Abstract
Arabidopsis thaliana UVR8 (AtUVR8) is a long-sought-after photoreceptor that undergoes dimer dissociation in response to UV-B light. Crystallographic and mutational studies have identified two crucial tryptophan residues for UV-B responses in AtUVR8. However, the mechanism of UV-B perception and structural events leading up to dimer dissociation remain elusive at the molecular level. We applied dynamic crystallography to capture light-induced structural events in photoactive AtUVR8 crystals. Here we report two intermediate structures at 1.67Å resolution. At the epicenter of UV-B signaling, concerted motions associated with Trp285/Trp233 lead to ejection of a water molecule, which weakens an intricate network of hydrogen bonds and salt bridges at the dimer interface. Partial opening of the β-propeller structure due to thermal relaxation of conformational strains originating in the epicenter further disrupts the dimer interface and leads to dimer dissociation. These dynamic crystallographic observations provide structural insights into the photo-perception and signaling mechanism of UVR8.
Keywords: photoreceptor, light signaling, dynamic crystallography, UVR8, photo-perception mechanism
Ultraviolet-B (280–315nm) is an intrinsic band of the solar spectrum when the sunlight reaches the surface of the earth. UV-B has significant biological effects arising from DNA damage. At low fluence rate, however, UV-B serves as an environmental cue that mediates important physiological responses ranging from photomorphogenesis, photo-protection to circadian rhythm in plants1–4. Although UV-B photoreceptors have been suggested for decades5, it was until very recently that UVR8 (UV RESISTANCE LOCUS8) was identified as a distinct photoreceptor responsible for UV-B responses in plants4,6–8. The 440-residue UVR8 from Arabidopsis forms a stable homodimer that readily dissociates upon UV-B irradiation6,9,10(Fig. 1, Fig. S1). The C-terminal segment in the AtUVR8 monomer is thought to be accessible for direct interactions with COP1 (constitutively photomorphogenesis 1), a master regulator of light signaling in plants7,11–13. Such light-dependent interactions seem crucial for UV-B responses and for cross-talks with other light-mediated signaling pathways6–8,13.
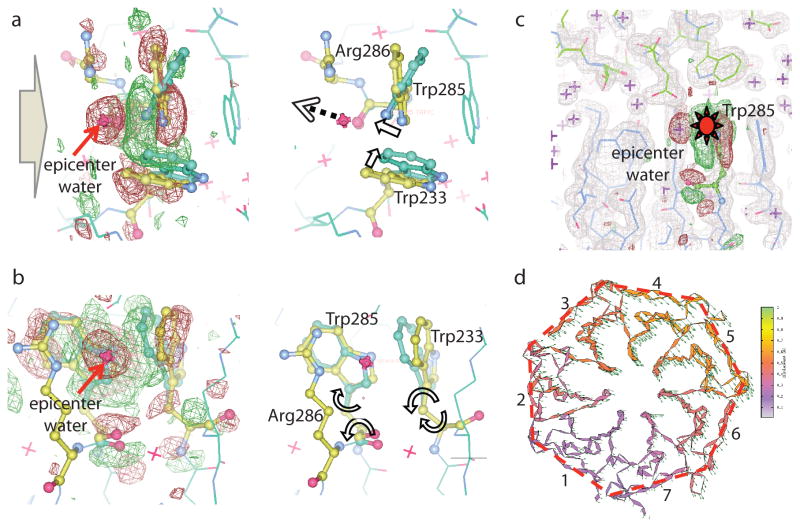
Fig. 1. Epicenter of UV-B signaling in AtUVR8.
a) Raw FUV-FDark difference map at 120 K for the entire dimer structure (contoured at +/−5σ with no mask applied) reveals two clusters of strong positive (red) and negative (green) difference densities at the dimer interface. b) A top-down view of one subunit from the dimer interface shows that light-induced structural changes are localized in blades 5 and 6. c) A zoom-in view of difference densities (contoured at +/−3.5σ) reveals structural rearrangements near the epicenter. Gray arrows mark the directions of light-induced motions from negative to positive densities.
Two AtUVR8 crystal structures, both with truncated C-termini, have been determined at high resolution9,10. Although they differ in space group and crystallization conditions, both structures show nearly identical dimer arrangements. As predicted, the AtUVR8 monomer adopts a 7-blade β-propeller structure14. Deletion of ~60 residues at the C-terminus of AtUVR8 did not seem to affect dimer formation nor UV-induced dimer dissociation9,10. The dimer interface, flat in shape and symmetrical in structure, buries significant surface area where a network of hydrogen bonds and salt bridges form extensive inter-subunit interactions9,10. UVR8 is also unique among all known photoreceptors - it does not use any non-protein cofactors as chromophore9,10,15. Instead, AtUVR8 is highly rich in UV-absorbing aromatic residues and comprises a total of 14 tryptophans in a monomer. Aside from the C-terminal Trp400, seven Trp residues from each monomer are found at the dimer interface while others are distributed among six blades at equivalent positions with their indole rings are buried between blades9,10. Trp285 and Trp233 at the dimer interface have been identified as potential chromophore based on structural analysis and mutational studies6,9–11. However, key questions concerning the photo-perception mechanism of UVR8 remain unanswered. Specifically, what is the structural nature of the triggering event? Are there any light-induced conformational changes associated with Trp285/Trp233? What is the sequence of molecular events leading up to dimer dissociation? Why does AtUVR8 evolve to comprise 14 Trp residues when only two of them are essential?
Charge neutralization of inter-subunit salt bridges has been proposed to be responsible for dimer dissociation in AtUVR89,10. It has been suggested that such charge neutralization is directly resulted from proton/electron transfers from the Trp285/Trp233 dyad due to light-induced charge separation within the dyad16–18, and that early signaling events in AtUVR8 may not involve conformational changes17. To address these questions, we applied dynamic crystallography to directly probe UV-B induced structural changes by initiating photoreactions in photoactive crystals. In this work, we present two intermediate structures of AtUVR8, which unambiguously establish the epicenter of UV-B signaling associated with concerted light-induced structural changes. Based on spatial organization and absorption properties of 13 Trp residues, we further propose an antenna mechanism for the UVR8 photoreceptor.
Results
The AtUVR8 construct used in this study comprises residues 12–381 and was crystallized under optimized conditions based on published protocols9. The dark structures of both wild type (WT) and a single mutant (W285F) were determined in space group C2 using molecular replacement (Table S1). The WT crystals visibly cracked upon UV-B irradiation at room temperature9, indicating photo-activity but also rendering the photoreaction irreversible in the AtUVR8 crystals.
To probe light-induced structural changes in photoactive AtUVR8 crystals, we adopted a temperature-scan strategy19. We first collected a complete reference dark dataset (FDark) from one segment of each mounted crystal at 100 K. We then initiated photoreaction by illuminating the same crystal for 10–15 minutes using two UV-B lamps at an elevated temperature. Light-induced structural changes were cryo-trapped by lowering the temperature of the illuminated crystal to 100 K, at which we collected a light dataset (FUV) from a different volume of the same crystal. By varying the temperature at which the reaction was initiated, we aimed to follow the progression of light-induced structural events by crystallography.
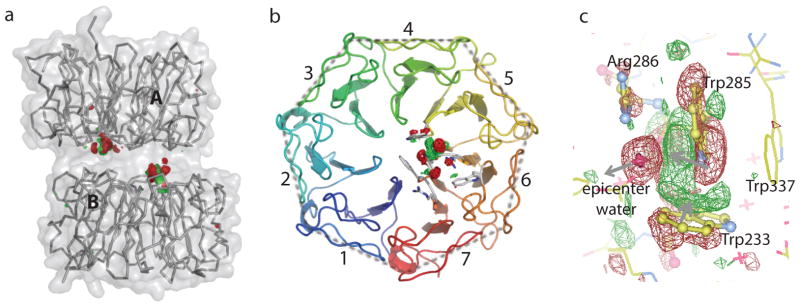
We calculated (FUV-FDark) difference Fourier maps using the phases from the dark structure. Such difference maps between paired dark and light datasets from a single crystal are highly sensitive to light-induced structural changes even when the extent of reaction initiation is as low as 10–15%20. We first tested our experimental protocol with the (FUV-FDark) difference map at 120 K using a WT AtUVR8 crystal. Two clusters of strong positive and negative difference electron densities are found at the dimer interface (Fig. 1). These map features are not detectable in a control experiment where we raised the crystal temperature to 120 K but did not apply UV-B irradiation (Fig. S1, Fig. S2). Nor did we observe any comparable signals in the FUV-FDark map from crystals of the W285F mutant that is able to form dimers but does not undergo UV-B induced dimer dissociation9,10 (Fig. S3, Fig. S4). These results confirmed that 1) difference densities in the (FUV-FDark) map at 120 K are indeed induced by UV-B irradiation; and 2) our experimental strategy is effective for capturing light-induced structural changes in the WT AtUVR8 crystals.
We scanned the temperature range between 100 and 180 K under the same data collection protocol on seven WT crystals (Table S2). No significant difference densities were observed when the photoreaction was initiated at 100 K (SI Fig. S1), while some weak difference densities are detected at 110 K. At 120 K, difference densities of significant signal-to-noise (S/N) ratios are clustered near two distinct yet structurally equivalent locations at the dimer interface (Fig. 1a,b), where they are specifically associated with Trp285/Trp233 and a water molecule (Fig. 1c). In all four UVR8 subunits in the asymmetric unit, similar difference density features are observed at 120 K and higher temperatures (Fig. S1). Not surprisingly, the FUV-FDark map at 180 K revealed more extensive structural changes beyond the epicenter. Using a WT crystal with slightly different cell parameters (Tables S1, S2; Fig. S4c), we were able to observe pronounced signals in subunit B (Fig. 2a, b). These extensive and pair-wise difference densities sandwich consecutive peptide planes and bulky side chains in the affected blades 4, 5 and 6, suggesting coordinated and large-scale motions along the same direction (Fig. 2c).
Fig. 2. Light-induced difference densities at 180 K.
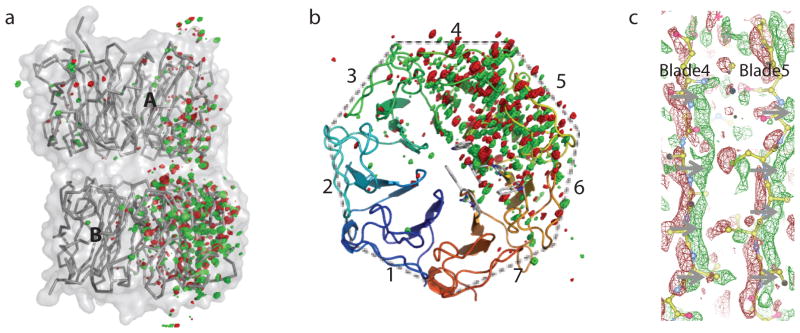
a) Raw FUV-FDark difference map (contoured at +/−4σ) in the entire dimer shows asymmetric distribution of difference densities. b) A top-down view of subunit B from the dimer interface shows widespread structural changes in blades 4, 5 and 6. Trp233 and Trp285 at the epicenter are located in blades 5 and 6, respectively. c) Difference densities (contoured at +/−2.5σ) that sandwich the peptide planes in affected blades suggest significant and concerted motions (indicated by gray arrows) from negative (red) to positive (green) densities. Two strands in blades 4 and 5 (ball-and-stick in yellow) are shown.
Given four independent crystallographic observations in the asymmetric unit, we obtained a total of 32 difference maps over seven temperatures and subjected them to singular value decomposition (SVD) analysis21. The scatter plots derived from the SVD decomposition show clear evidence for temperature-dependency of light-induced structural changes (Fig. S5). Furthermore, two subunits of the same AtUVR8 dimer seem to differ slightly at low temperatures, and further diverge when the crystal temperature rises above 160 K (Fig. S5b). As higher temperatures permit further progression along a reaction pathway, large-amplitude conformational changes are inevitably influenced by crystal lattice constraints. We hence attribute asymmetric behaviors among chemically identical subunits at higher temperatures to subtle differences in molecular packing (Fig. 2a; Fig. S5).
Since the difference maps between 120 and 160 K exhibit comparable features, we hereafter use the FUV-FDark map at 120 K for illustration and denote the corresponding intermediate structure I120K. Real-space refinement of I120K was carried out against difference electron densities within a 5-Å radius around residues at the epicenter21. The I120K structure represents an early signaling event (Fig. 1b), in which the indole rings of Trp285 and Trp233 are displaced as if they were about to collide with each other due to strong attraction (Fig. 3a,b). As a result, the indole ring of Trp233 rotates about 10° as it shifts towards Trp285. And the indole ring of Trp285 tilts about 30° with associated positive and negative difference densities forming a crossover pattern (Fig. 3a). Such concerted motions of the indole rings are concomitant with torsional motions about the Cα-Cβ and Cβ-Cγ bonds in Trp285 and Trp233 (Fig. 3a), leading to the strained backbone conformations in the vicinity of Trp285 and Trp233. In I120K, the carbonyl oxygen atoms of Trp285 and Gly284 move away from the dimer interface by ~0.5 Å (Fig. 3b and Fig. S1).
Fig. 3. Light-induced structural changes.
a,b) Superposition of the dark (yellow) and I120K (green) structures, with (left) and without (right) FUV-FDark difference densities at 120K (contoured at +/− 3.5σ). The red cross marks the location of the epicenter water in the dark structure. The gray block arrow indicates the viewpoint of b) in respect to a). c) A region of the 2Fo-Fc map in the dark structure (grey mesh contoured at 1σ) is superimposed with the FUV-FDark difference map at 120 K. Two subunits (stick models) are colored in green and blue, respectively; water molecules in purple crosses. Trp285 is shown in ball-and-stick. d) Light-induced motions at 180 K in subunit B. Directions and lengths of green arrows correspond to directions and amplitudes (3×) of motions in each blade. The structure is represented by consecutive peptide planes and colored according to the amplitude of motions (color bar; right).
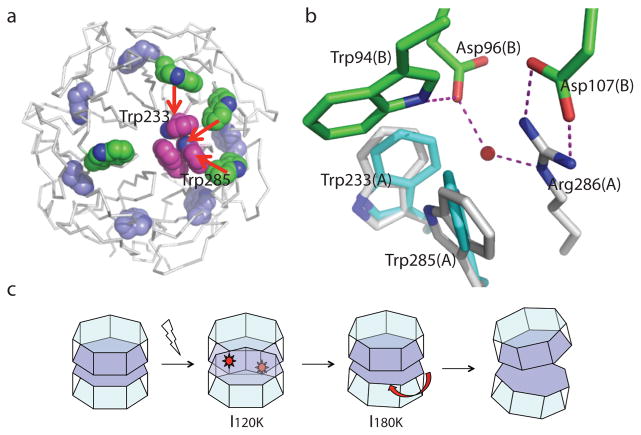
The most striking feature in the FUV-FDark difference map at 120 K is the significant negative density sitting on top of a water molecule in the dark structure (Fig. 3a). This water molecule, denoted “epicenter water”, is part of a well-knit hydrogen-bonding network at the dimer interface that involves Trp285/Arg286 in subunit A and Asp96/Trp94/Asp107 in subunit B (Fig. 4b, Fig. S6a). Such strong negative density (−14σ at the peak) clearly indicates that the epicenter water is kicked away like a “soccer ball” in the I120K structure when the indole ring of Trp285 rotates. Given its strategic location, departure of this epicenter water is expected to profoundly weaken inter-subunit interactions at the dimer interface (Fig. 3c). Not surprisingly, mutations that alter the network of hydrogen bonds and salt bridges (e.g. Arg286, Arg338, Asp96, Asp107 and Asp129) near the epicenter are more prone to monomerization compared to WT9,10,22,23 (Fig. S6).
Fig. 4. Photoreaction and dimer dissociation mechanism of UVR8.
a) Excitation energy captured by Trp pigments is funneled to the epicenter to generate a structural signal. Red arrows indicate putative directions of energy flow. b) The Coulomb force arising from light-induced charge separation drives the observed motions in Trp285 and Trp233. This leads to ejection of the epicenter water (red sphere), which weakens the network of hydrogen bonds and salt bridges at the dimer interface. Two subunits of the dark structure are colored in gray and green, respectively. The main features of I120K are highlighted in cyan. c) Schematic illustration of light-induced structural events captured by dynamic crystallography.
Dimer dissociation clearly requires more extensive motions than ejection of a water molecule. This is evident by large-scale motions shown in the FUV-FDark difference map at 180K (Fig. 2). The corresponding structure, denoted I180K, was refined against difference densities in the entire subunit B. I180K reveals remarkable backbone motions consistent with partial opening or unwinding of the circular β-propeller structure (Fig. 3d). The most prominent structural changes occur in blades 5 and 6, where Trp285 and Trp233 are respectively located. Other significant difference densities are found in blades 1 and 7 in subunit A across the dimer interface (Fig. 2a). We speculate that I180K is resulted from thermal relaxation of backbone conformational strains in Trp285 and Trp233 observed in I120K (Fig. 1c, Fig. 3a). It is plausible that such large-scale structural rearrangements originating from the epicenter would further disrupt hydrogen bonds and salt bridges at the dimer interface and lead to the eventual dimer dissociation (Fig. 4).
Are these cryo-trapped UVR8 structures relevant to light-induced molecular events at physiological temperatures? To address this, we conducted time-resolved fluorescence spectroscopic experiments at room temperature in solution with both WT and W285F24. Upon excitation by sub-nanosecond pulsed UV light25, we recorded time-courses of fluorescence decay, from which the time constants associated with distinct decay species were determined. The excitation wavelength was selected at 320nm, 1) to preferentially excite the Trp triad at the epicenter that is expected to absorb at longer UV wavelengths due to exciton coupling; and 2) to reduce excitation by tyrosine that absorbs at shorter wavelengths. Three major decay species were obtained for WT (Table S3), which correspond to three distinct states associated with the time constants of 70-ps (78%), 1.5-ns (16%) and 8.5-ns (6%), respectively. Clearly, the 70-ps species represents a dominant quenching event in WT. Consistently, the overall fluorescence emission is significantly enhanced in W285F compared to WT18 (Fig. S9). We attributed it to the absence of light-induced conformational changes in W285F (Fig. S4d). W285F exhibited three decay species in fluorescence quenching dynamics – 55-ps (4%), 3-ns (2%) and 13-ns (94%) - that are similar to those of WT. However, the long-lived ns species dominates in W285F (Table S3). These time-resolved results are in agreement with the findings of Liu et al18. Minor discrepancies might be attributed to potential sample heterogeneity and/or different excitation wavelengths.
Although the dimerization and crystal structure of W285F are largely unaffected, W285F does not undergo dimer dissociation in response to UV-B9. The absence of detectable light-induced structural changes in W285F (Fig. S3b, Fig. S4b) is consistent with significantly enhanced fluorescence compared to WT (Fig. S9). We thus speculate that the fluorescence-quenching event on the 100-ps timescale may correspond to formation of the I120K structure; and light energy quenched on the ~100-ps scale is converted to kinetic energy to drive motions in Trp285/Trp233 (Fig. 4; Table S3).
Discussion
Our dynamic crystallographic observations confirm that Trp285 and Trp233 are indeed at the epicenter of UV-B signaling9,10,26 and function as chromophores to generate light-induced structural changes9,11. But what triggers the observed motions in Trp233/Trp285? In the dark structure, the indole rings of Trp285 and Trp233 adopt a nearly perpendicular arrangement via a N-H…π interaction (Fig. 1c). Voityuk et al proposed that charge separation upon UV-B excitation occurs within the Trp285/Trp233 dyad, where 0.96e is transferred from one indole ring to the other17. We further estimated that such charge separation over a short distance (~4.7 Å – an average distance between two indole rings) would generate a considerable electrostatic attraction force between Trp285 and Trp233, as large as 1 nN according to the Coulomb’s Law. The Coulomb force brings together Trp285 and Trp233 such that their indole rings must rotate to avoid steric clashes. Two effects are directly resulted from rotation of the indole rings. First, the indole ring of Trp285 moves towards the epicenter water by ~1.7 Å, which ejects the water and subsequently perturbs inter-subunit interactions including a key salt bridge (Arg286-Asp107) in close proximity. This water ejection event, however, seems to precede the rupturing event of this salt bridge since there is no significant difference densities associated with Asp107 and Arg286 at 120 K (Fig. 1c). Second, the backbone structures of Trp285 and Trp233 are evidently strained; subsequent thermal relaxation partially opens up the circular β-propeller fold (I180K). We postulate that disintegration of an extensive dimer interface as in UVR8 would require both local and large-scale structural rearrangements as depicted in the I120K and I180K structures. Our findings from UVR8 also suggest a general mechanism how other signaling proteins in the WD40 family e.g. the β-subunit in the heterotrimeric G-proteins may transmit and amplify small-amplitude structural signals via the widespread β-propeller scaffold27.
We then ask why UVR8 comprise 14 Trp residues when only two of them are required for action? What is the role of “non-action” Trp pigments in UV-B signaling? The spatial and conformational distributions of Trp residues in the AtUVR8 dimer structure (Fig. S7) hints at a possible antenna model reminiscent of plant light-harvesting complexes and phycobilisomes in cyanobacteria28,29. This hypothesis implies that 1) all Trp residues contribute to the UVR8 function; 2) excitation energy flow is directed by energy gradient between pigments, and is eventually funneled to the epicenter; 3) energy transfer efficiency is governed by both distance and spectral overlap between donor and acceptor pigments.
To explore this, we calculated individual absorption spectrum for each Trp residue in the AtUVR8 dark structure using a time-dependent density functional tight-binding approach (TD-DFTB)30–33. Two interfacial Trp residues (Trp233, 94) partner with a nearby Asp residue while others (Trp337, 302, 250, 198) form hydrogen bonds with water molecules (Fig. S8a). Not surprisingly, six distal Trp residues (Trp39, 92, 144, 196, 300 and 352) exhibit largely similar optical properties, given their nearly identical conformations and protein environment34,35 (Fig. S8). In contrast, seven interfacial Trp residues (Trp94, 198, 250, 302, 233, 285 and 337) in diverse rotamer conformations (Fig. S7, S8; Table S4) give rise to a much wider absorption coverage that extends to the longer wavelength range of UV-B (Fig. S8c). We also note that all 13 Trp residues except Trp285 are stabilized by an in-plane hydrogen bond via indole nitrogen, although with different partners. Specifically, the indole nitrogen in a distal Trp residue forms hydrogen bond with the main chain carbonyl group. If the exciton coupling effect is considered, the effective spectra of closely clustered pigments (Trp285, 233, 337 and Trp94B) would be significantly shifted to longer wavelengths, which renders the energy sink of this putative UV-B antenna right at the epicenter (Fig. S7b). Consistent results from the TD-DFTB (Fig. S8c) and MS-CASP2 calculations17 suggest that the Trp pigments collectively convey much broader UV-B sensitivity than any single Trp alone in UVR8. If the “action” pigments (Trp233/Trp285) at the epicenter are able to receive UV-B signals detected by other “non-action” pigments via this proposed antenna mechanism (Fig. 4a), the UV-B sensitivity range of UVR8 would be significantly extended.
It is indeed intriguing that the overall dimer scaffolds in the crystal structures of WT, W285A and W285F are largely similar given their distinct phenotypes in vivo and in vitro9,11. Altered conformations of Trp233 and Trp337 in W285A (PDB ID: 4DNU9) due to the absence of the bulky indole ring evidently render a much weaker dimer similar to the disarrayed epicenter in I120K. W285F, on the other hand, exhibits a minimally altered dimer interface compared to WT. The benzene ring of Phe in general absorbs at shorter wavelength compared to the indole ring of a Trp residue. As a result, W285F is unable to directly respond to a UV-B signal9 (Fig. S3). Directional energy flow from UV-B-absorbing antenna pigments to the epicenter might be also affected. Indeed, the W285F dimer dissociates in response to UV-C instead of UV-B10.
The temperature-scan method is effective in probing early signaling events with small-amplitude structural changes tolerated by crystal lattices19. Compared to room-temperature methods20,36, reaction initiation at cryogenic temperatures has advantages for studying irreversible systems. The UVR8 crystals cracked under UV-B irradiation at room temperature9, thus they are not amenable to time-resolved crystallography that requires repetitive “pump-probe”. One cannot over-emphasize the importance of collecting dark and light datasets from the same crystal in achieving the best possible difference signals in dynamic crystallography19 (Fig. S2). It is also noteworthy that subtle differences due to non-crystallography symmetry can be exploited to explore structural events more advanced along the reaction trajectory.
In summary, this work provides structural insights into how light-induced dimer dissociation of UVR8 is triggered at the molecular level (Fig. 4). Excitation energy captured by UV-B-absorbing pigments are funneled to the epicenter (Fig. 4a), where charge separation between Trp285 and Trp233 occurs17. The resulting Coulomb force drives concerted motions in both indole rings and backbones of Trp285 and Trp233 (I120K). Consequently, the epicenter water is ejected (Fig. 4b) and inter-subunit interactions at the dimer interface are perturbed. Subsequent thermal relaxation of the backbone strains in Trp285/Trp233 results in partial unwinding of the 7-blade β-propeller structure (I180K). Synergistic actions at the epicenter and across the circular β-propeller scaffold lead to extensive disarray at the dimer interface, which eventually breaks the dimer apart (Fig. 4c).
Methods
Temperature-scan cryo-crystallography is a form of dynamic crystallography. It employs thermal constraints imposed by cryogenic temperatures to control the reaction progression and is able to capture early signaling events before extensive structural changes inevitably destroy the crystal lattices 19. Difference signals are so subtle that they are susceptible to non-isomorphism and random noises from crystal to crystal (Table S4). Difference R-factors between two dark datasets from different crystals can be much larger than those between the light and dark datasets from the same crystal (Table S1; Fig. S2). So in a dynamic crystallographic experiment, it is critically important to collect both the light and reference datasets from the same crystal in order to obtain the best possible difference densities in real space. We adopt real space analysis because it offers several advantages in dealing with dynamic crystallographic datasets from multiple crystals 21. First, difference electron densities are largely independent of the extent of reaction initiation; and they are very sensitive to structural changes even when light-induced populations are low20. Second, difference signals (FUV-FDark) are scattered in the entire reciprocal space (Fig. S2). In contrast, structural signals in real space are often concentrated around the action center such as the chromophore binding pocket and the catalytic site 19 (Fig. 1a). Such locality permits the use of a map mask, which greatly alleviates the computation burden in the dynamic crystallography data analysis. Third, difference densities derived from two datasets collected from the same crystal (Fig. S2) are much less susceptible to crystal-to-crystal variations. Therefore, they are more suitable for joint data analysis from multiple sources in a dynamic crystallographic experiment 21.
Supplementary Material
Acknowledgments
We thank L.C. Peng for providing Arabidopsis thaliana cDNA library. We thank the LS-CAT staff at Advanced Photon Source, Argonne National Laboratory for beamline support. XY acknowledges support from National Institute of Health grants GM036452 and EY024363. KHZ acknowledges support from National Natural Science Foundation of China (Grants 31270893 and 31110103912) and State Key Laboratory of Agricultural Microbiology of China. The TD-DFTB calculation work was supported by the Research Grants Council of Hong Kong SAR grant (project No. CityU 103812 to RZ) and grants from City University of Hong Kong (project No. 7200350 and 9610291 to JF). Use of the Advanced Photon Source was supported by the US Department of Energy (Office of Science and Office of Basic Energy Sciences) under Contract No. DE-AC02-06CH11357.
Footnotes
Author Contributions
X.Y. conceived and designed research, initiated and coordinated collaborations; X.Z. cloned, purified, crystallized wild type and mutant proteins, conducted mutagenesis and spectroscopic work; Z.R. carried out data analysis, refined and interpreted structures; Q.W., J.F. and R.Z. carried out TD-DFTB calculations; P.P.P. and K.T. contributed to mutagenesis and protein purification; K.H.Z. supervised and designed research at HZAU; X.Y. carried out X-ray experiments and data analysis; X.Y. and Z.R. wrote manuscript.
References
- 1.Heijde M, Ulm R. UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci. 2012;17:230–237. doi: 10.1016/j.tplants.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Fehér B, et al. Functional interaction of the circadian clock and UV RESISTANCE LOCUS 8-controlled UV-B signaling pathways in Arabidopsis thaliana. Plant J. 2011;67:37–48. doi: 10.1111/j.1365-313X.2011.04573.x. [DOI] [PubMed] [Google Scholar]
- 3.Kolowrat C, et al. Ultraviolet stress delays chromosome replication in light/dark synchronized cells of the marine cyanobacterium Prochlorococcus marinus PCC9511. BMC Microbiol. 2010;10:204. doi: 10.1186/1471-2180-10-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jenkins GI. The UV-B Photoreceptor UVR8: From Structure to Physiology. Plant Cell Online. 2014;26:21–37. doi: 10.1105/tpc.113.119446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein RM. Plants and near-ultraviolet radiation. Bot Rev. 1978;44:1–127. [Google Scholar]
- 6.Rizzini L, et al. Perception of UV-B by the Arabidopsis UVR8 Protein. Science. 2011;332:103–106. doi: 10.1126/science.1200660. [DOI] [PubMed] [Google Scholar]
- 7.Favory JJ, et al. Interaction of COP1 and UVR8 regulates UV B induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 2009;28:591–601. doi: 10.1038/emboj.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown BA, et al. A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci U S A. 2005;102:18225–18230. doi: 10.1073/pnas.0507187102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu D, et al. Structural basis of ultraviolet-B perception by UVR8. Nature. 2012;484:214–219. doi: 10.1038/nature10931. [DOI] [PubMed] [Google Scholar]
- 10.Christie JM, et al. Plant UVR8 Photoreceptor Senses UV-B by Tryptophan-Mediated Disruption of Cross-Dimer Salt Bridges. Science. 2012;335:1492–1496. doi: 10.1126/science.1218091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Hara A, Jenkins GI. In Vivo Function of Tryptophans in the Arabidopsis UV-B Photoreceptor UVR8. Plant Cell Online. 2012;24:3755–3766. doi: 10.1105/tpc.112.101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu M, Strid Å, Eriksson LA. Interactions and Stabilities of the UV RESISTANCE LOCUS8 (UVR8) Protein Dimer and Its Key Mutants. J Chem Inf Model. 2013;53:1736–1746. doi: 10.1021/ci4001822. [DOI] [PubMed] [Google Scholar]
- 13.Cloix C, et al. C-terminal region of the UV-B photoreceptor UVR8 initiates signaling through interaction with the COP1 protein. Proc Natl Acad Sci. 2012;109:16366–16370. doi: 10.1073/pnas.1210898109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kliebenstein DJ, Lim JE, Landry LG, Last RL. Arabidopsis UVR8 Regulates Ultraviolet-B Signal Transduction and Tolerance and Contains Sequence Similarity to Human Regulator of Chromatin Condensation 1. Plant Physiol. 2002;130:234–243. doi: 10.1104/pp.005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofmann NR. The Molecular Mechanism of the UVR8 UV-B Photoreceptor. Plant Cell Online. 2012;24:3485–3485. doi: 10.1105/tpc.112.240910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu M, Strid Å, Eriksson LA. Photochemical Reaction Mechanism of UV-B-Induced Monomerization of UVR8 Dimers as the First Signaling Event in UV-B-Regulated Gene Expression in Plants. J Phys Chem B. 2014;118:951–965. doi: 10.1021/jp4104118. [DOI] [PubMed] [Google Scholar]
- 17.Voityuk AA, Marcus RA, Michel-Beyerle M-E. On the mechanism of photoinduced dimer dissociation in the plant UVR8 photoreceptor. Proc Natl Acad Sci. 2014:201402025. doi: 10.1073/pnas.1402025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z, et al. Quenching Dynamics of Ultraviolet-Light Perception by UVR8 Photoreceptor. J Phys Chem Lett. 2014;5:69–72. doi: 10.1021/jz402396k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang X, Ren Z, Kuk J, Moffat K. Temperature-scan cryocrystallography reveals reaction intermediates in bacteriophytochrome. Nature. 2011;479:428–432. doi: 10.1038/nature10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren Z, et al. A Molecular Movie at 1.8 Å Resolution Displays the Photocycle of Photoactive Yellow Protein, a Eubacterial Blue-Light Receptor, from Nanoseconds to Seconds†. Biochemistry (Mosc) 2001;40:13788–13801. doi: 10.1021/bi0107142. [DOI] [PubMed] [Google Scholar]
- 21.Ren Z, et al. Resolution of structural heterogeneity in dynamic crystallography. Acta Crystallogr Sect D. 2013;69:946–959. doi: 10.1107/S0907444913003454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang X, Yang P, Ouyang X, Chen L, Deng XW. Photoactivated UVR8-COP1 Module Determines Photomorphogenic UV-B Signaling Output in Arabidopsis. PLoS Genet. 2014;10:e1004218. doi: 10.1371/journal.pgen.1004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heilmann M, Christie JM, Kennis JTM, Jenkins GI, Mathes T. Photoinduced transformation of UVR8 monitored by vibrational and fluorescence spectroscopy. Photochem Photobiol Sci. 2014 doi: 10.1039/C4PP00246F. [DOI] [PubMed] [Google Scholar]
- 24.McGuinness CD, Sagoo K, McLoskey D, Birch DJS. Selective excitation of tryptophan fluorescence decay in proteins using a subnanosecond 295nm light-emitting diode and time-correlated single-photon counting. Appl Phys Lett. 2005;86:261911. [Google Scholar]
- 25.Peng H, et al. Ultraviolet light-emitting diodes operating in the 340nm wavelength range and application to time-resolved fluorescence spectroscopy. Appl Phys Lett. 2004;85:1436–1438. [Google Scholar]
- 26.Heijde M, et al. Constitutively active UVR8 photoreceptor variant in Arabidopsis. Proc Natl Acad Sci. 2013;110:20326–20331. doi: 10.1073/pnas.1314336110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neer EJ, Schmidt CJ, Nambudripad R, Smith TF. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 28.Glazer AN. Light guides. Directional energy transfer in a photosynthetic antenna. J Biol Chem. 1989;264:1–4. [PubMed] [Google Scholar]
- 29.Liu Z, et al. Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution. Nature. 2004;428:287–292. doi: 10.1038/nature02373. [DOI] [PubMed] [Google Scholar]
- 30.Porezag D, Frauenheim T, Köhler T, Seifert G, Kaschner R. Construction of tight-binding-like potentials on the basis of density-functional theory: Application to carbon. Phys Rev B. 1995;51:12947–12957. doi: 10.1103/physrevb.51.12947. [DOI] [PubMed] [Google Scholar]
- 31.Elstner M, et al. Self-consistent-charge density-functional tight-binding method for simulations of complex materials properties. Phys Rev B. 1998;58:7260–7268. [Google Scholar]
- 32.Niehaus TA, et al. Tight-binding approach to time-dependent density-functional response theory. Phys Rev B. 2001;63:085108. [Google Scholar]
- 33.Niehaus TA. Approximate time-dependent density functional theory. J Mol Struct THEOCHEM. 2009;914:38–49. [Google Scholar]
- 34.Rogers DM, Besley NA, O’Shea P, Hirst JD. Modeling the Absorption Spectrum of Tryptophan in Proteins. J Phys Chem B. 2005;109:23061–23069. doi: 10.1021/jp053309j. [DOI] [PubMed] [Google Scholar]
- 35.Reshetnyak YK, Koshevnik Y, Burstein EA. Decomposition of Protein Tryptophan Fluorescence Spectra into Log-Normal Components. III Correlation between Fluorescence and Microenvironment Parameters of Individual Tryptophan Residues. Biophys J. 2001;81:1735–1758. doi: 10.1016/S0006-3495(01)75825-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wöhri AB, et al. Light-Induced Structural Changes in a Photosynthetic Reaction Center Caught by Laue Diffraction. Science. 2010;328:630–633. doi: 10.1126/science.1186159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.