Abstract
Type 1 diabetes is a T cell-mediated autoimmune disease. Environmental factors play an important role in the initiation of the disease in genetically predisposed individuals. With the improved control of infectious disease, the incidence of autoimmune diseases, particularly type 1 diabetes, has dramatically increased in developed countries. Increasing evidence suggests that gut microbiota are involved in the pathogenesis of type 1 diabetes. Here we focus on recent advances in this field and provide a rationale for novel therapeutic strategies targeting gut microbiota for the prevention of type 1 diabetes.
Type 1 diabetes (T1D) is an autoimmune disease characterized by the immune cell-mediated destruction of insulin-secreting pancreatic beta cells in genetically predisposed individuals upon environmental stimulation. The interaction between pancreatic β-cells and immune cells leads to the development of T1D (1). Strategies targeting cells or signaling pathways of immune system have been proven effective in preventing and reversal of T1D (2–7). Nevertheless, over the past few decades, there has been a steady 3~4% increase in the incidence of T1D, particularly in young children, in developed countries (8). Although genetic factors, especially genes in the HLA region, can predispose an individual to T1D, twin and family studies show that only a fraction of those genetically predisposed individuals will develop the disease (9–11). The cumulative incidence among monozygotic co-twins of persons with T1D is less than 50 % (11). Comparison of the frequency of HLA class II haplotypes in patients diagnosed more than 50 years ago with age and sex-matched patients between 1985 and 2002 suggests that the impact of environment on children with lower-risk HLA class II genes accounts for the rising incidence and decreasing age at diagnosis of T1D (12, 13). Thus, it is strongly believed that environmental factors are important for the development of T1D (14). Furthermore, environmental factors participate in the initiation, as well as various stages of the natural history of the T1D (15, 16).
There have been a variety of environmental factors, including viral infection and diet, suggested to promote T1D. A recent study provides evidence for the presence of enterovirus in pancreatic islets of newly diagnosed patients with type 1 diabetes (17), suggesting that a low-grade enteroviral infection in the pancreatic islets contributes to disease progression in humans. Consistent with the detection of virus in islets, virus-responsive interferon responsive factor 7 (IRF7) network genes and their regulatory locus are implicated in the pathogenesis of T1D by integrated genome-wide approaches (18). Coxsackie B virus (CVB) infected engrafted human islets in mice contain viral RNA, express viral protein, and show reduced insulin production compared to the grafts from uninfected mice (19). These observations imply that viral infection may trigger islet autoimmunity. In addition to virus, it has also been shown that various foods or food components such as cow’s milk and gluten affect the development of T1D (20–23). The insulin from cow’s milk may activate insulin-specific autoimmunity before the establishment of oral tolerance (22, 23). Other components from cow’s milk, such as casein, can alter gut permeability and potentially promote the incidence of T1D (22, 23). Food-derived gluten has also been shown to modify gut permeability and again potentially affect the development of T1D (20, 21). Interestingly, mice treated with a gluten-free diet showed modifications in gut microbiota and protection from T1D (20, 21). Consistently, accumulating studies showed in recent years the involvement of environment factors, particularly an association with gut microbiota, in the pathogenesis of T1D.
In this review, we will focus on the possible roles of gut microbiota in the development of T1D and provide the notion of using prebiotics, probiotics or even antibiotics as a potential strategy for the prevention of T1D.
Understanding the Gut microbiota
Gut microbiota and host have a symbiotic relationship due to their co-existence and co-evolution. On one hand, gut microbiota depend on the host for their growth and survival. On the other hand, most of the gut microbiota are non-pathogenic and can benefit the host in many ways: 1) extraction of nutrients and energy from diet intake (24–26), 2) protection from enteropathogen invasion (27), 3) contribution to the development of a normal immune system or function (28–30). In contrast, the imbalance between the gut microbiota and the host has been associated with many diseases including malnutrition (26), obesity (31–34), autoimmune disorders (35–40) and neurological diseases (41, 42). Thus, an understanding of how the gut flora may affect health and disease will provide an insight into how to promote health by the modification of gut microbiota.
The inability to culture the majority of the gut microbiota in the past had hindered our understanding of the microbial communities. With the advent of high-throughput sequencing technology, it has dramatically speeded up the dissection of the symbiotic relationship between gut microbes and their host. Several large-scale projects such as the US Human Microbiome Project and the European Metagenomics of the Human Intestinal Tract have substantially contributed to the understanding of healthy composition and functional states of gut microbiota (43–47). The analysis of 16S rRNA gene sequences of bacteria from human stool samples has identified that the majority of gut microbial populations comprise bacteria from four main phyla: Bacteriodetes, Firmicutes, Actinobacteria and Proteobacteria (48). The results from metagenomics analysis also provided information about the functional properties of gut bacteria, particularly the important function of low-abundance gut microbes, involved in an array of physiological processes (47, 49). Furthermore, the metabolomics analysis revealed the effect of metabolites derived from microbiome on their host (50). Equipped with knowledge at different levels, we are gaining better recognition about the importance of gut microbiota in our health.
Gut microbiota and type 1 diabetes
During the last decade, there has been considerable research activity and a sharp increase in the amount of experimental data on the role of gut microbiota in health and disease. Of note, gut microbiota play an important role in the regulation of autoimmunity and tolerance (36, 39, 40, 51). However, the contribution of gut microbiota to the development of T1D remains limited, although involvement of the microbiota has been suggested in the development of T1D as early as 1987 (52). Following on from this, experiments using NOD mice that were transferred from specific pathogen-free (SPF) conditions to germ free (GF)-conditions showed a marked change in insulitis and the incidence confirmed the role of gut microbiota as a regulator of islet-specific autoimmunity (53, 54).
The first gut microbiota study in humans for T1D compared the microbiome between 4 Finnish children with T1D and 4 age- and HLA-DQ-matched healthy children (55, 56). By employing the16S rRNA pyrosequencing method, lower diversity and stability of the fecal microbiome was identified in the children in their first year of life who later went on to develop T1D when compared with the healthy control subjects (55). Recent data from the Human Microbiome Project also imply that higher diversity and stability of the gut microbiome is associated with health (46). Furthermore, in the follow-up study, those Finnish children who developed T1D had a decreased ratio of Firmicutes vs Bacteroidetes, supporting a cross-sectional study showing that Bacteroidetes were more abundant in islet-specific autoantibody-positive children than in autoantibody-negative children (57, 58). All the evidence from both animal and human studies thus far further supports the involvement of gut microbiota in the development of T1D. There are several mechanisms by which gut microbiota could affect the development of T1D as discussed below.
Alteration of intestinal permeability
Heightened gut permeability has been demonstrated to be one of the phenomena that precede the clinical onset of T1D in both animal models of autoimmune diabetes, as well as in patients with T1D and prediabetic individuals (59–63). Evidence from animal studies has been largely derived from two rodent models: NOD mice and the BioBreeding diabetes-prone (BBDP) rat. It has been suggested that the imbalance of bacteria, such as Bacteroidetes, which ferment short-chain fatty acid (SCFA), can affect the gut permeability. Indeed, in parallel to the changed gut permeability, BBDP rats, before clinic onset, have a different gut bacterial composition from that of diabetes-resistant (BBDR) rats, with relatively higher abundance of Bacterioides sp in diabetic rat (64, 65). At disease onset, the gut bacterial profile was also different between BBDP and BBDR rats (66). Specifically, the BBDP rats had a lower proportion of the probiotic-like bacteria, such as Bifidobacterium and Lactobacillus, but had higher numbers of Bacteroides, Ruminococcus and Eubacterium (66). At the cellular level, there were also structural changes in the intestinal morphology, such as greater percentage of goblet cells and mucosal crypt depth, accompanying the increased permeability in BBDP rats (59, 62, 67). At the molecular level, the expression of multiple tight junction proteins was down- or up-regulated, in both BBDP rats and T1D patients, thus affecting the gut permeability, including occludin, members of claudin family and zonulin (61, 67, 68).
However, there is not much evidence, thus far, suggesting that the gut microbiota are actually responsible for the cellular and molecular changes in gut. Nevertheless, a study has shown that the metabolites of gut microbiota, such as butyrate, an anti-inflammatory factor, can affect gut permeability by enhancing the gut barrier function via tight junctions (69). In the children with beta cell autoimmunity, there was a low abundance of butyrate-producing bacteria including Clostridium clusters XIVa and IV (55–57). Butyrate can be metabolized from lactate. Children with T1D also have low numbers of lactate-producing bacteria, such as Bifidobacterium adolescentis (57). Those studies provided supporting evidence that gut microbiota could affect gut permeability through their metabolites. Nevertheless, gut permeability is only an index for the later possible beta cell autoimmunity. Although both BBDP and BBDR rats showed transient increases in gut permeability during early life, only BBDP rats that exhibited morphological changes and inflammation in intestine developed the disease (59), which implies that the enteropathy is fundamentally linked to the disease development. Thus, we have to be cautious in interpreting the gut permeability data in this setting.
Modification of intestinal immunity
Gut microbiota are essential for healthy development of the mammalian immune system (70). Direct evidence comes from germ-free mice, in which multiple defects in the gut immune system have been noted, including impaired development of gut-associated lymphoid tissue (GALT) (71), generation of colonic regulatory T cells (72) and production of IgA (73). Importantly, the profound effects that commensal microbiota have on immunity is not limited to the gut immune system, but extends to the systemic immune response (74). Germ-free mice have an elevated IgE level and overall are skewed toward Th2 immune responses, which can be normalized by exposure to a diverse microbiota during early life (75). Interestingly, administration of Bacteroides fragilis-derived polysaccharide A (PSA) to GF mice can correct the Th1/Th2 imbalance in the spleen (74).
Although the various mechanisms by which gut microbiota regulate host immunity are, as yet poorly elucidated, a couple of mechanisms have been proposed: 1) directly activating the innate immune response through Toll-like receptors (TLR) by molecular patterns from gut bacteria (76–79); 2) modulating immune responses via G-protein-coupled receptor (GPCR) by bacterially-derived metabolites (80, 81). The activation of innate immune responses by gut microbiota-derived molecular patterns that are mostly bacteria cell wall components such as flagellin (78, 82) and PSA (79), as well as commensal genomic DNA (77). For example, mice that are deficient in the flagellin receptor, TLR5, develop metabolic syndrome in a gut microbiota-dependent manner (82). The transfer of gut bacteria from TLR5-deficient mice into germ-free mice leads to development of many features of the metabolic syndrome in the recipients (82). This suggests that malfunction of the innate immune system may promote the development of metabolic syndrome through modification of the gut bacterial profile. On the other hand, Bacteroides fragilis-derived PSA, a TLR2 ligand, can induce IL-10-producing CD4 T cells and reciprocally suppress Th17 responses, thus protecting against experimental colitis (79). Similarly, TLR9-deficient mice display an elevated frequency of Foxp3+ Treg at intestinal effector sites and suppressed constitutive IL-17- and IFN-γ-producing effector T cells (77). Further mechanistic studies indicate that DNA from gut flora plays a major role in intestinal homeostasis through TLR9 engagement (77). In the NOD mouse model of T1D, we also demonstrated that a deficiency of the master adaptor protein MyD88 led to resistance to the development of T1D through the modification of gut bacteria (54). All the observations imply that different innate immune-activating components of gut bacterial origin have a different role in the regulation of gut immune homeostasis and our results provide the first evidence of the “missing” link between gut microbiota and innate immunity with T1D development.
Similar to the diet-independent components, diet-dependent gut bacteria-derived metabolites, such as short-chain fatty acid (SCFA) and vitamins, have far-reaching effects on the immune responses (70, 83). SCFAs are produced through fermentation of dietary fiber by gut microbiota, which bind the G-protein-coupled receptor 43 (Gpr43), also called free fatty acid receptor 2 (FFA2/FFAR2). Studies have demonstrated the anti-inflammatory role of SCFAs on immune cells (80, 84, 85). Several SCFAs, including acetate (27, 80, 84), butyrate(84), and propionate (84), have been well characterized as playing a role in gut immune homeostasis. Mice that take in acetate through drinking water display suppressed dextran sulfate sodium (DSS)-induced colitis, inflammatory arthritis and asthma in a Gpr43-dependent manner (80). The oral administration of acetate, butyrate or propionate not only augments population size but also enhances the suppressive function of colonic Treg in SPF mice (84). Among these three SCFAs, butyrate is the most potent inducer of the differentiation of naïve T cells into Treg, while acetate and propionate are important for the migration of Treg to intestine (86, 87). On the other hand, certain Bifidobacteria produce B-group vitamins that can activate mucosa-associated invariant T cells and the Jurkat T cell line (88, 89). Although it has been shown that vitamin D plays a role in the development of T1D (90), studies identifying how SCFAs directly affect islet-specific autoimmunity are still lacking.
In humans, more males develop diabetes than females develop T1D after puberty. However, in NOD mice, females develop diabetes earlier and in a greater proportion than the male mice. In NOD mice, the gut microbiota were shown to be involved in the gender bias of T1D and there is sex hormones play a role in altering commensal gut microbiota which influence the development of diabetes through the IFN-γ signaling pathway (35).
It is clear that gut bacteria affect systemic immunity. Mounting evidence also suggests that gut microbiota have profound effects on autoimmunity. Several studies in animal models implied that alterations in the gut microbiota are associated with the development of T1D (54, 59, 91, 92). During early life, BBDP rats have impaired intestinal barrier function that may be the underlying cause for the altered response to luminal antigens, but the intestinal inflammation might be a trigger that leads ultimately to diabetes development (59, 62). Studies have confirmed the role of gut microbiota as a regulator/facilitator of inflammation in the pancreatic islets. Antibiotic treatment partially protects against T1D in BBDP rats (64, 65). Germ-free NOD mice display an imbalance between Th1, Th17 and Treg differentiation in the intestine (53). This imbalance is associated with accelerated insulitis, which can be interpreted by the shared immune cell homing receptors, such as α4β7-integrin, in the gut and inflamed pancreas (93). Compared with BBDP rats, BBDR rats carry more probiotic-like bacteria in stool, such as Lactobacillus johnsonii (66). Lactobacillus johnsonii can induce Th17 responses in mesenteric lymph nodes and spleen, thus, affecting the development of T1D (76, 94, 95), although the role of Th17 immunity in T1D has been controversial (96–102). The transfer of gut bacteria from diabetes-resistant MyD88 deficient NOD mice can reduce insulitis and significantly delay the onset of diabetes through the upregulation of IgA and TGF-β production in the intestine (92).
Modifying gut flora to prevent T1D
Existing evidence has suggested the role of dysbiosis in T1D development, including reduced bacterial and functional diversity, which are accompanied by impaired gut barrier function and elevated inflammation due to decreased induction of Treg (55–58). With the improvement in our understanding of the role of gut microbiota in autoimmunity, we can develop therapies targeting intestinal immunity by modification of gut microbiota to prevent T1D development. It has been shown that experimental manipulation of gut microbiota in young NOD mice can significantly protect them from T1D development, which provides proof of concept that therapy targeting gut microbiota is effective in genetically predisposed individuals (38, 92). During infancy, the intestinal environment undergoes major developmental changes and the gut microbiome is extensively remodeled; it later becomes relatively resistant to variation after puberty, due to the regulation by intestinal immunity (25, 103). Therefore, minor modifications at the early stages in life would have profound effects on normal intestinal immune homeostasis in adulthood (104). Previous studies also suggested that neonatal gut immunity plays an important role in controlling the development of diabetes (105, 106). Thus, early life treatment with the antimicrobial drug vancomycin can expand Akkermansia muciniphila and reduce diabetes incidence in the NOD mouse (107). Our recent study also demonstrated that the offspring from NOD mothers treated with antibiotics that target gram-negative bacteria had reduced and delayed T1D development (Hu, et al., unpublished data). Neonatal oral administration of DiaPep2, an analogue of HSP60 peptide p277, in combination with hydrolyzed casein diet can protect against T1D in BBDP rats (105). These treatments during early life have a crucial effect on the intestinal barrier function, cytokine production and the development of diabetes (106). Long term administration of “friendly” gut bacteria or the probiotic compound VSL#3 to NOD mice starting from 4 weeks of age could also prevent the NOD mice from T1D development in regulatory cytokine IL-10 or TGF-β dependent mechanism (92, 108). Furthermore, administration of genetically modified gut bacteria can even reverse diabetes. Oral delivery of genetically-modified Lactococcus lactis alone or in combination with low dose of systemic anti-CD3 can reverse new-onset T1D in NOD mice (109, 110).
Although administration of gut microbiota can successfully prevent and reverse T1D in animal models, the application of this therapeutic strategy in humans has not yet been tested. One of the obstacles is the lack of reproducible development and manufacture of microbial mixtures with well-defined genetic content and metabolic output such as the one used for Clostridium difficile infection treatment (111, 112). Nevertheless, there are studies manipulating microbiota-induced immunoregulation by non-bacterial strategies including diet and/or bacterial metabolites. A gluten-free diet could affect gut microbiota and thus reduce the incidence of diabetes (20, 21). It has been shown that gut microbiota-derived metabolites, such as acetate, butyrate, propionate, can modulate intestinal immunity through induction or recruitment of Treg (80, 84–87). Regulatory T cells are essential for the controlling of islet autoimmunity. Thus, testing the effect of those SCFAs on the development of T1D would be important in future preclinical studies.
Conclusion
A better understanding of how gut bacteria-induced immunoregulation contributes to the pathogenesis of T1D is necessary. The existing evidence is exciting and encouraging in that modulation of gut microbiota can affect the progress of diabetes in preclinical studies. It is likely that the beneficial effects of gut microbes come from both the live bacteria and their metabolites (Figure 1). As an ecosystem, the gut microbiome is a community in which the components affect each other and a balance is important for the health of host. Although “omics” analyses can significant improve our understanding of the profile of gut flora and their metabolites, we have to be cautious in thinking that a single bacterial strain or molecule may be useful as therapeutics. Nevertheless, the advantages of microbial therapies are obvious: less expensive, less invasive and potentially long-lasting beneficial effects. Once we gain a better knowledge of specific host and gut microbial functional pathways involved in the development of T1D, direct or indirect microbiota-based therapies can be developed to prevent or cure T1D.
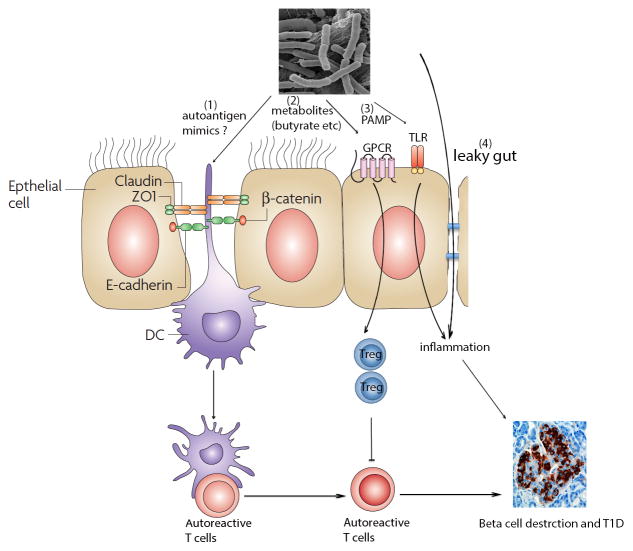
Figure 1.
The role of gut microbiota in the development of T1D. Gut flora can affect islet autoimmunity through mechanisms: 1) expression of autoantigen mimicry to activate autoreactive T cells by antigen-presenting cells to destruct islet beta cells; 2) generating metabolites, such as acetate, butyrate etc, to induce the differentiation or migration of regulatory T cells to control autoreactivity through GPCR signaling pathway (such as Gpr43); 3) gut bacteria-derived pathogen-associated molecular patterns (PAMP) activate TLR signaling pathway to initiate the inflammation, which activates autoreactive T cells and/or directly cause injury to beta cells through inflammatory cytokines; 4) gut bacteria can penetrate the leaky gut and cause inflammation to destruct beta cells.
Table 1.
Role of gut microbiota in T1D
| Function | Phenotype | References |
|---|---|---|
| Alteration of intestinal permeability | Increased ratio of Bacteroidetes vs Firmicutes | Ref 64, 65f, 66 |
| Modified tight junction | Ref 61, 67, 68, 69 | |
| Modified mucosal immunity | Impaired GALT development Modified innate immunity: | Ref 71 |
| 1) TLR2 | Ref 79 | |
| 2) TLR5 | Ref 78, 82 | |
| 3) TLR9 | Ref 77 | |
| Modified adaptive immunity: | ||
| 1) Affecting T cells: | ||
| i) Treg | Ref 27, 53, 72, 80, 84, 86, 87 | |
| ii) Th1 vs Th2 | Ref 53, 74, 75 | |
| iii) Th17 | Ref 53, 76, 77, 79, 94, 95 | |
| 2) Affecting B cell function | Ref 73, 75, 92 | |
| Therapies targeting gut microbiota | Manipulation gut flora at early life prevents T1D; | Ref 38, 92, 104, 106, 107 |
| Long-term supplement of probiotics prevents T1D; | Ref 92, 108 | |
| Administration of genetically modified bacteria reverses T1D. | Ref 109, 110 |
GALT: Gut-associated lymphoid tissue
Acknowledgments
This work was supported by NIH DK-088181, DK092882, JDRF and IACOCCA family foundation.
Footnotes
Conflict of Interest: the authors declare no conflict of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lehuen A, Diana J, Zaccone P, Cooke A. Immune cell crosstalk in type 1 diabetes. Nat Rev Immunol. 2010;10:501–513. doi: 10.1038/nri2787. [DOI] [PubMed] [Google Scholar]
- 2.Ansari MJ, Fiorina P, Dada S, Guleria I, Ueno T, Yuan X, Trikudanathan S, Smith RN, Freeman G, Sayegh MH. Role of ICOS pathway in autoimmune and alloimmune responses in NOD mice. Clin Immunol. 2008;126:140–147. doi: 10.1016/j.clim.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 3.Guleria I, Gubbels Bupp M, Dada S, Fife B, Tang Q, Ansari MJ, Trikudanathan S, Vadivel N, Fiorina P, Yagita H, et al. Mechanisms of PDL1-mediated regulation of autoimmune diabetes. Clin Immunol. 2007;125:16–25. doi: 10.1016/j.clim.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 4.Ben Nasr M, D’Addio F, Usuelli V, Tezza S, Abdi R, Fiorina P. The rise, fall, and resurgence of immunotherapy in type 1 diabetes. Pharmacol Res. 2014 doi: 10.1016/j.phrs.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Herold KC, Bluestone JA, Montag AG, Parihar A, Wiegner A, Gress RE, Hirsch R. Prevention of autoimmune diabetes with nonactivating anti-CD3 monoclonal antibody. Diabetes. 1992;41:385–391. doi: 10.2337/diab.41.3.385. [DOI] [PubMed] [Google Scholar]
- 6.Herold KC, Hagopian W, Auger JA, Poumian-Ruiz E, Taylor L, Donaldson D, Gitelman SE, Harlan DM, Xu D, Zivin RA, et al. Anti-CD3 monoclonal antibody in new-onset type 1 diabetes mellitus. N Engl J Med. 2002;346:1692–1698. doi: 10.1056/NEJMoa012864. [DOI] [PubMed] [Google Scholar]
- 7.Hu CY, Rodriguez-Pinto D, Du W, Ahuja A, Henegariu O, Wong FS, Shlomchik MJ, Wen L. Treatment with CD20-specific antibody prevents and reverses autoimmune diabetes in mice. J Clin Invest. 2007;117:3857–3867. doi: 10.1172/JCI32405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tuomilehto J. The emerging global epidemic of type 1 diabetes. Curr Diab Rep. 2013;13:795–804. doi: 10.1007/s11892-013-0433-5. [DOI] [PubMed] [Google Scholar]
- 9.Todd JA, Walker NM, Cooper JD, Smyth DJ, Downes K, Plagnol V, Bailey R, Nejentsev S, Field SF, Payne F, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–864. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Redondo MJ, Yu L, Hawa M, Mackenzie T, Pyke DA, Eisenbarth GS, Leslie RD. Heterogeneity of type I diabetes: analysis of monozygotic twins in Great Britain and the United States. Diabetologia. 2001;44:354–362. doi: 10.1007/s001250051626. [DOI] [PubMed] [Google Scholar]
- 12.Gillespie KM, Bain SC, Barnett AH, Bingley PJ, Christie MR, Gill GV, Gale EA. The rising incidence of childhood type 1 diabetes and reduced contribution of high-risk HLA haplotypes. Lancet. 2004;364:1699–1700. doi: 10.1016/S0140-6736(04)17357-1. [DOI] [PubMed] [Google Scholar]
- 13.Fourlanos S, Varney MD, Tait BD, Morahan G, Honeyman MC, Colman PG, Harrison LC. The rising incidence of type 1 diabetes is accounted for by cases with lower-risk human leukocyte antigen genotypes. Diabetes Care. 2008;31:1546–1549. doi: 10.2337/dc08-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. 2002;347:911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 15.Trucco M. Gene-environment interaction in type 1 diabetes mellitus. Endocrinol Nutr. 2009;56(Suppl 4):56–59. [PubMed] [Google Scholar]
- 16.Todd JA. Etiology of type 1 diabetes. Immunity. 2010;32:457–467. doi: 10.1016/j.immuni.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Krogvold L, Edwin B, Buanes T, Frisk G, Skog O, Anagandula M, Korsgren O, Undlien D, Eike M, Richardson SJ, et al. Detection of a low-grade enteroviral infection in the islets of Langerhans of living patients newly diagnosed with type 1 diabetes. Diabetes. 2014 doi: 10.2337/db14-1370. [DOI] [PubMed] [Google Scholar]
- 18.Heinig M, Petretto E, Wallace C, Bottolo L, Rotival M, Lu H, Li Y, Sarwar R, Langley SR, Bauerfeind A, et al. A trans-acting locus regulates an anti-viral expression network and type 1 diabetes risk. Nature. 2010;467:460–464. doi: 10.1038/nature09386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallagher GR, Brehm MA, Finberg RW, Barton BA, Shultz LD, Greiner DL, Bortell R, Wang JP. Viral infection of engrafted human islets leads to diabetes. Diabetes. 2014 doi: 10.2337/db14-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marietta EV, Gomez AM, Yeoman C, Tilahun AY, Clark CR, Luckey DH, Murray JA, White BA, Kudva YC, Rajagopalan G. Low incidence of spontaneous type 1 diabetes in non-obese diabetic mice raised on gluten-free diets is associated with changes in the intestinal microbiome. PLoS One. 2013;8:e78687. doi: 10.1371/journal.pone.0078687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen CH, Krych L, Buschard K, Metzdorff SB, Nellemann C, Hansen LH, Nielsen DS, Frokiaer H, Skov S, Hansen AK. A maternal gluten-free diet reduces inflammation and diabetes incidence in the offspring of NOD mice. Diabetes. 2014;63:2821–2832. doi: 10.2337/db13-1612. [DOI] [PubMed] [Google Scholar]
- 22.Vaarala O. Is type 1 diabetes a disease of the gut immune system triggered by cow’s milk insulin? Adv Exp Med Biol. 2005;569:151–156. doi: 10.1007/1-4020-3535-7_22. [DOI] [PubMed] [Google Scholar]
- 23.Kolb H, Pozzilli P. Cow’s milk and type I diabetes: the gut immune system deserves attention. Immunol Today. 1999;20:108–110. doi: 10.1016/s0167-5699(98)01425-x. [DOI] [PubMed] [Google Scholar]
- 24.Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 25.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 28.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahern PP, Faith JJ, Gordon JI. Mining the human gut microbiota for effector strains that shape the immune system. Immunity. 2014;40:815–823. doi: 10.1016/j.immuni.2014.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moran CP, Shanahan F. Gut microbiota and obesity: role in aetiology and potential therapeutic target. Best Pract Res Clin Gastroenterol. 2014;28:585–597. doi: 10.1016/j.bpg.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 33.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 34.Bekkering P, Jafri I, van Overveld FJ, Rijkers GT. The intricate association between gut microbiota and development of type 1, type 2 and type 3 diabetes. Expert Rev Clin Immunol. 2013;9:1031–1041. doi: 10.1586/1744666X.2013.848793. [DOI] [PubMed] [Google Scholar]
- 35.Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, Antonopoulos D, Umesaki Y, Chervonsky AV. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39:400–412. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu HJ, Wu E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes. 2012;3:4–14. doi: 10.4161/gmic.19320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sorini C, Falcone M. Shaping the (auto)immune response in the gut: the role of intestinal immune regulation in the prevention of type 1 diabetes. Am J Clin Exp Immunol. 2013;2:156–171. [PMC free article] [PubMed] [Google Scholar]
- 38.Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, von Bergen M, McCoy KD, Macpherson AJ, Danska JS. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 39.Longman RS, Yang Y, Diehl GE, Kim SV, Littman DR. Microbiota: host interactions in mucosal homeostasis and systemic autoimmunity. Cold Spring Harb Symp Quant Biol. 2013;78:193–201. doi: 10.1101/sqb.2013.78.020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chervonsky AV. Microbiota and autoimmunity. Cold Spring Harb Perspect Biol. 2013;5:a007294. doi: 10.1101/cshperspect.a007294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sherman MP, Zaghouani H, Niklas V. Gut microbiota, the immune system, and diet influence the neonatal gut-brain axis. Pediatr Res. 2014 doi: 10.1038/pr.2014.161. [DOI] [PubMed] [Google Scholar]
- 42.Gonzalez A, Stombaugh J, Lozupone C, Turnbaugh PJ, Gordon JI, Knight R. The mind-body-microbial continuum. Dialogues Clin Neurosci. 2011;13:55–62. doi: 10.31887/DCNS.2011.13.1/agonzalez. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Backhed F, Fraser CM, Ringel Y, Sanders ME, Sartor RB, Sherman PM, Versalovic J, Young V, Finlay BB. Defining a healthy human gut microbiome: current concepts, future directions, and clinical applications. Cell Host Microbe. 2012;12:611–622. doi: 10.1016/j.chom.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 44.Cantarel BL, Lombard V, Henrissat B. Complex carbohydrate utilization by the healthy human microbiome. PLoS One. 2012;7:e28742. doi: 10.1371/journal.pone.0028742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gevers D, Knight R, Petrosino JF, Huang K, McGuire AL, Birren BW, Nelson KE, White O, Methe BA, Huttenhower C. The Human Microbiome Project: a community resource for the healthy human microbiome. PLoS Biol. 2012;10:e1001377. doi: 10.1371/journal.pbio.1001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu GD, Lewis JD, Hoffmann C, Chen YY, Knight R, Bittinger K, Hwang J, Chen J, Berkowsky R, Nessel L, et al. Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol. 2010;10:206. doi: 10.1186/1471-2180-10-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Atarashi K, Honda K. Microbiota in autoimmunity and tolerance. Curr Opin Immunol. 2011;23:761–768. doi: 10.1016/j.coi.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki T, YT, Takao T, Fujimura T, Kawamura E, Shimizu ZM, Tamashita R, Nomoto K. Diabetogenic effects of lymphocyte transfusion on the NOD or NOD nude mice. In: Rygaard MBJ, Graem N, Sprang-Thomsen M, editors. Immune-deficient animals in biomedical research. Basel: Karger; 1987. pp. 112–116. [Google Scholar]
- 53.Alam C, Bittoun E, Bhagwat D, Valkonen S, Saari A, Jaakkola U, Eerola E, Huovinen P, Hanninen A. Effects of a germ-free environment on gut immune regulation and diabetes progression in non-obese diabetic (NOD) mice. Diabetologia. 2011;54:1398–1406. doi: 10.1007/s00125-011-2097-5. [DOI] [PubMed] [Google Scholar]
- 54.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giongo A, Gano KA, Crabb DB, Mukherjee N, Novelo LL, Casella G, Drew JC, Ilonen J, Knip M, Hyoty H, et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011;5:82–91. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, Casella G, Drew JC, Ilonen J, Knip M, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One. 2011;6:e25792. doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Goffau MC, Luopajarvi K, Knip M, Ilonen J, Ruohtula T, Harkonen T, Orivuori L, Hakala S, Welling GW, Harmsen HJ, et al. Fecal microbiota composition differs between children with beta-cell autoimmunity and those without. Diabetes. 2013;62:1238–1244. doi: 10.2337/db12-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Goffau MC, Fuentes S, van den Bogert B, Honkanen H, de Vos WM, Welling GW, Hyoty H, Harmsen HJ. Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia. 2014;57:1569–1577. doi: 10.1007/s00125-014-3274-0. [DOI] [PubMed] [Google Scholar]
- 59.Neu J, Reverte CM, Mackey AD, Liboni K, Tuhacek-Tenace LM, Hatch M, Li N, Caicedo RA, Schatz DA, Atkinson M. Changes in intestinal morphology and permeability in the biobreeding rat before the onset of type 1 diabetes. J Pediatr Gastroenterol Nutr. 2005;40:589–595. doi: 10.1097/01.mpg.0000159636.19346.c1. [DOI] [PubMed] [Google Scholar]
- 60.Bosi E, Molteni L, Radaelli MG, Folini L, Fermo I, Bazzigaluppi E, Piemonti L, Pastore MR, Paroni R. Increased intestinal permeability precedes clinical onset of type 1 diabetes. Diabetologia. 2006;49:2824–2827. doi: 10.1007/s00125-006-0465-3. [DOI] [PubMed] [Google Scholar]
- 61.Sapone A, de Magistris L, Pietzak M, Clemente MG, Tripathi A, Cucca F, Lampis R, Kryszak D, Carteni M, Generoso M, et al. Zonulin upregulation is associated with increased gut permeability in subjects with type 1 diabetes and their relatives. Diabetes. 2006;55:1443–1449. doi: 10.2337/db05-1593. [DOI] [PubMed] [Google Scholar]
- 62.Graham S, Courtois P, Malaisse WJ, Rozing J, Scott FW, Mowat AM. Enteropathy precedes type 1 diabetes in the BB rat. Gut. 2004;53:1437–1444. doi: 10.1136/gut.2004.042481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vaarala O. Leaking gut in type 1 diabetes. Curr Opin Gastroenterol. 2008;24:701–706. doi: 10.1097/MOG.0b013e32830e6d98. [DOI] [PubMed] [Google Scholar]
- 64.Schwartz RF, Neu J, Schatz D, Atkinson MA, Wasserfall C. Comment on: Brugman S et al. (2006) Antibiotic treatment partially protects against type 1 diabetes in the Bio-Breeding diabetes-prone rat. Is the gut flora involved in the development of type 1 diabetes? Diabetologia. 2007;49:2105–2108. doi: 10.1007/s00125-006-0334-0. [DOI] [PubMed] [Google Scholar]; Diabetologia. 50:220–221. doi: 10.1007/s00125-006-0526-7. [DOI] [PubMed] [Google Scholar]
- 65.Brugman S, Klatter FA, Visser JT, Wildeboer-Veloo AC, Harmsen HJ, Rozing J, Bos NA. Antibiotic treatment partially protects against type 1 diabetes in the Bio-Breeding diabetes-prone rat. Is the gut flora involved in the development of type 1 diabetes? Diabetologia. 2006;49:2105–2108. doi: 10.1007/s00125-006-0334-0. [DOI] [PubMed] [Google Scholar]
- 66.Roesch LF, Lorca GL, Casella G, Giongo A, Naranjo A, Pionzio AM, Li N, Mai V, Wasserfall CH, Schatz D, et al. Culture-independent identification of gut bacteria correlated with the onset of diabetes in a rat model. ISME J. 2009;3:536–548. doi: 10.1038/ismej.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Watts T, Berti I, Sapone A, Gerarduzzi T, Not T, Zielke R, Fasano A. Role of the intestinal tight junction modulator zonulin in the pathogenesis of type I diabetes in BB diabetic-prone rats. Proc Natl Acad Sci U S A. 2005;102:2916–2921. doi: 10.1073/pnas.0500178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vaarala O, Atkinson MA, Neu J. The “perfect storm” for type 1 diabetes: the complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes. 2008;57:2555–2562. doi: 10.2337/db08-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hague A, Butt AJ, Paraskeva C. The role of butyrate in human colonic epithelial cells: an energy source or inducer of differentiation and apoptosis? Proc Nutr Soc. 1996;55:937–943. doi: 10.1079/pns19960090. [DOI] [PubMed] [Google Scholar]
- 70.Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14:676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bouskra D, Brezillon C, Berard M, Werts C, Varona R, Boneca IG, Eberl G. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–510. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- 72.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moreau MC, Ducluzeau R, Guy-Grand D, Muller MC. Increase in the population of duodenal immunoglobulin A plasmocytes in axenic mice associated with different living or dead bacterial strains of intestinal origin. Infect Immun. 1978;21:532–539. doi: 10.1128/iai.21.2.532-539.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 75.Cahenzli J, Koller Y, Wyss M, Geuking MB, McCoy KD. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe. 2013;14:559–570. doi: 10.1016/j.chom.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kingma SD, Li N, Sun F, Valladares RB, Neu J, Lorca GL. Lactobacillus johnsonii N6.2 stimulates the innate immune response through Toll-like receptor 9 in Caco-2 cells and increases intestinal crypt Paneth cell number in biobreeding diabetes-prone rats. J Nutr. 2011;141:1023–1028. doi: 10.3945/jn.110.135517. [DOI] [PubMed] [Google Scholar]
- 77.Hall JA, Bouladoux N, Sun CM, Wohlfert EA, Blank RB, Zhu Q, Grigg ME, Berzofsky JA, Belkaid Y. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29:637–649. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oh JZ, Ravindran R, Chassaing B, Carvalho FA, Maddur MS, Bower M, Hakimpour P, Gill KP, Nakaya HI, Yarovinsky F, et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity. 2014;41:478–492. doi: 10.1016/j.immuni.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thangaraju M, Cresci GA, Liu K, Ananth S, Gnanaprakasam JP, Browning DD, Mellinger JD, Smith SB, Digby GJ, Lambert NA, et al. GPR109A is a G-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res. 2009;69:2826–2832. doi: 10.1158/0008-5472.CAN-08-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Greer RL, Morgun A, Shulzhenko N. Bridging immunity and lipid metabolism by gut microbiota. J Allergy Clin Immunol. 2013;132:253–262. doi: 10.1016/j.jaci.2013.06.025. quiz 263. [DOI] [PubMed] [Google Scholar]
- 84.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Inan MS, Rasoulpour RJ, Yin L, Hubbard AK, Rosenberg DW, Giardina C. The luminal short-chain fatty acid butyrate modulates NF-kappaB activity in a human colonic epithelial cell line. Gastroenterology. 2000;118:724–734. doi: 10.1016/s0016-5085(00)70142-9. [DOI] [PubMed] [Google Scholar]
- 86.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 88.Rossi M, Amaretti A, Raimondi S. Folate production by probiotic bacteria. Nutrients. 2011;3:118–134. doi: 10.3390/nu3010118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, Bhati M, Chen Z, Kostenko L, Reantragoon R, et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature. 2012;491:717–723. doi: 10.1038/nature11605. [DOI] [PubMed] [Google Scholar]
- 90.Mathieu C, Badenhoop K. Vitamin D and type 1 diabetes mellitus: state of the art. Trends Endocrinol Metab. 2005;16:261–266. doi: 10.1016/j.tem.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 91.Visser JT, Lammers K, Hoogendijk A, Boer MW, Brugman S, Beijer-Liefers S, Zandvoort A, Harmsen H, Welling G, Stellaard F, et al. Restoration of impaired intestinal barrier function by the hydrolysed casein diet contributes to the prevention of type 1 diabetes in the diabetes-prone BioBreeding rat. Diabetologia. 2010;53:2621–2628. doi: 10.1007/s00125-010-1903-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Peng J, Narasimhan S, Marchesi JR, Benson A, Wong FS, Wen L. Long term effect of gut microbiota transfer on diabetes development. J Autoimmun. 2014;53:85–94. doi: 10.1016/j.jaut.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hanninen A, Nurmela R, Maksimow M, Heino J, Jalkanen S, Kurts C. Islet beta-cell-specific T cells can use different homing mechanisms to infiltrate and destroy pancreatic islets. Am J Pathol. 2007;170:240–250. doi: 10.2353/ajpath.2007.060142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Valladares R, Sankar D, Li N, Williams E, Lai KK, Abdelgeliel AS, Gonzalez CF, Wasserfall CH, Larkin J, Schatz D, et al. Lactobacillus johnsonii N6.2 mitigates the development of type 1 diabetes in BB-DP rats. PLoS One. 2010;5:e10507. doi: 10.1371/journal.pone.0010507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lau K, Benitez P, Ardissone A, Wilson TD, Collins EL, Lorca G, Li N, Sankar D, Wasserfall C, Neu J, et al. Inhibition of type 1 diabetes correlated to a Lactobacillus johnsonii N6.2-mediated Th17 bias. J Immunol. 2011;186:3538–3546. doi: 10.4049/jimmunol.1001864. [DOI] [PubMed] [Google Scholar]
- 96.Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci U S A. 2011;108:11548–11553. doi: 10.1073/pnas.1108924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Marwaha AK, Crome SQ, Panagiotopoulos C, Berg KB, Qin H, Ouyang Q, Xu L, Priatel JJ, Levings MK, Tan R. Cutting edge: Increased IL-17-secreting T cells in children with new-onset type 1 diabetes. J Immunol. 2010;185:3814–3818. doi: 10.4049/jimmunol.1001860. [DOI] [PubMed] [Google Scholar]
- 98.Honkanen J, Nieminen JK, Gao R, Luopajarvi K, Salo HM, Ilonen J, Knip M, Otonkoski T, Vaarala O. IL-17 immunity in human type 1 diabetes. J Immunol. 2010;185:1959–1967. doi: 10.4049/jimmunol.1000788. [DOI] [PubMed] [Google Scholar]
- 99.Li S, Joseph C, Becourt C, Klibi J, Luce S, Dubois-Laforgue D, Larger E, Boitard C, Benlagha K. Potential role of IL-17-producing iNKT cells in type 1 diabetes. PLoS One. 2014;9:e96151. doi: 10.1371/journal.pone.0096151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Boehm BO, Rosinger S, Sauer G, Manfras BJ, Palesch D, Schiekofer S, Kalbacher H, Burster T. Protease-resistant human GAD-derived altered peptide ligands decrease TNF-alpha and IL-17 production in peripheral blood cells from patients with type 1 diabetes mellitus. Mol Immunol. 2009;46:2576–2584. doi: 10.1016/j.molimm.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 101.Silva JA, Ferrucci DL, Peroni LA, Abrahao PG, Salamene AF, Rossa-Junior C, Carvalho HF, Stach-Machado DR. Sequential IL-23 and IL-17 and increased Mmp8 and Mmp14 expression characterize the progression of an experimental model of periodontal disease in type 1 diabetes. J Cell Physiol. 2012;227:2441–2450. doi: 10.1002/jcp.22979. [DOI] [PubMed] [Google Scholar]
- 102.Marwaha AK, Tan S, Dutz JP. Targeting the IL-17/IFN-gamma axis as a potential new clinical therapy for type 1 diabetes. Clin Immunol. 2014;154:84–89. doi: 10.1016/j.clim.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 103.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sudo N, Yu XN, Aiba Y, Oyama N, Sonoda J, Koga Y, Kubo C. An oral introduction of intestinal bacteria prevents the development of a long-term Th2-skewed immunological memory induced by neonatal antibiotic treatment in mice. Clin Exp Allergy. 2002;32:1112–1116. doi: 10.1046/j.1365-2222.2002.01430.x. [DOI] [PubMed] [Google Scholar]
- 105.Brugman S, Klatter FA, Visser J, Bos NA, Elias D, Rozing J. Neonatal oral administration of DiaPep277, combined with hydrolysed casein diet, protects against Type 1 diabetes in BB-DP rats. An experimental study. Diabetologia. 2004;47:1331–1333. doi: 10.1007/s00125-004-1452-1. [DOI] [PubMed] [Google Scholar]
- 106.Scott FW, Rowsell P, Wang GS, Burghardt K, Kolb H, Flohe S. Oral exposure to diabetes-promoting food or immunomodulators in neonates alters gut cytokines and diabetes. Diabetes. 2002;51:73–78. doi: 10.2337/diabetes.51.1.73. [DOI] [PubMed] [Google Scholar]
- 107.Hansen CH, Krych L, Nielsen DS, Vogensen FK, Hansen LH, Sorensen SJ, Buschard K, Hansen AK. Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia. 2012;55:2285–2294. doi: 10.1007/s00125-012-2564-7. [DOI] [PubMed] [Google Scholar]
- 108.Calcinaro F, Dionisi S, Marinaro M, Candeloro P, Bonato V, Marzotti S, Corneli RB, Ferretti E, Gulino A, Grasso F, et al. Oral probiotic administration induces interleukin-10 production and prevents spontaneous autoimmune diabetes in the non-obese diabetic mouse. Diabetologia. 2005;48:1565–1575. doi: 10.1007/s00125-005-1831-2. [DOI] [PubMed] [Google Scholar]
- 109.Robert S, Gysemans C, Takiishi T, Korf H, Spagnuolo I, Sebastiani G, Van Huynegem K, Steidler L, Caluwaerts S, Demetter P, et al. Oral delivery of glutamic acid decarboxylase (GAD)-65 and IL10 by Lactococcus lactis reverses diabetes in recent-onset NOD mice. Diabetes. 2014;63:2876–2887. doi: 10.2337/db13-1236. [DOI] [PubMed] [Google Scholar]
- 110.Takiishi T, Korf H, Van Belle TL, Robert S, Grieco FA, Caluwaerts S, Galleri L, Spagnuolo I, Steidler L, Van Huynegem K, et al. Reversal of autoimmune diabetes by restoration of antigen-specific tolerance using genetically modified Lactococcus lactis in mice. J Clin Invest. 2012;122:1717–1725. doi: 10.1172/JCI60530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Allen-Vercoe E, Reid G, Viner N, Gloor GB, Hota S, Kim P, Lee C, O’Doherty K, Vanner SJ, Weese JS, et al. A Canadian Working Group report on fecal microbial therapy: microbial ecosystems therapeutics. Can J Gastroenterol. 2012;26:457–462. doi: 10.1155/2012/213828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Petrof EO, Claud EC, Gloor GB, Allen-Vercoe E. Microbial ecosystems therapeutics: a new paradigm in medicine? Benef Microbes. 2013;4:53–65. doi: 10.3920/BM2012.0039. [DOI] [PubMed] [Google Scholar]