Abstract
In vivo delivery of siRNAs designed to inhibit genes important in cancer and other diseases continues to be an important biomedical goal. We now describe a new nanoparticle construct that has been engineered for efficient delivery of siRNA to tumors. The construct is comprised of a 47-nm mesoporous silica nanoparticle (MSNP) core coated with a cross-linked PEI-PEG copolymer, carrying siRNA against the HER2 oncogene, and coupled to the anti-HER2 monoclonal antibody (trastuzumab). The construct has been engineered to increase siRNA blood half-life, enhance tumor-specific cellular uptake, and maximize siRNA knockdown efficacy. The optimized anti-HER2-nanoparticles produced apoptotic death in HER2 positive (HER2+) breast cancer cells grown in vitro, but not in HER2 negative (HER2−) cells. One dose of the siHER2-nanoparticles reduced HER2 protein levels by 60% in trastuzumab-resistant HCC1954 xenografts. Multiple doses administered intravenously over 3 weeks significantly inhibited tumor growth (p < 0.004). The siHER2-nanoparticles have an excellent safety profile in terms of blood compatibility and low cytokine induction, when exposed to human peripheral blood mononuclear cells. The construct can be produced with high batch-to-batch reproducibility and the production methods are suitable for large-scale production. These results suggest that this siHER2-nanoparticle is ready for clinical evaluation.
Keywords: mesoporous silica nanoparticles, siRNA, HER2+ breast cancer, cancer nanomedicine
1. Introduction
The Cancer Genome Atlas project, the International Cancer Genome Consortium and other large-scale genomics projects are identifying genomic aberrations and affected regulatory networks that enable aspects of cancer progression including proliferation, angiogenesis, invasion, drug resistance, and metastasis.[1] These discovery efforts and associated large scale functional studies[2] are guiding the development of a wide range of therapeutic agents designed to inhibit the genes and pathways on which cancers depend. Unfortunately, many of the most attractive therapeutic targets currently are not druggable using small molecule inhibitors or antibodies.[3] RNA interference using siRNA is an attractive alternative to inhibiting otherwise intractable therapeutic targets.[4] This strategy has been effective in vitro. However, delivery of siRNAs to tumors in patients is still challenging.
Several strategies to deliver siRNAs in vivo have involved packing siRNAs into nanoparticle constructs designed to increase siRNA half-life in the blood,[5] allow escape from the reticuloendothelial system (RES) recognition that rapidly causes nanoparticles to accumulate in the liver and spleen,[6] and enhance tumor specific cellular uptake. A wide range of organic and inorganic nanoparticle materials have been evaluated as siRNA carriers to achieve these goals. These include viral-capsids, cyclodextrin, cationic polymers, gold nanoparticles, peptides (see reviews),[7] and mesoporous silica nanoparticles (MSNP, see reviews).[8]
Several organic nanoparticles developed for anticancer agent delivery show promise but limitations have been identified. For example, viral-based carriers sometimes induce adverse immune responses. Cationic lipid nanoparticles have shown efficacy in treating liver cancer[9] since they home to the liver and spleen via RES recognition. Unfortunately, they did not show objective efficacy when used to treat tumors at other anatomic sites.[10] In addition, they elicited hematological toxicity in some cases[11] and some have been difficult to manufacture reproducibly at large scale. A cyclodextrin-based nanoparticle targeted to the human transferrin protein (hTf) was the first targeted siRNA delivery system to demonstrate anti-cancer efficacy at sites other than the liver (i.e., melanoma). A Phase 1 trial showed that this construct successfully silenced the target gene, RRM2, in tumors of three patients.[12] However, a subsequent report found that particle instability in kidneys reduced siRNA half-life.[13] In particular, only 1.4% of the injected siRNA remained in blood at 1 hour after injection into non-human primates without tumors.[14] SiRNA complexed with tumor penetrating peptides also have shown some efficacy.[15] Interestingly, a siRNA-peptide complex against PLK1 coupled to a HER2 scFv for targeted delivery to HER2+ breast cancer showed efficacy against HER2+ BT474 xenografts.[15b]
Inorganic nanoparticles such as gold, mesoporous silica, and iron oxide have been tested as siRNA carriers. These typically are easier to synthesize reproducibly at large scale. Spherical nucleic acid-gold nanoparticle conjugates[16] have been developed to deliver siRNA against Bcl2Like12 for treating glioblastoma. In vivo protein knockdown (~40%) and anti-tumor efficacy were achieved in the orthotopically-implanted tumor after 7 doses administered every other day. Several MSNP-based platforms for siRNA delivery have been tested.[17] These have exploited passive delivery to areas of tumors that have abnormal molecular and fluid transport dynamics due to abnormal vasculature and lymphatic structure – termed Enhanced Permeability and Retention (EPR).[18] These MSNPs were coated with cationic polymers including PEI,[17a, 17b] PEI-cyclodextrin,[17c] and PDMAEMA[17d] for cellular entrance and hence had no cancer cells specificity. Three of the four platforms[17a, 17c, 17d] did not have a steric layer such as PEG to shield them from RES recognition.[19] One PEI-modified MSNP platform without PEG or a targeting agent was employed to deliver siRNA against vascular endothelial growth factor in tumors upon intratumoral injection and shown to inhibit tumor growth.[17a] Two of the four platforms were loaded with siRNA (siRNA against the M2 isoform of the glycolytic enzyme pyruvate kinase (PKM2)[17c] or siRNA against polo-like kinase 1 (PLK-1))[17d] inside the pores, requiring large pore size, and in turn resulting in large particle sizes (80-150 nm as the MSNP core size). While promising, significant anti-tumor activity in vivo has not been reported for these constructs.[17c, 17d] Meng et. al.[17b] reported PEG-PEI-MSNP platform, but it had no targeting component and it showed tumor inhibition only upon combination with a chemotherapy drug (doxorubicin) due to the choice of targeted gene. Overall, MSNP remains attractive as a core material for siRNA delivery in vivo due its low toxicity, large pore volume),[20] large surface area, ease of controlling size, and high synthesis scalability.
Following on these reports, we developed a new MSNP construct to deliver a siRNA against the oncogenic human epidermal growth receptor type 2 (HER2) gene. We used a small diameter (~50 nm) rigid MSNP as the core. We coated the MSNP core with a PEI polymer to overcome delivery barriers, enable scale-up production and minimize toxicity compared to whole PEI-siRNA complex. PEI was also cross-linked by bioreducible crosslinkers to enhance its buffering capacity. We incorporated a PEG layer to protect the siRNA against blood enzymes, to prevent aggregation of cationic nanoparticles, to enhance blood safety and to prevent adverse immune response to the nanoparticles. We attached the antibody trastuzumab to the nanoparticle surface to target the particles to cells that overexpress the HER2 protein and to provide independent therapeutic benefit. Lastly, we loaded siRNA on the external surface of the MSNP (but protected under the PEG layer) to allow easy siRNA escape from the endosome prior to degradation by lysosomes.
We chose HER2 as the initial siRNA target because it is a particularly strong and well validated therapeutic target in breast cancer. Amplification of this gene occurs in 15-25% of diagnosed breast cancers[21] and is linked to aggressiveness and poor prognosis.[22] First line HER2+ targeted therapies, trastuzumab and lapatinib, demonstrate significant clinical efficacy, thereby providing validation of HER2 as a therapeutic target. However, up to 70% of patients with advanced disease demonstrate intrinsic or acquired resistance to trastuzumab within one year.[23] A combination of trastuzumab and lapatinib provided additional benefits to patients, but 50% of patients still did not respond to the treatment.[24] Combinations with newer therapeutic agents including trastuzumab emtansine, pertuzumab and neratinib are promising and some will likely become part of the standard of care for HER2+ tumors. However, early studies of these agents also show that many tumors will eventually progress on treatment.[25] Thus, additional therapeutic options are needed. By silencing HER2 in the tumors with siRNA in combination with simultaneous trastuzumab delivery, we provide a new strategy that may increase efficacy in tumors that continue to depend on HER2, thereby advancing treatment of HER2+ breast cancers.
2. Results and Discussion
2.1. Synthesis, characterization and optimization of siRNA-nanoparticles
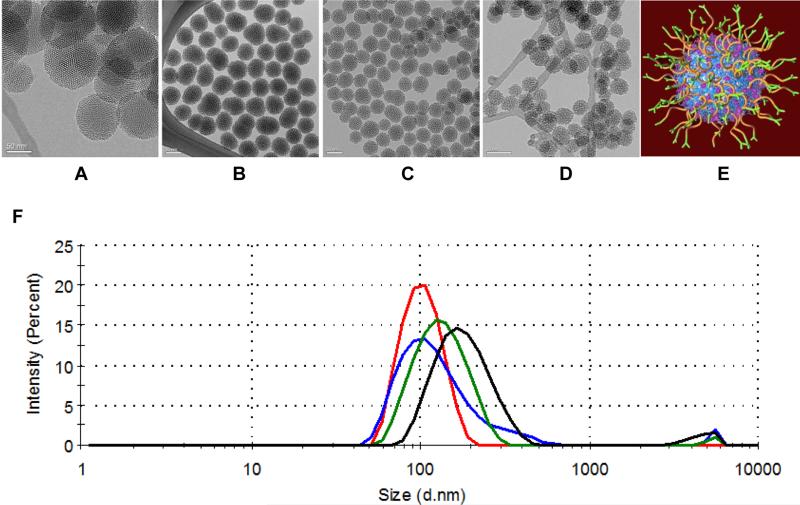
Our siRNA delivery construct is built around a MSNP core. MSNP size is an important consideration in siRNA-nanoparticle design. Several studies suggest that nanoparticles should be within the size range of 20 – 200 nm in order to enable EPR mediated delivery.[26] We used MSNPs at the smaller end of this size range since we expect that smaller nanoparticles will extravasate best into tumor tissues.[27] We compared two synthetic methods to produce the MSNP core; one yielded uniform sizes of 34 ± 3 nm, 47 ± 4 nm, or 61 ± 7 nm (by varying the ratios of two surfactants, see Experimental section) and the other[28] yielded particles that were larger and less uniform in size of 87 ± 14 nm. These results are illustrated in the TEM images in Figure 1A-D and summarized in Table 1. We analyzed these MSNPs using FT-IR (Figure S1A) to make sure there were no remaining surfactants (pore templates) in the NP cores.
Figure 1. Characterization of mesoporous silica nanoparticle (MSNP) cores and schematic of siRNA-nanoparticles.
(A-D) TEM images of four batches of mesoporous silica nanoparticle (MSNP) cores: non-uniform-sized MSNP with an average size of 87 nm (O-87, A), and uniform-sized MSNP with an average size of 61 nm (S-61, B), 47 nm (S-47, C), and 34 nm (S-34, D), respectively (Scale bar = 50 nm). (E) Schematic illustration of the nanoparticles with exterior modified layer-by-layer with cationic polymer (cross-linked PEI, blue), PEG (orange), antibody (green) and siRNA (magenta). (F) Hydrodynamic size distribution of S-34 (blue), S-47 (red), S-61 (green) and O-87 (black) cores after surface modification as in (E) except O-87, of which PEI was coated without cross-linking.
Table 1.
Hydrodynamic size, zeta potential, and silencing efficacy of six different nanoparticles.
| Material (MSNP core) | MSNP core size by TEMa) [nm] | Surface modificationb) | Hydrodynamic size (DLS) | Zeta charge [mV] | |
|---|---|---|---|---|---|
| Size [nm]c) | PDId) | ||||
| O-87 | 87 ± 14 | T-NP10 | 214 ± 22 | 0.22 | 22 ± 0.5 |
| S-61 | 61 ± 7 | T-NP10 | 113 ± 1.0 | 0.20 | 18 ± 0.4 |
| T-NP10C | 131 ± 0.3 | 0.20 | 19 ± 3.7 | ||
| S-47 | 47 ± 4 | T-NP10C | 117 ± 0.5 | 0.19 | 25 ± 0.1 |
| T-NP1.8C | 117 ± 2.4 | 0.20 | 19 ± 4.0 | ||
| S-34 | 34 ± 3 | T-NP10C | 133 ± 4.1 | 0.37 | 19 ± 4.0 |
core size measured in dry state, average size of 50 particles.
“10” stands for 10-kDa PEI; “1.8C” and “10C” stand for cross-linked 1.8-kDa and cross-linked 10-kDa PEI, respectively. All PEI-MSNP were then conjugated with PEG, and trastuzumab (T).
Average of three measurements; the z-average diameter and polydispersity index (PDI) values were defined according to International Standard on DLS (ISO13321).
PDI ranges from 0 to 1; smaller number indicates narrower size distribution; e.g., PDI <0.05 is considered monodisperse (one size only), while PDI >0.5 indicates a broad distribution of particle sizes.
We modified the MSNP core surface as illustrated in Figure 1E by adding polyethyleneimine (PEI), polyethyleneglycol (PEG), the targeting antibody, and siRNA. We included PEI in these constructs to promote endosomal escape of nucleic acids.[29] However, the toxicity of PEI is of concern. We attempted to reduce this by adding a PEG layer.[30] Nanoparticles were further conjugated with trastuzumab (designated as T) to target cells expressing HER2 and with rituximab targeting CD20 (designated as R) as a negative control. These nanoparticle constructs will be referred to hereafter as T-NP or R-NP, designating trastuzumab-conjugated PEG-PEI-MSNP or rituximab-conjugated PEG-PEI-MSNP, respectively. We employed several siRNAs during the course of our studies including a scrambled siRNA control designated siSCR, a siRNA against luciferase designated siLUC, and a siRNA against HER2 designated siHER2. Their specific sequences are described in Supporting Information.
The nanoparticles after surface modification had a hydrodynamic size of ~100 nm for the three uniform-sized core materials (S-34, S-47, S-61) and 200 nm for the non-uniformsized core material (O-87) in PBS. All materials were also positively charged after the modification due to the cationic PEI. The hydrodynamic sizes and zeta potentials of these materials after surface modification are summarized in Table 1 and hydrodynamic size histograms are shown in Figure 1F. Composition of the optimized nanoparticles (also see next section) was analyzed by TGA analysis and BCA assay, as reported in Figure S1B and Table 2.
Table 2.
Composition of T-siRNA-NP (all reported as percent by mass of the whole construct).
| Materials | Surface modification | PEI by TGA [%] | PEG by TGA [%] | Antibody by BCA [%] | NP/siRNA mass ratio (fluorescent method) |
|---|---|---|---|---|---|
| S-47 | T-NP10C | 13.5 | 18.2 | 3 | Complete at 25 and 50 |
| S-47 | T-NP1.8C | 15.9 | 6.1 | 3 | Complete at 25 and 50 |
The siHER2 was selected from 76 potential sequences by measuring the efficacy and specificity with which these siRNAs reduced mRNA levels and growth in 4 HER2+ cell lines and 2 HER2− cell lines. The best siHER2 from these studies was further confirmed for the efficacy and specificity in 20 additional HER2+ cell lines and 2 HER2− cell lines as shown in Figure S2A. The dose of this siRNA required to inhibit growth by 50% (GI50) was <5 nM in 19 of 20 HER2+ cell lines including 13 that did not respond to trastuzumab (30 μg mL−1) (Figure S2B).
2.2. Engineering endo-lysosomal vesicle escape and in vitro gene knockdown by siRNA-nanoparticles
Internalized nanoparticles ultimately end up in perinuclear lysosomal vesicles. siRNAs must escape from this environment early to be effective since the nucleases and acidic pH in the lysosomal vesicles will destroy the entrapped complexes. Polymers that exhibit high transfection efficiencies, such as PEI,[31] have buffering capacity in the endo-lysosomal pH range of 5–7 due to presence of unprotonated secondary and tertiary amines. This buffering is thought to increase influx of hydrated protons and chloride ions, thereby causing the vesicles to swell, rupture and release the siRNAs into cytosol.[32] This phenomenon is referred to as the proton sponge effect.
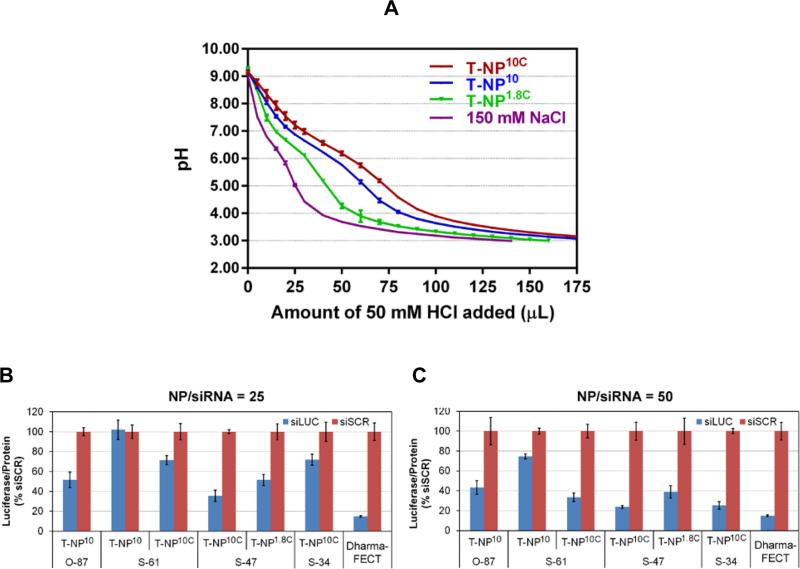
We tested the possibility that endo-lysosomal escape and gene silencing efficacy of our siRNA-nanoparticles could be increased by increasing buffering capacity of the nanoparticles. First, we measured the buffering capacities of nanoparticle platforms with cross-linked 1.8-kDa PEI (T-NP1.8C), cross-linked 10-kDa PEI (T-NP10C), and non-cross-linked 10-kDa PEI (T-NP10) in 150 mM NaCl (see Figure 2A). We found that the nanoparticle platforms had buffering capacity in the order of T-NP10C > T-NP10 > T-NP1.8C. Thus, the cross-linking of the PEI creates more secondary and tertiary amines yielding greater buffering capacity than primary amines.[33] We expected that gene silencing efficacy would be in the same order based on the proton sponge mediated siRNA release concept.
Figure 2. Buffering capacity and luciferase silencing efficacy of nanoparticles.
(A) Buffering capacity of three nanoparticle platforms with cross-linked 1.8-kDa PEI (T-NP1.8C), and non-cross-linked (T-NP10) and cross-linked 10-kDa PEI (T-NP10C) measured in NaCl (150 mM). (B-C) Silencing of luciferase in MDA-MB-231-H2N-luc (high HER2, high luciferase) upon treatment with 30 nM siLUC on nanoparticles (NP) at NP/siRNA weight ratio of 25 (B) and 50 (C), measured at 48 hours post transfection (with overnight media change).
We tested this by comparing the gene silencing ability of test siLUC on various nanoparticles (four core sizes, loaded with PEI of 1.8-kDa or 10-kDa, cross-linked or no cross-linked). Specifically, we assessed luciferase silencing at 24 hours post exposure of the siLUC-nanoparticles on a breast cancer cell line, MDA-MB-231-H2N-luc, that was genetically modified to overexpress both HER2 and luciferase.[34] Figure 2B-C show the luciferase silencing (vs. siSCR) of all nanoparticles. Complete siRNA binding was achieved for all materials at NP/siRNA mass ratio of 25 and above (confirmed by no remaining unbound siRNA in the solution phase after the loading step). However, materials with NP/siLUC of 50 offered better gene silencing efficacy (per same dose of siLUC) (Figure 2C) than those with NP/siLUC of 25 (Figure 2B) due to the greater number of nanoparticles to which the cells were exposed. We used the 50 mass ratio throughout this study unless specified otherwise. We found that smaller particles had reduced silencing efficacy compared to larger particles (see S-61 vs. O-87, both were modified with 10-kDa PEI, designated as TNP10 in Figure 2B). This is likely due to poorer endo-lysosomal escape of the siLUC from the smaller particles. However, large particles are less desirable for tumor delivery using EPR effects. We tested the possibility that the siRNA endosomal release from smaller particles could be increased by PEI cross-linking to increase the buffering capacity as shown in Figure 2A. Figure 2B-C shows that the silencing efficacy was indeed improved with cross-linked materials compared to the non-cross-linked material (see T-NP10C vs. T-NP10, from S-61 core). The highest silencing efficacy (76%) was achieved with the nanoparticles that were developed from S-47 core. This S-47 material also yielded the best size distribution without large aggregates (Figure 1F). The S-47 modified with cross-linked 10-kDa PEI was more effective than that modified with cross-linked 1.8-kDa PEI (76% vs. 60% silencing efficacy). Their compositions are reported in Table 2 (see also Figure. S1B for TGA analysis for PEI and PEG loading characterization). The S-47 core material with cross-linked PEI was used for all subsequent experiments.
2.3. Protection of siRNA against blood enzymatic degradation
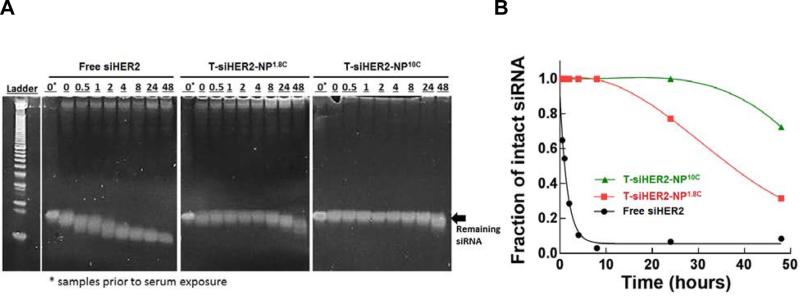
We assessed the ability of MSNP constructs (T-siHER2-NP1.8C vs. T-siHER2-NP10C) to protect siHER2 from degradation by blood enzymes by measuring the amount of siHER2 remaining after incubation of siHER2-nanoparticles for 0 to 48 hours in human serum (50% in PBS) at 37 °C. We compared these results to those obtained for free siHER2 without nanoparticles. Figure 3A shows the amount of intact siHER2 that survived enzymatic degradation as measured by gel electrophoresis. The corresponding siHER2 quantification based on the band intensity and location is shown in Figure 3B. Without the nanoparticles, naked siHER2 was degraded within 0.5 hour (observed as bands shifted toward lower molecular weight) and its half-life was about 1 hour, in agreement with previous reports for other siRNAs.[35] T-siHER2-NP1.8C fully protected siHER2 up to 8 hours, while T- siHER2-NP10C fully protected siHER2 up to 24 hours. The siRNA on both of our nanoparticle platforms experienced much less degradation than has been reported for the cyclodextrin-based nanoparticles that have already shown clinical antitumor efficacy.[12b] Those cyclodextrin-protected siRNAs were reported to experience 50% degradation within 12 hours, and 70% within 24 hours under 50% serum conditions.[36]
Figure 3. SiRNA protection from serum degradation.
(A) Residual siRNA against HER2 (siHER2) band after contact with 50% human serum after specified periods of time; conducted with free siHER2, or siHER2 loaded on two nanoparticle platforms with cross-linked 1.8-kDa (T-siHER2-NP1.8C) and cross-linked 10-kDa PEI (T-siHER2-NP10C), all at 37 °C with shaking, and (B) the corresponding siHER2 quantification using ImageJ software.
2.4. Importance of PEG
The higher siRNA protection for T-siHER2-NP10C compared to T-siHER2-NP1.8C is likely due to the increased PEG content for the higher-molecular-weight PEI of T-siHER2-NP10C. The PEG contents of T-siHER2-NP10C and T-siHER2-NP1.8C were 18.2% and 6.1%, respectively (Table 2). Higher PEG content is expected for T-siHER2-NP10C since it contains more reactive amine groups for PEG binding than T-siHER2-NP1.8C. PEG is known to provide a steric blocking effect[37] that reduces enzymatic degradation of siRNA.[38] It also reduces binding of blood proteins to the nanoparticles.[37b] We found in a separate experiment (Figure S3A) that siRNA on PEI-MSNP without PEG degraded faster than naked siRNA since positively charged PEI recruited more negatively charged siRNA degrading enzymes. In addition, we found significant aggregation of nanoparticles without PEG upon siRNA loading (Figure S3B). The PEG layer also improved blood compatibility and reduced immune response as described later in the paper.
2.5. Cellular uptake of the siRNA-nanoparticles
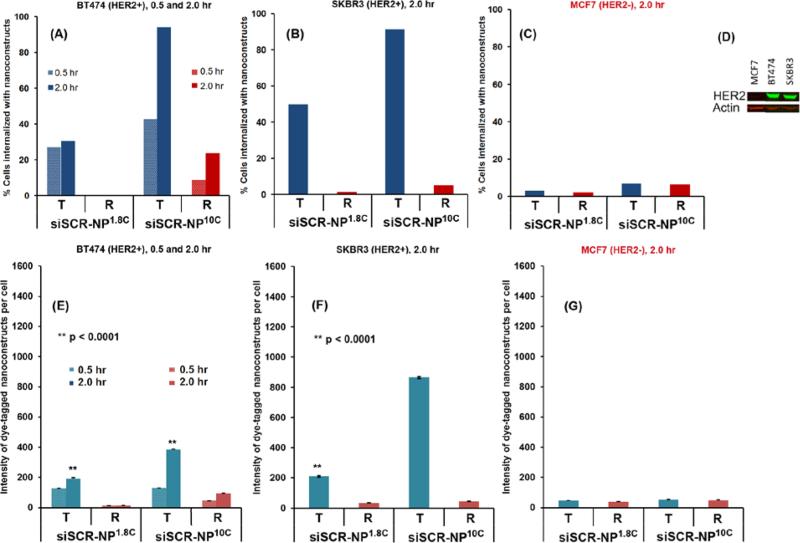
Cellular uptake relies on HER2 antibody (trastuzumab) conjugated on the nanoparticles, which was found to be optimal at 3% by mass of trastuzumab per nanoparticles as shown in Figure S4. This 3% loading was used throughout the manuscript. Without trastuzumab (Figure S4), the nanoparticles could be taken up by cells due to their positive charge, which is not specific to target cells only. Next, we assessed the specificity with which trastuzumab targeted siRNAs were taken up by cells that over express the HER2 protein by measuring the uptake of nanoparticles carrying a scrambled siRNA and coupled with trastuzumab (designated hereafter as T-siSCR-NP1.8C and T-siSCR-NP10C) and with rituximab targeting CD20 (designated as R-siSCR-NP1.8C and R-siSCR-NP10C). We measured cellular uptake of T-siSCR-NP10C and T-siSCR-NP1.8C in HER2+ breast cancer cells, BT474 and SKBR3, and the HER2− cell line MCF-7 at 0.5 or 2.0 hours post exposure to the nanoparticles. The siSCR was tagged with the fluorescent reporter, Alexa 488, for these experiments to enable quantitative analysis of siSCR uptake. R-siSCR-NP10C and R-siSCR-NP1.8C nanoparticles served as a negative control since BT474, SKBR3 and MCF-7 cells weakly express CD20. We measured the amount of Alexa 488-tagged siSCR in the interior of individual cells using flow cytometry after quenching fluorescence from Alexa 488-tagged siSCR on the external cell membrane using Trypan blue. Figure 4A-C shows that T-siSCR-NP10 were taken up effectively (>90%) into HER2+ cells (BT474 and SKBR3), but not HER2− cells (MCF7) and that uptake increased by extending the exposure time from 0.5 hr to 2 hr. Furthermore, uptake of T-siSCR-NP10C was greater than T-siSCR-NP1.8C. In addition, R-siSCR-NP10C and R-siSCR-NP1.8C nanoparticles were not taken up efficiently by any of the cell lines. Figure 4D illustrates HER2 protein expression in the three cell lines being evaluated. Figure 4E-G show the average intensity of Alexa 488-tagged siSCR signal per cell and the same trend can be observed. Fluorescence distributions of Alexa 488-siSCR uptake are reported in Figure S5. This confirms that nanoparticles enter cells primarily by a HER2-receptor mediated endocytosis mechanism and not by adsorptive endocytosis of positively charged particles as reported for PEI-MSNP.[39]
Figure 4. Cellular uptake of siRNA-nanoparticles.
Percentage of BT474 (HER2+) (A), SKBR3 (HER2+) breast cancer cells (B), and MCF7 (HER2−) breast cancer cells (C), that were internalized with fluorescent dye-tagged scrambled siRNA (siSCR)-nanoparticles having either cross-linked 1.8-kDa (NP1.8C) or cross-linked 10-kDa PEI (NP10C), and conjugated with either trastuzumab (T) or rituximab (R), (D) western blot confirming HER2 content of these 3 cell lines, and (E-G) the corresponding intensity (per cell) of dye-tagged siSCR-nanoparticles internalized into the cells. Data were presented as mean ± SEM. All were performed with 1 million cells and 100 μg of nanoparticles in 0.3 mL of cell culture media and exposure time of 0.5 or 2 hours. Also see histogram in Figure S5.
2.6. HER2 knockdown efficacy and therapeutic effects
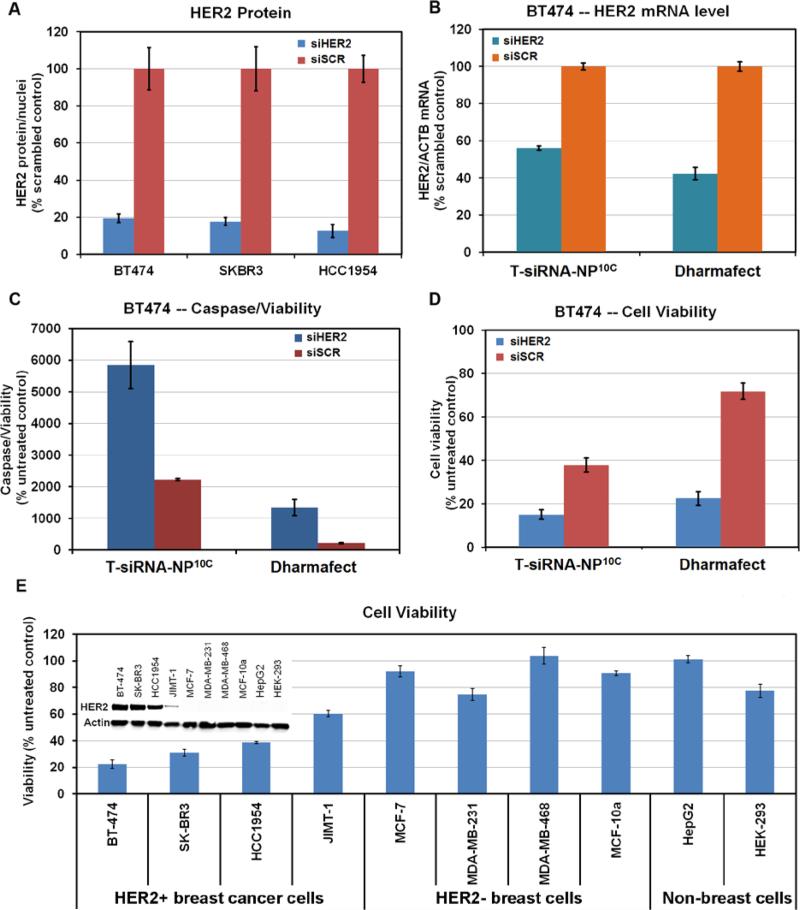
We assessed the efficiency of T-siHER2-NP10C and T-siHER2-NP1.8C in inhibiting HER2 mRNA levels and HER2 protein expression in the HER2+ breast cancer cell lines, BT474, SKBR3, and HCC1954. We used quantitative immunofluorescent imaging (IF) to assess HER2 protein levels (Figure 5A), the Quantigene RNA assay for HER2 mRNA level (Figure 5B), cleaved Caspase 3 and 7 assay for apoptotic markers (Figure 5C), and cellular ATP level assay for cell viability (Figure 5D-E). Figure 5A shows that T-siHER2-NP10C reduced HER2 levels by 81-93% compared to T-siSCR-NP10C. Figure S6A shows that the T-siHER2-NP10C were more effective than T-siHER2-NP1.8C at equivalent siRNA dose. Also, Figure S6B shows that doubling the dose of T-siHER2-NP1.8C did reduce HER2 protein levels by 79-83% in SKBR3 and HCC1954 but even that was not effective in BT474 cells. Overall, T-siHER2-NP10C demonstrated better anti-HER2 efficacy than T-siHER2-NP1.8C. Encouragingly, the T-siHER2-NP10C outperformed siHER2 with DharmaFECT in all cell lines as shown in Figure S6C.
Figure 5. HER2 knockdown by siHER2-nanoparticles and therapeutic responses.
(A) HER2 expression of three HER2+ breast cancer cells at 72 hours post treatment with siHER2 or siSCR (60 nM) on T-NP10C. (B) HER2 mRNA level (48 hours), (C) apoptotic activity (4 days) and (D) cell viability (4 days) of BT474 cells treated the same way as (A). (E) Cell viability after treatment with T-siHER2-NP10C for four days in various cell lines. All cells exposed to siRNA-nanoparticles overnight and media changed. Inset of (E) shows HER2 levels of all cells tested.
Quantitative interpretation of these results is complicated by the fact that treatment with T-siSCR-NP10C also reduced HER2 levels and killed HER2+ breast cancer cells (Figure S6D). We attribute this to the high levels of trastuzumab on the nanoparticles (3% by weight) since trastuzumab by itself is known to impact HER2 expression and cell viability independent of the siHER2.[40] Figure S6D, for example, shows that HER2 levels in BT474 were reduced by 41% with T-siSCR-NP10C and by 87% with the T-siHER2-NP10C compared to untreated controls. Likewise, Figure 5D shows that cell viability was reduced 59% by T-siSCR-NP10C and 86% by T-siHER2-NP10C. Hence, to evaluate the effect of siRNA, we compared results to the siSCR control instead of the untreated control. We also assessed T-siHER2-NP10C induced changes in HER2 mRNA levels in BT474 at 48 hours post treatment. Figure 5B shows a 44% reduction in HER2 mRNA relative to siSCR control. This compares with a 58% reduction using DharmaFECT (positive control). These results may be affected by the high cell death induced by the T-siHER2-NP10C since cells that are most strongly affected will be preferentially lost. We gained some insight into this by comparing knockdown efficiency in cells that are more resistant to trastuzumab, such as JIMT1 and HCC1954. In Figure S7, for example, we found that the mRNA reduction induced by T-siHER2-NP10C and DharmaFECT were more comparable; 69% vs. 72% in JIMT1 cells and 57% vs. 63% in HCC1954 cells.
Figure 5C shows that apoptotic activity was three-fold greater after treatment with T-siHER2-NP10C than with T-siSCR-NP10C, consistent with the reduced cell viability in Figure 5D. We also measured cell viability after treatment with T-siHER2-NP10C in a panel of HER2+ cells and HER2− cells. Figure 5E shows that treatment with T-siHER2-NP10C reduced viability more strongly in HER2+ breast cancer cells (BT474, SKBR3, HCC1954 and JIMT-1) than in HER2− breast cancer cells (MCF-7, MDA-MB-231, MDA-MB-468), HER2− breast epithelial cells (MCF-10a), HER2− human liver carcinoma cells (HepG2), and HER2− human embryonic kidney cells (HEK-293). The HER2 expression levels of these cells are included as an inset of Figure 5E.
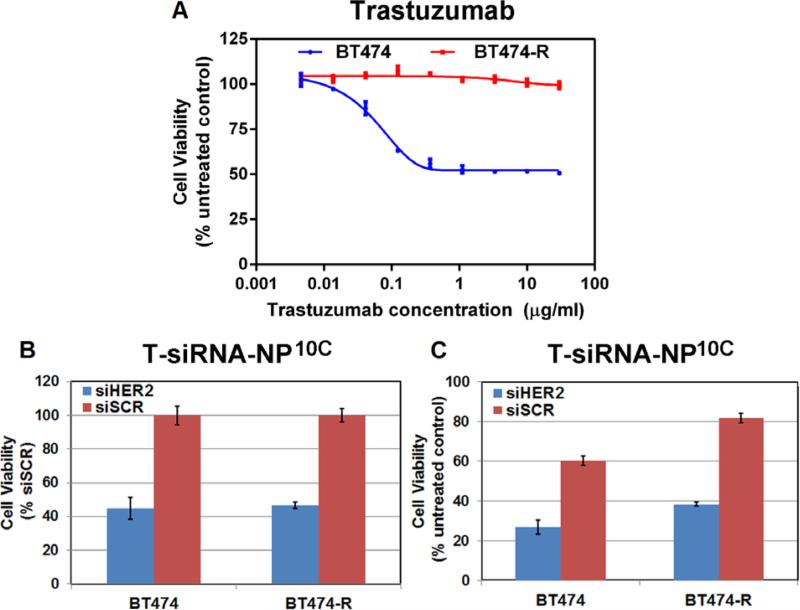
2.7. Impact of T-siHER2-NP10C on trastuzumab-resistant cells
We assessed the efficacy of T-siHER2-NP10C in the intrinsically trastuzumab-resistant HER2+ cell lines, HCC1954 and JIMT1 (Figure S2B), in the HER2+ cell line, BT474, that responds to trastuzumab and lapatinib, and in BT474-R, a derivative cell line that was made lapatinib-resistant by long-term treatment with 1 μM lapatinib. Figure 6A shows that the BT474-R cells were also less responsive to trastuzumab compared to parental BT474. However, Figure 6B shows that both cell lines were responsive to T-siHER2-NP10C compared to the T-siSCRNP10C control. This suggests a similar response to siRNA treatment in both cell lines. Meanwhile, Figure 6C shows that T-siHER2-NP10C reduced the viability of BT474 and BT474-R to 26.9% and 38.3% of the untreated control, respectively. We attribute the reduced efficacy (in terms of cell death compared to the untreated control) in BT474-R to the fact that trastuzumab on the nanoparticles does not elicit any therapeutic effect on BT474-R.
Figure 6. In vitro evaluation of siHER2-nanoparticles on HER2 silencing and ability to overcome trastuzumab resistance.
(A) Trastuzumab dose response (as 5-day cell viability) of BT474 and BT474-R (trastuzumab and lapatinib-resistant cell line derived from prolonged treatment of BT474 cells with lapatinib(1 μM)). (B-C) BT474 and BT474-R were treated with one dose of T-NP10C loaded with siHER2 or siSCR (60 nM siRNA) and cell viability was monitored at 5 days post treatment and reported as a percentage of siSCR control (B) or untreated control (C).
2.8. Blood compatibility
It is important that nanoparticle constructs intended for use systemically in vivo do not cause hemolysis, thrombogenesis, and platelet aggregation. We assessed these endpoints for T-siHER2-NP1.8C and T-siHER2-NP10C, and compared the results to those for the FDA-approved nanoparticle products: Abraxane (Paclitaxel-albumin nanoparticles) and Feraheme (iron oxide nanoparticles used as a MRI contrast agent). Nanoparticles were tested at 1X and 5X of the intended human blood level. Figure S8A shows that T-siHER2-NP1.8C and T-siHER2-NP10C did not cause hemolysis of red blood cells at either dose, while complete blood lysis was achieved with Triton-X (0.025%), the positive control. Figure S8B shows that these nanoparticles also did not affect the coagulation time of platelet poor plasma since all took about 37 s, while Feraheme prolonged the coagulation time, in agreement with its known side effects related to abnormal clotting.[41] Figure S8C shows that our nanoparticles and Abraxane did not trigger platelet aggregation while a collagen related peptide used as a positive control triggered aggregation immediately.
2.9. Immune response
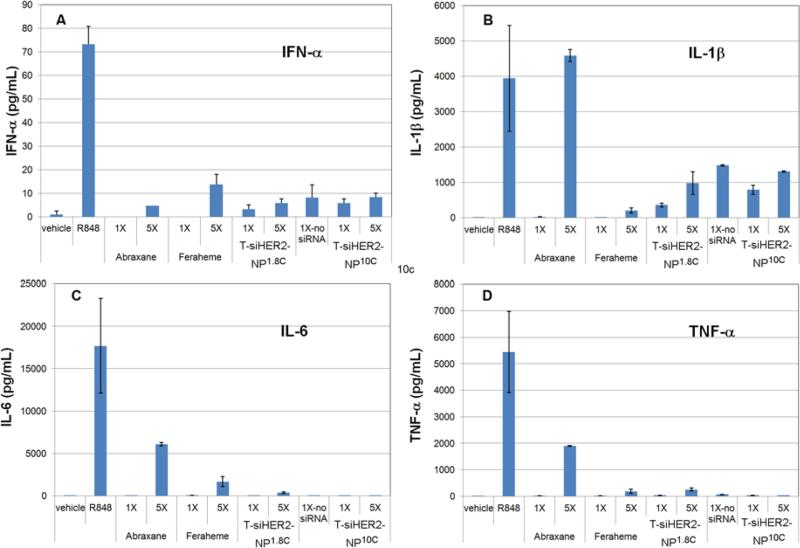
Induction of an adverse immune response is one of the major causes of failure of drug candidates during preclinical and clinical studies. Nanoparticles may elicit an inflammatory response in immune cells via toll-like receptor activation. We evaluated the effect of T-siHER2-NP1.8C and T-siHER2-NP10C on immune response by treating peripheral blood mononuclear cells (PBMCs) isolated from human blood with these nanoparticles. We measured induction of the cytokines IL-1β, IL-6, IFN-α, and TNF-α because their production is associated with Toll-like receptor activation on the surface of the cell membrane and on the endosomes.[42] PBMCs have been reported to respond to siRNA transfection with a sequence-specific TLR 7/8 dependent induction of IFN-α and TNF-α.[43] We used the TLR7/8 agonist (R848) as a direct positive control since TLR7 and TLR8 are located within the endosomes[44] where nanoparticles and siRNA are expected to reside. We compared results for our nanoparticles to those obtained for the FDA-approved nanoparticle-based drugs, Abraxane and Feraheme. Figure 7 shows that neither T-siHER2-NP1.8C nor T-siHER2-NP10C increased the levels of IL-6 and TNF-α at either the 1X or 5X level, while Abraxane significantly increased both cytokines at the 5X level. Both nanoparticles increased the levels of IFN-α and IL-1β somewhat, but not to the extent observed for Abraxane for IL-1β and Feraheme for IFN-α. The immune response was not significantly different for nanoparticles with or without siRNA, suggesting that the response was not siRNA specific. Lastly, the PBMC immunological response to T-siHER2-NP10C was not worse than T-siHER2-NP1.8C. This may be because the higher PEG content of T-siHER2-NP10C (Table 2) compensates for its higher charge of higher-molecular-weight PEI. We tested our nanoparticles for lipopolysaccharides or LPS, produced by gram-negative bacterial contamination since this might also trigger an adverse immune response. About 35% of clinically relevant nanoparticles have been found to carry this contaminant.[45] Figure S9 shows that T-siHER2-NP1.8C and T-siHER2-NP10C were not contaminated.
Figure 7. Cytokine induction in PBMCs.
24-hour exposure with various nanoparticles, T-siHER2-NP1.8C, T-NP10C (no-siRNA), T-siHER2-NP10C, Abraxane, and Feraheme. 1X = estimated human blood levels of the materials (i.e., 94 μg mL−1 for Abraxane, 102 μg mL−1 for Feraheme, and 70 μg mL−1 for the two nanoparticles), 5X = five-fold of such levels, Vehicle = PBS, R848 = TLR7/8 agonist (10 μM).
2.10. In vivo HER2 silencing efficacy and tumor growth inhibition in an orthotopic mouse tumor model
We chose the T-siHER2-NP10C (with S-47 MSNP core) over T-siHER2-NP1.8C because it yielded higher siHER2 protection, better cellular uptake, and higher gene knock-down and cell killing efficacy, without greater toxicity. Accordingly, we evaluated T-siHER2-NP10C for in vivo gene knockdown efficacy in HCC1954 xenografts grown orthotopically in mammary fad pads. Tumors were allowed to grow to 250 mm3 before treatment (n = 4 per group). We administered T-siHER2-NP10C and T-siSCR-NP10C as a single intravenous tail vein injection in order to deliver 1.25 mg siRNA kg−1 body weight. Tumors were harvested at 4 days post injection and analyzed. Figure 8A-B shows the HER2 protein levels in the HCC1954 tumors were reduced by 58.6% compared to saline control (p < 0.0013) and by 46.5% compared to treatment with the T-siSCR-NP10C control (p < 0.015). It should be noted that 22.7% (p = 0.27 vs. saline control) of the HER2 reduction in the siSCR control is likely due to trastuzumab on the nanoparticles. Similar results were obtained with orthotopic JIMT-1 tumor model as shown in Figure S10, but to a lesser extent (e.g., 38.2% HER2 reduction versus saline control (p < 0.0012)). This is perhaps due to lower level of HER2 expression in JIMT1 compared to HCC1954, resulting in reduced nanoparticle delivery and correspondingly reduced knock-down efficacy.
Figure 8. In vivo HER2 reduction and growth inhibition of orthotopic HCC1954 tumors.
(A) Representative immunofluorescent images of tumor tissues collected from mice (n = 4/group) at 4 days post i.v. injection with one dose of T-NP10C loaded with siHER2 or siSCR (1.25 mg siRNA kg−1, NP/siRNA of 50) or PBS control. (B) Quantitative HER2 levels of the tissues. Images were analyzed by CellProfiler; red = HER2 protein; green = CD31 endothelial marker; blue = DAPI staining cell nuclei. (C) Tumor growth in mice bearing orthotopic HCC1954 tumor xenografts (n = 5/group) receiving the same treatments as (A) but multiple doses (days of injection are indicated by arrows). All data are presented as means ± SEM. Specified p-values are against the saline control.
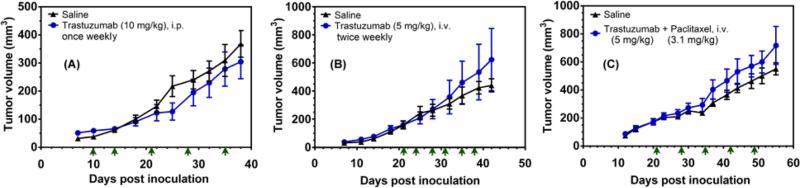
We assessed the ability of T-siHER2-NP10C (with S-47 MSNP core) to inhibit tumor growth in the same HCC1954 mouse model. Tumors were allowed to grow to ~100-150 mm3 in size prior to group randomization (n = 5 per group). Figure 8C shows that 5 intravenous tail vein injections of 1.25 mg siRNA kg−1, over a period of three weeks significantly inhibited tumor growth, while T-siSCR-NP10C produced little effect. This response is noteworthy since HCC1954 has been established as resistant to cisplatin,[46] trastuzumab,[47] and pertuzumab)[48] in vitro and/or in mice. We confirmed its resistance in vivo to trastuzumab (Figure 9A-B) and trastuzumab and paclitaxel combination (Figure 9C). We also found that T-siRNA-NP10 (with O-87 core) also inhibited ectopic HCC1954 xenograft growth (Figure S11A), while the siSCR control did not. In orthotopic tumors, however, T-siHER2-NP10C (with S-47 MSNP core) showed better efficacy than the larger particles, even at half the dose of siRNA (i.e., 1.25 mg siRNA kg−1 in Figure 8C vs. 2.5 mg siRNA kg−1 in Figure S11B).
Figure 9. Drug resistance in HCC1954 in vivo.
Tumor growth in mice bearing orthotopic HCC1954 tumor xenografts. (A) Mice (n=7/group) were injected intraperitonially with trastuzumab (10 mg kg−1) or saline. (B) Mice (n=5/group) were injected via tail vein with trastuzumab (5 mg kg−1). (C) Mice (n=9/group) were injected via tail vein with trastuzumab (5 mg kg−1) and paclitaxel (3.1 mg kg−1). Arrows indicate days of injection.
2.11 Reproducibility of nanoparticle synthesis
It is often difficult to achieve high batch-to-batch reproducibility during nanoparticle synthesis. We achieved manufacturing control of particle size and surface modification by combining inorganic and polymeric materials. Our sol-gel chemistry affords high batch-to-batch reproducibility of the MSNP core with 2.4% relative standard deviation (RSD) of particle sizes from 6 batches (Figure S12A-B). The layer-by-layer surface modification was also reproducible as shown in Figure S12A, C. Washing after each of the loading steps facilitates impurity removal. This is difficult to accomplish with one-pot synthesis methods. Based on these results, we anticipate that it will be straightforward to scale the sol-gel chemistry and the layer-by-layer modification manufacturing processes to the level needed for clinical studies.
3. Conclusion
We have produced a new generation siRNA in vivo delivery construct comprised of a MSNP core coated with PEI polymer to enhance delivery, a PEG layer to protect the siRNA and attached to an antibody for targeted delivery. We have demonstrated that our most effective construct, T-siHER2-NP10C (with S-47 core) was effectively taken up by HER2+ cells in a specific manner via antibody-receptor interaction and effectively silenced HER2 expression in vitro and in vivo. It did not appear to elicit unacceptable immune responses. This construct also produced excellent HER2 knockdown and growth inhibition of drug-resistant HER2+ tumor xenografts grown orthotopically. Manufacture of the construct is straightforward, reproducible and can be scaled to the level needed for clinical use.
HER2 oncogene/protein was chosen as the initial therapeutic target for siRNA (siHER2). Our data show that cancer resistant to current HER2 targeted therapies (e.g., trastuzumab and lapatinib) still responds well to siHER2 treatment. In addition to serving as a homing target, HER2 Ab (trastuzumab) also displayed therapeutic effect when conjugated on our nanoparticles. This provides a strong motivation to have both HER2 Ab and siHER2 on the same nanoparticle. With regard to the potential self-defeating issue that HER2 proteins might be decreased in the treated cells, rendering the targeting by HER2 Ab ineffective, we have two hypotheses: (1) not all cells within the tumors will receive T-siHER2-NP at once and hence some cells will over-express HER2 for HER2 Ab assisted delivery in subsequent doses, and (2) cells that receive sufficient T-siHER2-NP will undergo apoptosis (hence no longer needing T-siHER2-NP delivery) and the surviving cells will replenish HER2 proteins for HER2 Ab assisted delivery in subsequent doses. We found that cells that survived long term continuous treatment with siHER2, due to either not receiving sufficient siHER2 or not having siHER2 delivery at all, could replenish their HER2 level once relieved of siHER2 treatment to the same level with naïve cells. This indicates that they still can be the target for delivery by HER2 Ab. Our data also showed that long-term siHER2 treated cells remained sensitive to siHER2 treatment (same percent cell death as the parental or naïve cells). These finding will be reported in due course.
To move this technology forward to clinics, in addition to HER2, we are also actively working on finding other gene targets amenable to HER2+ breast cancer and other cancer types. We expect that this construct can be adapted to silence any gene deemed important in cancer growth. The siRNA-nanoparticle has the potential to impart both delivery specificity and therapeutic specificity via the RNAi mechanism, potentially providing a greater therapeutic index than conventional chemotherapeutics or small molecule inhibitors. In our platform, siRNA loading was accomplished last using a simple and quick mixing method. The loading employs electrostatic interaction, relying on negatively charged phosphodiester siRNA backbones. Therefore, the loading efficiency is independent of siRNA sequences. Our platform thus affords loading of different siRNAs or a cocktail of siRNAs without difficulty. In addition, since siRNA is loaded outside, our platform permits loading of drugs or imaging agents inside the pores, if needed. Co-delivery of drugs or imaging agents with siRNA has been investigated in our lab, but is outside the scope of this manuscript.
4. Experimental Section
Materials, siRNA, and cell lines
Detailed information on reagents and siRNA used is provided in the Supporting Information. Human breast cancer cell lines (BT474, JIMT1, SKBR3, HCC1954, MCF7, MDAMB231, and MDAMB468), breast epithelial cells (MCF-10a), human liver carcinoma (HepG2), and human embryonic kidney cells (HEK-293) were obtained from American Type Culture Collection. MDAMB231-H2N-luc was obtained as a gift from Prof. Robert Kerbel (University of Toronto, Canada). Cell media recipe for each cell line and siRNA sequence description are provided in the Supporting Information.
Synthesis and characterization of nanoparticles
The sol-gel synthesis of mesoporous silica nanoparticle cores (MSNPs) was modified from previous reports.[49] For 47-nm NP (S-47), CTAC (0.15 M) and TEA (350 μL) were mixed in water (125 mL) at 95 °C. Then, TEOS (3 mL) was added and the mixture was stirred for one hour. Afterwards, the pellets were recovered from suspension by centrifugation, washed with a copious amount of ethanol, and dried overnight. The particles were then resuspended and refluxed in acidic methanol (0.6 M HCl in methanol) overnight to remove CTAC and TEA. Bare MSNPs were then washed with ethanol and dried in a desiccator. TEA was also varied from 200 to 450 μL to achieve the MSNP sizes of 60 nm and 30 nm, respectively. MSNP (dry) size was measured with TEM (Phillips/FEI CM120/Biotwin TEM, Hillsboro, OR) and hydrodynamic size with Zetasizer (ZS-90/Malvern, Westborough, MA).
Non-uniform MSNPs (O-87) were synthesized by base-catalyzed synthesis. CTAB (6 mM) was dissolved in aqueous solution of pH 11.0 (240 mL, adjusted by 2 M NaOH). When the temperature stabilized at 80°C, TEOS (2.5 mL) was added. The reaction continued for 2 hours and particles were processed for surfactant removal in the same fashion as explained above.
Coating of PEI on the exterior of MSNP was carried out in ethanol by shaking MSNP (10 mg) and PEI (2.5 mg) in ethanol solution for 3 hours at room temperature. Next, PEIMSNP was cross-linked with DSP crosslinker (0.2 mg) for 40 minutes. The particles were pelleted down, washed, and resuspended in PBS (pH 7.2). For PEG loading, mal-PEG-5kDa-NHS (50 mg) was conjugated to the primary amine of MSNP-PEI (10 mg) in the PBS buffer under shaking (20 hours, RT, 300 rpm). The PEI and PEG loading was analyzed by thermogravimetric analysis (TGA Q50/ TA Instruments, New Castle, DE).
Antibody conjugation of MSNP-PEI-PEG utilized a thiol-maleimide reaction modified from literature.[50] First, antibody (trastuzumab (T) or rituximab (R)) was thiolated with Traut's reagent in PBS (pH 8.0) by 50-fold molar excess of Traut's reagent for 2 hours and then purified by Zeba spin column – MW-40,000 (Thermo Fisher Scientific, Waltham, MA). Thiolated antibody was mixed with MSNP-PEI-PEG at varied mass ratio from 0:1 to 1:1. The reaction was completed overnight at 4° C under shaking conditions (300 rpm). The material was pelleted down, resuspended in PBS, and washed with copious amount of PBS. Pierce Micro BCA kit (Thermo Fisher Scientific, Waltham, MA) was used to quantify the antibody loading of the nanoparticles. Lastly, the loading of siRNA was achieved by mixing MSNP-PEI-PEG-T (designated as T-NP) and siRNA (at nanoparticle/siRNA mass ratio of 25 or 50) in PBS solution (1 hour, room temp, 200 rpm shaking). The loading of siRNA was monitored by the fluorescent dyes tagged on siRNA.
Luciferase knockdown efficacy
The MDAMB231-H2N-luc cell line (over-expressing luciferase and HER2) was used for initial gene silencing efficacy assessment of the nanoparticles loaded with siRNA against luciferase (siLUC). Cells were plated at 3000 cells/well in a 96-well plate, maintained in corresponding cell media, as reported in Supporting Information. One day after seeding, cells were treated with siRNA-nanoparticles. The nanoparticles were loaded with siRNA at NP/siRNA ratio of 25 or 50 by mass. They were applied to each well at a fixed dose of 30 nM siRNA. The commercially available transfection agent, DharmaFECT (GE Dharmacon, Lafayette, CO), with the same siRNA dose (following manufacturer's recommended protocol) served as a positive control. After overnight incubation (~20 hours), cells were washed once and replenished with a complete media. At 48 hours post treatment, cells were lysed and analyzed for luciferase activity by Luciferase Glow Assay Kit (Thermo Fisher Scientific, Waltham, MA) and protein concentration by BCA protein assay kit (Thermo Fisher Scientific, Waltham, MA), following manufacturer's protocols. Luciferase activity of the lysate was normalized with the corresponding protein concentration in the same well and reported as a percentage of the untreated control. All treatments were performed in quadruplicate.
Buffering capacity measurement
Nanoparticles were suspended at 0.2 mg mL−1 in NaCl (150 mM, pH 9 -- pH adjusted with 0.05 M NaOH). Upon stabilization at pH 9.0, HCl (5 μL, 0.05 M) was added and solution was continuously stirred. When reaching steady state, the pH was recorded and the acid was added again. The process was repeated until the pH plateaued at around 3.0. The solution pH was then reported as a function of the amount of acid added.
Serum enzymatic protection assay
SiRNA-nanoparticles were incubated with human serum in PBS (50 v/v%) for a specified period of time (0, 0.5, 1, 2, 4, 8, 24, and 48 hours) at 37 °C under continuous shaking. At the end of each time point, the sample was mixed with proteinase K (200 μg mL−1) and frozen at -80 °C to stop the enzymatic reaction. For the analysis, samples were thawed and mixed with SDS (1.0 wt.%) in order to release siRNA from the nanoparticles. The sample was then mixed with an equal amount of 2X loading buffer and loaded into 15% TBE-urea gel (BioRad, Hercules, CA). The gel ran at 100 V for the first 20 minutes, and 150 V for another hour. The gel was then stained with SyBR Gold (Life Technologies, Eugene, OR) following manufacturer's protocol, and viewed under the UV chamber. The band intensity was analyzed by ImageJ software (NIH, Bethesda, MD). The fraction of intact siRNA was reported as a function of time that the siRNA-nanoparticles were in 50% serum.
Cellular uptake analysis by flow cytometry
We conducted the cell uptake studies in cell suspension where cells were exposed to nanoparticles in three dimensions. We also used short exposure time of 0.5 and 2 hours (rather than overnight exposure time as in routine transfection studies) since shorter time may better mimic in vivo exposure. However, the ratio of nanoparticle per cell was in the same par as in the routine transfection studies. Cells were harvested and resuspended in 1 million cells per 150 μL per tube. Each tube was mixed with 150 μL of siSCR (tagged with Alexa-488)-nanoparticles in PBS (containing 100 μg nanoparticles). Upon siSCR-nanoparticle addition, cells were placed on a rocker in the cell incubator (37 °C, 5% CO2) for 0.5 or 2.0 hours. After the specified incubation time, cells were washed (centrifuge at 115g, 5 min) with FACS buffer (1 mL, 1X Phosphate Buffered Saline (Ca/Mg++ free) + 1mM EDTA + 25mM HEPES pH 7.0 + 1% Fetal Bovine Serum (Heat-Inactivated)) three times. Cells were then resuspended in FACS buffer (550 μL) and transferred to 5-mL BD FACS tube. Cells were kept on ice until analysis. For cells stained with free antibody (for gating purpose), antibody labeling was performed on ice and under rocking conditions. Cells were stained with primary antibody (trastuzumab or rituximab: 2 μg per tube) for an hour, washed (centrifuge at 115g, 5 min) with PBS one time, stained with secondary antibody (Anti-human Alexa 488: 2 μg per tube) for 45 minutes, then washed 2 times with PBS, and resuspended in FACS buffer (550 μL) before analysis. All tubes were counter-stained for cellular DNA with DRAQ5 (2 μL, 5 mM) (Cell Signaling, Danvers, MA) for 15 minutes on ice. Then all tubes (except antibody-labeled cells for gating purpose) were incubated with Trypan Blue (500 μL, 0.4% in PBS) to quench fluorescence outside of the cells, and subjected to flow cytometry analysis. 10,000 events (cells) were analyzed for each sample. The intensity was processed with FlowJo software (FlowJo LLC, Ashland, OR).
In vitro efficacy: HER2 protein knockdown and cell viability
BT474, SKBR3 and HCC1954 were seeded in a 96-well plate for 24 hours prior to treatment. Nanoparticles were loaded with siHER2 or siSCR at NP/siRNA 50. siRNA dose was fixed at 60 nM. Media were switched to complete media after overnight incubation. Three days after treatment with siRNA-nanoparticles, cells were fixed and analyzed for HER2 protein expression by immunofluorescence method (see details in the Supplemental Materials). HER2 mRNA and β-actin mRNA levels were analyzed at 48 hours post treatment using Quantigene 2.0 Reagent System (Affymetrix Panomics, Santa Clara, CA) following the manufacturer's protocol. The HER2 mRNA level was then normalized with β-actin mRNA (housekeeping gene) and reported as the percentage of the untreated control. Cell viability and apoptosis were analyzed four days post treatment using CellTiter-Glo® Luminescent Assay (Promega, Madison, WI) and Caspase-Glo® 3/7 Assay Systems (Promega, Madison, WI), respectively. Caspase activity was normalized with the cell viability. Both were reported as a percentage of the untreated control.
Blood compatibility--hemolysis, coagulation, and platelet aggregation
Studies of blood compatibility were performed following or with minor modification from the Nanotechnology Characterization Laboratory (NCL)'s published protocols. The detailed protocols can be found in Supplemental Methods.
Immune response--peripheral blood mononuclear cells (PBMCs) cytokine release assay
A PBMC cytokine release assay was conducted according to the recommendations and method by the Nanotechnology Characterization Lab (NCL) of NCI for immunological studies of nanoparticles. The in vitro cell based assay evaluated cytokine production by PBMCs (200,000 cells/well) following a 24-hour exposure to the test materials. Test materials included nanoparticles with and without siHER2 to investigate the potential impact of siRNA mediated immune response. Following incubation, cell culture supernatants were collected and analyzed for IL-1β, IL-6, IFN-α, and TNF-α by a cytometry bead array (Milliplex Magnetic Bead/ Millipore, Billerica, MA) following manufacturer's protocol. Abraxane (Celgene, NJ) and Feraheme (AMAG Pharmaceuticals, MA) were used as FDA approved nanoparticle based drug benchmarks since there is no siRNA based nanoparticle drug in the market.
Animal studies--mouse tumor models and in vivo efficacy studies
All animals were recruited and used under an approved protocol of the Institutional Animal Care and Use Committee (IACUC) of Oregon Health and Science University. All animal experiments were carried out under the auspices of the OHSU Department of Comparative Medicine. In vivo gene silencing studies were performed in orthotopic mouse tumor models; HCC1954 cells (4 x 106) or JIMT-1 cells (1 x 106) unless otherwise specified were implanted into the mammary fat pads of 5-week-old SCID mice (Charles River, Wilmington, MA) and allowed to grow to an average size of ~250 and ~150 mm3, respectively. Mice were then grouped and proceeded to receive a single injection (tail vein) of the nanoparticles (T-siHER2-NP10C or T-siSCR-NP10C, S-47 core, 1.25 mg siRNA kg−1), or the PBS control. The tumors were harvested four days after treatment and analyzed for HER2 protein expression by immunofluorescence (Supplemental Methods). Subsequent study on tumor growth reduction study was also performed with the orthotopic HCC1954 tumor model. Tumor were allowed to grow to 150 mm3 prior to treatments (by tail vein injections) with siHER2 or siSCR (1.25 mg siRNA kg−1 body weight) delivered with T-NP10C (S-47 core) over a period of three weeks.
Separate tumor growth studies were performed with free trastuzumab treatments as follows (A) Mice (n=7/group) were injected intraperitonially with trastuzumab (10 mg kg−1) or saline. (B) Mice (n=5/group) were injected via tail vein with trastuzumab (5 mg kg−1) or saline. (C) Mice (n=9/group) were injected via tail vein with trastuzumab (5 mg kg−1) + paclitaxel (3.1 mg kg−1) or saline. Days of injection and doses were specified in Figure 9.
Statistical analysis
Pairwise statistical comparisons were performed using unpaired, two-tailed Student's t test. Tumor growth curve was analyzed with two-way ANOVA with post-hoc Dunnett's multiple comparison test against the saline group. Statistical significance was established at p ≤ 0.05. Graphpad Prism 6.0 software (GraphPad software Inc., San Diego, CA) was utilized for statistical analyses.
Supplementary Material
Acknowledgements
This work was funded by NCI/SBIR (Contract No. HHSN261201300078C), NIH/NIGMS (Grant No. R01GM089918), NIH/NIDDK (Grant No. R41DK094571), NIH/NCI (Grant No. U54CA112970), the Prospect Creek Foundation, OHSU's KCI pilot award, and OHSU's VPR fund. The authors are grateful to Prof. Rosalie Sears of OHSU for independent reviewing of the data in this paper, Dr. Lev Federov of OHSU's transgenic core for his expertise on mouse tumor models, Dr. Robert Kerbel of University of Toronto for breast cancer cell lines, Dr. Josephrajan Thainashmuthu for TEM imaging and FT-IR analysis, and Ethan Muldoon for his contribution to Figure S3A, Supporting Information. OHSU, W.N., D.J.C., J.W.G., and W.Y. have a significant financial interest in PDX Pharmaceuticals, LLC, a company that may have a commercial interest in the results of this research and technology. This potential personal and institutional conflict of interest has been reviewed and managed by OHSU.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Worapol Ngamcherdtrakul, Department of Biomedical Engineering, Oregon Health & Science University 3303 SW Bond Ave, Portland, OR 97239..
Jingga Morry, Department of Biomedical Engineering, Oregon Health & Science University 3303 SW Bond Ave, Portland, OR 97239..
Shenda Gu, Department of Biomedical Engineering, Oregon Health & Science University 3303 SW Bond Ave, Portland, OR 97239..
Dr. David J. Castro, Department of Biomedical Engineering, Oregon Health & Science University 3303 SW Bond Ave, Portland, OR 97239. PDX Pharmaceuticals 24 Independence Ave, Lake Oswego, OR 97035
Dr. Shaun M. Goodyear, Department of Biomedical Engineering, Oregon Health & Science University 3303 SW Bond Ave, Portland, OR 97239.
Thanapon Sangvanich, Department of Biomedical Engineering, Oregon Health & Science University 3303 SW Bond Ave, Portland, OR 97239..
Moataz M. Reda, Department of Biomedical Engineering, Oregon Health & Science University 3303 SW Bond Ave, Portland, OR 97239.
Richard Lee, Department of Biomedical Engineering, Oregon Health & Science University 3303 SW Bond Ave, Portland, OR 97239..
Samuel A. Mihelic, Department of Biomedical Engineering, Oregon Health & Science University 3303 SW Bond Ave, Portland, OR 97239.
Brandon L. Beckman, Department of Biomedical Engineering, Oregon Health & Science University 3303 SW Bond Ave, Portland, OR 97239.
Dr. Zhi Hu, Department of Biomedical Engineering, Oregon Health & Science University 3303 SW Bond Ave, Portland, OR 97239.
Prof. Joe W. Gray, Department of Biomedical Engineering, Oregon Health & Science University 3303 SW Bond Ave, Portland, OR 97239..
Prof. Wassana Yantasee, Department of Biomedical Engineering, Oregon Health & Science University 3303 SW Bond Ave, Portland, OR 97239.; PDX Pharmaceuticals 24 Independence Ave, Lake Oswego, OR 97035
References
- 1.a Cancer Genome Atlas Network Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Hanahan D, Weinberg Robert A. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.a Singel SM, Cornelius C, Batten K, Fasciani G, Wright WE, Lum L, Shay JW. Clinical Cancer Research. 2013;19:2061–2070. doi: 10.1158/1078-0432.CCR-13-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Bauer JA, Ye F, Marshall CB, Lehmann BD, Pendleton CS, Shyr Y, Arteaga CL, Pietenpol JA. Breast cancer research : BCR. 2010;12:R41. doi: 10.1186/bcr2595. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Ashworth A, Bernards R. Cold Spring Harbor perspectives in biology. 2010;2:a003327. doi: 10.1101/cshperspect.a003327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hopkins AL, Groom CR. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 4.Pecot CV, Calin GA, Coleman RL, Lopez-Berestein G, Sood AK. Nature reviews. Cancer. 2011;11:59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitehead KA, Langer R, Anderson DG. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo S, Huang L. Journal of Nanomaterials. 2011;2011:12. [Google Scholar]
- 7.a Kanasty R, Dorkin JR, Vegas A, Anderson D. Nat Mater. 2013;12:967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]; b Haussecker D. Mol Ther Nucleic Acids. 2012;1:e8. doi: 10.1038/mtna.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a Argyo C, Weiss V, Bräuchle C, Bein T. Chemistry of Materials. 2013;26:435–451. [Google Scholar]; b Tang F, Li L, Chen D. Advanced Materials. 2012;24:1504–1534. doi: 10.1002/adma.201104763. [DOI] [PubMed] [Google Scholar]
- 9.a Tabernero J, Shapiro GI, LoRusso PM, Cervantes A, Schwartz GK, Weiss GJ, Paz-Ares L, Cho DC, Infante JR, Alsina M, Gounder MM, Falzone R, Harrop J, White ACS, Toudjarska I, Bumcrot D, Meyers RE, Hinkle G, Svrzikapa N, Hutabarat RM, Clausen VA, Cehelsky J, Nochur SV, Gamba-Vitalo C, Vaishnaw AK, Sah DWY, Gollob JA, Burris HA. Cancer Discovery. 2013;3:406–417. doi: 10.1158/2159-8290.CD-12-0429. [DOI] [PubMed] [Google Scholar]; b Ramanathan RK, Hamburg SI, Borad MJ, Seetharam M, Kundranda MN, Lee P, Fredlund P, Gilbert M, Mast C, Semple SC, Judge AD, Crowell B, Vocila L, MacLachlan I, Northfelt DW. AACR 104th Annual Meeting 2013. Washington, DC: 2013. [Google Scholar]
- 10.Rudin CM, Marshall JL, Huang CH, Kindler HL, Zhang C, Kumar D, Gokhale PC, Steinberg J, Wanaski S, Kasid UN, Ratain MJ. Clinical Cancer Research. 2004;10:7244–7251. doi: 10.1158/1078-0432.CCR-04-0642. [DOI] [PubMed] [Google Scholar]
- 11.Fein DE, Bucki R, Byfield F, Leszczynska K, Janmey PA, Diamond SL. Molecular Pharmacology. 2010;78:402–410. doi: 10.1124/mol.110.066670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a Davis ME, Zuckerman JE, Choi CHJ, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Davis ME. Molecular pharmaceutics. 2009;6:659–668. doi: 10.1021/mp900015y. [DOI] [PubMed] [Google Scholar]
- 13.Zuckerman JE, Choi CHJ, Han H, Davis ME. Proceedings of the National Academy of Sciences. 2012;109:3137–3142. doi: 10.1073/pnas.1200718109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heidel JD, Yu Z, Liu JY-C, Rele SM, Liang Y, Zeidan RK, Kornbrust DJ, Davis ME. Proceedings of the National Academy of Sciences. 2007;104:5715–5721. doi: 10.1073/pnas.0701458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a Ren Y, Cheung HW, von Maltzhan G, Agrawal A, Cowley GS, Weir BA, Boehm JS, Tamayo P, Karst AM, Liu JF, Hirsch MS, Mesirov JP, Drapkin R, Root DE, Lo J, Fogal V, Ruoslahti E, Hahn WC, Bhatia SN. Science Translational Medicine. 2012:4. doi: 10.1126/scitranslmed.3003778. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Yao Y.-d., Sun T.-m., Huang S.-y., Dou S, Lin L, Chen J.-n., Ruan J.-b., Mao C.-q., Yu F.-y., Zeng M.-s., Zang J.-y., Liu Q, Su F.-x., Zhang P, Lieberman J, Wang J, Song E. Science Translational Medicine. 2012;4 doi: 10.1126/scitranslmed.3003601. [DOI] [PubMed] [Google Scholar]
- 16.Jensen SA, Day ES, Ko CH, Hurley LA, Luciano JP, Kouri FM, Merkel TJ, Luthi AJ, Patel PC, Cutler JI, Daniel WL, Scott AW, Rotz MW, Meade TJ, Giljohann DA, Mirkin CA, Stegh AH. Science Translational Medicine. 2013:5. doi: 10.1126/scitranslmed.3006839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.a Li X, Chen Y, Wang M, Ma Y, Xia W, Gu H. Biomaterials. 2013;34:1391–1401. doi: 10.1016/j.biomaterials.2012.10.072. [DOI] [PubMed] [Google Scholar]; b Meng H, Mai WX, Zhang H, Xue M, Xia T, Lin S, Wang X, Zhao Y, Ji Z, Zink JI, Nel AE. ACS Nano. 2013;7:994–1005. doi: 10.1021/nn3044066. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Shen J, Kim HC, Su H, Wang F, Wolfram J, Kirui D, Mai J, Mu C, Ji LN, Mao ZW, Shen H. Theranostics. 2014;4:487–497. doi: 10.7150/thno.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Lin D, Cheng Q, Jiang Q, Huang Y, Yang Z, Han S, Zhao Y, Guo S, Liang Z, Dong A. Nanoscale. 2013;5:4291–4301. doi: 10.1039/c3nr00294b. [DOI] [PubMed] [Google Scholar]
- 18.Maeda H, Wu J, Sawa T, Matsumura Y, Hori K. Journal of controlled release : official journal of the Controlled Release Society. 2000;65:271–284. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- 19.Roberts MJ, Bentley MD, Harris JM. Advanced drug delivery reviews. 2002;54:459–476. doi: 10.1016/s0169-409x(02)00022-4. [DOI] [PubMed] [Google Scholar]
- 20.Chung TH, Wu SH, Yao M, Lu CW, Lin YS, Hung Y, Mou CY, Chen YC, Huang DM. Biomaterials. 2007;28:2959–2966. doi: 10.1016/j.biomaterials.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 21.Murphy CG, Modi S. Biologics : targets & therapy. 2009;3:289–301. [PMC free article] [PubMed] [Google Scholar]
- 22.a Wang Y-C, Morrison G, Gillihan R, Guo J, Ward R, Fu X, Botero M, Healy N, Hilsenbeck S, Phillips G, Chamness G, Rimawi M, Osborne CK, Schiff R. Breast Cancer Research. 2011;13:R121. doi: 10.1186/bcr3067. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Ritter CA, Perez-Torres M, Rinehart C, Guix M, Dugger T, Engelman JA, Arteaga CL. Clinical Cancer Research. 2007;13:4909–4919. doi: 10.1158/1078-0432.CCR-07-0701. [DOI] [PubMed] [Google Scholar]; c Cooke T, Reeves J, Lanigan A, Stanton P. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2001;12(Suppl 1):S23–28. doi: 10.1093/annonc/12.suppl_1.s23. [DOI] [PubMed] [Google Scholar]
- 23.a Ahmad S, Gupta S, Kumar R, Varshney GC, Raghava GPS. Sci. Rep. 2014:4. doi: 10.1038/srep04483. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Nahta R, Esteva FJ. Breast cancer research : BCR. 2006;8:215. doi: 10.1186/bcr1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoeferlin LA, Park ECC,MA. Journal of surgery and science. 2013;1:3–7. [PMC free article] [PubMed] [Google Scholar]
- 25.a Barok M, Joensuu H, Isola J. Breast Cancer Research. 2014;16:1–12. doi: 10.1186/bcr3621. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Seyhan AA, Varadarajan U, Choe S, Liu W, Ryan TE. Molecular bioSystems. 2012;8:1553–1570. doi: 10.1039/c2mb05512k. [DOI] [PubMed] [Google Scholar]
- 26.a Allen TM, Cullis PR. Science. 2004;303:1818–1822. doi: 10.1126/science.1095833. [DOI] [PubMed] [Google Scholar]; b Kobayashi H, Watanabe R, Choyke PL. Theranostics. 2013;4:81–89. doi: 10.7150/thno.7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petros RA, DeSimone JM. Nat Rev Drug Discov. 2010;9:615–627. doi: 10.1038/nrd2591. [DOI] [PubMed] [Google Scholar]
- 28.Lu J, Liong M, Sherman S, Xia T, Kovochich M, Nel AE, Zink JI, Tamanoi F. Nanobiotechnology : the journal at the intersection of nanotechnology, molecular biology, and biomedical sciences. 2007;3:89–95. doi: 10.1007/s12030-008-9003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. Proceedings of the National Academy of Sciences. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.a Bieber T, Meissner W, Kostin S, Niemann A, Elsasser HP. Journal of controlled release : official journal of the Controlled Release Society. 2002;82:441–454. doi: 10.1016/s0168-3659(02)00129-3. [DOI] [PubMed] [Google Scholar]; b Kunath K, von Harpe A, Fischer D, Petersen H, Bickel U, Voigt K, Kissel T. Journal of controlled release : official journal of the Controlled Release Society. 2003;89:113–125. doi: 10.1016/s0168-3659(03)00076-2. [DOI] [PubMed] [Google Scholar]; c Breunig M, Lungwitz U, Liebl R, Fontanari C, Klar J, Kurtz A, Blunk T, Goepferich A. The journal of gene medicine. 2005;7:1287–1298. doi: 10.1002/jgm.775. [DOI] [PubMed] [Google Scholar]
- 31.Behr J. Chimia. 1997;51:34–36. [Google Scholar]
- 32.Eliyahu H, Barenholz Y, Domb AJ. Molecules (Basel, Switzerland) 2005;10:34–64. doi: 10.3390/10010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.a Ketola T-M, Hanzlíková M, Leppänen L, Raviña M, Bishop CJ, Green JJ, Urtti A, Lemmetyinen H, Yliperttula M, Vuorimaa-Laukkanen E. The Journal of Physical Chemistry B. 2013;117:10405–10413. doi: 10.1021/jp404812a. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Aravindan L, Bicknell KA, Brooks G, Khutoryanskiy VV, Williams AC. Macromolecular bioscience. 2013;13:1163–1173. doi: 10.1002/mabi.201300103. [DOI] [PubMed] [Google Scholar]
- 34.du Manoir JM, Francia G, Man S, Mossoba M, Medin JA, Viloria-Petit A, Hicklin DJ, Emmenegger U, Kerbel RS. Clinical Cancer Research. 2006;12:904–916. doi: 10.1158/1078-0432.CCR-05-1109. [DOI] [PubMed] [Google Scholar]
- 35.a Mantei A, Rutz S, Janke M, Kirchhoff D, Jung U, Patzel V, Vogel U, Rudel T, Andreou I, Weber M, Scheffold A. European journal of immunology. 2008;38:2616–2625. doi: 10.1002/eji.200738075. [DOI] [PubMed] [Google Scholar]; b Dykxhoorn DM, Palliser D, Lieberman J. Gene therapy. 2006;13:541–552. doi: 10.1038/sj.gt.3302703. [DOI] [PubMed] [Google Scholar]
- 36.Bartlett DW, Su H, Hildebrandt IJ, Weber WA, Davis ME. Proceedings of the National Academy of Sciences. 2007;104:15549–15554. doi: 10.1073/pnas.0707461104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.a Zhang Z, Berns AE, Willbold S, Buitenhuis J. Journal of colloid and interface science. 2007;310:446–455. doi: 10.1016/j.jcis.2007.02.024. [DOI] [PubMed] [Google Scholar]; b Gref R, Luck M, Quellec P, Marchand M, Dellacherie E, Harnisch S, Blunk T, Muller RH. Colloids and surfaces. B, Biointerfaces. 2000;18:301–313. doi: 10.1016/s0927-7765(99)00156-3. [DOI] [PubMed] [Google Scholar]
- 38.Mao S, Neu M, Germershaus O, Merkel O, Sitterberg J, Bakowsky U, Kissel T. Bioconjugate chemistry. 2006;17:1209–1218. doi: 10.1021/bc060129j. [DOI] [PubMed] [Google Scholar]
- 39.Zhang H, Xia T, Meng H, Xue M, George S, Ji Z, Wang X, Liu R, Wang M, France B, Rallo R, Damoiseaux R, Cohen Y, Bradley KA, Zink JI, Nel AE. ACS Nano. 2011;5:2756–2769. doi: 10.1021/nn200328m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nahta R. ISRN oncology. 2012;2012:428062. doi: 10.5402/2012/428062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwenk MH. Pharmacotherapy. 2010;30:70–79. doi: 10.1592/phco.30.1.70. [DOI] [PubMed] [Google Scholar]
- 42.Takeda K, Akira S. International Immunology. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 43.a Broering R, Real CI, John MJ, Jahn-Hofmann K, Ickenstein LM, Kleinehr K, Paul A, Gibbert K, Dittmer U, Gerken G, Schlaak JF. International Immunology. 2014;26:35–46. doi: 10.1093/intimm/dxt040. [DOI] [PubMed] [Google Scholar]; b Zamanian-Daryoush M, Marques JT, Gantier MP, Behlke MA, John M, Rayman P, Finke J, Williams BR. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research. 2008;28:221–233. doi: 10.1089/jir.2007.0090. [DOI] [PubMed] [Google Scholar]
- 44.Chaturvedi A, Pierce SK. Traffic (Copenhagen, Denmark) 2009;10:621–628. doi: 10.1111/j.1600-0854.2009.00899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crist RM, Grossman JH, Patri AK, Stern ST, Dobrovolskaia MA, Adiseshaiah PP, Clogston JD, McNeil SE. Integrative biology : quantitative biosciences from nano to macro. 2013;5:66–73. doi: 10.1039/c2ib20117h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beyer I, Cao H, Persson J, Song H, Richter M, Feng Q, Yumul R, van Rensburg R, Li Z, Berenson R, Carter D, Roffler S, Drescher C, Lieber A. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:3340–3351. doi: 10.1158/1078-0432.CCR-11-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beyer I, Li Z, Persson J, Liu Y, van Rensburg R, Yumul R, Zhang XB, Hung MC, Lieber A. Mol Ther. 2011;19:479–489. doi: 10.1038/mt.2010.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henjes F, Bender C, von der Heyde S, Braun L, Mannsperger HA, Schmidt C, Wiemann S, Hasmann M, Aulmann S, Beissbarth T, Korf U. Oncogenesis. 2012;1:e16. doi: 10.1038/oncsis.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.a Pan L, He Q, Liu J, Chen Y, Ma M, Zhang L, Shi J. Journal of the American Chemical Society. 2012;134:5722–5725. doi: 10.1021/ja211035w. [DOI] [PubMed] [Google Scholar]; b Slowing II, Trewyn BG, Giri S, Lin VSY. Advanced Functional Materials. 2007;17:1225–1236. [Google Scholar]
- 50.a Yousefpour P, Atyabi F, Vasheghani-Farahani E, Movahedi AA, Dinarvand R. International journal of nanomedicine. 2011;6:1977–1990. doi: 10.2147/IJN.S21523. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Koopaei MN, Dinarvand R, Amini M, Rabbani H, Emami S, Ostad SN, Atyabi F. International journal of nanomedicine. 2011;6:1903–1912. doi: 10.2147/IJN.S23211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.a Slowing II, Wu C-W, Vivero-Escoto JL, Lin VSY. Small. 2009;5:57–62. doi: 10.1002/smll.200800926. [DOI] [PubMed] [Google Scholar]; b Choi J, Reipa V, Hitchins VM, Goering PL, Malinauskas RA. Toxicological sciences : an official journal of the Society of Toxicology. 2011;123:133–143. doi: 10.1093/toxsci/kfr149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.