Abstract
IMPORTANCE
The core clinical and neuropathological feature of the autosomal dominant spinocerebellar ataxias (SCAs) is cerebellar degeneration. Mutations in the known genes explain only 50% to 60% of SCA cases. To date, no effective treatments exist, and the knowledge of drug-treatable molecular pathways is limited. The examination of overlapping mechanisms and the interpretation of how ataxia genes interact will be important in the discovery of potential disease-modifying agents.
OBJECTIVES
To address the possible relationships among known SCA genes, predict their functions, identify overlapping pathways, and provide a framework for candidate gene discovery using whole-transcriptome expression data.
DESIGN, SETTING, AND PARTICIPANTS
We have used a systems biology approach based on whole-transcriptome gene expression analysis. As part of the United Kingdom Brain Expression Consortium, we analyzed the expression profile of 788 brain samples obtained from 101 neuropathologically healthy individuals (10 distinct brain regions each). Weighted gene coexpression network analysis was used to cluster 24 SCA genes into gene coexpression modules in an unsupervised manner. The overrepresentation of SCA transcripts in modules identified in the cerebellum was assessed. Enrichment analysis was performed to infer the functions and molecular pathways of genes in biologically relevant modules.
MAIN OUTCOMES AND MEASURES
Molecular functions and mechanisms implicating SCA genes, as well as lists of relevant coexpressed genes as potential candidates for novel SCA causative or modifier genes.
RESULTS
Two cerebellar gene coexpression modules were statistically enriched in SCA transcripts (P = .021 for the tan module and P = 2.87 × 10−5 for the light yellow module) and contained established granule and Purkinje cell markers, respectively. One module includes genes involved in the ubiquitin-proteasome system and contains SCA genes usually associated with a complex phenotype, while the other module encloses many genes important for calcium homeostasis and signaling and contains SCA genes associated mostly with pure ataxia.
CONCLUSIONS AND RELEVANCE
Using normal gene expression in the human brain, we identified significant cell types and pathways in SCA pathogenesis. The overrepresentation of genes involved in calcium homeostasis and signaling may indicate an important target for therapy in the future. Furthermore, the gene networks provide new candidate genes for ataxias or novel genes that may be critical for cerebellar function.
Autosomal dominant cerebellar ataxias, also referred to as spinocerebellar ataxias (SCAs), are clinically heterogeneous, with an onset usually in adulthood. The SCAs are characterized by progressive cerebellar dysfunction, manifesting as progressive gait and limb incoordination, and can be associated with a wide range of neurological and nonneurological manifestations, including peripheral neuropathy, ophthalmoplegia, retinopathy, pyramidal and extrapyramidal signs, dementia, and epilepsy.1,2
Approximately 35 SCA loci have been mapped (Table), but only 24 causative genes have been identified to date. The SCAs can be classified into the following 3 major categories according to the mutation type: (1) polyglutamine ataxias caused by exonic (CAG)n expansions encoding for polyglutamine tracts, (2) noncoding repeat ataxias caused by expansions of repeat motifs located in noncoding regions, and (3) ataxias caused by conventional mutations and copy number variants, including missense, nonsense, and splice-site mutations, deletions, and duplications.3 However, 40% to 50% of patients with SCAs do not have mutations in the known genes.4
Table. Spinocerebellar Ataxia Loci, Genes, and Mutations.
| Ataxia | Locus | Gene | Type of Mutation | Protein or Complex | OMIM Accession No. |
|---|---|---|---|---|---|
| Coding Repeat Expansions a | |||||
| DRPLA | 12p13.31 | ATN1 | (CAG)n | Atrophin 1 | 125370 |
| SCA1 | 6p23 | ATXN1 | (CAG)n | Ataxin 1 | 164400 |
| SCA2 | 12q24 | ATXN2 | (CAG)n | Ataxin 2 | 183090 |
| SCA3/MJD | 14q32.1 | ATXN3 | (CAG)n | Ataxin 3 | 109150 |
| SCA6 | 19p13 | CACNA1A | (CAG)n | Calcium channel, voltage-dependent, P/Q-type, α-1A subunit | 183086 |
| SCA7 | 3p14 | ATXN7 | (CAG)n | Ataxin 7 | 164500 |
| SCA17 | 6q27 | TBP | (CAG)n | TATA box binding protein | 607136 |
|
| |||||
| Noncoding Repeat Expansions b | |||||
| SCA8 | 13q21.33 |
ATXN8,
ATXN8OS |
(CTG*CAG)n | Ataxin 8 and ATXN8 opposite strand (nonprotein coding) | 608768 |
| SCA10 | 22q13.31 | ATXN10 | (ATTCT)n | Ataxin 10 | 603516 |
| SCA12 | 5q32 | PPP2R2B | (CAG)n | Protein phosphatase 2, regulatory subunit B, β | 604326 |
| SCA31 | 16q21 | BEAN1 | (TGGAA)n | Brain expressed, associated with NEDD4, 1 | 117210 |
| SCA36 | 20p13 | NOP56 | (GGCCTG)n | NOP56 ribonucleoprotein homologue (yeast) | 614153 |
|
| |||||
| Other Types of Mutations c | |||||
| SCA5 | 11q13.2 | SPTBN2 | Point mutations | Spectrin, β, nonerythrocytic 2 | 600224 |
| SCA11 | 15q15.2 | TTBK2 | Point mutations | Tau tubulin kinase 2 | 604432 |
| SCA13 | 19q13.33 | KCNC3 | Point mutations | Potassium voltage-gated channel, Shaw-related subfamily, member 3 | 605259 |
| SCA14 | 19q13.42 | PRKCG | Point mutations | Protein kinase C, λ | 605361 |
| SCA15/16/29 | 3p26.1 | ITPR1 | Point mutations, large deletions | Inositol 1,4,5-trisphosphate receptor, type 1 | 606658, 117360 |
| SCA18 | 7q31.1 | IFRD1 | Point mutations | Interferon-related developmental regulator 1 | 607458 |
| SCA19/22 | 1p13.2 | KCND3 | Point mutations, small deletions | Potassium voltage-gated channel, Shal-related subfamily, member 3 | 607346 |
| SCA20 | 11p11.2-q13.3 | Not applicable | Genomic duplication | Region with ≥12 genes | 608687 |
| SCA23 | 20p13 | PDYN | Point mutations | Prodynorphin | 610245 |
| SCA26 | 19p13.3 | EEF2 | Point mutations | Eukaryotic translation elongation factor 2 | 609306 |
| SCA27 | 13q34 | FGF14 | Point mutations | Fibroblast growth factor 14 | 609307 |
| SCA28 | 18p11.21 | AFG3L2 | Point mutations | AFG3 adenosine triphosphatase family gene 3–like 2 (Saccharomyces cerevisiae) | 610246 |
| SCA35 | 20p13 | TGM6 | Point mutations | Transglutaminase 6 | 613908 |
|
| |||||
| Unknown Mutations | |||||
| SCA4 | 16q22.1 | ? | ? | ? | 600223 |
| SCA21 | 7p21.3-p15.1 | ? | ? | ? | 607454 |
| SCA25 | 2p21-p13 | ? | ? | ? | 608703 |
| SCA30 | 4q34.3-q35.1 | ? | ? | ? | 613371 |
| SCA32 | 7q32-q33 | ? | ? | ? | 613909 |
| SCA34 | 6p12.3-q16.1 | ? | ? | ? | 133190 |
| SCA37 | 1p32 | ? | ? | ? | HGNC 43726 |
Abbreviations: DRPLA, dentatorubral-pallidoluysian atrophy; HGNC, HUGO Gene Nomenclature Committee; MJD, Machado-Joseph disease; OMIM, Online Mendelian Inheritance in Man; SCA, spinocerebellar ataxia.
Polyglutamine ataxias.
Noncoding repeat ataxias.
Ataxias caused by conventional mutations.
The paradigm to investigate rare mendelian disorders is evolving as a result of the availability of next-generation sequencing, which has facilitated the identification of causal genes. Several SCA genes were recently identified using next-generation sequencing (eg, SCA19/22 and SCA35).5-7 The interpretation of next-generation sequencing data represents a major ongoing challenge, particularly in distinguishing causal mutations from the thousands of benign variants present in every exome (often >20 000 variants). Therefore, it is important to develop frameworks to prioritize candidate mutations of monogenic diseases in these large data sets.
One way forward is to use the expression and function of known SCA genes to predict novel genes. Although neuronal loss can be widespread in some SCAs (eg, SCA3), certain brain regions (eg, cortical regions) are usually less affected (reviewed by Seidel et al8). The pathological features of SCAs are most prominent in the cerebellum, which is characteristically atrophic despite the fact that SCA genes are ubiquitously expressed in the brain. This would suggest that, in common with many other neurodegenerative disorders, regional differences in gene expression within the human central nervous system are insufficient to explain the observed selective vulnerability of neurons. Furthermore, SCA genes have a wide range of functions, including ion transport, deubiquitination, dephosphorylation, phosphorylation, transcriptional regulation, translational elongation, and others, rendering it surprising that mutations in this diverse set of genes can give rise to a consistent phenotype. A study by Lim et al9 revealed that some ataxia-associated proteins share interacting partners, suggesting that phenotypes shared among ataxias may arise from their involvement in common molecular pathways. We hypothesized that expression patterns of 24 known SCA-associated genes in the human brain and relevant molecular pathways may be used to prioritize novel candidate genes and identify disease modifiers. In addition, this strategy may be used to identify potential therapeutic targets, as was found for the α-subunit of eukaryotic translation initiation factor (eIF2) in prion neurodegeneration.10
To date, limited information exists regarding the expression and function of SCA genes within the human brain. We performed expression profiling of 788 brain samples obtained from 101 neuropathologically healthy individuals as part of the United Kingdom Brain Expression Consortium and used weighted gene coexpression network analysis (WGCNA) to group genes into modules in an unsupervised manner.11-14 This approach has proven useful in identifying modules of biologically related genes that are not only coexpressed but also coregulated.13,15-17 We used a systems biology approach18,19 based on whole-transcriptome gene expression analysis to address possible relationships among known SCA genes, predict their functions, and propose novel candidate genes to aid the discovery of new SCA genes. Our data revealed 2 significant SCA transcript–enriched coexpression modules that provide testable hypotheses about the function of proteins that may interact with known SCA genes.
Methods
Human Brain Samples and Analysis of Gene Expression Arrays
All 101 samples had received fully informed consent and were authorized for ethically approved scientific investigation by the National Hospital for Neurology and Neurosurgery and Institute of Neurology Research Ethics Committee. Brain samples were collected by the Medical Research Council Sudden Death Brain and Tissue Bank.20 The 101 individuals had been neuropathologically healthy. Total RNA was isolated and processed for array analysis (Exon 1.0 ST; Affymetrix UK Ltd) as previously described (National Center for Biotechnology Gene Expression Omnibus GSE46706).21,22 Samples were randomized for all experimental procedures, from RNA extraction to array hybridization, and array data were thoroughly investigated for technical quality as detailed in the eMethods on the authors’ website (http://www.ucl.ac.uk/ukpdc/publications/data/Bettencourt_et_al_2014_JAMA_Neurol_Supplement.docx).
Gene expression patterns throughout development and aging were assessed using data from the Human Brain Transcriptome project (http://hbatlas.org).23,24 The Biological General Repository for Interaction Datasets, version 3.2.106 (http://thebiogrid.org),25 was used for the analysis of SCA interactors.
Weighted Gene Coexpression Network Analysis
The SCA genes and transcripts were assigned to modules (arbitrary colors) identified through WGCNA on whole-transcriptome gene expression. In total, 15 409 transcripts (13 706 genes) passing quality control were used to identify modules as previously described,22 and 3743 additional transcripts (3541 genes) were assigned to modules based on their highest module membership (MM).
Briefly, the WGCNA network11,14 was constructed for each tissue using a signed network with β power of 12 to achieve a scale-free topology. A dissimilarity matrix based on topological overlap measure, a pairwise measure of node similarity, was used to identify gene modules (ie, densely interconnected and coexpressed genes) through a dynamic tree-cutting algorithm.
Module preservation statistics26 were calculated (z score) to assess how well modules from one tissue are reproducible (or preserved) in another tissue. Previously proposed thresholds26 were considered (z score of <2 indicates no evidence of module preservation, z score between 2 and <10 indicates weak to moderate evidence, and z score of ≥10 indicates strong evidence).
The hypergeometric distribution was used to evaluate SCA transcript–enrichedmodules(P < .05 was considered significant). Only modules with at least 2 SCA transcripts were considered.
Gene Enrichment Analysis
DAVID Bioinformatics Resources, version 6.7 (http://david.abcc.ncifcrf.gov/home.jsp),27,28 was used to evaluate the biological and functional relevance among SCA genes and genes within SCA transcript–enriched modules. The overrepresentation of gene ontology (GO) categories and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways was examined (Bonferroni-corrected P < .05 was considered significant). oPOSSUM, version 3.0 (http://opossum.cisreg.ca/oPOSSUM3),29-31 was used for enrichment analysis on conserved transcription factor binding sites (TFBSs) in SCA genes (cutoffs were a z score of ≥10 and a Fisher exact test score of ≥7).
An overview of the coexpression analysis work flow and associated data sets is shown in eFigure 1 on the authors’ website. Further details are given in the eMethods.
Results
Forty Percent of SCA Genes Are Most Highly Expressed in the Cerebellum
Globally, SCA genes are highly expressed in the human brain throughout development and aging. We found that approximately 40% of SCA genes (SPTBN2, CACNA1A, ATXN7, KCNC3, ITPR1, TBP, KCND3, FGF14, NOP56, and ATN1) have the highest expression in the cerebellum, but 2 genes (PPP2R2B and PDYN) exhibit among the lowest levels in this region (eTable 1 on the authors’ website). Several genes have highly variable expression levels across brain regions, including all SCA genes with ion channel activity (CACNA1A, KCNC3, ITPR1, and KCND3) and modulators of ion channel activity (SPTBN2 and FGF14),32,33 which exhibit the highest expression levels in the cerebellum, followed by cortical regions, and the lowest expression in the white matter, suggesting that expression patterns may relate to gene function.
NR4A2 TFBSs Are Enriched Around SCA Genes
We had hypothesized that a feature common to SCA genes might be the coregulation of gene expression. Analysis of conserved TFBSs showed shared TFBSs across many SCA genes (up to 21 of 23 genes), suggesting that common transcription factors may be involved in the coregulation of these genes. We found that only TFBSs for NR4A2 are significantly overrepresented nearby SCA genes (z score, 10.4; P = .0003), with a mean of 7 TFBSs per gene (21 of 23 genes, absent only around AFG3L2 and BEAN1).
WGCNA Identifies 2 SCA Transcript–Enriched Modules in the Cerebellum
To gain insights into the functional organization of the brain transcriptome, we used WGCNA22 focusing on SCA genes (Table). Except for TGM6 (SCA35), which was not robustly detected, all other SCA genes were assigned to gene coexpression modules in 10 distinct brain regions (eTable 2 on the authors’ website). We were particularly interested in cerebellar gene MM because this region is most affected in these conditions.
Twenty-three gene coexpression modules were identified in the cerebellum, 14 of which contain at least 1 SCA transcript. Two of these modules include 4 SCA genes (eTable 2 on the authors’ website), namely, the tan module (ATXN3, ATXN10, TTBK2, and AFG3L2) and the light yellow module (ATXN8OS, PRKCG, ITPR1, and KCND3). This clustering was statistically significant (P = .021 for the tan module and P = 2.87 × 10−5 for the light yellow module).
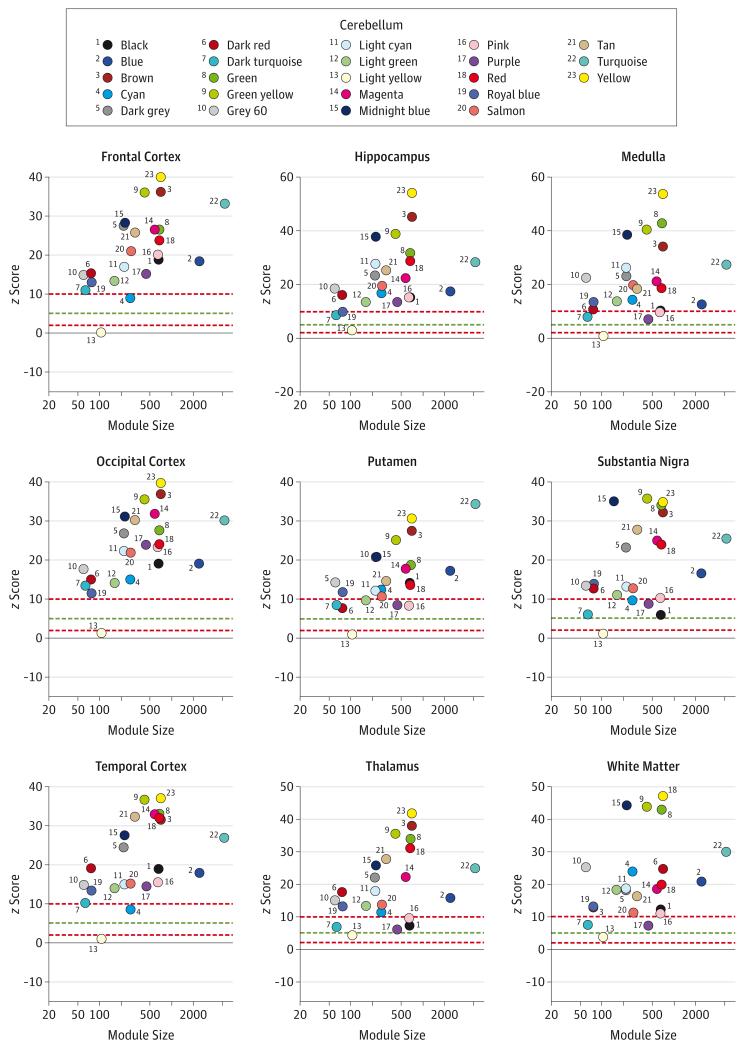
Whereas the tan module seems to be preserved across all brain regions (mean z score, 23.19), the light yellow module is poorly preserved and seems to be unique to the cerebellum (mean z score, 2.26) (Figure 1). Consistent with these findings, we note that SCA genes contained within the light yellow module give rise to more pure ataxia phenotypes despite being ubiquitously expressed in the human brain, whereas those in the tan module give rise to more complex phenotypes.
Figure 1. z Scores Indicating Whether Genes in the Coexpression Modules Identified in the Cerebellum Cluster in Other Brain Regions.
z Scores of less than 2 (bottom red line) indicate no evidence of preservation (the case for the cerebellar light yellow module in most brain regions), while scores exceeding 5 (green line) and exceeding 10 (upper red line) indicate moderate and strong module preservation across brain regions, respectively.
SCA Genes Have High MM Within SCA Transcript–Enriched Modules
To determine the relevance of each gene in the SCA transcript–enriched modules, we estimated the MM for all genes within these modules (eTable 3 and eTable 4 on the authors’ website). The MM measures how well the expression pattern of each gene within the module correlates with the eigengene (first principal component of gene expression for the module). Except for KCND3 (17th quantile), all SCA genes have an MM above the median in the tan module (TTBK2 [78th quantile], AFG3L2 [75th quantile], ATXN10 [61st quantile], and ATXN3 [61st quantile]) and in the light yellow module (ITPR1 [96th quantile], PRKCG [83rd quantile], and ATXN8OS [79th quantile]), with the ITPR1 gene being a hub gene. These data suggest that genes with high MM may constitute good candidate genes for ataxias of yet unknown cause. For example, these data indicate that 2 members of the light yellow module, C7orf16 (7p15 [94th quantile]) and GPR63 (6q16.1-q16.3 [78th quantile]), which we note are located within the SCA21 and SCA34 loci, respectively, are particularly promising candidate genes.
SCA Transcript–Enriched Modules Contain Additional Ataxia-Related Genes
The tan module contains genes involved in other autosomal dominant and recessive ataxia syndromes, including PRNP (prion disease gene, OMIM 176640), SACS (autosomal recessive spastic ataxia of Charlevoix-Saguenay, OMIM 604490), AFG3L2 (both SCA28 and autosomal recessive spastic ataxianeuropathy syndrome,34 OMIM 604581), and MTPAP (autosomal recessive spastic ataxia 4, OMIM 613669). Also present in the tan module are genes associated with other neurodegenerative disorders (eg, APP [OMIM 104760], VAPB [OMIM 605704], and VCP [OMIM 601023]), some of which exhibit cerebellar degeneration as a pathological feature (eg, POLR3A [OMIM 614258], POLR3B [OMIM 614366], EIF2B1 [OMIM 606686], and EIF2B2 [OMIM 606454]). The light yellow module also includes genes associated with additional ataxia syndromes (eg, CA8 [OMIM 114815] and TRPC3 [OMIM 602345]) and with other neurological diseases (eg, LARGE [OMIM 603590] and KCNMA1 [OMIM 600150]).
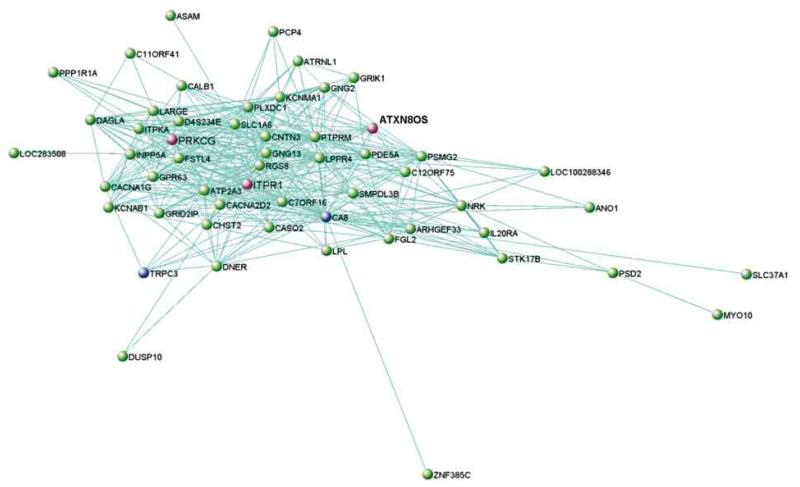
We investigated gene interconnections within SCA transcript–enriched modules using VisANT (http://visant.bu.edu).35 Genes with topological overlap measure values exceeding 0.01 were considered connected. For the tan module, no SCA genes passed this cutoff. For the light yellow module, among the genes in the network (Figure 2) are CA8 (gene with the top MM) and TRPC3 (57th quantile), both previously known to interact with ITPR1 and associated with ataxia syndromes.36,37
Figure 2. Network Representation of the Light Yellow Module in the Cerebellum.
Shown are all genes connected with a topological overlap measure exceeding 0.01. The spinocerebellar ataxia genes in this module are highlighted in pink, together with all genes that are directly connected to it based on the topological overlap measure cutoff used. Genes previously reported as interactors of spinocerebellar ataxia genes are highlighted in blue.
SCA Transcript–Enriched Modules Contain Established Granule and Purkinje Cell Markers
We used previously published cell-specific expression signatures38 to obtain insights into the cell types relating to SCA transcript–enriched modules in the human cerebellum. These signatures indicated that only genes specifically associated with granule cells (C12orf24 [91st quantile], THYN1 [86th quantile], ZDHHC13 [85th quantile], BLOC1S2 [66th quantile], PARP2 [57th quantile], and C14orf101 [53rd quantile]) show an MM above the median in the tan module. On the other hand, only genes specifically associated with Purkinje cells show high MM in the light yellow module (SMPDL3B [89th quantile], LARGE [87th quantile], and PDE5A [73rd quantile]), and the enrichment in these cell markers was statistically significant (P = 3.752 × 10−4). No astrocyte-associated or oligodendrocyte-associated genes were present in the 2 SCA transcript–enriched modules. Therefore, while the tan module is more closely related to gene expression signatures of granule cells, the light yellow module is associated with Purkinje cells (eFigure 2 on the authors’ website) and may contribute to the specificity of the neuropathology.
SCA Interactome Network vs SCA Transcript–Enriched Expression Modules Were Searched
We next searched whether genes in these SCA transcript–enriched modules have already been deposited in the Biological General Repository for Interaction Datasets25 as interactors of SCA genes. Several genes in the modules are previously known SCA interactors (eTable 5 on the authors’ website), some of which are also associated with neurodegenerative diseases, including ataxias (eg, APP, VCP, CA8, and TRPC3).
Some of these interactors (eg, BECN1 and PICK1)39,40 are established modulators of SCA-related phenotypes, which suggests that the SCA interactome network may aid the identification of genetic modifiers and candidate genes for SCAs.
SCA Transcript–Enriched Modules Implicate the Ubiquitin-Proteasome System and Calcium Signaling in Disease Pathogenesis
Enrichment analysis for the tan module shows an overrepresentation of multiple GO biological processes mostly related to protein catabolic processes, the regulation of ubiquitin protein ligase activity, and intracellular transport (eTable 6 on the authors’ website). Two KEGG pathways, proteasome and ubiquitin-mediated proteolysis, are overrepresented. The latter pathway is also enriched in previously reported SCA interactors (eTable 5 on the authors’ website). To date, no SCA genes have been assigned to these pathways. However, ATXN3 is known to act as a deubiquitinating enzyme in the ubiquitinproteasome pathway,41 and other ataxia genes are involved in this pathway (eFigure 3 on the authors’ website).
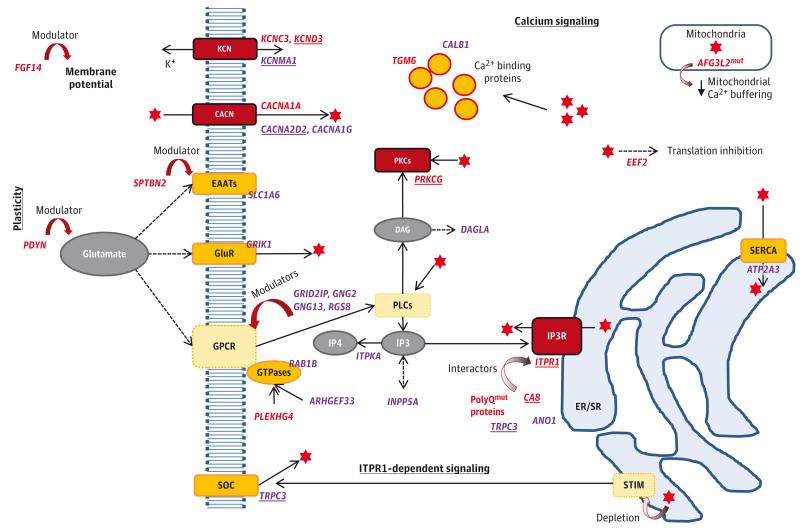
In the light yellow module, neuron projection (GO cellular component) and gated channel activity (GO molecular function) are overrepresented. The KEGG calcium signaling pathway is also significantly enriched. The KEGG long-term depression includes the same SCA genes but did not reach statistical significance (Bonferroni-corrected P = .051). However, many SCA genes have not been assigned to these canonical pathways (only 6 of 24 genes according to results from DAVID Bioinformatics Resources). Based on cerebellar light yellow module genes and their functions, our data suggest that a single signaling pathway interconnects many SCA genes and other ataxia-related genes (Figure 3).
Figure 3. Proposed Molecular Mechanisms Contributing to Intracellular Calcium Homeostasis and Signaling in Neurons Depicting the Involvement of Genes Known to Cause Ataxia and Genes in the Cerebellar Light Yellow Coexpression Module.
Genes are shown in italics: red indicates human ataxia genes (underlined if in the light yellow module) and purple indicates additional relevant genes in the light yellow module (underlined if involved in ataxia phenotypes in mice). The red star represents calcium ions (Ca2+). Red boxes highlight proteins encoded by SCA genes; yellow boxes represent proteins encoded by other genes in the light yellow module; pale yellow boxes show relevant proteins although not found in our data. KCN indicates Potassium channels; CACN, Voltage-gated calcium channels; EAATs, Excitatory amino-acid transporters; GluR, Glutamate receptors; GPCR, G protein-coupled receptors; SOC, Store-operated calcium channels; PLC, Phospholipase C; PKC, Protein kinase C; IP3, Inositol 1,4,5-triphosphate; IP4, Inositol 1,3,4,5-tetrakisphosphate; IP3R, Inositol 1,4,5-triphosphate receptors; DAG, diacylglycerol; STIM, Stromal interaction molecule; SERCA, Sarcoendoplasmic reticulum (SR) calcium transport ATPase; ER/SR, Endo/sarcoplasmic reticulum; mut, mutated; PolyQmut, proteins with expanded polyglutamine tracts, namely those encoded by mutated ATXN1, ATXN2, ATXN3, CACNA1A, ATXN7, TBP, and ATN1.
Discussion
Different SCA subtypes share a core set of clinical and pathological features. To determine whether molecular pathways can explain such similarities, we have performed the first comprehensive transcriptomic analysis to date of 24 SCA genes in the human brain. We have shown that SCA genes have specific patterns of expression among brain regions, with those involved in ion channel activity having the highest expression in the cerebellum.
Using genome-wide expression data and WGCNA, we assigned all known SCA genes to expression modules without making a priori assumptions regarding their functions. Our results show that 2 gene coexpression modules are enriched for SCA genes and relate to expression profiles within granule and Purkinje cells of the cerebellum. These modules contain known SCA interactors, validating the importance of these expression modules in SCAs.
Because little is known about the function of most SCA genes, many have not been yet assigned to canonical pathways. The present gene coexpression network analysis using a hypothesis-free approach implicates roles for the ubiquitinproteasome system (UPS) in granule cells and calcium homeostasis and signaling in Purkinje cells. The significant enrichment of TFBSs for NR4A2 (or NURR1) near SCA genes also links to calcium regulation. NR4A2 encodes for an orphan nuclear receptor that is regulated by neural activity through voltage-dependent calcium channels and calcineurin and has been implicated in the pathogenesis of Parkinson disease.42,43 In addition to data revealed on the known SCA genes, this work is important to future studies identifying new variants and proving their pathogenicity.
The UPS is involved in the turnover of multiple cellular proteins, enabling cells to dispose of biologically nonuseful proteins (eg, mutant, misfolded, and overaccumulated proteins). In addition, the UPS is implicated in controlling gene transcription and protein expression in signal transduction systems as well as in neural synapse (reviewed by Lehman 44). Our network analysis revealed that biological processes related to the regulation of transcription and protein catabolic process are relevant for SCAs. Protein misfolding is recognized as a key feature of neurodegenerative diseases. Alzheimer disease, Parkinson disease, amyotrophic lateral sclerosis, prion disease, polyglutamine diseases (including polyglutamine SCAs), and possibly others (eg, SCA11) are proteinopathies, in which a particular protein or set of proteins misfolds and aggregates. In such aggregates, besides the mutated protein, additional proteins are sequestered, including ubiquitin, proteasome components, and transcription factors.45 This suggests that, in these diseases, the UPS function may be compromised in the presence or absence of mutations in its components. The phenotype of UPS-associated ataxia genes is of a complex cerebellar ataxia with additional features, such as prominent cognitive dysfunction or pyramidal signs.
Disruption of calcium signaling has been proposed as a common mechanism in the pathogenesis of SCAs46-48 and other neurodegenerative diseases.49-51 Our data support this hypothesis, showing that this pathway is important in the SCA transcript–enriched coexpression module relevant to Purkinje cells (light yellow module) and may explain selective neuropathology. Purkinje cells are important target cells in the pathogenesis of many SCAs, and calcium signaling is crucial for their normal cellular function. Abnormal calcium levels in Purkinje cells are thought to uncouple plasticity and activate toxic cascades, resulting in cell death.47 In this module, ITPR1 is a hub gene, and ITPR1-dependent signaling has been previously proposed as an important link among several ataxias.48 The CA8 (quadruple-gait ataxia gene) product, also a highly interconnected gene in the same module, is thought to be an ITPR1 antagonist. Evidence also exists that the PRKCG product may be activated by robust activation of ITPR1.48 Although located in a different coexpression module, the CACNA1A gene, which encodes P/Q-type calcium channel, also modulates ITPR1-dependent plasticity.48 It has further been shown that mutant ataxin 1 (ATXN1), ataxin 2 (ATXN2), ataxin 3 (ATXN3), and possibly other pathogenically expanded polyglutamine proteins interact with ITPR1,47 suggesting that proteins not normally involved in this pathway can also disrupt ITPR1-dependent signaling. The phenotype of the ataxias associated with calcium signaling abnormalities is usually a pure ataxia, with a few exceptions (eg, CA8 mutations, which are associated with mild mental retardation and an unusual gait). Drugs targeting the stabilization of calcium levels, the ITPR1 directly, or even the trial of calcium agonists in cell models may be useful therapies and might represent potential modifiers of this disease group regardless of the genetic cause.
Conclusions
Normal expression of SCA genes in the human brain reveals pathways linking SCA genes and other ataxia genes. Some of these links have been previously proposed, while others suggest potential new targets for genetic and functional testing. An important next step will be to analyze the expression in different brain regions from individuals affected with the common SCA subtypes. These networks provide valuable lists of candidate genes for diseases with overlapping phenotypes, which are good candidates for novel genetic modifiers.
Supplementary Material
Acknowledgments
Funding/Support: The United Kingdom Brain Expression Consortium is supported by the United Kingdom Medical Research Council through the Sudden Death Brain and Tissue Bank (Dr Smith) and by project grant G0901254 (Dr Hardy) and training fellowship grant G0802462 (Dr Ryten). This study was further supported by the Brain Research Trust, The Royal Society, award WT089698 from the Medical Research Council/Wellcome Trust Joint Call in Neurodegeneration, and the National Institute for Health Research UCL Hospitals/UCL Biomedical Research Centre.
Role of the Sponsor: The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Footnotes
Conflict of Interest Disclosures: None reported.
REFERENCES
- 1.Schöls L, Bauer P, Schmidt T, Schulte T, Riess O. Autosomal dominant cerebellar ataxias: clinical features, genetics, and pathogenesis. Lancet Neurol. 2004;3(5):291–304. doi: 10.1016/S1474-4422(04)00737-9. [DOI] [PubMed] [Google Scholar]
- 2.Dueñas AM, Goold R, Giunti P. Molecular pathogenesis of spinocerebellar ataxias. Brain. 2006;129(pt 6):1357–1370. doi: 10.1093/brain/awl081. [DOI] [PubMed] [Google Scholar]
- 3.Soong BW, Paulson HL. Spinocerebellar ataxias: an update. Curr Opin Neurol. 2007;20(4):438–446. doi: 10.1097/WCO.0b013e3281fbd3dd. [DOI] [PubMed] [Google Scholar]
- 4.Durr A. Autosomal dominant cerebellar ataxias: polyglutamine expansions and beyond. Lancet Neurol. 2010;9(9):885–894. doi: 10.1016/S1474-4422(10)70183-6. [DOI] [PubMed] [Google Scholar]
- 5.Wang JL, Yang X, Xia K, et al. TGM6 identified as a novel causative gene of spinocerebellar ataxias using exome sequencing. Brain. 2010;133(pt 12):3510–3518. doi: 10.1093/brain/awq323. [DOI] [PubMed] [Google Scholar]
- 6.Lee YC, Durr A, Majczenko K, et al. Mutations in KCND3 cause spinocerebellar ataxia type 22. Ann Neurol. 2012;72(6):859–869. doi: 10.1002/ana.23701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duarri A, Jezierska J, Fokkens M, et al. Mutations in potassium channel kcnd3 cause spinocerebellar ataxia type 19. Ann Neurol. 2012;72(6):870–880. doi: 10.1002/ana.23700. [DOI] [PubMed] [Google Scholar]
- 8.Seidel K, Siswanto S, Brunt ER, den Dunnen W, Korf HW, Rüb U. Brain pathology of spinocerebellar ataxias. Acta Neuropathol. 2012;124(1):1–21. doi: 10.1007/s00401-012-1000-x. [DOI] [PubMed] [Google Scholar]
- 9.Lim J, Hao T, Shaw C, et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125(4):801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 10.Moreno JA, Radford H, Peretti D, et al. Sustained translational repression by eIF2α-P mediates prion neurodegeneration. Nature. 2012;485(7399):507–511. doi: 10.1038/nature11058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. doi:10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oldham MC, Horvath S, Geschwind DH. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc Natl Acad Sci U S A. 2006;103(47):17973–17978. doi: 10.1073/pnas.0605938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oldham MC, Konopka G, Iwamoto K, et al. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11(11):1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- 15.Konopka G. Functional genomics of the brain: uncovering networks in the CNS using a systems approach. Wiley Interdiscip Rev Syst Biol Med. 2011;3(6):628–648. doi: 10.1002/wsbm.139. [DOI] [PubMed] [Google Scholar]
- 16.Rosen EY, Wexler EM, Versano R, et al. Functional genomic analyses identify pathways dysregulated by progranulin deficiency, implicating Wnt signaling. Neuron. 2011;71(6):1030–1042. doi: 10.1016/j.neuron.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winden KD, Oldham MC, Mirnics K, et al. The organization of the transcriptional network in specific neuronal classes. Mol Syst Biol. 2009;5:291. doi: 10.1038/msb.2009.46. doi:10.1038/msb.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P. Coexpression analysis of human genes across many microarray data sets. Genome Res. 2004;14(6):1085–1094. doi: 10.1101/gr.1910904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302(5643):249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 20.Millar T, Walker R, Arango JC, et al. Tissue and organ donation for research in forensic pathology: the MRC Sudden Death Brain and Tissue Bank. J Pathol. 2007;213(4):369–375. doi: 10.1002/path.2247. [DOI] [PubMed] [Google Scholar]
- 21.Trabzuni D, Ryten M, Walker R, et al. Quality control parameters on a large dataset of regionally dissected human control brains for whole genome expression studies. J Neurochem. 2011;119(2):275–282. doi: 10.1111/j.1471-4159.2011.07432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forabosco P, Ramasamy A, Trabzuni D, et al. Insights into TREM2 biology by network analysis of human brain gene expression data. Neurobiol Aging. 2013;34(12):2699–2714. doi: 10.1016/j.neurobiolaging.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang HJ, Kawasawa YI, Cheng F, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478(7370):483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson MB, Kawasawa YI, Mason CE, et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62(4):494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34(database issue):D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langfelder P, Luo R, Oldham MC, Horvath S. Is my network module preserved and reproducible? PLoS Comput Biol. 2011;7(1):e1001057. doi: 10.1371/journal.pcbi.1001057. doi:10.1371/journal.pcbi.1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho Sui SJ, Mortimer JR, Arenillas DJ, et al. oPOSSUM: identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res. 2005;33(10):3154–3164. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho Sui SJ, Fulton DL, Arenillas DJ, Kwon AT, Wasserman WW. oPOSSUM: integrated tools for analysis of regulatory motif over-representation. Nucleic Acids Res. 2007;35(web server issue):W245–W252. doi: 10.1093/nar/gkm427. doi:10.1093/nar/gkm427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwon AT, Arenillas DJ, Worsley Hunt R, Wasserman WW. oPOSSUM-3: advanced analysis of regulatory motif over-representation across genes or ChIP-Seq datasets. G3 (Bethesda) 2012;2(9):987–1002. doi: 10.1534/g3.112.003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perkins EM, Clarkson YL, Sabatier N, et al. Loss of β-III spectrin leads to Purkinje cell dysfunction recapitulating the behavior and neuropathology of spinocerebellar ataxia type 5 in humans. J Neurosci. 2010;30(14):4857–4867. doi: 10.1523/JNEUROSCI.6065-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lou JY, Laezza F, Gerber BR, et al. Fibroblast growth factor 14 is an intracellular modulator of voltage-gated sodium channels. J Physiol. 2005;569(pt 1):179–193. doi: 10.1113/jphysiol.2005.097220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pierson TM, Adams D, Bonn F, et al. Whole-exome sequencing identifies homozygous AFG3L2 mutations in a spastic ataxia-neuropathy syndrome linked to mitochondrial m-AAA proteases. PLoS Genet. 2011;7(10):e1002325. doi: 10.1371/journal.pgen.1002325. doi:10.1371/journal.pgen.1002325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu Z, Mellor J, Wu J, DeLisi C. VisANT: an online visualization and analysis tool for biological interaction data. BMC Bioinformatics. 2004;5:17. doi: 10.1186/1471-2105-5-17. doi:10.1186/1471-2105-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Türkmen S, Guo G, Garshasbi M, et al. CA8 mutations cause a novel syndrome characterized by ataxia and mild mental retardation with predisposition to quadrupedal gait. PLoS Genet. 2009;5(5):e1000487. doi: 10.1371/journal.pgen.1000487. doi:10.1371/journal.pgen.1000487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Becker EB, Oliver PL, Glitsch MD, et al. A point mutation in TRPC3 causes abnormal Purkinje cell development and cerebellar ataxia in moonwalker mice. Proc Natl Acad Sci U S A. 2009;106(16):6706–6711. doi: 10.1073/pnas.0810599106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuhn A, Kumar A, Beilina A, Dillman A, Cookson MR, Singleton AB. Cell population–specific expression analysis of human cerebellum. BMC Genomics. 2012;13:610. doi: 10.1186/1471-2164-13-610. doi:10.1186/1471-2164-13-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nascimento-Ferreira I, Santos-Ferreira T, Sousa-Ferreira L, et al. Overexpression of the autophagic beclin-1 protein clears mutant ataxin-3 and alleviates Machado-Joseph disease. Brain. 2011;134(pt 5):1400–1415. doi: 10.1093/brain/awr047. [DOI] [PubMed] [Google Scholar]
- 40.McGurk L, Bonini NM. Protein interacting with C kinase (PICK1) is a suppressor of spinocerebellar ataxia 3-associated neurodegeneration in Drosophila. Hum Mol Genet. 2012;21(1):76–84. doi: 10.1093/hmg/ddr439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bettencourt C, Lima M. Machado-Joseph disease: from first descriptions to new perspectives. Orphanet J Rare Dis. 2011;6:35. doi: 10.1186/1750-1172-6-35. doi:10.1186/1750-1172-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tokuoka H, Hatanaka T, Metzger D, Ichinose H. Nurr1 expression is regulated by voltage-dependent calcium channels and calcineurin in cultured hippocampal neurons. Neurosci Lett. 2014;559:50–55. doi: 10.1016/j.neulet.2013.11.033. [DOI] [PubMed] [Google Scholar]
- 43.Decressac M, Volakakis N, Björklund A, Perlmann T. NURR1 in Parkinson disease: from pathogenesis to therapeutic potential. Nat Rev Neurol. 2013;9(11):629–636. doi: 10.1038/nrneurol.2013.209. [DOI] [PubMed] [Google Scholar]
- 44.Lehman NL. The ubiquitin proteasome system in neuropathology. Acta Neuropathol. 2009;118(3):329–347. doi: 10.1007/s00401-009-0560-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paulson HL. Protein fate in neurodegenerative proteinopathies: polyglutamine diseases join the (mis)fold. Am J Hum Genet. 1999;64(2):339–345. doi: 10.1086/302269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carlson KM, Andresen JM, Orr HT. Emerging pathogenic pathways in the spinocerebellar ataxias. Curr Opin Genet Dev. 2009;19(3):247–253. doi: 10.1016/j.gde.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kasumu A, Bezprozvanny I. Deranged calcium signaling in Purkinje cells and pathogenesis in spinocerebellar ataxia 2 (SCA2) and other ataxias. Cerebellum. 2012;11(3):630–639. doi: 10.1007/s12311-010-0182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schorge S, van de Leemput J, Singleton A, Houlden H, Hardy J. Human ataxias: a genetic dissection of inositol triphosphate receptor (ITPR1)–dependent signaling. Trends Neurosci. 2010;33(5):211–219. doi: 10.1016/j.tins.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garwood C, Faizullabhoy A, Wharton SB, et al. MRC Cognitive Function and Ageing Neuropathology Study Group Calcium dysregulation in relation to Alzheimer-type pathology in the ageing brain. Neuropathol Appl Neurobiol. 2013;39(7):788–799. doi: 10.1111/nan.12033. [DOI] [PubMed] [Google Scholar]
- 50.Grosskreutz J, Van Den Bosch L, Keller BU. Calcium dysregulation in amyotrophic lateral sclerosis. Cell Calcium. 2010;47(2):165–174. doi: 10.1016/j.ceca.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 51.Hurley MJ, Brandon B, Gentleman SM, Dexter DT. Parkinson’s disease is associated with altered expression of CaV1 channels and calcium-binding proteins. Brain. 2013;136(pt 7):2077–2097. doi: 10.1093/brain/awt134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.