Abstract
Although Bmp2 is essential for tooth formation, the role of Bmp2 during enamel formation remains unknown in vivo. In this study, the role of Bmp2 in regulation of enamel formation was investigated by the Bmp2 conditional knock out (Bmp2 cKO) mice. Teeth of Bmp2 cKO mice displayed severe and profound phenotypes with asymmetric and misshaped incisors as well as abrasion of incisors and molars. Scanning electron microscopy analysis showed that the enamel layer was hypoplastic and enamel lacked a typical prismatic pattern. Teeth from null mice were much more brittle as tested by shear and compressive moduli. Expression of enamel matrix protein genes, amelogenin, enamelin, and enamel-processing proteases, Mmp-20 and Klk4 was reduced in the Bmp2 cKO teeth as reflected in a reduced enamel formation. Exogenous Bmp2 up-regulated those gene expressions in mouse enamel organ epithelial cells. This result for the first time indicates Bmp2 signaling is essential for proper enamel development and mineralization in vivo.
Introduction
Tooth development is a highly organized process involving sequential and reciprocal interaction between epithelial and mesenchymal cells. The enamel formation results from the differentiation of the dental inner enamel epithelium into functional ameloblasts in a distinct spatial-temporal pattern during amelogenesis (Linde and Goldberg, 1993; Bartlett, 2013). The ameloblasts synthesize and secrete the enamel matrix proteins including amelogenin (AMEL) and non-amelogenin matrix proteins. AMEL is the most abundant enamel matrix protein (Termine et al., 1980; Salido et al., 1992; Smith, 1998). AMEL mutations in humans and mice cause amelogenesis imperfecta (AI), (Lagerstrom et al., 1991; Ravassipour et al., 2000; Gibson et al., 2001).
Non-amelogenin matrix proteins include enamelin (ENAM) and ameloblastin (AMBN). ENAM is the largest enamel matrix protein, accounting for 3–5% of enamel matrix proteins (Fukae et al., 1996; Hu et al., 1997). Human ENAM gene mutations cause autosomal dominant forms of AI (Rajpar et al., 2001; Mardh et al., 2002). Enam null mice exhibited abnormal enamel structures with a rough and pitted enamel surface (Hu et al., 2008). AMBN is suggested to be a cell adhesion molecule that regulates proliferation and differentiation of ameloblastic cells as well as that of other cells (Cerny et al., 1996; Krebsbach et al., 1996; Sonoda et al., 2009). Ambn ablation in mice and humans causes severe enamel abnormalities (Fukumoto et al., 2004; Poulter et al., 2014).
During enamel formation, these enamel matrix proteins are processed by several proteases to produce active molecules following their secretion into the developing enamel matrix (Bartlett, 2013). Matrix metalloprotein-20 (MMP-20) and kallikrein-related peptidase-4 (KLK4) are expressed in ameloblasts and secreted into the enamel layer. They have been shown to cleave these enamel matrix proteins including Amel, Ambn, and Enam (Li et al., 1999; Iwata et al., 2007; Yamakoshi et al., 2006; Chun et al., 2010; Nagano et al., 2009). Mutations of MMP-20 and KLK4 in humans cause autosomal recessive hypomaturation AI (Hart et al., 2004; Kim et al., 2005; Ozdemir et al., 2005b). In mice lacking Mmp-20 the enamel forms as a bilayer without rod-interrod organization (Caterina et al., 2002). In Klk4 null mice the enamel layer obtains normal thickness and organization, but is hypomineralized (Simmer et al., 2009).
Amelogenesis is a complex process controlled by many growth factors and transcriptional factors (Thesleff, 2003). Members of the bone morphogenetic protein (BMP) family have diverse biological functions during osteogenesis and embryonic development (Hogan, 1996; Ducy and Karsenty, 2000; Chen et al., 2004; Rosen, 2009). Among the BMP family members, BMP2 has been extensively studied for its various biological functions during chondrogenic and osteogenic differentiation as well as organ development (Zhang and Bradley, 1996; Ma et al., 2005; Lee et al., 2007; Singh et al., 2008; Feng et al., 2011; Yang et al., 2013). Bmp2 expression has been observed in odontoblasts and ameloblasts during tooth cytodifferentiation from mouse embryonic and postnatal stages (Aberg et al., 1997; Chen et al., 2008). Bmp2 is able to induce ameloblast differentiation and enamel-related gene expression in vitro (Miyoshi et al., 2008). These results indicate that Bmp2 is important for ameloblast differentiation and enamel formation. We previously showed that Bmp2 plays a critical role in postnatal tooth development when Bmp2 gene was conditionally deleted (Feng et al., 2011; Yang et al., 2012). Bmp2 null mice display abnormal tooth phenotypes with asymmetric and open forked incisors. However, it remains unclear how Bmp2 controls enamel development. In this study, we reported that teeth of Bmp2 conditional knock-out (cKO) mice exhibited similar symptoms to AI and effect of Bmp2 on enamel formation regulates expression of enamel matrix protein and enamel-processing protease genes.
Materials and Methods
Animals
All animal protocols were reviewed and approved by the Institute Animal Care at the University of Texas Health Science Center at San Antonio. All animals were fed with a soft diet (ClearH2O, Portland, ME). Bmp2 cKO mice and Bmp2 control mice (wild type and heterozygotes Bmp2) were used for this study. A conditional allele of the mouse Bmp2 gene was created by introducing Cre recombinase recognition sites (loxP) flanking upstream and downstream of exon 3 of the mouse Bmp2 gene (Feng et al., 2011). The floxed Bmp2 mice were crossed with Osx-Cre mice (Maes et al., 2010) to generate Bmp2 cKO mice. For tissue-specific genotyping, tail lysates were prepared by adding tail snips to the tissue lysis buffer and proteinase K according to manufacture protocol (Promega, Madison, WI). Genomic DNA was isolated from mouse tails using a DNA purification kit (Promega) and performed for PCR analysis. The primers used for detecting the wild and mutant alleles were as follows: forward 5′-AGGGTTTCAGGTCAGTTTCCG-3′ and reverse 5′-GATGATGAGGTTCTTGGCGG-3′ (floxed); forward 5′-AGCATGAACCCTCATGTGTTG-3′ and reverse 5′-GCTGTTTGTGTTTGGCTTGA-3′ (recombinant).
Microscopic analysis of teeth
Animals were anesthetized using Ketamine (Sigma-Aldrich, St Louis, MO). Tooth morphology was recorded under light microscopy. For histological examination, mandibles were fixed in 4% paraformaldehyde overnight. The samples were decalcified, embedded in paraffin, sectioned and stained with hematoxylin/ eosin.
X-ray analysis
Radiography was used to measure changes in teeth and bones. Maxillae and mandibles were radiographed using a Faxitron radiograph inspection unit (Field X-ray Corporation, Lincolnshire, IL). Digitized images were analyzed using the AnalySIS software to measure size and width of selected components in the incisors and molars.
Micro-computer tomography (Micro-CT)
Hemi-mandibles were scanned in a desktop SkyScan 1172 system (Bruker SkyScan, Aartselaar, Belgium). Each specimen was positioned with the incisal edge pointing superiorly, and the tube was sealed with Parafilm (American National Can, Greenwich, CT). Samples were scanned at 60 kV, 167 μA beam intensity, 5-μm image pixel size, a 0.35° rotation step, 7-frame averaging, and a 1,090-millisecond exposure time at each step. A 0.5-mm aluminum filter was used during scanning (Kovacs et al., 2009). A polynomial correction was also used to reduce beam-hardening effects during reconstructions (Kovacs et al., 2009; Zou et al., 2011). The images were reconstructed with NRecon (Bruker SkyScan) with a Feldkamp cone-beam algorithm. Successive 1-mm–long volumes of interest were created. Enamel volumes were determined from the manually drawn. HA phantoms (0.25 and 0.75 g/cm3 (SkyScan), and 2.927 g/cm3 (Himed, Bethpage, NY) were used to calibrate mineral density analysis. The various tissue types (enamel, dentin, and pulp chamber) were analyzed and quantified. The datasets were re-orientated so the axes of the first molar approached the image x, y and z planes. The division between the enamel and dentin was manually isolated, but an automated script was used to isolate the various tissues types within the two regions. Hemi-mandibles from different animals were used for micro-CT (n = 4, two males and 2 females/per group).
Scanning electron microscopy (SEM)
Mandibular incisors from 1-, 3-month-old Bmp2 cKO and wild-type mice were fractured at the level of the labial alveolar crest and mounted on metallic stubs using conductive carbon cements. The surface was washed in 0.1 M sodium cacodylate buffer and fixed for 20 min in 2.5% (w/v) glutaraldehyde (Sigma-Aldrich) in 0.1 M cacodylate buffer. The surface was washed in sodium cacodylate buffer and dehydrated in an ascending alcohol series. Finally, the surface was fixed in hexamethydioslazane (Sigma-Aldrich) allowed to air dry and then sputter-coated with gold. The samples were examined by SEM at 20 kV (JEOLJSM 6610 LV; JEOL, Inc., Peabody, MA).
RNA extraction and quantitative real time PCR (qRT-PCR)
The mandibles and maxillae from the Bmp2 cKO and control mice at postnatal days (PN) 2, PN4 and PN6 were homogenized with a homogenizer (Bertin Technology, Rockville, MD). Total RNA was isolated using TRIZOL reagent (Qiagen Inc. Valencia, CA), treated with DNase I (Promega), and purified with the RNeasy Mini kit (Qiagen Inc.). RNA concentration was determined by UV spectroscopy at 260 nm. Complementary DNA synthesis and PCR amplification were performed using standard protocols. Bmp2 primer set used for RT-PCR was as follows: forward 5′-CGGGAACAGATACAGGAAGC-3′ and reverse 5′-GCTGTTTGTGTTTGGCTTGA-3′. qRT-PCR amplification reaction was analyzed in real time on an ABI 7500 (Applied Biosystems, Foster City, CA) using SYBR Green chemistry, and threshold values were calculated using SDS2 software (Applied Biosystems). qRT-PCR was performed with Amel, Ambn, Enam, Mmp-20, Klk4 and cyclophilin A. Primers used for qRT-PCR were as follows: Amel, forward 5′-TGAAGTGGTACCAGAGCATGA-3′ and reverse 5′-ACAGGGATGATTTGGTGGTG-3′; Ambn, forward 5′-TTCCCATGGATAGGACCAAG-3′ and reverse 5′-ATCAGCTCTCCTTCCTGCAA-3′; Enam, forward 5′-GGAACCACCAAATGAAGCAG-3′ and reverse 5′-CCAAAGCCGTGATATCCAAA-3′; Mmp-20, forward 5′-CTCGTCCTTTGATGCAGTGA-3′ and reverse 5′-AAGAAAGCAATGCCTCGTTC-3′; Klk4, forward 5′-ATGATGGTCACTGCACGAAC-3′ and reverse 5′-CAAGACTCCCGAGCAGAAA-3′; cyclophilin A, forward 5′-GGTGACTTCACACGCCATAA-3′ and reverse 5′-CATGGCCTCCACAATATTCA-3′. The ΔΔCt method was used to calculate gene expression levels normalized to cyclophilin A value. The results were performed in triplicate of three separate experiments and expressed as a relative fold change in gene expression compared to the control.
Mechanical property measurements of teeth
Mandibular incisors were dissected from 4 week-old control and Bmp2 cKO mice (n = 6 for each genotype). The teeth were embedded in Acrymount embedding resin (Electron Microscopy Sciences, Hatfield, PA) with an iron holder. Mechanical property of shear and compression was conducted with a testing machine (ReNew Model 1125 Upgrade Package, MTS Systems Corporation, Eden Prairie, MN) using a 4448 N load endpoint and an initial speed of 1 mm/min. Data were analyzed using Image-Pro Plus Software (Media Cybernetics, Inc., Rockville, MD).
In situ hybridization
Mouse mandibles were fixed and processed. Serial sections were mounted on saline-treated slides. Representative sections from each block were stained with hematoxylin. The mouse cDNA corresponding to coding region of Bmp2 gene was generated by PCR (Chen et al., 2008). The amplified PCR product was subcloned into pCRII vectors containing Sp6 and T7 promoters (Invitrogen, Carlsbad, CA). Labeled 32P-rUTP antisense and sense Bmp2 probes were generated using Sp6 or T7 RNA polymerases after linearization with appropriate restriction enzymes, respectively. The method of in situ hybridization was performed as described earlier (Chen et al., 2008). Briefly, hybridization was performed at 55 °C overnight in a solution containing 50% formamide, 20 mM Tris-HCl (pH 8.0), 1 mM EDTA, 0.3 M NaCl, 10% dextran sulfate, 1 x Denhardt’s solution, 100 μg/ml denatured SS-DNA, 500 units/ml tRNA, and 1 μg/ml of 32P-rUTP labeled RNA probe. After hybridization, the cover slips were removed in 2 x SSC at room temperature, and sections were washed in RNase-free buffer (0.3 M NaCl, 10 mM Tris-HCl, 5 mM EDTA) at 37 °C for 10 min. The sections were incubated with RNases in the RNase-free buffer at 37 °C for 1 h, followed by incubation in the RNase-free buffer for 30 min. Consecutive 5-minute washes at 57 °C were done twice with 2 x SSC, four times in 0.5 x SSC, and three times in 0.1 x SSC. After washing, the sections were dehydrated using ethanol containing 0.3 M ammonium acetate. For autoradiography, slides were dipped in photographic emulsion (Kodak Scientific Imaging, Rochester, NY) diluted 1:1 with 0.6 M ammonium acetate at 42 °C. After drying at room temperature, the slides were exposed in the presence of desiccant for 3 days to 3 weeks and developed in a Kodak D-19 developer. The slides were counter-stained with hematoxylin, dehydrated through ethanol, cleared in xylene, and mounted with Permount (Fisher Scientific, Pittsburgh, PA).
Immunohistochemistry
Rabbit polyclonal anti-Amel, rabbit polyclonal anti-Ambn, goat polyclonal anti-Enam, rabbit polyclonal anti-Klk4 antibodies (Santa Cruz Biotechnology, Inc. Santa Cruz, CA) and rabbit monoclonal anti-Mmp-20, rabbit polyclonal anti-Bmp2 antibodies (Abcam, Cambridge, MA) were used as primary antibodies. For immunohistochemistry, tissue sections were dewaxed with xylene, rehydrated with a series of ethanol. To identify the distribution of Bmp2 in mouse tooth tissues, anti-Bmp2 antibody was used as the first antibody. In brief, the rehydrated tissue sections were incubated with 3% H2O2 to remove the endogenous peroxidase activity. For antigen retrieval, the sections were incubated in 10 mM sodium citrate solution for 20 min at 92 °C and in blocking agent for 20 min, followed by overnight incubated at 4 °C with 10 μg/ml of Bmp2 antibody. Those slides incubated with control IgG instead of the first antibody were used for negative control (Dako Carpinteria, CA). The sections were then incubated with biotinylated secondary antibody (anti-rabbit IgG, Vector Laboratory Inc., Burlingame, CA) and Vectastain Elite ABC reagent (Vector Laboratory Inc.). Subsequently, the tissue sections were stained with DAB and counterstained with Mayer’s hematoxylin and examined by light microscopy. For fluorescent immunohistochemistry, the tissue sections were dewaxed with xylene, rehydrated with ethanol and treated with H2O2. Then, the tissue sections were blocked with 10% normal donkey serum (Sigma–Aldrich) and incubated with 10 μg/ml of either the primary goat or rabbit antibody, which was recognized by the donkey anti-goat or anti-rabbit secondary antibody conjugated with Alexa Fluor ® 488 (Molecular Probes, Eugene, OR) overnight at 4 °C. The sections were rinsed with phosphate buffered saline, incubated with secondary antibody for 90 min, rinsed in H2O. For the nucleus staining, the tissue sections were incubated with Hoechst (Sigma–Aldrich) for 5 min at room temperature. After washing, the tissue sections were mounted using vectashield mounting medium (Vector Laboratory Inc.). For negative control, the primary antibody was replaced by mouse IgG I (Dako). Images of Alexa Fluor ® 488 staining of the various proteins were captured by a Nikon Eclipse TE2000S microscope with a filter (B-2E/C, C84941, a 450 ± 490 nm excitation filter) using a digital cooled camera connected to a PC computer and analyzed with NIS-Elements3.2 software. Image of nuclear staining with Hoechst was caught with a filter UV-2E/C, C86826. For each experiment, all slides were simultaneously processed for a specific antibody, so that homogeneity in the staining procedure was ensured between the samples. After the capture of these images at the same magnification, the threshold was set with the same resolution [fast (focus)], quality (capture), exposure, gain, and contrast, and maintained for each slide in the experiment. The optical density was calculated by use of the morphometric analysis within the software package.
Western blot analysis
The mouse enamel organ epithelial (EOE-2M) cells were described previously (Feng et al., 2012). Briefly, the EOE-2M cells were maintained in Dulbecco’s modified eagle medium (DMEM) (Invitrogen) with 10% fetal calf serum (FCS), 100 units/ml of penicillin/streptomycin, 50 μg/ml ascorbic acid. The EOE-2M cells were treated with or without 300 ng/ml of recombinant Bmp2 (Invitrogen) in DMEM medium with 0.5% FCS and 100 units/ml of penicillin/streptomycin at 37 °C in a 5% CO2 atmosphere for 24-,48- and 72-h, respectively. The cells were then washed with 1 x cold PBS and lysed with RIPA buffer (1 x PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 10 mg/ml phenylmethylsulfonyl fluoride (PMSF), 50 KIU/ml aprotinin, 100 mM sodium orthovanadate; Santa Cruz Biotechnology, Inc.). Whole cell lysates were resolved by 7% SDS-PAGE gels and transferred to Trans-Blot membranes (Bio-Rad, Hercules, CA). For the detection of mouse Amel, Ambn, Enam, Mmp-20 and Klk4, these antibodies described above were used as primary antibodies. The membranes were blocked with 5% non-fat milk in TBST buffer (10 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.1% Tween-20) for 60 min at room temperature. After washing, the membranes were incubated with primary antibodies against Amel, Ambn, Enam, Mmp-20 and Klk4 with appropriate dilution (1:500–1,000) overnight at 4 °C, respectively. The secondary antibody (horseradish peroxidase-conjugated anti-rabbit or anti-goat IgG) were diluted to 1:5,000–10,000 at room temperature for 60 min.
Immunoreactivity was determined using ECL chemiluminescence reagent (Thermo Scientific, Pittsburgh, PA). As a control, goat polyclonal anti-mouse β-actin antibody was used (Santa Cruz Biotechnology, Inc.). The band intensity was measured using ImageJ software (ImageJ, NIH./gov/iJ). Enamel protein expression level of each sample was normalized to β-actin value. The Bmp2 untreated enamel protein was used as control and acts as 1-fold increase. The fold increase in the Bmp2 treated enamel protein was calculated by dividing the control group.
Statistical analysis
Quantitative data were presented as means ± S.D. from three independent experiments and compared with the results of one-way ANOVA using GraphPad Prism 5 (GraphPad Software, Inc. La Jolla, CA). The differences between groups were statistically significant at *P <0.05 and **P <0.01.
Results
Expression of Bmp2 gene in ameloblasts during amelogenesis
To determine whether Bmp2 is expressed in ameloblasts during mouse enamel formation, we studied expression patterns of Bmp2 at postnatal stages of tooth development, using an in situ hybridization assay. At PN 1, expression of Bmp2 was observed in ameloblasts besides its expression in odontoblasts and osteoblasts in alveolar bones (Fig. 1b). At PN 5, Bmp2 expression pattern was similar to those at PN 1 (Fig. 1f). To further assess Bmp2 expression at translational levels in ameloblasts, immunohistochemistry was performed to analyze Bmp2 protein expression. Bmp2 signal was detected within the nucleus and cytoplasm in ameloblasts at PN1 and PN5 while Bmp2 expression was also observed in odontoblasts (Fig. 1d, h). However, Bmp2 protein expression was more intense within the nuclei than that of the cytoplasm. This indicates that Bmp2 is expressed in ameloblastic cells during amelogenesis.
Fig. 1.
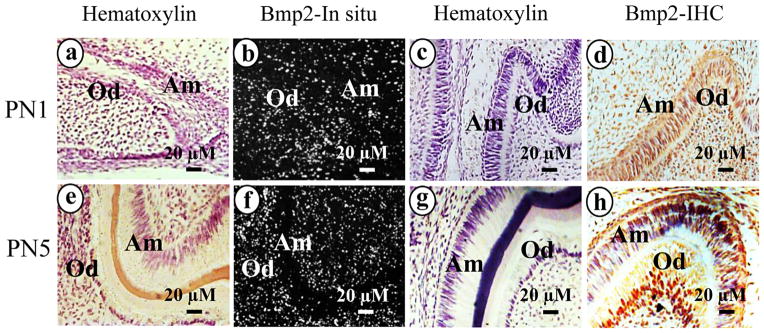
Bmp2 expression in ameloblasts during enamel formation. b and f. In situ hybridization of mouse tooth development at PN1 and PN5 with Bmp2 antisense probe. Bmp2 expression was observed in ameloblasts as well as in dental pulp, odontoblasts and osteoblasts. Hematoxylin staining is shown in a and e. Bmp2 protein expression in PN1 and PN5 mice was detected within the nucleus and cytoplasm in ameloblasts, odontoblasts and osteoblasts using immunohistochemistry (d and h). Panels, c and g are control tissue sections incubated with normal IgG instead of the primary antibodies. Abbreviations: Am, ameloblasts; Od, odontoblasts.
Bmp2 cKO mice analyzed by gross tooth morphology and radiography
To identify Bmp2 cKO mice, genomic DNA was isolated from the wild type and Bmp2 cKO mice.
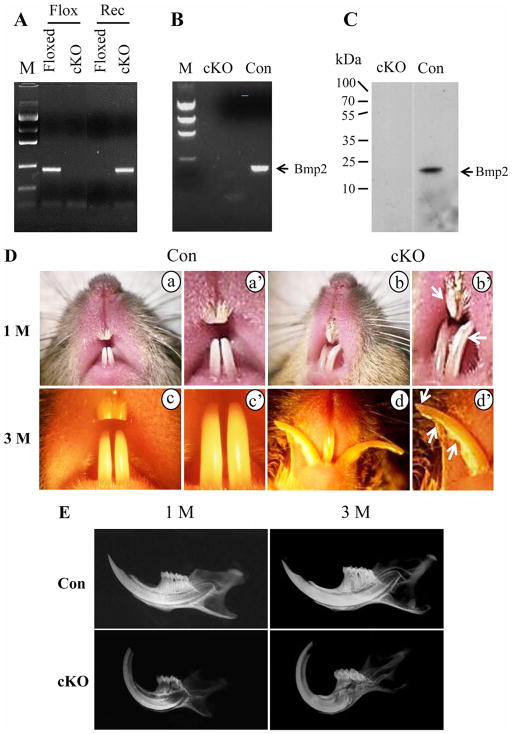
The genotyping of the Bmp2 cKO mice was identified using PCR assay (Fig. 2A). Bmp2 gene expression was not detected in the null teeth using RT-PCR and Western blot assays (Fig. 2B–C). Teeth with the Bmp2 cKO mice displayed abnormalities with asymmetric and uneven incisors (Fig. 2D). Noticeably, the mandibular incisors of the null mice were chalky white and the mandibular incisor enamel at the functional edge was apparently abraded and exhibited a prominent wear facet on the buccal surface. Also, mandibular incisor tips of the Bmp2 ablated mice had uneven, chisel chipping off irregular shape rather than the evenly smooth shape of the control incisors. Similarly, molars of the Bmp2 cKO mice were rough and rugged with wearing or abrasion of the cusps (data not shown). Dental radiography analysis revealed that the mandibular incisors of the Bmp2 cKO mice had abnormal morphology as well as the radio-opacity of the mandibles and the dental enamel mineral density was decreased in 1- and 3-month-old null mice compared to the control mice (Fig. 2E). Also, the dentin-enamel junction (DEJ) contrast in the Bmp2 mutant mice was reduced and tips of the mandibular incisors were abraded and dental pulp chambers exposed. The enamel layer of the mandibular incisors of the Bmp2 cKO mice was thin.
Fig. 2.
Gross morphology of teeth. A. Genotyping and PCR strategy. Tail genomic DNA was isolated from floxed Bmp2 and Bmp2 cKO mice and amplified by PCR using Bmp2 specific primers described as “Materials and Methods”. Floxed and Rec are floxed and recombinant primers used for PCR, respectively. B. RNA was isolated from the Bmp2 floxed and cKO teeth. RT-PCR results showed that Bmp2 mRNA is not detected in the Bmp2 null teeth using Bmp2 specific primer set. C. Proteins isolated from the Bmp2 floxed and cKO mandibles were detected by Western blot assay using the Bmp2 antibody. Bmp2 expression was seen in the Bmp2 floxed tissues, but not in the Bmp2 null mice. D (a–d). In the mutant mice, both sides of up and lower incisors are asymmetric with open forked (b, d). The incisors appear chalky white (arrows, b′). The incisor edge is jagged with chipping and tip incisor wearing (arrows, d′). a and c. In the normal mice, the enamel appears smooth, opalescent and incisors are symmetric. a′, b′, c′ and d′ indicate higher magnification of a, b, c and d. E. X-ray analysis showed that teeth in the Bmp2 cKO mice are abnormal. Incisor tip is abraded and dental pulp cavity exposed. Enamel layer is thin and the contrast of dentin-enamel junction (DEJ) reduced. M, DNA ladder marker; cKO, Bmp2 conditional knock out; Con, control; 1M, 3M, 1 and 3 months.
Abnormal teeth from Bmp2 cKO mice assayed by microcomputered tomography
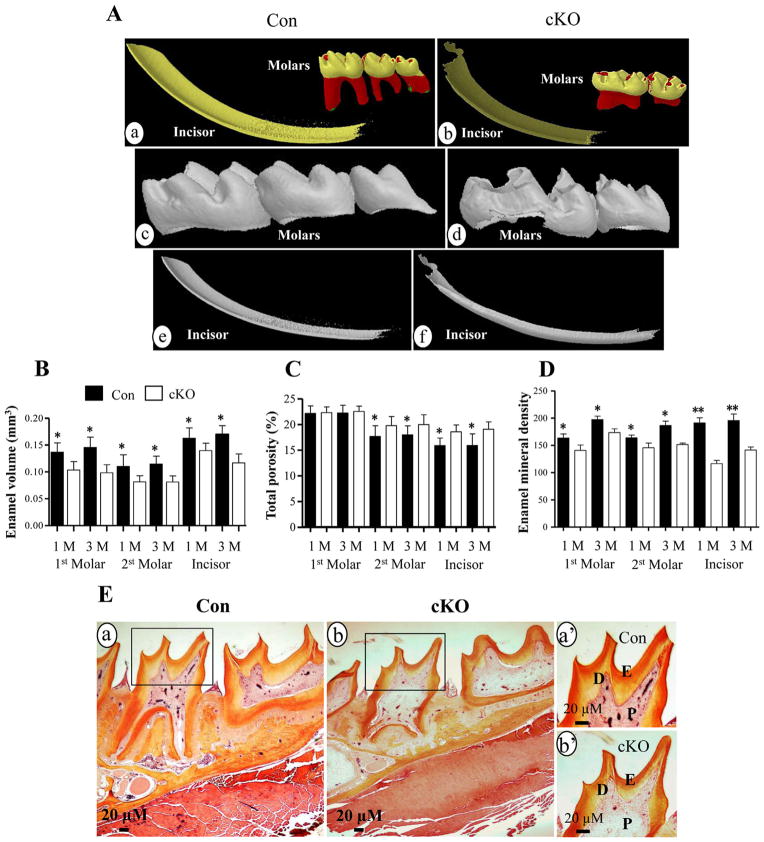
To further determine whether the X-ray data were correlated with Micro-CT study, Micro-CT assay was used to exam tooth changes. Micro-CT analysis demonstrated that the third molar in the null mouse was missing (Fig. 3Ab,d) and the enamel volume of the mandibular incisors, first and secondary molars of 1- and 3-month-old null mice was decreased compared to the control groups (Fig. 3Ab,d,f; 3B). In contrast, the porosity in sizes of the mandibular secondary molars and incisors were larger than that of the control mice (Fig. 3C). Enamel mineral density of incisors and molars in the Bmp2 cKO teeth was also decreased (Fig. 3D). Histological study showed that enamel layer of the Bmp2 null mice at 1 month-old Bmp2 null mice was thin compared to that of 1 month-old control mice besides thin dentin, enlarged dental pulp chamber and delayed root formation (Fig. 3E).
Fig. 3.
Micro-computer tomography and histology of teeth. A. Micro-CT analysis of teeth of the Bmp2 cKO (b, d, f) and control (a, c, e) mice. The molars (a, b, c, d) and incisors (a, b, e, f) from 1 month old mice were subjected to Micro-CT analysis and showed that enamel layer in the null mice is thinner than that of the control mice. Mineral density of the incisors and molars of the Bmp2 cKO mice is decreased versus the control groups. The third molar of the null mice is missing. B. Enamel volume of the first, second molars and incisors in the null mice is reduced in the 1- and 3-month-old mice. C. Porosity of the second molars and incisors of the Bmp2 mutant mice is increased in size. D. Enamel mineral density of the first, second molars and incisors is decreased in the 1- and 3-month-old cKO mice. E is representative images, and histological staining showed that enamel layer and dentin of the molars are thinner (b) than the control teeth (a) in 1 mouth-old mice. a′ and b′ show higher magnification of a and b. *(P <0.05) and **(P <0.01) indicate significant differences between the control and Bmp2 cKO groups. Con, control; cKO, Bmp2 conditional knock out; D, dentin; E, enamel; P, dental pulp. Yellow and red colors of molars in panels Aa and Ab show tooth crown and root, respectively. Dark and light yellow colors of incisors in panels Aa and Ab indicate enamel and dentin.
Characterization of null mouse enamel by SEM
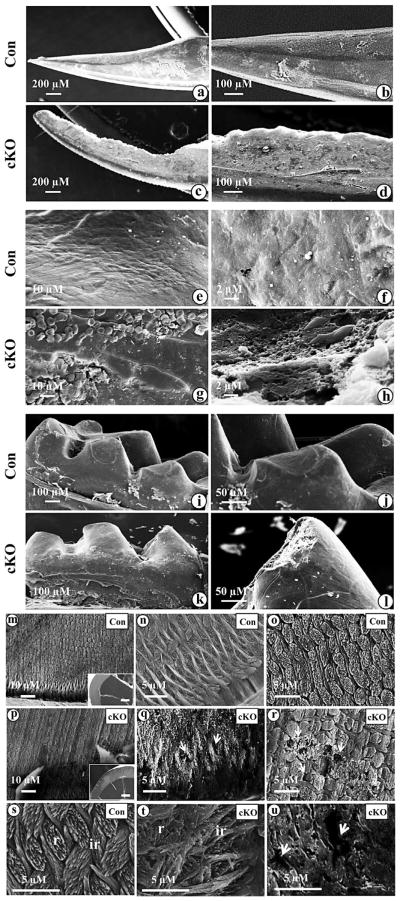
SEM analysis demonstrated that the mandibular incisor surface in 3 month-old Bmp2 null mice was rough with an abnormal angle and the incisor edges were jagged with rounded tips (Fig. 4c,d,g,h). Similar phenotype was seen in the molars of the Bmp2 null mice (Fig. 4k,l). Higher magnification showed that incisor surface appeared obviously uneven in the Bmp2 null mice (Fig. 4p–r,t,u) compared to the control group (Fig. 4m,n,o, s). Furthermore, in the Bmp2 null mice, the prismatic architecture was notably uneven and abraded and lost the typical decussating rod and inter-rod structures (Fig. 4q,r,t,u). Also, the broken surface in enamel contains numerous holes ranging from approximately 1 to 10 μM, whereas in the control mice, the rods and inter-rods of the enamels were well aligned, showing a distinctively prismatic pattern, which is the hallmark of organized mineral (Fig. 4n,o,s). In this study, we only observed transverse sections of incisor enamels between the wild type and Bmp2 cKO mice using SEM. We will study the transverse sections of incisor and molar enamels from these mice as mouse enamel of incisors and molars presents several structural differences (Goldberg et al., 2014).
Fig. 4.
SEM of the mandibular incisors and molars from Bmp2 null mice. a. Enamel layer of the incisor of a 3 month-old control mouse is smooth. b, e and f. Higher magnification of a. c. Enamel layer of the incisor at a 3 month-old Bmp2 cKO mouse is rough. The buccal surface is jagged and tip of the incisor is abraded on the enamel layer. d, g and h. Higher magnification of c. i. Molar of a 3 month-old control mouse. k. Molar cusps in the null mouse are rugged and abraded. j and l. Higher magnification of i and k. m, n, o and s. Enamel in the control mice shows a typical prismatic pattern of rod (r) and inter-rod (ir) structures. Rods and inter-rods are well aligned. q, r, t and u. Enamel in the out layer of the null mice is abraded and has irregular architectures of rods and inter-rods. The rods and inter-rods have disorganized prismatic patterns. The broken surface of the enamel between the rods and inter-rods forms numerous “holes” (arrows) ranging from about 1 to 10 μM in sizes. The transverse section from the control mice in the panel m is shown in inset. The transverse fracture from Bmp2 cKO mice in the panel p is shown in inset. Bar, 100 μM.
Mechanical properties of teeth from wild-type and Bmp2 cKO mice
In order to determine whether morphological change of teeth in the cKO mice causes alteration of mechanical property of the teeth, we tested shear and compressive modulus of the incisors from the Bmp2 cKO mice compared to the control. The results showed that mechanic property of shear and compression of the control groups is 1.45- and 1.88-fold strengths compared to that of the Bmp2 cKO teeth, respectively (Fig. 5). As the Bmp2 cKO teeth are less resistant to shear and compressive forces, the teeth are much more brittle.
Fig. 5.
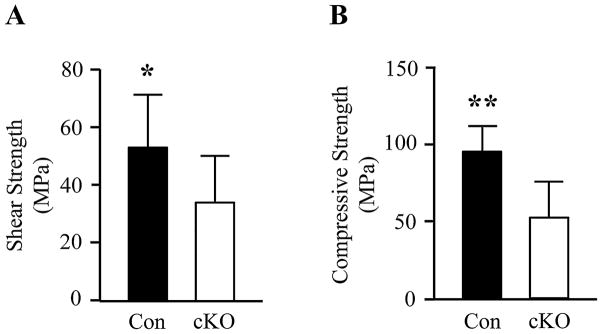
Mechanical properties of control and Bmp2 cKO incisors. A. Tooth shear strength between the control and Bmp2 cKO mice. B. Tooth compressive strength from the control and Bmp2 cKO mice at aged 4 weeks. * and ** indicate statistically significant differences by ANOVA (P <0.05, P <0.01). Con, control; cKO, Bmp2 cKO.
Altered expression of enamel matrix and enamel-processing protease genes in Bmp2 cKO enamel
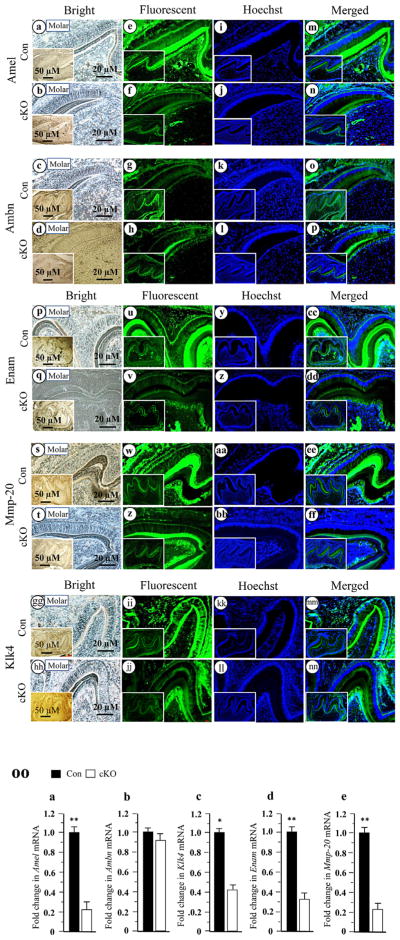
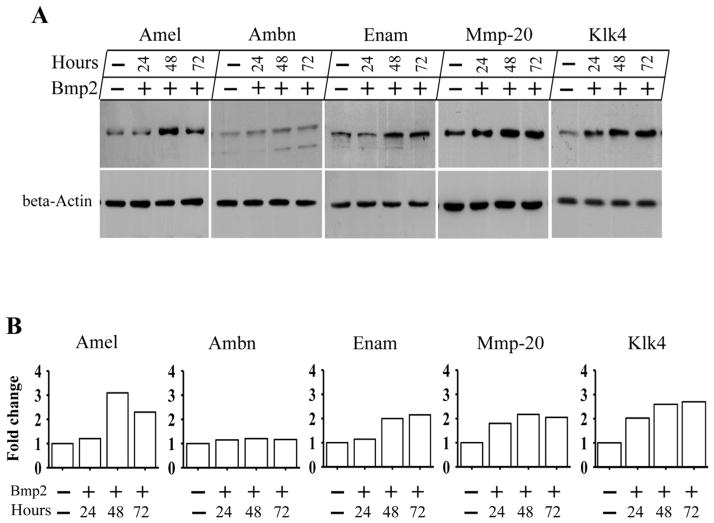
To investigate whether enamel defects are related to altered expression of enamel matrix and enamel-specific protease genes in the cKO enamel, we studied expression of enamel matrix proteins and enamel-processing proteins using fluorescent immunohistochemistry with specific antibodies. At PN6, expression of Amel, Enam, Mmp-20 and Klk4 proteins except for Ambn in the Bmp2 cKO enamel was less intense compared to control mice (Fig. 6). To further determine how deletion of Bmp2 signaling affects the ameloblasts during amelogenesis, we performed qRT-PCR analysis specifically for these genes including Amel, Ambn, Enam, Mmp-20 and Klk4 at PN6 teeth. Similar to translational levels, mRNA expressional levels of Amel, Enam, Mmp-20 and Klk4 were dramatically decreased in the Bmp2 cKO enamel, but Ambn expression in the null enamel was unaffected (Fig. 6OO). Then, we examined whether forced expression of Bmp2 is able to induce those gene expressions in mouse enamel organ epithelial cells. The cells were treated with exogenous Bmp2 for up to 72 hours. This result showed that Bmp2 is able to stimulate expression of Amel, Enam, Mmp-20 and Klk4 proteins compared to the control groups by Western blotting assay (Fig. 7), indicating that dental enamel defects are relevant to expression of enamel matrix and enamel-processing protease genes in the Bmp2 cKO mice during mouse amelogenesis.
Fig. 6.
Altered expression of mouse enamel matrix and enamel-processing protein genes in the Bmp2 cKO teeth. Tissue sections at PN6 were photographed under a light microscope using a digital cooled camera (a–d, q–t, gg–hh). The expression of enamel matrix and enamel processing proteins were analyzed and quantified by fluorescent immunohistochemistry assay with antibodies specific to Amel, Ambn, Enam, Mmp-20 and Klk4 (e–h, u–x, ii–jj). The tissue sections were stained with Hoechst for the nucleus (i–l, y–bb, kk–ll). Images were merged (m–p, cc–ff, mm–nn). Lower magnifications are shown in insets. Protein expression of Amel, Enam, Mmp-20 and Klk4 were reduced in the Bmp2 cKO enamels, whereas Ambn expression was unaffected. OO. Total RNAs were isolated from the wild-type and Bmp2 cKO teeth at PN6. The mRNA levels of Amel, Ambn, Enam, Mmp-20 and Klk4 were measured by qRT-PCR. Cyclophilin A was used as an internal control. Expression of these mRNAs in the teeth of the control mice acts as a 1.0-fold increase. The bar graphs show mean ± S.D. (n = 3) from three independent experiments. qRT-PCR shows a significant decrease of Amel (a), Klk4 (c), Enam (d) and Mmp-20 (e) mRNAs and no difference in Ambn (b) mRNA expression at PN6 in the Bmp2 cKO mice. Asterisks indicate significant differences between the control and Bmp2 cKO groups (*P <0.05, **P <0.01). Con, control; cKO, Bmp2 cKO.
Fig. 7.
Effect of Bmp2 on expression of enamel matrix and enamel processing proteins in mouse enamel organ epithelial cells. A. The cells were treated with or without 300 ng/ml of recombinant Bmp2 for 24-, 48- and 72-h. The cells were lysed and protein expressional levels were detected by Western blot assay using antibodies specific to Amel, Ambn, Enam, Mmp-20 and Klk4, respectively. Beta-actin was used as an internal control. Bmp2 induces protein expression of Amel, Enam, Mmp-20 and Klk4 at given time points in the mouse enamel organ epithelial cells, but there is no effect on Ambn protein expression. B. The enamel protein band intensity was qualified by ImageJ software. The Bmp2 untreated enamel protein was normalized to β actin as control. The fold increase in the Bmp2 treated enamel protein expression level was calculated by dividing the Bmp2 untreated group. This result demonstrates that exogenous Bmp2 was able to induce expression of Amel, Enam, Mmp-20 and Klk4 proteins at given time periods.
Discussion
Bmp2 is a multiple-functional growth factor and involved in many organ development and formation (Zhang and Bradley, 1996; Ma et al., 2005; Singh et al., 2008; Feng et al., 2011; Yang et al., 2012; Yang et al., 2013). In this study, we observed that teeth with Bmp2 cKO mice displayed abnormal phenotypes and enamel layer of incisors and molars was thin as well as the enamel surface was rough with chipping that are consistent with our previous observations (Feng et al., 2011). We further found that enamel volume and mineral density are decreased whereas tooth porosity of the secondary molars and incisors in size is increased in the Bmp2 null mice (Fig. 3). Also, enamel in the Bmp2 cKO mice loses the prismatic patterns with alteration of rod and inter-rod architecture (Fig. 4). Teeth of the Bmp2 null mice are much more brittle (Fig. 5). These Bmp2 cKO mice developed tooth phenotype that resembles AI (Gibson et al., 2001; Caterina et al., 2002; Hu et al., 2008; Simmer et al., 2009). In the present study, we found that the expression of Amel, Enam, Mmp-20, and Klk4 genes in the Bmp2 cKO teeth was reduced, and exogenous Bmp2 was able to induce these gene expressions in mouse enamel organ epithelial cells, indicating the dental enamel defects in the null mice are related to precise expression of the enamel matrix protein and enamel-processing protease genes tested.
Mmp-20 is primarily expressed in ameloblasts during the secretory stage of enamel formation (Bartlett, 2013) and is to initiate hydrolysis of three major enamel matrix proteins, amelogenin, ameloblastin and enamelin (Yamakoshi et al., 2006; Iwata et al., 2007; Chun et al., 2010). MMP-20 mutations in humans and mice exhibit hypoplastic enamel, altered rod-interrod structures and enamel matrix proteins (Caterina et al., 2002; Kim et al., 2005; Ozdemir et al., 2005b). Interestingly, the changes in enamel crystallites found in the Bmp2 null mice are similar to those observed in MMP-20 mutations in humans and mice (Figs. 2–4). Our data showed that expression of Mmp-20 was reduced at the transcriptional and translational levels in the cKO mice, suggesting that there might be associations between Bmp2 signaling pathways and Mmp-20. Lee et al. identified that p.Ala304Thr MMP-20 mutation in human is localized at the hemopexin domain and further found that expression level of the mutant MMP-20 protein was significantly decreased compared to the wild type MMP-20 protein, but the mutant MMP-20 has catalytic activity (Lee et al., 2010). Recently, Shin et al. have generated Mmp-20 transgenic founder mouse lines with low, medium and high Mmp-20 expression levels driven by mouse amelogenin gene promoter (Shin et al., 2014). These Mmp-20 transgenic mice were crossed to either Mmp-20 −/− null or Mmp-20 +/+ normal background mice. They found that each transgene of the low, medium or high levels of Mmp-20 expression rescued the defective enamel back nearly full thickness in the Mmp-20 null background mice. In contrast, the transgenic mice with either the medium or high Mmp-20 expression resulted in dysplastic enamel in the Mmp-20 +/+ background mice. These data demonstrate that Mmp-20 expression levels must be within a specific range for normal enamel development. Herein, the reduction of Mmp-20 expression in the Bmp2 cKO mice might be related to enamel defects during amelogenesis.
On the other hand, we observed that expression of Klk4 in the Bmp2 cKO mice was lower than the control. Like Mmp-20, Klk4 digests Amel, Enam and Ambn into small fragments (Yamakoshi et al., 2006; Nagano et al., 2009). Klk4 mutations cause autosomal recessive hypomaturation AI. Interestingly, the changes in enamel morphology seen in our cKO mice had some similarities to those observed in Klk4 mutations in mice and humans (Hart et al., 2004; Simmer et al., 2009). It showed that teeth in the Bmp2 cKO mice were chalky white in the erupted portion and enamel layer in the mandibular incisors was abraded and the enamel volume was decreased. Contrast of the density was reduced between enamel and dentin (Fig. 2–4).
However, the Klk4 null mice showed several differences from the Bmp2 cKO mice. In the Bmp2 cKO mice, enamel layer appeared thin and displayed loss of typical prismatic patterns, and Amel expression was decreased. In contrast, in the Klk4 null mice, enamel attained normal thickness, the enamel layer retained decussating enamel rods although it was rapidly abraded away following weaning. The Amel expression in the Klk4 null mice was more intense at the enamel surface and in enamel matrix than that of the control mice (Simmer et al., 2009). It is raised questions whether reduced Klk4 expression in the Bmp2 cKO mice is sufficient to interrupt enamel development. What is the threshold of Klk4 expression required for correct enamel formation? It leaves this question unresolved at this time.
In the current study, we also found that expression of Amel in the null mice was reduced, and the gene expression was induced by Bmp2 treatment in mouse enamel organ epithelial cells. In Amel null mice, enamel layer appeared thin, hypoplastic and lacked a discernible enamel rod and inter-rod structures of the mineral (Gibson et al., 2001). The enamel phenotype with amelogenin mutations resembles those found in the Bmp2 cKO mice, suggesting that reduction of Amel gene expression in the Bmp2 null mice is related to enamel defects. Gibson et al. reported that expression of Amel gene in Amel null mice rescues the enamel phenotype (Gibson et al., 2011). This indicates that the amelogenin is required for normal enamel development.
Furthermore, we observed that the Enam expression was decreased in the Bmp2 cKO mice. Enam gene mutations in humans and mice cause autosomal dominant AI and show a dose-dependent effect on mice and human beings (Masuya et al., 2005; Ozdemir et al., 2005a; Hu et al., 2008) A single mutant allele exhibits a mild form of AI while mutations of both alleles have severe enamel malformation with little or no mineral covering dentin, Teeth with the heterozygous Enam mutant mice were chalky white in the erupted portion and enamel appeared rough and abraded in the surface (Hu et al., 2008). Noteworthy, some changes in enamel phenotypes found in the Bmp2 null mice resemble those found in the Enam heterozygous mutant mice. Therefore, enamel defects observed in the Bmp2 null mice are due in part to a reduction of Enam expression.
To correlate Bmp2 signaling and the ameloblast-specific downstream targets, we analyzed mRNA expression of representative enamel matrix protein and enamel-specific protease genes using qRT-PCR. These results revealed that in the absence of Bmp2 signaling, the mRNA expression levels of Amel, Enam, Mmp-20 and Klk4 were dramatically decreased in the cKO mice. The data are consistent with immunohistochemistry assay. It is proposed that loss of Bmp2 signal causes abnormal enamel development duo to down-regulation of expression of Amel, Enam, Mmp-20 and Klk4 genes.
Although many Bmp molecules besides Bmp2 including Bmp4 and Bmp7 are expressed during the amelogenesis (Aberg et al., 1997; Miyoshi et al., 2008), our data suggest that these Bmp molecules cannot compensate for loss of Bmp2 in enamel formation although we did not evaluate expression of Bmp4 and Bmp7 genes in this model. It needs to be further investigated in the future. It has been reported that a number of factors regulates enamel-related gene expression in ameloblasts, including β-catenin, C/EBP, DLX3, LEF-1, MSX2, Notch and Smad1/5/8 (Price et al., 1999; Zhou et al., 2000; Mitsiadis et al., 2010; Molla et al., 2010; Tian et al., 2010). Amelogenin gene transcription is up-regulated by C/EBP and MSX2 (Zhou et al., 2000). In addition, DLX3 is homeodomain transcription factor necessary for tooth development (Dong et al., 2005; Duverger et al., 2008; Price et al., 1999). However, molecular mechanisms of Bmp2 signaling transduction pathways in enamel development have not been completely understood in particular Enam, Mmp-20 and Klk4 in vivo. This study demonstrates that Bmp2 has diverse biological functions in controlling enamel gene expression, matrix formation and mineralization. Understanding of the Bmp2 signaling pathways will lead to a greater understanding of how Bmp2 controls expression of individual enamel matrix protein and enamel-specific protease genes during amelogenesis.
In summary, the present study showed that the Bmp2 deletion causes enamel defects via controlling expression of enamel matrix protein and enamel-specific protease genes. We demonstrate that proper Bmp2 signaling during mouse amelogenesis plays an important role in intricately orchestrated synthesis and processing of enamel matrix proteins and enamel-processing proteases for correct enamel development, formation and mineralization.
Acknowledgments
Contract grant sponsor: National Institute of Health (NIH); Contract grant number: DE019892.
Contract grant sponsor: San Antonio Area Foundation.
We thank Ms. Sonya Marie Lopez and Melissa Garcia for critical reading of the manuscript and Dr. Roberto Fajardo and Mr. James Schmitz for help with micro-CT analysis. This work was supported by NIH grant DE019892 and San Antonio Area Foundation to SC.
Literature Cited
- Aberg T, Wozney J, Thesleff I. Expression patterns of bone morphogenetic proteins (Bmps) in the developing mouse tooth suggest roles in morphogenesis and cell differentiation. Dev Dyn. 1997;210:383–396. doi: 10.1002/(SICI)1097-0177(199712)210:4<383::AID-AJA3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Bartlett JD. Dental Enamel Development: Proteinases and Their Enamel Matrix Substrates. ISRN Dent. 2013;2013:684607. doi: 10.1155/2013/684607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caterina JJ, Skobe Z, Shi J, Ding Y, Simmer JP, Birkedal-Hansen H, Bartlett JD. Enamelysin (matrix metalloproteinase 20)-deficient mice display an amelogenesis imperfecta phenotype. The J Biol Chem. 2002;277:49598–49604. doi: 10.1074/jbc.M209100200. [DOI] [PubMed] [Google Scholar]
- Cerny R, Slaby I, Hammarstrom L, Wurtz T. A novel gene expressed in rat ameloblasts codes for proteins with cell binding domains. J Bone Miner Res. 1996;11:883–891. doi: 10.1002/jbmr.5650110703. [DOI] [PubMed] [Google Scholar]
- Chan HC, Estrella NM, Milkovich RN, Kim JW, Simmer JP, Hu JC. Target gene analyses of 39 amelogenesis imperfecta kindreds. Eur J Oral Sci. 2011;119:311–323. doi: 10.1111/j.1600-0722.2011.00857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors. 2004;22:233–241. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- Chen S, Gluhak-Heinrich J, Martinez M, Li T, Wu Y, Chuang HH, Chen L, Dong J, Gay I, MacDougall M. Bone morphogenetic protein 2 mediates dentin sialophosphoprotein expression and odontoblast differentiation via NF-Y signaling. J Biol Chem. 2008;283:19359–19370. doi: 10.1074/jbc.M709492200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Gluhak-Heinrich J, Wang YH, Wu YM, Chuang HH, Chen L, Yuan GH, Dong J, Gay I, MacDougall M. Runx2, osx, and dspp in tooth development. J Dental Res. 2009;88:904–909. doi: 10.1177/0022034509342873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho A, Haruyama N, Hall B, Danton MJ, Zhang L, Arany P, Mooney DJ, Harichane Y, Goldberg M, Gibson CW, Kulkarni AB. TGF-ss regulates enamel mineralization and maturation through KLK4 expression. PLoS ONE. 2013;8:e82267. doi: 10.1371/journal.pone.0082267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun YH, Yamakoshi Y, Yamakoshi F, Fukae M, Hu JC, Bartlett JD, Simmer JP. Cleavage site specificity of MMP-20 for secretory-stage ameloblastin. J Dent Res. 2010;89:785–790. doi: 10.1177/0022034510366903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Amor D, Aldred MJ, Gu T, Escamilla M, MacDougall M. DLX3 mutation associated with autosomal dominant amelogenesis imperfecta with taurodontism. Am J Med Genet A. 2005;133A:138–141. doi: 10.1002/ajmg.a.30521. [DOI] [PubMed] [Google Scholar]
- Ducy P, Karsenty G. The family of bone morphogenetic proteins. Kidney Int. 2000;57:2207–2214. doi: 10.1046/j.1523-1755.2000.00081.x. [DOI] [PubMed] [Google Scholar]
- Duverger O, Lee D, Hassan MQ, Chen SX, Jaisser F, Lian JB, Morasso MI. Molecular consequences of a frameshifted DLX3 mutant leading to Tricho-Dento-Osseous syndrome. J Biol Chem. 2008;283:20198–20208. doi: 10.1074/jbc.M709562200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, McDaniel JS, Chuang HH, Huang O, Rakian A, Xu X, Steffensen B, Donly KJ, MacDougall M, Chen S. Binding of amelogenin to MMP-9 and their co-expression in developing mouse teeth. J Mol Histol. 2012;43:473–485. doi: 10.1007/s10735-012-9423-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Yang G, Yuan G, Gluhak-Heinrich J, Yang W, Wang L, Chen Z, McDaniel Schulze, Donly J, Harris KJ, Macdougall SE, Chen M. Abnormalities in the enamel in bmp2-deficient mice. Cells Tissues Organs. 2011;194:216–221. doi: 10.1159/000324644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukae M, Tanabe T, Murakami C, Dohi N, Uchida T, Shimizu M. Primary structure of the porcine 89-kDa enamelin. Adv Dent Res. 1996;10:111–118. doi: 10.1177/08959374960100020201. [DOI] [PubMed] [Google Scholar]
- Fukumoto S, Kiba T, Hall B, Iehara N, Nakamura T, Longenecker G, Krebsbach PH, Nanci A, Kulkarni AB, Yamada Y. Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J Cell Biol. 2004;167:973–983. doi: 10.1083/jcb.200409077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson CW, Li Y, Suggs C, Kuehl MA, Pugach MK, Kulkarni AB, Wright JT. Rescue of the murine amelogenin null phenotype with two amelogenin transgenes. Eur J Oral Sci. 2011;119:70–74. doi: 10.1111/j.1600-0722.2011.00882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson CW, Yuan ZA, Hall B, Longenecker G, Chen E, Thyagarajan T, Sreenath T, Wright JT, Decker S, Piddington R, Harrison G, Kulkarni AB. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J Biol Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- Goldberg M, Kellermann O, Dimitrova-Nakov S, Harichane Y, Baudry A. Comparative studies between mice molars and incisors are required to draw an overview of enamel structural complexity. Front Physiol. 2014;5:359. doi: 10.3389/fphys.2014.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart PS, Hart TC, Michalec MD, Ryu OH, Simmons D, Hong S, Wright JT. Mutation in kallikrein 4 causes autosomal recessive hypomaturation amelogenesis imperfecta. J Med Genet. 2004;41:545–549. doi: 10.1136/jmg.2003.017657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan BL. Bone morphogenetic proteins: Multifunctional regulators of vertebrate development. Genes dev. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- Hu CC, Fukae M, Uchida T, Qian Q, Zhang CH, Ryu OH, Tanabe T, Yamakoshi Y, Murakami C, Dohi N, Shimizu M, Simmer JP. Cloning and characterization of porcine enamelin mRNAs. J Dent Res. 1997;76:1720–1729. doi: 10.1177/00220345970760110201. [DOI] [PubMed] [Google Scholar]
- Hu JC, Hu Y, Smith CE, McKee MD, Wright JT, Yamakoshi Y, Papagerakis P, Hunter GK, Feng JQ, Yamakoshi F, Simmer JP. Enamel defects and ameloblast-specific expression in Enam knock-out/lacz knock-in mice. J Biol Chem. 2008;283:10858–10871. doi: 10.1074/jbc.M710565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata T, Yamakoshi Y, Hu JC, Ishikawa I, Bartlett JD, Krebsbach PH, Simmer JP. Processing of ameloblastin by MMP-20. J Dent Res. 2007;86:153–157. doi: 10.1177/154405910708600209. [DOI] [PubMed] [Google Scholar]
- Jernvall J, Aberg T, Kettunen P, Keranen S, Thesleff I. The life history of an embryonic signaling center: BMP-4 induces p21 and is associated with apoptosis in the mouse tooth enamel knot. Development. 1998;125:161–169. doi: 10.1242/dev.125.2.161. [DOI] [PubMed] [Google Scholar]
- Kim JW, Simmer JP, Hart TC, Hart PS, Ramaswami MD, Bartlett JD, Hu JC. MMP-20 mutation in autosomal recessive pigmented hypomaturation amelogenesis imperfecta. J Med Genet. 2005;42:271–275. doi: 10.1136/jmg.2004.024505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs M, Danyi R, Erdelyi M, Fejerdy P, Dobo-Nagy C. Distortional effect of beam-hardening artefacts on microCT: A simulation study based on an in vitro caries model. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2009;108:591–599. doi: 10.1016/j.tripleo.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Krebsbach PH, Nakata K, Bernier SM, Hatano O, Miyashita T, Rhodes CS, Yamada Y. Identification of a minimum enhancer sequence for the type II collagen gene reveals several core sequence motifs in common with the link protein gene. J Biol Chem. 1996;271:4298–4303. doi: 10.1074/jbc.271.8.4298. [DOI] [PubMed] [Google Scholar]
- Lagerstrom M, Dahl N, Nakahori Y, Nakagome Y, Backman B, Landegren U, Pettersson U. A deletion in the amelogenin gene (AMG) causes X-linked amelogenesis imperfecta (AIH1) Genomics. 1991;10:971–975. doi: 10.1016/0888-7543(91)90187-j. [DOI] [PubMed] [Google Scholar]
- Lee KY, Jeong JW, Wang J, Ma L, Martin JF, Tsai SY, Lydon JP, DeMayo FJ. Bmp2 is critical for the murine uterine decidual response. Mol Cell Biol. 2007;27:5468–5478. doi: 10.1128/MCB.00342-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Seymen F, Kang HY, Lee KE, Gencay K, Tuna B, Kim JW. MMP20 hemopexin domain mutation in amelogenesis imperfecta. J Dent Res. 2010;89:46–50. doi: 10.1177/0022034509352844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Machule D, Gao C, DenBesten PK. Activation of recombinant bovine matrix metalloproteinase-20 and its hydrolysis of two amelogenin oligopeptides. Eur J Oral Sci. 1999;107:352–359. doi: 10.1046/j.0909-8836.1999.eos107506.x. [DOI] [PubMed] [Google Scholar]
- Linde A, Goldberg M. Dentinogenesis. Crit Rev Oral Biol Med. 1993;4:679–728. doi: 10.1177/10454411930040050301. [DOI] [PubMed] [Google Scholar]
- Ma L, Lu MF, Schwartz RJ, Martin JF. Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development. 2005;132:5601–5611. doi: 10.1242/dev.02156. [DOI] [PubMed] [Google Scholar]
- Maes C, Kobayashi T, Selig MK, Torrekens S, Roth SI, Mackem S, Carmeliet G, Kronenberg HM. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev Cell. 2010;19:329–344. doi: 10.1016/j.devcel.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardh CK, Backman B, Holmgren G, Hu JC, Simmer JP, Forsman-Semb K. A nonsense mutation in the enamelin gene causes local hypoplastic autosomal dominant amelogenesis imperfecta (AIH2) Hum Mol Genet. 2002;11:1069–1074. doi: 10.1093/hmg/11.9.1069. [DOI] [PubMed] [Google Scholar]
- Masuya H, Shimizu K, Sezutsu H, Sakuraba Y, Nagano J, Shimizu A, Fujimoto N, Kawai A, Miura I, Kaneda H, Kobayashi K, Ishijima J, Maeda T, Gondo Y, Noda T, Wakana S, Shiroishi T. Enamelin (Enam) is essential for amelogenesis: ENU-induced mouse mutants as models for different clinical subtypes of human amelogenesis imperfecta (AI) Hum Mol Genet. 2005;14:575–583. doi: 10.1093/hmg/ddi054. [DOI] [PubMed] [Google Scholar]
- Mitsiadis TA, Graf D, Luder H, Gridley T, Bluteau G. BMPs and FGFs target Notch signalling via jagged 2 to regulate tooth morphogenesis and cytodifferentiation. Development. 2010;137:3025–3035. doi: 10.1242/dev.049528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Nagata H, Horiguchi T, Abe K, Wahyudi Arie, Baba I, Harada Y, Noma H. BMP2-induced gene profiling in dental epithelial cell line. J Med Invest. 2008;55:216–226. doi: 10.2152/jmi.55.216. [DOI] [PubMed] [Google Scholar]
- Molla M, Descroix V, Aioub M, Simon S, Castaneda B, Hotton D, Bolanos A, Simon Y, Lezot F, Goubin G, Berdal A. Enamel protein regulation and dental and periodontal physiopathology in MSX2 mutant mice. Am J Pathol. 2010;177:2516–2526. doi: 10.2353/ajpath.2010.091224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Kakegawa A, Yamakoshi Y, Tsuchiya S, Hu JC, Gomi K, Arai T, Bartlett JD, Simmer JP. Mmp-20 and Klk4 cleavage site preferences for amelogenin sequences. J Dent Res. 2009;88:823–828. doi: 10.1177/0022034509342694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdemir D, Hart PS, Firatli E, Aren G, Ryu OH, Hart TC. Phenotype of ENAM mutations is dosage-dependent. J Dent Res. 2005a;84:1036–1041. doi: 10.1177/154405910508401113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdemir D, Hart PS, Ryu OH, Choi SJ, Ozdemir-Karatas M, Firatli E, Piesco N, Hart TC. MMP20 active-site mutation in hypomaturation amelogenesis imperfecta. J Dent Res. 2005b;84:1031–1035. doi: 10.1177/154405910508401112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulter JA, Murillo G, Brookes SJ, Smith CE, Parry DA, Silva S, Kirkham J, Inglehearn CF, Mighell AJ. Peletion of ameloblastin exon 6 is associated with amelogenesis imperfecta. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JA, Wright JT, Walker SJ, Crawford PJ, Aldred MJ, Hart TC. Tricho-dento-osseous syndrome and amelogenesis imperfecta with taurodontism are genetically distinct conditions. Clin Genet. 1999;56:35–40. doi: 10.1034/j.1399-0004.1999.550105.x. [DOI] [PubMed] [Google Scholar]
- Rajpar MH, Harley K, Laing C, Davies RM, Dixon MJ. Mutation of the gene encoding the enamel-specific protein, enamelin, causes autosomal-dominant amelogenesis imperfecta. Hum Mol Genet. 2001;10:1673–1677. doi: 10.1093/hmg/10.16.1673. [DOI] [PubMed] [Google Scholar]
- Ravassipour DB, Hart PS, Hart TC, Ritter AV, Yamauchi M, Gibson C, Wright JT. Unique enamel phenotype associated with amelogenin gene (AMELX) codon 41 point mutation. J Dent Res. 2000;79:1476–1481. doi: 10.1177/00220345000790070801. [DOI] [PubMed] [Google Scholar]
- Rosen V. BMP2 signaling in bone development and repair. Cytokine Growth Factor Rev. 2009;20:475–480. doi: 10.1016/j.cytogfr.2009.10.018. [DOI] [PubMed] [Google Scholar]
- Salido EC, Yen PH, Koprivnikar K, Yu LC, Shapiro LJ. The human enamel protein gene amelogenin is expressed from both the X and the Y chromosomes. Am J Hum Genet. 1992;50:303–316. [PMC free article] [PubMed] [Google Scholar]
- Shin M, Hu Y, Tye CE, Guan X, Deagle CC, Antone JV, Smith CE, Simmer JP, Bartlett JD. Matrix metalloproteinase-20 over-expression is detrimental to enamel development: A Mus musculus model. PLoS ONE. 2014;9:e86774. doi: 10.1371/journal.pone.0086774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer JP, Hu Y, Lertlam R, Yamakoshi Y, Hu JC. Hypomaturation enamel defects in Klk4 knockout/LacZ knockin mice. J Biol Chem. 2009;284:19110–19121. doi: 10.1074/jbc.M109.013623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AP, Castranio T, Scott G, Guo D, Harris MA, Ray M, Harris SE, Mishina Y. Influences of reduced expression of maternal bone morphogenetic protein 2 on mouse embryonic development. Sex Dev. 2008;2:134–141. doi: 10.1159/000143431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE. Cellular and chemical events during enamel maturation. Crit Rev Oral Biol Med. 1998;9:128–161. doi: 10.1177/10454411980090020101. [DOI] [PubMed] [Google Scholar]
- Sonoda A, Iwamoto T, Nakamura T, Fukumoto E, Yoshizaki K, Yamada A, Arakaki M, Harada H, Nonaka K, Nakamura S, Yamada Y, Fukumoto S. Critical role of heparin binding domains of ameloblastin for dental epithelium cell adhesion and ameloblastoma proliferation. J Biol Chem. 2009;284:27176–27184. doi: 10.1074/jbc.M109.033464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Termine JD, Belcourt AB, Miyamoto MS, Conn KM. Properties of dissociatively extracted fetal tooth matrix proteins. II. Separation and purification of fetal bovine dentin phosphoprotein. The J Biol Chem. 1980;255:9769–9772. [PubMed] [Google Scholar]
- Thesleff I. Epithelial-mesenchymal signalling regulating tooth morphogenesis. J Cell Sci. 2003;116:1647–1648. doi: 10.1242/jcs.00410. [DOI] [PubMed] [Google Scholar]
- Tian H, Lv P, Ma K, Zhou C, Gao X. Beta-catenin/LEF1 activated enamelin expression in ameloblast-like cells. Biochem Biophys Res Commun. 2010;398:519–524. doi: 10.1016/j.bbrc.2010.06.111. [DOI] [PubMed] [Google Scholar]
- Wang F, Wu LA, Li W, Yang Y, Guo F, Gao Q, Chuang HH, Shoff L, Wang W, Chen S. Immortalized mouse dental papilla mesenchymal cells preserve odontoblastic phenotype and respond to bone morphogenetic protein 2. In vitro Cell Dev Biol Anim. 2013;49:626–637. doi: 10.1007/s11626-013-9641-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakoshi Y, Hu JC, Fukae M, Yamakoshi F, Simmer JP. How do enamelysin and kallikrein 4 process the 32-kDa enamelin. Eur J Oral Sci. 2006;114:379–380. doi: 10.1111/j.1600-0722.2006.00281.x. [DOI] [PubMed] [Google Scholar]
- Yang W, Harris MA, Cui Y, Mishina Y, Harris SE, Gluhak-Heinrich J. Bmp2 is required for odontoblast differentiation and pulp vasculogenesis. J Dent Res. 2012;91:58–64. doi: 10.1177/0022034511424409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Guo D, Harris MA, Cui Y, Gluhak-Heinrich J, Wu J, Chen XD, Skinner C, Nyman JS, Edwards JR, Mundy GR, Lichtler A, Kream BE, Rowe DW, Kalajzic I, David V, Quarles DL, Villareal D, Scott G, Ray M, Liu S, Martin JF, Mishina Y, Harris SE. Bmp2 in osteoblasts of periosteum and trabecular bone links bone formation to vascularization and mesenchymal stem cells. J Cell Sci. 2013;126:4085–4098. doi: 10.1242/jcs.118596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Bradley A. Mice deficient for BMP2 are nonviable and have defects in amnion/ chorion and cardiac development. Development. 1996;122:2977–2986. doi: 10.1242/dev.122.10.2977. [DOI] [PubMed] [Google Scholar]
- Zhou YL, Lei Y, Snead ML. Functional antagonism between Msx2 and CCAAT/ enhancer-binding protein alpha in regulating the mouse amelogenin gene expression is mediated by protein-protein interaction. J Biol Chem. 2000;275:29066–29075. doi: 10.1074/jbc.M002031200. [DOI] [PubMed] [Google Scholar]
- Zou W, Hunter N, Swain MV. Application of polychromatic microCT for mineral density determination. J Dent Res. 2011;90:18–30. doi: 10.1177/0022034510378429. [DOI] [PMC free article] [PubMed] [Google Scholar]