Abstract
Singleminded-2s (SIM2s) is a member of the bHLH/PAS family of transcription factors and a key regulator of mammary epithelial cell differentiation. SIM2s is highly expressed in mammary epithelial cells and down regulated in human breast cancer. Loss of Sim2s causes aberrant mouse mammary ductal development with features suggestive of malignant transformation, whereas over-expression of SIM2s promotes precocious alveolar differentiation in nulliparous mouse mammary glands, suggesting that SIM2s is required for establishing and enhancing mammary gland differentiation. To test the hypothesis that SIM2s regulates tumor cell differentiation, we analyzed SIM2s expression in human primary breast ductal carcinoma in situ (DCIS) samples and found that SIM2s is lost with progression from DCIS to invasive ductal cancer (IDC). Utilizing a MCF10DCIS.COM progression model, we have shown that SIM2s expression is decreased in MCF10DCIS.COM cells compared to MCF10A cells and reestablishment of SIM2s in MCF10DCIS.COM cells significantly inhibits growth and invasion in vitro and in vivo. Analysis of SIM2s-MCF10DCIS.com tumors showed that SIM2s promoted a more differentiated tumor phenotype including the expression of a broad range of luminal markers (CSN2 (β-casein), CDH1 (E-cadherin), and KER18 (keratin-18)) and suppressed genes associated with stem cell maintenance and a basal phenotype (SMO (smoothened), p63, SLUG (snail-2), KER14 (keratin-14) and VIM (vimentin)). Furthermore, loss of SIM2s expression in MCF10DCIS.COM xenografts resulted in a more invasive phenotype and increased lung metastasis likely due to an increase in hedgehog signaling and matrix metalloproteinase expression. Together, these exciting new data support a role for SIM2s in promoting human breast tumor differentiation and maintaining epithelial integrity.
Keywords: SIM2s, Breast Cancer, DCIS, Differentiation
Introduction
Ductal Carcinoma In Situ (DCIS) has been shown to be a precursor to invasive ductal cancer (IDC) (1) with 20–30% of DCIS showing evidence of invasion upon diagnosis (2, 3). Though the progression of DCIS to IDC is believed to be an important aspect of tumor aggressiveness, prognosis and molecular markers that can predict progression are poorly understood. Analysis of biomarkers and molecular profiles of IDC and DCIS have failed to identify progression-specific pathways (4–8). Therefore, determining the mechanisms by which some DCIS progress is critical for future breast cancer diagnostics and treatment.
There is increasing evidence that DCIS are heterogeneous tumors, which enhances complexity when attempting to define the mechanisms that promote progression to IDC in in vivo models. Recent studies utilizing the MCF10DCIS.COM cell line, which was derived from the non-cancerous MCF10A cell line, have shown that these cells contain a unique bipotent progenitor ability that forms a myoepithelial cell layer in addition to luminal-type cells in vivo, which results in basal-like DCIS with high similarities to human DCIS samples (9–13). Intraductal and flank injections have shown that MCF10DCIS.COM cells not only form DCIS like structures, but also spontaneously progress to invasive breast cancer (9, 11). These observations suggest that MCF10DCIS.COM cells are a unique model to study DCIS, as well as the role of different factors in regulating the progression to IDC.
We have previously shown that the basic helix-loop-helix/PER-ARNT-SIM (bHLH/PAS) transcription factor Singleminded-2s (SIM2s) plays a role in normal mammary gland development as well as in promoting tumor cell differentiation (14–18). Loss of Sim2s expression in the mouse mammary gland and in normal breast and breast cancer cell lines is associated with an epithelial mesenchymal transition(EMT), whereas over-expression of Sim2s under the Mouse Mammary Tumor Virus (MMTV) promoter induces precocious alveolar differentiation in nulliparous mice and delayed forced involution (14–18). SIM2s is down-regulated in primary human breast cancer samples, and re-establishment of SIM2s in human breast cancer cell lines inhibits cell proliferation and invasion (15, 17). Moreover, we have found that SIM2s mRNA gene expression is inhibited by activation of C/EBPβ and NOTCH signaling, two known EMT promoters, in RAS-transformed MCF10A cells (15). Both C/EBPβ and NOTCH expression have been shown to have to play a role in breast cancer progression through mediation of breast cancer stem cells, cellular proliferation, and oncogenesis (19–23). Together, these observations suggest that SIM2s is a tumor suppressor gene that is required to maintain epithelial integrity by inhibiting EMT-like pathways and promoting differentiation.
In the studies here, we examined the role of SIM2s in regulating the progression of DCIS to IDC and metastasis, while promoting the less aggressive, luminal-like breast cancer subtype. Based on our studies of SIM2s as a breast cancer tumor suppressor, we hypothesize that SIM2s expression will decrease spontaneous metastasis seen in the MCF10DCIS.COM model, and play a role in the inhibition of DCIS progression.
Results
SIM2s is lost during DCIS progression
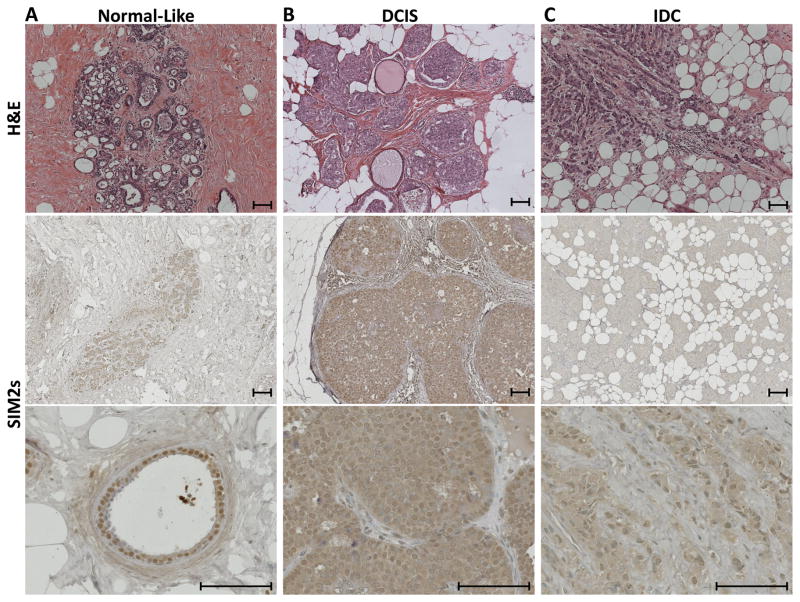
To determine the role of SIM2s in progression from DCIS to IDC, fourteen human primary DCIS and IDC samples were analyzed for SIM2s expression by immunohistochemistry. We have previously shown that SIM2s is localized in the nuclei of human breast and mouse mammary ductal epithelial cells (16, 17), and similar punctate staining was observed in normal ductal structures surrounding the tumors analyzed (Fig 1A). In these studies, we found that SIM2s expression is prominent in both the nucleus and cytoplasm in over 75% of DCIS samples (Fig. 1B), suggesting a loss in localization at the onset of progression. In contrast, as DCIS progresses to IDC, SIM2s staining is dramatically down-regulated with no evidence of nuclear expression in over 80% of IDC samples (Fig. 1C), supporting a role for loss of SIM2s in breast cancer progression.
Figure 1. SIM2s expression is progressively lost in human ductal carcinoma in situ (DCIS) transition to invasive ductal cancer (IDC).
A – Human Normal-like tissue, B – Human DCIS, C – Human IDC. Top Row – H&E staining of Normal, DCIS, and IDC samples. Bottom Rows - SIM2s immunohistochemistry of Normal, DCIS and IDC samples. Normal-like structures show clean, punctate nuclear staining. DCIS samples show nuclear and cytoplasmic SIM2s staining in over 75% of samples, and this expression is lost in over 80% of IDC samples. Images were taken at 10x and 40x objective (6.3x and 25.2x), scale bars represent 100μm. An n=14 was used for each tumor classification (DCIS and IDC).
Re-establishment of SIM2s in the MCF10.DCIS.COM cell line induces genetic and morphologic changes in vitro
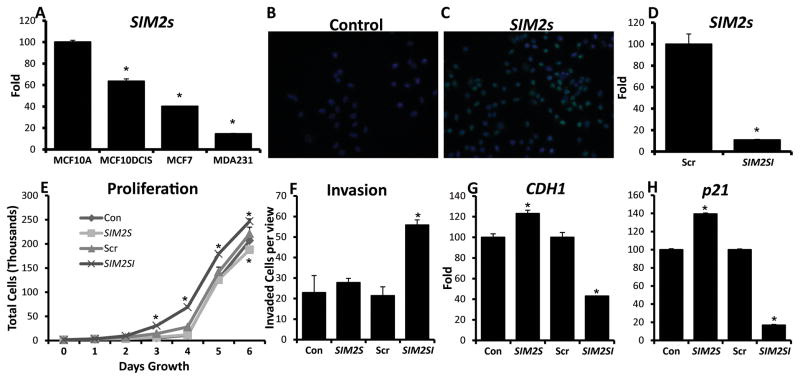
MCF10DCIS.COM cells are a unique human breast cancer cell line which form DCIS-like lesions in in vivo mouse models, similar to primary human DCIS lesions, and spontaneously progress to invasive cancer (13). In addition, MCF10DCIS.COM cells are thought to contain a bipotent progenitor population that generate both myoepithelial and luminal cells, mimicking the heterogeneity observed in human DCIS tumors (9, 11). Quantitative real-time PCR (Q-PCR) analysis of SIM2s levels showed that SIM2s is significantly down-regulated in MCF10DCIS.COM cells compared to parent MCF10A cells, however, SIM2s levels are still higher in MCF10DCIS.COM cells than levels found in luminal MCF7 and basal MDA.MB.231 cell lines, suggesting that SIM2s expression is lost with progression (Fig. 2A). To determine the effect of SIM2s loss and gain of function, we stably transduced MCF10DCIS.COM cells with SIM2s and previously validated SIM2s-shRNA (SIM2si) lentiviruses (15, 16). SIM2s levels and localization were confirmed using Q-PCR and immunofluorescence (Fig. 2B, C, & D) (Supplemental Fig. 1). Q-PCR analysis of SIM2s mRNA levels show an approximate 80% loss of expression in SIM2si cells compared to scrambled controls (Fig. 2D). In growth assays, SIM2s inhibited cell proliferation, whereas loss of SIM2s led to a significant increase in proliferation as compared to scrambled controls (Fig. 2E). We observed no change in invasive potential with SIM2s over-expression in Boyden chamber assays; however, there was a significant increase in invasion in the SIM2si cells (Fig. 2F). Q-PCR analysis also showed a significant increase in E-Cadherin (CDH1) expression with SIM2s expression, as well as a decrease with SIM2si (Fig. 2G). Similarly, p21, an important senescence and cell cycle regulator, was also significantly altered in response to SIM2s (Fig. 2H).
Figure 2. Analysis of MCF10DCIS cell transductions in vitro show changes in proliferation, invasion, and differentiation markers.
A – Q-PCR analysis of SIM2s mRNA levels in MCF10DCIS.COM and parent MCF10A cells, as well as commonly used breast cancer cell lines MCF7 and MDA.MB.231. B, C - Immunofluorescent staining of SIM2s to confirm nuclear SIM2s overexpression. D – Q-PCR analysis of SIM2s in the MCF10DCIS.com cell line confirming an approximate 80% loss of expression in adhered SIM2si cells. E – Proliferation assays confirm that SIM2s expression inhibits breast cancer cell proliferation, while loss of SIM2s increases growth. The values shown are the mean ± SE of triplicate samples. F - Boyden chamber invasion assay shows significantly more SIM2si cells were able to invade and migrate compared to controls. Values are the average number of cells per five fields per membrane of three separate plates. G & H - Q-PCR analysis of differentiation markers CDH1 and p21. * = p-value < .05.
In vivo analysis of transduced MCF10DCIS.com cell xenografts show changes in growth and morphology
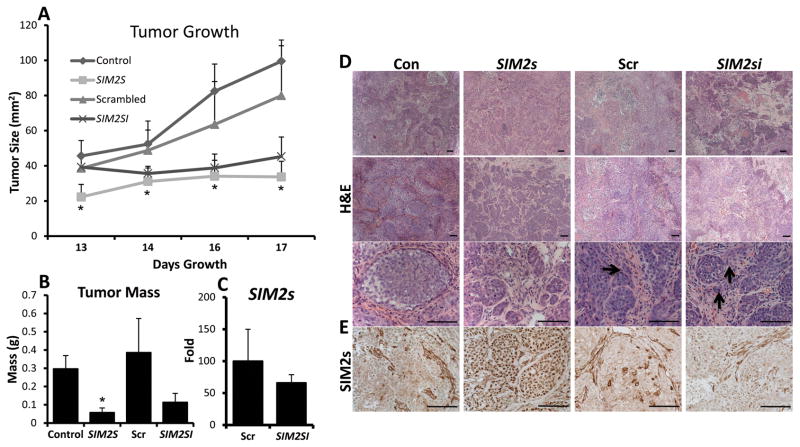
Similar to the in vitro results, we found that SIM2s xenograft tumors grew significantly slower than controls (Fig. 3A & B). In contrast to our in vitro observations, we were surprised to find that down-regulation of SIM2s led to a decreased trend in tumor size and weight as compared to scrambled controls; however statistical significance was not obtained (Fig. 3A & B). To determine if SIM2s expression affected changes in tumor morphology, we analyzed H&E sections from SIM2s and SIM2si tumors. Histological analysis confirmed a distinct phenotypic differences with SIM2s expression (Fig. 3D): SIM2s tumors had a more differentiated phenotype including lobular-like structures with intact myoepithelial layers whereas SIM2si tumors, despite growing at a slower rate, were more invasive and had large necrotic areas as compared to scrambled controls (Fig. 3D, invasion shown by arrows). These observations are similar to previous studies that found no correlation between MCF10DCIS.COM growth in vivo and invasive potential and support the hypothesis that SIM2s inhibits DCIS progression by promoting and maintaining a luminal phenotype (9, 11, 12). Immunohistological analysis of tumors confirmed that SIM2s and SIM2si tumors continued to overexpress or knock down SIM2s protein levels in vivo (Fig. 3E). Q-PCR analysis of SIM2s levels confirmed an approximate 50% knockdown of SIM2s expression in SIM2si xenografts (Fig. 3C).
Figure 3. Differential SIM2s expression regulates growth in vivo.
A & B – Analysis of xenograft tumor growth and mass shows that over-expression and loss of SIM2s expression inhibits xenograft growth. C – Q-PCR analysis of SIM2s mRNA expression in SIM2si tumors shows an approximate 50% loss of expression in vivo. D – H&E histological analysis shows that SIM2s expressing tumors exhibit smaller, more lobulo-like structures throughout the tumor, with less necrosis and inflammation. Images were taken using a 5x objective (5x) 10x objective (6.3x) and a 40x objective (25.2x). Scale bars represent 100μm. Arrows indicate areas of invasion. D – Histological analysis of SIM2s expression in xenografts confirms SIM2s overexpression and loss of expression. Images were taken using a 40x objective (25.2x). Scale bars represent 100μm. * = p-value < .05.
SIM2s inhibits expression of basal breast cancer markers
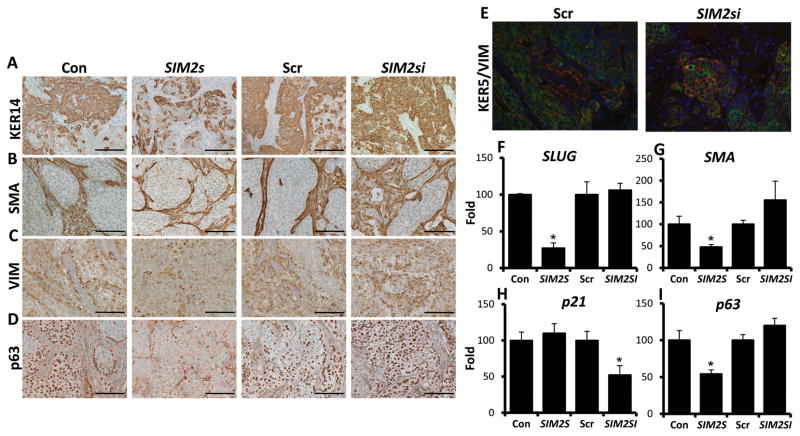
We have previously established that SIM2s is a negative regulator of EMT and promotes mammary gland differentiation in vitro and in vivo (14, 17, 18). To determine if the morphological changes associated with SIM2s expression in MCF10DCIS.COM xenografts are similar to our previous gain and loss of function studies in the mouse mammary gland, we examined basal markers involved in breast cancer progression and EMT via Q-PCR and immunohistochemical analysis. Immunostaining for basal markers including keratin 14 (KER14), alpha-smooth muscle actin (αSMA), vimentin (VIM) and p63 show a decrease in staining in SIM2s over-expressing MCF10DCIS.COM tumors, and up-regulation with loss of SIM2s (Fig. 4A, B, C, & D). Moreover, indicative of enhanced tumor aggressiveness and progression, we observed an increase in co-localization of KER5 and VIM in SIM2si tumors (Fig. 4E, Supplemental Fig. 2). Further analysis of EMT and basal markers, including the EMT transcription factor, SLUG, which we have previously shown is directly regulated and suppressed by SIM2s, was significantly decreased in SIM2s tumors along with SMA and p63 (Fig. 4F, G, & I). These results are consistent with SIM2s’s role in mammary gland differentiation as p63, SMO and SLUG regulate cell differentiation and stem cell maintenance, which suggests that re-establishment of SIM2s is sufficient to promote a decrease in basal breast cancer markers (24–30). Analysis of p21 mRNA levels also showed a decrease in SIM2si xenografts, similar to what was seen in vitro (Fig. 4H). p21 is an important cell cycle regulator and is involved in the p53 stress response pathway as well as senescence (31–33).
Figure 4. SIM2s decreases markers associated with basal breast cancer in MCF10DCIS.COM xenografts.
A, B, C, & D – Immunohistochemical staining for basal markers including KER14, SMA, VIM, and p63. Images were taken using a 40x objective (25.2x). Scale bars represent 100μm. E – KER5/VIM immunofluorescence shows increased overlap of KER5 and VIM in SIM2si xenografts, which is associated with enhanced invasive potential and aggressiveness. F, G, H, & I – Q-PCR analysis of basal markers SLUG, SMA, and p63, as well as cell cycle regulator p21. * = p-value < .05.
SIM2s promotes expression of luminal markers
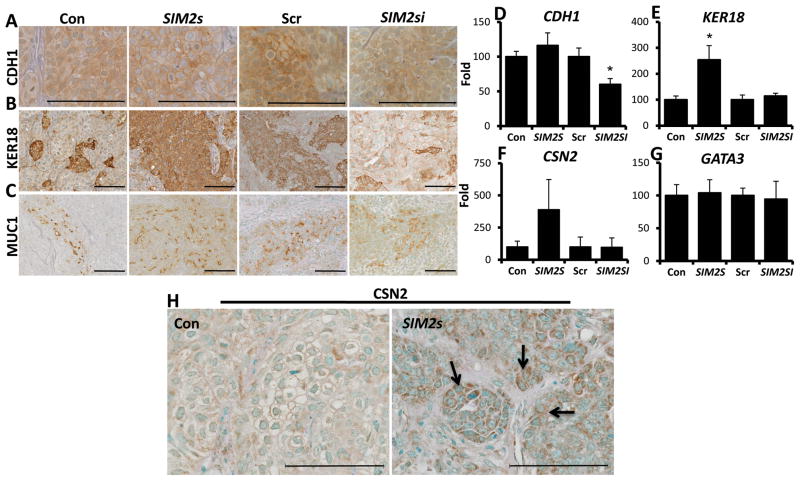
To determine whether differential expression of SIM2s regulates prominent luminal markers in DCIS xenografts, we evaluated the expression of E-cadherin (CDH1), keratin-18 (KER18), and Mucin-1 (MUC1). Immunostaining results for CDH1 showed increased trends and localization to the cellular membrane in the SIM2s over-expressing tumors, and a decrease in localized staining with SIM2si tumors (Fig. 5A). Interestingly, Q-PCR analysis of CDH1 showed no significant changes with SIM2s overexpression; however SIM2si tumors had significantly lower levels of CDH1 mRNA compared to controls (Fig. 5D). Analysis of keratin 18 (KER18) showed a significant positive relationship with SIM2s expression; KER18 protein and mRNA levels were elevated in SIM2s tumors and protein appeared decreased with loss of SIM2s (Fig. 5B & E). MUC1, an apical luminal marker that is often mis-localized in cancer, showed an increase in apical staining in SIM2s xenografts, while a loss of localization is seen in SIM2si tumors (Fig. 5C). In addition, we also examined changes in the transcription factor GATA3, a prognostic factor associated with positive breast cancer outcome and regulator of breast tumor cell differentiation (34, 35). Analysis of GATA3 mRNA expression showed no differences in with SIM2s expression, indicating that the differentiation phenotype associated with SIM2s expression appears to be GATA3 independent in this model (Fig. 5G). To determine whether SIM2s tumors undergo partial lactogenic differentiation, we also examined the expression of the milk protein β-casein (CSN2). Immunostaining for β-casein in SIM2s tumors not only showed increased protein expression over controls, but also secretory globule formation indicative of milk protein expression as seen in the lactating mammary gland (Fig. 5H, arrows) and a distinct trend in CSN2 mRNA gene expression in SIM2s tumors (Fig. 5F & H). Overall these data support the hypothesis that expression of SIM2s induces a luminal phenotype, including expression of milk proteins, while loss of SIM2s significantly decreases the expression of luminal markers.
Figure 5. SIM2s xenografts have increased levels of luminal markers and express β-Casein.
A, B, & C –Re-establishment of SIM2s promotes apical localization of CDH1 and increased KER18 expression, while SIM2s loss results in a decrease in expression. SIM2s expression also promotes apical expression of luminal marker MUC-1, whereas loss of SIM2s causes a loss of localization. D, E, F, & G – Q-PCR analysis of luminal markers CDH1, KER18, CSN2, and GATA3. H – Immunohistochemical analysis for CSN2 shows elevated levels in SIM2s tumors (see arrows). Images were taken using a 40x objective (25.2x) and 63x objective (63x). Scale bars of images represent 100μm. * = p-value < .05.
Angiogenesis and metastasis are inhibited by SIM2s
We have shown that gain and loss of SIM2s expression in MCF10DCIS.COM correlates with phenotypic changes in invasive behavior and expression of luminal and basal differentiation markers. To investigate if these phenotypic differences affect cancer progression and metastasis, we analyzed lungs from tumor bearing mice for vimentin (VIM), which is expressed in MCF10DCIS.COM cells, but not in normal lung tissue (36). The results showed positive VIM staining in lungs from control and scrambled tumors; however, we did not detect VIM expression in lungs from mice with SIM2s-expressing tumors (Fig. 6A & C). To confirm SIM2s-dependent changes in progression, we performed Q-PCR analysis for human specific β-2-globulin (β2M) gene expression in mouse lungs, which has been previously shown as an indicator of human cells in mouse lung tissues either due to metastasis or circulating tumor cells (36). While moderate levels of β2M expression were observed in control tissues, we did not detect β2M expression in lung tissue from mice with SIM2s over-expressing tumors (Fig. 6B). In contrast, the majority of the lungs from mice with SIM2si tumors had high levels of β2M expression (Fig. 6B). Consistent with differences in metastatic potential and SIM2s expression, we observed a decrease in angiogenesis in SIM2s tumors compared to controls (Fig. 6D and E) Interestingly, this difference is seen without taking into account the drastic change in tumor size (Fig. 6E). In contrast, no significant change in angiogenesis was seen with loss of SIM2s, indicating that the increase in metastasis is due to other metastatic processes.
Figure 6. SIM2s inhibits metastasis and alters angiogenesis.
A – Immunohistochemical analysis of lung tissue for vimentin (VIM) positive micrometastases showed decreased staining in SIM2s tumors, whereas loss of SIM2s enhanced lung metastasis. Images were taken with a 40x objective (25.2x). B – Q-PCR analysis for human β-2-Globulin as an indicator of lung metastasis confirmed the effect of SIM2s on metastasis with a decreased in β-2-Globulin expression in SIM2s tumors and increased expression with loss of SIM2s. Data is shown as the number of β-2G positive samples out of the total number of samples analyzed. C – Increased magnification (63x) of VIM staining to indicate the presence of vimentin positive cells in Scrambled controls and SIM2si tumors. D – Immunostaining for CD31 expression showed that SIM2s tumors have smaller blood vessels that remained on the outer perimeter of the tumors, whereas SIM2si tumors had an increased trend in angiogenesis. E – Quantification of CD31 staining by measuring blood vessel length confirms trends seen with CD31 immunohistochemistry. Images were taken using a 10x objective (6.3x) and a 40x objective (25.2x). Scale bars in images represent 100μm. *= p-value < .05
SIM2s-dependent regulation of matrix metalloproteinase and hedgehog signaling
We have previously shown cell type specific SIM2s-dependent regulation of matrix metalloproteinase (MMP). These studies found SIM2s binds to the MMP3 promoter and inhibits MMP3 expression in MDA.MB.435 cells (17), while MMP2 expression is increased in MCF7-SIM2si cells and mouse Sim2s knockout mammary glands (16). Using Q-PCR, we examined MMP gene expression in SIM2s over and under-expressing tumors (Fig. 7). The results show that SIM2s xenografts have decreased MMP expression, whereas SIM2si tumors have increased MMP levels (Fig. 7A, B, C, & D). In addition, the hedgehog signaling pathway has been shown to be overexpressed in breast cancer and play a role in proliferation and differentiation (27, 28). Analysis of Indian Hedgehog (iHH) and SMO by Q-PCR showed a drastic increase in iHH and SMO mRNA levels with loss of SIM2s (Fig. 7E & F).
Figure 7. Loss of SIM2s increases tumor invasiveness through MMP expression and Hedgehog signaling.
A, B, C & D – Q-PCR analysis of various MMPs showing a decrease in expression with SIM2s tumors and an increase in SIM2si tumors, a likely mechanism for increased tumor invasiveness and metastasis. E & F – Q-PCR analysis of Indian Hedgehog (IHH) and Smoothened (SMO) show elevated levels of expression in SIM2si tumors. * - p-value < .05.
Discussion
There is significant evidence that relates the differentiation status of a tumor with its metastatic potential (37–39). Analysis of luminal markers by microarray analysis and immunohistochemistry have confirmed that loss of epithelial characteristics correlates with an increase in cancer progression (40–43). Though pathways have been identified that promote differentiation, few molecules have been identified that maintain and enhance differentiation potential. We have previously shown that SIM2s is a negative regulator of EMT in normal breast, breast cancer cell lines and the mouse mammary gland by suppressing SLUG and MMP2 gene transcription (16, 17). In contrast, overexpression of Sim2s in the mouse mammary gland under the MMTV promoter induces precocious lactogenic differentiation in virgin mice and delayed involution following forced weaning (14, 18). Together, these observations led us to hypothesize that expression of SIM2s in breast cancer would inhibit tumor growth by regulating differentiation potential. We report here that SIM2s expression is lost in human DCIS progression to invasive breast cancer and, utilizing the MCF10DCIS.com progression model, we demonstrate that reestablishment of SIM2s promotes a more luminal-like phenotype, whereas down-regulation of SIM2s leads to an increase in invasive potential. These new data support a role for SIM2s in regulating epithelial identity and a potential novel molecular target for differentiation therapy.
At the onset of xenograft studies, as expected we observed that tumors over-expressing SIM2s grew at a slower rate, as compared to controls, with lower amounts of necrosis. Surprisingly, we found that SIM2si tumors exhibited a decreased trend in growth compared to scrambled controls. However, upon histological analysis, we observed a more invasive phenotype and large necrotic areas in the SIM2si tumors. In comedo DCIS, necrotic centers have been implicated as a more rapidly progressing DCIS with a worse prognosis compared to DCIS lacking necrosis (12). Further analysis of luminal and basal-like breast cancer markers showed distinct trends regarding SIM2s gain and loss of function. We observed that basal markers were inhibited in SIM2s tumors, with upward trends occurring in SIM2si tumors. Conversely, when examining luminal markers, we found an increase either in the appropriate localization (CDH1 and MUC1) or increased expression with SIM2s tumors and a loss of localization and expression with SIM2si. Another unique expression pattern was the presence of αSMA in the SIM2s over-expressing tumors. We anticipated that re-establishment of SIM2s in MCF10DCIS.COM cells would inhibit the bipotent progenitor capabilities of the cell line, and thus prevent the development of a myoepithelial layer in vivo. Surprisingly, however, SIM2s did not affect the bipotent progenitor ability. The significant decrease in SMA mRNA expression seen in SIM2s tumors could possibly be due to the smaller size of the tumors and lobular units rather than a biologically significant change in SMA expression. Finally, the expression of CSN2 in SIM2s tumors further indicates that not only does reestablishment of SIM2s maintain epithelial integrity, but also promotes functional differentiation.
Recent studies have identified a number of transcription factor cascades that control key events in regulating mammary epithelial differentiation including GATA3, ELF5, NOTCH and C/EBPβ (21, 22, 35, 44). For instance, analysis of Gata3 conditional knockout mammary glands found an increase in luminal progenitor cells during alveolar differentiation and a defect in virgin ductal morphogenesis as a result of compromised estrogen responsiveness (34), whereas loss of Elf5 has no effect on virgin development and exclusively regulates alveolar cell fate (45). Although these factors play key roles in promoting differentiation along the luminal lineage, it begs the question: what maintains mammary ductal epithelial cells in a differentiated state and keeps them from de-differentiating and acquiring stem cell characteristics? It can be hypothesized that this/these factor(s) may target pathways regulating tumor initiating cell self-renewal by blocking induction of EMT and maintaining epithelial integrity. Interestingly, studies with GATA3 have shown that while overexpression is sufficient to promote epithelial differentiation, forced loss of GATA3 is not tolerated (35). With SIM2s we observed that while overexpression is sufficient to induce differentiation, loss of expression promotes malignant transformation. Moreover, the lack of change in GATA3 in the SIM2s tumors, which has been shown to promote differentiation and inhibit breast cancer growth and metastasis, indicates that SIM2s is operating independently of GATA3. This has significant implications for our understanding of normal mammary development and breast cancer progression by introducing a novel pathway that may play a role in maintaining tumor initiating cells or in promoting functional differentiation.
One of the most interesting phenotypes of this study was the dramatic increase in lung metastasis with loss of SIM2s. MMPs have long been attributed as playing a key role in breast cancer invasion and (46–50) are integral to the degradation of the basement membrane during normal biology functions including mammary gland involution and lobular development (14, 50–56). Previous work in our lab has shown that SIM2s differentially regulates MMP2 and MMP3 gene expression (16, 17). In the study here, we observed a SIM2s-dependent regulation of MMP gene expression in SIM2s over and under-expressing DCIS xenografts. We showed that MMP1 and MMP10 were significantly decreased by SIM2s expression, whereas MMP3 was significantly increased by loss of SIM2s, accompanied by increased trends in other MMPs analyzed. This data indicates that a likely mechanism for SIM2s’ effect on metastasis may be through a global regulation of MMPs in a complex and varied manner. Hedgehog signaling has also been implicated in breast cancer progression and invasion as well as the maintenance of cancer stem cells (27, 28, 57, 58). In our study the decrease in SMO expression in SIM2s tumors is unique since SMO up-regulation in tumors causes a phenotype similar to that seen in SIM2si tumors and Sim2s knockout mammary glands, including increased proliferation and altered differentiation, possibly indicating interaction between these genes (28). Studies in gastric and ovarian cancer have also connected hedgehog signaling with invasion and MMP expression (59, 60), suggesting a potential mechanism of action by which SIM2s inhibits invasion and metastasis in vivo.
These observations provide a possible mechanism for the promotion of tumor differentiation through re-establishment of SIM2s, as well as possible roles for SIM2s in breast cancer progression. Determining what point of the metastatic cascade that SIM2s functions will be key in understanding SIM2s’ role in breast cancer progression. Moreover, the use of established metastatic models and the impact of SIM2s on breast cancer subtypes will help elucidate the mechanism by which SIM2s affects tumor progression as well as its possible effect on tumor initiating cells. Together, these results suggest that SIM2s has the potential to be a novel target for differentiation therapy by inhibiting or reversing breast cancer progression.
Materials and Methods
Cell Culture
MCF10DCIS cells were generously provided by Dr. Dan Medina (Baylor College of Medicine, Houston, TX) and maintained in DMEM-F12 (Invitrogen, Carlsbad, CA) with 10% horse serum (Atlanta Biologicals). HEK-293T Ampho-Phoenix packaging cells were obtained with permission from Gary Nolan at Stanford University and maintained as recommended.
Plasmids and Lentiviral Transductions
Lentiviral transduction of MCF10DCIS.com cells was performed as previously described (17). Lentiviral transduction, SIM2s shRNA, and SIM2s over-expression plasmids have been previously described (15–17).
Invasion assays
Invasion was measured using control and Matrigel-coated invasion chambers (Falcon BD, Franklin Lakes, NJ). A total of 12,500 cells were seeded in serum-free Dulbecco’s modified Eagle’s medium (DMEM-F12) Invitrogen, Carlsbad, CA) in the upper chamber, with serum-containing medium in the lower chamber as a chemo attractant. After 18 h at 37°C, cells were scraped from the upper chamber with a cotton swab, and the undersides of the membranes were fixed in 3.8% paraformaldehyde (Sigma, St. Louis, MO), stained with DAPI (4′,6′-diamidino-2-phenylindole)(Invitrogen), and counted. The percent invasion was calculated according to the manufacturer’s instructions.
Proliferation Assays
Proliferation was measured using a Coulter particle counter. 15,000 cells of each transduction were plated in triplicates on 6 well plates, then every 24 hours cells were trypsinized and counted in triplicate. The procedure continued for 6 days, or until the cells reached 100% confluency.
Xenograft Studies
For xenograft studies, MCFDCIS.com cells (50,000) were injected subcutaneously into 8 to 14-week-old female nude mice in 50% Matrigel (BD Biosciences, Bedford, MA). Tumors were allowed to grow for 18 days, and were measured using calipers starting on day 13. Xenografts were weighed at harvest and either snap frozen in liquid nitrogen and stored at −80°C for DNA/RNA purification or formalin fixed and paraffin embedded. Animal experiments were conducted following protocols approved by the Texas A&M Animal Care and Use Committee.
Immunostaining
Immunostaining was carried out as previously described (17). Samples were incubated in blocking solution for one hour, followed by incubation in primary antibody overnight at 4°C. Antibodies used were SIM2s (Santa Cruz, Santa Cruz, CA), SMA (Sigma, St. Louis, MO), Keratin 14 (Covance, Princeton, NJ), Vimentin (Sigma), p63 (NeoMarkers, Freemont, CA), Keratin 5 (Covance), E-Cadherin (Cell Signaling, Beverly, MA), Keratin 18 (NeoMarkers), Mucin-1 (NeoMarkers), β-Casein (Santa Cruz), and PECAM-1 (Santa Cruz). Tissue preparation and Hematoxylin and Eosin (H&E) staining were done by the Histology Core Facility at Texas A&M University College of Veterinary Medicine & Biomedical Sciences. Immunostaining for SIM2s was performed on ductal carcinoma in situ and invasive breast cancer tissue paraffin sections provided by the Pathology Department at the University of Kansas Medical Center per human subjects approved protocols. CD31 quantification was performed using ImageJ Software (NIH) and has been previously described (61, 62).
RNA Isolation and Reverse Transcription
RNA was isolated from tissues using Trizol Reagent (Invitrogen, Carlsbad, CA), followed by purification using a Qiagen RNEasy Mini Kit (Qiagen, Valencia, CA) as previously described (17). 2μg total RNA was reverse-transcribed into cDNA using oligo(dT) and Superscript II Reverse transcriptase (Invitrogen). Reverse Transcription (RT) reactions were performed on tissues as previously described (63).
Quantitative PCR
Quantitative PCR was performed as previously described (17). QPCR was performed using SYBR Green Master Mix (Applied Biosystems, Foster City, CA). Primers (IDT, Coralville, IA) for analysis of SIM2s, and CSN2 mRNA levels have been previously described (18) as well as ECadherin (CDH1), SLUG, and Keratin 18 (Ker18)(16). Primers used for further mRNA analysis include p21(FP: CCT-AAT-CCG-CCC-ACA-GGA-A; RP: AAG-ATG-TAG-AGC-GGG-CCT-TTG), SMA (FP: CAA-GTG-ATC-ACC-ATC-GGA-AAT-G; RP: AGC-AGA-CTC-CAT-CCC-GAT-GA), Smoothened (SMO) (FP: CAC-CCT-GGC-CAC-ATT-CGT; RP: CGC-ATT-GAC-GTA-GAA-GAG-AAT-AAC-A), p63 (FP: CCT TCT GTG AGC CAG CTT ATC A; RP: CAT CAG GAA TGG TTG TAG GAG TGA), GATA3 (FP: CTG-GCT-CGC-AGA-ATT-GCA; RP: AAC-TGG-GTA-TGG-CAG-AAT-AAA-ACG), and β-2-Globulin (36). GAPDH and TBP were used as the normalizing genes, and data was analyzed using the ΔΔCT method (64).
Supplementary Material
Supplemental Figure 1. Channel images of the SIM2s fluorescence previously shown (Figure 2). Adhered cells were stained with SIM2s (green) and DAPI (blue). Images were taken using a 40x objective (25.2x).
Supplemental Figure 2. Channel images of the KER5/VIM fluorescence previously shown (Figure 4). Tumors were stained with KER5 (green), VIM (red), and DAPI (blue). Images were taken using a 40x objective (25.2x).
Acknowledgments
Grants: National Cancer Institute (NCI) R01CA111551 (W.W.P.)
Department of Defense (DOD-CDMRP) W81XWH-11-1-0158 (K.C.S.)
NCI 5R00CA127462-06 (F.B.)
We would like to thank Dr. Daniel Medina (Baylor College of Medicine, Houston, TX) for providing the MCF10DCIS.com cell line and the Histology Core Facility at Texas A&M University College of Veterinary Medicine & Biomedical Sciences for tissue preparation and H&E staining. We would thank the University of Kansas Medical Center for providing the human DCIS and IDC samples.
Footnotes
Conflict of Interest
The authors have nothing to disclose.
References
- 1.Burstein HJ, Polyak K, Wong JS, Lester SC, Kaelin CM. Ductal carcinoma in situ of the breast. N Engl J Med. 2004;350(14):1430–41. doi: 10.1056/NEJMra031301. Epub 2004/04/09. [DOI] [PubMed] [Google Scholar]
- 2.Cody HS., 3rd Sentinel lymph node biopsy for DCIS: are we approaching consensus? Ann Surg Oncol. 2007;14(8):2179–81. doi: 10.1245/s10434-006-9300-9. Epub 2007/02/01. [DOI] [PubMed] [Google Scholar]
- 3.Maffuz A, Barroso-Bravo S, Najera I, Zarco G, Alvarado-Cabrero I, Rodriguez-Cuevas SA. Tumor size as predictor of microinvasion, invasion, and axillary metastasis in ductal carcinoma in situ. J Exp Clin Cancer Res. 2006;25(2):223–7. Epub 2006/08/22. [PubMed] [Google Scholar]
- 4.Yao J, Weremowicz S, Feng B, Gentleman RC, Marks JR, Gelman R, et al. Combined cDNA array comparative genomic hybridization and serial analysis of gene expression analysis of breast tumor progression. Cancer Res. 2006;66(8):4065–78. doi: 10.1158/0008-5472.CAN-05-4083. Epub 2006/04/19. [DOI] [PubMed] [Google Scholar]
- 5.Chin K, de Solorzano CO, Knowles D, Jones A, Chou W, Rodriguez EG, et al. In situ analyses of genome instability in breast cancer. Nat Genet. 2004;36(9):984–8. doi: 10.1038/ng1409. Epub 2004/08/10. [DOI] [PubMed] [Google Scholar]
- 6.Ma XJ, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, et al. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci U S A. 2003;100(10):5974–9. doi: 10.1073/pnas.0931261100. Epub 2003/04/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Porter D, Lahti-Domenici J, Keshaviah A, Bae YK, Argani P, Marks J, et al. Molecular markers in ductal carcinoma in situ of the breast. Mol Cancer Res. 2003;1(5):362–75. Epub 2003/03/26. [PubMed] [Google Scholar]
- 8.Cocker R, Oktay MH, Sunkara JL, Koss LG. Mechanisms of progression of ductal carcinoma in situ of the breast to invasive cancer. A hypothesis. Med Hypotheses. 2007;69(1):57–63. doi: 10.1016/j.mehy.2006.11.042. Epub 2007/01/30. [DOI] [PubMed] [Google Scholar]
- 9.Behbod F, Kittrell FS, LaMarca H, Edwards D, Kerbawy S, Heestand JC, et al. An intraductal human-in-mouse transplantation model mimics the subtypes of ductal carcinoma in situ. Breast Cancer Res. 2009;11(5):R66. doi: 10.1186/bcr2358. Epub 2009/09/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller FR, Santner SJ, Tait L, Dawson PJ. MCF10DCIS.com xenograft model of human comedo ductal carcinoma in situ. J Natl Cancer Inst. 2000;92(14):1185–6. doi: 10.1093/jnci/92.14.1185a. Epub 2000/07/25. [DOI] [PubMed] [Google Scholar]
- 11.Hu M, Yao J, Carroll DK, Weremowicz S, Chen H, Carrasco D, et al. Regulation of in situ to invasive breast carcinoma transition. Cancer Cell. 2008;13(5):394–406. doi: 10.1016/j.ccr.2008.03.007. Epub 2008/05/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shekhar MP, Tait L, Pauley RJ, Wu GS, Santner SJ, Nangia-Makker P, et al. Comedo-ductal carcinoma in situ: A paradoxical role for programmed cell death. Cancer Biol Ther. 2008;7(11):1774–82. doi: 10.4161/cbt.7.11.6781. Epub 2008/09/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tait LR, Pauley RJ, Santner SJ, Heppner GH, Heng HH, Rak JW, et al. Dynamic stromal-epithelial interactions during progression of MCF10DCIS.com xenografts. Int J Cancer. 2007;120(10):2127–34. doi: 10.1002/ijc.22572. Epub 2007/02/03. [DOI] [PubMed] [Google Scholar]
- 14.Scribner KC, Wellberg EA, Metz RP, Porter WW. Singleminded-2s (Sim2s) Promotes Delayed Involution of the Mouse Mammary Gland through Suppression of Stat3 and NF{kappa}B. Mol Endocrinol. 2011 doi: 10.1210/me.2010-0423. Epub 2011/02/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gustafson TL, Wellberg E, Laffin B, Schilling L, Metz RP, Zahnow CA, et al. Ha-Ras transformation of MCF10A cells leads to repression of Singleminded-2s through NOTCH and C/EBPbeta. Oncogene. 2009 doi: 10.1038/onc.2008.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laffin B, Wellberg E, Kwak HI, Burghardt RC, Metz RP, Gustafson T, et al. Loss of singleminded-2s in the mouse mammary gland induces an epithelial-mesenchymal transition associated with up-regulation of slug and matrix metalloprotease 2. Mol Cell Biol. 2008;28(6):1936–46. doi: 10.1128/MCB.01701-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwak HI, Gustafson T, Metz RP, Laffin B, Schedin P, Porter WW. Inhibition of breast cancer growth and invasion by single-minded 2s. Carcinogenesis. 2007;28(2):259–66. doi: 10.1093/carcin/bgl122. [DOI] [PubMed] [Google Scholar]
- 18.Wellberg E, Metz RP, Parker C, Porter WW. The bHLH/PAS transcription factor singleminded 2s promotes mammary gland lactogenic differentiation. Development. 2010;137(6):945–52. doi: 10.1242/dev.041657. Epub 2010/02/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LaMarca HL, Visbal AP, Creighton CJ, Liu H, Zhang Y, Behbod F, et al. CCAAT/enhancer binding protein beta regulates stem cell activity and specifies luminal cell fate in the mammary gland. Stem Cells. 2010;28(3):535–44. doi: 10.1002/stem.297. Epub 2010/01/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stylianou S, Clarke RB, Brennan K. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 2006;66(3):1517–25. doi: 10.1158/0008-5472.CAN-05-3054. Epub 2006/02/03. [DOI] [PubMed] [Google Scholar]
- 21.Grimm SL, Rosen JM. The role of C/EBPbeta in mammary gland development and breast cancer. J Mammary Gland Biol Neoplasia. 2003;8(2):191–204. doi: 10.1023/a:1025900908026. Epub 2003/11/26. [DOI] [PubMed] [Google Scholar]
- 22.Politi K, Feirt N, Kitajewski J. Notch in mammary gland development and breast cancer. Semin Cancer Biol. 2004;14(5):341–7. doi: 10.1016/j.semcancer.2004.04.013. Epub 2004/08/04. [DOI] [PubMed] [Google Scholar]
- 23.Kiaris H, Politi K, Grimm LM, Szabolcs M, Fisher P, Efstratiadis A, et al. Modulation of notch signaling elicits signature tumors and inhibits hras1-induced oncogenesis in the mouse mammary epithelium. Am J Pathol. 2004;165(2):695–705. doi: 10.1016/S0002-9440(10)63333-0. Epub 2004/07/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du Z, Li J, Wang L, Bian C, Wang Q, Liao L, et al. Overexpression of DeltaNp63alpha induces a stem cell phenotype in MCF7 breast carcinoma cell line through the Notch pathway. Cancer Sci. 2010;101(11):2417–24. doi: 10.1111/j.1349-7006.2010.01700.x. Epub 2010/10/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yalcin-Ozuysal O, Fiche M, Guitierrez M, Wagner KU, Raffoul W, Brisken C. Antagonistic roles of Notch and p63 in controlling mammary epithelial cell fates. Cell Death Differ. 2010;17(10):1600–12. doi: 10.1038/cdd.2010.37. Epub 2010/04/10. [DOI] [PubMed] [Google Scholar]
- 26.de Biase D, Morandi L, Degli Esposti R, Ligorio C, Pession A, Foschini MP, et al. p63 short isoforms are found in invasive carcinomas only and not in benign breast conditions. Virchows Arch. 2010;456(4):395–401. doi: 10.1007/s00428-010-0900-1. Epub 2010/03/13. [DOI] [PubMed] [Google Scholar]
- 27.Visbal AP, LaMarca HL, Villanueva H, Toneff MJ, Li Y, Rosen JM, et al. Altered differentiation and paracrine stimulation of mammary epithelial cell proliferation by conditionally activated Smoothened. Dev Biol. 2011;352(1):116–27. doi: 10.1016/j.ydbio.2011.01.025. Epub 2011/02/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moraes RC, Zhang X, Harrington N, Fung JY, Wu MF, Hilsenbeck SG, et al. Constitutive activation of smoothened (SMO) in mammary glands of transgenic mice leads to increased proliferation, altered differentiation and ductal dysplasia. Development. 2007;134(6):1231–42. doi: 10.1242/dev.02797. [DOI] [PubMed] [Google Scholar]
- 29.Kallergi G, Papadaki MA, Politaki E, Mavroudis D, Georgoulias V, Agelaki S. Epithelial to mesenchymal transition markers expressed in circulating tumour cells of early and metastatic breast cancer patients. Breast Cancer Res. 2011;13(3):R59. doi: 10.1186/bcr2896. Epub 2011/06/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dubois-Marshall S, Thomas JS, Faratian D, Harrison DJ, Katz E. Two possible mechanisms of epithelial to mesenchymal transition in invasive ductal breast cancer. Clin Exp Metastasis. 2011 doi: 10.1007/s10585-011-9412-x. Epub 2011/07/27. [DOI] [PubMed] [Google Scholar]
- 31.Bond J, Haughton M, Blaydes J, Gire V, Wynford-Thomas D, Wyllie F. Evidence that transcriptional activation by p53 plays a direct role in the induction of cellular senescence. Oncogene. 1996;13(10):2097–104. Epub 1996/11/21. [PubMed] [Google Scholar]
- 32.Zuo S, Liu C, Wang J, Wang F, Xu W, Cui S, et al. IGFBP-rP1 induces p21 expression through a p53-independent pathway, leading to cellular senescence of MCF-7 breast cancer cells. Journal of cancer research and clinical oncology. 2012 doi: 10.1007/s00432-012-1153-y. Epub 2012/03/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malkin D, Li FP, Strong LC, Fraumeni JF, Jr, Nelson CE, Kim DH, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250(4985):1233–8. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 34.Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127(5):1041–55. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9(2):201–9. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- 36.McDaniel SM, Rumer KK, Biroc SL, Metz RP, Singh M, Porter W, et al. Remodeling of the mammary microenvironment after lactation promotes breast tumor cell metastasis. Am J Pathol. 2006;168(2):608–20. doi: 10.2353/ajpath.2006.050677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu R, Wang X, Chen GY, Dalerba P, Gurney A, Hoey T, et al. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356(3):217–26. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 38.Bloom HJ, Richardson WW. Histological grading and prognosis in breast cancer; a study of 1409 cases of which 359 have been followed for 15 years. Br J Cancer. 1957;11(3):359–77. doi: 10.1038/bjc.1957.43. Epub 1957/09/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–6. doi: 10.1038/415530a. Epub 2002/02/02. [DOI] [PubMed] [Google Scholar]
- 40.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. doi: 10.1038/35021093. Epub 2000/08/30. [DOI] [PubMed] [Google Scholar]
- 41.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–74. doi: 10.1073/pnas.191367098. Epub 2001/09/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zajchowski DA, Bartholdi MF, Gong Y, Webster L, Liu HL, Munishkin A, et al. Identification of gene expression profiles that predict the aggressive behavior of breast cancer cells. Cancer Res. 2001;61(13):5168–78. Epub 2001/06/30. [PubMed] [Google Scholar]
- 43.West M, Blanchette C, Dressman H, Huang E, Ishida S, Spang R, et al. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc Natl Acad Sci U S A. 2001;98(20):11462–7. doi: 10.1073/pnas.201162998. Epub 2001/09/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou J, Chehab R, Tkalcevic J, Naylor MJ, Harris J, Wilson TJ, et al. Elf5 is essential for early embryogenesis and mammary gland development during pregnancy and lactation. Embo J. 2005;24(3):635–44. doi: 10.1038/sj.emboj.7600538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oakes SR, Hilton HN, Ormandy CJ. The alveolar switch: coordinating the proliferative cues and cell fate decisions that drive the formation of lobuloalveoli from ductal epithelium. Breast Cancer Res. 2006;8(2):207. doi: 10.1186/bcr1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walsh LA, Cepeda MA, Damjanovski S. Analysis of the MMP-dependent and independent functions of tissue inhibitor of metalloproteinase-2 on the invasiveness of breast cancer cells. Journal of cell communication and signaling. 2012 doi: 10.1007/s12079-011-0157-8. Epub 2012/01/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu D, Guo H, Li Y, Xu X, Yang K, Bai Y. Association between Polymorphisms in the Promoter Regions of Matrix Metalloproteinases (MMPs) and Risk of Cancer Metastasis: A Meta-Analysis. PLoS One. 2012;7(2):e31251. doi: 10.1371/journal.pone.0031251. Epub 2012/02/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mendes O, Kim HT, Stoica G. Expression of MMP2, MMP9 and MMP3 in breast cancer brain metastasis in a rat model. Clin Exp Metastasis. 2005;22(3):237–46. doi: 10.1007/s10585-005-8115-6. [DOI] [PubMed] [Google Scholar]
- 49.Pellikainen JM, Ropponen KM, Kataja VV, Kellokoski JK, Eskelinen MJ, Kosma VM. Expression of matrix metalloproteinase (MMP)-2 and MMP-9 in breast cancer with a special reference to activator protein-2, HER2, and prognosis. Clin Cancer Res. 2004;10(22):7621–8. doi: 10.1158/1078-0432.CCR-04-1061. Epub 2004/12/01. [DOI] [PubMed] [Google Scholar]
- 50.Ioachim EE, Athanassiadou SE, Kamina S, Carassavoglou K, Agnantis NJ. Matrix metalloproteinase expression in human breast cancer: an immunohistochemical study including correlation with cathepsin D, type IV collagen, laminin, fibronectin, EGFR, c-erbB-2 oncoprotein, p53, steroid receptors status and proliferative indices. Anticancer Res. 1998;18(3A):1665–70. [PubMed] [Google Scholar]
- 51.Schedin P, Mitrenga T, McDaniel S, Kaeck M. Mammary ECM composition and function are altered by reproductive state. Mol Carcinog. 2004;41(4):207–20. doi: 10.1002/mc.20058. Epub 2004/10/07. [DOI] [PubMed] [Google Scholar]
- 52.Witty JP, Wright JH, Matrisian LM. Matrix metalloproteinases are expressed during ductal and alveolar mammary morphogenesis, and misregulation of stromelysin-1 in transgenic mice induces unscheduled alveolar development. Mol Biol Cell. 1995;6(10):1287–303. doi: 10.1091/mbc.6.10.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141(1):52–67. doi: 10.1016/j.cell.2010.03.015. Epub 2010/04/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sternlicht MD, Lochter A, Sympson CJ, Huey B, Rougier JP, Gray JW, et al. The stromal proteinase MMP3/stromelysin-1 promotes mammary carcinogenesis. Cell. 1999;98(2):137–46. doi: 10.1016/s0092-8674(00)81009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang C, Hu F, Guo S, Mi D, Shen W, Zhang J, et al. BMP-6 inhibits MMP-9 expression by regulating heme oxygenase-1 in MCF-7 breast cancer cells. Journal of cancer research and clinical oncology. 2011;137(6):985–95. doi: 10.1007/s00432-010-0963-z. Epub 2010/12/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sims JD, McCready J, Jay DG. Extracellular heat shock protein (Hsp)70 and Hsp90alpha assist in matrix metalloproteinase-2 activation and breast cancer cell migration and invasion. PLoS One. 2011;6(4):e18848. doi: 10.1371/journal.pone.0018848. Epub 2011/05/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kasperczyk H, Baumann B, Debatin KM, Fulda S. Characterization of sonic hedgehog as a novel NF-kappaB target gene that promotes NF-kappaB-mediated apoptosis resistance and tumor growth in vivo. FASEB J. 2009;23(1):21–33. doi: 10.1096/fj.08-111096. Epub 2008/09/06. [DOI] [PubMed] [Google Scholar]
- 58.Kasper M, Jaks V, Fiaschi M, Toftgard R. Hedgehog signalling in breast cancer. Carcinogenesis. 2009;30(6):903–11. doi: 10.1093/carcin/bgp048. Epub 2009/02/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoo YA, Kang MH, Lee HJ, Kim BH, Park JK, Kim HK, et al. Sonic hedgehog pathway promotes metastasis and lymphangiogenesis via activation of Akt, EMT, and MMP-9 pathway in gastric cancer. Cancer Res. 2011;71(22):7061–70. doi: 10.1158/0008-5472.CAN-11-1338. Epub 2011/10/07. [DOI] [PubMed] [Google Scholar]
- 60.Liao X, Siu MK, Au CW, Wong ES, Chan HY, Ip PP, et al. Aberrant activation of hedgehog signaling pathway in ovarian cancers: effect on prognosis, cell invasion and differentiation. Carcinogenesis. 2009;30(1):131–40. doi: 10.1093/carcin/bgn230. Epub 2008/11/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koodie L, Ramakrishnan S, Roy S. Morphine suppresses tumor angiogenesis through a HIF-1alpha/p38MAPK pathway. Am J Pathol. 2010;177(2):984–97. doi: 10.2353/ajpath.2010.090621. Epub 2010/07/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stockmann C, Doedens A, Weidemann A, Zhang N, Takeda N, Greenberg JI, et al. Deletion of vascular endothelial growth factor in myeloid cells accelerates tumorigenesis. Nature. 2008;456(7223):814–8. doi: 10.1038/nature07445. Epub 2008/11/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Metz RP, Kwak HI, Gustafson T, Laffin B, Porter WW. Differential transcriptional regulation by mouse single-minded 2s. J Biol Chem. 2006;281(16):10839–48. doi: 10.1074/jbc.M508858200. [DOI] [PubMed] [Google Scholar]
- 64.Hettinger AM, Allen MR, Zhang BR, Goad DW, Malayer JR, Geisert RD. Presence of the acute phase protein, bikunin, in the endometrium of gilts during estrous cycle and early pregnancy. Biol Reprod. 2001;65(2):507–13. doi: 10.1095/biolreprod65.2.507. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Channel images of the SIM2s fluorescence previously shown (Figure 2). Adhered cells were stained with SIM2s (green) and DAPI (blue). Images were taken using a 40x objective (25.2x).
Supplemental Figure 2. Channel images of the KER5/VIM fluorescence previously shown (Figure 4). Tumors were stained with KER5 (green), VIM (red), and DAPI (blue). Images were taken using a 40x objective (25.2x).