Abstract
Co-speciation is a fundamental concept of evolutionary biology and intuitively appealing, yet in practice hard to demonstrate as it is often blurred by other evolutionary processes. We investigate the phylogeographic history of the monogenean ectoparasites Gyrodactylus teuchis and G. truttae on European salmonids of the genus Salmo. Mitochondrial cytochrome oxidase subunit 1 and the nuclear ribosomal internal transcribed spacer 2 were sequenced for 189 Gyrodactylus individuals collected from 50 localities, distributed across most major European river systems, from the Iberian- to the Balkan Peninsula. Despite both anthropogenic and naturally caused admixture of the principal host lineages among major river basins, co-phylogenetic analyses revealed significant global congruence for host and parasite phylogenies, providing firm support for co-speciation of G. teuchis and its salmonid hosts brown trout (S. trutta) and Atlantic salmon (S. salar). The major split within G. teuchis, coinciding with the initial divergence of the hosts was dated to ~1.5 My BP, using a Bayesian framework based on an indirect calibration point obtained from the host phylogeny. The presence of G. teuchis in Europe thus predates some of the major Pleistocene glaciations. In contrast, G. truttae exhibited remarkably low intraspecific genetic diversity. Given the direct life cycle and potentially high transmission potential of gyrodactylids, this finding is interpreted as indication for a recent emergence (<60 ky BP) of G. truttae via a host-switch. Our study thus suggests that instances of two fundamentally different mechanisms of speciation (co-speciation vs. host-switching) may have occurred on the same hosts in Europe within a time span of less than 1.5 My in two gyrodactylid ectoparasite species.
Introduction
Co-evolution is a fundamental concept in evolutionary biology and in very general terms refers to “the change of a biological object triggered by the change of a related object" [1]. Co-evolution in host-parasite- or mutualistic systems is a special case defined as the reciprocal adaptive change of two antagonists through reciprocal selective pressures [2]. Another closely related phenomenon is co-speciation, whose characteristics can but need not fit the above definition of co-evolution. In host-parasite systems, co-speciation is thought to occur when the association between host and parasite species is close and speciation of the host results in parallel speciation of the parasite. The concept of co-speciation was originally proposed for avian parasites and their hosts [3,4], and the expectation that hosts and their parasites should evolve in synchrony is known as Farenholz’s rule [5]. However, host-parasite co-speciation is often difficult to demonstrate as it may be blurred by other evolutionary processes (reviewed e.g. in [6]), including (i) host switching, i.e. a parasite species partly or fully switches to another host species, (ii) duplication, i.e. parasite speciation without host speciation (sometimes also referred to as intra-host speciation [7]), and (iii) lineage sorting, i.e. the extinction or the absence of a parasite lineage in the founding population of a new host species (“missing the boat”). Parasites may also fail to speciate in response to host speciation due to increased gene flow among parasite populations relative to the host populations [8]. In a recent review [9] only 7% of all surveyed claims of co-speciation provided convincing evidence. Nonetheless a major prerequisite for tracking co-speciation events is a solid understanding of the phylogenies of the involved host and parasite species [10].
Gyrodactylid monogeneans (Platyhelminthes) are for various reasons fascinating model systems for studying the processes of speciation in host-parasite associations. The hyperdiverse genus Gyrodactylus currently includes about 400 ectoparasitic species [11] primarily infecting teleost fish, but this number poorly reflects the diversity of the radiation, estimated to be at least ~ 20,000 species [12]. Many species of Gyrodactylus have a direct life cycle and are hyperviviparous, a rare but highly efficient reproductive mode [13], which gives them the potential for rapid population growth [14]. Additionally, species of Gyrodactylus are capable of both sexual and asexual reproduction and new parasite populations may frequently originate from single founder individuals [14]. Given these unique reproductive features it is not surprising that host switching is believed to be the most abundant mode of speciation in Gyrodactylus [15]. Paradoxically, many Gyrodactylus species are highly host specific [12,16], a trait which is considered to result from the continuous adaptation inherent in a co-evolutionary arms race as first envisioned by Darwin [17].
However, in-depth studies on the co-evolution of gyrodactylids and their hosts are scarce. Huyse & Volckaert [18] investigated the evolutionary associations between European gobies (Gobiidae) and their Gyrodactylus parasites. They concluded that the radiation of highly host specific gill parasites was largely driven by host switching from the three-spined stickleback (Gasterosteus arcuatus) onto two gobiid genera Pomatoschistus and Gobius, followed by phylogenetically conserved host-switching among various goby hosts. In contrast, they suggested co-speciation for the less host specific fin-parasites resulting in several host-associated species complexes [18]. Another case of possible host-parasite co-speciation has been reported for the Central American guppies Poecilia reticulate and P. picta and their respective gyrodactylid parasites G. turnbulli and G. pictae [19].
The brown trout (Salmo trutta) is one of the best-studied European freshwater fish species with a complex, yet well-resolved phylogeographic structure across its wide European range. At least five distinct major lineages are distinguished for S. trutta in Europe [20–26] although a purely river-basin based phylogeographic view has been called into question in recent years [22,25,27,28]. Brown trout is an economically important species, and translocation of fish stocks has been common across large geographical distances. Only in recent years has the preservation of the genetic integrity of local brown trout populations become the objective of modern management strategies. However, while possible introgression into the gene pool of locally adapted fish populations is an important issue in conservation genetics, the effects of stocking on the integrity of host-parasite interactions, which have potentially evolved over a long period of time, have not yet been studied in detail for monogeneans. Brown trout is a suitable host for several species of Gyrodactylus, the three most commonly found species in Europe being G. derjavinoides, G. teuchis, and G. truttae. Parasite transmission occurs directly from host to host (no free-living stages) and detachment is usually fatal for the parasite within less than 24h (reviewed in [16]). Range expansion in gyrodactylids is thus entirely dependent on the host, and co-phylogeographic patterns might be expected.
The current study aims at identifying signatures of co-speciation in the widely distributed host-parasite system of ectoparasitic gyrodactylids and their European hosts brown trout and Atlantic salmon (Salmo salar). Co-phylogenetic methods are applied to evaluate phylogeographic congruence and potential allopatric co-divergence as initial steps of co-speciation.
Materials and Methods
Sampling
Gyrodactylus parasites were collected from the skin and fins of ethanol preserved specimens of primarily salmonid host species: brown trout, Atlantic salmon, brook trout (Salvelinus fontinalis), rainbow trout (Oncorhynchus mykiss), and European grayling (Thymallus thymallus). Fish were screened for gyrodactylids using a stereomicroscope. Parasites were removed using watchmaker forceps and stored in 96% ethanol individually until further processing. No specific sampling permits were required for the collection of flatworm parasites. None of the species (fish or parasites) sampled is endangered or protected. The sampling of gyrodactylids was purely opportunistic, in that only host (fish) samples collected as part of independent routine monitoring and/or scientific collection operations were used. These operations were conducted with certified electric fishing generators, licensed operators, and with sampling permission from the local (district) level authorities, as required. Gyrodactylus spp. specimens were collected from 50 localities including three hatcheries (see Fig 1 and Table 1). We reiterate that the authors of this study were not responsible for, nor did they carry out fish sampling, rather they were simply given access to previously collected material.
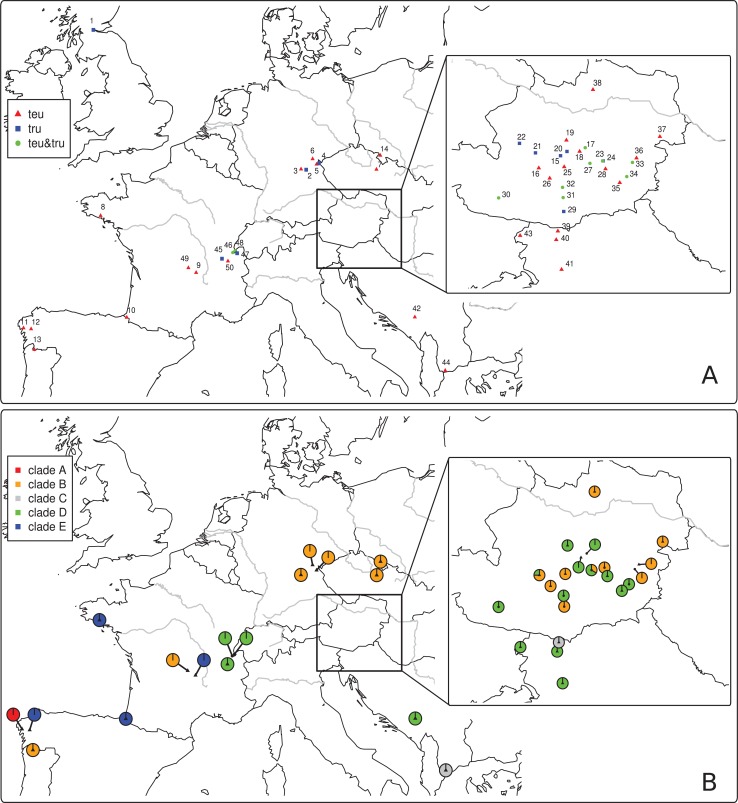
Fig 1. Maps illustrating (A) the principal sampling locations of the study (location numbers as in Table 1) as well as the occurrence of G. teuchis (red triangle), G. trutta (blue square), or both species (green circle) in a given location, and (B) depicting the geographic distribution of the five mtDNA clades (A-E, see Fig 2) of G. teuchis throughout the study area.
The enlarged boxes represent sampling locations or mtDNA clade data in Austria and Slovenia.
Table 1. Summary of sampling locations.
| river | drainage | ID | host | coordinates | date | N | ||
|---|---|---|---|---|---|---|---|---|
| Gteu | Gtru | |||||||
| Bannock Burn | Forth | Atlantic | 1 | St | 56.1004°N, | 2006 | - | 1 |
| 3.9085°W | ||||||||
| Aufsess | Regnitz → Main → Rhine | Atlantic | 2 | St | 49.8797°N, | 25.10.2011 | - | 2 |
| Tt | 11.2293°E | - | 1 | |||||
| Regnitz | Main → Rhine | Atlantic | 3 | St | 49.9053°N, | 21.10.2011 | 2 | - |
| 10.8660°E | ||||||||
| Selb | Eger → Elbe | Atlantic | 4 | St | 50.1648°N | 24.10.2011 | - | 2 |
| 12.1450°E | ||||||||
| Eger | Elbe | Atlantic | 5 | St | 50.1203°N | 24.10.2011 | 2 | - |
| 11.9711°E | ||||||||
| Selbitz | Saale → Elbe | Atlantic | 6 | St | 50.3605°N | 24.10.2011 | 2 | - |
| 11.6903°E | ||||||||
| Loučná | Elbe | Atlantic | 7 | St | 49.8935°N, | 05.12.2011 | 2 | - |
| 16.2311°E | ||||||||
| Scorff | Blavet | Atlantic | 8 | Ss | 47.8351°N, | 19.09.2011 | 1 | - |
| St | 3.3941°W | 2 | - | |||||
| Allier | Loire | Atlantic | 9 | Ss | 45.3020°N, | 15.03.2011 | 1 | - |
| St | 3.4033°E | 2 | - | |||||
| Nivelle | - | Atlantic | 10 | St | 43.3070°N, | 15.09.2011 | 2 | - |
| 1.5297°W | ||||||||
| Donas | Tambre | Atlantic | 11 | St | 42.8353°N, | 07.2012 | 2 | - |
| 8.8752°W | ||||||||
| Ulla | - | Atlantic | 12 | St | 42.7946°N | 07.2012 | 2 | - |
| 8.3362°W | ||||||||
| Vilamea | Caldo | Atlantic | 13 | St | 41.8755°N, | 07.2012 | 2 | - |
| 8.1069°W | ||||||||
| Scinawka | Nysa Kłodzka → Oder | Baltic Sea | 14 | Tt | 50.5141°N, | 20.05.2011 | 2 | - |
| 16.5018°E | ||||||||
| Palten | Enns → Danube | Black Sea | 15 | St | 47.5245°N, | 17.07.2009 | - | 10 |
| Tt | 14.3887°E | - | 1 | |||||
| Schwarzenseebach | Enns → Danube | Black Sea | 16 | Sf | 47.3390°N, | 16.11.2009 | 2 | - |
| St | 13.9164°E | 2 | - | |||||
| Hinterwildalpenbach | Enns → Danube | Black Sea | 17 | St | 47.6455°N, | 21.09.2009 | 2 | 1 |
| Sf | 14.9239°E | - | 2 | |||||
| Erzbach | Enns → Danube | Black Sea | 18 | St | 47.5886°N, | 07.10.2009 | 2 | - |
| 14.8024°E | ||||||||
| Leerensackbach | Enns → Danube | Black Sea | 19 | St | 47.7583°N, | 12.09.2006 | 1 | - |
| 14.5158°E | ||||||||
| Enns | Danube | Black Sea | 20 | St | 47.5887°N, | 18.10.2008 | - | 2 |
| 14.528°E | ||||||||
| Riedlbach | Traun → Danube | Black Sea | 21 | St | 47.5678°N, | 28.09.2011 | - | 2 |
| 13.8451°E | ||||||||
| Weissenbach | Traun → Danube | Black Sea | 22 | St | 47.7112°N, | 22.09.2009 | - | 2 |
| 13.4977°E | ||||||||
| Mürz Hafendorf | Mur → Drau → Danube | Black Sea | 23 | St | 47.4467°N, | 11.10.2008 | - | 2 |
| Tt | 15.312°E | - | 1 | |||||
| Mürz Kapfenberg | Mur → Drau → Danube | Black Sea | 24 | St | 47.445°N | 06.10.2008 | 2 | - |
| Tt | 15.3022°E | - | 1 | |||||
| Pöls | Mur → Drau → Danube | Black Sea | 25 | St | 47.3565°N, | 05.10.2009 | 1 | - |
| Sf | 14.4680°E | 1 | - | |||||
| Katschbach | Mur → Drau → Danube | Black Sea | 26 | Sf | 47.1853°N | 23.07.2009 | 2 | - |
| St | 14.1537°E | 8 | - | |||||
| Vordernbergerbach | Mur → Drau → Danube | Black Sea | 27 | St | 47.4100°N, | 23.07.2009 | 2 | 1 |
| Om | 15.0264°E | 1 | - | |||||
| Mur Mixnitz | Drau → Danube | Black Sea | 28 | St | 47.3258°N | 03.10.2009 | 2 | - |
| 15.3642°E | ||||||||
| Gurk | Drau → Danube | Black Sea | 29 | Tt | 46.6848°N, | 21.10.2008 | - | 2 |
| 14.4577°E | ||||||||
| Möll | Drau → Danube | Black Sea | 30 | St | 46.8895°N, | 27.11.2008 | 2 | 2 |
| Tt | 13.0455°E | - | 1 | |||||
| Metnitz | Gurk → Drau → Danube | Black Sea | 31 | St | 46.8942°N, | 17.11.2009 | 2 | 1 |
| 14.4447°E | ||||||||
| Olsa | Gurk → Drau → Danube | Black Sea | 32 | St | 47.0481°N, | 23.09.2009 | 5 | 10 |
| 14.4321°E | ||||||||
| Lafnitz | Raab → Danube | Black Sea | 33 | Tt | 47.4237°N, | 17.10.2008 | 1 | 1 |
| 15.9487°E | ||||||||
| Feistritz | Raab → Danube | Black Sea | 34 | St | 47.2105°N, | 08.10.2008 | 2 | 2 |
| Tt | 15.8255°E | - | 1 | |||||
| Kötschmanngrabenbach | Raab → Danube | Black Sea | 35 | St | 47.1181°N, | 22.09.2011 | 2 | - |
| 15.6731°E | ||||||||
| Pinka | Raab → Danube | Black Sea | 36 | St | 47.4894°N, | 08.09.2011 | 2 | - |
| 16.0361°E | ||||||||
| Wulka | Neusiedlersee | - | 37 | St | 47.8118°N, | 15.10.2009 | 10 | - |
| 16.5437°E | ||||||||
| Kleiner Kamp | Kamp → Danube | Black Sea | 38 | St | 48.5192°N, | 20.11.2008 | 10 | - |
| 15.0922°E | ||||||||
| Bistrica | Sava → Danube | Black Sea | 39 | St | 46.3893°N, | 15.06.2011 | 2 | - |
| 14.3304°E | ||||||||
| Besnica | Sava → Danube | Black Sea | 40 | St | 46.2594°N, | 15.10.2011 | 2 | - |
| 14.2949°E | ||||||||
| Mahnečica | Sava → Danube | Black Sea | 41 | St | 45.8106°N, | 21.10.2011 | 2 | - |
| 14.4107°E | ||||||||
| Tara | Sava → Danube | Black Sea | 42 | St | 43.3231°N, | 05.06.2010 | 3 | - |
| 18.9610°E | ||||||||
| Kranska | Lake Prespa → Drin | Adriatic | 43 | Stp | 40.9465°N, | 27.10.2010 | 2 | - |
| 21.1219°E | ||||||||
| Gljun | Soča | Adriatic | 44 | Stm | 46.3217°N, | 01.12.2011 | 10 | - |
| 13.5116°E | ||||||||
| Ain | Rhone | Mediterranean | 45 | St | 45.9196°N, | 16.03.2011 | - | 2 |
| 5.2418°E | ||||||||
| Allondon | Rhone | Mediterranean | 46 | St | 46.1934°N, | 28.09.2010 | 7 | 10 |
| 6.0029°E | ||||||||
| Foron de Fillinge | Arve → Rhone | Mediterranean | 47 | St | 46.1638°N, | 15.06.2010 | - | 1 |
| Tt | 6.3206°E | - | 1 | |||||
| Versoix | Rhone | Mediterranean | 48 | St | 46.2763°N, | 08.10.2010 | 1 | 2 |
| Tt | 6.1672°E | 1 | - | |||||
| hatchery | Loire | Atlantic | 49 | St | - | 15.03.2011 | 2 | - |
| hatchery | Rhone | Mediterranean | 50 | St | - | 16.03.2011 | 2 | - |
| fish farm 1 | Vistula | Balitc | 51 | St | - | 27.03.2002 | - | ? |
Shown are the river names, the associated drainage, an ID number that corresponds to sites shown in Fig 1, the host species (Om—O. mykiss, Sf—S. fontinalis, Ss—S. salar, St—S. trutta, Tt—T. thymallus), the sites coordinates, collection date, and the number of individuals (N) of the two study species, G. teuchis and G. truttae, analysed at each site.
1Genbank accession EU304826
Molecular methods
Total genomic DNA was extracted from individual worms as described earlier [29]. Only one parasite specimen per fish was processed to minimize the likelihood of analysing directly related Gyrodactylus individuals. The internal transcribed spacer 2 (ITS2) of the nuclear ribosomal gene cluster was amplified and sequenced as described previously [29] using published primers ITS4.5 (5'-CATCGGTCTCTCGAACG-3') and ITS2 (5'-TCCTCCGCTTAGTGATA-3') [30]. Obtained ribosomal sequences were subjected to a BLAST search [31] against GenBank for species identification. For the amplification of a ~1,350 bp fragment of the mitochondrial cytochrome oxidase I gene (COI) we used the forward primer teutruforw (5'-TTGTTTTCAAAACAAAAAGTGC-3') with the reverse primer teurev (5'-TACTACTTTCGTTTCATAGCCC-3') or trurev (5'-TACTTCTTTCACTCCATAACCC-3'), newly designed primer pairs for species-specific amplification in G. teuchis or G. truttae, respectively. The COI fragments were likewise amplified under standard PCR conditions as previously described [29] and sequenced (both strands) using the PCR primers and BigDye chemistry (Applied Biosystems). Sequences were quality checked and edited manually using the Sequencher 4.1.4 software (Gene Codes Corporation).
In addition to the novel COI sequences of G. teuchis and G. truttae generated in this study sequences for G. derjavinoides (EU293891), G. lavareti (EF446765), G. salaris (DQ988931), G. thymalli (EF527269), and G. truttae (EU304826) were retrieved from GenBank and included in subsequent analyses. Additionally, a dataset from previously published sequences of the mitochondrial control region (CR) of the salmonid hosts Atlantic salmon and brown trout (see Table 2) was compiled. The control region was chosen due to the extensive number of available sequences and a reasonably acceptable calibrated divergence rate in salmonid fishes. Brown trout sequences from five major geographical mtDNA lineages (Adriatic, Atlantic, Danubian, Duero, Mediterranean) were carefully selected to capture a wide range of the reported European diversity. Alignments were generated using MAFFT [32] and for parasite sequences manually edited and analyzed (p-distances, dS/dN ratio) in MEGA5 [33] and DNAsp [34].
Table 2. General information on the host sequences used in the current study.
| species | GenBank | mtDNA lineage | mt haplotype |
|---|---|---|---|
| St | AY185571 1 | Danube | Da3 |
| St | AY185572 | Danube | Da9 |
| St | AY185575 | Danube | Da23b |
| St | AF273086 1 | Atlantic | H1 |
| St | AF273087 | Atlantic | H2 |
| St | EF530495 | Atlantic | ATcs33 |
| St | EF530513 1 | Duero | DUcs1 |
| St | EF530514 | Duero | DUcs2 |
| St | EF530515 | Duero | DUcs3 |
| St | AY836350 1 | Mediterranean | MEcs1 |
| St | AY836351 | Mediterranean | MEcs2 |
| St | AY836352 | Mediterranean | MEcs3 |
| St | AY8363301 | Adriatic | ADcs1 |
| St | AY836331 | Adriatic | ADcs2 |
| St | AY836332 | Adriatic | ADcs3 |
| Ss | U12143 1 | - | - |
| Ss | JQ390056 | - | - |
| Ss | AF133701 | - | - |
St—S. trutta, Ss—S. salar. The mtDNA haplotypes refer to the haplotype names given in the initial publications.
1 sequence used in host-parasite association analyses
Phylogenetic analyses and species delimitation
The best fitting substitution models were determined using MrModeltest2 [35]. Phylogenetic relationships were inferred using Bayesian inference (BI) with MrBayes 3.2.1 [36] as well as Maximum likelihood (ML) with RAxML 7.8.3 [37]. On each alignment BI was run for four analyses with four Metropolis-coupled Markov chain Monte Carlo (MCMC) chains with an incremental heating temperature of 0.1 for 50,000,000 generations and sampled every 1,000 generations. The first 25% of the generations were discarded as “burn-in”, and posterior probabilities were estimated for the remaining generations. ML tree robustness was assessed by >100 bootstrap pseudo replicates. Molecular species delimitation was subsequently performed on the ML trees inferred from the COI sequences using the recently proposed Poisson Tree Processes (PTP) model [38] as implemented on the PTP-webserver (http://species.h-its.org/ptp/, last accessed February, 2015).
Co-phylogenetic analyses of G. teuchis and Salmo spp.
To assess topological congruence of phylogenetic trees and to test for signatures of co-divergence between G. teuchis and Salmo spp. the programs ParaFit [39] and PACo [40] were used. Prior to analyses both the parasite and the host datasets were reduced to retain only one representative sequence for each of the major lineages. For G. teuchis five lineages were defined according to the least conservative species delimitation result (see Fig 2 and results section below). For lineages comprising more than one haplotype the most abundant haplotype was chosen as representative. For Salmo spp. we randomly chose one of the previously published haplotypes for each of the relevant mtDNA trout lineages (see Table 2) and one haplotype for Salmo salar. Subsequently, two host-parasite association matrices (HPM) were constructed to evaluate geographical congruence of parasite and host lineages. HPM-1 (see Fig 3A) allowed for ambiguity in the geographical associations between host and parasite lineages. That is, parasite haplotypes could be assigned to more than one host lineage, thus taking into consideration that parasite haplotypes were detected in more than one distinct river basin, associated with major host lineages, or that parasite haplotypes originated from regions of known admixture between major host lineages.
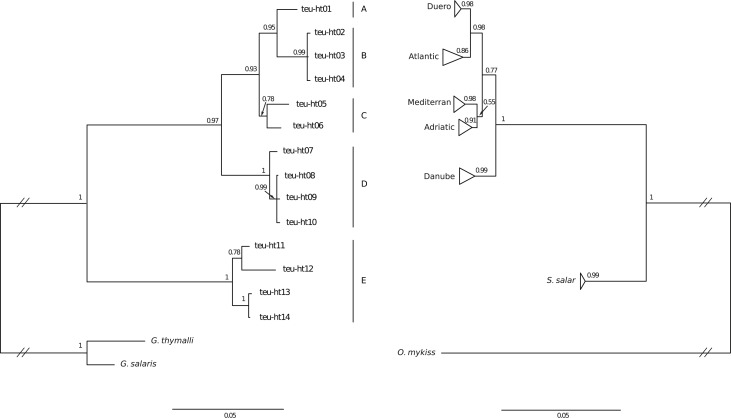
Fig 2. Results of Bayesian phylogenetic inference (GTR+Γ) for G. teuchis (tree on the left) based on COI gene, and for Salmo sp.
(tree on the right) based on mitochondrial control region. The labels in the upper part of the Salmo tree refer to the five major lineages of S. trutta.
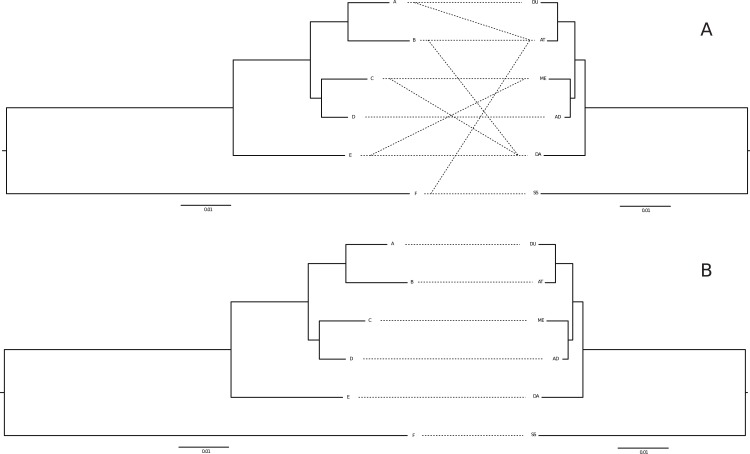
Fig 3. Two alternative host parasite associations between G. teuchis (left) and Salmo sp.
(right), whereby (A) represents the HPM-1 model allowing for some ambiguity due to shared clades within drainages, and B is the most parsimonious model based on the most frequent or presumed native lineages (see methods for details). G. teuchis lineages A-E refer to the major lineages recovered in the Bayesian analysis illustrated in Fig 2. The major lineages for Salmo sp. are abbreviated as follows: DU—Duero, AT—Atlantic, AD—Adriatic, DA—Danube, and SS—S. salar.
HPM-2 (see Fig 3B), in contrast, is based on the most parsimonious host association for each parasite lineage. Here, meta-data on the sampling locations, e.g., information about earlier introduction of non-native fish stocks, were taken into consideration.
For example, G. teuchis haplotypes assigned to clade D were identified in both the Danubian and Adriatic drainages (see results section below), which harbour distinct Danubian (DA) and Adriatic (AD) trout lineages, respectively. Thus clade D is connected to lineage DA and AD in HPM-1. However, the Adriatic river in question (Soca drainage) is known to have been heavily stocked with Danubian trout (see Discussion below) and HPM-2 thus only takes into account the most parsimonious host-parasite association of G. teuchis clade D to trout lineage DA.
PACo and ParaFit tests for each HPM were conducted in R [41] and run for 100,000 permutations based on pairwise genetic (TN93 evolutionary model of substitution) and patristic distances (computed from ML trees inferred with MEGA5 [33]), respectively.
Estimating the nucleotide substitution rate for the mitochondrial COI gene in G. teuchis
The flatworm fossil record is rather poor and hence direct calibration points are scarce [42]. Based on topological congruence between the molecular phylogenies of G. teuchis and its hosts S. truttae and S. salar the nucleotide substitution rate for the COI sequences of G. teuchis was inferred using an indirect calibration, i.e. the previously inferred divergence time of approx. 0.6 My for the Atlantic and Danubian lineages of S. trutta (see e.g. [20,25]). Accordingly the tMRCA prior for the corresponding node in G. teuchis was imposed as a lognormal distribution with a mean of 0.6 My (SD = 0.1 My). We applied a speciation Yule tree prior and the defaults of BEAUTi v. 1.8.0 [43] for all other parameters. Initial BEAST runs using a relaxed molecular clock with uncorrelated lognormal distributed substitution rates revealed a rate variation with a standard deviation close to zero, which indicates that a clock-like mode of substitution cannot be rejected. Consequently, all analyses were repeated under a strict molecular clock model with otherwise identical settings. To compare the inferred node ages between host and parasite the same procedure was applied to the host dataset. For each analysis, three independent Markov chains were run for 20 million generations and parameters were sampled every 1,000 generations. Parameter estimates of the independent analyses were checked for convergence using Tracer v. 1.5 [44] and combined with LogCombiner v. 1.8.0 [43].
Results
A total of 39 of 50 sampled salmonid populations (78%) were infected with G. teuchis, while G. truttae occurred in 22 populations (42%) (Table 1). Co-infection with both species was observed in 10 populations (20%). G. teuchis was thus significantly more prevalent than G. truttae (p < 0.001, Fisher’s exact test). The 189 Gyrodactylus individuals (nteu = 122; ntru = 67) analysed in the current study were collected from five salmonid host species: Salmo trutta (nteu = 110; ntru = 55), Salmo salar (nteu = 2), Salvelinus fontinalis (nteu = 5; ntru = 2), Oncorhynchus mykiss (nteu = 1), Thymallus thymallus (nteu = 4; ntru = 10). In a few brown trout and grayling populations, we also detected G. derjavinoides and G. thymalli, respectively.
Gyrodactylus truttae
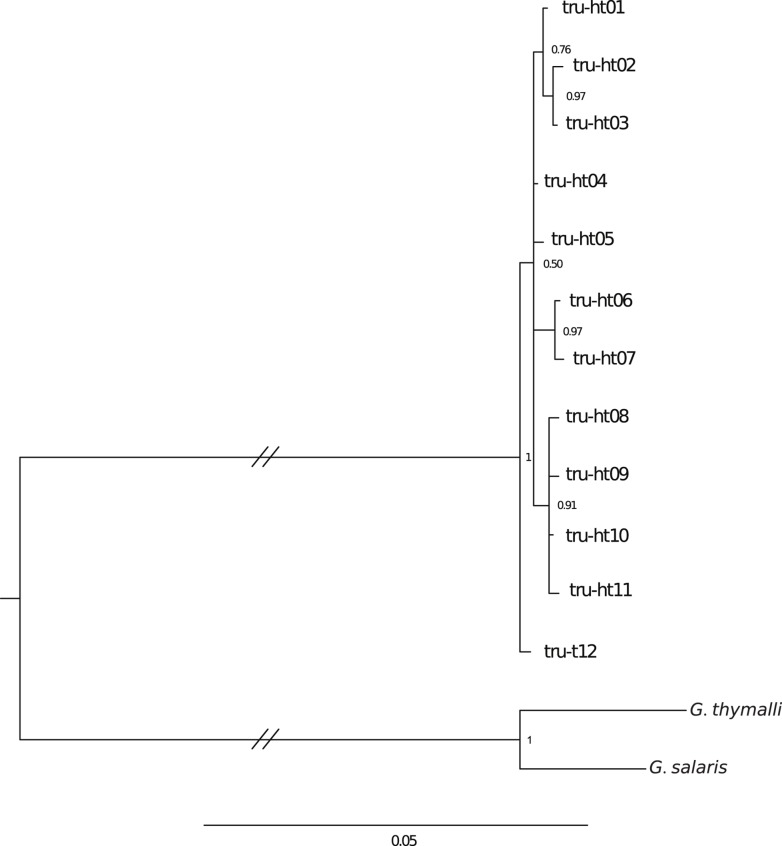
In total, we detected 11 novel mitochondrial haplotypes across the 67 analyzed G. truttae specimens from the 22 infected populations. None were identical to the previously published sequence of G. truttae from a Polish fish farm (GenBank entry EU304826). Nucleotide sequences of the new haplotypes were submitted to GenBank (accessions: KR080729—KR080739). The final alignment of 1,349 bp contained 14 variable sites (dS/dN ratio = 12/2), of which seven were parsimony informative. The average GC-content was 35.7%, and the transition/transversion ratio was 4.02. The average K2P-distance between any two COI haplotypes was 0.003 (range 0.001–0.005). Bayesian inference and ML methods recovered identical tree topologies. Automated species delimitation based on the PTP model assigned all G. truttae haplotypes to one single species (p<0.001). Table 3 summarizes the geographic distribution of individual haplotypes and Fig 4 presents the phylogeographic hypothesis for the species. Additionally, the ITS2 region was sequenced in 16 individuals from 14 sampling sites (see Table 3). BLAST search of the ITS2 sequences in all cases returned G. truttae as the best hit with > 99% identity. Only one variable site was detected at ITS2 position 86, which resulted in three variants (see Table 3). Variant A was identical to previously published sequences for G. truttae and was found in 11 populations (Table 3) and on two host species (S. trutta, T. thymallus). Variant B was found in two localities and two host species (S. trutta, T. thymallus). Variant C was only detected on S. trutta at one single site. No obvious congruence in relationships between COI haplotypes and ITS2 sequences could be identified. The novel ITS2 sequences obtained in the current study were submitted to GenBank (accessions: KR080740—KR080742).
Table 3. Geographical distribution of mitochondrial haplotypes and ITS sequences for G. truttae.
| ID | host | COI haplotypes | ITS2 variant | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |||
| 1 | St | 1 | - | |||||||||||
| 2 | St | 2 | A | |||||||||||
| 2 | Tt | 1 | - | |||||||||||
| 4 | St | 2 | - | |||||||||||
| 15 | St | 10 | A | |||||||||||
| 15 | Tt | 1 | - | |||||||||||
| 17 | Sf | 2 | - | |||||||||||
| 17 | St | 1 | - | |||||||||||
| 20 | St | 1 | 1 | B | ||||||||||
| 21 | St | 2 | - | |||||||||||
| 22 | St | 2 | - | |||||||||||
| 23 | St | 2 | A | |||||||||||
| 23 | Tt | 1 | - | |||||||||||
| 24 | Tt | 1 | A | |||||||||||
| 27 | St | 1 | - | |||||||||||
| 29 | Tt | 2 | A | |||||||||||
| 30 | Tt | 1 | A | |||||||||||
| 30 | St | 2 | A | |||||||||||
| 31 | St | 1 | A | |||||||||||
| 32 | St | 4 | 6 | A | ||||||||||
| 33 | Tt | 1 | - | |||||||||||
| 34 | Tt | 1 | - | |||||||||||
| 34 | St | 2 | A | |||||||||||
| 45 | St | 2 | - | |||||||||||
| 46 | St | 3 | 7 | C | ||||||||||
| 47 | Tt | 1 | B | |||||||||||
| 47 | St | 1 | B | |||||||||||
| 48 | St | 2 | A | |||||||||||
| 51 | St | ? | A | |||||||||||
Fig 4. Bayesian inference (GTR+Γ) of the intraspecific phylogenetic relationships within G. truttae based on COI.
Gyrodactylus teuchis
Among the 122 analyzed G. teuchis individuals we identified 14 novel mitochondrial haplotypes (GenBank accessions: KR080710—KR080723). The final alignment of 1,355 bp contained 187 variable sites (dS/dN ratio = 196/9) of which 160 were parsimony informative. The average GC-content was 36.3%, and the transition/transversion ratio was 5.98. The average K2P-distance between any two COI haplotypes was 0.061 (range 0.001–0.11). BI and ML methods yielded identical tree topologies (Fig 2). The PTP model delimited (p < 0.001) a total of two species in the G. teuchis dataset when using G. salaris and G. thymalli as outgroups (species 1—ht01-10; species 2—ht11-14). However, when using G. lavareti (GenBank EF446765) as an outgroup five species were delimited (species 1—ht01; species 2—ht02-ht-4; species 3—ht05-ht06; species 4—ht07-ht10; species 5—ht11-14). The genetic divergence estimates between the two outgroup species and G. teuchis were similar (G. lavareti vs. G. teuchis K2P = 0.186; G.salaris/thymalli vs. G. teuchis K2P = 0.218). Sequencing of the ITS2 region in 24 G. teuchis individuals from 22 sampling localities revealed five distinct ITS2 variants (see Table 4 and Table 5). Variant A was identical to the ITS2 sequence previously reported in the re-descriptions of G. teuchis [29,45] and was detected at 18 localities in parasites isolated from four host species (S. fontinalis, S. salar, S. trutta, T. thymallus), while variant D was found at two locations in parasites from two host species (S. fontinalis, S. trutta). In contrast, variants B, C, and E were each found at only one location in parasites from S. trutta (see Table 5). Overall there were seven variable sites, none of which were parsimony informative (Table 4). BLAST searches of the ITS2 sequences always returned G. teuchis as the best match with more than 99% identity. The novel ITS2 sequences obtained in this study were submitted to GenBank (accessions: KR080724—KR080728).
Table 4. Summary of ITS2 variants observed in G. teuchis and G. truttae.
| G. teuchis | ITS2 position | G. truttae | ITS2 position | ||||||
|---|---|---|---|---|---|---|---|---|---|
| variant | 58 | 68 | 100 | 127 | 173 | 180 | 255 | variant | 86 |
| A | A | T | T | G | A | G | C | A | A |
| B | G | A | C | R | A | G | C | B | T |
| C | R | W | Y | R | A | G | C | C | W |
| D | R | T | Y | G | R | R | Y | ||
| E | A | T | T | G | R | R | Y | ||
Table 5. Geographical distribution of mitochondrial haplotypes and ITS sequences for G. teuchis.
| ID | host | COI haplotype | ITS2 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | variant | |||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |||
| 3 | St | 2 | - | |||||||||||||
| 5 | St | 2 | - | |||||||||||||
| 6 | St | 2 | - | |||||||||||||
| 7 | St | 2 | - | |||||||||||||
| 8 | Ss | 1 | A | |||||||||||||
| 8 | St | 2 | A | |||||||||||||
| 9 | Ss | 1 | A | |||||||||||||
| 9 | St | 2 | A | |||||||||||||
| 49 | St | 2 | - | |||||||||||||
| 10 | St | 1 | 1 | B | ||||||||||||
| 11 | St | 2 | C | |||||||||||||
| 12 | St | 2 | A | |||||||||||||
| 13 | St | 2 | A | |||||||||||||
| 14 | Tt | 2 | A | |||||||||||||
| 16 | Sf | 1 | 1 | D | ||||||||||||
| 16 | St | 2 | E | |||||||||||||
| 17 | St | 2 | - | |||||||||||||
| 18 | St | 2 | D | |||||||||||||
| 19 | St | 1 | A | |||||||||||||
| 24 | St | 2 | A | |||||||||||||
| 25 | St | 1 | - | |||||||||||||
| 25 | Sf | 1 | - | |||||||||||||
| 26 | Sf | 2 | A | |||||||||||||
| 26 | St | 8 | - | |||||||||||||
| 27 | St | 1 | 1 | - | ||||||||||||
| 27 | Om | 1 | - | |||||||||||||
| 28 | St | 2 | - | |||||||||||||
| 30 | St | 2 | A | |||||||||||||
| 31 | St | 2 | A | |||||||||||||
| 32 | St | 5 | - | |||||||||||||
| 33 | Tt | 1 | - | |||||||||||||
| 34 | St | 2 | A | |||||||||||||
| 35 | St | 2 | - | |||||||||||||
| 36 | St | 2 | - | |||||||||||||
| 37 | St | 10 | A | |||||||||||||
| 38 | St | 10 | - | |||||||||||||
| 39 | St | 2 | - | |||||||||||||
| 40 | St | 2 | - | |||||||||||||
| 41 | St | 2 | - | |||||||||||||
| 42 | St | 3 | A | |||||||||||||
| 43 | St | 10 | A | |||||||||||||
| 44 | St | 2 | A | |||||||||||||
| 46 | St | 7 | A | |||||||||||||
| 48 | St | 1 | A | |||||||||||||
| 48 | Tt | 1 | - | |||||||||||||
| 50 | St | 2 | - | |||||||||||||
Co-phylogenetic analyses
The phylogenetic tree based on mitochondrial COI haplotypes of G. teuchis was compared with the phylogenetic tree of brown trout and Atlantic salmon based on sequences of the mitochondrial control region (see Fig 2). At first glance the topologies of both trees appear congruent. The two alternative host-parasite associations (HPM-1 and HPM-2) tested using co-phylogenetic methods are illustrated in Fig 3. For the HPM-1 association model, global congruence between host and parasite tree topologies was significant or of borderline significance (0.06) in three of the four tests using pairwise genetic (PPACo = 0.06; PParaFit = 0.089) or patristic distances (PPACo = 0.04; PParaFit = 0.061). However, more convincing significant global congruence was found for HPM-2 model using both genetic (PPACo = 0.035; PParaFit = 0.046) and patristic distances (PPACo = 0.019; PParaFit = 0.039).
The nucleotide substitution rate for the mitochondrial COI gene in G. teuchis
Applying the divergence time of ~ 0.6 My between the Atlantic and Danubian trout lineages [20,25] as an indirect calibration for the corresponding node in the G. teuchis phylogeny of mitochondrial haplotypes, BEAST inferred a mean substitution rate of 5.1% (95% HPD 2.9–7.7%) per million years using a strict molecular clock, which corresponds to an age of 1.46 My (95% HPD 0.8–2.2) for the most recent common ancestor of G. teuchis. The 95% confidence intervals for the inferred mean node ages for G. teuchis and its hosts overlapped in all cases. However, the mean age of nodes for G. teuchis tended to be lower than for the host trees.
Discussion
Despite both anthropogenic and naturally caused admixture of major host lineages among major river basins, the most parsimonious host-parasite association model revealed significant global congruence in tree topologies, providing support of co-speciation of G. teuchis and its salmonid hosts (S. trutta and S. salar). The anthropogenic introduction of brown trout stocks from various Atlantic basin sources into the Danube basin is well documented, but strong evidence has also been presented for a natural postglacial north-south range expansion [22,23,25]. Recently, the genetic distinction of anthropogenically and naturally expanding Atlantic lineages of brown trout into the Danube basin has been analysed in detail, and natural admixture is at least regionally dominant over human-mediated expansion [28]. This postulated unidirectional expansion of the host S. trutta is congruent with our parasite data, reflected in the occurrence of the putative Atlantic haplotype teu-ht03 in both Atlantic and Danubian watercourses, while haplotypes associated with Danubian brown trout lineage (teu-ht07-10) were absent from rivers of the Atlantic basin. The apparently stable co-occurrence of Atlantic and Danubian G. teuchis haplotypes in some watercourses of the Austrian Danube (e.g. Schwarzenseebach, Vordernbergerbach) is likewise congruent with gradual secondary postglacial contact rather than invasion of Atlantic haplotypes due to human mediated translocation. The absence of Danubian haplotypes in some Austrian rivers most likely reflects the limited sampling. However, local replacement of Danubian haplotypes via introgression and/or subsequent selective sweeps cannot be ruled out. In contrast, the occurrence of teu-ht08 in Rhone tributaries is most likely the result of human mediated translocation of parasites along with their fish hosts as this putative Danubian haplotype was also detected in a local Rhone basin fish farm. Currently only two trout populations have been sampled for Gyrodactylus in the Rhone catchment and it is expected that more extensive sampling is likely to also reveal distinct Rhone haplotypes of G. teuchis corresponding to the Rhone trout lineage. The detection of the common Danubian haplotype teu-ht08 in the Adriatic Soca system is easily explained by extensive stocking of Danubian trout in Soca tributaries (A. Snoj personal communication). The trout population of the river Kranska (sometimes referred to as S. peristericus [46]) was recently genotyped [47], and the authors reported that the population is fixed for a specific Adriatic trout haplotype. There is therefore little doubt that the G. teuchis haplotype collected from this very population (teu-ht06) indeed represents a novel Adriatic haplotype, an interpretation that is also supported by co-phylogenetic analyses. The Balkans are believed to be a centre of diversity for the genus Salmo [46], and an extensive exploration of the associated Gyrodactylus species is likely to reveal further cryptic diversity. Although found in a Danube tributary, haplotype teu-ht05 was assigned to the Adriatic lineage based on the results of the PTP species delimitation. Taking into consideration the topological congruence of the host and parasite trees as illustrated in Fig 2 one might expect an association of teu-ht05 with the Mediterranean trout lineage. There have been no reports of introgression of Adriatic or Mediterranean trout into the Bistrica trout population (A. Snoj personal communication). However, it should be kept in mind that the reproductive strategy of gyrodactylids facilitates rapid population growth and population expansion starting from a very low number of founder individuals.
The G. teuchis haplotype teu-ht01 was sufficiently divergent from the remaining Atlantic haplotypes (teu-ht02-04) to be delimited as separate species by the PTP algorithm (using G. lavareti as an outgroup) and topological congruence with the Salmo tree indicates an association with the Duero trout lineage. We emphasize that this choice of outgroup and application of the PTP algorithm alone does not provide a definitive answer of species delineation. Rather, we apply it as an objective approach for at least delineating clades or generating hypotheses concerning species diversity. The respective sampling locality for teu-ht01 (Donas) is not part of the Duero system and the local trout population has never been genotyped. Individual trout carrying Duero clade haplotypes, however, are not limited to the Duero system [24] and have been reported to occur in the relatively nearby north-flowing Mandeo River system [48]. In the absence of a reference haplotype from an unambiguously identified Duero trout location we speculate that teu-ht01 represents a parasite lineage specific to, or at least originating with the Duero trout lineage.
Co-phylogenetic analyses clearly indicate an association between Atlantic salmon and the highly divergent clade E of G. teuchis. The haplotypes within this clade (teu-ht11-14) were collected exclusively from rivers that support sympatric populations of brown trout and Atlantic salmon. Although restocking programs aiming to restore the dwindling salmon populations in these rivers have been carried out, the native salmon populations have not gone extinct [49,50], and the results of the co-phylogenetic analyses corroborate an ancient association of G. teuchis with Atlantic salmon in these rivers. In contrast, the contemporary salmon population of Tambre River was shown to be the result of stray individuals from neighbouring rivers [51], and the traces of the ancient association of G. teuchis with salmon in this river might have disappeared with the local extinction of the native salmon stock. The observed G. teuchis haplotype, associated with the Atlantic mtDNA lineage of brown trout, may reflect a secondary occupation of an open ecological niche.
Two recent studies attempted to date various splits within Salmonidae but did not specifically address the splits within the genus Salmo [52,53]. Nevertheless, the divergence time between the Atlantic and Danubian mtDNA brown trout lineages of approx. 0.6 My [20,25] is largely accepted. Using this calibration point as a proxy for the split of the respective mitochondrial lineages of G. teuchis yielded a nucleotide substitution rate of 5.1% (2.9–7.7%) per million years. This estimate is substantially lower than the 13.7–20.3% per million years reported for the COI genes of G. salaris and G. thymalli [54]. When comparing both estimates it is important to note that the latter was also derived indirectly. The authors interpreted the phylogeographic pattern of the COI haplogroups recovered for G. salaris and G. thymalli as indicative of simultaneous isolation of mitochondrial lineages and the separation of the hosts into four refugia situated around the Continental Ice Cap during the beginning of the last major glaciation cycle approx. 130,000 years ago [54]. While some key parameters such as the dS/dN ratio are very similar for the data presented here and that from [54] (21.7:1 for G. teuchis and 17:1 for G. salaris and G. thymalli) other characteristics such as the GC-content (36.3% for G. teuchis and 46% for G. salaris and G. thymalli) and the transition/transversion ratio (5.98 for G. teuchis and 1.1 for G. salaris and G. thymalli) differ significantly. It remains thus open to what extent the differences in the estimated substitution rates reflect (i) lineage differences, (ii) sampling bias, and/or (iii) uncertainties in the assumptions that the estimates rest on. Nevertheless, the substitution rate determined here for the mitochondrial COI gene of G. teuchis is very close to the estimate of 5.5% suggested for the ITS region of the nuclear rDNA cluster of Gyrodactylus, an estimate derived from vicariance of G. aphyae on the Eurasian minnow (Phoxinus phoxinus) on opposite sides of the Baltic–White Sea watershed [15]. This seems surprising as mitochondrial DNA in animals generally evolves substantially faster than nuclear loci, but the variation among taxonomic groups is large, and for some basal groups (e.g. corals) the opposite is true (see [55,56], and references therein). Huyse & Volckaert [18] reported the ITS region to evolve at a rate of only 0.07% per My in G. branchialis, an estimate in the same order of magnitude as the low genetic diversity of the ITS2 region observed in G. teuchis.
The current study not only suggests co-speciation of G. teuchis and its salmonids hosts, but also documents pronounced differences in the phylogeographic structure and the evolution of the host parasite systems between G. teuchis and G. truttae. While the association of G. teuchis with its host lineages seems to result from co-divergence, the relatively low genetic diversity and the lack of phylogeographic structure in G. truttae might indicate a very recent host switch onto brown trout (or a historical bottleneck) and a subsequent rapid range expansion; however, based on the limited sampling, the host-parasite association of G. truttae and Salmo spp. appears considerably more recent than for G. teuchis. Human mediated transfer of trout stocks may have significantly enhanced the spread of G. truttae. Applying the nucleotide substitution rate determined for COI of G. teuchis, the observed genetic diversity implies an age of <60,000 years for the most recent common ancestor of all currently identified G. truttae haplotypes, significantly younger than the splits among major mtDNA lineages of S. trutta.
Taxonomic implications
Species delimitation in Gyrodactylus is a difficult issue. Morphological species identification is usually based on only a few characters and is far from straightforward as it requires sophisticated morphometric techniques [57]. Molecular species identification in Gyrodactylus was therefore highly welcomed, and currently relies largely on sequencing of the ribosomal internal transcribed spacers (ITS). However, the current study reveals substantial difference in genetic diversity between ITS and the mtDNA COI. The PTP algorithm splits G. teuchis into two to five putative species based on COI data while the ITS data are in favour of G. teuchis being only one species. It is particularly interesting that the COI haplotypes of clade E, which were the only ones detected in parasites infecting Atlantic salmon differ from all other haplotypes by roughly 10%. Neither the original description of G. teuchis [58] nor the re-descriptions [29,45] were accompanied by COI sequences and genetic information was until now restricted to the ITS sequences produced by these studies. However, the river Scorff, sampled in the current study, was one of the localities where G. teuchis had initially been reported [58]. The COI data obtained for the river Kleiner Kamp in the current study is based on the same material that was used in the re-description of G. teuchis [29].
A similar pattern of differential resolution between ITS and COI sequence diversity has been reported for the notorious salmon parasite G. salaris and its benign sibling species G. thymalli infecting European grayling (T. thymallus). Mitochondrial COI haplogroups in G. salaris and G. thymalli differ by ~2–3% while the ribosomal ITS sequences are identical [15], reflecting a considerably lower substitution rate. The ITS data were taken as an indication of conspecificity by some authors [15] (see also [59] for review), while others reject this conclusion based on experimental evidence showing differences in host specificity and possibly co-evolutionary adaptations to the respective hosts [60,61]. In a recent study, Fromm et al. [62] investigated microRNA loci in G. salaris and G. thymalli and detected very low sequence diversity, which was interpreted as further support for the hypothesis of conspecificity initially drawn from the ITS data. However, in G. thymalli the various mitochondrial haplogroups seem to largely correspond to different European drainage systems [63]. The sister species G. salmonis and G. salvelini infecting North American Salvelinus fontinalis and European S. alpinus, respectively, represent a further example of a taxonomic dilemma. While the species show a > 2.5% mitochondrial divergence [64] they likewise share identical ITS sequences.
The current study emphasizes once more that taxonomic conclusions based on single molecular markers can be ambiguous. The application of high throughput genome-wide analyses will certainly add a new dimension to understanding the short-term evolutionary history in Gyrodactylus species and is likely to challenge our current view of species boundaries in this highly diverse, yet relatively understudied genus. However, we adhere to a view that molecular distances alone should not be used to revise taxonomy. Nonetheless, the molecular divergence for clade E within the G. teuchis complex is approximately 10% and far exceeds distances typically associated with species delineation, and further, this clade is associated with a shift in the host species, altogether supporting the main thesis that our study demonstrates co-speciation. However, this is not a taxonomic study, and in the absence of morphological data and experimental evaluations of potential reproductive barriers we render a taxonomic revision of the G. teuchis complex premature. Once more data is available, G. teuchis may become an ideal model species for exploring fine scale host-parasite interactions of monogenean fish parasites.
Acknowledgments
The authors are grateful to H. Hansen, R. Haunschmid, G. Holzer, E. Lorenz, S. Marić, G. Parthl, H. Persat, M. Popiolek, E. Prévost, J. Sanchez-Hernandez, A. Snoj, T. Speierl, V. Svinger, and G. Woschitz for providing samples and/or the opportunity to join electro-fishing operations. The assistance of J. Grond with DNA sequencing is also kindly acknowledged.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The study was financed exclusively by the Natural History Museum, University of Oslo, in the form of a PhD fellowship including material costs to CH.
References
- 1. Yip KY, Patel P, Kim PM, Engelman DM, McDermott D, Gerstein M (2008) An integrated system for studying residue coevolution in proteins. Bioinformatics 24: 290–292. [DOI] [PubMed] [Google Scholar]
- 2. Woolhouse ME, Webster JP, Domingo E, Charlesworth B, Levin BR (2002) Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat Genet 32: 569–577. [DOI] [PubMed] [Google Scholar]
- 3. Kellogg VL (1913) Distribution and species-forming of ecto-parasites. Am Nat 47: 129–158. [Google Scholar]
- 4. Fahrenholz H (1913) Ectoparasiten und Abstammungslehre. Zoologischer Anzeiger 41: 371–374. [Google Scholar]
- 5. Eichler W (1948) Some rules in ectoparasitism. Ann Mag Nat Hist (Ser 12) 1: 588–589. [Google Scholar]
- 6. Page RD, Charleston MA (1998) Trees within trees: phylogeny and historical associations. Trends Ecol Evol 13: 356–359. [DOI] [PubMed] [Google Scholar]
- 7. Simkova A, Morand S, Jobet E, Gelnar M, Verneau O (2004) Molecular phylogeny of congeneric monogenean parasites (Dactylogyrus): a case of intrahost speciation. Evolution 58: 1001–1018. [DOI] [PubMed] [Google Scholar]
- 8. Johnson KP, Adams RJ, Page RD, Clayton DH (2003) When do parasites fail to speciate in response to host speciation? Syst Biol 52: 37–47. [DOI] [PubMed] [Google Scholar]
- 9. de Vienne DM, Refregier G, Lopez-Villavicencio M, Tellier A, Hood ME, Giraud T (2013) Cospeciation vs host-shift speciation: methods for testing, evidence from natural associations and relation to coevolution. New Phytol 198: 347–385. 10.1111/nph.12150 [DOI] [PubMed] [Google Scholar]
- 10. Page RD, Clayton DH, Paterson AM (1996) Lice and cospeciation: a response to Barker. Int J Parasitol 26: 213–218. [DOI] [PubMed] [Google Scholar]
- 11. Harris PD, Shinn AP, Cable J, Bakke TA (2004) Nominal species of the genus Gyrodactylus von Nordmann 1832 (Monogenea: Gyrodactylidae), with a list of principal host species. Syst Parasitol 59: 1–27. [DOI] [PubMed] [Google Scholar]
- 12. Bakke TA, Harris PD, Cable J (2002) Host specificity dynamics: observations on gyrodactylid monogeneans. Int J Parasitol 32: 281–308. [DOI] [PubMed] [Google Scholar]
- 13. Cohen J (1977) Reproduction London, UK: Butterworth. [Google Scholar]
- 14. Cable J, Harris PD (2002) Gyrodactylid developmental biology: historical review, current status and future trends. Int J Parasitol 32: 255–280. [DOI] [PubMed] [Google Scholar]
- 15. Zietara MS, Lumme J (2002) Speciation by host switch and adaptive radiation in a fish parasite genus Gyrodactylus (Monogenea, Gyrodactylidae). Evolution 56: 2445–2458. [DOI] [PubMed] [Google Scholar]
- 16. Bakke TA, Cable J, Harris PD (2007) The biology of gyrodactylid monogeneans: the "Russian-doll killers". Adv Parasitol 64: 161–376. [DOI] [PubMed] [Google Scholar]
- 17. Darwin CR (1862) On the various contrivances by which British and foreign orchids are fertilized London, UK: Murray. [PMC free article] [PubMed] [Google Scholar]
- 18. Huyse T, Volckaert FA (2005) Comparing host and parasite phylogenies: gyrodactylus flatworms jumping from goby to goby. Syst Biol 54: 710–718. [DOI] [PubMed] [Google Scholar]
- 19. Cable J, van Oosterhout C, Barson N, Harris PD (2005) Gyrodactylus pictae n. sp. (Monogenea: Gyrodactylidae) from the Trinidadian swamp guppy Poecilia picta Regan, with a discussion on species of Gyrodactylus von Nordmann, 1832 and their poeciliid hosts. Syst Parasitol 60: 159–164. [DOI] [PubMed] [Google Scholar]
- 20. Bernatchez L (2001) The evolutionary history of brown trout (Salmo trutta L.) inferred from phylogeographic, nested clade, and mismatch analyses of mitochondrial DNA variation. Evolution 55: 351–379. [DOI] [PubMed] [Google Scholar]
- 21. Cortey M, Pla C, Garcia-Marin JL (2004) Historical biogeography of Mediterranean trout. Mol Phylogenet Evol 33: 831–844. [DOI] [PubMed] [Google Scholar]
- 22. Weiss S, Schlotterer C, Waidbacher H, Jungwirth M (2001) Haplotype (mtDNA) diversity of brown trout Salmo trutta in tributaries of the Austrian Danube: massive introgression of Atlantic basin fish—by man or nature? Mol Ecol 10: 1241–1246. [DOI] [PubMed] [Google Scholar]
- 23. Duftner N, Weiss S, Medgyesy N, Sturmbauer C (2003) Enhanced phylogeographic information about Austrian brown trout populations derived from complete mitochondrial control region sequences. Journal of Fish Biology 62: 427–435. [Google Scholar]
- 24. Cortey M, Vera M, Pla C, Garcia-Marin JL (2009) Northern and Southern expansions of Atlantic brown trout (Salmo trutta) populations during the Pleistocene. Biological Journal of the Linnean Society 97: 904–917. [Google Scholar]
- 25. Lerceteau-Kohler E, Schliewen U, Kopun T, Weiss S (2013) Genetic variation in brown trout Salmo trutta across the Danube, Rhine, and Elbe headwaters: a failure of the phylogeographic paradigm? BMC Evol Biol 13: 176 10.1186/1471-2148-13-176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cortey M, Garcia-Marin JL (2002) Evidence for phylogeographically informative sequence variation in the mitochondrial control region of Atlantic brown trout. Journal of Fish Biology 60: 1058–1063. [Google Scholar]
- 27. Susnik S, Snoj A, Wilson IF, Mrdak D, Weiss S (2007) Historical demography of brown trout (Salmo trutta) in the Adriatic drainage including the putative S. letnica endemic to Lake Ohrid. Mol Phylogenet Evol 44: 63–76. [DOI] [PubMed] [Google Scholar]
- 28.Schenekar T, Lerceteau-Kohler E, Weiss S (2014) Fine-scale phylogeographic contact zone in Austrian brown trout Salmo trutta reveals multiple waves of post-glacial colonization and a pre-dominance of natural versus anthropogenic admixture. Conservation Genetics.
- 29. Hahn C, Bakke TA, Bachmann L, Weiss S, Harris PD (2011) Morphometric and molecular characterization of Gyrodactylus teuchis Lautraite, Blanc, Thiery, Daniel & Vigneulle, 1999 (Monogenea: Gyrodactylidae) from an Austrian brown trout population. Parasitol Int 60: 480–487. 10.1016/j.parint.2011.08.016 [DOI] [PubMed] [Google Scholar]
- 30. Matejusova I, Gelnar M, McBeath AJ, Collins CM, Cunningham CO (2001) Molecular markers for gyrodactylids (Gyrodactylidae: Monogenea) from five fish families (Teleostei). Int J Parasitol 31: 738–745. [DOI] [PubMed] [Google Scholar]
- 31. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33: 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R (2003) DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497. [DOI] [PubMed] [Google Scholar]
- 35. Nylander JAA (2004) MrModeltest v2 Program distributed by the author. Evolutionary Biology Centre, Uppsala University. [Google Scholar]
- 36. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 37. Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- 38. Zhang J, Kapli P, Pavlidis P, Stamatakis A (2013) A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29: 2869–2876. 10.1093/bioinformatics/btt499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Legendre P, Desdevises Y, Bazin E (2002) A statistical test for host-parasite coevolution. Syst Biol 51: 217–234. [DOI] [PubMed] [Google Scholar]
- 40. Balbuena JA, Miguez-Lozano R, Blasco-Costa I (2013) PACo: a novel procrustes application to cophylogenetic analysis. PLoS One 8: e61048 10.1371/journal.pone.0061048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. R-Core-Team (2013) R: A language and environment for statistical computing Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 42. Poinar G (2003) A rhabdocoel turbellarian (Platyhelminthes, Typhloplanoida) in Baltic amber with a review of fossil and sub-fossil platyhelminths. Invertebrate Biology 122: 308–312. [Google Scholar]
- 43. Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rambaut A, Drummond AJ (2007) Tracer (http://beast.bio.ed.ac.uk/Tracer).
- 45. Cunningham CO, Mo TA, Collins CM, Buchmann K, Thiery R, Blanc G, et al. (2001) Redescription of Gyrodactylus teuchis Lautraite, Blanc, Thiery, Daniel & Vigneulle, 1999 (Monogenea: Gyrodactylidae); a species identified by ribosomal RNA sequence. Syst Parasitol 48: 141–150. [DOI] [PubMed] [Google Scholar]
- 46.Kottelat M, Freyhof J (2007) Handbook of European freshwater fishes; Kottelat C, Switzerland and Freyhof, Berlin, Germany, editor.
- 47. Snoj A, Maric S, Berrebi P, Crivelli AJ, Shumka S, Susnik S (2009) Genetic architecture of trout from Albania as revealed by mtDNA control region variation. Genet Sel Evol 41: 22 10.1186/1297-9686-41-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bouza C, Vilas R, Castro J (2008) Mitochondrial Haplotype variability of brown trout populations from north-western Iberian Peninsula, a secondary contact area between lineages. Conservation Genetics 9: 917–920. [Google Scholar]
- 49. Descroix A, Desvilettes C, Martin P, Anneville O, Bec A, Bourdier G (2009) Feeding, growth and nutritional status of restocked salmon parr along the longitudinal gradient of a large European river: the Allier. Ecology of Freshwater Fish 18: 282–296. [Google Scholar]
- 50. Martinez JL, Dumas J, Beall E, Garcia-Vazquez E (2001) Assessing introgression of foreign strains in wild Atlantic salmon populations: variation in microsatellites assessed in historic scale collections. Freshwater Biology 46: 835–844. [Google Scholar]
- 51. Saura M, Caballero P, Moran P (2008) Are there Atlantic salmon in the River Tambre? Journal of Fish Biology 72: 1223–1229. [Google Scholar]
- 52. Shedko SV, Miroshnichenko IL, Nemkova GA (2013) [Phylogeny of salmonids (Salmoniformes, Salmonidae) and molecular dating: analysis of mtDNA data]. Genetika 49: 718–734. [DOI] [PubMed] [Google Scholar]
- 53. Crete-Lafreniere A, Weir LK, Bernatchez L (2012) Framing the Salmonidae family phylogenetic portrait: a more complete picture from increased taxon sampling. PLoS One 7: e46662 10.1371/journal.pone.0046662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Meinila M, Kuusela J, Zietara MS, Lumme J (2004) Initial steps of speciation by geographic isolation and host switch in salmonid pathogen Gyrodactylus salaris (Monogenea: Gyrodactylidae). Int J Parasitol 34: 515–526. [DOI] [PubMed] [Google Scholar]
- 55. Willett CS (2012) Quantifying the elevation of mitochondrial DNA evolutionary substitution rates over nuclear rates in the intertidal copepod Tigriopus californicus. J Mol Evol 74: 310–318. 10.1007/s00239-012-9508-1 [DOI] [PubMed] [Google Scholar]
- 56. Lynch M, Koskella B, Schaack S (2006) Mutation pressure and the evolution of organelle genomic architecture. Science 311: 1727–1730. [DOI] [PubMed] [Google Scholar]
- 57. Vignon M (2011) Putting in shape: towards a unified approach for the taxonomic description of monogenean haptoral hard parts. Syst Parasitol 79: 161–174. 10.1007/s11230-011-9303-1 [DOI] [PubMed] [Google Scholar]
- 58. Lautraite A, Blanc G, Thiery R, Daniel P, Vigneulle M (1999) Gyrodactylids parasitizing salmonids in Brittany and Western Pyrénées water basins: Epidemiological features of infection and species composition. Bulletin Français de Pêche et de la Pisciculture 355: 305–325. [Google Scholar]
- 59. Hansen H, Bakke TA, Bachmann L (2007) DNA taxonomy and barcoding of monogenean parasites: lessons from Gyrodactylus. Trends Parasitol 23: 363–367. [DOI] [PubMed] [Google Scholar]
- 60. Bakke TA, Soleng A, Harris PD (1999) The susceptibility of Atlantic salmon (Salmo salar L.) x brown trout (Salmo trutta L.) hybrids to Gyrodactylus salaris Malmberg and Gyrodactylus derjavini Mikailov. Parasitology 119 (Pt 5): 467–481. [DOI] [PubMed] [Google Scholar]
- 61. Soleng A, Bakke TA (2001) The susceptibility of grayling (Thymallus thymallus) to experimental infections with the monogenean Gyrodactylus salaris. Int J Parasitol 31: 793–797. [DOI] [PubMed] [Google Scholar]
- 62. Fromm B, Burow S, Hahn C, Bachmann L (2014) MicroRNA loci support conspecificity of Gyrodactylus salaris and Gyrodactylus thymalli (Platyhelminthes: Monogenea). Int J Parasitol 44: 787–793. 10.1016/j.ijpara.2014.05.010 [DOI] [PubMed] [Google Scholar]
- 63. Hansen H, Bakke TA, Bachmann L (2007) Mitochondrial haplotype diversity of Gyrodactylus thymalli (Platyhelminthes; Monogenea): extended geographic sampling in United Kingdom, Poland, and Norway reveals further lineages. Parasitol Res 100: 1389–1394. [DOI] [PubMed] [Google Scholar]
- 64. Gilmore SR, Abbott CL, Cone DK (2010) The placement of Gyrodactylus salmonis (Yin & Sproston) in the molecular phylogeny of studied members of the Gyrodactylus wageneri-group parasitizing salmonids. J Fish Dis 33: 461–467. 10.1111/j.1365-2761.2010.01154.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.