentially pathogenic microorganism; −PPM, PPM-negative; +PPM, PPM-positive; qPCR, quantitative real-time polymerase chain reaction; SA, Staphylococcus aureus; SGRQ, St George’s Respiratory Questionnaire; SP, Streptococcus pneumoniae; VAS, visual analog scale.
The mean sputum neutrophil count in patients with positive pathogen-specific qPCR (ie, bacterial loads above and below 106 copies/mL) was 75% compared with 64% in those with negative pathogen-specific qPCR (mean difference 12%, 95% CI, 5% to 18%; P<0.001). There were no differences in age, smoking pack-years, FEV1, or FEV1% predicted, in subjects that were culture-negative and qPCR-positive compared with the remainder of the group. Pathogen-specific qPCR identified subjects with neutrophilic airway inflammation, whether associated with a positive culture or not (Figure 1). Only qPCR pathogen-specific H. influenzae bacterial load correlated with percentage sputum neutrophils and sputum total cell count (r=0.37, P<0.001 and r=0.26, P=0.017, respectively). A dose-response sputum neutrophil relationship was seen with H. influenzae only (Table 2). The sensitivity and specificity of qPCR to detect neutrophilic inflammation was 75% and 34% in comparison with that of the standard culture technique, which was 37% and 87%, respectively.
Table 2.
Clinical and inflammatory characteristics of COPD patients with positive qPCR categorized into absent (<106), present (between 106 to 107), and highly present (greater than 108 copies/mL)
|
Haemophilus influenzae
|
Moraxella catarrhalis
|
Streptococcus pneumoniae
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Absent n=30 |
Present (106–107) |
High present, (>108) |
P-value | Absent n=30 |
Present (106–107) |
High present, (>108) |
P-value | Absent n=30 |
Present (106–107) |
High present, (>108) |
P-value | |
| n | 30 | 30 | 22 | 6 | 2 | 5 | 6 | 3 | 2 | |||
| Any +PPM sputum culture | 13% | 23% | 91% | <0.01 | 17% | 50% | 60% | 0.38 | 67% | 0% | 50% | 0.20 |
| Log CFU/mL* | 7.0 (6.6–7.4) | 7.1 (6.7–7.4) | 7.7 (7.2–8.3) | 0.04 | 7.0 (5.1–8.8) | 7.8 (7.7–8.0) | 8.7 (6.8–10.6) | 0.19 | 7.2 (6.6–7.8) | 6.6 (4.1–9.1) | 5.5 (5.3–5.6) | 0.05 |
| Log total 16S genome copies/mL* | 8.1 (7.7–8.4) | 8.5 (8.2–8.8) | 9.2 (8.8–9.6) | <0.01 | 8.1 (6.8–9.4) | 9.4 (5.6–13.2) | 9.7 (8.9–10.4) | 0.06 | 9.3 (8.8–9.7) | 8.4 (7.7–9.0) | 8.4 (5.5–11.4) | 0.01 |
| Sputum neutrophils, % | 66 (22) | 72 (21) | 84 (16) | 0.01 | 65 (25) | 72 (25) | 74 (25) | 0.86 | 66 (30) | 69 (30) | 33 (28) | 0.45 |
| CRQ dyspnea, units | 3.5 (1.3) | 2.7 (0.9) | 3.1 (1.2) | 0.04 | 3.6 (0.8) | 2.7 (0.7) | 4.0 (0.9) | 0.23 | 3.7 (1.2) | 2.7 (0.6) | 2.7 (0.4) | 0.33 |
| CRQ fatigue, units | 3.8 (1.3) | 3.2 (1.1) | 3.8 (1.2) | 0.13 | 4.1 (1.2) | 3.4 (0.9) | 4.5 (0.7) | 0.45 | 4.3 (1.3) | 4.4 (1.1) | 3.4 (2.3) | 0.70 |
| CRQ emotion, units | 4.5 (1.3) | 4.2 (1.3) | 4.6 (1.5) | 0.58 | 4.3 (1.4) | 3.9 (1.3) | 5.5 (0.9) | 0.24 | 5.1 (1.4) | 5.6 (0.1) | 4.1 (1.3) | 0.39 |
| CRQ mastery, units | 4.9 (1.5) | 4.5 (1.6) | 4.7 (1.5) | 0.61 | 4.2 (1.5) | 3.6 (1.2) | 5.3 (0.9) | 0.24 | 5.9 (0.8) | 6.2 (1.0) | 3.5 (2.1) | 0.06 |
| VAS cough, mm | 37 (25) | 39 (27) | 39 (25) | 0.94 | 30 (27) | 57 (14) | 49 (24) | 0.34 | 33 (13) | 42 (38) | 44 (42) | 0.83 |
| VAS dyspnea, mm | 47 (25) | 45 (26) | 51 (25) | 0.65 | 43 (31) | 76 (13) | 39 (25) | 0.29 | 44 (19) | 61 (29) | 37 (20) | 0.36 |
| VAS sputum purulence, mm | 39 (24) | 40 (26) | 48 (26) | 0.51 | 20 (25) | 68 (28) | 42 (26) | 0.69 | 28 (12) | 59 (41) | 41 (35) | 0.90 |
| VAS sputum volume, mm | 33 (26) | 35 (26) | 42 (32) | 0.40 | 18 (22) | 26 (16) | 27 (14) | 0.11 | 37 (20) | 53 (18) | 31 (2) | 0.22 |
Notes: Data for Staphylococcus aureus not analyzed or presented here. Data are presented as mean (SD) unless stated.
Mean (95% CI).
Abbreviations: CFU, colony-forming unit; CRQ, Chronic Respiratory Disease Questionnaire; +PPM, potentially pathogenic microorganism-positive; qPCR, quantitative real-time polymerase chain reaction; VAS, visual analog scale.
In 65 subjects, sputum was available at every time point over 1 year (four visits). Repeatability of pathogen identification in these subjects, analyzed by the ICC (95% CI), for CFU and total 16S was 0.80 (0.68 to 0.88) and 0.61 (0.37 to 0.77), respectively. The mean within-subject log SD for CFU and 16S was 0.62 and 0.76, respectively. Repeatability was greatest for H. influenzae pathogen-specific qPCR (Table 3). Persistence of pathogen, defined as the occurrence of three or more samples positive for the same pathogen, was only observed in H. influenzae culture (n=8) and pathogen-specific H. influenzae qPCR (n=19). Persistence of H. influenzae, whether defined by culture, qPCR detection, or both, was associated with lower FEV1, increased neutrophilic airway inflammation, and worse symptoms, both at baseline and over time (Table 4A and B).
Table 3.
Frequency of sputum PPM detection in culture or qPCR, in 65 COPD subjects with repeated sampling over 12 months
| Number of times PPM detected | 0, n (%) | 1, n (%) | 2, n (%) | 3/4, n (%) | Repeatability index | Mean within-subject log SD |
|---|---|---|---|---|---|---|
| +PPM culture | 30 (46.2%) | 16 (24.6%) | 10 (15.4%) | 9 (13.8%) | 0.74 | NA |
| HI culture | 35 (54.8%) | 16 (24.6%) | 6 (9.2%) | 8 (12.3%) | 0.51 | NA |
| MC culture | 59 (90.8%) | 4 (6.2%) | 2 (3.0%) | 0 (0.0%) | 0.37 | NA |
| SP culture | 57 (87.7%) | 3 (4.6%) | 5 (7.8%) | 0 (0.0%) | 0.49 | NA |
| SA culture | 64 (98%) | 1 (1.5%) | 0 (0.0%) | 0 (0.0%) | 0.00 | NA |
| CFU | NA | NA | NA | 65 (100%) | 0.80 | 0.62 |
| Total 16S | NA | NA | NA | 65 (100%) | 0.61 | 0.76 |
| HI qPCR | 7 (11%) | 10 (15%) | 16 (25%) | 32 (49%) | 0.84 | 1.39 |
| MC qPCR | 14 (22%) | 19 (29%) | 13 (20%) | 19 (29%) | 0.75 | 1.46 |
| P qPCR | 23 (35%) | 15 (23%) | 15 (23%) | 12 (18%) | 0.62 | 1.26 |
| SA qPCR | 49 (75%) | 11 (17%) | 2 (3%) | 3 (5%) | 0.14 | 0.36 |
Notes: The qPCR detection threshold was >106 genome copies/mL of sputum. The repeatability index was a measure of the repeatability of a positive detection of pathogen.
Abbreviations: CFU, colony-forming unit; HI, Haemophilus influenzae; MC, Moraxella catarrhalis; NA, not applicable; PPM, potentially pathogenic microorganism; +PPM, PPM-positive; qPCR, quantitative real-time polymerase chain reaction; SA, Staphylococcus aureus; SD, standard deviation; SP, Streptococcus pneumoniae.
Table 4.
Clinical and inflammatory baseline characteristics in COPD subjects, with sputum sampling over 12 months
| Categorized according to qPCR threshold above 106 for HI-detection subgroups
| ||||
|---|---|---|---|---|
| HI −/−, n=25 | HI +/−, n=21 | HI +/+, n=19 | P-value | |
| Exacerbations/yr during study* | 2.6 (1.7) | 2.6 (1.5) | 2.5 (1.2) | 0.975 |
| Proportion of frequent exacerbations‡ | 36% | 38% | 53% | 0.514 |
| Sputum total cell count, ×106/g¥ | 3.1 (1.7–5.4) | 2.4 (1.4–3.9) | 6.9 (3.7–12.8) | 0.026 |
| Sputum neutrophils (%)* | 67 (21) | 64 (22) | 79 (22) | 0.070 |
| Total sputum neutrophils, ×106/g¥ | 2.1 (1.1–3.9) | 1.5 (0.8–2.7) | 5.2 (2.4–11.2) | 0.023 |
| Δ sputum neutrophils | −2.4 (−9.9 to 5.1) | −12.2 (−22.7 to −1.7) | 0.6 (−11.6 to 12.7) | 0.160 |
| Δ FEV1 (L)# | −0.06 (−1.5 to 0.04) | −0.12 (−0.21 to −0.03) | −0.03 (−0.13 to 0.08) | 0.437 |
| Δ FVC# | −0.25 (−0.45 to −0.05) | −0.40 (−0.58 to −0.23) | −0.00 (−0.25 to 0.25) | 0.029 |
| Δ VAS cough (mm) | 2 (−6 to 9) | −2 (−17 to 13) | 3 (−9 to 15) | 0.809 |
| Δ VAS dyspnea (mm) | −2 (−15 to 11) | 23 (9 to 36) | 2 (−10 to 14) | 0.016 |
| Δ VAS production (mm) | 6 (−4 to 15) | −4 (−16 to 8) | −10 (−23 to 3) | 0.122 |
| Δ VAS purulence (mm) | 12 (3 to 21) | 0 (−14 to 15) | 2 (−13 to 17) | 0.307 |
| Δ SGRQ symptoms | −5.02 (−13.15 to 3.11) | 3.15 (−7.30 to 13.61) | 6.39 (−0.90 to 13.7) | 0.134 |
| Δ SGRQ activity | 1.63 (−5.00 to 8.26) | 5.63 (−3.87 to 15.13) | 6.09 (0.18 to 12.0) | 0.599 |
| Δ SGRQ impacts | −4.13 (−9.95 to 1.68) | 2.53 (−7.04 to 12.11) | 0.90 (−3.48 to 5.28) | 0.288 |
| Δ SGRQ total | −2.60 (−7.70 to 2.49) | 5.95 (−1.00 to 12.89) | 3.22 (−1.09 to 7.54) | 0.060 |
| Categorized according to detection of any HI PPM on standard culture subgroups
| ||||
|---|---|---|---|---|
| HI PPM −/−, n=35 | HI PPM +/−, n=22 | HI PPM +/+, n=8 | P-value§ | |
| Exacerbations/yr during study* | 2.5 (1.6) | 2.6 (1.5) | 2.5 (1.1) | 0.997 |
| Proportion of frequent exacerbations‡ | 34% | 45% | 63% | 0.310 |
| Sputum total cell count, ×106/g¥ | 2.6 (1.6–4.1) | 4.4 (2.6–7.6) | 8.8 (3.2–24.1) | 0.037 |
| Sputum neutrophils (%)* | 66 (21) | 71 (25) | 81 (20) | 0.262 |
| Total sputum neutrophils, ×106/g¥ | 1.7 (1.0–2.9) | 2.9 (1.5–5.7) | 6.9 (2.1–22.7) | 0.058 |
| Δ sputum neutrophils | −2.0 (−8.4 to 4.3) | −12.3 (−22.9 to −1.73) | 8.0 (−13.9 to 30.0) | 0.055 |
| Δ FEV1 (L)# | −0.10 (−0.18 to −0.02) | −0.07 (−0.15 to 0.01) | 0.12 (−0.11 to 0.35) | 0.071 |
| Δ FVC# | −0.31 (−0.47 to −0.15) | −0.20 (−0.41 to 0.02) | 0.06 (−0.32 to 0.44) | 0.132 |
| Δ VAS cough (mm) | 2 (−7 to 11) | 4 (−8 to 15) | −9 (−26 to 9) | 0.484 |
| Δ VAS dyspnea (mm) | 6 (−5 to 17) | 15 (1 to 29) | −6 (−27 to 15) | 0.242 |
| Δ VAS production (mm) | 6 (−3 to 14) | −10 (−21 to 2) | −16 (−32 to 0) | 0.024 |
| Δ VAS purulence (mm) | 8 (−1 to 17) | 5 (−9 to 19) | −4 (−22 to 15) | 0.552 |
| Δ SGRQ symptoms | −1.26 (−7.84 to 5.31) | 1.11 (−8.33 to 10.56) | 8.15 (−7.81 to 24.11) | 0.476 |
| Δ SGRQ activity | −0.51 (−5.94 to 4.92) | 9.94 (2.25 to 17.64) | 7.15 (−1.86 to 16.16) | 0.050 |
| Δ SGRQ impacts | −3.38 (−8.48 to 1.73) | 2.13 (−5.19 to 9.46) | 1.91 (−3.60 to 7.43) | 0.337 |
| Δ SGRQ total | −1.28 (−5.50 to 2.95) | 4.21 (−2.09 to 10.51) | 4.35 (−1.29 to 10.00) | 0.213 |
Notes: N=65. Data were presented as mean difference (95% confidence interval) unless stated.
Mean (SD),
Geometric mean (95% confidence interval).
Frequent exacerbations was defined as the occurrence of three or more exacerbations during 12 months of study.
Postbronchodilator.
ANOVA or chi-square, as appropriate. Δ represents change from baseline over 12 months of study.
Abbreviations: −/−, never detected; +/−, sometimes detected; +/+, always detected +/+; ANOVA, analysis of variance; FEV1, forced expiratory volume in 1 second; FVC, forced vital capacity; HI, Haemophilus influenzae; PPM, potentially pathogenic microorganisms; qPCR, quantitative real-time polymerase chain reaction; SD, standard deviation; SGRQ, St George’s Respiratory Questionnaire; VAS, visual analog scale.
Discussion
Our findings indicate that sputum qPCR is a sensitive measure of airway bacterial presence in patients with stable COPD and identifies patients with neutrophilic inflammation. Bacteria qPCR copy numbers were repeatable within subjects and were particularly important in detecting a group of patients with persistent bacterial presence, severe neutrophilic airway inflammation, and poor clinical outcomes. qPCR has the additional advantages of providing a rapid and quantifiable result. Collectively, these properties suggest important advantages over conventional culture-based methods for the assessment of airway bacteria in patients with airway disease.
H. influenzae was the most prevalent airway pathogen detected by qPCR and the only pathogen to be identified persistently in patients with stable COPD, in keeping with earlier, culture-based studies in patients with stable COPD.1,2 Our study extends these earlier studies by showing that bacterial presence, whether assessed by qPCR, culture, or both, is associated with increased neutrophilic airway inflammation and, when persistent, with worse clinical outcomes measured by the SGRQ, with clinical but not statistically significant deterioration. Furthermore, our study is consistent with a more recent study investigating qPCR pathogen detection in patients with COPD,18 although we extended this finding to assess airway inflammation and outcomes longitudinally. A noninterventional study such as this cannot establish a causal link between the presence of bacteria and increased neutrophilic airway, but such a relationship is likely, as short-term antibiotic treatment reduces both bacterial load and the sputum neutrophil count in patients with stable COPD.9,19 We cannot exclude the possibility that the relationships seen are a function of the presence of H. influenzae alone. The dose-response relationship between H. influenzae and the sputum neutrophil count identified by qPCR, and the demonstration that persistent presence of H. influenzae is associated with particularly high sputum neutrophil counts, would be consistent with this. A causal link with this pathogen and neutrophilic airway inflammation is also biologically plausible since patients with COPD have been shown to have impaired innate immune response and a reduced phagocytic response to H. influenza.20 Moreover, nontypeable H. influenzae, in contrast to other respiratory PPMs, has been found to have the ability to invade lung parenchyma and can be identified intracellularly.21,22 Once established in the airway, H. influenzae might directly drive neutrophilic airway inflammation since it increases airway mucosal TNF production and has a marked cytotoxic T cell response;23 we recently observed that H. influenzae presence is an independent predictor of airway production of TNF and IL1.24
The most noteworthy and important finding of our study is that a significant number of sputum samples were culture-negative but contained high copy numbers for a PPM, using qPCR. This pattern of pathogen-specific PPM isolation was particularly seen with H. influenzae; it was associated with increased neutrophilic airway inflammation and, when present repeatedly, with worse lung function and quality of life. One potential explanation is that nonviable pathogens contribute to neutrophilic airway inflammation, as has been shown for H. Influenzae in animal models.25 Alternatively, pathogens might exist in a quiescent state, contributing to airway inflammation and only detectable using molecular methods. An earlier longitudinal study repeatedly assessing airway bacteria by culture methods strongly supports such behavior for H. influenza.26 Gaps in H. influenzae detection by sputum culture over 1 to 6 months were invariably associated with molecular evidence of persistence of the same strain of H. influenza.26 Long-term treatment with prophylactic antibiotics, in particular low-dose macrolides, is associated with a reduction in symptoms and exacerbation frequency in subjects with COPD,27,28 an effect specific to subjects with chronic bronchitis and purulent sputum.29 Although it remains to be established whether this effect is mediated by a reduction in airway bacteria, repeated small, randomized, placebo-controlled trials have shown parallel reductions in bacterial numbers and neutrophilic airway inflammation during antibiotic treatment.19,28 Future stud ies should investigate whether the response to long-term low-dose macrolides is associated with traditional and molecular measures of airway bacteria. Targeting therapy to a particularly responsive population would be an advance, as long-term macrolide therapy has been associated with hearing decrements and an increased rate of macrolide-resistant infections, including nontuberculous mycobacteria infection.27
A limitation of our study includes our failure to use molecular techniques to identify P. aeruginosa, or to investigate variation between the sampling position and site and the absence of information on change in bacterial strain. Further studies to evaluate these potentially important aspects are thus required. We have also been unable to assess the functional properties of neutrophils in these subjects, to determine whether there is differential expression, migration, or function. However, recent experimental assays have revealed that neutrophil phagocytosis of bacteria is similar between healthy controls and subjects with COPD,30 although this has yet to be studied in airway neutrophils. A final limitation is that computed tomography was not undertaken as part of the study protocol, so we are not in a position to determine whether the presence of bronchiectasis was an important cofactor in these subjects. We have previously shown that in a subgroup of this cohort, emphysema is the most common radiological finding, and airway inflammation is similar in patients with emphysema or bronchiectasis,31 suggesting that the presence of bronchiectasis alone is unlikely to be an important factor. We were unable to show any differences in exacerbation frequency in the population with higher bacterial burden as measured by qPCR or culture. Potentially, this may reflect selection bias, as subjects were selected on the basis of a prior history of exacerbations. Exacerbations are recognized to be heterogeneous in mechanism and pathogenesis,32 and another possibility is that bacterial presence is associated with an increased rate of a particular type of event.
To conclude, we have shown that a significant number of patients with stable COPD have evidence of airway PPMs, that qPCR is a more sensitive measure of this than is traditional culture, and that patients with a bacteria-associated disease have increased neutrophilic airway inflammation. The presence of a positive correlation between qPCR copy numbers and the sputum neutrophil count, and the finding that patients with persistently positive sputum samples have particularly severe neutrophilic airway inflammation and poor clinical outcomes, would be consistent with a causal link. H. influenzae was the most commonly isolated pathogen, the only one to be identified repeatedly, and was most closely associated with neutrophilic airway inflammation and poor clinical outcomes, suggesting a more important role for this pathogen in stable COPD. Further work is warranted to investigate whether subjects with this pattern of disease benefit most from prophylactic antimicrobial therapy and to further define the role of H. influenzae in the pathogenesis of COPD.
Acknowledgments
The study was funded by the Medical Research Council (UK) and the Wellcome Trust (CEB), and the research was performed in laboratories partly funded by the European Regional Development Fund (grant number ERDF 05567). This article presents independent research funded by the National Institute for Health Research (NIHR). The views expressed are those of the authors and not necessarily those of the National Health Service (NHS), the NIHR, or the Department of Health. The Medical Research Council, Wellcome Trust, and the NIHR had no involvement in the design of the study, data collection, analyses, data interpretation, writing of the manuscript, or the decision to submit the manuscript.
The authors would like to thank all the volunteers that took part in the study and Mrs B Hargadon, Mrs S McKenna, Mrs C Whitehead, Miss A Singapuri, and Mrs S Terry for their assistance with volunteer characterization.
Footnotes
Disclosure
MB has received conference travel support from Boehringer Ingelheim, GlaxoSmithKline, and Almirall, and consultancy fees from AstraZeneca, Almirall, Chiesi, and GlaxoSmithKline. IDP has received consultancy and honoraria fees from GlaxoSmithKline, AstraZeneca, Novartis, Boehringher Ingelheim, and Aerocrine, and received travel support from GlaxoSmithKline and Boerhingher Ingelheim. CEB has received grant support and consultancy fees from AstraZeneca, MedImmune, Novartis, Roche/Genentech, Chiesi, Boehringher Ingelheim, and GlaxoSmithKline, and biomarker assays provided by Genentech.
The authors report no other conflicts of interest in this work.
Author contributions
HP, BB, VM, and KH were involved in data collection and data interpretation. IDP and MRB were involved in study design, data collection, and interpretation. MB and CEB were involved in the design of the study, volunteer recruitment, data collection, interpretation, and analysis, and were responsible for the integrity of the data and the final decision to submit. All authors contributed to the writing of the manuscript and approved the final version for submission.
References
- 1.Monsó E, Rosell A, Bonet G, et al. Risk factors for lower airway bacterial colonization in chronic bronchitis. Eur Respir J. 1999;13(2):338–342. doi: 10.1034/j.1399-3003.1999.13b20.x. [DOI] [PubMed] [Google Scholar]
- 2.Cabello H, Torres A, Celis R, et al. Bacterial colonization of distal airways in healthy subjects and chronic lung disease: a bronchoscopic study. Eur Respir J. 1997;10(5):1137–1144. doi: 10.1183/09031936.97.10051137. [DOI] [PubMed] [Google Scholar]
- 3.Murphy TF, Sethi S, Klingman KL, Brueggemann AB, Doern GV. Simultaneous respiratory tract colonization by multiple strains of nontypeable haemophilus influenzae in chronic obstructive pulmonary disease: implications for antibiotic therapy. J Infect Dis. 1999;180(2):404–409. doi: 10.1086/314870. [DOI] [PubMed] [Google Scholar]
- 4.Hill AT, Campbell EJ, Hill SL, Bayley DL, Stockley RA. Association between airway bacterial load and markers of airway inflammation in patients with stable chronic bronchitis. Am J Med. 2000;109(4):288–295. doi: 10.1016/s0002-9343(00)00507-6. [DOI] [PubMed] [Google Scholar]
- 5.Sethi S, Maloney J, Grove L, Wrona C, Berenson CS. Airway inflammation and bronchial bacterial colonization in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;173(9):991–998. doi: 10.1164/rccm.200509-1525OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerjee D, Khair OA, Honeybourne D. Impact of sputum bacteria on airway inflammation and health status in clinical stable COPD. Eur Respir J. 2004;23(5):685–691. doi: 10.1183/09031936.04.00056804. [DOI] [PubMed] [Google Scholar]
- 7.Patel IS, Seemungal TA, Wilks M, Lloyd-Owen SJ, Donaldson GC, Wedzicha JA. Relationship between bacterial colonisation and the frequency, character, and severity of COPD exacerbations. Thorax. 2002;57(9):759–764. doi: 10.1136/thorax.57.9.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bafadhel M, McKenna S, Terry S, et al. Acute exacerbations of chronic obstructive pulmonary disease: identification of biologic clusters and their biomarkers. Am J Respir Crit Care Med. 2011;184(6):662–671. doi: 10.1164/rccm.201104-0597OC. [DOI] [PubMed] [Google Scholar]
- 9.Siva R, Bafadhel M, Monteiro W, Brightling CE, Pavord ID. Effect of levofloxacin on neutrophilic airway inflammation in stable COPD: a randomized, double-blind, placebo-controlled trial. Int J Chron Obstruct Pulmon Dis. 2014;9:179–186. doi: 10.2147/COPD.S55419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bafadhel M, McKenna S, Terry S, et al. Blood eosinophils to direct corticosteroid treatment of exacerbations of chronic obstructive pulmonary disease: a randomized placebo-controlled trial. Am J Respir Crit Care Med. 2012;186(1):48–55. doi: 10.1164/rccm.201108-1553OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller MR, Hankinson J, Brusasco V, et al. ATS/ERS Task Force Standardisation of spirometry. Eur Respir J. 2005;26(2):319–338. doi: 10.1183/09031936.05.00034805. [DOI] [PubMed] [Google Scholar]
- 12.Brightling CE, Monterio W, Green RH, et al. Induced sputum and other outcome measures in chronic obstructive pulmonary disease: safety and repeatability. Respir Med. 2001;95(12):999–1002. doi: 10.1053/rmed.2001.1195. [DOI] [PubMed] [Google Scholar]
- 13.Guyatt G. Measuring health status in chronic airflow limitation. Eur Respir J. 1988;1(6):560–564. [PubMed] [Google Scholar]
- 14.Jones PW, Quirk FH, Baveystock CM. The St George’s Respiratory Questionnaire. Respir Med. 1991;85(Suppl B):S25–S31. doi: 10.1016/s0954-6111(06)80166-6. discussion S33–S37. [DOI] [PubMed] [Google Scholar]
- 15.Health Protection Agency . UK Standards for Microbiology Investigations. Investigation of bronchoalveolar lavage, sputum and associated specimens. London: Health Protection Agency; 2009. [Accessed March 9, 2015]. (Bacteriology; B 57). Available from: http//www.hpa.org.uk/webc/HPAwebFile/HPAweb_C/1317132860548. [Google Scholar]
- 16.Pye A, Stockley RA, Hill SL. Simple method for quantifying viable bacterial numbers in sputum. J Clin Pathol. 1995;48(8):719–724. doi: 10.1136/jcp.48.8.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belda J, Leigh R, Parameswaran K, O’Byrne PM, Sears MR, Hargreave FE. Induced sputum cell counts in healthy adults. Am J Respir Crit Care Med. 2000;161(2 Pt 1):475–478. doi: 10.1164/ajrccm.161.2.9903097. [DOI] [PubMed] [Google Scholar]
- 18.Garcha DS, Thurston SJ, Patel AR, et al. Changes in prevalence and load of airway bacteria using quantitative PCR in stable and exacerbated COPD. Thorax. 2012;67(12):1075–1080. doi: 10.1136/thoraxjnl-2012-201924. [DOI] [PubMed] [Google Scholar]
- 19.Banerjee D, Honeybourne D, Khair OA. The effect of oral clarithromycin on bronchial airway inflammation in moderate-to-severe stable COPD: a randomized controlled trial. Treat Respir Med. 2004;3(1):59–65. doi: 10.2165/00151829-200403010-00007. [DOI] [PubMed] [Google Scholar]
- 20.Taylor AE, Finney-Hayward TK, Quint JK, et al. Defective macrophage phagocytosis of bacteria in COPD. Eur Respir J. 2010;35(5):1039–1047. doi: 10.1183/09031936.00036709. [DOI] [PubMed] [Google Scholar]
- 21.Möller LV, Timens W, van der Bij W, et al. Haemophilus influenzae in lung explants of patients with end-stage pulmonary disease. Am J Respir Crit Care Med. 1998;157(3 Pt 1):950–956. doi: 10.1164/ajrccm.157.3.9707010. [DOI] [PubMed] [Google Scholar]
- 22.Drömann D, Rupp J, Rohmann K, et al. The TGF-beta-pseudoreceptor BAMBI is strongly expressed in COPD lungs and regulated by nontypeable Haemophilus influenzae. Respir Res. 2010;11:67. doi: 10.1186/1465-9921-11-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King PT, Lim S, Pick A, et al. Lung T-cell responses to nontypeable Haemophilus influenzae in patients with chronic obstructive pulmonary disease. J Allergy Clin Immunol. 2013;131(5):1314–1321. e14. doi: 10.1016/j.jaci.2012.09.030. [DOI] [PubMed] [Google Scholar]
- 24.Barker BL, Haldar K, Patel H, et al. Association between pathogens detected using quantitative polymerase chain reaction with airway inflammation in COPD at stable state and exacerbations. Chest. 2015;147(1):46–55. doi: 10.1378/chest.14-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Essilfie AT, Simpson JL, Dunkley ML, et al. Combined Haemophilus influenzae respiratory infection and allergic airways disease drives chronic infection and features of neutrophilic asthma. Thorax. 2012;67(7):588–599. doi: 10.1136/thoraxjnl-2011-200160. [DOI] [PubMed] [Google Scholar]
- 26.Murphy TF, Brauer AL, Schiffmacher AT, Sethi S. Persistent colonization by Haemophilus influenzae in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2004;170(3):266–272. doi: 10.1164/rccm.200403-354OC. [DOI] [PubMed] [Google Scholar]
- 27.Albert RK, Connett J, Bailey WC, et al. COPD Clinical Research Network Azithromycin for prevention of exacerbations of COPD. N Engl J Med. 2011;365(8):689–698. doi: 10.1056/NEJMoa1104623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He ZY, Ou LM, Zhang JQ, et al. Effect of 6 months of erythromycin treatment on inflammatory cells in induced sputum and exacerbations in chronic obstructive pulmonary disease. Respiration. 2010;80(6):445–452. doi: 10.1159/000321374. [DOI] [PubMed] [Google Scholar]
- 29.Sethi S. Moxifloxacin for the treatment of acute exacerbations of chronic obstructive pulmonary disease. Clin Infect Dis. 2005;41(Suppl 2):S177–S185. doi: 10.1086/428058. [DOI] [PubMed] [Google Scholar]
- 30.Walton GM, Purvis T, Chadwick C, Stockley RA, Sapey E. S46 Phagocytosis by blood neutrophils is not attenuated in patients with chronic obstructive pulmonary disease. Thorax. 2014;69(Suppl 2):A26–A27. [Google Scholar]
- 31.Bafadhel M, Umar I, Gupta S, et al. The role of CT scanning in multidimensional phenotyping of COPD. Chest. 2011;140(3):634–642. doi: 10.1378/chest.10-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han MK, Agusti A, Calverley PM, et al. Chronic obstructive pulmonary disease phenotypes: the future of COPD. Am J Respir Crit Care Med. 2010;182(5):598–604. doi: 10.1164/rccm.200912-1843CC. [DOI] [PMC free article] [PubMed] [Google Scholar]