Abstract
This paper presents a novel method for Glioblastoma (GBM) feature extraction based on Gaussian mixture model (GMM) features using MRI. We addressed the task of the new features to identify GBM using T1 and T2 weighted images (T1-WI, T2-WI) and Fluid-Attenuated Inversion Recovery (FLAIR) MR images. A pathologic area was detected using multithresholding segmentation with morphological operations of MR images. Multiclassifier techniques were considered to evaluate the performance of the feature based scheme in terms of its capability to discriminate GBM and normal tissue. GMM features demonstrated the best performance by the comparative study using principal component analysis (PCA) and wavelet based features. For the T1-WI, the accuracy performance was 97.05% (AUC = 92.73%) with 0.00% missed detection and 2.95% false alarm. In the T2-WI, the same accuracy (97.05%, AUC = 91.70%) value was achieved with 2.95% missed detection and 0.00% false alarm. In FLAIR mode the accuracy decreased to 94.11% (AUC = 95.85%) with 0.00% missed detection and 5.89% false alarm. These experimental results are promising to enhance the characteristics of heterogeneity and hence early treatment of GBM.
1. Introduction
Providing quantitative and accurate information for medical diagnosis, Magnetic Resonance Imaging (MRI) plays an essential role in medical imaging [1]. MRI has several advantages over other medical imaging techniques regarding its multiple applications, namely, for cardiovascular, musculoskeletal, and, in particular, for imaging of the brain and neurological systems [2, 3]. However, a bottleneck of MR image processing arises from variations in intensity due to B1 and B0 field inhomogeneity [4, 5]. This is manifested by the nonuniform appearance even of a single tissue which may mislead image analysis algorithms, which enhance abnormality area detection by a segmentation model [2, 6].
In the last decade, MR imaging established itself as key imaging modality in diagnosis and follow-up of brain tumors including Glioblastoma (GBM) [7]. GBM is the most common primary malignant brain tumor in adults [8]. It is characterized by abnormal and uncontrolled cell proliferation, necrosis, and vascular proliferation [9]. Despite the ongoing research and clinical trials, GBM remains one of the most aggressive malignant tumors with less than 5% of patients surviving five years after diagnosis [10]. This is attributed to the highly infiltrative nature and the heterogeneity that Glioblastoma exhibits on molecular and genomic levels which lead to differences in individual treatment response and prognosis [11].
Accordingly, research has focused on exploring associations between certain imaging features and the underlying genomic profiles of GBM in a new branch in clinical radiology known as “imaging genomics” [12, 13]. Using a GMM, Simon et al. recently showed that delineation and quantification of apparent diffusion coefficient in gliomas can be performed reliably and fast and demonstrated how thereby user-dependent variability can automatically be removed [14]. Consequently, recent work has focused on developing robust methods for reading and imaging features extraction from such MR images.
Automatic reading algorithms can foster faster and more precise readings of MR images as well as segmenting the abnormal imaging areas to classify them as GBM or not. Robust reading of MR images includes several consecutive steps. The system must first segment the image by detecting and extracting the abnormal area from their surrounding medium using multithresholding segmentation and morphological image processing. This step requires careful selection of the appropriate segmentation methodology for processing of high resolution grayscale MR images. While several segmentation methods based on MR images have been proposed using filtering to remove noise, these techniques are not generally applicable to automated detection of GBM as the tumor can be unintentionally eliminated during the process of noise reduction. Segmentation methods based on thresholding or multithresholding are thus preferred.
That way, it is likely that GBM and the normal brain tissue “survive” the thresholding. This method divides an image into several regions using multithresholding [15–18]. The second step following the detection of area of imaging abnormality, representing GBM, involves extraction of some characteristic parameters and texture features that are specific for GBM [19–26]. Plurals based on the texture features were proposed where the visual analysis of texture is a difficult task, particularly with GBM. The texture analysis based on gray level cooccurrence matrix (GLCM) determines neighborhoods around pixels (texture elements) where the GLCM is counted using the specific offset and phases [27]. Also, shape and texture feature were used to classify the brain tumor type and grade using SVM model; however the classifier accuracy was limited by 85% [28].
Moreover, the feature quality is essential to improve the classifier accuracy and accordingly the applications. For example, wavelet based classification has proven to be a powerful technique [29–31]. However, due to its comparatively low classification accuracy this approach was not promising to follow in our MRI data. We therefore aimed to investigate GBM tumor features that may have the potential to measure specific GBM characteristics. To achieve this, we focused on features derived from Gaussian mixture model (GMM) analysis on both weighted T1 and T2 and FLAIR sequences.
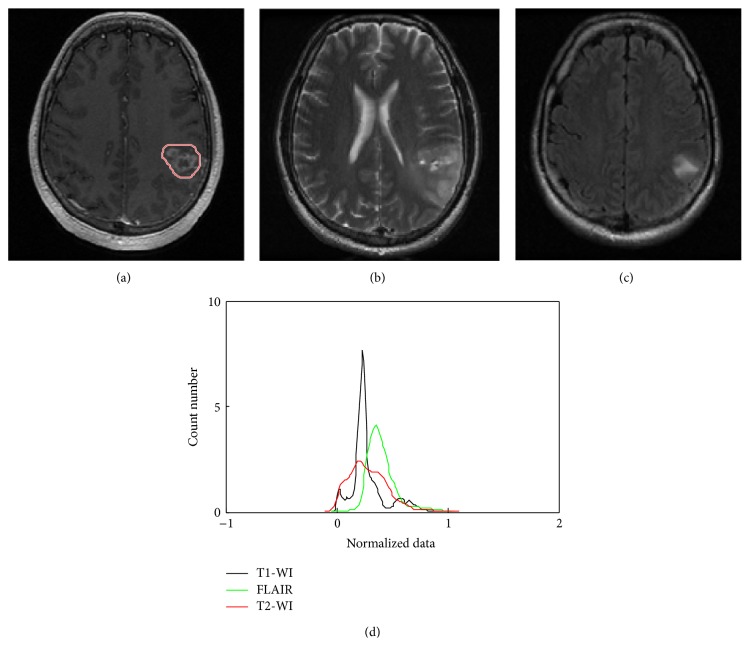
Figure 1 shows 2D axial image of brain within GBM region indicated by the red line in Figure 1(a). Clearly, GBM area has higher intensity on the grayscale level brain MR image, but some pixels of normal brain share the same intensity values as the GBM pixels. These pixels closely resemble the GBM pixels in terms of their intensity, rendering GBM detection a difficult task. Also, histogram of GBM area is not similar in the three MRI sequences.
Figure 1.
Analysis of GBM schema: (a) brain tumor image on axial T1-WI, (b) axial T2-WI, (c) axial FLAIR sequence, and (d) GBM data fitting in three MR sequences.
2. Materials and Methods
The schematic of the proposed method is shown in Figure 2: (1) preprocessing to normalize grayscales and filtering to remove the noise from images in the three MRI sequences, T1-WI, T2-WI, and FLAIR; (2) tumor (GBM) areas detection by multithresholding segmentation and normal areas determined from the normal brain material; (3) feature extraction from the GMM curve fitting of the grayscale histogram on T1-WI, T2-WI, and FLAIR images; (4) applying three classifier techniques to discriminate between the tumor areas and normal areas based on GMM features; and (5) validating the effect of GMM features by comparative study with PCA and wavelet features. The details of the schematic are given below.
Figure 2.

Schematic diagram of the proposed method for automatic feature extraction.
2.1. Data Acquisition
A data set of 17 patients was collected by November 2013 from the publicly available Cancer Imaging Archive (http://www.cancerimagingarchive.net/) database for our preliminary study. We excluded patients with incomplete imaging data set. All of the images had 512 × 512 pixels acquisition matrices and were converted into grayscale before further processing. MRI raw data were filtered to remove noise and standardized by the linear normalization, followed by multithreshold-based segmentation. This technique was applied to determine the tumor position and was successfully applied for the GBM data collection process. Note that preprocessing of skull stripping is required; it is necessary to obtain only the brain material without the skull bone; however, multithresholding segmentation with morphological operation filter may be detecting GBM area in two-dimension axial image. In this context, automated operation can be a difficult task if the GBM area is smaller than skull thickness.
2.2. Multithresholding Segmentation
Single threshold segmentation for GBM region pixels may resemble normal brain pixels. Segmentation based on multithresholding resolved this problem. Accordingly, we carried out an initial estimation of GBM localization by using multithresholding (multilevel image thresholds) segmentation method proposed by Otsu [32]. This approach enabled the definition of thresholds that maximize the interclass variances, thus also minimizing the intraclass variances. It can offer multilevel image thresholds in order to segment the desired object (brain tumor). In our case, we adjusted the multithresholding of an image for skull stripping and tumor detection. In order to robustly detect GBM, we had to resolve the problem arising from resembling pixels spots. This could be easily resolved by median filter depending on the window size.
2.3. GMM Feature Extracted
Many GMM had previously been considered in the literature for face identification [27], which was found to offer the best trade-off in terms of complexity, robustness, and discrimination. It has also been used for voice identification based on the feature and score normalization techniques [33, 34]. Also, GMM based features show promise accuracy classifier to distinguish between target and ghost/clutter regions [35].
GMM is a parametric probability density function represented as a weighted sum of K Gaussian component densities according to
| (1) |
where x represented the N dimensional continuous valued data vector, w i the mixture weight, and g the component Gaussian densities.
Each component density was controlled by the N-variate Gaussian function according to
| (2) |
where μ i was the average of a vector, T is the transpose, and Σi is the covariance matrix.
The complete Gaussian mixture model was parameterized by the mean vectors, covariance matrices, and mixture weights from all component densities. These parameters can be expressed according to
| (3) |
The variance (v i) of components was represented by the diagonal of the covariance matrix Σi. We extracted then a feature vector R from three components of GMM according to
| (4) |
where w, µ, and v are the weight, average, and variance of GMM components (indexes 1, 2, and 3 are the first, second, and third component of GMM).
Each segmented GBM area could be represented by the feature vector R GBM that is of size 1 by 9 elements. A similar feature vector size (R N) for normal area was computed.
For n GBM areas, we had Rt GBM matrix that was of size n by 9 elements, meaning n samples. Similar matrix for the area of normal brain was Rt N. When computing the matrixes Rt GBM and Rt N, the classification operation became ready.
2.4. Principal Component Analysis Applied on GMM Features
In the following, we present a principal component analysis technique to reduce the data and to get the appropriate feature from each vector feature.
Each feature vector of GBM and of normal brain was extracted from several Gaussian distributions which were represented by the average, standard deviation, and weight. Concatenating the parameters of GMM, this technique could show the correlation between the features extracted. Further, it could have been a good factor classifier to distinguish between GBM and normal brain tissue. Two matrixes (Rt GBM) and (Rt N) of n GBM and normal samples were n by 9, where each feature row concerns 9 elements. GBM and normal area samples of n = 17 patients were arranged into data matrixes Rt GBM and Rt N according to
| (5) |
where [R 1G ⋯ R nG] and [R 1N ⋯ R nN] were the GMM features of GBM and normal area, respectively.
Training data were received by Rt GBM and Rt N. PCA was employed, where the covariance of Rt GBM and Rt N was computed. The covariance matrix could be found according to
| (6) |
where cov was the covariance. C GBM and C N are the same size 9 by 9.
According to the following equation, the eigenvalues and eigenvectors could be computed according to
| (7) |
where V was the matrix of principal component and each column in V was an eigenvector. Λ was the diagonal matrix where the diagonal elements were the values of the eigenvalues.
We organized the eigenvectors by their corresponding eigenvalues and we retained three eigenvectors as the PCs of the data from C GBM and C N, respectively, where the higher variance represented the three largest eigenvalues. Figure 3 shows the variance of eigenvalues of three MRI modes. Clearly, the maximum variances common between T1-WI, T2-WI, and FLAIR were located in the first three indexes of eigenvalues. The matrix dimension of Rt GBM and Rt N was reduced by the projection of each row according to
| (8) |
where { }T was the transpose indicator, PCGBM T and PCN T were the transpose of principal components of GBM and normal area, respectively, and i was the index of row in GBM matrix Rt GBM and normal area matrix Rt N, respectively.
Figure 3.

Principal components based on higher variance of GBM and normal areas: (black curve) variance magnitude of 17 normal areas from T1-WI, T2-WI, and FLAIR, (blue curve) variance magnitude of 17 GBMs chosen from T1-WI, (red curve) variance magnitude of 17 GBMs chosen from T2-WI, and (green curve) variance magnitude of 17 chosen GBMs chosen from FLAIR mode of MRI.
Using 3 PCs, a new matrix P GBM of GBM that was of size 17 by 3 and matrix P N of normal area had a similar size. We considered then three classifier models to evaluate GBM and normal areas discrimination based on GMM features and their three principal components, respectively.
2.5. Classifier Setting
In general, the goal of a learning/classification algorithm is to build a set of training examples with class labels. In this context, we implemented three classifier techniques, namely, naïve Bayes (NB) [36], support vector machine (SVM) [37], and probabilistic neural network (PNN) [38]. The implementation of NB is performed using a kernel estimation method which approximated the complex distributions of the data. Then, SVM was implemented using the Gaussian radial basis function, and radial basis network based PNN was employed which is a fast classifier technique. The reason for using these specific classifier methods is to achieve the trade-off performance which is reported.
Due to the limited data available (17 patients), validation data sets were performed based on leave-one-out cross-validation [39]. Performance metrics are expressed by the following equations:
| (9) |
Moreover, receiver operating characteristic (ROC) curves and the associated area under the curve (AUC) values were computed to assess the discrimination between GBM and normal areas [40]. The results of the performance metrics reflected the succeeding GMM features for discrimination between GBM and normal area. Note that the training data set of the normal brain tissue regions represent different normal regions within the MR image.
3. Experimental Results and Discussions
3.1. Segmentation of the GBM
GBM tumor tissue was detected using the multithresholding segmentation based on Otsu's technique and the morphology operation to obtain only the abnormal brain regions in robust term. Figure 4 shows GBM tumor segmented using several steps. The process of tumor detection from MR images may appear to be a difficult task as MR images contained some areas which have a similar range of gray color (Figure 4(b)). Morphology operators or filtering was necessary to remove noise like the boundary of skull and brain (Figures 4(c) and 4(d)). Then GBM was detected and located (Figures 4(e) and 4(f)).
Figure 4.
GBM detection by segmentation and morphology operations: (a) T1-MR image, (b) image segmented by four levels, (c) range of GBM gray level conserve, (d) filtering of (c), (e) raw GBM data detected, and (f) GBM located on the brain image.
3.2. GMM Feature Extraction and Classification
Three GMM curve fittings based on the histogram analysis showed three components of GBM (see Figure 5). Three Gaussian components were chosen based on the empirical metrics which showed three components of GBM. GMM features were shown to be feasible for discriminating between GBM and normal brain.
Figure 5.

GMM curve fitting: example of GBM based GMM features.
Table 1 shows a comparative study between the three modes of MR images based on the classifier accuracy, false alarm, and missed detection. These metrics represented the highest performance using NB classifier with the classification accuracy range between 94.00 and 97.00%, false alarm range between 0.00 (which means that the normal area samples were correctly classified without error) and 5.89%, and missed detection range between 0.00 (which means that the GBM samples were correctly classified without error) and 2.95%. This latter value of missed detection represented the one GBM sample from 17 that was incorrectly classified (or classified as normal area).
Table 1.
Performance metrics (%) based on the GMM features.
| Classifier | Sequence | Accuracy | False alarm | Missed detection |
|---|---|---|---|---|
| NB | T1-WI | 97.05 | 2.95 | 0 |
| T2-WI | 97.05 | 0 | 2.95 | |
| FLAIR | 94.11 | 5.89 | 0 | |
| ∗Entire GBM | 86.27 | 2.94 | 10.78 | |
|
| ||||
| SVM | T1-WI | 70.58 | 0 | 29.41 |
| T2-WI | 64.70 | 5.88 | 29.41 | |
| FLAIR | 67.64 | 2.94 | 29.41 | |
| ∗Entire GBM | 66.66 | 4.90 | 28.43 | |
|
| ||||
| PNN | T1-WI | 94.11 | 5.89 | 0 |
| T2-WI | 70.58 | 11.76 | 17.64 | |
| FLAIR | 94.11 | 2.94 | 2.94 | |
| ∗Entire GBM | 86.27 | 2.94 | 10.78 | |
∗Entire GBM refers to T1-WI, T2-WI, and FLAIR features combined together.
GMM features were reduced with a PCA, which accounted for 97% of the cumulative variance from these features. Table 2 shows the performance metrics of the classifier accuracy based on the PCA. Clearly, the accuracy was decreased in the two MRI sequences T1-WI and T2-WI with the best performance achieved using BN classifier, where the accuracy ranged between 73.52 and 79.41%, false alarm ranged between 2.94 and 8.82%, and missed detection is 17.64%. In FLAIR sequence, PNN model showed highest value (94.11%, 0.00%, and 5.88%) of accuracy, false alarm, and missed detection, respectively.
Table 2.
Performance metrics (%) based on the PCA features.
| Classifier | Sequence | Accuracy | False alarm | Missed detection |
|---|---|---|---|---|
| NB | T1-WI | 73.52 | 8.82 | 17.64 |
| T2-WI | 79.41 | 2.94 | 17.64 | |
| FLAIR | 82.35 | 5.88 | 11.76 | |
| Entire GBM | 68.62 | 15.68 | 15.68 | |
|
| ||||
| SVM | T1-WI | 55.88 | 0 | 44.11 |
| T2-WI | 61.76 | 8.82 | 29.41 | |
| FLAIR | 85.29 | 0 | 14.70 | |
| Entire GBM | 35.29 | 20.58 | 44.11 | |
|
| ||||
| PNN | T1-WI | 61.76 | 8.82 | 29.41 |
| T2-WI | 52.94 | 14.70 | 32.35 | |
| FLAIR | 94.11 | 0 | 5.88 | |
| Entire GBM | 51.96 | 9.80 | 38.23 | |
Clearly, the accuracy decreased in T1-WI and T2-WI which reflected the lack of PCA features. In other words, GBM features provided from GMM were likely independent which was represented by the decrease accuracy value when we applied the PCA, while FLAIR sequence showed a similar value of 94.11% with GMM and PCA features which represent the correlation between the features. This is also represented by the highest correlation value of GMM features in the FLAIR sequence (Figure 6), while the heat map of correlation shows a less value in T1-WI and T2-WI sequences.
Figure 6.
Heat map with correlation coefficients between GMM features: w, μ, and v are the weight, average, and variance, respectively; N and T are the index of normal and tumor (GBM) areas, respectively.
Another aspect of the classification was considered by using all data based on 51 images (entire GBM) including 17 images T1-WI, T2-WI, and FLAIR. Table 1 shows the classifier accuracy decrease to 86.27% with 2.94% false alarm and 10.78% missed detection using BN classifier. Clearly, the discrimination between GBM and normal brain tissue using single MR sequence was better than using all together. The classifier accuracy decreased to 68.62% (see Table 2) when we applied the PCA which proved again that the GMM features had no redundancy information in T1-WI and T2-WI and are better to be used for the discrimination of GBM from normal brain based on single MRI sequence. Note that BN classifier showed a better performance than SVM and PNN classifier model.
Moreover, ROC curves and the associated AUC have been computed. Figure 7 shows the ROC curves to evaluate the quality of a classifier. Table 4 shows that AUC values of 95.85% based on GMM features are better than PCA (AUC = 86.16%) using FLAIR sequence. Clearly, the AUC obtained by using GMM features were better than those from PCA.
Figure 7.
ROC curves of GBM and normal area discrimination based on T1-WI, T2-WI, FLAIR, and entire MR mode (T1-WI, T2-WI, and FLAIR, 51 images): (a) GMM features, (b) PCA features, and (c) Daubechies (db1) and Coiflets (coif1) wavelet features.
Table 4.
Comparison of area under (%) ROC curve between three feature types.
| Feature | T1-WI | T2-WI | FLAIR | |
|---|---|---|---|---|
| Wavelets | coif1 | 91.35 | 94.46 | 74.5 |
| db1 | 68.51 | 87.54 | 73.7 | |
|
| ||||
| GMM | 92.73 | 91.70 | 95.85 | |
|
| ||||
| PCA | 67.82 | 75.43 | 86.16 | |
3.3. Comparatives and Discussions
A comparative study was employed using wavelet based feature [29–31]. Two wavelets, namely, Daubechies (db1) and Coiflets (coif1), were considered [41, 42]. Three quantified functions were computed, namely, average, standard deviation, and entropy. Classifier accuracy based on the wavelet features showed the highest accuracy value of 88.23% (coif1) in T2-WI using SVM and PNN, 79.41% (coif1) in T1-WI, and 70.58% (db1) in FLAIR sequence using PNN and NB (see Table 3). ROC curves were associated with AUC based on wavelet feature (coif1) with range value 74.50−94.46% better than those based on wavelet feature (db1) which showed values of 68.51–87.54% (Figure 7(c) and Table 4). We note that the main goal of the feature extraction using discrete wavelet transform technique is that the approximation coefficients usually contain the most important information (low frequency). Hence they constitute one part of the extracted features and another represented by the critical information from the high-frequency part (details). In comparison, GBM identification is more promising using the GMM features with highest accuracy classifier.
Table 3.
Performance metrics (%) based on the wavelets.
| Classifier | Metrics | T1-WI | T2-WI | FLAIR | |||
|---|---|---|---|---|---|---|---|
| db1 | coif1 | db1 | coif1 | db1 | coif1 | ||
| NB | Accuracy | 67.64 | 79.41 | 82.35 | 85.29 | 70.58 | 64.70 |
| False alarm | 17.64 | 8.82 | 8.82 | 8.88 | 8.82 | 14.70 | |
| Missed detection | 14.70 | 8.8235 | 8.82 | 8.82 | 20.58 | 20.58 | |
|
| |||||||
| SVM | Accuracy | 50 | 76.47 | 85.29 | 88.23 | 58.82 | 55.88 |
| False alarm | 20.58 | 14.70 | 2.94 | 0 | 8.82 | 8.82 | |
| Missed detection | 29.41 | 8.82 | 11.76 | 11.76 | 32.35 | 35.29 | |
|
| |||||||
| PNN | Accuracy | 70.58 | 79.41 | 82.35 | 88.23 | 70.588 | 58.82 |
| False alarm | 20.58 | 20.58 | 11.76 | 5.88 | 20.58 | 29.41 | |
| Missed detection | 8.82 | 0 | 5.88 | 5.88 | 8.82 | 11.76 | |
∗ db1 and coif1 are the first order of Daubechies and Coiflet wavelet, respectively.
This work is the first focusing on the robust GBM characteristics using the GMM features, while previous literature showed the efficiency of the texture and statistical features analysis to discover brain tumors heterogeneity [24–27, 38]. Through the texture analysis we find that this changes depending on the size and number of the pixels in the determined region. For example, cooccurrence matrix provides valuable information about the relative position of the neighboring pixels in an image. It has been proved that the texture descriptors improve performance of diagnosis where texture is an important source of image characteristics [24, 25]. However, GBM region can be easily distinguished from normal brain areas using the GMM features because we determined the number of Gaussian components (three components in this work [43]), and the number or swap of pixels did not affect the accuracy result. In other words, the number of features is independent of the pixels in the brain tumor image. All GBM tumor regions being diagnosed following MR imaging, the big advantage of this technique derives from the robust processing of MRI data to the final decision.
Obviously, neuroradiologists are becoming more and more important players to early diagnose GBM. Our vision is to integrate engineering based methods as described in daily practice to enhance radiologists' performance beyond their routine “vision.” In particular in utterly devastating diseases, like GBM, improvement in any medical specialty involved is of utmost essence.
As a limitation, this study is based on a single cohort design and subject to its respective limitations. Second, the number of 17 patients limited our analysis of the GBM heterogeneity. However, there is currently no consensus on how to assess GBM heterogeneity. Third, the goal of this study was to analyze the number of images features to determine the efficiency of the GMM features as prognostic indicator. And last, the entire algorithm worked and collected data automatically.
4. Conclusions
This paper implements the GMM features extracted from GBM tumor using MR images to assist in radiologic GBM heterogeneity recognition. By a comparative study of other features types as PCA and wavelet, this technique resides in its ability to detect GBM automatically with high accuracy performance which was difficult previously. Turning to the future, it is the authors' intentions to extend this work to implement a full algorithm integrated on MRI equipment in order to identify the GBM robustly using GMM features.
Ethical Approval
The materials are in compliance with all applicable laws, regulations, and policies for the protection of medical data, and any necessary approvals, authorizations, and informed consent documents were obtained.
Consent
Informed consent was obtained from all patients for being included in the study.
Conflict of Interests
The author declares that there is no conflict of interests regarding the publication of this paper.
References
- 1.Chen C.-C., Wan Y.-L., Wai Y.-Y., Liu H.-L. Quality assurance of clinical MRI scanners using ACR MRI phantom: preliminary results. Journal of Digital Imaging. 2004;17(4):279–284. doi: 10.1007/s10278-004-1023-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sachdeva J., Kumar V., Gupta I., Khandelwal N., Ahuja C. K. Segmentation, feature extraction, and multiclass brain tumor classification. Journal of Digital Imaging. 2013;26(6):1141–1150. doi: 10.1007/s10278-013-9600-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patriarche J. W., Erickson B. J. Part 1. Automated change detection and characterization in serial MR studies of brain-tumor patients. Journal of Digital Imaging. 2007;20(3):203–222. doi: 10.1007/s10278-006-1038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Likar B., Viergever M. A., Pernuš F. Retrospective correction of MR intensity inhomogeneity by information minimization. IEEE Transactions on Medical Imaging. 2001;20(12):1398–1410. doi: 10.1109/42.974934. [DOI] [PubMed] [Google Scholar]
- 5.Yang F., Thomas M. A., Dehdashti F., Grigsby P. W. Temporal analysis of intratumoral metabolic heterogeneity characterized by textural features in cervical cancer. European Journal of Nuclear Medicine and Molecular Imaging. 2013;40(5):716–727. doi: 10.1007/s00259-012-2332-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caban J. J., Yao J., Mollura D. J. Enhancing image analytic tools by fusing quantitative physiological values with image features. Journal of Digital Imaging. 2012;25(4):550–557. doi: 10.1007/s10278-011-9449-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nachimuthu D. S., Baladhandapani A. Multidimensional texture characterization: on analysis for brain tumor tissues using MRS and MRI. Journal of Digital Imaging. 2014;27(4):496–506. doi: 10.1007/s10278-013-9669-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamstra D. A., Galbán C. J., Meyer C. R., et al. Functional diffusion map as an early imaging biomarker for high-grade glioma: correlation with conventional radiologic response and overall survival. Journal of Clinical Oncology. 2008;26(20):3387–3394. doi: 10.1200/jco.2007.15.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helseth E., Unsgaard G., Dalen A., Vik R. Effects of type beta transforming growth factor in combination with retinoic acid or tumor necrosis factor on proliferation of a human glioblastoma cell line and clonogenic cells from freshly resected human brain tumors. Cancer Immunology Immunotherapy. 1988;26(3):273–279. doi: 10.1007/BF00199941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ostrom Q. T., Gittleman H., Farah P., et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro-Oncology. 2013;15(supplement 2):ii1–ii56. doi: 10.1093/neuonc/not151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verhaak R. G. W., Hoadley K. A., Purdom E., et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zinn P. O., Colen R. R. Imaging genomic mapping in glioblastoma. Neurosurgery. 2013;60(1):126–130. doi: 10.1227/01.neu.0000430773.18220.3f. [DOI] [PubMed] [Google Scholar]
- 13.Zinn P. O., Majadan B., Sathyan P., et al. Radiogenomic mapping of edema/cellular invasion MRI-phenotypes in glioblastoma multiforme. PLoS ONE. 2011;6(10) doi: 10.1371/journal.pone.0025451.e25451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simon D., Fritzsche K. H., Thieke C., et al. Diffusion-weighted imaging-based probabilistic segmentation of high- and low-proliferative areas in high-grade gliomas. Cancer Imaging. 2012;12(1):89–99. doi: 10.1102/1470-7330.2012.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balla-Arabé S., Gao X. Image multi-thresholding by combining the lattice Boltzmann model and a localized level set algorithm. Neurocomputing. 2012;93:106–114. doi: 10.1016/j.neucom.2012.04.019. [DOI] [Google Scholar]
- 16.Rojas Domínguez A., Nandi A. K. Detection of masses in mammograms via statistically based enhancement, multilevel-thresholding segmentation, and region selection. Computerized Medical Imaging and Graphics. 2008;32(4):304–315. doi: 10.1016/j.compmedimag.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 17.Hertz L., Schafer R. W. Multilevel thresholding using edge matching. Computer Vision, Graphics, & Image Processing. 1988;44(3):279–295. doi: 10.1016/0734-189x(88)90125-9. [DOI] [Google Scholar]
- 18.Otsu N. A threshold selection method from Gray-Level histograms. IEEE Transactions on Systems, Man, and Cybernetics. 1979;9(1):62–66. [Google Scholar]
- 19.Chaddad A., Tanougast C., Golato A., Dandache A. Carcinoma cell identification via optical microscopy and shape feature analysis. Journal of Biomedical Science and Engineering. 2013;6(11):1029–1033. doi: 10.4236/jbise.2013.611128. [DOI] [Google Scholar]
- 20.Castellano G., Bonilha L., Li L. M., Cendes F. Texture analysis of medical images. Clinical Radiology. 2004;59(12):1061–1069. doi: 10.1016/j.crad.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 21.Mahmoud-Ghoneim D., Alkaabi M. K., de Certaines J. D., Goettsche F.-M. The impact of image dynamic range on texture classification of brain white matter. BMC Medical Imaging. 2008;8, article 18 doi: 10.1186/1471-2342-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasikala M., Kumaravel N. A wavelet-based optimal texture feature set for classification of brain tumours. Journal of Medical Engineering and Technology. 2008;32(3):198–205. doi: 10.1080/03091900701455524. [DOI] [PubMed] [Google Scholar]
- 23.Yu O., Mauss Y., Zollner G., Namer I. J., Chambron J. Distinct patterns of active and non-active plaques using texture analysis on brain NMR images in multiple sclerosis patients: preliminary results. Magnetic Resonance Imaging. 1999;17(9):1261–1267. doi: 10.1016/s0730-725x(99)00062-4. [DOI] [PubMed] [Google Scholar]
- 24.Dettori L., Semler L. A comparison of wavelet, ridgelet, and curvelet-based texture classification algorithms in computed tomography. Computers in Biology and Medicine. 2007;37(4):486–498. doi: 10.1016/j.compbiomed.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 25.Georgiadis P., Cavouras D., Kalatzis I., et al. Improving brain tumor characterization on MRI by probabilistic neural networks and non-linear transformation of textural features. Computer Methods and Programs in Biomedicine. 2008;89(1):24–32. doi: 10.1016/j.cmpb.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 26.Chaddad A., Tanougast C., Dandache A., Al Houseini A., Bouridane A. Improving of colon cancer cells detection based on Haralick's features on segmented histopathological images. Proceedings of the IEEE Conference on Computer Applications and Industrial Electronics (ICCAIE '11); December 2011; Penang, Malaysia. IEEE; pp. 87–90. [DOI] [Google Scholar]
- 27.Cardinaux F., Sanderson C., Bengio S. User authentication via adapted statistical models of face images. IEEE Transactions on Signal Processing. 2006;54(1):361–373. doi: 10.1109/TSP.2005.861075. [DOI] [Google Scholar]
- 28.Zacharaki E. I., Wang S., Chawla S., et al. Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magnetic Resonance in Medicine. 2009;62(6):1609–1618. doi: 10.1002/mrm.22147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dash J. K., Sahoo L. Wavelet based features of circular scan lines for mammographic mass classification. Proceedings of the 1st International Conference on Recent Advances in Information Technology (RAIT '12); March 2012; pp. 58–61. [DOI] [Google Scholar]
- 30.Dua S., Rajendra Acharya U., Chowriappa P., Vinitha Sree S. Wavelet-based energy features for glaucomatous image classification. IEEE Transactions on Information Technology in Biomedicine. 2012;16(1):80–87. doi: 10.1109/TITB.2011.2176540. [DOI] [PubMed] [Google Scholar]
- 31.Li S., Liao C., Kwok J. T. Wavelet-based feature extraction for microarray data classification. Proceedings of the International Joint Conference on Neural Networks (IJCNN '06); July 2006; pp. 5028–5033. [Google Scholar]
- 32.Otsu N. A threshold selection method from gray-level histograms. IEEE Transactions on Systems, Man and Cybernetics. 1979;9(1):62–66. doi: 10.1109/tsmc.1979.4310076. [DOI] [Google Scholar]
- 33.Kinnunen T., Li H. An overview of text-independent speaker recognition: from features to supervectors. Speech Communication. 2010;52(1):12–40. doi: 10.1016/j.specom.2009.08.009. [DOI] [Google Scholar]
- 34.Barras C., Gauvain J. Feature and score normalization for speaker verification of cellular data. Proceedings of the IEEE International Conference on Acoustics, Speech, and Signal Processing (ICASSP '03); April 2003; IEEE; pp. II49–II52. [DOI] [Google Scholar]
- 35.Kilaru V., Amin M., Ahmad F., Sévigny P., DiFilippo D. Gaussian mixture modeling approach for stationary human identification in through-the-wall radar imagery. Journal of Electronic Imaging. 2015;24(1) doi: 10.1117/1.jei.24.1.013028.013028 [DOI] [Google Scholar]
- 36.Aggarwal C. C. Data Classification: Algorithms and Applications. CRC Press; 2014. [Google Scholar]
- 37.Hearst M., Dumais S., Osman E., Platt J., Scholkopf B. Support vector machines. IEEE Intelligent Systems and their Applications. 1998;13(4):18–28. doi: 10.1109/5254.708428. [DOI] [Google Scholar]
- 38.Specht D. F. Probabilistic neural networks. Neural Networks. 1990;3(1):109–118. doi: 10.1016/0893-6080(90)90049-q. [DOI] [PubMed] [Google Scholar]
- 39.Stone M. Cross-validatory choice and assessment of statistical predictions. Journal of the Royal Statistical Society. Series B. Methodological. 1974;36:111–147. [Google Scholar]
- 40.Park S. H., Goo J. M., Jo C.-H. Receiver operating characteristic (ROC) curve: practical review for radiologists. Korean Journal of Radiology. 2004;5(1):11–18. doi: 10.3348/kjr.2004.5.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mallat S. G. Theory for multiresolution signal decomposition: the wavelet representation. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1989;11(7):674–693. doi: 10.1109/34.192463. [DOI] [Google Scholar]
- 42.Daubechies I. Ten Lectures on Wavelets. Vol. 61. Philadelphia, Pa, USA: SIAM; 1992. (CBMS-NSF Regional Conference Series in Applied Mathematics). [DOI] [Google Scholar]
- 43.Chaddad A., Zinn P. O., Colen R. R. Brain tumor identification using Gaussian Mixture Model features and Decision Trees classifier. Proceedings of the 48th Annual Conference on Information Sciences and Systems (CISS '14); March 2014; Princeton, NJ, USA. IEEE; pp. 1–4. [DOI] [Google Scholar]