Abstract
This manuscript presents the latest algorithmic and methodological developments to the structure-based design program DOCK 6.7 focused on an updated internal energy function, new anchor selection control, enhanced minimization options, a footprint similarity scoring function, a symmetry-corrected RMSD algorithm, a database filter, and docking forensic tools. An important strategy during development involved use of three orthogonal metrics for assessment and validation: pose reproduction over a large database of 1043 protein-ligand complexes (SB2012 test set), cross-docking to 24 drug-target protein families, and database enrichment using large active and decoy data sets (DUD-E test set) for 5 important proteins including HIV protease and IGF-1R. Relative to earlier versions, a key outcome of the work is a significant increase in pose reproduction success in going from DOCK 4.0.2 (51.4%) → 5.4 (65.2%) → 6.7 (73.3%) as a result of significant decreases in failure arising from both sampling 24.1% → 13.6% → 9.1% and scoring 24.4% → 21.1% → 17.5%. Companion cross-docking and enrichment studies with the new version highlight other strengths and remaining areas for improvement, especially for systems containing metal ions. The source code for DOCK 6.7 is available for download and free for academic users at http://dock.compbio.ucsf.edu/.
Keywords: docking, pose reproduction, cross-docking, enrichment, virtual screening, ligand flexibility, DOCK
Graphical Abstract
DOCK is a structure-based design program developed over the past 30+ years. The current performance of DOCK version 6.7, made possible by new advances to the codebase, algorithmic updates, and optimized input parameters, is presented. The effectiveness of DOCK is demonstrated in pose reproduction, cross-docking (pictured), and enrichment experiments to systems that are of interest as drug targets. The current release is available for download at: http://dock.compbio.ucsf.edu/.
Introduction
Computer-based molecular docking can facilitate the early stages of drug discovery through systematic pre-screening of ligands (i.e. small molecules) for shape and energetic compatibility with a receptor (i.e. protein) prior to experimental evaluation.[1–3] The inaugural molecular docking program, DOCK, developed by Kuntz and coworkers, has a long history of new advances and accomplishments in the field of structure-based design. At its first inception, DOCK version 1 (DOCK 1), was fundamentally a geometric shape-matching algorithm[4] that evaluated the quality of ligand-receptor complexes on steric overlap.[5] Later, DOCK 2 provided users more sophisticated control over the thoroughness of the sampling algorithm, which in turn provided control over the timing and accuracy of docking calculations.[6,7] DOCK 3 included more rigorous parameterization of the input ligands and receptors, as well as the introduction of a physics-based scoring function that utilized grids,[8,9] and it is this version of DOCK that was used to perform the first documented virtual screen[10] to the enzyme thymidylate synthase. Importantly, the DOCK 3 branch is still actively developed, and it is one of the most rigorously validated molecular docking programs in terms of correspondence between predictions and high-throughput screening or X-ray crystallographic experiments.[11] DOCK 4 was released with an improved graph-matching algorithm for ligand orienting,[12] as well as ligand flexibility based on an incremental construction algorithm (termed anchor-and-grow) and random search.[13] A major re-write into object-oriented C++, including MPI parallelization, led to the release of DOCK 5, which further improved the accuracy of flexible-ligand docking and made the code more modular, which in turn facilitated the implementation of new scoring functions and clustering features.[14,15] Finally, DOCK 6 (versions 6.0 to 6.7) is an extension of DOCK 5 with improved sampling and scoring capabilities, optimization and testing for compatibility with RNA, numerous bug fixes, and a close integration with the AmberTools package[16,17] to allow for implicit solvent, conjugate gradient minimizations, and molecular dynamics simulations.[18–20]
The molecular docking approach has precipitated a number of compounds with experimental activity; a thorough, but not comprehensive, list of successes over the past five years was recently compiled by Coleman et al.[11] Recent representative examples of these successes using DOCK include discoveries to the targets HIVgp41,[21] fatty acid binding protein,[22] β-lactamase,[23] a previously unknown amidohydrolase,[24] thiamine phosphate synthase,[25] tyrosine phosphatase,[26] prostaglandin E2 synthase 1,[27] and thymidine monophosphate kinase,[28] among others. In addition to direct empirical confirmation, the performances of DOCK and other docking programs are occasionally evaluated in formal docking competitions and special symposia.[29–32] A brief list of widely used docking programs, primarily selected from a survey of published literature from 2010–2011,[33] includes: AutoDock,[34] AutoDock Vina,[35] GOLD,[36,37] Glide,[38,39] Surflex,[40] FlexX,[41] ICM,[42] FRED,[43] MOE,[44] CDOCKER,[45] and eHITS.[46] A detailed summary of the latest developments in these programs and others can be found in a recent review by Yuriev and Ramsland.[33]
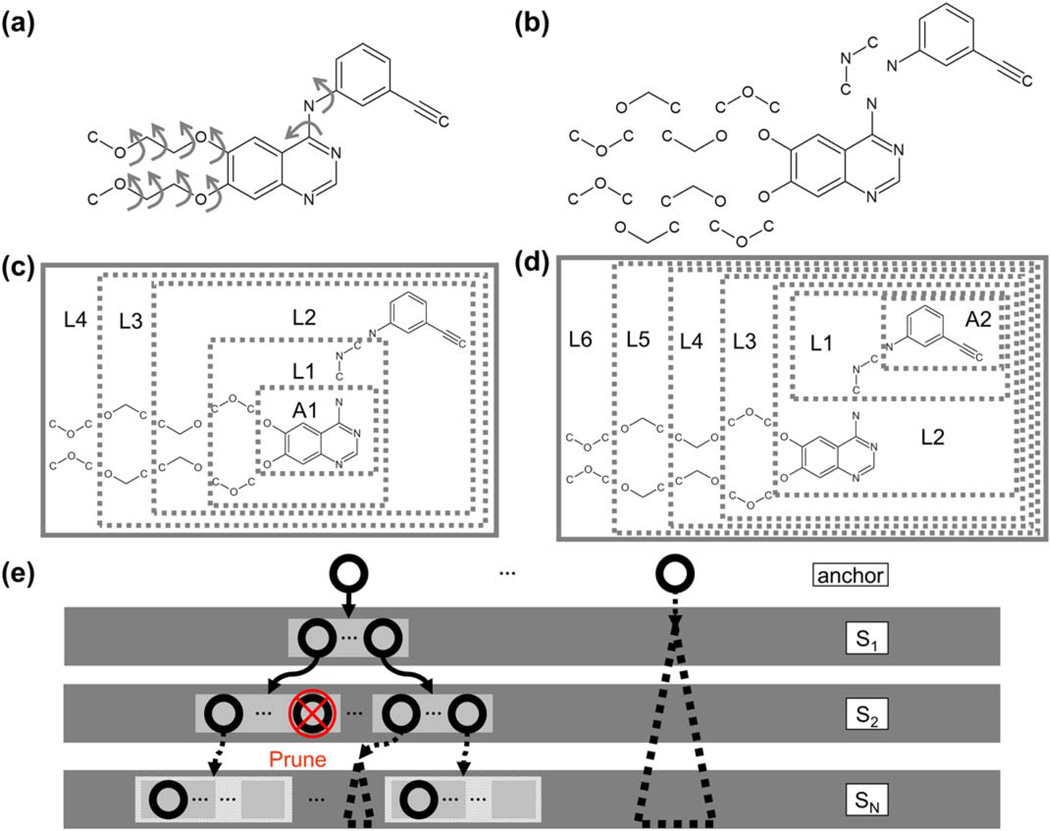
Contributing to the success of DOCK is the open-source codebase (free for registered academic users) and the active community of developers and users over the past three decades. One of the core features that distinguishes DOCK 6 from other docking programs is the anchor-and-grow search algorithm (Figure 1),[12] a breadth-first method for small molecule conformational sampling. In brief, a candidate ligand is disassembled into rigid segments connected by rotatable bonds (RB), for example see Figure 1a, b. A number of the largest segments, termed here the anchors (labeled A1 and A2 in Figure 1), are oriented to the binding site using binding site spheres and a graph-matching algorithm.[12] Next, the rigid segments are flexibly grown in layers (labeled L1–L6) radiating from the anchor until the full molecule is restored (Figure 1c, d).[13] A key advantage of this approach is that the ligand internal conformation remains within the constraints of the receptor binding site, drastically reducing the conformational space sampled. As each ligand segment is grown, conformers are energy minimized and pruned before proceeding to the next step (Figure 1e).
Figure 1.
(a) Structure of prescription drug erlotinib is shown with arrows indicating rotatable bonds. (b) The molecule is broken into rigid segments at rotatable bonds. (c–d) Layers for the specific molecule (L1, L2, … LN) are defined for two different anchors: A1 and A2. (e) Cartoon representation of the growth tree is shown. Circles represent partially or fully grown ligand conformers, which are energy minimized and pruned following each addition of a segment (S1, S2, … SN).
The purpose of this manuscript is to introduce and discuss several important updates to the DOCK 6 codebase, focused on version 6.7, that improve docking success rates and run times dramatically compared to previous versions. First, in Methods and Details, we present the SB2012 test set – an expanded version of the SB2010 test set[47] – that incorporates 1043 crystallographic receptor-ligand complexes for large-scale validation and testing of algorithmic developments. Further, we discuss the protocols and input file parameters recommended for using DOCK in either a pose reproduction or virtual screening (enrichment) capacity. In Results I: Impact of New Features, we discuss in detail new features including: (1) an updated internal energy function, (2) new anchor selection control, and (3) enhanced minimization options. Other new features, including a footprint similarity scoring function, a symmetry-corrected RMSD algorithm, a database filter, and docking forensic tools are briefly summarized, then described in more detail in Supporting Information. In Results II: Current Docking Performance, we discuss the latest performance of the program DOCK in terms of pose reproduction, cross-docking, and enrichment. In addition, we assess the performance of DOCK relative to previous versions.[13,14] Finally, in Summary and Future Perspectives, we summarize the results and briefly introduce some upcoming and in-development features planned for future releases of the DOCK program. Overall, these works represent as yet unreported novel docking results, obtained with newly added DOCK functionality, over a significantly large validation test set (1043 systems).
Three key outcomes from this work include: (1) A steady progression of increased pose reproduction success rates from DOCK version 4.0.2 (51.4%) → 5.4 (65.2%) → 6.7 (73.3%) with the most dramatic improvements for large and flexible ligands. (2) Cross-docking success for many protein families is not majorly dependent on the specific receptor structure used, and for some structurally important cases (i.e. active vs. inactive, wild-type vs. mutant) DOCK can discriminate between different receptor forms. (3) DOCK generally performs well in both early and overall enrichment, and tuning the number of ligand anchors improves run times without negatively impacting performance. The overall major advance in the docking algorithm described in this work was not due to any one change, but rather a series of complimentary and independent incremental improvements that employed a range of orthogonal test sets and tools.
Methods and Details
SB2012 Test Set
The SB2010 test set of 780 crystallographic receptor-ligand complexes described in Mukherjee et al.[47] was expanded to 1043 systems in a new test set termed SB2012. Receptor and ligand preparation protocols were updated, and a considerable number of new systems were introduced. New entries include 72 additional systems from the Astex diverse set,[37] 29 additional systems from the Directory of Useful Decoys[48] (DUD, discussed in Brozell et al.),[29] and 169 new systems from the Protein Data Bank (PDB),[49] with an emphasis on kinases corresponding to receptors in the Astex set. Specifically, we added systems representing insulin-like growth factor 1 receptor (IGF-1R), Abelson tyrosine-protein kinase 1 (ABL1), peroxisome proliferator-activated receptor gamma (PPAR-γ), and epidermal growth factor receptor (EGFR), among others. From the original SB2010 test set, five systems with semi-covalent boron-containing ligands and two other aberrant systems were removed.
A thorough description of the test set preparation protocols can be found in the original SB2010 paper.[47] Here, we will briefly discuss the most important preparation steps, focusing on new or updated steps. Beginning with SB2012, protocols were revised for compatibility with Amber 11,[50] and to support systems with small-molecule cofactors. In SB2012, ligands were protonated and initially assigned empirical Gasteiger-Marsili charges[51] with the program MOE,[44] taking every effort to match the protonation state and net charge described in the literature for the crystallographic complex. Ligands were then assigned more rigorous AM1-BCC charges[52,53] using antechamber (AmberTools). Small-molecule cofactors, when present, follow the same preparation protocols as the ligands. Receptors were protonated and assigned ff99SB parameters[54] in tLEaP (AmberTools), then subjected to short energy minimizations with heavy restraints on non-hydrogen atoms using sander (Amber 11). Importantly, the new protocols enable the energy minimization of newly-added receptor hydrogen atoms in the presence of the bound cognate ligand (and cofactor, if applicable), allowing for physically reasonable positioning. In addition, the new top2mol2 tool allows for facile conversion between Amber parm and crd files and separate receptor, ligand, and cofactor mol2 files (Tripos Sybyl conventions[55]) while retaining the atomic partial charges, bond types, and atom types as assigned by tLEaP (receptor) or antechamber (ligand, cofactors).
Pose Reproduction Methods
The first key experiment, pose reproduction, involves taking a known (i.e. crystallographic) receptor-ligand complex, removing the ligand from the system, then docking the ligand back into its original binding site (Figure 2). The test is a docking success if the top-scoring pose is within 2.0 Å RMSD from the crystallographic position (termed here the correct pose), a scoring failure if a correct pose was sampled but not scored as the best pose, or a sampling failure if a correct pose was never sampled. Across a large test set, e.g. SB2012, the sum of successes + scoring failures + sampling failures equals 100%. Here, we briefly present methods for preparing the system and performing pose reproduction experiments in the program DOCK, including input file parameters, which together are the generally recommended protocols for this type of experiment. Additional details can be found in the DOCK User’s Manual.[56]
Figure 2.
Standard pose reproduction experiment following a flexible docking protocol. (a) Crystal ligand conformation shown in blue. (b) DOCK sampling space shown as overlay of magenta molecules. (c) Top-ranked pose (magenta) relative to crystal pose (blue), 1.35 Å RMSD.
First, the molecular surface of the receptor absent hydrogen atoms and the ligand was determined using the DMS program[57] with a probe radius of 1.4 Å. Then, the DOCK accessory program sphgen[4] was used to generate spheres within the ligand binding site. A maximum of 75 spheres that were within 8.0 Å of the crystallographic ligand position were retained for docking. Following sphere generation, the DOCK accessory program showbox was used to construct a box around the spheres plus an 8.0 Å margin in all directions. Finally the DOCK accessory program GRID[8] was used to pre-compute energy interactions between a dummy probe atom and all receptor atoms on a 0.3 Å resolution grid within the box. For these experiments, the Lennard-Jones potential with 6–9 attractive and repulsive exponents was used to model the van der Waals interactions, and the Coulomb potential with a distance-dependent dielectric coefficient of ε=4r was used to model the electrostatic interactions. This resulting grid formed the basis for computing single grid energy (SGE) score, which was used in all tests in this manuscript unless otherwise noted.
For pose reproduction experiments, the ligands were treated as flexible based on the the FLX protocol in Mukherjee et al.[47] unless otherwise specified. Although a complete example input file is distributed with the test set, some of the important parameters are as follows: All rigid segments of the molecule with at least five atoms were treated as anchors, and the maximum number of anchor orientations attempted (max_orientations) was 1000. In addition, a maximum of 1000 anchor orients (pruning_max_orients) were retained for anchor-and-grow search following a pruning heuristic[56] that combines conformer rank and root-mean-square deviation (RMSD) using a pruning clustering cutoff of 100. During ligand growth, a repulsive van der Waals term with an exponent of 12 (referred to as the repulsive-only internal energy) was used to alleviate internal clashes. A simplex minimizer[58] was employed for one cycle of minimization per ligand conformation per stage of growth, and the score was considered to be converged if the changes in energy between steps were less than 0.1 kcal/mol. The translational, rotational, and torsional step sizes for the simplex minimizer were 1.0 Å, 0.1π radians (18°), and 10.0°, respectively. One cycle of the simplex minimizer constituted a maximum of 500 iterations for the anchor (simplex_anchor_max_iterations), and 500 iterations for each subsequent partially grown molecule (simplex_grow_max_iterations). Any conformer with a score greater than 100.0 kcal/mol (pruning_conformer_score_cutoff) following energy minimization was rejected. Finally, a maximum number of 5000 scored conformers were retained for examination, following best-first clustering with a 2.0 Å RMSD threshold. Together, these parameters are referred to in this manuscript as the “standard flexible docking (FLX) protocol” or the “standard protocol”.
Cross-docking Methods
The second key experiment, cross-docking, is an additional method for evaluating the pose reproduction ability of docking programs under different conditions. Here, multiple receptor-ligand crystallographic complexes are aligned into a common reference frame. An aligned group of systems, also called a cross-docking family, facilitates the experiment of docking all ligands into all receptors, forming a matrix of N×N docking calculations where N is the number of systems in the family. As part of the SB2012 release, 24 cross-docking families were prepared (ranging from 6 to 59 systems per family) and made available for download. Membership in a cross-docking family was initially determined according to the “Molecule Name” record in the PDB[49] and the Enzyme Commission number (EC#) when available. The receptors in each family were aligned on the alpha carbon atoms using the matchmaker tool in the Chimera program.[59] The first PDB code in each family was arbitrarily chosen as the master reference frame for alignment. During the alignment step, PDB codes with a low number of backbone matches (<50% alpha carbon matches) or high backbone RMSD (>2.0 Å) to the reference structure were rejected from the set. In addition, systems with ligands that bound in a disparate binding site were removed. For some particular families, however, models with larger-than-expected backbone RMSD were deliberately retained for biological motives. As an example, active and inactive conformations of EGFR were retained in the same family. Finally, each group was visually inspected to further ensure that all structures were well aligned.
An important assumption in cross-docking is that the aligned ligand pose in each receptor provides a physically reasonable RMSD reference for evaluating success or failure of off-diagonal elements. However, some aligned ligand poses may not fit in a non-native binding site due to, for example, induced fit effects or receptor mutations, and thus should not be used as a docking reference. To assess compatibility, every ligand from the aligned set was energy minimized in the context of every receptor from the same aligned set, prior to any cross-docking calculations (1000 iterations of Cartesian space minimization with an RMSD restraint of 10 kcal mol−1 Å−2 and van der Waals exponents of 6–12). If the RMSD of the minimized crystallographic ligand exceeded 2.0 Å, or if it had a positive energy, then it was considered a non-viable reference and that pairing was not included in the test. Stated another way, if any particular crystallographic ligand pose was incompatible with an off-diagonal receptor, that pair was not considered when computing docking statistics.
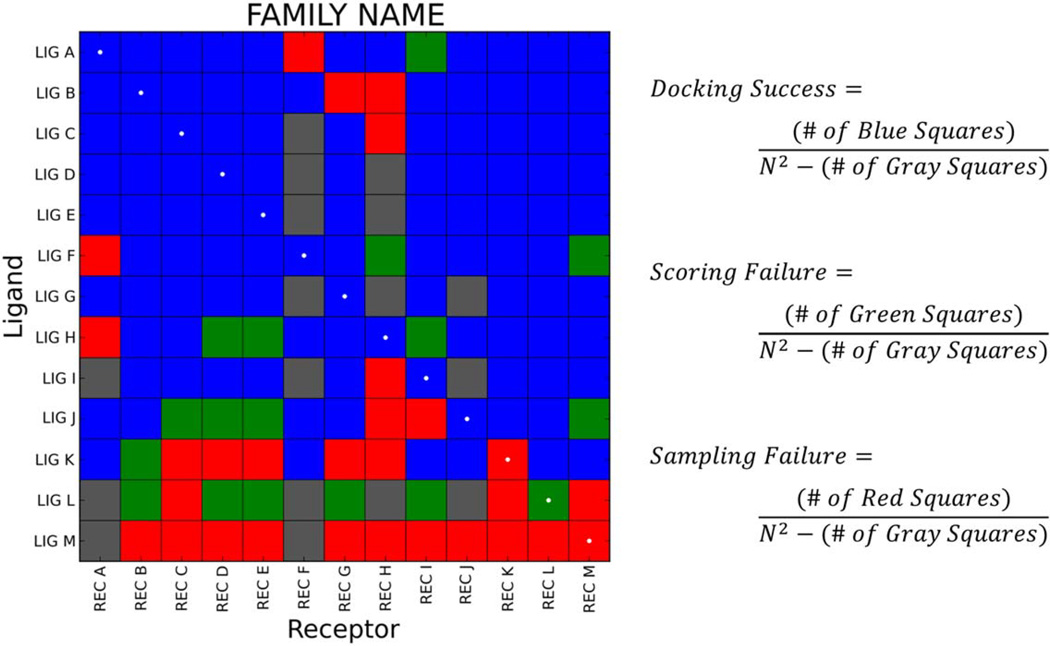
A particularly useful way to evaluate cross-docking performance is to employ heatmaps, as illustrated for a representative family in Figure 3. Here, ligands are labeled along the y-axis, and receptors are labeled in the same order along the x-axis. The diagonal of the matrix, or those cells representing the cognate (native) receptor-ligand pairs, is marked with white dots. Cells in the matrix are color-coded using the docking outcome definitions previously described for pose reproduction experiments: docking success (blue), scoring failure (green), sampling failure (red), and the addition of a non-viable pairing (gray). Ligand order along the y-axis is determined lexicographically first by success rate (number of blue squares), then by scoring failures (number of green squares), then by sampling failures (number of red squares), and finally by the number of non-viable pairings (number of gray squares). The overall matrix docking success is computed by dividing the number of docking successes (blue squares) by the number of viable docking calculations (N2 minus the number of gray squares). As an example, Figure 3 shows cross-docking to a family of size N=13 which required 13×13=169 docking calculations. The matrix contains 17 gray squares denoting non-viable pairings, thus the denominator in the matrix success calculations is 169−17=152. There are 106 blue squares denoting successful dockings, therefore, the matrix docking success is 106/152=69.7%. Similar calculations can be made to determine the matrix scoring failure rate (green squares), the matrix sampling failure rate (red squares), or the success and failure rates pertaining to the diagonal.
Figure 3.
Sample cross-docking heatmap (N=13) color-coded as: blue=docking success, green=scoring failure, red=sampling failure, or gray=non-viable pairing. The diagonal is marked with white dots. In this matrix, there are 169 total squares: 106 blue, 17 green, 29 red, and 17 gray. Thus, the matrix docking success is 106/152=69.7%, the matrix scoring failure is 17/152=11.2%, and the matrix sampling failure is 29/152=19.1%.
It is important to emphasize that the protocol decisions defined here for crossdocking and elsewhere in the manuscript are an informed design choice, and we recognize that other researchers may have different ways of approaching system preparation and docking protocols. Our protocol for classifying theoretical complexes as viable or non-viable is based on the simple premise that if the off-diagonal reference ligand itself is not energetically and/or sterically compatible when overlaid with a rigid receptor (in which it was not co-crystallized), then docking to reproduce that reference pose cannot (a priori) be expected to work. Stated another way, some structures when overlaid produce un-physical complexes and the standard 2.0 Å RMSD definition for docking success could not possibly be achieved with those pairings. On-the-fly docking protocols that includes both receptor and ligand flexibility in DOCK are under investigation. For the present study, however, receptor flexibility tests are not presented.
Enrichment Methods
The third key experiment, enrichment, provides a means to assess the performance of DOCK in a virtual screening capacity (see Figure 4). Enrichment involves rank-ordering a database of compounds containing a set of known actives seeded among a large number of decoys according to their predicted binding affinity to a target. Ideally, known actives are ranked higher than the decoys by the docking program or method used. One common metric for evaluating the outcome of an enrichment experiment is to use receiver operating characteristic (ROC) curves. These curves plot the true positive rate (true positives divided by the total number of positives, also called sensitivity) against the false positive rate (false positives divided by the total number of decoys, also described as [1-specificity]) for a rank-ordered list.[60] Subsequently, ROC curves can be evaluated on the total area under the curve (AUC), or smaller subsets such that early enrichment can be evaluated (e.g. enrichment after 1% or 10% of the database has been screened). Good early enrichment is considered to be important for virtual screening where only a small number of compounds might be purchased for experimental testing. Enrichment studies using an earlier version of DOCK (version 6.6) were recently performed on all 40 DUD systems[48] and the results were reported in a manuscript by Brozell et al.[29] Here, we performed enrichment on five systems from the expanded DUD-E set which includes larger active and decoy sets, as well as more targets.[61] The systems used for enrichment in this work include estrogen receptor α (PDB code 1SJ0),[62] human immunodeficiency virus type 1 (HIV) protease (PDB code 1XL2),[63] thrombin (PDB code 1YPE),[64] insulin-like growth factor 1-receptor (IGF-1R, PDB code 2OJ9),[65] and 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase (PDB code 3CCW).[66]
Figure 4.
Schematic protocol for enrichment experiment. (a) Lists of known active compounds and decoy compounds are prepared for a given receptor. In this specific example, five actives (represented by black circles) and ten decoys (represented by open circles) are shown. (b) Following docking of all actives and decoys to a receptor, outcomes are ranked based on score. Here, two different outcomes are described: good enrichment (blue box) and poor enrichment (red box). (c) Data are plotted as receiver operating characteristic (ROC) curves: true positive rate on the y-axis, false positive rate on the x-axis. The blue line represents a good enrichment outcome with an area-under-the-curve (AUC) of >0.5; the red line represents a poor enrichment outcome with an AUC of <0.5; the dashed line represents random enrichment with an AUC equal to 0.5.
In the initial DOCK preparation steps, the receptor is treated the same as in the pose reproduction experiments. The ligands originate from databases such as ZINC,[67,68] DUD,[48] and DUD-E.[61] Protonation states and tautomeric forms for a range of pH values are predicted, then partial atomic charges using semi-empirical methods in AMSOL are computed.[69,70] Importantly, this approach generates molecules in a format compatible with current DOCK protocols.[71] During docking, the standard FLX protocol was used with one major difference in the input file: the number of scored conformers retained is set to 1 rather than 5,000. Note that prior DOCK enrichment studies employed 100 or 1000 as the parameter for maximum number of orients (pruning_max_orients).[29,72] Here, we employ 1000 for all experiments, which is the generally recommended setting for this parameter.
Computational Aspects
All tests described in this manuscript were run on a 240-CPU Linux cluster consisting of 20 Dell PowerEdge C6100 servers with dual Intel Xeon x5660 2.80 GHz hexa-core Nehalem processors. It is necessary to note that, in some cases, different compilers, environments, or computer architectures can influence docking speed. Thus, while useful as a general guide, timings presented in this manuscript are highly specific to our infrastructure and should be viewed in that light. The most recent version of DOCK is available to registered users online at http://dock.compbio.ucsf.edu. In addition, the SB2012 test set, including example input file parameters, is available for download at www.rizzolab.org.
Results I: Impact of New Features
Ligand Conformational Sampling and Orienting
Internal Energy Function
Previous versions of DOCK (5.4–6.3) employed a three-term (attractive van der Waals, repulsive van der Waals, electrostatic) internal energy function that was used only to evaluate the final docked pose. To alleviate potential internal clashes among ligand atoms during torsional sampling, users could specify an optional clash overlap function. Briefly, for every non-bonded pair of ligand atoms, the clash overlap function sums the two van der Waals radii and multiplies that value by a user-specified clash overlap parameter ranging from 0 to 1. If the result is greater than the distance between the pair of atoms, a clash is registered and the conformer is rejected from the ensemble. A high clash overlap parameter (near 1) can negatively impact sampling, while a low parameter (near 0) can allow too many unphysical ligand conformers to appear. By default, DOCK uses a value of 0.5, which provides a reasonable balance. However, upon close examination, this practice did not always prevent significant internal ligand clashes from forming during anchor-and-grow conformational sampling. To correct this behavior we modified the internal energy function to employ solely the repulsive van der Waals term, and to function at all stages of growth. Note that the repulsive-only component of the van der Waals term is always unfavorable, so it will serve only to prevent internal clashes between ligand heavy atoms, which is the primary goal of the function. Without an explicit torsional energy term, the attractive van der Waals and electrostatic terms could favor highly-strained ligand conformations where atoms are tightly clustered, giving rise to unfavorable and unphysical results.
A substantial speed-up of the new internal energy code was accomplished by building a pair-list of atoms that could form a van der Waals interaction internal to the ligand. This pair-list ignored 1–2, 1–3, and 1–4 atom pairs, then pre-computed van der Waals repulsive coefficients for all other pairs of atoms separated by at least four bonds. Using this approach, the internal energy function could be called efficiently within the energy minimizer at every stage during anchor-and-grow. A beneficial consequence of this protocol is that the anchor-and-grow process was made more effective in that partially-grown conformers with internal clashes are pruned early during growth (see Figure 1). This dramatically improves docking proficiency (compared to using no internal energy function), as well as overall sampling.
Figure 5a demonstrates that significant internal clashes can occur if the internal energy and clash overlap functions are not applied during growth. Unlike the clash overlap function, the energy minimizer can utilize the new internal energy function to relax torsions and gradually ameliorate internal clashes instead of discarding such poses outright. It should be emphasized that problems with ligand conformational expansion did not emerge in earlier versions of DOCK because development was largely limited to testing relatively small, rigid molecules with less than seven rotatable bonds,[14] and partially-grown conformers at each stage of growth were not rigorously examined.
Figure 5.
(a) Top-scoring ligand pose with internal energy and clash overlap functions turned off (top) vs. only the internal energy function turned on (the standard FLX protocol, bottom) using DOCK 6.7 (PDB code 1M17).[73] Internal clashes in top molecule indicated by red arrows. (b) Histograms of ligand internal energy (repulsive van der Waals term only) for native crystallographic ligands from the SB2012 test set following no minimization (black line) or 100 (red), 1000 (blue), and 5000 (green) iterations of flexible minimization.
In order to choose a hard cutoff parameter for the internal energy, thus setting a baseline for testing, the repulsive-only internal energy was measured for all crystallographic poses in the SB2012 test set following several different energy minimization protocols. As shown in Figure 5b, the large majority of crystallographic conformations have a repulsive-only internal energy score between 0 and 40 kcal/mol. Thus, employing a maximum allowable unfavorable score of +100.0 kcal/mol provides an upper bound consistent with experimental observations and will ensure that only severe clashes are penalized, and that smaller unfavorable interactions occurring during ligand growth will not result in outright rejection of the conformation. During energy minimization, this unfavorable score is added to the intermolecular interaction energy (computed, for example, on the grid), and the sum of the two energies is minimized. It should be noted that the parameter pruning_conformer_score_cutoff now depends on both protein-ligand interaction energy and ligand internal energy. As a consequence, we found that one member of the SB2012 test set (PDB code 3FEI)[74] is unable to complete growth due to a relatively high ligand internal energy, and slightly unfavorable docked poses early in growth. Increasing the score cutoff parameter from +100.0 kcal/mol to +200.0 kcal/mol rescues this system. Table 1 compares the results of the standard docking protocol without any consideration of internal energies or clashes (none), the clash overlap feature from earlier versions (clash-ovr), and the new internal energy function (int-ener).
Table 1.
Comparison of internal energy protocols to assess pose reproduction in SB2012.
| Internal Energy Method |
# RB | Subset Size |
Success # (%) |
Sampling Failure # (%) |
Scoring Failure # (%) |
Avg. Time (min) |
|
|---|---|---|---|---|---|---|---|
| A | nonea | <8 | 542 | 417 (76.9) | 18 (3.3) | 107 (19.7) | 0.8 |
| B | none | 8–15 | 403 | 163 (40.4) | 93 (23.1) | 147 (36.5) | 2.7 |
| C | none | >15 | 98 | 9 (9.2) | 66 (67.3) | 23 (23.5) | 8.7 |
| D | none | all | 1043 | 589 (56.5) | 177 (17.0) | 277 (26.6) | 2.3 |
| E | clash-ovrb | <8 | 542 | 417 (76.9) | 17 (3.1) | 108 (19.9) | 0.8 |
| F | clash-ovr | 8–15 | 403 | 164 (40.7) | 85 (21.1) | 154 (38.2) | 2.7 |
| G | clash-ovr | >15 | 98 | 9 (9.2) | 62 (63.3) | 27 (27.6) | 8.5 |
| H | clash-ovr | all | 1043 | 590 (56.6) | 164 (15.7) | 289 (27.7) | 2.3 |
| I | int-enerc | <8 | 542 | 453 (83.6) | 13 (2.4) | 76 (14.0) | 1.2 |
| J | int-ener | 8–15 | 403 | 264 (65.5) | 53 (13.2) | 86 (21.3) | 5.2 |
| K | int-ener | >15 | 98 | 48 (49.0) | 29 (29.6) | 21 (21.4) | 22.8 |
| L | int-ener | all | 1043 | 765 (73.3) | 95 (9.1) | 183 (17.5) | 4.8 |
none refers to the standard docking method without the internal energy function or the clash-overlap function.
clash-ovr refers to the standard docking method with the clash overlap function turned on and the internal energy function turned off.
int-ener refers to the standard docking method with the internal energy function turned on and the clash-overlap function turned off (equivalent to the standard FLX protocol).
Notably, the docking success rate across all 1043 systems is 73.3% when the internal energy function is used (Table 1, row L), which is a substantial improvement when compared to 56.5% success without the internal energy function (row D), or 56.6% success with only the clash overlap function (row H). The higher success rate with the internal energy function can be attributed to a substantial decrease in both the sampling failures (from ~17% to ~9%) and the scoring failures (from ~27% to ~18%). Looking at specific subsets, the overall greatest improvement using the internal energy function is for the group of molecules with >15 rotatable bonds. Here, the success rate is 49.0% using the standard internal energy (Table 1, row K), vs. 9.2% without using the internal energy and with or without the clash overlap function (row C, G). Conceptually, this is consistent with greater opportunity for internal clashes among ligands that have a large number of rotatable bonds. An examination of the results for smaller, more rigid ligands shows less dramatic improvement. For example, for all systems with <8 rotatable bonds, the sampling failure rates are all low (between 2.4–3.3%) irrespective of whether the internal energy or clash overlap functions were used (Table 1, rows A, E, I). A consequence of better sampling and scoring using internal energy is an increase in average docking time from 2.3 min (Table 1, rows D, H) to 4.8 min (row L). This increase, however, is not due to the time required for internal energy calculations alone, rather it is primarily due to the larger number of anchor-and-grow conformer branches (see Figure 1) retained. Specifically, in the absence of the internal energy function, many growth tree branches are terminated prematurely due to internal clashes and unfavorable energies. With the internal energy function, more branches contain physically acceptable conformations, resulting in improved sampling.
Anchor Selection and Limits
Two important considerations in the anchor-and-grow strategy are the identities and the number of anchors used to seed growth. In earlier versions of DOCK, the user could specify a minimum anchor size (e.g. five heavy atoms or greater), and all rigid segments of the molecule that matched the criteria would be used as anchors. Recently, options were added to allow users to specify one specific rigid segment of their choice as the anchor, or to allow users to specify a certain number of rigid segments to be used as anchors. By limiting the number of anchors, or by beginning growth from a specific anchor, the anchor-and-grow strategy will have improved efficiency (i.e. faster time) without negatively impacting success rates. As an example of the utility of manually choosing a specific anchor, given a hypothetical binding site with high propensity to bind aromatic rings in a specific location, a user could specify an aromatic ring in a candidate ligand as the anchor, then limit the anchor-placement search space to the expected binding location. As described in Mukherjee et al,[47] DOCK success rates are typically higher when the exact anchor position is known, as is the case for fixed anchor docking (FAD) and the ligand atom spheres protocol (LAS) discussed in Minimization Options below.
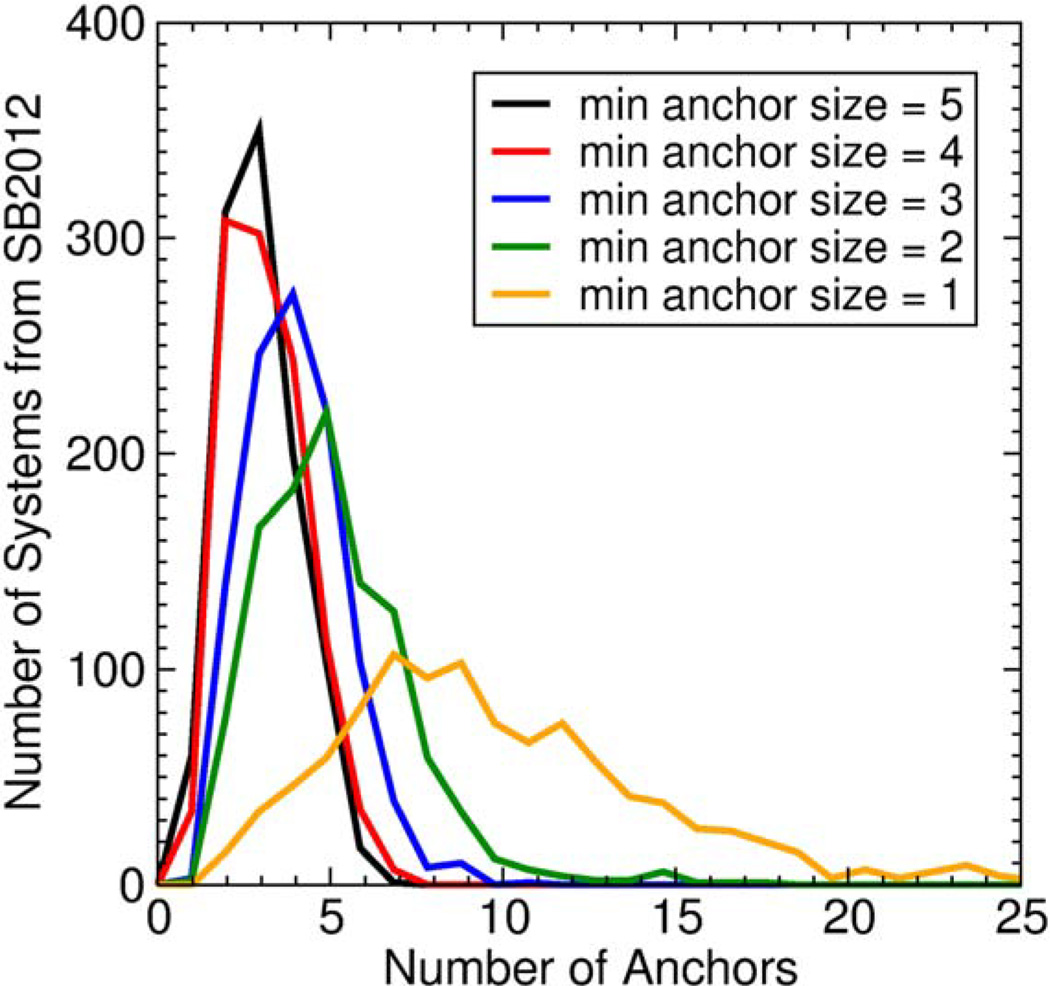
In the case of limiting the number of anchors, the DOCK anchor-and-grow search algorithm is proficient enough that it may find the correct binding orientation of a ligand beginning from any one anchor. For example, if a molecule contains five anchors that fit the user-defined criteria, the search algorithm might find a correct pose five times – once for each anchor – but it would require five times the computation cost as using a single anchor. Thus, one potential strategy to reduce calculation time without reducing the success rate would be to use fewer anchors. Figure 6 plots histograms of the number of anchors identified in each ligand from the SB2012 test set using different minimum number of atoms per anchor criteria. The plot can be interpreted as follows: if the user specifies a minimum anchor size of “5” (five or more heavy atoms), then the majority of ligands in the test set will have three anchors, but a large fraction will have four or more anchors (Figure 6, black line). To test the effects of different anchor strategies, we performed docking using the standard FLX protocol and varying the number of anchors between 1, 3, or all anchors that fit the criteria of five or greater heavy atoms. The results are summarized in Table 2.
Figure 6.
Histograms of the number of anchors for five different thresholds for minimum anchor size. These data are from the SB2012 test set, N=1043.
Table 2.
Comparison of different numbers of anchors to assess pose reproduction in SB2012.
| # Anchors |
# RB | Subset Size |
Success # (%) |
Sampling Failure # (%) |
Scoring Failure # (%) |
Avg. Time (min) |
|
|---|---|---|---|---|---|---|---|
| A | 1a | <8 | 542 | 433 (80.0) | 47 (8.7) | 62 (11.4) | 0.4 |
| B | 1 | 8–15 | 403 | 226 (56.1) | 122 (30.3) | 55 (13.6) | 1.5 |
| C | 1 | >15 | 98 | 37 (37.8) | 53 (54.1) | 8 (8.2) | 7.0 |
| D | 1 | all | 1043 | 696 (66.7) | 222 (21.3) | 125 (12.0) | 1.5 |
| E | 3b | <8 | 542 | 453 (83.6) | 13 (2.4) | 76 (14.0) | 1.1 |
| F | 3 | 8–15 | 403 | 260 (64.5) | 57 (14.1) | 86 (21.3) | 4.1 |
| G | 3 | >15 | 98 | 49 (50.0) | 31 (31.6) | 18 (18.4) | 17.2 |
| H | 3 | all | 1043 | 762 (73.1) | 101 (9.7) | 180 (17.3) | 3.8 |
| I | allc | <8 | 542 | 453 (83.6) | 13 (2.4) | 76 (14.0) | 1.2 |
| J | all | 8–15 | 403 | 264 (65.5) | 53 (13.2) | 86 (21.3) | 5.2 |
| K | all | >15 | 98 | 48 (49.0) | 29 (29.6) | 21 (21.4) | 22.8 |
| L | all | all | 1043 | 765 (73.3) | 95 (9.1) | 183 (17.5) | 4.8 |
Only 1 anchor is chosen.
No more than 3 anchors that have five or more heavy atoms are chosen. If there is no anchor bigger than five atoms, then the biggest with the most branching points is chosen.
All rigid segments with five atoms or greater are used as anchors.
As shown in Table 2, performing flexible ligand docking beginning from only 1 anchor (largest rigid anchor) results in a reasonable docking success rate of 66.7% across all systems (Table 2, row D). A significant improvement, however, is observed when the number of anchors is increased to 3 (73.1%, row H) or when all anchors are used (73.3%, row L). Importantly, limiting sampling from all to 3 anchors decreases average run time from 4.8 min to 3.8 min, a substantial speed up without negatively impacting docking success. It is noteworthy that the largest increase in success rate when using multiple anchors is observed in the >15 subset. Specifically, using 3 anchors (row G) or all anchors (row K) yields ~50% docking success on the >15 rotatable bond subset, compared to 1 anchor at 37.8% (row C). The difference is primarily due a decrease in sampling failures from ~54% when using 1 anchor (row C) to ~30–32% for all anchors or 3 anchors, respectively (rows G, K). Overall, for large-scale virtual screening applications where timing may be important, the current results suggest limiting orienting to only the 3 largest rigid segments may be appropriate (further discussed in Enrichment Results).
Minimization Options
The primary goals of energy minimization in DOCK are to resolve minor clashes between the ligand and protein, and to relax the conformation of the ligand into a reasonable internal geometry. Larger conformational sampling involving rigid-body ligand rotations and translations, as well as ligand dihedral angle rotations, should be tasked to the anchor-and-grow engine in DOCK, not the minimizer. Here, we present improvements to the simplex minimizer (torsion pre-minimizer and RMSD restraint minimizer) that have been designed to address larger-than-expected ligand movements observed during energy minimization.
Torsion Pre-minimizer
By default, the simplex minimizer in DOCK treats the six rigid degrees of freedom (3 translational, 3 rotational) and each torsion equally. However, it is possible that a newly grown torsion coarsely sampled at the angles specified in the DOCK flex definition file[56] could introduce clashes with the receptor. One strategy for such cases is to resolve the clash by minor adjustment of the newly-added torsion angle without altering any other part of the molecule that already has a favorable interaction with the protein (e.g. from previous rounds of minimization). However, if all ligand degrees of freedom are treated equally, the minimizer may prefer the entire ligand pose to be rigidly translated or rotated away from the source of the clash, rather than adjusting the torsion. To resolve this problem, we implemented a torsion pre-minimizer that disregards all degrees of freedom except the dihedral angle of the newly added fragment. Then, a small number of iterations of energy minimization are performed (typically 10 to 50), prior to a full energy minimization with the standard degrees of freedom. Because there is only one degree of freedom in the torsion pre-minimization stage, a small number of iterations still provides the desired result.
RMSD Restraint Minimizer
Our initial testing demonstrated that the torsion pre-minimization cannot always satisfactorily resolve clashes between a ligand and receptor. In such cases, the ligand may still undergo large translations or rotations to resolve a clash when full energy minimization is initiated. To address this, we also implemented an RMSD restraint minimizer into DOCK. The goal of the RMSD restraint minimizer is resolution of clashes between the ligand and receptor, or internal clashes, by moving the position of the ligand heavy atoms as little as possible, thus preserving the pose of the current branch of anchor-and-grow. This is achieved through Hooke’s law:
| (Eq. 1) |
Where Erestraint is the potential energy in the RMSD restraint, and k is a user-specified spring constant in kcal mol−1 Å−2. Simplex minimization energy evaluation then attempts to minimize the sum:
| (Eq. 2) |
Where Einteraction is the interaction energy between the ligand and receptor, and Einternal is the internal energy of the ligand. The default value of the parameter k is 10.0 kcal mol−1 Å−2. As an example, for a 0.5 Å RMSD change during minimization, Erestraint=10(0.5)2=2.5 kcal mol−1, which represents a minor penalty for a small conformational change. For a larger conformational change of 2.0 Å, Erestraint=10(2.0)2=40 kcal mol−1. In most cases, such a large penalty would offset any favorable intermolecular score achieved and disallow such moves during minimization, effectively preventing the minimizer from behaving as a sampling engine.
Comparison of Minimization Methods
To test the efficacy of these two new minimization methods, we performed pose reproduction experiments over the SB2012 test set using the standard flexible ligand protocol (FLX, see Methods and Details), as well as two additional protocols. Fixed-anchor docking (FAD) does not use the orient function of DOCK, rather growth is initiated from crystallographic anchor positions. Ligand-atom spheres (LAS) is the same as FLX, with the exception that only the positions of heavy atoms from the crystallographic ligand are used as spheres and, subsequently, for orienting the anchors. Thus, in LAS, the anchor will have a high propensity for finding the correct anchor position, but will also find several decoy positions, making the problem slightly more challenging than FAD. The results are summarized in Table 3.
Table 3.
Torsion pre-minimizer and RMSD restrained minimization statistics in SB2012.
| Minim. Method |
Docking Protocol |
Success # (%) |
Sampling Failure # (%) |
Scoring Failure # (%) |
Avg. Time (min) |
|
|---|---|---|---|---|---|---|
| A | pre-mina | FAD | 854 (81.9) | 34 (3.3) | 155 (14.9) | 2.9 |
| B | pre-min | LAS | 757 (72.6) | 83 (8.0) | 203 (19.5) | 4.8 |
| C | pre-min | FLX | 737 (70.7) | 100 (9.6) | 206 (19.8) | 4.9 |
| D | restraintb | FAD | 909 (87.2) | 17 (1.6) | 117 (11.2) | 2.3 |
| E | restraint | LAS | 780 (74.8) | 71 (6.8) | 192 (18.4) | 4.2 |
| F | restraint | FLX | 721 (69.1) | 128 (12.3) | 194 (18.6) | 4.3 |
| G | standardc | FAD | 854 (81.9) | 30 (2.9) | 159 (15.2) | 3.0 |
| H | standard | LAS | 774 (74.2) | 75 (7.2) | 194 (18.6) | 4.7 |
| I | standard | FLX | 765 (73.3) | 95 (9.1) | 183 (17.5) | 4.8 |
pre-min refers to the torsion pre-minimizer with 50 iterations.
restraint refers to the RMSD restraint minimizer with a spring constant of 10.0 kcal mol−1 Å−2.
standard refers to the standard minimization protocol.
With regards to the FAD protocol, the RMSD restraint method decreases sampling failures by ~1.5% (Table 3, row D vs. rows A, G) and scoring failures by ~4.0% (row D vs. rows A, G), improving overall success rate to 87.2% (row D) compared to the torsion pre-minimizer alone or the standard protocol, both at 81.9% (rows A, G). In addition, use of the restraint reduces average run times from ~3 minutes (torsion pre-minimizer or standard) to 2.3 minutes (RMSD restraint minimizer). A similar but less drastic trend is observed for the LAS docking protocol, where the RMSD restraint minimizer has a slightly higher docking success at 74.8% (Table 3, row E) when compared to the torsion pre-minimizer at 72.6% (row B) or the standard protocol at 74.2% (row H). Interestingly however, the RMSD restraint minimizer and the torsion pre-minimizer both negatively impact success when using the FLX docking protocol. The standard protocol yields 73.3% docking success (Table 3, row I), whereas the torsion pre-minimizer and the RMSD restraint minimizer reduce success rates to 70.7% (row C) and 69.1% (row F), respectively. The majority of the decrease in success can be attributed to an increase in sampling failures. These data taken together indicate that these modified minimization tools should be used only in scenarios where the anchor position is known with high confidence.
Despite this somewhat surprising negative result with respect to the FLX docking protocol, examination of the growth steps for docking with and without the RMSD restraint shows that the algorithm produces the expected behavior. As one representative example from a large sampling ensemble, Figure 7 shows that, when the RMSD restraint minimizer is used, ligand growth proceeds in a highly methodical fashion and without sudden changes in RMSD. Here, the largest rigid ligand segment begins 2.6 Å RMSD from the crystal pose (Figure 7a), and in subsequent steps, the partially grown molecule moves incrementally (Figure 7b→k), ultimately converging on a position 0.4 Å RMSD from the crystal pose. An overlay of all stages of growth (Figure 7l) highlights the incremental movement, wherein the average deviation between any two frames was 0.4 Å. In contrast to Figure 7, Figure 8 shows that, without the RMSD restraint, a more sporadic ligand motion occurs. Following initial minimized anchor placement 3.2 Å RMSD from the crystal pose (Figure 8a), the ligand scaffold appears to jump during growth, particularly between frames a→b (1.8 Å RMSD) and c→d (1.4 Å RMSD). Growth continues until the complete conformer is obtained (Figure 8k), which, in this example is also 0.4 Å away. An overlay of all stages of growth (Figure 8l) shows that the motion of the ligand is less predictable and more sporadic when compared to Figure 7 with the RMSD restraint applied. Although the final conformer is a docking success, the sporadic behavior is less desirable because energy minimization calculations performed in previous growth stages are essentially wasted when the ligand undergoes large, drastic movements arising from clashes with receptor atoms. The more slow and deliberate sampling motion shown in Figure 7 would be preferable, provided that equal or better success rates can be achieved, which may be possible through more accurate initial anchor placements.
Figure 7.
Stages of growth for flexible docking (FLX) of 2-deoxy-2,3-dehydro-N-acetyl-neuraminic acid ligand to neuraminidase receptor (PDB code 1F8B)[75] using the RMSD restraint minimizer with spring constant 10.0 kcal mol−1 Å−2. Crystal structure pose shown as gray sticks, and partially grown conformers shown as orange sticks. (a) Initial minimized ligand anchor position (beginning 2.6 Å RMSD from crystal). (b→k) Incremental growth stages for all rigid segments (ultimately 0.4 Å RMSD from crystal). (l) Overlay of all stages of growth colored sequentially in white to black gradient.
Figure 8.
Stages of growth for flexible docking (FLX) of 2-deoxy-2,3-dehydro-N-acetyl-neuraminic acid ligand to neuraminidase receptor (PDB code 1F8B)[75] absent the RMSD restraint minimizer. Crystal structure pose shown as light gray sticks, and partially grown conformers shown as magenta sticks. (a) Initial minimized ligand anchor position (beginning 3.2 Å RMSD from crystal). (b→k) Incremental growth stages for all rigid segments (ultimately 0.4 Å RMSD from crystal). (l) Overlay of all stages of growth colored sequentially in white to black gradient.
An additional, potentially useful application of the RMSD restraint minimizer is to perform hydrogen optimization. Typically, X-ray crystal structures from the PDB do not contain hydrogen atoms, and as a result, they must be added to the model prior to docking. By using a modified torsion library and restraints on the ligand heavy atoms, it is now possible to sample only the polar ligand hydrogen atoms without altering heavy atom positions. When subsequently minimized on the receptor coordinates, with the RMSD restraint enabled, this feature is particularly useful in generating a reference ligand energy footprint for use with the footprint scoring function, as described in Balius et al.[76] and further discussed below.
Ligand Scoring and Assessment
Several recent additions to the DOCK codebase represent significant advances to the manner by which ligand poses are evaluated and scored. First, the new footprint similarity scoring function[72,76] is an effective method for characterizing and quantifying ligand binding on a per-residue basis (Figure S1, Table S1). The method has been shown to be a powerful approach for applications in virtual screening[21,22] and in understanding important drug targets.[77,78] Second, a new symmetry-corrected RMSD function[79] based on the Hungarian algorithm[80,81] has been implemented to accurately evaluate ligand binding pose RMSD in cases of symmetric molecules or partially symmetric substructures. The new RMSD metric contributes to the accurate reporting of DOCK successes and failures (Figure S2), and it has also been co-opted to serve as a scoring function or applied to molecular dynamics simulations.[79] And finally, a database filter has been implemented to calculate various ligand properties including molecular weight, number of rotatable bonds, and net charge. These enhancements are described in further detail in the Supporting Information.
Docking Forensic Tools
Careful analysis of the docking process and outcomes, particularly the docking failures, is a means of discovering problems in or limitations of the DOCK codebase, input parameters, or structure preparation protocols. In response to problematic behaviors observed by the developers or reported by users to the DOCK-Fans mailing list (http://dock.compbio.ucsf.edu/), a set of forensic tools were implemented which enabled in many cases discovery of the root causes underlying aberrant sampling and scoring failures in DOCK. Two primary forensics tools added to the program include growth trees (Figure S3) and verbose statistics (Figure S4). The impact of these new features and various other bug fixes are described in detail in the Supporting Information.
Results II: Current Docking Performance
Pose Reproduction Results
The first set of key experiments, pose reproduction calculations, were performed using the standard FLX protocol with the SGE scoring function over the SB2012 test set and three different versions of DOCK including the current version (6.7), and two previous release versions (4.0.2 and 5.4). Success rates, sampling failure rates, scoring failure rates, and run times for sets of ligands of varying size are summarized in Table 4. Note that the input parameter options have evolved between versions of DOCK, but every effort was made to use the same or comparable parameters in terms of sampling, scoring, and energy minimization, to match the standard protocol of version 6.7. In addition, earlier versions of DOCK (4.0.2 and 5.4) do not contain the Hungarian-algorithm based symmetry correction method for RMSD calculations.[79] Thus, to provide a fair comparison all docking outcomes using earlier versions were re-evaluated in DOCK 6.7 to obtain symmetry-corrected RMSDs without any additional sampling or scoring.
Table 4.
Comparison of different DOCK versions using the SB2012 test set (N=1043) to assess pose reproduction.
| DOCK Version |
# RB | Subset Size |
Success # (%) |
Sampling Failure # (%) |
Scoring Failure # (%) |
Avg. Time (min) |
|
|---|---|---|---|---|---|---|---|
| A | 4.0.2 | <8 | 542 | 360 (66.4) | 66 (12.2) | 116 (21.4) | 0.1 |
| B | 4.0.2 | 8–15 | 403 | 166 (41.2) | 118 (29.3) | 119 (29.5) | 0.5 |
| C | 4.0.2 | >15 | 98 | 11 (11.2) | 67 (68.4) | 20 (20.4) | 1.6 |
| D | 4.0.2 | all | 1043 | 537 (51.4) | 251 (24.1) | 255 (24.4) | 0.4 |
| E | 5.4 | <8 | 542 | 436 (80.4) | 26 (4.8) | 80 (14.8) | 12.0 |
| F | 5.4 | 8–15 | 403 | 220 (54.6) | 70 (17.4) | 113 (28.0) | 86.8 |
| G | 5.4 | >15 | 98 | 24 (24.5) | 46 (46.9) | 28 (28.6) | 376.6 |
| H | 5.4 | all | 1043 | 680 (65.2) | 142 (13.6) | 221 (21.1) | 74.0 |
| I | 6.7 | <8 | 542 | 453 (83.6) | 13 (2.4) | 76 (14.0) | 1.2 |
| J | 6.7 | 8–15 | 403 | 264 (65.5) | 53 (13.2) | 86 (21.3) | 5.2 |
| K | 6.7 | >15 | 98 | 48 (49.0) | 29 (29.6) | 21 (21.4) | 22.8 |
| L | 6.7 | all | 1043 | 765 (73.3) | 95 (9.1) | 183 (17.5) | 4.8 |
Overall, DOCK 6.7 significantly outperforms the two previous DOCK versions in terms of increased docking success and decreased sampling and scoring failures across all ligand subsets. Specifically, DOCK 6.7 successfully reproduces 765/1043, or 73.3% of experimental crystal poses (Table 4, row L), compared to DOCK 5.4 at 680/1043 or 65.2% (row H) and DOCK 4.0.2 at 537/1043 or 51.4% (row D). The increased success rate can be attributed to dramatic decreases in both sampling failures and scoring failures across all ligand subsets. Notably, the largest success rate increase is observed for the set of ligands with >15 rotatable bonds, which follows the series 49.0% (DOCK 6.7, row K) > 24.5% (DOCK 5.4, row G) > 11.2% (DOCK 4.0.2, row C). Average docking time in DOCK 4.0.2 is considerably faster than subsequent versions, but the speed is marred by lower success rates. It is important to emphasize that the correlation of low success rates with faster run times often indicates undersampling caused by early termination of docking calculations. In practice, a balance between high success and reasonable run times is desirable. DOCK 6.7 (4.8 min/ligand, row L) performs, on average, much faster than DOCK 5.4 (74.0 min/ligand, row H). As noted in Methods and Details, however, run times computed for various versions of DOCK depend on many factors including specific compiler options and features. Thus, the slower performance of DOCK 5.4 could likely be improved through various compiler optimization strategies, although it lacks the code enhancements, new features, and bug fixes that account for the speed of DOCK 6.7.
The principal challenge remaining in pose reproduction is a high instance in scoring failures. Progress toward improved scoring, however, is observed with each new release of DOCK, as scoring failure rates follow the series 24.4% (DOCK 4.0.2, row D) > 21.1% (DOCK 5.4, row H) > 17.5% (DOCK 6.7, Table 4, row K). More advanced scoring methods, including the footprint similarity function discussed above, will continue to diminish scoring failure rates.[72,76] Another challenge that remains is the sampling failure rate, particularly for very large ligands of >15 rotatable bonds. DOCK 6.7 performs reasonably well with relatively low sampling failure rates for the <8 rotatable bonds subset (2.4%, row I) and the 8–15 rotatable bond subset (13.2%, row J), which are most representative of drug-like ligands. And, although the sampling failure rate for DOCK 6.7 on the >15 rotatable bonds subset is still relatively high (29.6%, row K), it significantly outperforms DOCK 5.4 (46.9%, row G) and DOCK 4.0.2 (68.4%, row C).
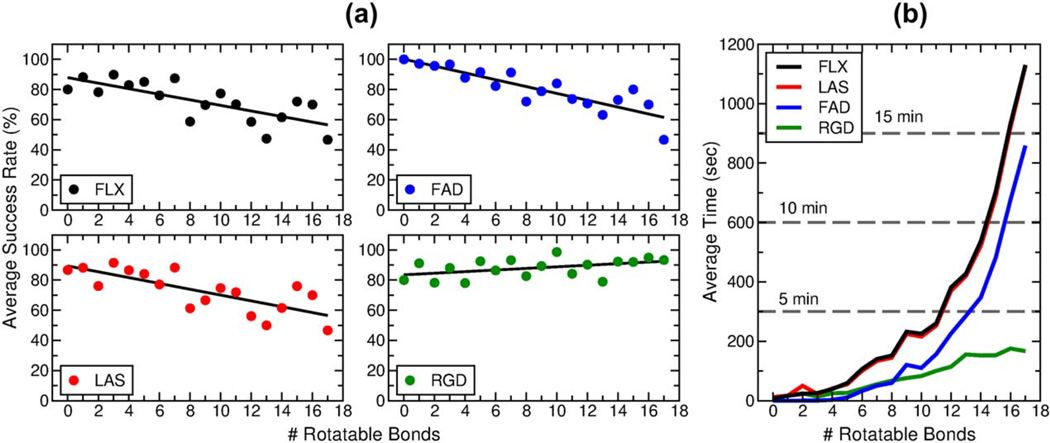
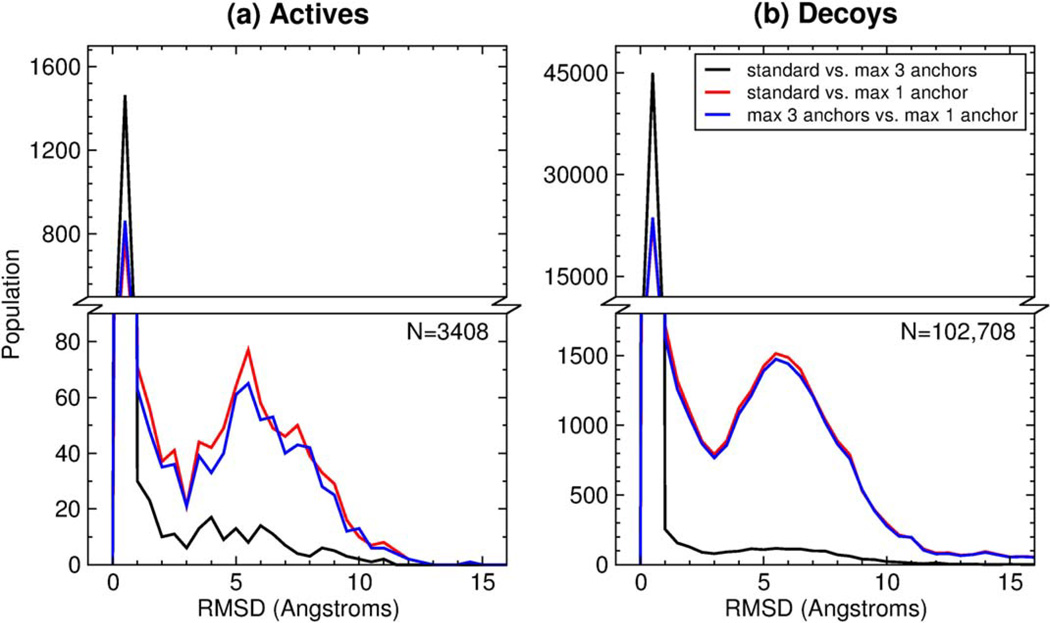
It is informative to closely examine the pose reproduction outcomes as a function of ligand flexibility. Figure 9 shows the variation of docking success rates and timings with the number of ligand rotatable bonds for four different docking protocols using DOCK 6.7. The standard FLX, LAS, and FAD protocols (described above) are accompanied here by the rigid docking protocol (RGD), wherein ligands are docked to the receptor rigidly constrained in the starting crystallographic conformation and without sampling any dihedral torsions. To compile this data, systems from the SB2012 test set were sub-divided into individual bins according to the number of ligand rotatable bonds. Bins for ligands with more than 17 rotatable bonds had fewer than ten members and thus were excluded from this plot. The FLX, LAS, and FAD protocols result in a decrease in docking success rate with increasing number of rotatable bonds, and the data fits show linear relationships with nearly identical slopes (Figure 9a, black, red, and blue circles). The RGD protocol, however, results in a slight increase in docking success rate with increasing number of rotatable bonds (Figure 9a, green circles), likely due to the fact that as ligand size increases, there are fewer feasible binding orientations without steric clashes.
Figure 9.
(a) Success rate as a function of the number of ligand rotatable bonds for the FLX (black), LAS (red), FAD (blue), and RGD (green) protocols over the SB2012 test set with DOCK 6.7. Colored circles represent the success rate for a particular bin, and black lines are the best-linear fit. (b) Average docking run time as a function of the number of ligand rotatable bonds for the FLX (black), LAS (red), FAD (blue), and RGD (green) protocols over the SB2012 test set. Subsets greater than 17 rotatable bonds were excluded from this analysis due to a small bin size of less than ten systems.
Each ligand rotatable bond adds a new degree of freedom to the standard six degrees of freedom (three translational, three rotational) leading to an exponential increase in the sampling search space. Figure 9b shows a corresponding exponential increase in docking run time as ligand flexibility increases for the FLX (black), LAS (red), and FAD (blue) protocols. On average, using the standard FLX protocol, ligands of 11 or fewer rotatable bonds complete docking in five minutes or less, and ligands of 14 or fewer rotatable bonds complete docking in ten minutes or less. The FAD protocol requires less time than the FLX or LAS protocols because anchor starting positions are restricted to the known crystallographic positions, thus fewer anchor starting orientations are used. The RGD protocol shows an approximately linear increase in docking run time as ligand flexibility increases, as expected, and the average docking run time for ligands of 17 or fewer rotatable bonds is three minutes or less. Taken together, the improvements made to DOCK 6.7 yield overall strong success rates and reasonable computation times over previous versions. The remainder of the manuscript focuses on DOCK version 6.7.
Cross-docking Results
The second key experiment, cross-docking, provides an additional metric for evaluating docking performance over a very large set of protein-ligand pairs. As described in Methods and Details, protein-ligand complexes from the SB2012 test set were divided into 24 cross-docking families (based on receptor identity) ranging in size from 6 to 59 systems. Prior to docking, systems in each family were aligned on backbone α-carbons. To illustrate the low variability in receptor backbone conformation, a representative overlay of 15 receptors from the EGFR family of proteins is shown in Figure 10. In this example, low backbone RMSDs of no greater than 0.6 Å and high backbone atom matches of approximately 80% or greater were achieved using PDB code 1M17[73] as the template. Cross-docking relies heavily on well-aligned protein structures, thus systems that did not satisfy the criteria described in Cross-docking Methods were removed from consideration. Ultimately, 541 total protein-ligand complexes were retained, constituting 17,457 total docking combinations across the 24 families.
Figure 10.
Representative alignment of a cross-docking family. Fifteen systems from the EGFR protein family were aligned on the backbone of PDB code 1M17.[73] Protein backbones are shown as gray ribbon, and bound ligands are shown as red (erlotinib-like), blue (lapatinib-like), green (ATP-like), or yellow (staurosporine-like) sticks. The conformational search and scoring space is shown as a magenta box.
Figure 11 shows cross-docking outcomes using the standard FLX protocol with the SGE scoring function plotted onto 24 separate heatmaps, sorted left-to-right, top-to-bottom by decreasing docking success rate. Full-size heatmaps for all families are also provided as Supporting Information (see Supporting Information Figures S5–S28). Analogous to the pose reproduction outcomes, cross-docking outcomes are categorized as success (blue), scoring failure (green), and sampling failure (red), with the addition of non-viable pairings (gray, see Methods and Details). In addition, Table 5 quantifies the results for each family including success and failure rates for both the entire matrix and for the diagonal alone, sorted by overall matrix success rate.
Figure 11.
Cross-docking outcomes for 24 families. Blue squares indicate docking success, green squares indicate scoring failure, red squares indicate sampling failure, and gray squares indicate a non-viable pairing. Panels are sorted left-to-right, top-to-bottom by overall docking success rate.
Table 5.
Cross-docking outcomes for protein families from SB2012 test set using DOCK 6.7.
| Success Rate (%) |
Sampling Failure Rate (%) |
Scoring Failure Rate (%) |
|||||
|---|---|---|---|---|---|---|---|
| Family Namea | # Sys |
Matrix | Diagonal | Matrix | Diagonal | Matrix | Diagonal |
| neuraminidase | 43 | 91.0 | 93.0 | 0.8 | 2.3 | 8.2 | 4.7 |
| cyclooxygenase | 6 | 88.5 | 100.0 | 7.7 | 0.0 | 3.8 | 0.0 |
| streptavidin | 8 | 82.8 | 87.5 | 1.7 | 0.0 | 15.5 | 12.5 |
| T4 lysozyme | 13 | 76.4 | 92.3 | 0.0 | 0.0 | 23.6 | 7.7 |
| estrogen receptor | 45 | 71.5 | 97.8 | 0.4 | 0.0 | 28.1 | 2.2 |
| HIV reverse transcriptase | 21 | 71.4 | 95.2 | 10.4 | 0.0 | 18.3 | 4.8 |
| HMG CoA reductase | 13 | 69.7 | 76.9 | 19.1 | 15.4 | 11.2 | 7.7 |
| β-trypsin | 29 | 64.8 | 72.4 | 6.5 | 3.4 | 28.7 | 24.1 |
| factor Xa | 41 | 60.6 | 95.1 | 9.8 | 0.0 | 29.6 | 4.9 |
| tyrosine phosphatase | 16 | 60.2 | 87.5 | 21.1 | 6.3 | 18.7 | 6.3 |
| ribonuclease T1 | 7 | 56.3 | 85.7 | 29.2 | 0.0 | 14.6 | 14.3 |
| trypsin | 46 | 53.9 | 63.0 | 24.7 | 15.2 | 21.4 | 21.7 |
| ribonuclease A | 14 | 45.0 | 50.0 | 18.3 | 14.3 | 36.6 | 35.7 |
| thymidylate synthase | 12 | 41.0 | 58.3 | 24.3 | 0.0 | 34.7 | 41.7 |
| carboxypeptidase A | 8 | 40.3 | 37.5 | 30.6 | 25.0 | 29.0 | 37.5 |
| thrombin | 37 | 38.5 | 70.3 | 24.5 | 10.8 | 37.1 | 18.9 |
| thermolysin | 26 | 37.6 | 46.2 | 26.5 | 15.4 | 35.9 | 38.5 |
| acetylcholinesterase | 18 | 30.6 | 50.0 | 21.1 | 5.6 | 48.3 | 44.4 |
| HIV protease | 59 | 26.8 | 57.6 | 48.0 | 37.3 | 25.2 | 5.1 |
| lysozyme | 14 | 26.0 | 35.7 | 13.3 | 0.0 | 60.7 | 64.3 |
| EGFR | 15 | 26.0 | 46.7 | 31.5 | 13.3 | 42.5 | 40.0 |
| IGF-1R | 7 | 21.9 | 71.4 | 25.0 | 0.0 | 53.1 | 28.6 |
| matrix metalloproteinase | 14 | 19.0 | 7.1 | 43.6 | 28.6 | 37.4 | 64.3 |
| carbonic anhydrase | 29 | 18.7 | 34.5 | 7.8 | 3.4 | 73.5 | 62.1 |
| average | - | 50.8 | 67.2 | 18.6 | 8.2 | 30.7 | 24.7 |
Families sorted by matrix success rate.
Overall, the diagonal success rate – derived from docking cognate receptor-ligand pairs – is consistently higher than the matrix success rate for any given family (Table 5). The average diagonal success rate is 67.2%, and the average matrix success rate is 50.8%. This observation follows our expectations, and it demonstrates that DOCK performs optimally when searching for cognate receptor-ligand binding poses. Interestingly, a linear fit between the diagonal and matrix success rates for each family yields a relatively high correlation (r2=0.704). This could suggest that the general characteristics of a certain protein family (including binding site shape, hydrophobic character, existence of cofactors or ions, etc.) play a larger role in docking success than specific differences between members in a family (e.g. point mutations, small binding site conformational differences, etc.). In addition, it should be noted that congeneric series of ligands will typically bind a given protein family in the same orientation and conformation, and will be inherently more amenable to cross-docking experiments. Table 5 also indicates that the diagonals show significantly lower sampling failure rates (average 8.2%) than the average for the total matrix (average 18.6%), suggesting that receptor flexibility plays an important role. The result serves as a reminder that careful consideration of the receptor structure used in a virtual screening capacity is paramount, and whenever possible, a ligand-bound receptor conformation should be used for virtual screening with DOCK 6.7.
To discuss some specific cross-docking outcomes in detail while maintaining brevity, the results below are arranged into four main categories: (1) families for which DOCK performs well, (2) families for which DOCK performs poorly, (3) DOCK performance in the presence of multiple receptor forms (e.g. isoforms or conformational states), and (4) DOCK performance in the presence of receptor mutations. Some specific families will not be discussed in detail, but are still presented here for completeness. These results may be useful as a reference to the reader to identify specific PDB structures that are amenable to docking or virtual screening experiments, or to quickly assess how DOCK 6.7 may perform on a given family in terms of success and failure rates.
Case 1: Excellent DOCK Performance
Focusing first on families for which DOCK performs well, we find neuraminidase (N=43 systems, 91.0% success rate), streptavidin (N=8 systems, 82.8% success rate), and model system T4 lysozyme (N=13 systems, 76.4% success rate) among the top of the list (Figure 11, Table 5). Docking successes dominate the cross-docking outcomes across all three of these families and, importantly, the incidence of sampling failures or non-viable pairings is very low. Cross-docking to the neuraminidase family results in 0.8% sampling failure rate, the streptavidin family 1.7% sampling failure rate, and the T4 lysozyme family 0% sampling failure rate. Low sampling failure rate and few non-viable pairings indicate that the receptors as a group are amenable to accommodating all ligands in the group.
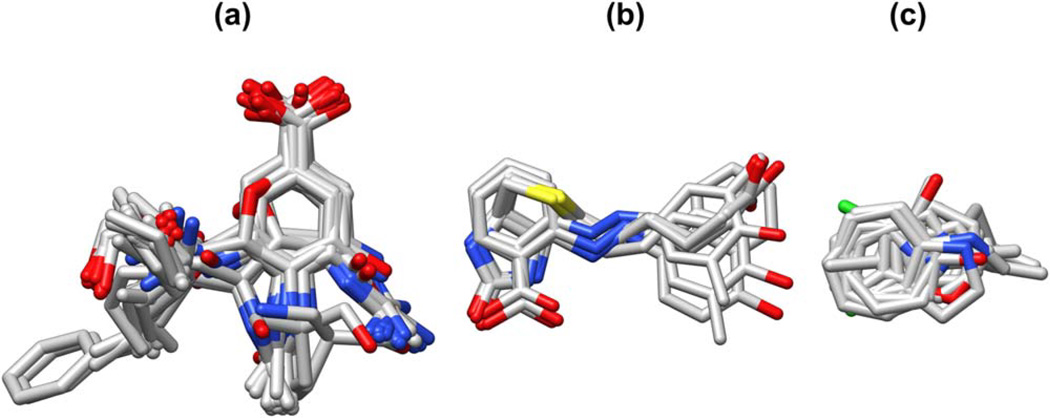
One key contributor to high success rate is the high degree of similarity among ligands in each of these three families (Figure 12). For example, the 43 neuraminidase ligands include the anti-influenza agent zanamivir[82] and only other close analogs of sialic acid. As a congeneric series (Figure 12a), all ligands in the group tend to be compatible with all receptors in the group (see Supporting Information Figure S19). Of the eight streptavidin ligands (Figure 12b), five are biotin or a biotin derivative [83–86] and three are azobenzene analogs.[87] The five biotin-like molecules tend to dock successfully into both the original biotin-containing receptors as well as the azobenzene-containing receptors. The azobenzene analogs, however, only dock well into the azobenzene-containing receptors, and attempts to dock them into the biotin-containing receptors yield several instances of non-viable pairings and other failures (see Supporting Information Figure S22). High success rates are also observed in T4 lysozyme. Ligands in this set are mostly small and inflexible (Figure 12c), a characteristic conducive to successful docking. Furthermore, ligands in this set are all benzene or pyrrole analogs,[71,88–90] thus receptors in the set are amenable to docking compounds that are similar to their cognate ligand (see Supporting Information Figure S23). In addition to these three examples, several other families representing important drug targets exhibited promising overall success rates including cyclooxygenase (88.5%), estrogen receptor (71.5%), HIV reverse transcriptase (71.4%), and HMG CoA reductase (69.7%), as summarized in Table 5.
Figure 12.
Overlay of ligands from (a) neuraminidase (N=43), (b) streptavidin (N=8), and (c) T4 lysozyme (N=13) cross-docking families. Hydrogens and protein atoms hidden for clarity. For complete PDB code lists, see Supporting Information Figures S19, S22, and S23.
Case 2: Poor DOCK Performance
Representative families for which DOCK yields poor cross-docking outcomes include carbonic anhydrase (N=29 systems, 18.7% success rate), matrix metalloproteinase (N=14 systems, 19.0% success rate), and lysozyme (N=14 systems, 26.0% success rate). In the case of carbonic anhydrase and lysozyme, the docking outcomes are dominated by scoring failures (73.5% for carbonic anhydrase; 60.7% for lysozyme). In the case of matrix metalloproteinase, the scoring and sampling failures are roughly equal (37.4% and 43.6%, respectively). One likely reason for such high scoring failure rates is the presence of metal ions in the binding site, as is the case for carbonic anhydrase and matrix metalloproteinase. Visual inspection of docking outcomes to carbonic anhydrase reveals that the zinc ion in the active site frequently coordinates the incorrect atom from the sulfonamide group[91] (present in all 29 ligands from this family), or coordinates atoms in the sulfonyl group[92] (present in 14 ligands from this family), all of which are highly polarized. The strong electrostatic interaction yields a large favorable DOCK score, but incorrect coordination of the zinc ion can prevent the remainder of the ligand from adopting the correct binding site orientation, resulting in many docking failures. Similar trends can be observed with coordination of the active site zinc ion in matrix metalloproteinase. Non-polarizable force fields in molecular mechanics rely on static partial charges, which is a known limitation, but an interesting area of active research.[93–95] Alternatively, use of different metal ion parameters based on coordination state has been shown to improve docking outcomes,[14,96] although this was not pursued here.
Another likely reason for the high incidence of scoring failures among this group is failure to account for water-mediated interactions or solvation effects, which are important to both carbonic anhydrase[97,98] and lysozyme.[99] In our current standard DOCK receptor preparation protocol, all waters are removed. Accounting for explicit water molecules as a component of the receptor, for example, can help improve accurate pose reproduction predictions. Prior tests in our laboratory demonstrated that accounting for crystallographic waters in the carbonic anhydrase family improves overall cross-docking success rate by 14% and success rate for cognate ligand-receptor pairs by 17%.[47] Ideally, not all water molecules that co-crystallize with the receptor need be retained, instead a limited number of hand-picked key water molecules should be chosen and modeled. Ongoing work in our group and by others [38,100–107] is seeking to address this challenge. Note that these are just a few representative examples of the known limitations to docking, and factors such as entropic effects, induced fit effects, and others also present challenges to successful docking.
Case 3: Multiple Receptor Isoforms or Conformations
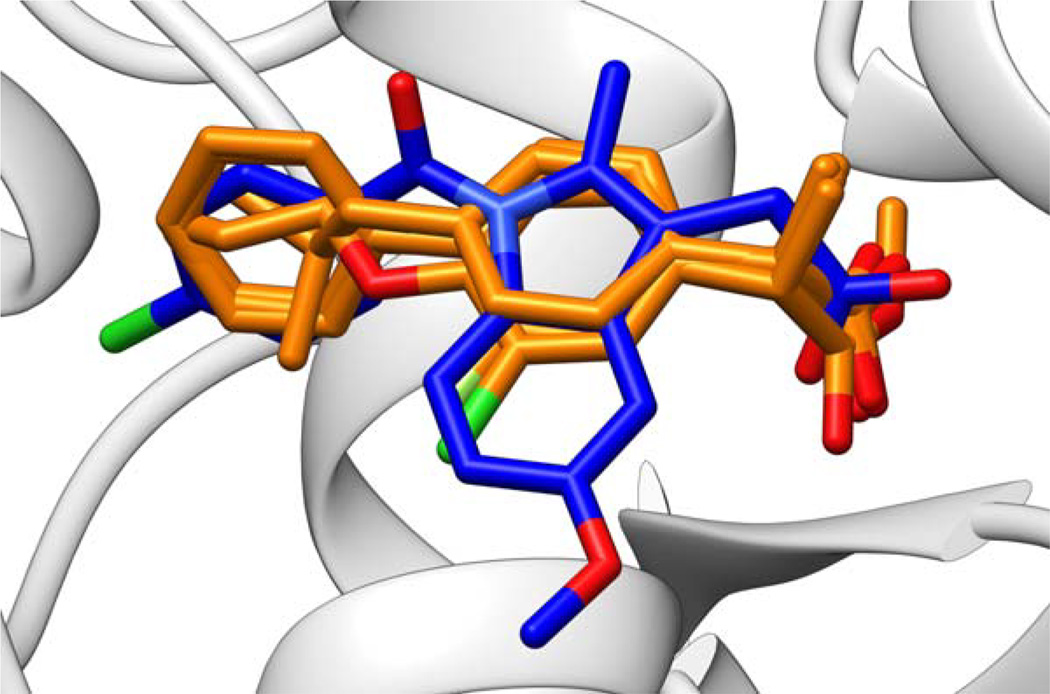
In some specific cases, we see interesting cross-docking outcomes that can be directly correlated with what is known about the structure and biology for a given family. Focusing on one particular example, the cyclooxygenase family (N=6 systems) performed quite well overall (Table 5, 88.5% success rate). The worst-performing ligand from the set originated from PDB code 4COX.[108] The ligand, indomethacin, successfully docked into its cognate receptor, but was incompatible with all five other receptors (see Supporting Information Figure S9). The other five ligands are ibuprofen and ibuprofen analogs,[109] and for the 4COX receptor, only one of these successfully docked. Overlay of the ligands from the cross-docking family highlight the differences in size and functionality for the ibuprofen and ibuprofen analogs vs. indomethacin (Figure 13). Of these six receptors, only 4COX is cyclooxygenase isoform 2 (COX-2), whereas the other five are cyclooxygenase isoform 1 (COX-1). The observed specificity of indomethacin for COX-2 and of ibuprofen analogs for COX-1 is consistent with the experimental specificity for inhibitors of the two cyclooxygenase isoforms.[108]
Figure 13.
Overlay of ligands from the cyclooxygenase family. Five ibuprofen or ibuprofen-analog ligands, specific to COX-1, shown in orange; one indomethacin ligand, specific to COX-2, shown in blue. Hydrogen atoms hidden for clarity. For complete PDB code lists, see Supporting Information Figure S9.
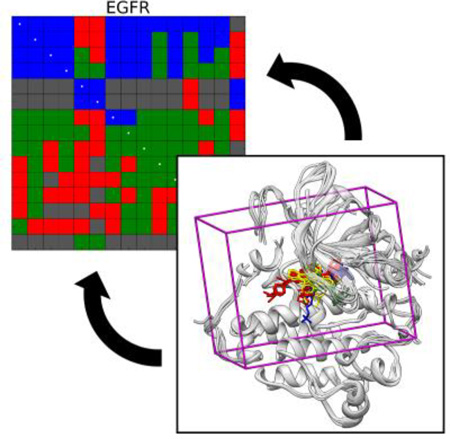
Focusing on a larger family, EGFR (N=15), we identify interesting trends across the various receptor conformations. Within this family, the receptors adopt local variations in conformation due in part to the identity of the co-crystallized ligands (see Figure 10; also discussed in Supporting Information, Figure S1), which fall into several classes: erlotinib-like (1M17, 2ITO, 2ITP, 2ITT, 2ITY, 2J6M),[73,110] lapatinib-like (1XKK, 2RGP, 3BEL),[111–113] ATP-like (2EB3, 2ITV, 2ITX),[110,114] and staurosporine-like (2ITQ, 2ITU, 2ITW).[110] The receptors also adopt more global conformational variations between the 3 inactive forms (those bound to lapatinib-like inhibitors) and 12 active forms (all others, Figure 14), defined principally by the conformation of the EGFR A-loop.[110] Overall, cross-docking performance to this family is poor at 26.0% matrix success rate (Table 5). However, if we consider sub-matrices defined by ligand classes, we observe improved outcomes. For example, cross-docking the lapatinib-like ligands into their cognate (inactive) receptors forms a 3×3 sub-matrix with an improved success rate of 66.7% and 33.3% scoring failures (Figure 14d). Similarly, cross-docking the erlotinib-like ligands into their cognate (active) receptors forms a 6×6 sub-matrix with an improved success rate of 55.6%, 41.7% scoring failures, and 2.8% sampling failures (Figure 14b). In both cases the individual success rates are higher than the overall success rate for this family, and there are no non-viable pairings within either sub-matrix. Docking the ATP-like ligands into their cognate receptors (a 3×3 sub-matrix), however, produces no successful docking outcomes (Figure 14c), likely owing to the fact that highly polarized and charged molecules are difficult to dock successfully (as discussed in the previous section). In preparation for cross-docking, we found that the three lapatinib-like ligands were incompatible with 11 of the 12 active forms of the receptor (Figure 14, also see Supporting Information Figure S10). In addition, the erlotinib-like, ATP-like, and staurosporine-like ligands were dominated by sampling failures (66.7%) and non-viable pairings (22.2%) when docked into the three inactive forms of the receptor. Both of these observations are consistent with the experimental specificity of lapatinib for inactive forms of EGFR and the other ligand classes for active forms of EGFR.[110] This highlights the importance of choosing the appropriate receptor form prior to a virtual screen, depending on the desired outcome.
Figure 14.
Heatmap of EGFR cross-docking outcomes. Ligand PDB codes and number of rotatable bonds listed on y-axis, receptor PDB codes listed on x-axis. Structures are grouped by the identity of the bound ligand: (a) staurosporine-like (N=3), (b) erlotinib-like (N=6), (c) ATP-like (N=3), and (d) lapatinib like (N=3). The first 12 columns are all active forms, the last three columns are inactive forms.
Case 4: Receptor Mutations
Finally, we examine cross-docking outcomes to the viral targets HIV reverse transcriptase (N=21 systems) and HIV protease (N=59 systems). Reverse transcriptase has a high matrix success rate (71.4%, Table 5), but with a high incidence of non-viable pairings (200 non-viable out of 441 pairings). Six mutations known to confer resistance to various non-nucleoside inhibitors are represented in this data set including L100I, K101E, K103N, E138K, Y181C, and Y188C.[115–119] Mutation at residue 138 causes resistance to the drug rilpivirine,[120] mutation at residues 103 or 188 causes resistance to the drugs efavirenz and nevirapine,[117,121] and mutation at residues 100, 101, or 181 causes resistance to all three.[119,122–125] One might reasonably expect docking outcomes to mirror these trends, i.e. lower docking success should be observed for a compound docked to receptors containing mutations that confer resistance (termed here “resistant pairings”), compared to wild type receptors. Counterintuitively, we observed an increase in docking success (84.2%) among resistant pairings, which is higher than the HIV reverse transcriptase family as a whole (71.4%, see Supporting Information Figure S14). Interestingly, upon closer examination, we identified that 54.3% of the possible resistant pairings were in fact classified as non-viable, which is somewhat higher than the number of non-viable elements from the entire matrix (45.4%). Taken together, these observations could suggest that mutations that confer drug resistance in HIV reverse transcriptase might be more easily identified from non-viable pairings than from cross-docking outcomes, although additional studies need to be performed. However, we would emphasize that these analyses are no substitution for experimental measurements or rigorous free energy of binding calculations.
Turning our attention to HIV protease, we observed a low matrix success rate (26.3%, Table 5) with a high incidence of sampling failures (48.7%). Unique to this family, less than half of the receptors (24 out of 59) contain the wild type sequence. Despite the frequency of mutations, the global backbone conformation is highly conserved. Following alignment using PDB code 1A8K as the reference,[126] the average α-carbon RMSD is 0.64 Å. To measure the effect of receptor mutations in DOCK, we consider the drug darunavir with both wild type and mutant protease. Darunavir effectively binds wild type protease; however V32I, I47V, I50V, V82F, or I84V mutations cause resistance to the drug and attenuated binding.[127–130] In this cross-docking family, the ligand darunavir is represented 7 times, and receptors with at least one of the above mutations are represented 16 times, providing 7×16=112 combinations of darunavir and darunavir-resistant receptors. The remaining 43 receptors provide 7×43=301 combinations of darunavir and wild type or non-resistant mutant receptors. In practice, we find essentially no difference in docking darunavir to any form of protease. When docking to darunavir-resistant receptors, we find 9 out of 112 non-viable pairings, and a 43.7% success rate. When docking to all other receptors, we find 19 out of 301 non-viable pairings, and a 41.5% success rate. For this particular family, the high incidence of sampling failures seems to be more dependent on ligand size and complexity rather than any specific mutation. As shown previously in Figure 9 and presented in Table 4, docking success rates decrease and sampling failure rates increase with the number of ligand rotatable bonds. The protease family is no exception to this trend, with an r2=0.620 correlation (see Supporting Information, Figure S29) between the number of ligand rotatable bonds and the number of sampling failures. With the current protocols, it is important to elaborate that docking may only be able to discriminate between strong and weak binders in cases where mutations directly interact with the ligand; docking should not be expected to be sensitive to mutations with indirect effects, including solvation or allosteric changes, for example. More rigorous calculations (e.g. free energy perturbation) or experiment would be required to determine quantitative free energy of binding differences between wild-type and mutant forms.
It should also be noted that the computational experiments described here all employ a rigid receptor. Ongoing work in our laboratory, as well as other groups, includes evaluating methods to effectively model receptor conformational variability or ensembles during docking.[34,131–135] Under ideal docking conditions, success should be independent of the actual starting receptor structure. In practice, however, the starting conditions (including different forms, mutations, variable induced fit effects, etc.) play a large role in influencing the outcome. The close correspondence (r2=0.704) between the matrix and diagonal success rates, however, suggest that, to a limited degree and for certain systems, docking outcomes can be independent of starting receptor conformation. Importantly, the family-based alignments shown here derived from the SB2012 test set provide a reasonable starting point for future development efforts that aim to improve matrix success rates by accounting for receptor flexibility and other factors.
Enrichment Results
The final key set of experiments, enrichment, serve to estimate how DOCK would perform in a real-world virtual screening application. Prior enrichment studies from our laboratory, reported in Brozell et al.,[29] evaluated the standard FLX docking protocol with the SGE scoring function across all 40 systems in the Directory of Useful Decoys (DUD) data set.[48] In the current work, we evaluate a smaller subset of systems, namely thrombin (PDB code 1YPE),[64] IGF-1R (PDB code 2OJ9),[65] HIV protease (PDB code 1XL2),[63] HMG CoA reductase (PDB code 3CCW),[66] and estrogen receptor (PDB code 1SJ0)[62] for the updated, significantly larger DUD-Enhanced (DUD-E) data set.[61] Enrichment outcomes under three different conditions were evaluated: (1) the standard FLX protocol, which orients all ligand anchors, (2) the FLX protocol with a maximum of 3 anchors, or (3) the FLX protocol with a maximum of 1 anchor. The primary objective of these tests was to determine if reducing the number of anchor seed positions during anchor-and-grow would yield similar success but with faster timings. Following standard practices, enrichment outcomes are plotted as ROC curves (Figure 15). The accompanying Table 6 presents the results numerically, including area under the curve (AUC) at 1% and 100% of the database screened.
Figure 15.
Enrichment curves using standard all anchor (red), max 3 anchors (blue), or max 1 anchor (green) protocols with the DUD-E database for five different receptors. For comparison, random results are shown (black) against the diagonal (dashed line). Note that the standard all anchors (red) and max 3 anchors (blue) curves are essentially indistinguishable due to high overlap.
Table 6.
Enrichment outcomes using DOCK 6.7, three different protocols, and five systems from DUD-E.
| 1% of the database | 100% of the database | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Targeta | Protocolb | # Mol | # Act | # Dec | AUCc | # Mol | # Act | # Dec | AUC |
| (Max) | (100.00) | (10000.00) | |||||||
| (Random) | (0.50) | (5000.00) | |||||||
| all anchors | 116 | 159 | 8.74 | 8663.35 | |||||
| THRB | 3 anchors | 275 | 112 | 163 | 8.92 | 27409 | 461 | 26948 | 8662.42 |
| 1 anchors | 95 | 180 | 8.41 | 8123.13 | |||||
| all anchors | 13 | 82 | 4.69 | 7375.85 | |||||
| IGF-1R | 3 anchors | 95 | 14 | 81 | 4.66 | 9439 | 148 | 9291 | 7391.56 |
| 1 anchors | 12 | 83 | 4.70 | 7409.44 | |||||
| all anchors | 70 | 293 | 6.94 | 7171.28 | |||||
| HIVPR | 3 anchors | 363 | 68 | 295 | 6.83 | 36224 | 536 | 35688 | 7131.71 |
| 1 anchors | 71 | 292 | 6.80 | 7096.54 | |||||
| all anchors | 34 | 56 | 9.10 | 6867.27 | |||||
| HMDH | 3 anchors | 90 | 35 | 55 | 8.99 | 8913 | 170 | 8743 | 6845.12 |
| 1 anchors | 35 | 55 | 9.96 | 6779.79 | |||||
| all anchors | 57 | 154 | 8.19 | 6009.76 | |||||
| ESR1 | 3 anchors | 211 | 55 | 156 | 8.40 | 21046 | 383 | 20663 | 5981.02 |
| 1 anchors | 64 | 147 | 8.57 | 6139.25 | |||||
THRB=thrombin, PDB code 1YPE; IGF-1R=insulin-like growth factor 1 receptor, PDB code 2OJ9; HIVPR=human immunodeficiency virus protease, PDB code 1XL2; HMDH=3-hydroxy-3-methyl-glutaryl-coenzyme A reductase, PDB code 3CCW; ESR1=estrogen receptor 1, PDB code 1SJ0.
all anchors=standard FLX protocol using all anchors; 3 anchors=FLX protocol using a maximum of 3 anchors; 1 anchors=FLX protocol using only 1 anchor.
AUC=area under the curve. All AUC values, including Max AUC and Random AUC, are multiplied by 104 for readability.
As a general observation for these five systems, the ROC curves (Figure 15) and the AUC values (Table 6) demonstrate good enrichment for all three DOCK protocols. As expected, scrambling the results by randomly choosing actives and decoys generates curves that lie on the ROC diagonals (black lines). In contrast, ROC curves derived using standard (red), max 3 anchors (blue), or max 1 anchor (green) protocols are all significantly above the diagonals. More importantly, the very steep upward slopes indicate significant early enrichment, which is the primary goal of virtual screening. For these five active / decoy data sets, 1% of the database examined is equivalent to 90 to 363 compounds which, depending on resources, would be a reasonable range for acquisition and experimental testing. Under the present conditions, enrichment at 1% equates to between 12 and 116 active compounds depending on the system and protocol (Table 6). In practice, however, enrichment results are highly system-dependent and subject not only to the specific computational protocols employed, but also the experimental assays used to evaluate activity. For more lengthy discussion, including additional examples of using DOCK to perform enrichment analysis, interested readers may consult Brozell et al.[29]
Relative to the standard all anchor protocol, the max 1 anchor protocol is considerably faster (240% speedup) and the max 3 anchor protocol is marginally faster (23% speedup). As the faster protocol does not lead to noteworthy changes in ROC curve shape (Figure 15) or AUC values (Table 6), with the possible exception of thrombin, a possible conclusion is that 1 anchor is in fact adequate for virtual screening. However, since the pose prediction tests discussed earlier (Table 2) clearly showed a significant decrease in success using only 1 anchor (66.7%) relative to max 3 anchors (73.1%) or all anchors (73.3%), this conclusion could be enigmatic. To explore this question in greater detail, we visually examined poses for top-scoring molecules obtained with the standard protocol (red), and compared those with the corresponding poses obtained by either the max 3 anchors (blue) or max 1 anchor (green) protocols. Figure 16 shows the top five pose overlays from each of the five enrichment studies.
Figure 16.
Ligand pose comparisons using standard (red), max 3 anchors (blue), or max 1 anchor (green) protocols. Molecules shown were selected based on the top 5 scored results obtained with the standard protocol. Results for which the max 1 anchor protocol yields substantially different poses from the other two protocols are highlighted with a gray box. Results for which all protocols yield discernibly different poses are highlighted with a dashed gray box. To facilitate colored visualization for identical poses, molecules were offset 0.2 Å in the x- or y-direction.
For these examples, the standard (Figure 16, red) and max 3 anchors (Figure 16, blue) poses visually show identical or substantially well-overlaid geometries in 22 out of 25 cases suggesting, on average, use of either protocol will yield consistent binding poses. In contrast, the 1 anchor protocol yields a similar pose in only 15 out of 25 cases. The ten cases wherein the 1 anchor protocol yielded a pose dissimilar from the standard protocol are highlighted by gray boxes (Figure 16, all gray boxes). In three instances, all three protocols yield discernibly different poses (Figure 16, dashed gray boxes). Importantly, the alternative poses determined by the 1-anchor or 3-anchor protocols always yielded less favorable energy scores than the poses found using the standard protocol, highlighting the improved sampling attained by using more anchors per molecule. Further, the trends observed visually from this small subset of compounds are reflective of the results as a whole, as shown in Figure 17, which plots symmetry-corrected RMSD histograms using all of the final ligand poses generated divided into active (N=3048) and decoy (N=102,708) groups. Despite the majority of poses being identical or nearly identical, as shown by the large population peaks near 0 Å RMSD, the standard vs. max 3 anchor results (Figure 17, black) show a much greater correspondence than do the standard vs. max 1 anchor results (Figure 17 red). As expected, RMSD differences for max 3 anchors vs. max 1 anchor (Figure 17 blue) are similar to standard vs. max 1 anchor. The significant second population peak in the 3 to 10 Å RMSD range in both histograms indicates a large number of predicted active and decoy poses that are geometrically different when using the max 1 anchor protocol.
Figure 17.
Differences in geometry for the sets of all (a) active (N=3408 ligands) and (b) decoy (N=102,708 ligands) poses sampled using different DOCK protocols across the five enrichment systems evaluated by symmetry-corrected RMSD. Standard vs. max 3 anchors (black), standard vs. max 1 anchor (red), and max 3 anchors vs. max 1 anchor (blue) are shown. Bin size is 0.5 Å RMSD.
It is important to emphasize that enrichment, per se, does not require that docked poses be correct in terms of their experimental binding geometry, only that actives be rank ordered earlier than decoys for any given protocol. The observation that the max 1 anchor protocol in many cases yielded different binding geometries yet similar ROC and AUC results highlights challenges associated with devising good enrichment tests. Reasonable explanations for the similarity of ROC and AUC results could be related to the alternative poses being nearly isoenergetic, or alternatively, scored less favorably by an approximately constant amount which, in both cases, would lead to similar stratification in terms of rank-ordering. Moreover, Ferrari et al.[136] examined the effect of varying van der Waals parameters on enrichment results. In this case, different van der Waals coefficients appeared to affect the actives more so than the decoys, ultimately leading to worse overall enrichment. In any event, these observations are likely not unique to DOCK and should be explored further. A key point from the present studies is that docking accuracy should be assessed in multiple ways (e.g. pose reproduction, cross-docking, and enrichment). Use of any one metric alone is not sufficient.
Summary and Future Perspectives
As the seminal docking program in continual development since the early 1980s, DOCK has evolved through major algorithmic additions, updates, and rewrites, and with considerable community input due to its widespread use. The primary goals of this current manuscript were to (1) highlight the impact of new and updated algorithms most recently implemented into the DOCK codebase, and (2) assess the performance of the most current release (version 6.7) for pose reproduction, cross-docking, and enrichment.
Regarding the impact of new features and algorithms, a modified internal energy function (Figure 5, Table 1) improves overall success rates by removing unphysical ligand geometries from the population early during growth. New input parameters have provided users more control over the number and identity of anchors that are used for docking (Figure 6, Table 2), which may lead to faster, more efficient docking under the present anchor-and-grow strategy. The new minimization options (Figures 7–8, Table 3) have proven useful in optimizing hydrogen positions and limiting the impact of clashes during sampling, and the RMSD restrained minimizer has strong potential to further increase docking performance provided more accurate initial anchor placement. Finally, the footprint similarity score provides an effective new way to assess and score ligand binding (Figure S1, Table S1), and the symmetry-corrected RMSD function removes many false sampling failures and contributes to the accurate reporting of docking success (Figure S2). These improvements and other DOCK developments were made possible with the introduction of user-friendly forensic tools including growth trees and expanded verbose statistics (Figures 7–8, S3–S4).
Currently, DOCK 6.7 is able to successfully reproduce 73.3% of crystallographic ligand poses to within 2.0 Å RMSD over a large test set of 1043 protein-ligand complexes. This marks an 8.1 to 21.9% improvement over previous versions, largely due to the substantial decrease in sampling failures going from DOCK 4.0.2 (24.1%) → 5.4 (13.6%) → 6.7 (9.1%). The present balance between docking performance and run time (approximately 4.8 min per ligand) is acceptable given the current standards and availability of high-performance computing. The variation in success rates, failures, and run time (Figure 9) as a function of the number of ligand rotatable bonds is useful in gauging sampling and tractability for screening very flexible small molecule libraries. Additional tests exploring cross-docking performance for 541 proteins arranged into 24 families (Figure 11, Table 5) reveal both strengths and limitations of DOCK, and provide a valuable starting point for future work aimed at improving success rates for particularly challenging groups (Figures 12–14). For example, four out of five of the worst performing families in terms of diagonal success rate have metal ions in the binding site (Table 5). On the other hand, the relatively high correlation between matrix and diagonal success rates (r2=0.704) suggests that, particularly for certain families, docking outcomes can be independent of the initial X-ray structure employed. Enrichment studies also demonstrate the ability of DOCK to rank order known active compounds seeded into a large group of decoys (Figure 15, Table 6), a capability that has proven successful in recent real-world applications.[21,22] The appropriate parameters to employ for virtual screening is an important consideration, and future studies that further investigate the balance between performance vs. time would be worthwhile (Figures 16–17).
Additionally, several new features not discussed in this manuscript are in development and planned for future releases of DOCK. Among several new scoring functions, a Boltzmann-weighted function is in development that employs multiple receptor conformations and / or homologs simultaneously. A pharmacophore matching scoring function has been developed[137] to account for hydrophobic, hydrogen bonding, charged, aromatic, and non-aromatic ring features in on-the-fly docking. And, a volume matching scoring function has been implemented that scores ligands based on their spatial overlap to a reference compound. A major update to the DOCK descriptor scoring function has been made that allows different functions to be combined in the form:
| (Eq 3.) |
Where C1…CN are user-input coefficients, and F1…FN are user-chosen scoring functions such as the grid energy score, footprint similarity score, pharmacophore matching score, volume matching score, and chemical fingerprint, among others. This combined approach is highly customizable and can be leveraged to improve scoring accuracy for diverse types of ligands and receptors. In addition to new scoring functions, new ligand sampling functions are also in development. Principally among them is a new de novo design strategy which co-opts the existing anchor-and-grow methodology to assemble new molecules from fragments of pre-existing molecules.
In summary, this manuscript highlights the size and scope of experiments including pose reproduction, cross-docking, and enrichment over systems with diverse structure and chemistry that can be performed using one standard system preparation protocol, one standard input parameter file, and one physics-based energy scoring function in DOCK. The steady progression of features and performance from version 4 to 5 to 6 is a testament to the importance of employing multiple metrics and large data sets for validation, to the advantages of an open source infrastructure, and to the benefits of a large community of users and developers, past and present. DOCK can be downloaded at http://dock.compbio.ucsf.edu/.
Supplementary Material
Acknowledgments
The authors thank Brian C. Fochtman and Yuchen Zhou for computational assistance. This research utilized computational facilities at the Ohio Supercomputer Center (SRB) and resources at the New York Center for Computational Sciences at Stony Brook University / Brookhaven National Laboratory which is supported by the U.S. Department of Energy under Contract No. DE-AC02-98CH10886 and by the State of New York. This work was funded in part by the Stony Brook University Office of the Vice President for Research, the New York State Office of Science Technology and Academic Research (NYSTAR), and NIH grants F32GM105400 (to WJA), F31CA134201 (to TEB), and R01GM083669 (to RCR).
Footnotes
Author Contributions: WJA, TEB, SM, and RCR conceived and designed the experiments. WJA, TEB, and SM performed the experiments and analysis. WJA, TEB, SM, SRB, DTM, and PTL contributed and implemented DOCK source code. DAC, IDK, and RCR provided mentoring and intellectual support. WJA authored the final version of the manuscript, TEB, SM, and RCR contributed to writing the manuscript. All authors read and reviewed the manuscript.
References
- 1.Kuntz ID. Science. 1992;257:1078–1082. doi: 10.1126/science.257.5073.1078. [DOI] [PubMed] [Google Scholar]
- 2.Shoichet BK. Nature. 2004;432:862–865. doi: 10.1038/nature03197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jorgensen WL. Science. 2004;303:1813–1818. doi: 10.1126/science.1096361. [DOI] [PubMed] [Google Scholar]
- 4.DesJarlais RL, Sheridan RP, Seibel GL, Dixon JS, Kuntz ID, Venkataraghavan R. J. Med. Chem. 1988;31:722–729. doi: 10.1021/jm00399a006. [DOI] [PubMed] [Google Scholar]
- 5.Kuntz ID, Blaney JM, Oatley SJ, Langridge R, Ferrin TE. J. Mol. Biol. 1982;161:269–288. doi: 10.1016/0022-2836(82)90153-x. [DOI] [PubMed] [Google Scholar]
- 6.Shoichet BK, Kuntz ID. J. Mol. Biol. 1991;221:327–346. doi: 10.1016/0022-2836(91)80222-g. [DOI] [PubMed] [Google Scholar]
- 7.Shoichet BK, Bodian DL, Kuntz ID. J. Comput. Chem. 1992;13:380–397. [Google Scholar]
- 8.Meng EC, Shoichet BK, Kuntz ID. J. Comput. Chem. 1992;13:505–524. [Google Scholar]
- 9.Goodford PJ. J. Med. Chem. 1985;28:849–857. doi: 10.1021/jm00145a002. [DOI] [PubMed] [Google Scholar]
- 10.Shoichet BK, Stroud RM, Santi DV, Kuntz ID, Perry KM. Science. 1993;259:1445–1450. doi: 10.1126/science.8451640. [DOI] [PubMed] [Google Scholar]
- 11.Coleman RG, Carchia M, Sterling T, Irwin JJ, Shoichet BK. PLoS One. 2013;8:e75992. doi: 10.1371/journal.pone.0075992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ewing TJA, Kuntz ID. J. Comput. Chem. 1997;18:1175–1189. [Google Scholar]
- 13.Ewing TJA, Makino S, Skillman AG, Kuntz ID. J. Comput.-Aided Mol. Des. 2001;15:411–428. doi: 10.1023/a:1011115820450. [DOI] [PubMed] [Google Scholar]
- 14.Moustakas DT, Lang PT, Pegg S, Pettersen E, Kuntz ID, Brooijmans N, Rizzo RC. J. Comput.-Aided Mol. Des. 2006;20:601–619. doi: 10.1007/s10822-006-9060-4. [DOI] [PubMed] [Google Scholar]
- 15.Zou X, Sun Y, Kuntz ID. J. Am. Chem. Soc. 1999;121:8033–8043. [Google Scholar]
- 16.Case DA, Cheatham TE, III, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 18.Lang PT, Brozell SR, Mukherjee S, Pettersen EF, Meng EC, Thomas V, Rizzo RC, Case DA, James TL, Kuntz ID. RNA. 2009;15:1219–1230. doi: 10.1261/rna.1563609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graves AP, Shivakumar DM, Boyce SE, Jacobson MP, Case DA, Shoichet BK. J. Mol. Biol. 2008;377:914–934. doi: 10.1016/j.jmb.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macke TJ, Case DA. ACS Symp. Ser. 1998;682:379–393. [Google Scholar]
- 21.Holden PM, Kaur H, Goyal R, Gochin M, Rizzo RC. Bioorg. Med. Chem. Lett. 2012;22:3011–3016. doi: 10.1016/j.bmcl.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berger WT, Ralph BP, Kaczocha M, Sun J, Balius TE, Rizzo RC, Haj-Dahmane S, Ojima I, Deutsch DG. PLoS One. 2012;7:e50968. doi: 10.1371/journal.pone.0050968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teotico DG, Babaoglu K, Rocklin GJ, Ferreira RS, Giannetti AM, Shoichet BK. Proc. Natl. Acad. Sci. U. S. A. 2009;106:7455–7460. doi: 10.1073/pnas.0813029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hermann JC, Marti-Arbona R, Fedorov AA, Fedorov E, Almo SC, Shoichet BK, Raushel FM. Nature. 2007;448:775–779. doi: 10.1038/nature05981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khare G, Kar R, Tyagi AK. PLoS One. 2011;6:e22441. doi: 10.1371/journal.pone.0022441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu Z-H, Chen L, Wu L, Liu S, Wang L, Zhang Z-Y. Bioorg. Med. Chem. Lett. 2011;21:4238–4242. doi: 10.1016/j.bmcl.2011.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He S, Li C, Liu Y, Lai L. J. Med. Chem. 2013;56:3296–3309. doi: 10.1021/jm301900x. [DOI] [PubMed] [Google Scholar]
- 28.Koseki Y, Kinjo T, Kobayashi M, Aoki S. Eur. J. Med. Chem. 2013;60:333–339. doi: 10.1016/j.ejmech.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 29.Brozell SR, Mukherjee S, Balius TE, Roe DR, Case DA, Rizzo RC. J. Comput.-Aided Mol. Des. 2012;26:749–773. doi: 10.1007/s10822-012-9565-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carlson HA, Dunbar JB. J. Chem. Inf. Model. 2011;51:2025–2026. doi: 10.1021/ci200398g. [DOI] [PubMed] [Google Scholar]
- 31.Damm-Ganamet KL, Smith RD, Dunbar JB, Stuckey JA, Carlson HA. J. Chem. Inf. Model. 2013;53:1853–1870. doi: 10.1021/ci400025f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunbar JB, Smith RD, Damm-Ganamet KL, Ahmed A, Esposito EX, Delproposto J, Chinnaswamy K, Kang Y-N, Kubish G, Gestwicki JE, Stuckey JA, Carlson HA. J. Chem. Inf. Model. 2013;53:1842–1852. doi: 10.1021/ci4000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuriev E, Ramsland PA. J. Mol. Recognit. 2013;26:215–239. doi: 10.1002/jmr.2266. [DOI] [PubMed] [Google Scholar]
- 34.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trott O, Olson AJ. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Proteins: Struct., Funct., Genet. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 37.Hartshorn MJ, Verdonk ML, Chessari G, Brewerton SC, Mooij WTM, Mortenson PN, Murray CW. J. Med. Chem. 2007;50:726–741. doi: 10.1021/jm061277y. [DOI] [PubMed] [Google Scholar]
- 38.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 39.Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. J. Med. Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 40.Spitzer R, Jain AN. J. Comput.-Aided Mol. Des. 2012;26:687–699. doi: 10.1007/s10822-011-9533-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rarey M, Kramer B, Lengauer T, Klebe G. J. Mol. Biol. 1996;261:470–489. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- 42.Abagyan R, Totrov M, Kuznetsov D. J. Comput. Chem. 1994;15:488–506. [Google Scholar]
- 43.McGann M. J. Chem. Inf. Model. 2011;51:578–596. doi: 10.1021/ci100436p. [DOI] [PubMed] [Google Scholar]
- 44.MOE. Montreal, Canada: Chemical Computing Group; 2009. http://www.chemcomp.com/, [Google Scholar]
- 45.Wu G, Robertson DH, Brooks CL, III, Vieth M. J. Comput. Chem. 2003;24:1549–1562. doi: 10.1002/jcc.10306. [DOI] [PubMed] [Google Scholar]
- 46.Zsoldos Z, Reid D, Simon A, Sadjad BS, Johnson AP. Curr. Protein Pept. Sci. 2006;7:421–435. doi: 10.2174/138920306778559412. [DOI] [PubMed] [Google Scholar]
- 47.Mukherjee S, Balius TE, Rizzo RC. J. Chem. Inf. Model. 2010;50:1986–2000. doi: 10.1021/ci1001982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang N, Shoichet BK, Irwin JJ. J. Med. Chem. 2006;49:6789–6801. doi: 10.1021/jm0608356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Wang B, Hayik S, Roitberg A, Seabra G, Kolossvary I, Wong KF, Paesani F, Vanicek J, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh M-J, Cui G, Roe DR, Mathews DH, Seetin MG, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. AMBER 11. San Francisco: University of California; 2010. http://ambermd.org/, [Google Scholar]
- 51.Gasteiger J, Marsili M. Tetrahedron. 1980;36:3219–3222. [Google Scholar]
- 52.Jakalian A, Bush BL, Jack DB, Bayly CI. J. Comput. Chem. 2000;21:132–146. [Google Scholar]
- 53.Jakalian A, Jack DB, Bayly CI. J. Comput. Chem. 2002;23:1623–1641. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 54.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Proteins: Struct., Funct., Bioinf. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tripos. St. Louis, MO: Tripos Mol2 File Format; 2009. http://www.tripos.com/, [Google Scholar]
- 56.Lang PT, Moustakas DT, Brozell SR, Carrascal N, Mukherjee S, Balius TE, Allen WJ, Holden PM, Pegg S, Raha K, Shivakumar D, Rizzo RC, Case DA, Shoichet BK, Kuntz ID. DOCK 6.6 User's Manual. 2013 http://dock.compbio.ucsf.edu/DOCK_6/dock6_manual.htm, [Google Scholar]
- 57.DMS. San Francisco, CA: UCSF Computer Graphics Laboratory; 2003. http://www.cgl.ucsf.edu/Overview/software.html. [Google Scholar]
- 58.Leach AR. Molecular Modelling: Principles and Applications. 2nd Ed. Chapter 5. Harlow, England: Prentice Hall; 2001. pp. 258–260. [Google Scholar]
- 59.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 60.Triballeau N, Acher F, Brabet I, Pin J-P, Bertrand H-O. J. Med. Chem. 2005;48:2534–2547. doi: 10.1021/jm049092j. [DOI] [PubMed] [Google Scholar]
- 61.Mysinger MM, Carchia M, Irwin JJ, Shoichet BK. J. Med. Chem. 2012;55:6582–6594. doi: 10.1021/jm300687e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim S, Wu JY, Birzin ET, Frisch K, Chan W, Pai L-Y, Yang YT, Mosley RT, Fitzgerald PMD, Sharma N, Dahllund J, Thorsell A-G, DiNinno F, Rohrer SP, Schaeffer JM, Hammond ML. J. Med. Chem. 2004;47:2171–2175. doi: 10.1021/jm034243o. [DOI] [PubMed] [Google Scholar]
- 63.Specker E, Boettcher J, Lilie H, Heine A, Schoop A, Mueller G, Griebenow N, Klebe G. Angew. Chem. Int. Ed. 2005;44:3140–3144. doi: 10.1002/anie.200462643. [DOI] [PubMed] [Google Scholar]
- 64.Fokkens J, Klebe G. Angew. Chem. Int. Ed. 2006;45:985–989. doi: 10.1002/anie.200502302. [DOI] [PubMed] [Google Scholar]
- 65.Velaparthi U, Wittman M, Liu P, Stoffan K, Zimmermann K, Sang X, Carboni J, Li A, Attar R, Gottardis M, Greer A, Chang CY, Jacobsen BL, Sack JS, Sun Y, Langley DR, Balasubramanian B, Vyas D. Bioorg. Med. Chem. Lett. 2007;17:2317–2321. doi: 10.1016/j.bmcl.2007.01.102. [DOI] [PubMed] [Google Scholar]
- 66.Sarver RW, Bills E, Bolton G, Bratton LD, Caspers NL, Dunbar JB, Harris MS, Hutchings RH, Kennedy RM, Larsen SD, Pavlovsky A, Pfefferkorn JA, Bainbridge G. J. Med. Chem. 2008;51:3804–3813. doi: 10.1021/jm7015057. [DOI] [PubMed] [Google Scholar]
- 67.Irwin JJ, Shoichet BK. J. Chem. Inf. Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. J. Chem. Inf. Model. 2012;52:1757–1768. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chambers CC, Cramer CJ, Truhlar DG. J. Phys. Chem. 1996;100:16385–16398. [Google Scholar]
- 70.Li J, Zhu T, Cramer CJ, Truhlar DG. J. Phys. Chem. A. 1998;102:1820–1831. [Google Scholar]
- 71.Wei BQ, Baase WA, Weaver LH, Matthews BW, Shoichet BK. J. Mol. Biol. 2002;322:339–355. doi: 10.1016/s0022-2836(02)00777-5. [DOI] [PubMed] [Google Scholar]
- 72.Balius TE, Allen WJ, Mukherjee S, Rizzo RC. J. Comput. Chem. 2013;34:1226–1240. doi: 10.1002/jcc.23245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stamos J, Sliwkowski MX, Eigenbrot C. J. Biol. Chem. 2002;277:46265–46272. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- 74.Grether U, Benardeau A, Benz J, Binggeli A, Blum D, Hilpert H, Kuhn B, Marki HP, Meyer M, Mohr P, Puntener K, Raab S, Ruf A, Schlatter D. ChemMedChem. 2009;4:951–956. doi: 10.1002/cmdc.200800425. [DOI] [PubMed] [Google Scholar]
- 75.Smith BJ, Colman PM, Von IM, Danylec B, Varghese JN. Protein Sci. 2001;10:689–696. doi: 10.1110/ps.41801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Balius TE, Mukherjee S, Rizzo RC. J. Comput. Chem. 2011;32:2273–2289. doi: 10.1002/jcc.21814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang Y, Rizzo RC. Biochemistry. 2012;51:2390–2406. doi: 10.1021/bi2016553. [DOI] [PubMed] [Google Scholar]
- 78.McGillick BE, Balius TE, Mukherjee S, Rizzo RC. Biochemistry. 2010;49:3575–3592. doi: 10.1021/bi901915g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Allen WJ, Rizzo RC. J. Chem. Inf. Model. 2014;54:518–529. doi: 10.1021/ci400534h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kuhn HW. Nav. Res. Logist. Q. 1955;2:83–97. [Google Scholar]
- 81.Munkres J. J. Soc. Indust. Appl. Math. 1957;5:32–38. [Google Scholar]
- 82.Varghese JN, Epa VC, Colman PM. Protein Sci. 1995;4:1081–1087. doi: 10.1002/pro.5560040606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weber PC, Ohlendorf DH, Wendoloski JJ, Salemme FR. Science. 1989;243:85–88. doi: 10.1126/science.2911722. [DOI] [PubMed] [Google Scholar]
- 84.Katz BA. J. Mol. Biol. 1997;274:776–800. doi: 10.1006/jmbi.1997.1444. [DOI] [PubMed] [Google Scholar]
- 85.Freitag S, Le Trong I, Chilkoti A, Klumb LA, Stayton PS, Stenkamp RE. J. Mol. Biol. 1998;279:211–221. doi: 10.1006/jmbi.1998.1735. [DOI] [PubMed] [Google Scholar]
- 86.Hyre DE, Le Trong I, Freitag S, Stenkamp RE, Stayton PS. Protein Sci. 2000;9:878–885. doi: 10.1110/ps.9.5.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Weber PC, Pantoliano MW, Simons DM, Salemme FR. J. Am. Chem. Soc. 1994;116:2717–2724. [Google Scholar]
- 88.Morton A, Matthews BW. Biochemistry. 1995;34:8576–8588. doi: 10.1021/bi00027a007. [DOI] [PubMed] [Google Scholar]
- 89.Graves AP, Brenk R, Shoichet BK. J. Med. Chem. 2005;48:3714–3728. doi: 10.1021/jm0491187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mobley DL, Graves AP, Chodera JD, McReynolds AC, Shoichet BK, Dill KA. J. Mol. Biol. 2007;371:1118–1134. doi: 10.1016/j.jmb.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chakravarty S, Kannan KK. J. Mol. Biol. 1994;243:298–309. doi: 10.1006/jmbi.1994.1655. [DOI] [PubMed] [Google Scholar]
- 92.Boriack-Sjodin PA, Zeitlin S, Chen H-H, Crenshaw L, Gross S, Dantanarayana A, Delgado P, May JA, Dean T, Christianson DW. Protein Sci. 1998;7:2483–2489. doi: 10.1002/pro.5560071201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ponder JW, Wu C, Ren P, Pande VS, Chodera JD, Schnieders MJ, Haque I, Mobley DL, Lambrecht DS, Di Stasio RA, Head-Gordon M, Clark GNI, Johnson ME, Head-Gordon T. J. Phys. Chem. B. 2010;114:2549–2564. doi: 10.1021/jp910674d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xiang JY, Ponder JW. J. Comput. Chem. 2013;34:739–749. doi: 10.1002/jcc.23190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu J, He X, Zhang JZH. J. Chem. Inf. Model. 2013;53:1306–1314. doi: 10.1021/ci400067c. [DOI] [PubMed] [Google Scholar]
- 96.Irwin JJ, Raushel FM, Shoichet BK. Biochemistry. 2005;44:12316–12328. doi: 10.1021/bi050801k. [DOI] [PubMed] [Google Scholar]
- 97.Snyder PW, Mecinovic J, Moustakas DT, Thomas SW, III, Harder M, Mack ET, Lockett MR, Heroux A, Sherman W, Whitesides GM. Proc. Natl. Acad. Sci. U. S. A. 2011;108:17889–17894. doi: 10.1073/pnas.1114107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mecinovic J, Snyder PW, Mirica KA, Bai S, Mack ET, Kwant RL, Moustakas DT, Heroux A, Whitesides GM. J. Am. Chem. Soc. 2011;133:14017–14026. doi: 10.1021/ja2045293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cheetham JC, Artymiuk PJ, Phillips DC. J. Mol. Biol. 1992;224:613–628. doi: 10.1016/0022-2836(92)90548-x. [DOI] [PubMed] [Google Scholar]
- 100.Forli S, Olson AJ. J. Med. Chem. 2012;55:623–638. doi: 10.1021/jm2005145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Huang N, Shoichet BK. J. Med. Chem. 2008;51:4862–4865. doi: 10.1021/jm8006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Young T, Abel R, Kim B, Berne BJ, Friesner RA. Proc. Natl. Acad. Sci. U. S. A. 2007;104:808–813. doi: 10.1073/pnas.0610202104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Abel R, Young T, Farid R, Berne BJ, Friesner RA. J. Am. Chem. Soc. 2008;130:2817–2831. doi: 10.1021/ja0771033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 105.Verdonk ML, Chessari G, Cole JC, Hartshorn MJ, Murray CW, Nissink JWM, Taylor RD, Taylor R. J. Med. Chem. 2005;48:6504–6515. doi: 10.1021/jm050543p. [DOI] [PubMed] [Google Scholar]
- 106.Lie MA, Thomsen R, Pedersen CNS, Schiott B, Christensen MH. J. Chem. Inf. Model. 2011;51:909–917. doi: 10.1021/ci100510m. [DOI] [PubMed] [Google Scholar]
- 107.Huggins DJ, Tidor B. Protein Eng., Des. Sel. 2011;24:777–789. doi: 10.1093/protein/gzr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kurumbail RG, Stevens AM, Gierse JK, McDonald JJ, Stegeman RA, Pak JY, Gildehaus D, Miyashiro JM, Penning TD, Seibert K, Isakson PC, Stallings WC. Nature. 1996;384:644–648. doi: 10.1038/384644a0. [DOI] [PubMed] [Google Scholar]
- 109.Selinsky BS, Gupta K, Sharkey CT, Loll PJ. Biochemistry. 2001;40:5172–5180. doi: 10.1021/bi010045s. [DOI] [PubMed] [Google Scholar]
- 110.Yun C-H, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, Eck MJ. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wood ER, Truesdale AT, McDonald OB, Yuan D, Hassell A, Dickerson SH, Ellis B, Pennisi C, Horne E, Lackey K, Alligood KJ, Rusnak DW, Gilmer TM, Shewchuk L. Cancer Res. 2004;64:6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- 112.Xu G, Searle LL, Hughes TV, Beck AK, Connolly PJ, Abad MC, Neeper MP, Struble GT, Springer BA, Emanuel SL, Gruninger RH, Pandey N, Adams M, Moreno-Mazza S, Fuentes-Pesquera AR, Middleton SA, Greenberger LM. Bioorg. Med. Chem. Lett. 2008;18:3495–3499. doi: 10.1016/j.bmcl.2008.05.024. [DOI] [PubMed] [Google Scholar]
- 113.Xu G, Abad MC, Connolly PJ, Neeper MP, Struble GT, Springer BA, Emanuel SL, Pandey N, Gruninger RH, Adams M, Moreno-Mazza S, Fuentes-Pesquera AR, Middleton SA. Bioorg. Med. Chem. Lett. 2008;18:4615–4619. doi: 10.1016/j.bmcl.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 114.Yoshikawa S, Kukimoto-Niino M, Parker L, Handa N, Terada T, Fujimoto T, Terazawa Y, Wakiyama M, Sato M, Sano S, Kobayashi T, Tanaka T, Chen L, Liu ZJ, Wang BC, Shirouzu M, Kawa S, Semba K, Yamamoto T, Yokoyama S. Oncogene. 2013;32:27–38. doi: 10.1038/onc.2012.21. [DOI] [PubMed] [Google Scholar]
- 115.Ren J, Nichols CE, Chamberlain PP, Weaver KL, Short SA, Stammers DK. J. Mol. Biol. 2004;336:569–578. doi: 10.1016/j.jmb.2003.12.055. [DOI] [PubMed] [Google Scholar]
- 116.Ren J, Nichols CE, Stamp A, Chamberlain PP, Ferris R, Weaver KL, Short SA, Stammers DK. FEBS J. 2006;273:3850–3860. doi: 10.1111/j.1742-4658.2006.05392.x. [DOI] [PubMed] [Google Scholar]
- 117.Ren J, Nichols CE, Chamberlain PP, Weaver KL, Short SA, Chan JH, Kleim J-P, Stammers DK. J. Med. Chem. 2007;50:2301–2309. doi: 10.1021/jm061117m. [DOI] [PubMed] [Google Scholar]
- 118.Ren J, Chamberlain PP, Stamp A, Short SA, Weaver KL, Romines KR, Hazen R, Freeman A, Ferris RG, Andrews CW, Boone L, Chan JH, Stammers DK. J. Med. Chem. 2008;51:5000–5008. doi: 10.1021/jm8004493. [DOI] [PubMed] [Google Scholar]
- 119.Das K, Bauman JD, Clark AD, Frenkel YV, Lewi PJ, Shatkin AJ, Hughes SH, Arnold E. Proc. Natl. Acad. Sci. U. S. A. 2008;105:1466–1471. doi: 10.1073/pnas.0711209105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Xu H-T, Colby-Germinario SP, Asahchop EL, Oliveira M, McCallum M, Schader SM, Han Y, Quan Y, Sarafianos SG, Wainberg MA. Antimicrob. Agents Chemother. 2013;57:3100–3109. doi: 10.1128/AAC.00348-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xu H-T, Oliveira M, Quan Y, Bar-Magen T, Wainberg MA. J. Antimicrob. Chemother. 2010;65:2291–2299. doi: 10.1093/jac/dkq338. [DOI] [PubMed] [Google Scholar]
- 122.Maga G, Amacker M, Ruel N, Hubscher U, Spadari S. J. Mol. Biol. 1997;274:738–747. doi: 10.1006/jmbi.1997.1427. [DOI] [PubMed] [Google Scholar]
- 123.Azijn H, Tirry I, Vingerhoets J, de Bethune M-P, Kraus G, Boven K, Jochmans D, Van Craenenbroeck E, Picchio G, Rimsky LT. Antimicrob. Agents Chemother. 2010;54:718–727. doi: 10.1128/AAC.00986-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Corbau R, Mori J, Phillips C, Fishburn L, Martin A, Mowbray C, Panton W, Smith-Burchnell C, Thornberry A, Ringrose H, Knochel T, Irving S, Westby M, Wood A, Perros M. Antimicrob. Agents Chemother. 2010;54:4451–4463. doi: 10.1128/AAC.01455-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Asahchop EL, Wainberg MA, Oliveira M, Xu H, Brenner BG, Moisi D, Ibanescu IR, Tremblay C. AIDS. 2013;27:879–887. doi: 10.1097/QAD.0b013e32835d9f6d. [DOI] [PubMed] [Google Scholar]
- 126.Weber IT, Wu J, Adomat J, Harrison RW, Kimmel AR, Wondrak EM, Louis JM. Eur. J. Biochem. 1997;249:523–530. doi: 10.1111/j.1432-1033.1997.00523.x. [DOI] [PubMed] [Google Scholar]
- 127.Delaugerre C, Pavie J, Palmer P, Ghosn J, Blanche S, Roudiere L, Dominguez S, Mortier, J.-M. Molina E, de Truchis P. AIDS. 2008;22:1809–1813. doi: 10.1097/QAD.0b013e328307f24a. [DOI] [PubMed] [Google Scholar]
- 128.Van Marck H, Dierynck I, Kraus G, Hallenberger S, Pattery T, Muyldermans G, Geeraert L, Borozdina L, Bonesteel R, Aston C, Shaw E, Chen Q, Martinez C, Koka V, Lee J, Chi E, de Bethune MP, Hertogs K. J. Virol. 2009;83:9512–9520. doi: 10.1128/JVI.00291-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rhee S-Y, Taylor J, Fessel WJ, Kaufman D, Towner W, Troia P, Ruane P, Hellinger J, Shirvani V, Zolopa A, Shafer RW. Antimicrob. Agents Chemother. 2010;54:4253–4261. doi: 10.1128/AAC.00574-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sterrantino G, Zaccarelli M, Colao G, Baldanti F, Giambenedetto S, Carli T, Maggiolo F, Zazzi M. Infection. 2012;40:311–318. doi: 10.1007/s15010-011-0237-y. [DOI] [PubMed] [Google Scholar]
- 131.Fischer M, Coleman RG, Fraser JS, Shoichet BK. Nat. Chem. 2014;6:575–583. doi: 10.1038/nchem.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Totrov M, Abagyan R. Curr. Opin. Struct. Biol. 2008;18:178–184. doi: 10.1016/j.sbi.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Durrant JD, McCammon JA. Curr. Opin. Pharmacol. 2010;10:770–774. doi: 10.1016/j.coph.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rueda M, Totrov M, Abagyan R. J. Chem. Inf. Model. 2012;52:2705–2714. doi: 10.1021/ci3001088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cosconati S, Marinelli L, Di Leva FS, La Pietra V, De Simone A, Mancini F, Andrisano V, Novellino E, Goodsell DS, Olson AJ. J. Chem. Inf. Model. 2012;52:2697–2704. doi: 10.1021/ci300390h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ferrari AM, Wei BQ, Costantino L, Shoichet BK. J. Med. Chem. 2004;47:5076–5084. doi: 10.1021/jm049756p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jiang L, Rizzo RC. J. Phys. Chem. B. 2015;119:1083–1102. doi: 10.1021/jp506555w. [PubMed: 25229837] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.