Abstract
All DNA replicases achieve high fidelity by a conserved mechanism, but each translesion polymerase carries out mutagenic DNA synthesis in its own way. Here we report crystal structures of human DNA polymerase ν (Pol ν), which is homologous to high-fidelity replicases and yet error-prone. Instead of a simple open-to-closed movement of the O helix upon binding of a correct incoming nucleotide, Pol ν has a different open state and requires the finger domain to swing sideways and undergo both opening and closing motions to accommodate the nascent base pair. A single amino acid substitution in the O-helix of the finger domain improves the fidelity of Pol ν nearly ten-fold. A unique cavity and the flexibility of the thumb domain allow Pol ν to generate and accommodate a looped-out primer strand. Primer loopout may be a mechanism for DNA trinucloetide-repeat expansion.
Introduction
Among 17 DNA polymerases identified in humans, Pol ν, Pol θ and Pol ζ stand out by sharing sequence homology with high-fidelity replicases but conducting mutagenic and translesion DNA synthesis (TLS) 1–5. The A-family Pol ν and Pol θ increase immunoglobulin gene diversity in chicken DT40 cells 6, and Pol θ plays a role in maintaining genome stability 7,8. The B-family Pol ζ is essential for normal cell proliferation 9–11. Based on the sequence conservation, Pol ν, Pol θ and Pol ζ are predicted to contain a three-domain catalytic core with the palm domain bearing the catalytic carboxylates, the thumb domain binding DNA duplex, and the finger domain interacting with the nascent base pair between a template and incoming nucleotide. However, the mechanism by which these polymerases carry out mutagenic DNA synthesis is unclear.
Two characteristics of replicases ensure high-fidelity DNA synthesis: proofreading by the 3′–5′ exonuclease 12–15 and discrimination against incorrect incoming nucleotides (dNTP) by the “open–and–closed” conformational change of the finger domain 16–18. Y-family DNA polymerases, specialized for translesion and error-prone DNA synthesis, lack these two features 19. Pol ν, Pol θ and Pol ζ are incapable of proofreading owing to mutations in the active site of the 3′–5′ exonuclease 1,20,21. Because of the absence of atomic resolution structures, how these specialized polymerases interact with DNA and the nascent base pair and the nature of conformational changes during a catalytic cycle are unknown.
The 900-residue human Pol ν 22 is the smallest among the three enigmatic TLS polymerases. The catalytic cores of Pol ν and Pol θ are homologous to mitochondrial Pol γ and E. coli Pol I 20,22,23, but each contains three insertions (Fig. 1a) 5,24,25. Pol ν has a unique error signature and frequently mis-incorporates dT opposite a template dG 1,2. To understand Pol ν and to shed light on the mutagenic properties of Pol θ and Pol ζ, we have determined the crystal structures of human Pol ν complexed with DNA and discovered unexpected conformational changes involving the finger and thumb domains.
Figure 1.
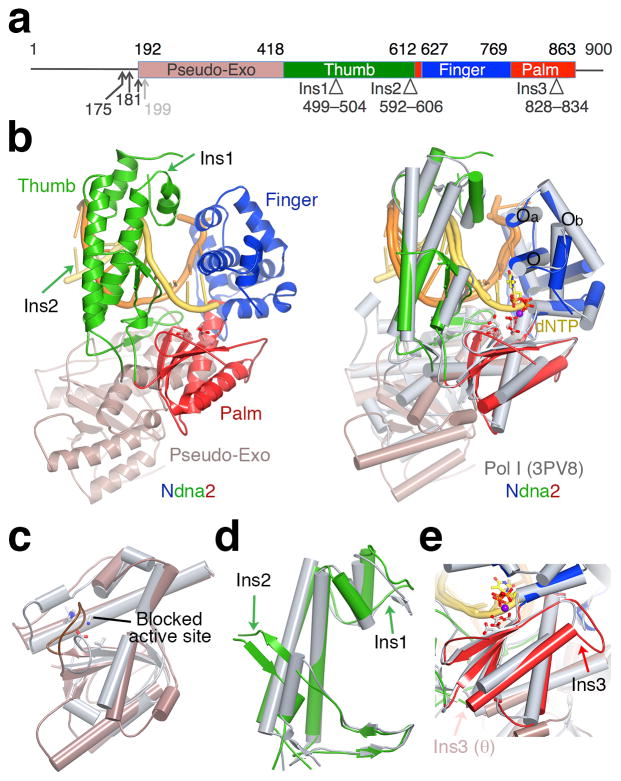
Structure of Pol ν. (a) The primary structure. The N-termini of four Pol ν variants are indicated by arrowheads. Structural domains and three insertions are marked with boundary residues. (b) Ndna2 structure (left) and its superposition with bacillus Pol I 33 (grey, right). DNA is colored yellow (primer) and orange (template). The catalytic residues and incoming nucleotide in Pol I are shown as ball–and–sticks. (c) A zoom-in view of the superimposed Exo domain. Pol ν Exo site is mutated (equivalent residues in bacillus Pol I are shown in sticks) and blocked by a long loop. (d) The Thumb domain. (e) The Palm domain. Ins1–3 are marked by arrowheads (d,e).
Results
A minimal active human Pol ν and four crystal structures
Human Pol ν is largely insoluble when expressed in E. coli 26,27. To improve its solubility, we expressed human Pol ν tagged with an N-terminal tandem repeat of maltose-binding protein (MBP) in HEK293 cells (Methods). We also serially deleted the N- and C-terminal unstructured regions to create Pol ν77 (E175–G863, 77 kDa) 27, Pol ν76, (A181–G863), Pol ν75 (A192–G863), and Pol ν74 (D199–G863) (Supplementary Fig. 1). All four Pol ν variants were soluble and readily purified. Except for Pol ν74, the N-terminal MBP tag was easily removed by PreScission cleavage. Tag-free Pol ν75, ν76 and ν77 were equally active in DNA binding and synthesis. Pol ν75 was purified to homogeneity and used for most of the following structural and biochemical studies.
DNA synthesis assays using Pol ν expressed in mammalian cells confirmed that it is an error prone DNA polymerase and preferentially misincorporates dT regardless of template sequence (Supplementary Fig. 2a,b). Using previously reported mutation hotspot and coldspot DNA substrates 26, the measured kcat and KM of Pol ν75 for incorporating correct (dC) and incorrect (dT) nucleotides (Supplementary Table 1) agreed well with the kinetic parameters obtained from E. coli expressed and refolded Pol ν77 26. In contrast to the majority of DNA polymerases, which carry out DNA synthesis with a similar efficiency and accuracy regardless of DNA sequence, Pol ν had up to a 20-fold difference in catalytic efficiency and fidelity depending on DNA sequence (Supplementary Table 1).
To understand Pol ν’s unusual error-prone DNA synthesis, we determined the crystal structure of a Pol ν75–DNA complex, Ndna1, at 3.1Å resolution (Methods) with a 14nt template, 11nt primer DNA and a non-reactive nucleotide analog dAMPNPP (Supplementary Fig. 3a,b). Due to crystal lattice interactions, the blunt end of each DNA was in the active site instead of the 3-nt 5′ overhang, and the incoming nucleotide was absent. After modifying the DNA substrate and changing the duplex length (Supplementary Fig. 3c,d), we obtained three additional crystals, Ndna2, Ndna3, and Ndna4 with Ndna2 and Ndna3 in the same space group as Ndna1. These three structures were solved by molecular replacement and refined at the highest resolution of 2.95Å (Ndna2) and lowest resolution of 3.3Å (Ndna3) (Table 1). Perhaps due to the high salt and low pH of the crystallization buffer (1.2M (NH4)2SO4 and pH 5.9–6.3), all four structures were protein–DNA binary complexes without dAMPNPP. Interestingly, in the first three structures (Ndna1–3), the finger domain changed from the open, ajar to a closed conformation correlating with the increases of the DNA length (Supplementary Fig. 3). In Ndna4 the finger domain was closed, and the thumb domain underwent a large rotation.
Table 1.
Data collection, SAD phasing and refinement statistics
| Ndna1 | Ndna2 | Ndna3 | Ndna4 | Ndna1 Br (SAD) | |
|---|---|---|---|---|---|
| Space group | H32 | H32 | H32 | I222 | H32 |
| Cell dimensions | |||||
| a, b, c (Å) | 292.7, 292.7, 110.7 | 290.9, 290.9, 110.2 | 292.1, 292.1, 108.1 | 98.0, 110.9, 277.4 | 294.1, 294.1, 110.8 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 120.0 |
| Wavelength (Å) | 0.9099 | 1.0000 | 1.0000 | 1.0000 | 0.9099 (Br peak) |
| Resolution (Å) a | 30.0 - 3.10 (3.15 - 3.10) | 30.0 - 2.95 (3.00 - 2.95) | 30.0 - 3.30 (3.36 - 3.30) | 30.0 - 3.20 (3.26 - 3.20) | 30.0 - 3.20 (3.26 - 3.20) |
| Rmerge (%) a | 11.9 (61.2) | 7.9 (63.7) | 13.2 (69.6) | 9.8 (61.9) | 9.3 (70.4) |
| I/σI a | 11.9 (2.1) | 16.7 (2.2) | 13.3 (2.5) | 13.0 (2.0) | 16.3 (2.8) |
| Completeness (%) a | 99.5 (98.2) | 99.0 (100.0) | 99.4 (98.6) | 98.9 (98.6) | 100.0 (100.0) |
| Redundancy a | 4.4 (3.6) | 4.7 (4.3) | 5.7 (5.3) | 5.2 (5.1) | 7.1 (7.1) |
| Refinement | |||||
| Resolution (Å) | 30.0 - 3.10 | 30.0 - 2.95 | 30.0 - 3.30 | 30.0 - 3.20 | |
| No. reflections | 32,894 | 37,333 | 26,294 | 25,063 | |
| Rwork/Rfree (%) | 21.3/23.7 | 22.2/24.1 | 20.3/22.3 | 24.2/28.1 | |
| No. atoms b | |||||
| Protein | 5,112 | 4,971 | 5,025 | 4,952 | |
| DNA | 505 | 550 | 489 | 449 | |
| ligand | 0 | 12 (MES) | 12 (MES) | 0 | |
| B factors | |||||
| Protein | 87.2 | 74.5 | 60.7 | 79.8 | |
| DNA | 85.9 | 82.7 | 60.2 | 88.9 | |
| ligand | 69.5 | 58.7 | |||
| r.m.s. deviations | |||||
| Bond lengths (Å) | 0.008 | 0.007 | 0.008 | 0.013 | |
| Bond angles (°) | 1.112 | 1.061 | 1.076 | 1.683 | |
Values in parenthesis are for highest-resolution shell. A single crystal was used for each data collection and structure refinement.
There is no water included in the structure refinement.
An unusual open state of Pol ν
The Ndna2 and Ndna4 structures resemble the finger-closed form of A-family polymerases (Fig. 1, 2a), which usually represents the reaction-ready state of a polymerase–DNA–dNTP ternary complex 16–18,28,29 and occasionally a post-reaction product state when DNA is not yet translocated for the next round of dNTP incorporation 30,31. The catalytic core of Pol ν75 superimposed well with that of bacillus and Taq DNA polymerase I (abbreviated as Pol I) in the ternary complexes 32,33, with 1.34 and 1.48 Å rmsd (root-mean-square deviation) over 380 pairs of Cα atoms, respectively. Residues 192–416 of Pol ν formed a degenerate 3′–5′ exonuclease domain, which deviates from the equivalent domain in E. coli and bacillus Pol I (2.6 Å over 145 pairs of Cα atoms) (Fig. 1b,c). The four catalytic essential carboxylates are replaced by T224, M226, F324 and L401 in the pseudo-Exo domain of Pol ν, and the mutated active site is further blocked by an elongated loop (229–241 aa) (Fig. 1c). The first two insertions (Ins1 and Ins2) in Pol ν and Pol θ are located at the tip and the base of the thumb domain (Fig. 1a,d) and likely influence DNA binding (see details below). The third insertion (Ins3) in Pol ν is on the backside of the palm domain distal from the DNA binding surface (Fig. 1e). Although Ins3 in Pol θ is further towards the C-terminus, modeling shows it is also on the backside of the palm domain and extends toward the pseudo-Exo domain (Fig. 1e) 24,25.
Figure 2.
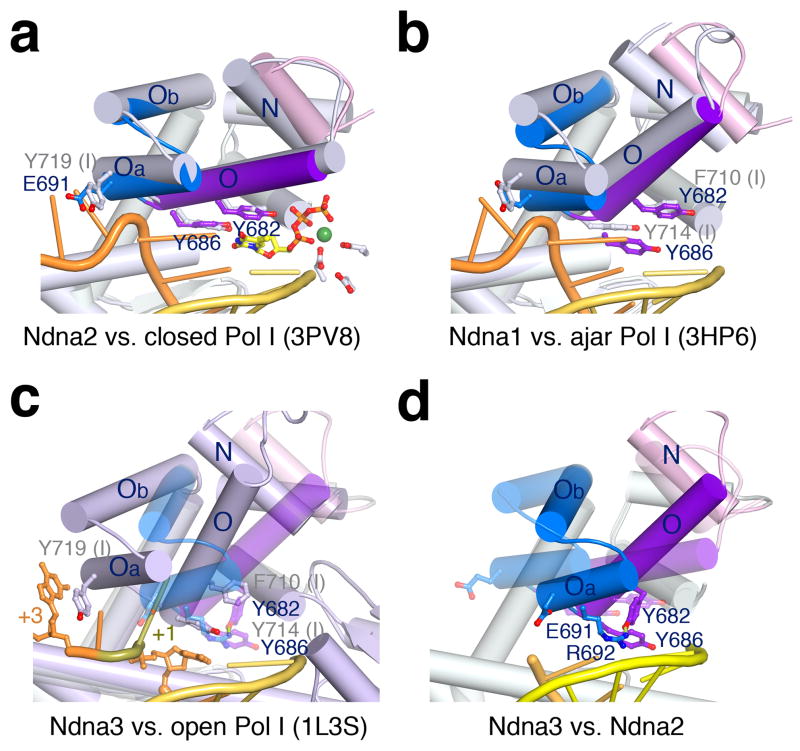
The unusual finger-open structure of Pol ν75. (a) Superposition of Ndna2 (protein only with helices N, O, Oa and Ob in different colors) and the finger-closed Pol I (3PV833, grey protein, yellow primer and orange template). Key residues are diagramed. (b) Superposition of Pol ν of Ndna1 and the finger-ajar Pol I with DNA (3HP635). (c) Superposition of Ndna3 (semi-transparent Pol ν) with the finger-open Pol I with DNA (1L3S34, light blue). (c) Superposition of Ndna3 (protein and DNA) and Ndna2 (semi-transparent). Rotations of helices in Ndna3 relative to Pol I and Ndna2 are indicated by red and yellow arrows.
In the absence of an incoming nucleotide, DNA Pol I of E. coli, bacillus and Taq assume an open state with the O helix in the finger domain rotating ~40° away from the catalytic center 16,34 (Video 1). The Ndna1 structure of Pol ν revealed a semi-open finger domain that resembled the ajar state of bacillus DNA Pol I (PDB: 3HP6), where the O helix is halfway between the open and closed state owing to a mismatched incoming nucleotide 35. The catalytic core of these two structures superimposed reasonably well except for the absence of dNTP in Ndna1 (Fig. 2b).
The O helix of Ndna3 was the most open among the four Pol ν75 structures, but Ndna3 was different from the open state of Pol I (Fig. 2c,d). The O helix in Ndna3 was rotated 25°, instead of 40°, away from the DNA substrate relative to that in Ndna2. Among the previously characterized A-family polymerases, helices Oa and Ob (following the O helix in the finger domain) move slightly outwards in the opposite direction of the O helix from the closed to the open state to accommodate the downstream (+1) template nucleotide (Fig. 2c). However, in Ndna3, Oa and Ob rotated 17.5° toward the DNA duplex in the same direction as the O helix and occluded both +1 and templating nucleotide binding site (Fig. 2c). Instead of the opening of O helix, a large portion of the finger domain (helices Oa, Ob, and O) swings over the DNA duplex (Video 2). In this unusual open state of Pol ν, binding of a templating base is blocked. Although binding of an incoming nucleotide is possible, without a template base dNTP binding has to be unstable and nonspecific. Pol ν needs to open the templating base binding site for a nascent base pair to form and close over the incoming dNTP for DNA synthesis to take place. The unusual open state of Pol ν may explain its weak binding of dNTP, reduced catalytic efficiency and low fidelity.
Unique residues in Pol ν and the error signature
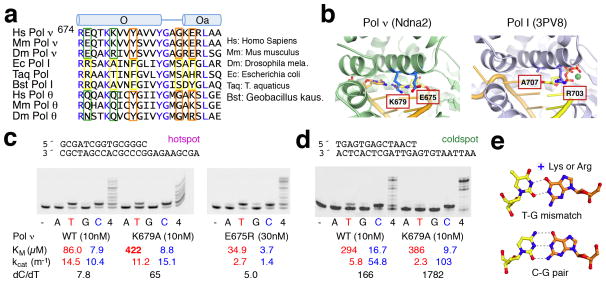
Several residues uniquely conserved in Pol ν appear to stabilize its unusual open state. Firstly, Y682 on the O helix of Pol ν replaces a phenylalanine residue in high-fidelity Pol I (F710 in the bacillus Pol I) (Fig. 3a) and favors the unusual open state by forming a hydrogen bond with R692 (on helix Oa). The Y682 and R692 pair closes off the binding site for the +1 template nucleotide, which is normally sandwiched between helices O and Oa in Pol I (Fig. 2c), and displaces the tyrosine (Y686 in Pol ν and Y714 in Pol I) that is a placeholder for the template base (Fig. 2c,d). Interestingly, the Y682F mutant Pol ν has greatly reduced catalytic activity but improved accuracy 5. Secondly, G689 of Pol ν replaces a conserved serine in Pol I and allows a sharp turn between helices O and Oa, thus keeping them together and preventing Pol ν from reaching the normal open state. Nearby E691 of Pol ν replaces the aromatic residue in Pol I (Y719), which stacks with the downstream template base (+3) and stabilizes the template strand (Fig. 2c). The negatively charged E691 of Pol ν cannot form favorable interaction with DNA. As a result, the downstream template is disordered in all Ndna structures.
Figure 3.
Unique residues of Pol ν for mutagenic synthesis. (a) Sequence alignment of helices O and Oa. Generally conserved residues are colored blue. Uniquely conserved residues are outlined in orange for stabilizing the open-state of Pol ν, and in green for dT misincorporation. (b) E675 and K679 and corresponding residues in Pol I. (c,d) Nucleotide incorporation in the mutation hotspot (c) and (d) coldspot by Klenow, WT, E675R and K679A Pol ν75 with each (A, G, T or C) or all four dNTPs (4) provided. Kinetic data and the preference of dC over dT (ratios of kcat/KM for dC over dT) are shown below. (e) A TG mismatch is stabilized by Lys or Arg in the major groove.
E675 and K679 are unique to Pol ν and located near the nascent base pair and thus may favor dT misincorporation (Fig. 3a,b) 5. We generated Pol ν mutants with these residues replaced by the corresponding amino acids in Pol I (Fig. 3a). The E675R mutation reduced polymerase activity but did not change nucleotide preference (Fig. 3c, Supplementary Table 1). In contrast, K679A mutant Pol ν75 was as active as the WT protein and exhibited 8 to 10-fold better accuracy in nucleotide selection than WT, regardless of whether the DNA substrate sequence was a “hotspot” or “coldspot” for mutation (Fig. 3c,d). The positively charged K679 potentially forms hydrogen bonds with O6 of dG (template) and O4 of the incoming dTTP, thus favoring the dT misincorporation opposite dG (Fig. 3e). This role would be similar to the positively charged R61 and R61K mutant of human Pol η, which promote G:T mismatches and A to G hypermutation in somatic cells 36.
The flexible thumb and primer strand loopout
Ins2 (592–606aa) in Pol ν forms an extended β hairpin replacing a short αhelix in replicative A-family polymerases (Fig. 4a,b). Interestingly, the addition of Ins2 creates a cavity in the DNA-binding surface of Pol ν. As a result, the primer strand upstream from its 3′ end (−2 to −4 nt) is exposed to solvent (Fig. 4a). Furthermore, the thumb domain undergoes a 29.3° rigid-body rotation towards the upstream DNA in the Ndna4 structure (Fig. 4c, Video 3). In other A-family polymerases, the α helix that is replaced by the β hairpin of Ins2 in Pol ν would prevent the thumb movement. While maintaining contacts with the template strand, the rotated thumb in Ndna4 leaves the primer strand solvent exposed from −2 nt upwards.
Figure 4.
Primer looped out by Pol ν. (a) Structural comparison of Pol ν75 (Ndna2) and bacillus Pol I. The proteins are shown as molecular surface and DNA as yellow tube-and-ladder. Ins2 (green) creates a cavity in Ndna2 and exposes the primer strand (indicated by the purple arrow). The cavity is absent in Pol I in the absence of Ins2, which is replace by an α helix (shown in cyan). (b) The Ndna2 and PolI structures are superimposed in ribbon diagrams. The cyan helix of Pol I and Ins2 in Pol ν75 occupy the same space. (c) The thumb rotation in Ndna4 (dark green) compared to Ndna2 (transparent green) further exposes the primer strand. (d) Fully base-paired (F-bp) and primer loopout P[x,y] DNA. (e) Primer extension by Klenow, Pol ν75 and Pol θ86 in the presence of each (A, G, T or C) or all four dNTPs (4). (f) Primer extension by WT and ΔIns2 mutant Pol ν75.
The cavity in the Pol ν thumb domain and exposed primer strand are reminiscent of the cavity on the template side of the DNA binding surface in E. coli DNA pol II that results in template strand loopout and deletional frameshifts 18. We suspect that Pol ν can accommodate looped-out primers in the cavity and lead to either insertional frameshifts or mutations if realignment ensues. To determine the size and location of primer loopouts, seven DNA substrates with 1 to 4 nucleotides looped out in the primer strand at the −2, −3 or −4 position were constructed (Supplemental Fig. 4). Indeed Pol ν75 could extend primers with a 1-nt loopout 3bp upstream from the 3′ end (P[3,1]) with 42% of the catalytic efficiency of normal DNA substrate, while the Klenow fragment of E. coli Pol I (Klenow) could not (Fig. 4d,e) (Supplementary Table 2). The loopout location at −3 position matches perfectly with the observed cavity in Pol ν. The nucleotide insertion efficiency of Pol ν75 inversely correlated with increase of the loop size from 1 to 3 nt at the −3 position (Supplemental Fig. 4). When the 1-nt loopout was 4 to 5 bp upstream (P[4 or 5,1]), Pol ν75 became quite efficient, and even Klenow showed weak activity.
Because Ins2 is conserved between Pol θ and Pol ν, we tested whether Pol θ could extend a loopout-containing primer. An active polymerase domain of Pol θ, Pol θ86 (Methods, Supplemental Fig. 5), was generated and purified. Pol θ86 was ~4 times more active than Pol ν75 and efficiently extended P[3,1] to full-length (25nt) and also 1-nt longer products (26 nt) (Fig. 4e). Interestingly, with normal DNA substrate Pol θ86 produced both the full-length (26 nt) and 1-nt shorter (25 nt) products (Fig. 4e), agreeing with the report that Pol θ86 is prone to generate both types of frameshift mutations 3.
The functional importance of the three inserted regions in Pol θ has been examined previously25. Deletion of Ins1 leads to reduced processivity of DNA synthesis only, but deletion of Ins2 or Ins3 appears to destabilize Pol θ and eliminate the translesion synthesis activities of Pol θ. To examine how Ins2 influences Pol ν’s tolerance of primer loopout, we replaced 25 residues of Pol ν (583–607 aa) including Ins2 with the corresponding 10 residues of E. coli Pol I (680–689 aa) (Methods). The resulting ΔIns2 Pol ν75 was stable and retained one-third of WT Pol ν75’s catalytic efficiency on normal DNA. But its catalytic efficiency with P[3,1] and P[3,2] loopout substrates was reduced to less than one-fifth of WT’s (Fig. 4f, Supplementary Tables 1, 2). It is not surprising that the ΔIns2 protein is not a perfect mimic of WT Pol ν or Pol I, but the differential reduction of its catalytic activity on normal versus loopout DNA is consistent with a role of the structural cavity and the primer-loopout model.
Discussion
Until now a simple open and closed rotation of the finger domain during each cycle of nucleotide incorporation has been universally observed in A-, B-, C-, and X-family DNA polymerases 16,18,28,29, reverse transcriptases, and RNA polymerases 37,38 (Fig. 5a). The Pol ν75 structures, however, reveal an unprecedented open state, which is open for an incoming nucleotide, but closed for template base binding. The unusual open state would change the kinetic and dynamic processes of nascent base pair formation, incoming nucleotide selection, and incorporation. A handful of residues unique to Pol ν (Fig. 3a) likely stabilize the unusual open state, and conservation of Y682 and G689 in Pol ν and Pol θ (Fig. 3a) indicates that the unusual open state is likely also present in Pol θ.
Figure 5.
Diagram of DNA synthesis by Pol ν and a primer-loopout model for trinucleotide-repeat (TNR) expansion. (a) The high-fidelity Pol I and TLS Pol ν differ in the open states, while identical in the closed states. When the O helix in Pol ν is open for dNTP binding, helices Oa and Ob exclude the template base from the active site. The unique K679 in Pol ν further promotes dTTP misincorporation. The 3′–5′ exonuclease (Exo, pink star), which proofreads and improves the accuracy of Pol I, is inactivated in Pol ν (pseudo-Exo, black star). (b) A primer–loopout model. When a downstream template base is unusable (indicated as a red dot), Pol ν can loop out 1–2 nucleotides of the primer strand at the −3 position to re-use the normal template base(s) for lesion-bypass DNA synthesis. The mobile thumb of Pol ν (shown as semi-transparent green) may facilitate DNA translocation and misalignment. (c) Repetitive DNA sequence such as trinucleotide repeats (CNG)n would ease loopout of repeat units, as diagramed here, and result in repeat expansion.
The error signature of Pol ν in misincorporating dTTP is also due to the unique K679 on the O helix (Fig. 3) Our analysis of K679A mutant Pol ν on mutation hotspot and coldspot confirmed the earlier finding 5 that replacement of one amino acid can substantially increase discrimination against dT misincorporation (Supplementary Table 1). Different amino-acid replacements at E675(Q), K679(Q) and E691(R) are found in Pol θ, which may result in mutation spectra and translesion properties different from Pol ν. The key feature of Y-family polymerases, which specialize in translesion synthesis, is the small and non-conserved finger domain that accommodates different DNA lesions in a preformed “closed” active site 39. The finger-closed conformation in the post-reaction product state was previously observed only among low-fidelity X- and Y-family polymerases 30,31. Whether the closed conformation revealed in Ndna2 and Ndna4 bears functional significance for Pol ν in the error-prone and translesion DNA synthesis awaits future studies. The dramatically different finger-open state of Pol ν and Pol θ with substitutions of amino acids that contact the nascent base pair (Fig. 5a) may provide the answer as to how these homologs of high-fidelity replicases carry out mutagenic and translesion DNA synthesis.
Primer loopout allows the template and primer to misalign and DNA synthesis to continue when the downstream template is not usable or blocked (Fig. 5b). Misalignment of primer and template, however, would depend on the local DNA sequence and require adjustment of the DNA position. The near 30° rotation of the thumb domain in Ndna4 (Fig. 4c) has not been observed among DNA polymerases, but it is reminiscent of the thumb movement observed with T7 RNA polymerase during transition from transcription initiation to elongation 40. The mobility and the cavity in the thumb domain, both of which appear to correlate with Ins2, may allow Pol ν to sample DNA for alternative template–primer alignments and accommodate nucleotide loopout in the primer strand (Fig. 5b). Removal of Ins2 does not alter the fidelity of Pol ν but decreases its tolerance for primer loopout (Supplementary Table 1, 2). Pol θ, which contains a larger Ins2 25 and is potentially more flexible, is more efficient than Pol ν at synthesizing DNA with a primer loopout (Fig. 4e).
Short DNA sequence repeats make it easy for primer and template to misalign by one-repeat unit (Fig. 5c). If not realigned, looping out of the primer strand would result in insertions during replication. Trinucleotide repeat (TNR) expansion, a common cause of a number of human diseases including Huntington disease, Fragile X syndrome, Friedreich’s ataxia and myotonic dystrophy, must involve DNA synthesis for these repeats to expand 41,42. Various models have been hypothesized to account for the expansions. Prior to Pol ν and Pol θ, no human DNA polymerase was known to loopout primer and generate expanded DNA products, however. The expansion has thus been attributed to formation of DNA secondary structure (hairpins or triplex) in the expanded strand outside of the DNA polymerase context 43,44. Because of its tendency to form secondary structures, trinucleotide repeats are inherently unstable and often require specialized TLS polymerases to replicate in normal cells. The ability of Pol ν and Pol θ to loopout primer strands raises the possibility that specialized TLS polymerases may also lead to repeat expansions when unrestrained or misguided.
Online Methods
Expression of human Pol ν in HEK293 cells
Human POLN cDNA was obtained from HEK293 cells by RT-PCR. Based on sequence-alignment with bacterial homologs and to remove non-conserved and predicted unstructured regions, four N-and C-terminal deletion clones POLN77, POLN76, POLN75 and POLN74, encoding residues from E175, A181, A192 or D199 to G863, respectively, were constructed (Supplemental Fig. 1a). POLN77 was previously reported 27. Amplified POLN variants were cloned into the mammalian expression vector pLEXm 45. To improve protein solubility and ease purification, a His8 tag and two tandem copies of maltose binding protein (2MBP) 46 followed by a Prescission protease site were fused to the N-terminus of Pol ν variants. The His8-2MBP-tagged Pol ν variants were overexpressed in HEK293GNT1 cells following the protocol described previously 21.
Generation of Pol ν mutants
Mutations of E675R, K679A, and ΔIns2, in which 25 residues in Pol ν (583–607aa) including Ins2 were replaced by the corresponding 10 residues in E. coli Pol I (680–689aa, PVRNEEGRRI), were introduced by a PCR-based mutagenesis using following primers (altered site are underlined), 5′-GACACACGCAGACAGACGGCAAACCAAGAAGGTG and its reverse complement for E675A, 5′-GAGAGCAAACCAAGGCGGTGGTGTACGCGG and its reverse complement for K679A, 5′-CCAGTGAGAAACGAGGAAGGCAGAAGGATCAGGGCCATGTTTGTTTCATC and 5′-GATCCTTCTGCCTTCCTCGTTTCTCACTGGGATACCTTGGATATTAGGAT for ΔIns2.
Protein purification
WT and mutant His8-2MBP-tagged Pol ν75 proteins were first purified using amylose resin (NEB) first. After removing the His8-2MBP tag by PreScission protease, Pol ν75 was purified over a HiTrap heparin HP column and Superdex 200 (GE Healthcare). Purified proteins were concentrated to ~2 mg/ml and stored at −25°C in 20 mM Tris (pH 8.0), 0.3 M NaCl, 2 mM TCEP and 30% glycerol. All purification steps were performed at 4°C.
Crystallization and structure determination
Pol ν75–DNA–dAMPNPP complexes with the Pol ν75 concentration at 5–6 mg/ml were crystallized by the hanging-drop vapor diffusion method at 20°C against a reservoir containing 1.2 ~ 1.5 M (NH4)2SO4, 100 mM MES (pH 5.9–6.3). The crystals were cryo-protected by the addition of 1.5 M sodium malonate (pH 6.0) to the mother liquor. For phase determination, four thymines in the DNA substrate were substituted by 5-bromo-deoxyuridines (Supplemental Fig. 3b). Diffraction data of native crystals and the single-wavelength anomalous diffraction (SAD) at the Br absorption edge were collected at the SER-CAT beam lines in Advanced Photon Source and processed using HKL2000 47 (Table 1). Phases were determined by combining SAD data and molecular replacement with Bacillus DNA Pol I (PDB: 3PV8) 33 as a search model. COOT 48 and PHENIX 49 were used for model building, phase determination and structure refinement. All protein residues are in the favored and allowed regions of Ramachandran plot and none is in disallowed regions. All structural figures and videos were made using PyMol (www.pymol.org).
DNA synthesis assay
DNA polymerase activities (Fig. 3c,d, 4e,f) were measured as previously described 21. Briefly, the DNA primer was 6-FAM labeled at the 5′-end. Reactions were carried out in 10 μL of reaction buffer (20 mM Tris (pH 8.3), 130 mM NaCl, 0.1 mg/ml BSA, 5 mM DTT, 3% glycerol and 5 mM MgCl2). The final concentrations of DNA substrates were 100 nM, Pol ν75 10 nM, and all four dNTPs or each dNTP 100 μM. After pre-incubation of the protein–DNA mixture at 37°C for 5 min, reactions were initiated by addition of desired nucleotides and MgCl2 and proceeded at 37°C for 10 min. Reactions shown in supplementary figures were carried out under similar conditions; differences (if any) are specified in figure legends. Primer extension products were resolved on 15% polyacrylamide – 7.5 M urea gel in 1 x TBE buffer, visualized by Typhon Trio (GE Healthcare), and quantified using ImageQuant TL (GE Healthcare) software.
Steady-state kinetic assay of single nucleotide incorporation
kcat and KM of single nucleotide incorporation were measured in 10 μl of the reaction buffer including 5 μM DNA substrates (1% primer was 6-FAM labeled), 10–200 nM Pol ν75, and a correct (0.1 to 250 μM) or an incorrect dNTP (1 to 1000 μM) for the template base. Reactions were initiated and terminated as describe above except for the 5 min duration of reaction instead of 10 min. Quantification and curve fitting to the Michaelis-Menten equation for calculating kcat and KM were carried out as previously described 21.
Preparation of active human Pol θ polymerase domain
Human POLQ cDNA was obtained from HEK293 cells by RT-PCR. Based on the previous publication 25 and predicted secondary structures, two N-terminal deletion clones POLQ90 (G1792–V2590, same as described in 25) and a smaller POLQ86 (S1822–V2590) were constructed and inserted into the pLEXm-His8-2MBP expression vector as for POLN75. Pol θ90 and Pol θ86 were over-expressed in HEK293 cells and purified as described for Pol ν75.
Supplementary Material
Acknowledgments
Funding: This work is funded by National Institutes of Health intramural program (DK036146-08, W.Y.).
We thank D. Leahy and R. Craigie for editing the manuscript. The research was supported by the intramural research program of US National Institutes of Health (DK036146-08, WY).
Footnotes
Accession Codes
Atomic coordinates and structure factors have been deposited with the Protein Data Bank with accession codes 4XVI, 4XVK, 4XVL and 4XVM for Ndna1–4. The authors declare no competing financial interests.
Author Contributions
Y-S.L. carried out most of experiments. Y.G. helped with kinetic measurement of K678A mutant Pol ν and with data deposition. Y-S.L. and W.Y. designed the project and prepared the manuscript.
References
- 1.Takata K, Shimizu T, Iwai S, Wood RD. Human DNA polymerase N (POLN) is a low fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J Biol Chem. 2006;281:23445–23455. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- 2.Arana ME, Takata K, Garcia-Diaz M, Wood RD, Kunkel TA. A unique error signature for human DNA polymerase nu. DNA Repair (Amst) 2007;6:213–223. doi: 10.1016/j.dnarep.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arana ME, Seki M, Wood RD, Rogozin IB, Kunkel TA. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 2008;36:3847–3856. doi: 10.1093/nar/gkn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seki M, Wood RD. DNA polymerase theta (POLQ) can extend from mismatches and from bases opposite a (6-4) photoproduct. DNA Repair (Amst) 2008;7:119–127. doi: 10.1016/j.dnarep.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takata K, Arana ME, Seki M, Kunkel TA, Wood RD. Evolutionary conservation of residues in vertebrate DNA polymerase N conferring low fidelity and bypass activity. Nucleic Acids Res. 2010;38:3233–3244. doi: 10.1093/nar/gkq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohzaki M, et al. DNA polymerases nu and theta are required for efficient immunoglobulin V gene diversification in chicken. J Cell Biol. 2010;189:1117–1127. doi: 10.1083/jcb.200912012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yousefzadeh MJ, Wood RD. DNA polymerase POLQ and cellular defense against DNA damage. DNA Repair (Amst) 2013;12:1–9. doi: 10.1016/j.dnarep.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koole W, et al. A Polymerase Theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat Commun. 2014;5:3216. doi: 10.1038/ncomms4216. [DOI] [PubMed] [Google Scholar]
- 9.Esposito G, et al. Disruption of the Rev3l-encoded catalytic subunit of polymerase zeta in mice results in early embryonic lethality. Curr Biol. 2000;10:1221–1224. doi: 10.1016/s0960-9822(00)00726-0. [DOI] [PubMed] [Google Scholar]
- 10.Lange SS, et al. Dual role for mammalian DNA polymerase zeta in maintaining genome stability and proliferative responses. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E687–696. doi: 10.1073/pnas.1217425110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood RD. DNA damage tolerance and a web of connections with DNA repair at Yale. Yale J Biol Med. 2013;86:507–516. [PMC free article] [PubMed] [Google Scholar]
- 12.Reha-Krantz LJ. DNA polymerase proofreading: Multiple roles maintain genome stability. Biochim Biophys Acta. 2010;1804:1049–1063. doi: 10.1016/j.bbapap.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 13.Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta. Nature structural & molecular biology. 2009;16:979–986. doi: 10.1038/nsmb.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hogg M, et al. Structural basis for processive DNA synthesis by yeast DNA polymerase varepsilon. Nature structural & molecular biology. 2014;21:49–55. doi: 10.1038/nsmb.2712. [DOI] [PubMed] [Google Scholar]
- 15.Lee YS, Kennedy WD, Yin YW. Structural insight into processive human mitochondrial DNA synthesis and disease-related polymerase mutations. Cell. 2009;139:312–324. doi: 10.1016/j.cell.2009.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doublie S, Sawaya MR, Ellenberger T. An open and closed case for all polymerases. Structure. 1999;7:R31–35. doi: 10.1016/S0969-2126(99)80017-3. [DOI] [PubMed] [Google Scholar]
- 17.Xia S, Konigsberg WH. RB69 DNA polymerase structure, kinetics, and fidelity. Biochemistry. 2014;53:2752–2767. doi: 10.1021/bi4014215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang F, Yang W. Structural insight into translesion synthesis by DNA Pol II. Cell. 2009;139:1279–1289. doi: 10.1016/j.cell.2009.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seki M, Marini F, Wood RD. POLQ (Pol theta), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 2003;31:6117–6126. doi: 10.1093/nar/gkg814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee YS, Gregory MT, Yang W. Human Pol zeta purified with accessory subunits is active in translesion DNA synthesis and complements Pol eta in cisplatin bypass. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:2954–2959. doi: 10.1073/pnas.1324001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marini F, Kim N, Schuffert A, Wood RD. POLN, a nuclear PolA family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J Biol Chem. 2003;278:32014–32019. doi: 10.1074/jbc.M305646200. [DOI] [PubMed] [Google Scholar]
- 23.Sharief FS, Vojta PJ, Ropp PA, Copeland WC. Cloning and chromosomal mapping of the human DNA polymerase theta (POLQ), the eighth human DNA polymerase. Genomics. 1999;59:90–96. doi: 10.1006/geno.1999.5843. [DOI] [PubMed] [Google Scholar]
- 24.Seki M, et al. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2004;23:4484–4494. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hogg M, Seki M, Wood RD, Doublie S, Wallace SS. Lesion bypass activity of DNA polymerase theta (POLQ) is an intrinsic property of the pol domain and depends on unique sequence inserts. J Mol Biol. 2011;405:642–652. doi: 10.1016/j.jmb.2010.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arana ME, Potapova O, Kunkel TA, Joyce CM. Kinetic analysis of the unique error signature of human DNA polymerase nu. Biochemistry. 2011;50:10126–10135. doi: 10.1021/bi201197p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arana ME, Powell GK, Edwards LL, Kunkel TA, Petrovich RM. Refolding active human DNA polymerase nu from inclusion bodies. Protein Expr Purif. 2010;70:163–171. doi: 10.1016/j.pep.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans RJ, et al. Structure of PolC reveals unique DNA binding and fidelity determinants. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20695–20700. doi: 10.1073/pnas.0809989106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moon AF, et al. The X family portrait: structural insights into biological functions of X family polymerases. DNA Repair (Amst) 2007;6:1709–1725. doi: 10.1016/j.dnarep.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Diaz M, Bebenek K, Krahn JM, Kunkel TA, Pedersen LC. A closed conformation for the Pol lambda catalytic cycle. Nature structural & molecular biology. 2005;12:97–98. doi: 10.1038/nsmb876. [DOI] [PubMed] [Google Scholar]
- 31.Zhao Y, et al. Structural basis of human DNA polymerase eta-mediated chemoresistance to cisplatin. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:7269–7274. doi: 10.1073/pnas.1202681109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang W, Hellinga HW, Beese LS. Structural evidence for the rare tautomer hypothesis of spontaneous mutagenesis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:17644–17648. doi: 10.1073/pnas.1114496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson SJ, Taylor JS, Beese LS. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3895–3900. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu EY, Beese LS. The structure of a high fidelity DNA polymerase bound to a mismatched nucleotide reveals an “ajar” intermediate conformation in the nucleotide selection mechanism. J Biol Chem. 2011;286:19758–19767. doi: 10.1074/jbc.M110.191130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y, et al. Mechanism of somatic hypermutation at the WA motif by human DNA polymerase eta. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:8146–8151. doi: 10.1073/pnas.1303126110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brueckner F, Ortiz J, Cramer P. A movie of the RNA polymerase nucleotide addition cycle. Curr Opin Struct Biol. 2009;19:294–299. doi: 10.1016/j.sbi.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 38.Steitz TA. Visualizing polynucleotide polymerase machines at work. EMBO J. 2006;25:3458–3468. doi: 10.1038/sj.emboj.7601211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang W. An overview of Y-Family DNA polymerases and a case study of human DNA polymerase eta. Biochemistry. 2014;53:2793–2803. doi: 10.1021/bi500019s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin YW, Steitz TA. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science. 2002;298:1387–1395. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- 41.Cleary JD, Nichol K, Wang YH, Pearson CE. Evidence of cis-acting factors in replication-mediated trinucleotide repeat instability in primate cells. Nature genetics. 2002;31:37–46. doi: 10.1038/ng870. [DOI] [PubMed] [Google Scholar]
- 42.Budworth H, McMurray CT. A brief history of triplet repeat diseases. Methods in molecular biology. 2013;1010:3–17. doi: 10.1007/978-1-62703-411-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mirkin EV, Mirkin SM. To switch or not to switch: at the origin of repeat expansion disease. Molecular cell. 2014;53:1–3. doi: 10.1016/j.molcel.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gacy AM, et al. GAA instability in Friedreich’s Ataxia shares a common, DNA-directed and intraallelic mechanism with other trinucleotide diseases. Molecular cell. 1998;1:583–593. doi: 10.1016/s1097-2765(00)80058-1. [DOI] [PubMed] [Google Scholar]
- 45.Aricescu AR, Lu W, Jones EY. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr. 2006;62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 46.Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 48.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 49.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.