Abstract
Formation of the muscular layer of the heart, the myocardium, involves the medial movement of bilateral progenitor fields; driven primarily by shortening of the endoderm during foregut formation. Using a combination of time-lapse imaging, microsurgical perturbations and computational modeling, we show that the speed of the medial-ward movement of the myocardial progenitors is similar, but not identical to that of the adjacent endoderm. Further, the extracellular matrix microenvironment separating the two germ layers also moves with the myocardium, indicating that collective tissue motion and not cell migration drives tubular heart assembly. Importantly, as myocardial cells approach the midline, they perform distinct anterior-directed movements relative to the endoderm. Based on the analysis of microincision experiments and computational models, we propose two characteristic, autonomous morphogenetic activities within the early myocardium: 1) an active contraction of the medial portion of the heart field and 2) curling - the tendency of the unconstrained myocardial tissue to form a spherical surface with a concave ventral side. In the intact embryo, these deformations are constrained by the endoderm and the adjacent mesoderm, nevertheless the corresponding mechanical stresses contribute to the proper positioning of myocardial primordia.
Keywords: myocardium, endoderm, mechanics, modeling
Introduction
In amniotes, the endoderm plays an important mechanical and signaling role in early heart development. As cardiac precursors in the splanchnic mesoderm move from bilateral heart fields to the embryonic midline, they are in close contact with the endoderm and intervening extracellular matrix. Myocardial precursor cells become epithelialized beginning at Hamburger and Hamilton (1951)/HH Stages 7–8 (Linask, 1992), and the myocardial tissue primordia fuse anterior to the opening of the foregut – the anterior intestinal portal (AIP) at HH9-10. The foregut lining is formed concomitantly via shortening of the endoderm, and the walls of the foregut are formed from splanchnic mesoderm adjacent to the myocardium (Bellairs and Osmond, 2005). The physical association between the mesoderm and endoderm (connected by extracellular matrix), as well as the forming heart and foregut (connected transiently via a mesentery) indicates the need to study tissue-level mechanical interactions during heart and foregut formation.
In avians, several studies have suggested that myocardial progenitors actively propel the midline-directed movement of cardiogenic fields (DeHaan, 1963; Rosenquist and DeHaan, 1966; Wiens 1996). Currently the most widely accepted (textbook) mechanism that attempts to explain these movements is the migration of myocardial cells (Gilbert, 2006). This migration is envisioned as taking place relative to the underlying endoderm of the forming foregut and the associated extracellular matrix (ECM). Furthermore, timely closure (regression) of the anterior intestinal portal (AIP) is critical for the midline directed myocardial precursor movements (reviewed in Brand, 2003), as perturbation of AIP regression or removal of the foregut endoderm results in cardia bifida (DeHaan, 1959; Rosenquist, 1970; Gannon and Bader, 1995). Varner and Taber (2012) provided additional evidence for a primary role of endoderm shortening (contraction) in driving convergence of the heart fields to the midline, and co-movement of labeled endodermal and myocardial tissue was demonstrated. In this study, we sought to determine if shortening of the endoderm was sufficient to form a tubular heart, or whether myocardial progenitors actively participated in driving the fusion of myocardial progenitor fields at the midline.
We demonstrate that in avians, myocardial precursors do not migrate substantially relative to their ECM microenvironment as has been suggested. Instead, in agreement with the results of Varner and Taber (2012), endodermal shortening during foregut morphogenesis predominantly drives the medial-ward displacement of the myocardial cells to the midline. However, here we show that in addition to the role of the endoderm - as the myocardial progenitor fields are moving towards the midline - they autonomously exert mechanical stresses within the tissue. These forces give rise to at least two distinct active autonomous deformations and propel the anterior displacement of the myocardium relative to the endoderm. Thus, our imaging and microincision studies as well as our computational models indicate that both endodermal contraction and autonomous myocardial deformations contribute to heart tube assembly.
Materials and Methods
Quail embryo preparation
Fertile wild type quail (Coturnix coturnix japonica) eggs (Ozark Egg Co., Stover, MO) were incubated for varying periods of time (from 20 to 36 hours) at 37°C to reach stages HH6 to HH11 (Hamburger and Hamilton, 1951). Embryos were then isolated and cultured as in Cui et al., 2009 (modified early chick “EC” culture; Chapman et al., 2001).
Plasmid generation and transfection for myocardial progenitor labeling
Genomic DNA was prepared from three HH12 chick embryos using Tri Reagent (Sigma) according to the manufacturer’s instructions. Primer sequences 5′-AAAAAATTAATCAAGGCTATGACTGCTGGAGTG-3′ and 5′-AAAAAGCTAGCCAGAGGTGCTGGTGGTGCTG-3′ were used to PCR amplify the region between −931 and +56 of the Cardiac Myosin Light Chain 2 (CMLC2/my17) gene. To generate pCMLC2::EGFP plasmid, the CMLC2 promoter was subcloned into the pEGFP-C1 vector (Clontech) between AseI and NheI restriction sites in place of the CMV promoter. To generate pCMLC2::MitoRFP, the CMLC2 promoter was similarly subcloned into the pEGFP-N1 vector (Clontech) between AseI and NheI sites. Next, the EGFP ORF was replaced by a cassette consisting of a mitochondrial localization signal fused to RFP (MitoRFP) using XhoI and NheI restriction sites. The MitoRFP cassette was derived from pCoxIV-RFP (a generous gift from Dr. Rusty Lansford, University of Southern California and Children’s Hospital Los Angeles, CA).
In vivo fluorescent labeling approaches
Monoclonal antibodies directed against fibrillin-2 (JB3) and fibronectin (B3D6) ECM proteins (Rongish et al., 1998, Czirok et al., 2006, Aleksandrova et al., 2012; DSHB, Iowa City, IA) or a quail endothelial cell surface marker (QH1; Pardanaud et al., 1987; DSHB) were directly conjugated to AlexaFluor 488, 555 or 647 (Molecular Probes) according to the manufacturer’s instructions. The direct conjugates were injected into the lateral plate mesoderm in 5–40 nl boluses using a PLI-100 (Harvard Instruments) microinjector as described in Little and Drake, 2000. Microinjections were performed 30–60 minutes prior to the beginning of the image acquisition to allow for antibody diffusion and antigen binding. We found that short exposure of the ventral side of the embryo to the MitoTracker Green dye solution, a dye retained in mitochondria, causes its retention only in the endoderm, if endodermal integrity was not compromised during treatment. The in vivo endoderm-labeling MitoTracker® Green FM dye (Invitrogen) was diluted in DMSO to 1 mM concentration according to the manufacturer’s recommendations. The obtained stock solution was further dissolved to 500 nM in PBS. HH8 stage embryos, placed on paper rings as described above, were submerged into the resultant solution with their ventral side down, and placed into a humidified incubator at 37°C for 30 min. Following three 5 min washes with PBS at room temperature, the quality of labeling and the integrity of the endoderm in each embryo was assessed under a Leica DM6000 epifluorescence microscope.
Post-fixation immunofluorescence labeling
Embryos were fixed and prepared for immunolabeling according to Little and Drake, 2000. Monoclonal antibodies against avian epitopes (JB3, B3D6, QH1; DSHB, Iowa City, IA), directly conjugated to AlexaFluor 488, 555 or 647 (Molecular Probes, Eugene, OR), were added at 1:1000 dilutions; rabbit polyclonal anti-GFP antibody, conjugated to AlexaFluor 488 – at 1:400, and mouse monoclonal antibody MF20 (DSHB, Iowa City, IA) – at 1:10 dilution in 3% BSA for overnight incubation. Goat anti-mouse secondary antibodies, conjugated to AlexaFluor 488, 555 or 647, were used to follow the MF20 staining at 1:10 dilution in 3% BSA. Nuclei were counterstained with 4′, 6-diamidino-2-phenylindole (DAPI) at 500 nM in PBS.
Preparation of transverse plastic sections
Embryos were dehydrated through a graded ethanol series, placed in JB4 infiltration medium (Electron Microscopy Sciences, Hatfield, PA) at 4°C overnight, and embedded in JB4 resin following the manufacturer’s protocol. Subsequently, 10 μm sections were prepared.
Wide-field time-lapse imaging
Automated microscopy of immunolabeled quail embryos was performed as described elsewhere (Czirok et al., 2002; Zamir et al., 2008). To enhance contrast, selected epifluorescence image stacks were deconvolved (Autoquant X, MediaCybernetics). Manual tracing of image details was performed using custom software (see, e.g., Czirok et al., 2004). Embryos stained with MitoTracker Green (as described above) were subsequently recorded in time lapse for 4–5 hours with 10–12 min intervals between frames. Fading of MitoTracker Green fluorescent signal prevented further imaging. Motion analysis of recorded image sequences were performed as described below and in Aleksandrova et al., 2012.
Confocal Imaging
Confocal imaging of whole-mount embryo specimens was performed using a Nikon 90i upright microscope with a Nikon C1 confocal scan head and Nikon Plan Apochromat 10x and 20x objectives.
Microincisions
50 – 800 μm incisions through the endoderm and adjacent splanchnic mesoderm of HH8 stage embryos were performed under a Leica stereomicroscope using tungsten needles. Embryos were then incubated at 37°C on a microscope stage (Czirok et al., 2002; Zamir, et al., 2008) for 0.5 – 1.5 h prior to initiation of time-lapse image acquisition.
Particle Image Velocimetry (PIV)
We used the two-step algorithm of Zamir et al. (2005); implemented in MatLab (Mathworks, Inc.) and as described in Aleksandrova et al., 2012. Briefly, images were divided into overlapping tiles, each 75 μm wide. The displacements of the tiles were determined by cross-correlation analysis; for each tile we searched the next image for the location that exhibited the most similar intensity pattern. The resulting displacement vectors were then interpolated and denoised by a thin-plate spline fit, yielding our coarse displacement map. This map was used to construct a second, higher resolution one. The cross-correlation procedure was repeated with tiles that were only 30 μm wide. To reduce ambiguities associated with smaller tiles (patterns within smaller tiles are less unique), the subsequent image was scanned only in the vicinity of positions predicted by the coarse map. In this study we did not apply a final smoothing (spline fitting) step, but kept the results of the second cross-correlation analysis.
Details on ECM tracking can be found in Czirok et. al., 2004 (Figure 3) and Aleksandrova et al., 2012 (methods and Supplemental Figure 1). Within the whole embryo, the ECM labeling was highly non-uniform, as some areas were exposed to more antibody than others. To account for the individual variability in ECM antibody staining, we analyzed 4–9 specimens for each developmental stage. Image tiles corresponding to areas where the immunofluorescence was weak were very noisy; hence the cross correlation analysis was more error-prone. To reflect this, for each PIV-derived displacement vector we assigned a weight, the local standard deviation of fluorescence intensity. Thus, displacement vectors characterizing a detail-rich region carry more weight in the subsequent analyses than those obtained in a weakly-labeled area. The PIV analysis comparing displacements of the endoderm and myocardial progenitors has been performed by identical methods. However, the images of the MitoTracker-mediated labeled endoderm were not suitable for deconvolution. Hence, we projected the maximal image intensities of the ‘z-stack’ into a single image for each frame, and used the resulting ‘z-projected’ image for PIV analysis. Thus, instead of performing the PIV analysis on each collected z-plane, it was performed only on a single collapsed image. To compare endoderm and myocardial movements, both optical channels were analyzed the same way.
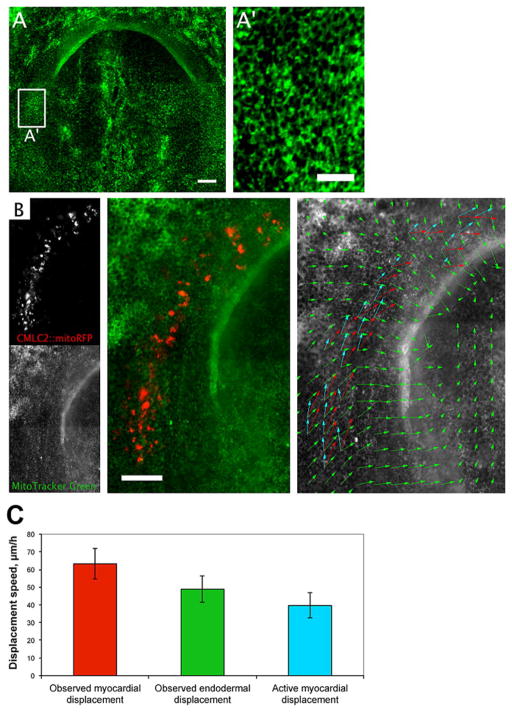
Figure 3. Analysis of myocardial progenitor movements relative to the endoderm.
A, A′ – HH8 embryo with the endoderm labeled with MitoTracker Green dye. Scale bars: A-100 μm, A′ − 40 μm; B – Localization of the myocardial CMLC2 and endodermal MitoTracker signals, and the corresponding vector field comparing the movements of the two tissue layers. Red and green vectors indicate myocardial and endodermal displacements, respectively. Blue vectors show myocardial movements relative to the local endoderm movement. Scale bar: 100 μm. C – observed myocardial, observed endodermal, and active myocardial (relative to the endoderm) displacement speeds, measured for the region outlined in B.
Motion analysis
To characterize ECM and cell rearrangements at various stages of heart development, pairs of deconvolved image stacks were selected. The images within the pairs were acquired on the average of 20 minutes apart. Each pair was analyzed through the following procedure. The corresponding optical sections and fluorescence channels were compared by PIV analysis as described above. Velocities (displacements normalized by the time lag between the images) were presented in a reference system co-moving with the somites. Thus, we use the term “velocity” to describe a vector value characterizing both the rate and direction of the displacements of selected objects. The term “speed” is used to describe the magnitude of the velocity vector. Observed velocity vectors represent total movement in a frame of reference where somites or the paraxial mesoderm is stationary. Active cell velocities were calculated as the local vectorial difference between the cell- and ECM-derived velocity vectors as described in Aleksandrova et al. (2012), and thus are indicative of relative movement. The “reliability” weight assigned to the vectorial difference is the lesser of the two weights assigned to the source vectors.
For statistical comparison of cell motion between embryos, three areas were selected around the anterior intestinal portal (AIP): one at the anterior aspect and one at each side. Velocity vectors from different embryos, but within the same location, were pooled. Data obtained from different embryos were considered statistically independent.
Quantitative measure of autonomous myocardial activity following tissue perturbation
To assess the movements of myocardial cells in embryos with bilateral microincisions, CMLC2::GFP or CMLC2::MitoRFP-expressing cells were tracked manually at the incision boundary. The achieved transfection efficiency, which we estimate at 10% of the total myocardial cell population, allows individual cells to be followed through space and time. Reproducible myocardial progenitor speeds were obtained despite labeling different myocardial cell populations in each embryo. To determine the local ECM movement, or that of the wound/incision edge, we implemented the following procedure: i) the center of mass of the tracked cells was calculated at both sides of the embryo. Since the tracked cells are in close proximity, the center of mass gives a convenient approximate location of the myocardial cells. ii) Centered around the position of the center of mass, a PIV cross-correlation calculation was performed on the ECM images, yielding the typical ECM displacements at those areas. As the most prominent feature on these images is the wound edge, the PIV analysis essentially returns the movement of the incision boundary. iii) Knowing the positions of the myocardial cells as well as the incision boundary, their relative speed, v, was calculated for each frame. iv) The autonomous myocardial activity was characterized as the magnitude of v, averaged over 10 frames following experimental microincision perturbations.
Computational modeling
Utilizing a recent particle-beam tissue model (Czirok and Isai, 2014) we have developed a model of heart development that reflects interactions between 2 germ layers. In the model, mechanical connectivity of the cells is explicitly represented as elastic beams connecting adjacent particles. The beams can be compressed and stretched, bent and twisted. Tissue movements, as driven by cell activities, are modeled using the following steps. First, an initial configuration is defined, which is in mechanical equilibrium. Second, cell activities are represented as changes in the neutral link lengths or orientation – parameters that characterize the link in a mechanical equilibrium. This approach is similar to prescribing growth laws in finite element models (Varner et al., 2010), but we prescribe the new equilibrium configuration on the cellular (particle) scale instead of the tissue scale. Finally, the new equilibrium configuration of the system is established using a 4th order Runge Kutta relaxation algorithm (Press et al., 1992). Since here we model only small deformations, we did not address changes in cell adjacency. Detailed model equations are provided as Supplemental Material.
Results
Myocardial progenitors move medially with their associated fibronectin-containing ECM to the anterior embryonic midline
To compare the movements of myocardial cells with their ECM environment, we visualized the fibronectin ECM in the myocardial fields in CMLC2::GFP or CMLC2::mitoRFP-transfected quail embryos at HH7-8 (n=21), and then recorded embryonic development until HH11-12 (Fig. 1, Supplemental Fig. 1, Movie 1). The population of myocardial cells transfected using this approach is estimated to be 10% of the total myocardial cell population, which allows the tracing of individual cells. Myocardial progenitor and fibronectin movements were compared in the same embryo using vector maps obtained by particle image velocimetry/PIV (Zamir et al., 2005; Aleksandrova et al., 2012). Movements are characterized both as observed (movement of myocardial progenitors and their local fibronectin ECM relative to the somites or paraxial mesoderm) and as active (relative movements between cells and the local ECM, calculated as the local vectorial difference between the velocity vectors of cell and ECM motion).
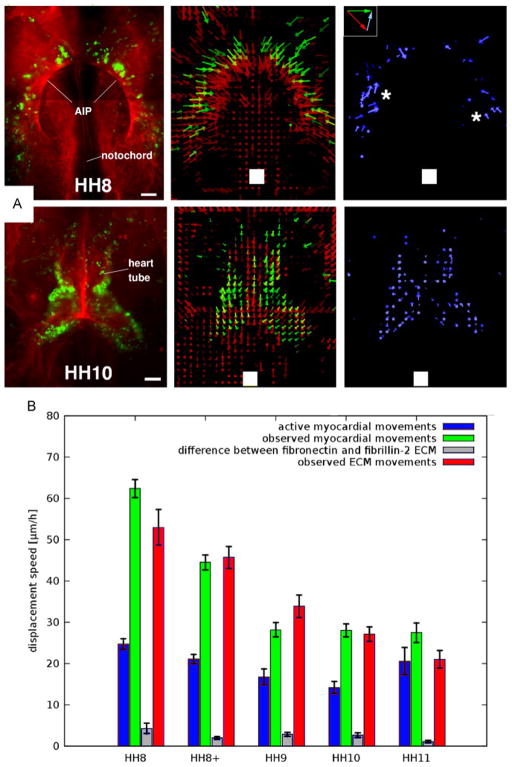
Figure 1. Characterization of myocardial progenitor movements relative to the fibronectin ECM.
A – representative epifluorescence images of embryos included in the PIV analysis at HH8 and HH10 (left panels, red: ECM immunolabeling, green: CMLC2::GFP labeled myocardial cells), with corresponding vector maps of observed myocardial (green) and ECM (red) velocities (middle panels) in a frame of reference anchored to the somites. The differences between the local myocardial and ECM movements, our measure of active myocardial cell movements (see inset), are plotted with blue vectors (right panels). While the overall (observed) motion of both the ECM and myocardium is medial and posterior, the active myocardial movement is mostly anteriorly-directed (asterisks). Red – B3D6 mAb-labeled fibronectin ECM, green – pCMLC2::GFP-transfected myocardial cells. Scale bars and white squares – 100 μm. The vectors represent speeds as extrapolated displacements during one hour. Dots indicate an absence of detectable movement, i.e., velocities smaller than 10 μm/hour. AIP – anterior intestinal portal
B – mean speeds of observed (green) and active (blue) myocardial progenitor movements, compared to the observed ECM (red) displacement speeds at progressive stages of heart tube assembly. Error bars represent SEM, the difference between ECM components (grey) is our estimate of the magnitude of the systematic error in our analysis.
Myocardial and ECM movements are similar in the vicinity of the AIP (Fig. 1A). To quantify the degree of co-movement, the typical magnitude of the myocardial and ECM movement vectors as well as of their difference was averaged at three locations close to the AIP in n=21 embryos. The observed movements of both the myocardium and ECM slow down from 60 μm/h to 30 μm/h as development progresses. While similar, ECM and myocardial movements are not identical; the magnitude of the vectorial difference between myocardial and ECM displacements (the measure of active movements) remains at 20 μm/h during the entire period. The distinct active myocardial cell movements exhibit predominantly anterior directionality (asterisks in Fig. 1A).
A similar relationship between observed and active cell movements, and ECM movements was previously characterized for the endocardial progenitor population (Aleksandrova et al., 2012). The calculated 20 μm/h autonomous speeds of myocardial progenitors are in the same range as those reported for endocardial cells in Aleksandrova et al., 2012, or endothelial cells moving in culture (10 μm/h: Szabo et al., 2010; 50 μm/h: Kouvroukoglou et al., 2000). Thus, cell repositioning (relative to its immediate environment) can be comparable to the size of the cell in approximately one hour. In comparison, the total distance myocardial progenitors moved (with their tissue environment) from their initial lateral location is around 1 mm.
Myocardial progenitors contract at the medial aspects of the heart field
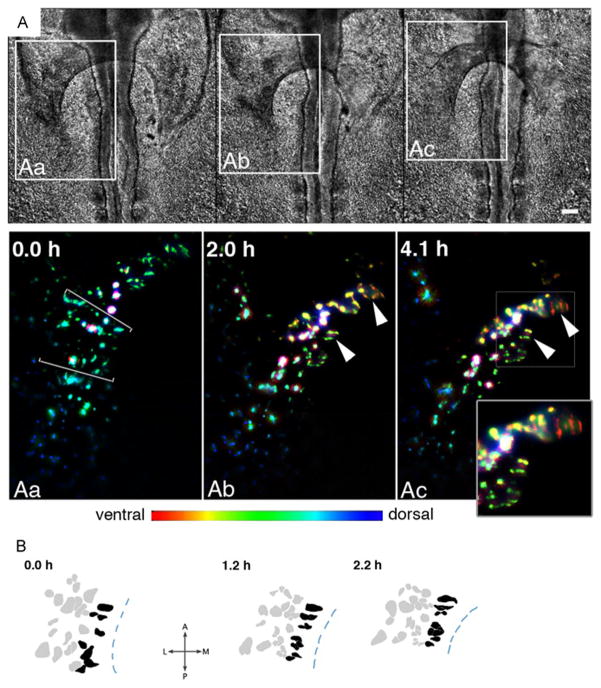
To investigate the behavior of individual myocardial progenitors, we subjected three CMLC2::GFP-transfected HH8 embryos to 1 frame/minute time-lapse imaging. Cells situated along the medial aspects of the bilateral heart fields undergo pronounced contraction along the edge of the AIP, becoming progressively more columnar, with their long axis perpendicular to the AIP arch (See cells marked by arrowheads in Fig. 2Aa-Ac, and Movie 2). In contrast, more laterally-positioned myocardial cells maintain their cuboidal/isotropic shapes. Note that contraction of individual cells appears to result in tissue-level contraction in the medial aspect of the heart field (See decrease in the anterior-posterior extent of the medial portion of the heart field in Fig. 2B at 0.0h compared to 2.2h). Simultaneously, the anterior heart fields turn ventrally (Fig. 2Aa-Ac), resulting in an inversion of the myocardium (Abu-Issa and Kirby, 2008).
Figure 2. Characteristic cell-shape changes in the medial aspect of the myocardial field.
A – DIC (top panels) and epifluorescence (GFP; bottom panels Aa-Ac) images of the right embryonic myocardial progenitor field in a pCMLC2::GFP-transfected embryo. Individual planes within the z-stack were color-coded to reflect the position of objects along the dorso-ventral embryonic axis as illustrated by the spectrum bar. Arrowheads mark two groupings of cells that manifest similar cell shape changes (See inset box, epifluorescence image Ac). In both cases, the labeled cells appear thinner (compressed), and elongate in the direction perpendicular to the anatomical margin of the AIP (shown in DIC images and depicted as segmented lines in B). Scale bar: 100 μm. B – outlines of the cells within the region demarcated by brackets in Aa. Segmented lines mark the AIP border. Elapsed time is shown in the top left corner of each image.
Myocardial tissue moves anteriorly relative to the endoderm
To compare the relative motion of the endoderm versus myocardial tissue, embryos were transfected with CMLC2::mitoRFP and their endoderm labeled with MitoTracker Green (n=11). This vital stain allows the observation of individual cells within the endoderm for up to 5 hours (Fig. 3A–A′). PIV analysis performed on sequential image pairs taken 15–24 minutes apart yields vector maps characterizing the co-movement of myocardial (red) and endodermal (green) tissues (Fig. 3B, Movie 3). In HH8-9 embryos, the overall movement of both tissues is directed towards the midline. However, the motion of the presumptive myocardial tissue relative to the endoderm has a pronounced anterior directionality (cyan vectors in Fig. 3B). Displacement speeds, calculated for both the myocardium and endoderm (Fig. 3C), demonstrate a greater degree of difference (i.e. exhibit greater relative motion) than the difference between the myocardial tissue and its ECM microenvironment (Fig. 1B).
Mechanical perturbations abrogate endoderm shortening and reveal autonomous myocardial movements
In order to identify the tissue-autonomous behavior of myocardial fields, we utilized microincisions (Fig. 4A–E) to alter the tissue mechanics of the myocardial and adjacent tissues. Incisions transecting the endoderm and underlying splanchnic mesoderm were conducted at HH8 (Fig. 4A). Prior to the microsurgeries, myocardial progenitors and fibronectin-ECM were labeled in each embryo.
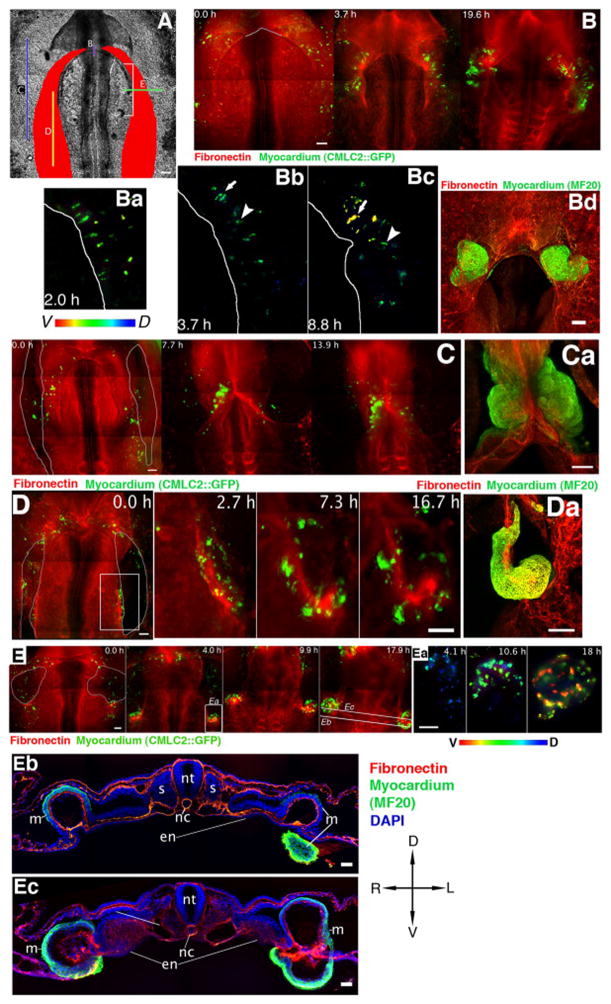
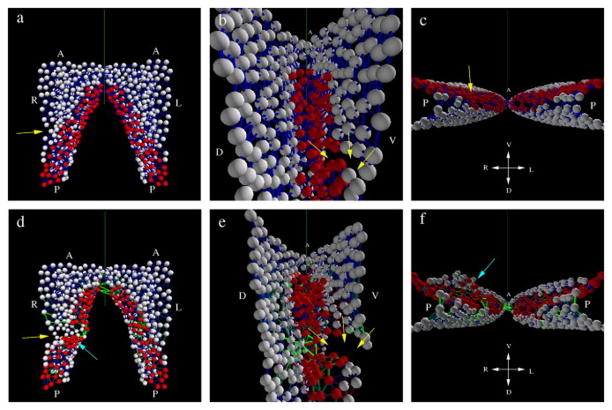
Figure 4. Typical phenotypes induced by localized incisions through the endoderm and splanchnic mesoderm of HH8 embryos.
A – location of incisions (labeled B–E) that result in phenotypes shown in panels B–E. Myocardial fields are schematically represented by red shading. B, C, D, E: Time-lapse image sequences of perturbed myocardial development. Red – B3D6 mAb-labeled fibronectin ECM, green – CMLC2::GFP or CMLC2::mitoRFP-expressing myocardial cells. Contour lines delineate the wound edges at the onset of recording, 0.5–1.5 hours post-incision. Time elapsed during recording is indicated on image panels. Scale bars: 100 μm unless otherwise indicated. Bd, Ca, Da: Myocardium visualized at the recording conclusion by post-fixation MF20 antibody labeling (green) and confocal imaging. B – An incision through the endoderm at the ventral midline leads to the failure of midline fusion and formation of two lateral “hearts” (cardia bifida). Ba-Bc – Images of the left myocardial field of a CMLC2::GFP-transfected embryo that underwent an incision through the endoderm at the ventral midline. Color-code indicates dorso-ventral position of the labeled cells. Contour lines illustrate the position of the AIP edge. The same cell groups are indicated by an arrow and an arrowhead. Note the elongated shapes of CMLC2::GFP-expressing cells in Bc, a contraction comparable to Fig. 2. Myocardial cells become increasingly ventral, as indicated by the change in color from blue/green to yellow/red. C – Bilateral incisions through the endoderm and the underlying splanchnic mesoderm lateral to the myocardial fields have minor effects on heart development and are compatible with myocardial fusion. D – Bilateral, longitudinal incisions through the myocardial fields and endoderm. The region marked by the white rectangle on the left panel is shown in detail in the adjacent panels. E – Bilateral, medio-lateral incisions, perpendicular to the AIP arch, through the endoderm and underlying myocardium and splanchnic mesoderm. Embryos shown are representative examples. The number of embryos that underwent the various types of surgical perturbations are as follows: n=15 (Fig. 4B), n=6 (Figs. 4C and 4D) and n=34 (Fig. 4E). Ea: Cellular resolution image sequence depicting CMLC2::GFP-expressing myocardial tissue curling ventrally at the wound edge, within the area marked by a white rectangle in panel E. The color code represents dorsal-ventral position. Eb and Ec are images of transverse sections taken post-fixation at the lines indicated in panel E. Note that the myocardium forms pronounced cup-like structures posterior to the incision site (Eb), whereas the endoderm remains relatively planar. At the incision site (Ec), deformed (“curled”) myocardium is found ventral to the endoderm. En – endoderm, mc – myocardium, nc – notochord, nt – neural tube, s – somite; D – dorsal, V - ventral, L – left, R – right. Scale bars in Eb and Ec: 50 μm.
Perturbations to the ventral midline endoderm induce separation of cardiac progenitor fields, termed cardia bifida (DeHaan, 1959; Glanzer and Peaslee, 1970; Rosenquist, 1970; Satin et al., 1988). This perturbation also slows foregut elongation, i.e., movement of the endoderm (Movie 4). Yet, despite the failure of the myocardial fields to converge, the bifid myocardial tissue continues to exhibit: 1) ventral cell repositioning and 2) cell contraction along the medial edge of the progenitor fields coupled with a columnar cell-shape change (Fig. 4B).
When 500–600 μm-long incisions, parallel to the anterior-posterior axis, were targeted either lateral (Fig. 4C, Movie 5) or medial (not shown) to the heart field, the perturbation had little or no effect on heart or AIP morphogenesis. In contrast, incisions within the myocardial field result in striking deformations (Fig. 4D, 4Da, Movie 6). The unconstrained wound edges of the myocardial tissue bend ventrally and medially yielding a “cylindrical” surface. Furthermore, myocardial tissue posterior to the incision advances in the anterior direction, past the wound edge. The morphology, and the processes within the perturbed myocardial tissue, is depicted in schematic form in Fig. 5a.
Figure 5. Schematic representations of tissue movements in a ventral view.
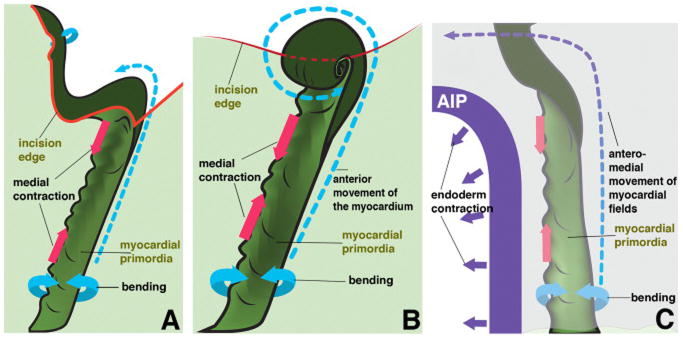
5a – longitudinally incised myocardial field, as shown in Fig. 4D (embryonic left side; boxed area). The differentiating myocardium (dark green), contiguous with the surrounding non-cardiac mesoderm (light green), is undergoing contraction along its medial edge (indicated by a decrease in distance between the magenta arrows). At the same time, the myocardium actively generates bending moments (forces) that generate a groove (lower cyan arrows). The incision (red contour line) introduces an unconstrained boundary of the myocardium, which in turn bends and rotates medially (upper cyan arrow). The forces generated within the myocardial field posterior to the incision propel the myocardium anteriorly (blue segmented arrow) past the wound edge. 5b – medio-laterally incised myocardial field, as shown in Fig. 4E, with the same notations used as in panel 5a. The incision separates the myocardium and the adjacent non-myocardial mesoderm from the anterior tissues. As in 5a, tissue forces generated by the myocardial field posterior to the incision propels the myocardial tissue anteriorly (blue segmented arrow), past the wound edge. Tissue-autonomous bending moments drive the formation of characteristic spheres of myocardium at the wound edge. 5c: normal heart tube assembly. The myocardial field is located in the close vicinity of the AIP (purple semi-arch), and the mesoderm is overlain by the endoderm (the top light purple tissue layer), continuous with the AIP. The AIP endoderm undergoes a uniform contraction that moves both itself and the mesoderm mediocaudally (purple arrows). At the same time, the medial portion of the myocardial field actively contracts (pink arrows) and develops bending moments within the tissue. The mechanical constraints of the adjacent tissue transform this activity into the observed ventral bending (cyan arrows). The combination of these activities leads to the net displacement of the myocardial field in the anterior and medial direction, illustrated by the segmented arrow.
Pronounced myocardial changes were also observable when 200–300 μm-long incisions were placed perpendicular to the anterior-posterior axis (white bracket, Fig. 4A). In perturbed embryos the myocardial tissue continues to move anteriorly. Specifically, it advances past the incision site and curls over the ventral surface of the endoderm yielding a spherical surface (Figure 4E, 4Ea, Fig. 5b and Movies 7–8). Figures 4Eb and 4Ec illustrate the cross-sectional anatomy of the perturbed embryo shown in Fig. 4E. The two physical sections were obtained caudal to the posterior wound edge and at the level of the incision site, respectively. These sections illustrate the cup-shaped myocardial structures that form by progressive ventral curling. Thus in the absence of contact with, or constraints due to the endoderm and the adjacent mesoderm, the myocardial tissue exhibits substantial autonomous deformations. We suggest that in the normal (constrained) condition, the identified autonomous tissue stresses (bending moments - a measure of spatially distributed internal forces associated with an imposed curvature on a structural element) contribute to tissue movements necessary for proper heart tube formation.
Computational modeling of myocardial and endodermal tissue behavior
Based on in vivo observations and experimental perturbations, we constructed a 3D tissue mechanics model utilizing a recent modeling framework (Czirok and Isai, 2014). This model represents the myocardial tissue, the adjacent splanchnic (ventral) and somatic (dorsal) mesodermal layers surrounding the coelom, and the endodermal layer comprising the foregut floor and lining the AIP (see Supplemental text and Supplemental Fig. 2). Intercellular mechanical connections are assumed within and between the germ layers. The left and right myocardial fields are kept distinct at the midline - linked only through their association with the subjacent endoderm. In this configuration, the tissue is at rest in the absence of cellular activities.
We consider three distinct cellular activities within the model: 1. Contraction of the medial aspect of the myocardial sheet - simulated by a 25% shortening of the interparticle bonds (green links in Suppl. Fig. 2C′). 2. A uniform and isotropic bending moment within a flat myocardium. In the absence of additional constraints, this distribution of cell-generated mechanical forces deforms the tissue to assume a ventrally concave spherical surface in mechanical equilibrium. 3. The endoderm moves medio-caudally to regress the AIP. We do not attempt to model the driving forces of the latter process. Instead, we prescribe a centripetally-directed displacement field along the AIP and at the ventral endodermal surface, which corresponds to our time-lapse data (Fig. 3).
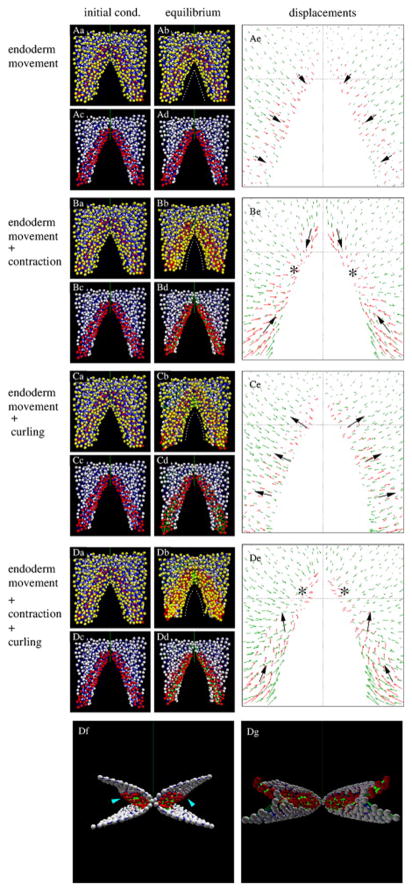
After imposing a combination of the above activities, by computer simulations we determine the new configuration in which the surrounding tissue accommodates these cellular activities and the whole system reaches a new mechanical equilibrium (Fig. 6). The simulations suggest that medio-caudal endodermal motion alone does not result in myocardial tissue-specific displacements. This lack of movement is reflected in an AIP with abnormally straight edges (Fig. 6A). Addition of myocardial contraction (Fig. 6B) pulls the myocardial sheet towards two, laterally-localized (instead of the normal medially-localized) contraction centers (asterisks), one on each side. As a result of these modeling conditions, the shape of the AIP resembles a triangle, instead of the characteristic arch-like shape observed in vivo. Endodermal motion with the presence of myocardial bending moments (Fig. 6C) moves the myocardial layer anteriorly and laterally (instead of medially), but creates a more normally shaped AIP. Finally, the addition of both myocardial tissue contraction and bending moments (Fig. 6D) moves the myocardial sheet anterio-medially. The centers of myocardial tissue contraction are also moved towards the midline. As a further relevant model outcome, the myocardial tissue and the surrounding endoderm (not shown) protrude into the coelom, thus forming a dorsal bulge (Fig. 6Df, 6Dg), as is observed in vivo. Finally, the shape of the AIP in embryos is assessed by establishing curvature radii at the lateral aspects of the AIP and these data are compared to the shape of the AIP obtained in the model. At the stage selected for modeling (“endocardial stage” 4 from Aleksandrova et al., 2012), we found that in normal embryos, the curvature radii are approximately equal to (100%) the lateral width of the AIP; i.e. the AIP has a substantial curvature at this stage. The model recapitulates this shape when all three driving mechanisms are considered.
Figure 6. Simulated tissue movements resulting from various prescribed (assigned) active cell behaviors.
A: A prescribed medio-caudal movement of the endoderm rolls the presumptive myocardium medially, contributing to the inversion of the myocardial field. B: Contraction of the medial aspect of the myocardial tissue (see green links in Suppl. Fig. 2C′) narrows the AIP and reduces its curvature. C: Uniform curling of the myocardium moves the myocardial field laterally and anteriorly, as well as widens the AIP and increases its curvature. D: The combination of the three effects moves the myocardial field antero-medially relative to the surrounding tissue. Panels a and b show all particles, while the endoderm and associated cell-cell connections are removed from panels c, d. The initial configuration (as in Suppl. Fig. 2) is shown in panels a, c. The mechanical equilibrium resulted by the prescribed cellular activity is shown in panels b, d. The shape of the AIP is indicated by white dotted lines in panels b. Panels e show the tissue displacements during the simulations. Red and green arrows indicate movements of the myocardial tissue and both surfaces of the folded mesoderm, respectively. Black arrows are guides for the eye, indicating the local direction of myocardial movements. Asterisks mark contraction centers. Panels Df and Dg show, from anterior and posterior aspects (ventral surface is at the top) the combined effects of all three cellular activities. Arrowheads indicate a developing bulge of the myocardium into the coelomic space.
These simulations demonstrate that due to the mechanical constraints imposed by adjacent tissues (endoderm and non cardiac mesoderm), simple myocardial cellular activities can produce tissue deformations that are markedly distinct from the prescribed behavior, and show a great degree of similarity with the empirically observed tissue movements in vivo. In particular, at the anterior AIP, the myocardial tissue dislocates anteriorly relative to the more ventral endoderm layer (Compare Fig. 3 to Fig. 6De). Furthermore, by moving the ventral (splanchnic) mesodermal layer medially, this myocardial activity can contribute to the regression of the AIP, by providing additional compression along the anterior intestinal portal.
Our model can also reproduce the myocardial tissue movements observed after mechanical perturbations when the initial condition of the simulation is a configuration where the links crossing the embryonic midline at the anterior-most aspect of the AIP were removed (Fig. 7). The simulated “wound” opens, and the lateral movement of the myocardial tissue (Supp. Fig 3E) is consistent with the presence of two contractile centers (bifid heart), one on each side (as in Fig. 4Bd). Similarly, a mediolateral “incision” in the myocardial tissue and the surrounding mesoderm yields a ventrally curled myocardial structure (Fig. 7), in accord with experimental observations (Fig. 4E). Thus, our computational model demonstrates that the proposed autonomous myocardial tissue activities, contraction and curling, are feasible mechanisms to drive early cardiac morphogenesis.
Figure 7. Computational simulations of microsurgical incisions.
A mechanical discontinuity was introduced into the initial configuration by removing interconnecting beams along a line at the embryonic right side (yellow arrows), shown from a ventral (a), lateral (b) and posterior (c) aspect. Myocardial activity results in a pronounced ventral curling of the myocardium, shown from a ventral (d), lateral (e) and posterior (f) aspect. A: anterior, P: posterior, V: ventral, D: dorsal, R: right, L: left.
Discussion
Myocardial progenitors and their local ECM move together as a tissue composite to the midline
Despite the prevalent literature postulating that cardiac progenitor movements to the midline occur relative to the ECM and/or the endoderm (classically defined as migration), we find that such movements are limited in scope, and are secondary to larger-scale tissue deformation events. Our in vivo fibronectin immunolocalization strategy labels both myocardial- and endodermal-associated ECM fractions (Supp. Fig 1). Hence, we can detect movement relative to both the endodermal- and myocardial-associated fibronectin. As shown in a number of recent studies by our group (Zamir et al., 2006; 2008; Aleksandrova et al., 2012), in avian embryos and by others (Sepich et al., 2005) in zebrafish embryos, cell populations at various stages of development can be moved passively with the surrounding tissue environment. Additionally, cells may move relative to their ECM micro-environment, which we refer to as “active” movement. In this study, we demonstrate that most of the midline-directed myocardial movement is shared with the surrounding fibronectin ECM (Fig. 1A). Hence, the observed medial movement of myocardial progenitors clearly cannot be interpreted as a migration process relative to their local ECM environment.
Wiens (1996) proposed that myocardial movement is driven by the contractile activity of the anterior-most cells within a cohesive epithelial sheet. These cells were suggested to interact with an anterior “patch” of fibronectin, activate their contractile mechanisms, and pull the posterior portion of the myocardial fields forward. As seen in Suppl. Fig. 1Aa, fibronectin is associated with both the presumptive myocardium and endoderm at both the anterior and posterior aspects of the heart fields, without the obvious presence of a mediolateral or anterio-posterior gradient.
Studies in which microincisions block foregut elongation also suggest that enhanced myocardial cell migration does not compensate for perturbed endodermal contraction; since in these experiments we do not observe a compensatory medial movement of the myocardial primordia.
Tissue mechanics underlying the microincision experiments
In microincision experiments, the endoderm adjacent to the wound does not exhibit substantial morphological changes. Thus, the myocardium likely responds to the incisions autonomously, without mechanical input from the adjacent endoderm. We therefore propose that the observed deformations are a cumulative result of forces acting within, and specific to the myocardial sheet. While long term effects of microincisions are more problematic to interpret than short term recoil over a time course of minutes, our experiments have reasonable controls: similar microincisions made at slightly different positions or at a different angle relative to the anterior-posterior axis. Most importantly, incisions placed through the endoderm and mesoderm in the regions outside of the heart fields – although eliciting a wound healing response – do not result in tissue curling or any other behaviors that we describe for incised myocardial primordia. The experimental incisions (red contour lines in Figs. 5a and 5b) introduce unconstrained (free) myocardial boundaries, which curl further ventrally (cyan arrows) in the absence of mechanical constraints.
Endodermal cell activities generate tissue movements necessary for early heart morphogenesis
The directionality and magnitude of the convective tissue movements studied in this work indicate endodermal contraction plays a primary role in drawing the heart fields toward the midline. In our experiments, as well as work reported previously (DeHaan, 1959; Rosenquist, 1970), when AIP regression is perturbed, myocardial field translocation to the midline fails. More recently, Varner and Taber (2012) implicated the endoderm, and not the myocardium, as the primary source of a midline-directed contractile force in chicken embryos. Their work proposes that a cytoskeletal contraction-driven shortening of the endoderm around the AIP actively pulls the heart primordia (splanchnic mesoderm) to the midline. While our work agrees with this important role for the endoderm in bringing the myocardial fields to the midline, our results also indicate that the endoderm motion alone is not sufficient for proper midline positioning of the myocardial primordia.
Autonomous behavior of the myocardial fields contributes to proper heart morphogenesis
Our data suggest a complementary, autonomous role for the myocardial primordia in heart tube morphogenesis not previously observed. We propose two myocardial tissue-specific cellular activities that contribute to early heart formation: 1) contraction of cells within the medial aspect of the heart field (magenta arrows in Fig. 5) and 2) exertion of bending moments within the myocardium that can give rise to a uniform ventral curling of the tissue in the absence of further constraints (Fig. 5c).
Contraction of the medial myocardial tissue boundary is visible at the cellular level: cells become progressively more columnar with their long axis perpendicular to the direction of tissue contraction (Fig. 2B). This observation is in accord with that of Auman et al. 2007, who demonstrated that regionally-confined cardiomyocyte cell shape changes underlie the emergence of ventricular chamber curvatures in the developing zebrafish heart. Likewise, in chicken embryos, Manasek et al. 1972 proposed regional changes in myocardial cell shape and alignment as a potential mechanism for cardiac looping.
The autonomous curling activity of the myocardium is readily apparent in the mechanical incision experiments, in which the free edge of the myocardial tissue bends/curls ventrally irrespective of the direction of the wound. Due to the constraints of adjacent (endodermal and mesodermal) tissues, in unperturbed embryos the myocardium does not assume a spherical shape. In particular, mechanical constraints at the long myocardial-mesodermal tissue boundary restrict the curvature along the A–P axis, while curvature still develops along a mediolateral axis - giving rise to a fold in the myocardium that protrudes dorsally (Supplemental Fig. 4B). We expect the exerted mechanical forces (bending moment) to contribute to the positioning of the heart fields and drive the observed anterior-directed movements of the myocardium relative to the endoderm (Fig. 3). Thus, in our proposed model, active contraction of the medial myocardial sheet is superimposed upon a more broadly-acting medio-caudal tissue movement that is driven by the regression of the AIP (Fig. 5c).
While our model cannot exclude alternative explanations, it demonstrates that our proposed mechanism is feasible, and - within the limits of our model - the three features considered (endoderm movement, myocardial contraction and bending) are both sufficient and necessary. Here we only explored the model in the elastic regime; future work will test how the proposed driving forces may contribute to sustained long-term elasto-plastic tissue flows generating a tubular heart from the lateral heart field.
Applicability of the mechanisms proposed in avians to zebrafish heart development
In contrast to the endoderm folding-based foregut morphogenesis of avians and mammals, in zebrafish the gut develops from a rod of cells that forms as a result of midline-directed movements of bilateral endoderm progenitor populations. These movements resemble those of the cardiac progenitors (Sakaguchi et al., 2006; Arrington and Yost, 2009), However, in natter/fibronectin mutants (Trinh and Stainier, 2004) cardia bifida is observed, yet foregut morphogenesis is not affected, suggesting that endodermal and cardiac progenitors may rely on distinct mechanisms for arrival at the midline. Likewise, Ye and Lin (2013) showed that endodermal S1pr2/Galpha13 signaling, working through a RhoGEF-dependent pathway, drives the convergence of the endoderm to the midline, but not the convergence of the mesoderm.
Conclusions
In conclusion, we have shown that the myocardial tissue is displaced to the embryonic midline via tissue motion (myocardial cells and their surrounding ECM moving together). These medial movements are driven primarily by endodermal contraction around the anterior intestinal portal. However, autonomous myocardial tissue deformations (contraction and curling) result in an anterior directed movement of the myocardial primordia relative to the endoderm. Our in vivo dynamic imaging and computational results are confirmed by our modeling approach and computer simulations. Thus, we conclude that the endoderm and myocardium join forces to drive early heart tube assembly. Collectively this results in the precise positioning of the myocardial primordia during tubular heart morphogenesis.
Supplementary Material
A - maximum intensity dorso-ventral projections of confocal stacks obtained at HH8 (top panels), and HH11 (bottom panels). Whole mount specimens were immunostained with antibodies MF20 (red), B3D6 anti-fibronectin (blue), and anti-GFP (green) to enhance pCMLC2-GFP expression-mediated labeling of transfected cells. A′, A″ -10 μm plastic sections taken at the levels indicated with horizontal white lines. Aa and Ab: areas marked by white rectangles that show the colocalization of MF20 and CMLC2::GFP signals in greater detail. Scale bars: 100 μm.
The model builds up the mesoderm (white), endoderm (yellow) and myocardium (red) using particles and their mechanical interconnections represented as links. Panels A, B and C show the initial configuration from a posterior, anterior and ventral aspect. The endoderm and its associated links are removed for a better view of the myocardium in the corresponding panels A′, B′ and C′. The green links in the medial aspect of the myocardium (magnified inset in C′) indicate spatially localized contractility, the effect of which on the surrounding tissue will be evaluated by simulations. A: anterior, P: posterior, V: ventral, D: dorsal, R: right, L: left.
Simulations with an “incision” (removed links) at the anterior aspect of the AIP yield a widening wound and AIP. The pronounced antero-lateral movement of the myocardium is consistent with the observations yielding bifidic hearts. The initial configuration (as in Suppl. Fig. 2) is shown in panels A and C. In the inset a white bracket indicates the area where cell-cell contacts were removed. The mechanical equilibrium resulted by the prescribed cellular activity is shown in panels B and D. Panels A and B show all particles, while the endoderm and associated cell-cell connections are removed from panels C and D. Panel E show the tissue displacements during the simulations. Red and green arrows indicate movements of the myocardium and both surfaces of the folded mesoderm, respectively. Black arrows are guides for the eye, indicating the local direction of myocardial movements.
Myocardial progenitor fields undergo a dramatic change in position along the dorso-ventral embryonic axis in caudal-to-cranial progression. A – a panel from Fig. 3 by van den Berg et al., 2009 (p. 182), to illustrate the anatomy of myocardial fields at HH8 and to indicate the cranio-caudal position of the sections shown in panel B. B - a series of 10 μm-thick sections (taken up to 200 μm apart from each other) through the cardiac fields of an HH8 embryo. The sections’ approximate positions, on the cranio-caudal axis, are indicated by lines connecting to A. Arrowheads point to the dorsal bends of the myocardium protruding into the coelom. Green – MF20 antibody-labeled myocardial progenitors, blue – DAPI. d – dorsal, and v - ventral side of the embryo. All sections are presented in the same orientation. Scale bar: 100 μm.
Heart tube assembly in a Tie1::H2B-YFP quail embryo during stages HH8-11 with endothelial/endocardial nuclei in green, CMLC2::MitoRFP-expressing myocardial progenitors shown in red and fluorescent B3D6 antibody-labeled fibronectin in blue.
Cellular behaviors of CMLC2::GFP-expressing myocardial progenitors within the embryonic right heart field. Original images were color-coded to reflect the position of cells located at different dz levels relative to the dorso-ventral axis of the embryo, with dorsal z-planes represented in blue, and ventral - in red portions of the spectrum.
Left panel: Movements of CMLC2::MitoRFP-expressing myocardial progenitors (red) relative to the MitoTracker Green-labeled endoderm (green). Right panel: PIV-derived vectors illustrating the magnitude and directions of observed myocardial (red vectors) and endodermal (green vectors) displacements that took place during the 15 min time period that separates the adjacent images. Vectorial differences between the above parameters, reflecting myocardial movements relative to the endoderm, are represented with cyan vectors.
Development of the cardia bifida phenotype in an embryo that underwent an incision through the AIP endoderm at the ventral midline at HH8. CMLC2::GFP-expressing myocardial progenitors are shown in green, and B3D6 mAb labeled fibronectin ECM in red.
Heart tube assembly in an embryo that underwent bilateral longitudinal incisions lateral to the heart field positions at HH8. CMLC2::GFP-expressing myocardial progenitors are shown in green, and B3D6 mAb labeled fibronectin ECM in red.
Deformations that take place within the embryonic left heart field in response to the incision placed within the cardiac field parallel to its anterior-posterior axis. CMLC2::GFP-expressing myocardial progenitors are shown in green, and B3D6 mAb-labeled fibronectin ECM in red.
Deformations taking place within heart fields in response to bilateral incisions across the cardiac fields perpendicular to their anterior-posterior axes. CMLC2::GFP-expressing myocardial progenitors are shown in green, and B3D6 mAb-labeled fibronectin ECM in red.
Deformations taking place within the embryonic left heart field (CMLC2::GFP-expressing) in response to an incision placed perpendicular to its anterior-posterior axis. Original images were colorcoded to reflect the position of cells located at different dz levels relative to the dorso-ventral axis of the embryo, with dorsal z-planes represented in blue, and ventral - in red portions of the spectrum.
Acknowledgments
We gratefully acknowledge Dr. Charles Little for critical reading of the manuscript, helpful comments and discussion. We are also grateful to Patricia St. John for advice on confocal microscopy, and to Phil Shafer for schematic illustration preparation.
MF20 and B3D6 antibodies were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242.
This work was supported by NIH grants HL085694 (BJR); HL087136 (AC); COBRE award P20 RR024214 (BJR); the Hungarian Research Fund OTKA K72664 (AC); the G. Harold & Leila Y. Mathers Charitable Foundation (BJR, AC), and the American Heart Association predoctoral fellowship 11PRE7580033 (AA).
Footnotes
Author Contributions
Anastasiia Aleksandrova – Conceptualized and performed experiments, analyzed data, prepared figures, prepared and edited manuscript
Andras Czirok – Conceptualized and performed modeling/simulations, analyzed computational data, prepared figures, prepared and edited manuscript
Edina Kosa – performed computational analyses and histological sample preparation
Oleksandr Galkin – performed experiments and molecular construct preparation
Tracey J. Cheuvront – histological sample preparation
Brenda J. Rongish – Conceptualized experiments, analyzed experimental data, prepared and edited manuscript
References
- Abu-Issa R, Kirby ML. Patterning of the heart field in the chick. Dev Biol. 2008;319:223–33. doi: 10.1016/j.ydbio.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleksandrova A, Czirók A, Szabó A, Filla MB, Hossain MJ, Whelan PF, Lansford R, Rongish B. Convective tissue movements play a major role in avian endocardial morphogenesis. Dev Biol. 2012;363:348–61. doi: 10.1016/j.ydbio.2011.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrington CB, Yost HJ. Extra-embryonic syndecan 2 regulates organ primordia migration and fibrillogenesis throughout the zebrafish embryo. Development. 2009;136:3143–52. doi: 10.1242/dev.031492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auman HJ, Coleman H, Riley HE, Olale F, Tsai HJ, Yelon D. Functional modulation of cardiac form through regionally confined cell shape changes. PLoS Biol. 2007;5:e53. doi: 10.1371/journal.pbio.0050053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellairs R, Osmond M. Atlas of Chick Development. Academic Press; 1998. [Google Scholar]
- Brand T. Heart development: molecular insights into cardiac specification and early morphogenesis. Dev Biol. 2003;258:1–19. doi: 10.1016/s0012-1606(03)00112-x. [DOI] [PubMed] [Google Scholar]
- Cui C, Cheuvront TJ, Lansford RD, Moreno-Rodriguez RA, Schultheiss TM, Rongish BJ. Dynamic positional fate map of the primary heart-forming region. Dev Biol. 2009;332:212–22. doi: 10.1016/j.ydbio.2009.05.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czirók A, Isai DG. Cell resolved, multiparticle model of plastic tissue deformations and morphogenesis. Physical Biology. 2014;12:016005. doi: 10.1088/1478-3975/12/1/016005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czirok A, Rongish BJ, Little CD. Extracellular matrix dynamics during vertebrate axis formation. Developmental Biology. 2004;268:111–122. doi: 10.1016/j.ydbio.2003.09.040. [DOI] [PubMed] [Google Scholar]
- Czirók A, Rupp PA, Rongish BJ, Little CD. Multi-field 3D scanning light microscopy of early embryogenesis. J Microsc. 2002;206:209–17. doi: 10.1046/j.1365-2818.2002.01032.x. [DOI] [PubMed] [Google Scholar]
- Czirok A, Zamir EA, Filla MB, Little CD, Rongish BJ. Extracellular matrix macro-assembly dynamics in early vertebrate embryos. Cur Topic Dev Biol. 2006;73:237–58. doi: 10.1016/S0070-2153(05)73008-8. [DOI] [PubMed] [Google Scholar]
- DeHaan RL. Cardia bifida and the development of pacemaker function in the early chick heart. Dev Biol. 1959;1:586–602. [Google Scholar]
- DeHaan RL. Migration patterns of the precardiac mesoderm in the early chick embryo. Exp Cell Res. 1963;29:544–60. doi: 10.1016/s0014-4827(63)80016-6. [DOI] [PubMed] [Google Scholar]
- Gannon M, Bader D. Initiation of cardiac differentiation occurs in the absence of anterior endoderm. Development. 1995;121:2439–50. doi: 10.1242/dev.121.8.2439. [DOI] [PubMed] [Google Scholar]
- Gilbert S, editor. Developmental biology. 8. Sunderland (Massachusetts); Sinauer Associates: 2006. p. 474. [Google Scholar]
- Glanzer ML, Peaslee MH. Inhibition of heart beat development by chloramphenicol in intact and Cardia bifida explanted chick embryos. Experientia. 1970;26:370–1. doi: 10.1007/BF01896893. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. J Morphol. 1951;88:49–92. [PubMed] [Google Scholar]
- Kouvroukoglou S, Dee KC, Bizios R, McIntire LV, Zygourakis K. Endothelial cell migration on surfaces modified with immobilized adhesive peptides. Biomaterials. 2000;21:1725–33. doi: 10.1016/s0142-9612(99)00205-7. [DOI] [PubMed] [Google Scholar]
- Linask KK. N-cadherin localization in early heart development and polar expression of Na+, K+-ATPase, and integrin during pericardial coelom formation and epithelialization of the differentiating myocardium. Dev Biol. 1992;151:213–224. doi: 10.1016/0012-1606(92)90228-9. [DOI] [PubMed] [Google Scholar]
- Little CD, Drake CJ. Whole-mount immunolabeling of embryos by microinjection. Increased detection levels of extracellular and cell surface epitopes. Methods Mol Biol. 2000;135:183–9. doi: 10.1385/1-59259-685-1:183. [DOI] [PubMed] [Google Scholar]
- Manasek FJ, Burnside MB, Waterman RE. Myocardial cell shape change as a mechanism of embryonic heart looping. Dev Biol. 1972;29:349–71. doi: 10.1016/0012-1606(72)90077-2. [DOI] [PubMed] [Google Scholar]
- Pardanaud L, Altmann C, Kitos P, Dieterlen-Lievre F, Buck CA. Vasculogenesis in the early quail blastodisc as studied with a monoclonal antibody recognizing endothelial cells. Development. 1987;100:339–49. doi: 10.1242/dev.100.2.339. [DOI] [PubMed] [Google Scholar]
- Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical Recipes. 2. Cambridge University Press; 1992. [Google Scholar]
- Rongish BJ, Drake CJ, Argraves WS, Little CD. Identification of the developmental marker, JB3-antigen, as fibrillin-2 and its de novo organization into embryonic microfibrous arrays. Dev Dyn. 1998;212:461–71. doi: 10.1002/(SICI)1097-0177(199807)212:3<461::AID-AJA13>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Rosenquist GC. Cardia bifida in chick embryos: anterior and posterior defects produced by transplanting tritiated thymidine-labeled grafts medial to the heart forming regions. Teratology. 1970;3:135–42. doi: 10.1002/tera.1420030205. [DOI] [PubMed] [Google Scholar]
- Rosenquist GC, DeHaan RL. Migration of precardiac cells in the chick embryo: a radioautographic study. Carnegie Inst. Wash. Publication no. 625 (Contrib. Embryol.) 1966;38:111–121. [Google Scholar]
- Sakaguchi T, Kikuchi Y, Kuroiwa A, Takeda H, Stainier DY. The yolk syncytial layer regulates myocardial migration by influencing extracellular matrix assembly in zebrafish. Development. 2006;133:4063–72. doi: 10.1242/dev.02581. [DOI] [PubMed] [Google Scholar]
- Satin J, Fujii S, DeHaan RL. Development of cardiac beat rate in early chick embryos is regulated by regional cues. Dev Biol. 1988;129:103–13. doi: 10.1016/0012-1606(88)90165-0. [DOI] [PubMed] [Google Scholar]
- Sepich DS, Solnica-Krezel L. Analysis of cell movements in zebrafish embryos: morphometrics and measuring movement of labeled cell populations in vivo. Methods Mol Biol. 2005;294:211–33. doi: 10.1385/1-59259-860-9:211. [DOI] [PubMed] [Google Scholar]
- Szabo A, Unnep R, Mehes E, Twal WO, Argraves WS, Cao Y, Czirok A. Collective cell motion in endothelial monolayers. Phys Biol. 2010;7:046007. doi: 10.1088/1478-3975/7/4/046007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh LA, Stainier DY. Fibronectin regulates epithelial organization during myocardial migration in zebrafish. Dev Cell. 2004;6:371–82. doi: 10.1016/s1534-5807(04)00063-2. [DOI] [PubMed] [Google Scholar]
- van den Berg G, Abu-Issa R, de Boer BA, Hutson MR, de Boer PA, Soufan AT, Ruijter JM, Kirby ML, van den Hoff MJ, Moorman AF. A caudal proliferating growth center contributes to both poles of the forming heart tube. Circ Res. 2009;104:179–88. doi: 10.1161/CIRCRESAHA.108.185843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varner VD, Taber LA. Not just inductive: a crucial mechanical role for the endoderm during heart tube assembly. Development. 2012;139:1680–90. doi: 10.1242/dev.073486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varner VD, Voronov DA, Taber LA. Mechanics of head fold formation: investigating tissue-level forces during early development. Development. 2010;137:3801–11. doi: 10.1242/dev.054387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiens DJ. An alternative model for cell sheet migration on fibronectin during heart formation. J Theor Biol. 1996;179:33–9. doi: 10.1006/jtbi.1996.0045. [DOI] [PubMed] [Google Scholar]
- Ye D, Lin F. S1pr2/Gα13 signaling controls myocardial migration by regulating endoderm convergence. Development. 2013;140:789–99. doi: 10.1242/dev.085340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir EA, Czirók A, Rongish BJ, Little CD. A digital image-based method for computational tissue fate mapping during early avian morphogenesis. Ann Biomed Eng. 2005;33:854–65. doi: 10.1007/s10439-005-3037-7. [DOI] [PubMed] [Google Scholar]
- Zamir EA, Czirók A, Cui C, Little CD, Rongish BJ. Mesodermal cell displacements during avian gastrulation are due to both individual cell-autonomous and convective tissue movements. Proc Natl Acad Sci USA. 2006;103:19806–11. doi: 10.1073/pnas.0606100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir EA, Rongish BJ, Little CD. The ECM moves during primitive streak formation—computation of ECM versus cellular motion. PLoS Biol. 2008;6:e247. doi: 10.1371/journal.pbio.0060247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A - maximum intensity dorso-ventral projections of confocal stacks obtained at HH8 (top panels), and HH11 (bottom panels). Whole mount specimens were immunostained with antibodies MF20 (red), B3D6 anti-fibronectin (blue), and anti-GFP (green) to enhance pCMLC2-GFP expression-mediated labeling of transfected cells. A′, A″ -10 μm plastic sections taken at the levels indicated with horizontal white lines. Aa and Ab: areas marked by white rectangles that show the colocalization of MF20 and CMLC2::GFP signals in greater detail. Scale bars: 100 μm.
The model builds up the mesoderm (white), endoderm (yellow) and myocardium (red) using particles and their mechanical interconnections represented as links. Panels A, B and C show the initial configuration from a posterior, anterior and ventral aspect. The endoderm and its associated links are removed for a better view of the myocardium in the corresponding panels A′, B′ and C′. The green links in the medial aspect of the myocardium (magnified inset in C′) indicate spatially localized contractility, the effect of which on the surrounding tissue will be evaluated by simulations. A: anterior, P: posterior, V: ventral, D: dorsal, R: right, L: left.
Simulations with an “incision” (removed links) at the anterior aspect of the AIP yield a widening wound and AIP. The pronounced antero-lateral movement of the myocardium is consistent with the observations yielding bifidic hearts. The initial configuration (as in Suppl. Fig. 2) is shown in panels A and C. In the inset a white bracket indicates the area where cell-cell contacts were removed. The mechanical equilibrium resulted by the prescribed cellular activity is shown in panels B and D. Panels A and B show all particles, while the endoderm and associated cell-cell connections are removed from panels C and D. Panel E show the tissue displacements during the simulations. Red and green arrows indicate movements of the myocardium and both surfaces of the folded mesoderm, respectively. Black arrows are guides for the eye, indicating the local direction of myocardial movements.
Myocardial progenitor fields undergo a dramatic change in position along the dorso-ventral embryonic axis in caudal-to-cranial progression. A – a panel from Fig. 3 by van den Berg et al., 2009 (p. 182), to illustrate the anatomy of myocardial fields at HH8 and to indicate the cranio-caudal position of the sections shown in panel B. B - a series of 10 μm-thick sections (taken up to 200 μm apart from each other) through the cardiac fields of an HH8 embryo. The sections’ approximate positions, on the cranio-caudal axis, are indicated by lines connecting to A. Arrowheads point to the dorsal bends of the myocardium protruding into the coelom. Green – MF20 antibody-labeled myocardial progenitors, blue – DAPI. d – dorsal, and v - ventral side of the embryo. All sections are presented in the same orientation. Scale bar: 100 μm.
Heart tube assembly in a Tie1::H2B-YFP quail embryo during stages HH8-11 with endothelial/endocardial nuclei in green, CMLC2::MitoRFP-expressing myocardial progenitors shown in red and fluorescent B3D6 antibody-labeled fibronectin in blue.
Cellular behaviors of CMLC2::GFP-expressing myocardial progenitors within the embryonic right heart field. Original images were color-coded to reflect the position of cells located at different dz levels relative to the dorso-ventral axis of the embryo, with dorsal z-planes represented in blue, and ventral - in red portions of the spectrum.
Left panel: Movements of CMLC2::MitoRFP-expressing myocardial progenitors (red) relative to the MitoTracker Green-labeled endoderm (green). Right panel: PIV-derived vectors illustrating the magnitude and directions of observed myocardial (red vectors) and endodermal (green vectors) displacements that took place during the 15 min time period that separates the adjacent images. Vectorial differences between the above parameters, reflecting myocardial movements relative to the endoderm, are represented with cyan vectors.
Development of the cardia bifida phenotype in an embryo that underwent an incision through the AIP endoderm at the ventral midline at HH8. CMLC2::GFP-expressing myocardial progenitors are shown in green, and B3D6 mAb labeled fibronectin ECM in red.
Heart tube assembly in an embryo that underwent bilateral longitudinal incisions lateral to the heart field positions at HH8. CMLC2::GFP-expressing myocardial progenitors are shown in green, and B3D6 mAb labeled fibronectin ECM in red.
Deformations that take place within the embryonic left heart field in response to the incision placed within the cardiac field parallel to its anterior-posterior axis. CMLC2::GFP-expressing myocardial progenitors are shown in green, and B3D6 mAb-labeled fibronectin ECM in red.
Deformations taking place within heart fields in response to bilateral incisions across the cardiac fields perpendicular to their anterior-posterior axes. CMLC2::GFP-expressing myocardial progenitors are shown in green, and B3D6 mAb-labeled fibronectin ECM in red.
Deformations taking place within the embryonic left heart field (CMLC2::GFP-expressing) in response to an incision placed perpendicular to its anterior-posterior axis. Original images were colorcoded to reflect the position of cells located at different dz levels relative to the dorso-ventral axis of the embryo, with dorsal z-planes represented in blue, and ventral - in red portions of the spectrum.