Abstract
Epigenetic regulation, including histone modification, is a critical component of gene regulation, although precisely how this contributes to the development of complex tissues such as the neural retina is still being explored. We show that during retinal development in mouse, there are dynamic patterns of expression of the polycomb repressive complex 2 (PRC2) catalytic subunit EZH2 in retinal progenitors and some differentiated cells, as well as dynamic changes in the histone modification H3K27me3. Using conditional knockout of Ezh2 using either Pax6-αCre or Six3-Cre, we find selective reduction in postnatal retinal progenitor proliferation, disruption of retinal lamination, and enhanced differentiation of several late born cell types in the early postnatal retina, including photoreceptors and Müller glia, which are ultimately increased in number and become reactive. RNA-seq identifies many non-retinal genes upregulated with loss of Ezh2, including multiple Hox genes and the cell cycle regulator Cdkn2a, which are established targets of EZH2-mediated repression. ChIP analysis confirms loss of the H3K27me3 modification at these loci. Similar gene upregulation is observed in retinal explants treated with an EZH2 chemical inhibitor. There is considerable overlap with EZH2-regulated genes reported in non-neural tissues, suggesting that EZH2 can regulate similar genes in multiple lineages. Our findings reveal a conserved role for EZH2 in constraining the expression of potent developmental regulators to maintain lineage integrity and retinal progenitor proliferation, as well as regulating the timing of late differentiation.
Keywords: histone modification, retina, neurogenesis, proliferation, PRC2
Introduction
During the development of complex tissues, the balance between proliferation and differentiation of progenitors, and the timing of differentiation is critical to ensure that various cell types are generated in appropriate numbers. The dramatic changes in gene expression driving these events are regulated primarily by transcription factors that are expressed in stage and cell-type dependent manner. This has been well defined in the developing neural retina, where specific sets of transcription factors define retinal progenitor identity and regulate the ordered genesis of distinct retinal cell types (Bassett and Wallace, 2012). But there is increasing evidence that in many systems epigenetic regulation, such as histone modification, also plays an important role. These modifications can serve as gatekeepers for the activity of transcription factors, and thus regulate gene activation or repression (Laugesen and Helin, 2014). While the molecular machinery involved in histone modifications have been well studied, their precise roles during in vivo development are still being explored.
One of the key repressive modifications is the trimethylation of lysine 27 on histone H3 (H3K27me3). The di-and tri-methylation of H3K27 is primarily catalyzed by polycomb repressive complex 2 (PRC2), with the enzymatic activity of the complex mediated by the subunit Enhancer of zeste 2 (EZH2), and in some instances by EZH1 (Margueron and Reinberg, 2011). Polycomb group proteins were originally identified in Drosophila for their role in silencing Hox genes (Schuettengruber et al., 2007), but are now recognized to have broader roles in both invertebrate and vertebrate development, including neural development (Aldiri and Vetter, 2012; Margueron and Reinberg, 2011). In embryonic stem cells (ESCs) PRC2 is required for neural differentiation (Pasini et al., 2007; Shen et al., 2008), and PRC2 targets many of the genes involved in neurogenesis for H3K27me3 modification (Boyer et al., 2006; Lee et al., 2006).
The in vivo role of Ezh2 during neural development is complex. During the late phase of neocortical development PRC2 cooperates with PRC1 to restrict the generation of neurons to allow astrocyte production, with Ezh2 conditional knockouts showing a prolonged neurogenic phase (Hirabayashi and Gotoh, 2010; Hirabayashi et al., 2009). In contrast, early deletion of Ezh2 in the developing cerebral cortex causes enhanced early neurogenesis, accompanied by earlier astrocyte production and a premature termination of cell proliferation (Pereira et al., 2010). Furthermore, knockdown of Ezh2 in neural stem cell cultures also leads to a reduction in their proliferation and self-renewal potential and enhanced production of astrocytes rather than oligodendrocytes upon differentiation (Sher et al., 2008). Interestingly, in the neural crest, Ezh2 is not required for neural development (Schwarz et al., 2014). Thus, Ezh2 appears to play complex roles in the developing nervous system, and can promote or block differentiation, maintain proliferation, and/or regulate the timing of cell fate acquisition. These diverse developmental roles have also been ascribed to Ezh2 function in other tissues, including skin, skeletal muscle, heart and lung (Ezhkova et al., 2009; He et al., 2012; Juan et al., 2011; Snitow et al., 2015; Woodhouse et al., 2013). However, it is unclear whether core molecular mechanisms are responsible for these functions in multiple tissues.
The developing neural retina is an excellent system to investigate this question, since progenitor properties and the transcriptional regulation of cell fate determination are well described (Bassett and Wallace, 2012). Furthermore, genome-wide H3K27me3 histone modification has been characterized at several key stages of mouse retinal development (Popova et al., 2012). In Xenopus, we previously found that knocking down or blocking the PRC2 core component Ezh2 reduces retinal neurogenesis and promotes Müller glial cell differentiation (Aldiri et al., 2013). We also observed reduced retinal progenitor proliferation, in part due to upregulation of the Cdk inhibitor p15 (Ink4b; Cdkn2b), which is a direct target of EZH2-mediated repression in multiple tissues (Popov and Gil, 2010). Conditional disruption of Ezh2 in the mouse retina using Dkk-Cre showed that Ezh2 is also required to repress cdkn2a and maintain progenitor proliferation and the normal timing of retinal differentiation (Aldiri et al., 2013; Iida et al., 2014).
However, we lack a detailed understanding of the molecular changes associated with loss of PRC2-mediated repression in the developing retina and how this relates to PRC2 function in other tissues. We performed conditional disruption of Ezh2 in the mouse retina using Pax6-αCre, and assessed effects on retinal development as well as gene expression changes by RNAseq. Our analysis shows that Ezh2 and its repressive histone modifications are necessary to maintain the transcriptional identity as well as proliferation state of retinal progenitors, to regulate proper timing of differentiation during early postnatal retinal development, and to constrain the generation of Müller glia.
Materials and Methods
Ezh2fl/fl, Pax6-αCre and R26EYFP mice and their genotyping have been described previously (Marquardt et al., 2001; Shen et al., 2008; Srinivas et al., 2001; Su et al., 2003). Mouse breeding details are provided in SI Materials and Methods. For immunostaining, retinal sections were generated as previously described (La Torre et al., 2013). Nuclear counterstain was done using either DAPI or Hoechst. Detailed methods and antibodies used are listed in SI Materials and Methods. In situ hybridization was performed on whole embryos and retinal sections using digoxigenin (DIG)-labeled riboprobe for mouse Ezh2, as previously described (Zhang et al., 2008). Hematoxylin and eosin staining of retinal sections was performed as previously described (Zhang et al., 2008). For explant culture, retinas from E14 mouse embryos were dissected and cultured as previously described (La Torre et al., 2013) with GSK126 (Xcess Biosciences) added at 2μM or 10μM, and corresponding concentrations of vehicle (DMSO) added to the control samples.
For RNAseq analysis the peripheral one-third of E16.5 retinas was dissected, left and right eyes combined, and total RNA isolated using RNeasy Plus Mini Kit according to manufacturer’s protocol (Qiagen). RNA integrity was confirmed using a Bioanalyzer RNA 6000 Nano Chip. Four biological replicates of Ezh2fl/− CKO animals and Ezh2fl/+ controls from the same litter were used for RNA sequencing on a 50 bp single-end run on Illumina HiSeq 2000. See SI Materials and Methods for details. RNAseq data have been deposited to the Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo/) under series accession number GSE65082.
Primers for PCR and ChIP are listed in Supplemental Table 7. Detailed methods for RNA isolation, RT-PCR and quantitative RT-PCR are described in SI Materials and Methods. For chromatin immunoprecipitation (ChIP) analysis, the peripheral one-third of postnatal day 0 retinas were dissected and processed, as described in SI Materials and Methods. Values were averaged from 3 biologically independent experiments and normalized for ChIP efficiency across biological replicates.
Results
Ezh2 is dynamically expressed during retinal development
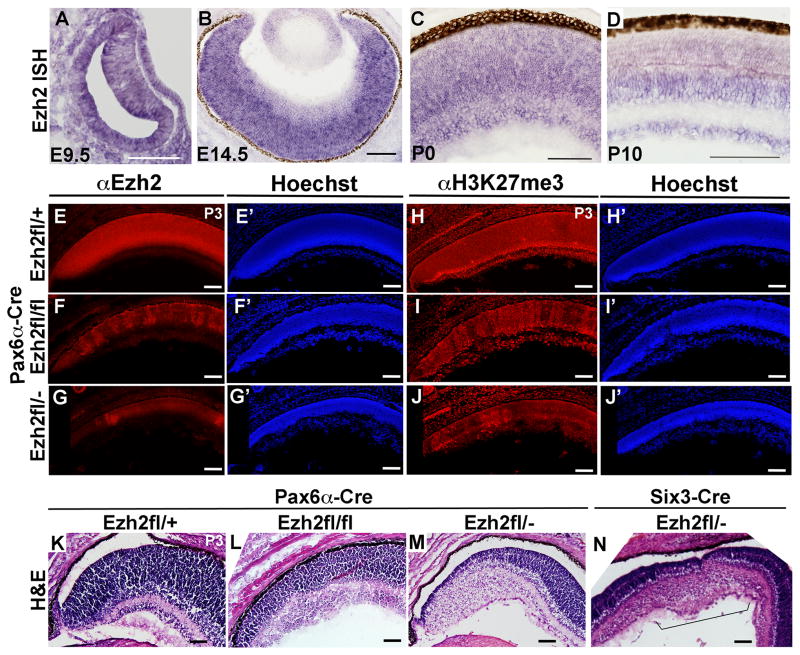
In the developing Xenopus retina, Ezh2 is enriched in retinal progenitors and downregulated in differentiated cells (Aldiri et al., 2013). To determine where Ezh2 is expressed during mouse retinal development we performed in situ hybridization and found expression in progenitor cells throughout the optic cup at e9.5 (Figure 1A), and in retinal progenitors at e12.5 (not shown). Expression was reduced in the emerging retinal ganglion cell layer (GCL) at e14.5, but maintained in retinal progenitors (Figure 1B). By P0, Ezh2 expression was evident in the GCL, as well as in the neuroblastic layer (Figure 1C), and by P10 Ezh2 expression was largely restricted to the GCL and inner nuclear layer (INL). Thus Ezh2 is enriched in both retinal progenitors and subsets of differentiating cells, with dynamic changes in expression over the course of development.
Figure 1. Ezh2 expression during retinal development and after Ezh2 conditional deletion.
(A–D) In situ hybridization (ISH) analysis. (E–G) EZH2 immunostaining shows increasing loss of protein in the peripheral retina with conditional deletion using Pax6-αCre in Ezh2fl/fl and Ezh2fl/− mice relative to control Ezh2fl/+ mice at P3. (E′–G′) Hoechst staining of the same sections. (H–J) Similarly H3K27me3 immunostaining shows increasing loss of this histone mark. (H′–J′) Hoechst staining of the same sections. (K–M) Hematoxylin and eosin staining shows increasing disruption to retinal lamination with conditional deletion using Pax6-αCre in Ezh2fl/fl and Ezh2fl/− mice relative to control Ezh2fl/+ mice at P3. (N) Similar disruption of retinal lamination in the central retina is observed after conditional deletion of Ezh2 using Six3-Cre in Ezh2fl/− mice. Scale bar = 100μm
Loss of Ezh2 results in impaired retinal development
To assess the function of Ezh2 during retinal development, we conditionally inactivated Ezh2 in retinal progenitors by crossing Pax6-αCre mice with either Ezh2fl/fl or Ezh2fl/− mice (Shen et al., 2008; Su et al., 2003). The Pax6-αCre retinal driver promotes recombination in the peripheral retina from E10.5 onwards, which is at the onset of retinal neurogenesis (Marquardt et al., 2001). By immunostaining retinal sections at P3, EZH2 protein was detected at high levels through the neuroblastic layer and at lower levels in the GCL in peripheral retina of control Ezh2fl/+::Pax6-αCre (Ezh2fl/+) mice, with mosaic loss of EZH2 protein in Ezh2fl/fl::Pax6-αCre (Ezh2fl/fl CKO) retina, that was more extensive in Ezh2fl/−::Pax6-αCre (Ezh2fl/− CKO) retina (Figure 1E–G). In addition, H3K27me3 labeling was enriched in the GCL and in the inner part of the neuroblastic layer at P3 in Ezh2fl/+ control retina (Popova et al., 2012), but showed mosaic loss in Ezh2fl/fl CKO retina, that was more extensive in Ezh2fl/− CKO retina (Figure 1H–J). When crossed with R26EYFP mice, the reporter showed mosaic expression that corresponded to the areas with loss of H3K27me3 (Supplemental Figure 1).
Since there was evidence for increasing penetrance of the phenotype with Ezh2fl/− versus Ezh2fl/fl CKO, we analyzed the phenotype in both to assess effects on retinal development. Hematoxylin and eosin staining of P3 retinal sections revealed disruption of retinal lamination and reduced thickness of the neuroblastic layer in Ezh2fl/fl CKO mice, that was more extensive in Ezh2fl/− CKO mice (Figure 1K–M). For Ezh2fl/fl CKO mice, the R26EYFP reporter confirmed that the phenotype corresponded to the areas of Cre-mediated recombination (Supplemental Figure 1). We also confirmed disruption of retinal lamination that was restricted to the central retina region in Ezh2fl/− CKO animals using Six3-Cre (Figure 1N), which drives recombination in retinal progenitors (Furuta et al., 2000). Thus, Ezh2 is required for normal retinal development.
Ezh2 is required to maintain late retinal progenitor proliferation
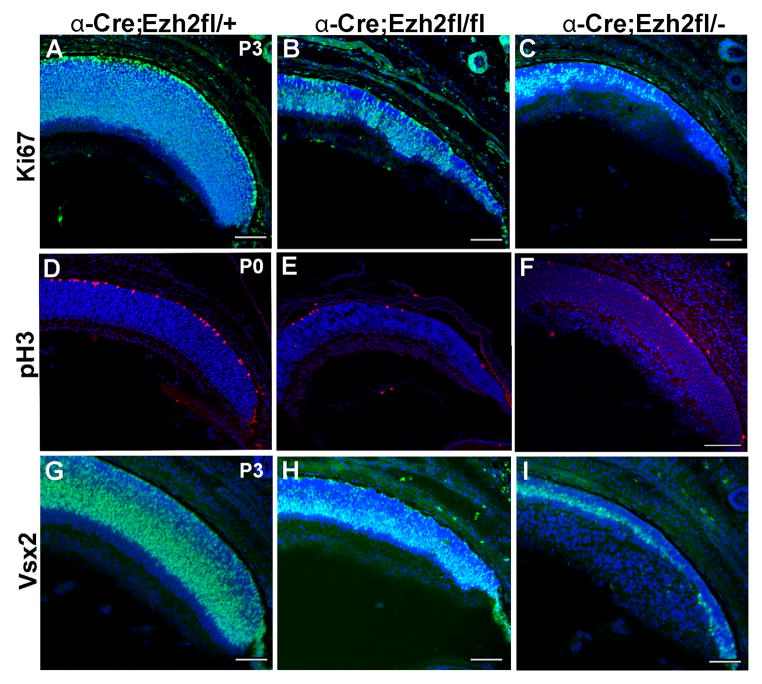
Reduced thickness of the neuroblastic layer suggested reduced retinal progenitor number in Ezh2 CKO retina. Labeling of progenitors with the proliferation marker Ki67 was reduced at P3 in Ezh2fl/fl CKO mice and to a greater extent in Ezh2fl/− CKO mice (Figure 2A–C). The reduction in proliferation was confirmed by a similar reduction of pH3 labeling, which marks progenitor cells at the G2-M transition (Figure 2D–F). We also found significant reduction at P3 in the numbers of cells labeled for the progenitor marker Vsx2 in Ezh2fl/fl CKO mice that was more pronounced in Ezh2fl/− CKO mice (Figure 2G–I).
Figure 2. Ezh2 deletion results in reduced retinal progenitor proliferation.
(A–C) Increasing loss of Ki67 labeling (green) in the peripheral retina in Ezh2fl/fl and Ezh2fl/− CKO mice relative to control Ezh2fl/+ mice at P3. (D–F) Similarly, there is increasing reduction of pH3 labeling (red), and (G–I) reduced labeling for the progenitor marker Vsx2 (green). Scale bar = 100μm
There was no effect on EdU labeling at e15 in Ezh2fl/fl CKO mice, suggesting that the proportion of cells in S-phase was unaltered (Supplemental Figure 1). In addition, the proportion of cycling cells appeared unchanged since there was no effect on PCNA labeling or Ki67 labeling at e15.5 in Ezh2fl/− CKO mice, although we cannot rule out subtle effects on cell cycle length (Supplemental Figure 1 and data not shown). Although there was no apparent effect on retinal progenitor proliferation, there was loss of EZH2 and H3K27me3 labeling of progenitors in the CKO region of the peripheral retina at this stage (Supplemental Figure 1). By P0 there was modest reduction in Ki67 labeling in Ezh2fl/− CKO mice, suggesting that premature exit of progenitors has begun by this stage (Supplemental Figure 1). Thus, we conclude that Ezh2 is required to maintain proliferating progenitors during postnatal development, but not embryonic retinal development.
Ezh2 deletion alters retinal ganglion cell and amacrine cell numbers
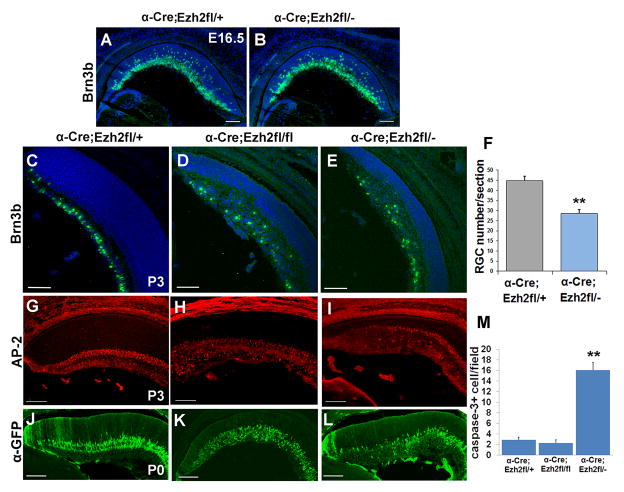
Since disruption of Ezh2 has an effect on retinal progenitor maintenance, we predicted that this would impact retinal cell fate specification. Retinal neurons in mouse are generated in a defined sequence beginning at e10.5 with retinal histogenesis largely complete by postnatal day 14. An early wave of differentiation during the embryonic period generates the majority of retinal ganglion cells, horizontal cells, cones and amacrine cells, while a later postnatal wave generates the bulk of rod photoreceptors, bipolar cells and Müller glia (Bassett and Wallace, 2012). We first assessed early born retinal neurons since we did not observe an effect on proliferation during the embryonic period of retinal neurogenesis. Consistent with this, there was no major effect on Brn3b-positive RGCs at e15.5 in Ezh2fl/− CKO mice (Figure 3A,B). But by P3 the numbers of Brn3b-labeled RGCs was significantly reduced in both Ezh2fl/− and Ezh2fl/fl CKO retina (Figure 3C–F). These results suggest that although the ganglion cells are generated in approximately normal numbers, these cells are reduced postnatally (see below).
Figure 3. Ezh2 deletion alters retinal ganglion cell and amacrine cell numbers.
(A,B) No change in Brn3b labeling at e16.5 (green). (C–E) Brn3b immunostaining (green) shows disruption of the RGC layer and reduced RGC number in Ezh2fl/fl and Ezh2fl/− CKO retina relative to control Ezh2fl/+ at P3. (F) There is a 36% reduction in Brn3b+ cells at P3 in Ezh2fl/− CKO retina. N= 45 sections from 6 retinas. Error bars represent the standard error, ** = P < 0.001 Student’s t-test. (G–I) Ap2+ amacrine cells are increased relative to control Ezh2fl/+ retina at P3 in Ezh2fl/fl CKO, but reduced in Ezh2fl/− CKO. (J–L) Similarly, at P0 the GFP reporter that is expressed from the Pax6-αCre transgene in amacrine cells is expanded in Ezh2fl/fl CKO, but reduced in Ezh2fl/− CKO retina relative to control Ezh2fl/+. (M) The number of cleaved caspase 3-positive cells is increased at P0 in Ezh2fl/− CKO but not Ezh2fl/fl CKO retina relative to control Ezh2fl/+. N=24, 24 and 27 sections from 6 retinas each, respectively for Ezh2fl/+, Ezh2fl/fl and Ezh2fl/−. Error bars represent the standard error, ** = P < 0.001 Student’s t-test. Scale bar = 100μm
Amacrine cells are also largely born during the embryonic period, so we investigated whether loss of Ezh2 affected this retinal cell population. We observed an increase in Ap2-labeled amacrine cells in Ezh2fl/fl CKO mice relative to Ezh2fl/+ controls at P3 (Figure 3G,H). This was confirmed by an increase in GFP-labeled cells in Ezh2fl/fl CKO mice, since at postnatal stages the GFP reporter from the Pax6-αCre transgene is expressed in amacrine cells (Figure 3J,K) (Marquardt et al., 2001). In contrast, there was a decrease in Ap2-labeled amacrine cells in Ezh2fl/− CKO mice, as well as a decrease in GFP-labeled cells at P3 (Figure 3I,L). By P10, Ap2-labeled amacrine cells were significantly reduced in Cre+ regions in Ezh2fl/fl CKO retina, largely due to reduced number of labeled cells in the INL (N= 3 retinas per genotype, 3 sections/retina, P < 0.01 by Student’s t-test), and were also clearly reduced at P14 in Ezh2fl/− CKO retina (data not shown). Thus we observe distinct effects on amacrine cell number in Ezh2fl/− versus Ezh2fl/fl CKO mice, but in both cases the amacrine and ganglion cell layers were disorganized and intermixed.
To determine whether the survival of the neurons was compromised from the loss in Ezh2, we labeled the retinas for cleaved caspase 3. We observed an increase in the numbers of cells labeled for cleaved caspase 3 at P0 in Ezh2fl/− CKO mice (Figure 3M). A small subset of cleaved caspase 3 positive cells colabeled for Brn3b (data not shown), indicating that an increase in cell death that may partially account for the reduced number of early born cell types in Ezh2fl/− CKO animals. This increase in cell death was not evident at P3 and was not significant at either stage in Ezh2fl/fl CKO mice. Thus, we conclude that Ezh2 is not required for the genesis of early born neurons during the embryonic period but is ultimately required for the normal complement of early retinal cell types.
Ezh2 disruption results in increased photoreceptor differentiation in the early postnatal retina
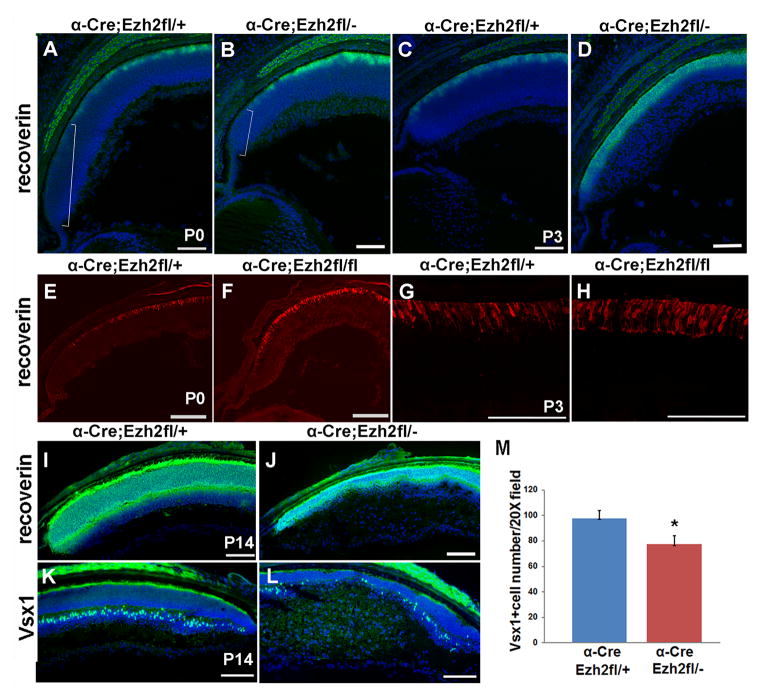
Since loss of Ezh2 results in reduced proliferation and progenitor depletion at early postnatal ages, we assessed whether this would affect the differentiation of later born cell types. We found that there was increased expression of photoreceptor differentiation markers in the peripheral retina of both Ezh2fl/− and Ezh2fl/fl CKO mice at early postnatal ages. At P0, recoverin labeling in the outer nuclear layer (ONL) extended more peripherally in both Ezh2fl/− and Ezh2fl/fl CKO retina, suggesting that more progenitors in this region were differentiating at this timepoint than normal (Figure 4A,B, E,F), with the average distance from the ciliary edge to the first recoverin labeling decreased by 40% in Ezh2fl/fl CKO retina (76uM) compared to Ezh2fl/+ controls (126uM) (N=3, P<0.01). Consistent with this, by P3 there was a thickened layer of recoverin-labeled cells in both Ezh2fl/− and Ezh2fl/fl CKO retina (Figure 4C,D,G,H). This was not simply due to derepression of recoverin expression, since we also observed increased labeling for the synaptic marker SV2 in the outer plexiform layer in Ezh2fl/fl CKO retina compared to Ezh2fl/+ controls, indicating that more functional photoreceptors are present in the P3 Ezh2 cKO retina (Supplemental Figure 2). This is likely due to more cells exiting and differentiating as photoreceptors at the earlier timepoint, but it is also possible that the process of photoreceptor differentiation is accelerated, or that retinal cell birthorder is altered.
Figure 4. Ezh2 deletion increases rod photoreceptor differentiation in the early postnatal retina, and affects overall numbers of later born retinal cell types.
(A,B) Conditional deletion in Ezh2fl/− retina results in increased photoreceptor differentiation relative to control Ezh2fl/+ at P0 as detected by recoverin labeling (green). Bracket indicates the extent of the undifferentiated region at the peripheral margin of the retina, which is reduced in CKO animals. (C,D) Increased photoreceptor differentiation is also evident at P3 as evident by the thickened ONL. (E,F) This is similarly observed in Ezh2fl/fl CKO retina at P0, and (G,H) at P3 (recoverin, red). (I,J) By P14 recoverin-positive photoreceptors are reduced in Ezh2fl/− CKO. (K,L) Bipolar cells are reduced in the Ezh2 CKO region at P14 as detected by Vsx1 labeling (green). (M) Cell counts show a significant reduction in Vsx1-postive cells at P14 in Ezh2fl/− CKO retina. N=10, 12 sections, 4 retinas each respectively for Ezh2fl/+ and Ezh2fl/−. Error bars represent the standard error. * = P < 0.01 Student’s t-Test. Scale bar = 100μm
However, by P14 the recoverin-labeled ONL was noticeably thinner in Ezh2fl/− CKO retina (Figure 4I,J). The reduction in proliferation in the postnatal retina, coupled with the reduced ONL thickness suggests that the progenitor depletion ultimately leads to an overall reduction in photoreceptor production. Consistent with this, the number of cells labeled with the photoreceptor progenitor marker Otx2 was reduced by P3 in Ezh2fl/fl CKO retina (data not shown).
Although the loss in Ezh2 leads to an acceleration in the timing of differentiation of photoreceptors in the postnatal retina, we did not observe a similar premature expression of the bipolar cell marker Vsx1 in Ezh2fl/− CKO retina at P3, prior to the onset of bipolar cell differentiation, or at P6, which is at the early stages of bipolar cell differentiation (data not shown). By P14, the number of Vsx1 labeled bipolar cells was significantly reduced in Ezh2fl/− CKO retina (Figure 4K,L). Thus, the requirement for Ezh2 in the normal timing of photoreceptor differentiation does not extend to all late generated cell types.
Ezh2 disruption results in increased early Müller glia differentiation
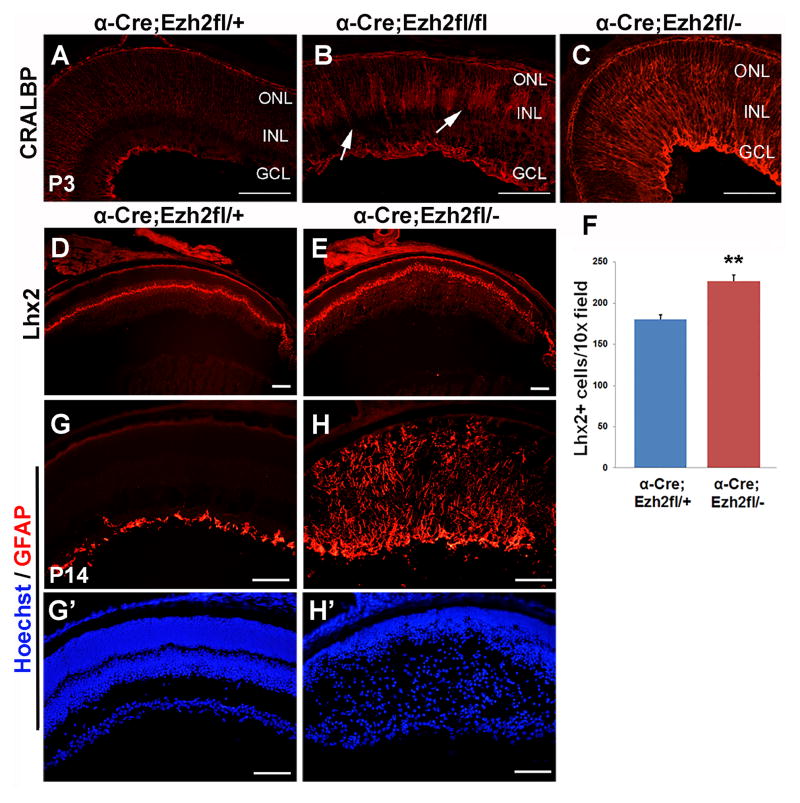
Since blocking Ezh2 in the Xenopus retina results in increased Müller glial cell differentiation (Aldiri et al., 2013), we assessed whether these cells would be affected with loss of Ezh2 in the mouse retina. We found that at P3 there was increased Müller glial differentiation in the peripheral retina of both Ezh2fl/− and Ezh2fl/fl CKO mice as detected by CRALBP expression (Figure 5A–C). This increase was already evident at P0 (data not shown). By P14 there was a 25% increase in Lhx2 positive Müller glia in the CKO region in Ezh2fl/− CKO retina as compared to Ezh2fl/+ retina (Figure 5D–F). Consistent with this, at P10 there was also an expansion of Sox2-labeled cells in the INL of Ezh2fl/fl CKO retina, suggesting an increase in Müller glia (data not shown). However, at P14 in Ezh2fl/− CKO retina Müller glia were reactive, and showed significant disorganization and upregulation of GFAP (Figure 5G,H). Glutamine synthase, was significantly downregulated, consistent with Müller glia reactivity (data not shown). At P10 we found no coexpression of NeuN, which labels neurons, and Id1, which labels Müller glia, in the CKO region of Ezh2fl/fl retina, indicating that the distinction between neuron versus glial identity is maintained (data not shown). Thus, we conclude that Ezh2 is required to constrain Müller glial cell differentiation, and to regulate the timing of their genesis.
Figure 5. Ezh2 deletion enhances Müller glia differentiation and leads to reactivity.
(A–C) Conditional deletion in Ezh2fl/fl and Ezh2fl/− retina results in increased Müller glia differentiation at P3 relative to control Ezh2fl/+ retina as detected by Cralbp immunstaining. (D,E) Lhx2 immunostaining at P14 shows an increase in Müller glia in Ezh2fl/− CKO retina. (F) Counts of Lhx2–labeled cells show a 25% increase in Müller glia in Ezh2fl/− CKO retina relative to Ezh2fl/+ controls. N=20, 24 sections, 6 retinas each respectively for Ezh2fl/+ and Ezh2fl/−. ** = P < 0.001, Student’s T-test. (G,H) Müller glia at P14 in the Ezh2fl/− CKO region become reactive and show increased GFAP expression. (G′,H′) Hoechst staining of the same sections. Scale bar = 100μm. GCL = ganglion cell layer, INL = inner nuclear layer, ONL = outer nuclear layer.
Ezh2 is required to inhibit the expression of many developmental regulators and non-retinal genes
To understand the mechanisms underlying this retinal phenotype we sought to determine which genes are deregulated by loss of Ezh2. We dissected peripheral retina tissue at e16.5 to enrich for regions with loss of Ezh2, and then performed RNA-seq. This age is prior to apparent changes in retinal development, since we sought to identify gene expression changes that are directly due to loss of Ezh2. A total of 232 genes were differentially expressed in Ezh2fl/− CKO versus Ezh2fl/+ control animals with a fold change cutoff of 2 (Abs log2 > 1, p < 0.05), with the majority of genes upregulated and only four significantly downregulated, which is consistent with the canonical function of EZH2 in promoting gene repression (Supplemental Table 1).
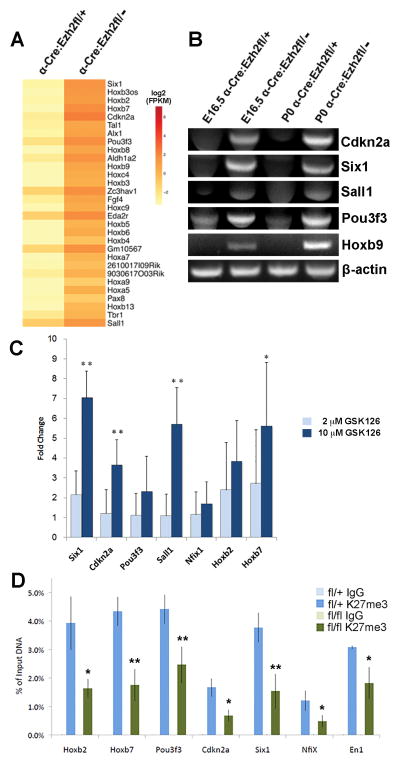
Examination of the top 30 most highly upregulated genes showed that most of them had either no or very low expression in the control retina, but were significantly upregulated in Ezh2fl/− CKO retina (Figure 6A). This included genes encoding many developmental regulators such as multiple Hox transcription factors (15/30 top upregulated genes), transcription factors (eg. Six1, Pou3f3), signaling factors (eg. Fgf4) and the cell cycle inhibitor Cdkn2a. Cdkn2b was also upregulated (Supplemental Table 1). To confirm the results of our RNA-seq analysis we performed RT-PCR for a subset of the most highly upregulated genes, Cdkn2a, Six1, Sall1, Pou3f3, and Hoxb9 using RNA isolated from peripheral retina tissue and found that these candidate genes were upregulated at e16.5, consistent with our RNA-seq timepoint, as well as at P0, when the retinal phenotype is apparent (Figure 6B). To further confirm these findings, we sought to independently reduce EZH2 activity. E14 retinal explants were treated with the EZH2 chemical inhibitor GSK126 at 2μM or 10μM for four days, then RNA isolated and quantitative RT-PCR performed to assess fold change in expression for Six1, Cdkn2a, Pou3f3, Sall1, Nfix, Hoxb2 and Hoxb7. There was significant upregulation of Six1, Cdkn2a, Sall1, and Hoxb7, particularly at the 10μM concentration, although Pou3f3, Nfix, and Hoxb2 were upregulated but not significantly (Figure 6C).
Figure 6. RNA-Seq reveals significant gene upregulation following conditional deletion of Ezh2.
(A) Heat map representation of log2 (avg FPKM) for the top thirty genes differentially expressed by at least twofold in peripheral retina at e16.5 in Ezh2fl/− CKO mice relative to control Ezh2fl/+ mice. Genes with no expression are assigned an FPKM value of 0.1. A color-coded log2 scale is shown at the side of the heat map. (B) RT-PCR for a subset of these genes confirms their upregulation following conditional deletion of Ezh2 at both e16.5 and P0. (C) Quantitative RT-PCR analysis following treatment of retinal explants with GSK126, a small molecule inhibitor of EZH2, also shows upregulation of many genes that increase in the Ezh2 CKO. (D) Chromatin immunoprecipitation (ChIP)-qPCR for H3K27me3 on a subgroup of genes that are upregulated after Ezh2 deletion confirms depletion of H3K27me3. The Engrailed1 promoter was also assayed as a gene that is not upregulated after Ezh2 deletion. * = p<0.05, ** = p<0.01, Students’ T-test.
Many of these most highly upregulated genes are repressed via EZH2 in other tissues, suggesting that EZH2 may target common genes regardless of cell lineage. To further investigate this, we compared our gene expression profiling data for e16.5 retina (GSE65082), which is ectodermal in origin, to data available in the GEO data repository for gene expression changes in other lineages after loss of Ezh2 (http://www.ncbi.nlm.nih.gov/geo/). For example, out of the 232 genes differentially expressed in our study, 40 were also significantly increased in e12.5 cardiomyocytes, which is mesodermal in origin (GSE29992; abs log2 > 0.585, p < 0.05; (He et al., 2012)). Similarly, 29 were also significantly increased in e14.5 lung, which is endodermal in origin (GSE66060; abs log2 > 0.585, p < 0.05; (Snitow et al., 2015)). Multiple genes or gene families were upregulated in all three datasets, including Cdkn2a, Hox genes and Pou3f3 (Supplemental Table 2). While some genes are upregulated in specific lineages, the fact that there is such overlap suggests that Ezh2 is required to repress similar classes of genes in multiple lineages, regardless of embryonic germ layer origin.
We also compared the gene expression changes we found in embryonic retina to those reported for adult tissues. An analysis of gene expression from right ventricles of adult wild type and Ezh2-deficient hearts showed up to 60 genes common with our dataset (GSE34274; Abs log2 > 0.585, p < 0.05; (Delgado-Olguin et al., 2012)). Of these, 18 genes were also upregulated in either e12.5 cardiomyocytes or e14.5 lung (Supplemental Table 3). This includes the skeletal muscle gene Six1, which was among the most highly upregulated in both lineages. This analysis strikingly shows that overlapping genes are repressed in an Ezh2-dependent manner regardless of lineage or developmental stage.
Many, but not all Ezh2 regulated genes have high levels of H3K27me3
To assess the link between H3K27me3 and retinal gene expression changes in the Ezh2 CKO, we examined the genes that are upregulated in the Ezh2 CKO and their levels of H3K27 trimethylation, as reported for the postnatal day 1 (P1) mouse retina (Popova et al., 2012). Among the genes that are upregulated in the Ezh2 CKO are those with the highest levels of H3K27me3 (Supplemental Table 4). In order to confirm that the conditional knockout of Ezh2 results in depletion of H3K27me3, we performed chromatin immunoprecipitation (ChIP) for this histone modification on a subgroup of these genes. The promoter regions (within 1kb upstream of the transcription start site) of the upregulated genes Hoxb2, Hoxb7, Pou3f3, Cdkn2a, Six1, Nfix were assayed for H3K27me3 enrichment by ChIP-qPCR from Ezh2fl/fl CKO and Ezh2fl/+ littermates. The Engrailed1 promoter was also assayed as a gene that is not upregulated after Ezh2 deletion. At all assayed gene promoters, H3K27me3 levels were reduced, confirming the predicted effect of Ezh2 deletion (Figure 6D). H3K27me3 levels were reduced, but not absent, at least in part due to the incomplete nature of the conditional deletion throughout the retina.
We found that H3K27me3 levels were reduced at the Engrailed1 promoter, although this gene is not upregulated following Ezh2 deletion. We identified additional genes that are most highly marked with H3K27me3 in the P1 mouse retina, but that are not significantly increased in the Ezh2 CKO (Supplemental Table 5), suggesting additional repressive mechanisms, or developmental gain of H3K27me3 at P1 as compared to e16.5. Conversely, many genes that increase in the Ezh2 CKO do not appear to have significant levels of H3K27me3 (Supplemental Table 6), suggesting indirect regulation, or developmental loss of H3K27me3 by P1. An important caveat is that different retinal cell types likely have different levels of these modifications (Popova et al., 2014; Popova et al., 2012), which may not be detected by bulk analysis. Overall, we conclude that EZH2 maintains repression of many potent developmental regulators, including genes not normally expressed in the developing retina, although clearly other repressive mechanisms are also involved in maintaining appropriate levels of gene expression in retinal progenitors.
Discussion
Development of the retina involves the ordered genesis of distinct retinal cell types from proliferating retinal progenitors, and by conditional deletion of Ezh2 in developing retina, we show that Ezh2 plays multiple roles in the development of this tissue. Ezh2 is required in postnatal retina to maintain progenitor proliferation, coordinate the timing of differentiation and generate the appropriate complement of late retinal cell types. In addition, RNAseq analysis revealed that loss of Ezh2 results in upregulation of cell cycle inhibitors and many non-retinal genes, indicating that Ezh2 also maintains transcriptional integrity of cells in this lineage. Genes upregulated after loss of Ezh2 in retina overlap significantly with those repressed by Ezh2 in other tissues of distinct developmental origin, suggesting that Ezh2 may have a core function in repressing the expression of potent developmental regulators and cell cycle genes regardless of lineage.
Loss of Ezh2 disrupts H3K27me3 in the developing retina
We observed that Ezh2 is highly expressed in retinal progenitors and weakly in postmitotic cells of the inner retina, consistent with previous studies showing significant expression in proliferating cells and embryonic tissues of multiple lineages, including retina (Aldiri et al., 2013; Aldiri and Vetter, 2012; Ezhkova et al., 2009; Iida et al., 2014; Margueron et al., 2008). Iida et al. (2014) showed EZH2 protein localized to Pax6-positive retinal progenitors and RGCs at embryonic stages, while postnatally EZH2 localized to glutamine synthase-positive Müller glia. In our study, conditional deletion of Ezh2 in retinal progenitors resulted in a large decrease in the level of H3K27me3 in the deleted region, particularly in progenitors. The presence of residual H3K27me3, particularly in some postmitotic cells, suggests potential contribution by Ezh1, which is generally more highly enriched in postmitotic cells and adult tissues than Ezh2 (Ezhkova et al., 2009; Margueron et al., 2008). However, the expression and function of Ezh1 in the developing retina remains to be determined.
We also noted increasing penetrance of phenotype, as well as loss of Ezh2 and H3K27me3, when comparing Ezh2fl/fl versus Ezh2fl/− CKO retina using the Pax6-αCre driver (Marquardt et al., 2001). Thus, the recombination efficiency and/or differences in the timing of deletion seem to impact the phenotype. We confirmed that the Ezh2fl/fl CKO phenotype was identical when generated by either lab. In addition, the Ezh2 CKO phenotype was also apparent using the Six3-Cre driver (Furuta et al., 2000), and has also recently been reported using Dkk-Cre (Iida et al., 2014). Together these findings demonstrate a consistent role for Ezh2 in the regulation of retinal development.
Ezh2 maintains retinal progenitor proliferation
One of the most highly conserved functions of Ezh2 in developing tissues is the maintenance of progenitor proliferation (Laugesen and Helin, 2014). Similar to the findings of Iida et al., 2014, we observed that loss of Ezh2 in retinal progenitors results in reduced proliferation at early postnatal ages, as well as upregulation of Cdkn2a (Arf/Ink4a) and Cdkn2b (Ink4b), which regulate the G1/S transition of the cell cycle and control of apoptosis. Both loci are not normally expressed in the developing retina, and have high levels of H3K27me3 histone modification in newborn mouse retina (Popova et al., 2012). It is likely that the proliferation defect is due to premature cell cycle exit of retinal progenitors, as demonstrated in the Xenopus retina (Aldiri et al., 2013), although changes in cell cycle kinetics or progenitor cell death may also contribute to progenitor depletion. In both Xenopus and mouse, loss or inhibition of Ezh2 leads to an increase in Arf/Ink4a or Ink4b, with ectopic expression of these genes causing reduced proliferation (Aldiri et al., 2013; Iida et al., 2014). Cell cycle genes are dysregulated in other regions of the CNS upon loss of Ezh2 (Pereira et al., 2010). Deficiency of Ink4a/Arf was sufficient to restore proliferation in Ezh2-deficient neural stem cells, demonstrating that it is the principal factor resulting in loss of proliferation (Hwang et al., 2014). Cdkn2a is a well-documented direct target for repression by Ezh2 in many other tissues, and is also associated with cancer (Popov and Gil, 2010). Thus maintaining proliferation by preventing the expression of cell cycle inhibitors is a core function of Ezh2.
Progenitor proliferation was unaffected by loss of Ezh2 in embryonic retina, suggesting that additional mechanisms regulate cell cycle gene expression. Consistent with this possibility, retinal progenitor proliferation and cell cycle genes are also regulated by G9a methyltransferase via H3K9 methylation (Katoh et al., 2012). It is also possible that distinct mechanisms function during embryonic versus postnatal development to regulate proliferation, since conditional knockout of the DNA methyltransferase gene Dnmt1 also alters cell cycle progression in postnatal but not embryonic retina (Rhee et al., 2012).
Ezh2 maintains the timing of late retinal cell differentiation
Ezh2 deletion and the concomitant loss of H3K27me3 in the developing retina caused striking alterations in histogenesis and retinal lamination. Differentiation of retinal ganglion cells, an early born cell type, was normal during the embryonic period, similar to the effect on proliferation, although numbers were reduced by early postnatal stages, potentially due to increased apoptosis. Although we confirmed loss of EZH2 and H3K27me3 embryonically, it is possible that other mechanisms act redundantly with Ezh2 to ensure normal differentiation during the embryonic period. We observed increased differentiation for several late born cell types at early postnatal ages, particularly rod photoreceptors and Müller glia, similar to Iida et al (2014) (Iida et al., 2014). Although generated early, by P14 rod photoreceptors were reduced in number, potentially due to early cell cycle exit and progenitor depletion. However, overall numbers of Müller glia were increased, suggesting a shift from neuron to glial fate. In Xenopus, Ezh2 knockdown also caused an increase in Müller glia differentiation at the expense of neurons (Aldiri et al., 2013), possibly due to reduced proneural gene expression, which we do not observe in our RNAseq data. Instead we find increased expression of the glial gene Nfix, and Ink4a/Arf, both of which have been shown to regulate glial fate acquisition (Price et al., 2014; Wilczynska et al., 2009).
Ezh2 regulates the timing of differentiation and the proportion of cell types generated in multiple developmental contexts. In the cerebral cortex after conditional Ezh2 deletion using Emx1-Cre, there is an acceleration of developmental timing, with early onset of gliogenesis, and a shift from self-renewal to differentiation in cortical progenitors (Pereira et al., 2010). In contrast, conditional deletion of Ezh2 during late corticogenesis results in an extended period of neurogenesis and failure to switch to astrocyte production (Hirabayashi et al., 2009). Thus the effects of Ezh2 loss on developmental timing are highly context-dependent (Aldiri and Vetter, 2012; Testa, 2011). Accelerated developmental timing has also been reported for non-neural tissues; basal cells in the developing lung epithelium appear earlier and in increased numbers upon loss of Ezh2 (Snitow et al., 2015). Thus Ezh2 may act to regulate the timing of differentiation and the proportion of cell types generated in multiple developmental contexts.
Not all late born cell types were affected, since there was no evidence for increased early bipolar cell differentiation (see also (Iida et al., 2014)). In addition, in Ezh2fl/fl CKO retina there was an increase in amacrine cell number, while in Ezh2fl/− CKO retina, amacrine cells were decreased, potentially due to increased penetrance of the phenotype, resulting in distinct effects on timing of differentiation or survival of these neurons. Overall, the gene expression changes induced by loss of Ezh2 may selectively affect the differentiation of a subset of retinal neurons, perhaps by mimicking the effects of related endogenous developmental regulators with shared binding specificity.
Upregulation of non-retinal genes with loss of Ezh2
Several classes of genes appear to be particularly susceptible to silencing by the H3K27me3 histone modification. In addition to cell cycle inhibitors, 24 Hox genes were upregulated in Ezh2fl/− CKO retina, including multiple genes from the Hoxa, Hoxb and Hoxc clusters, consistent with the Hox loci having some of the highest levels of H3K27me3 in the P1 retina (Popova et al., 2012). Hox genes are upregulated in many tissues upon loss of Ezh2, including tissues with very different embryonic origin such as cardiac tissue or lung endoderm (He et al., 2012; Snitow et al., 2015). Such deregulation can have significant developmental consequences. For example, derepression of Hox genes in neural crest cells, where they are not normally expressed, prevents craniofacial bone and cartilage formation (Schwarz et al., 2014). Polycomb genes were first identified for their role in the regulation of Hox genes in Drosophila, suggesting an ancient and strongly conserved relationship between Hox gene regulation and H3K27 trimethylation (Sparmann and van Lohuizen, 2006).
We confirmed that other potent developmental regulators are upregulated in the Ezh2 CKO retina, including genes with high levels of H3K27 trimethylation, such as Six1, Sall1, and Pou3f3, which showed reduced H3K27me3 by ChIP analysis in the Ezh2 CKO retina. For all these genes the H3K27me3 modification levels are much higher than the levels of the permissive mark H3K4me2 in the P1 retina (Popova et al., 2012). However, unlike Iida et al (2014), we did not observe changes in the expression of bHLHb4, Vsx1 or Isl1 (Iida et al., 2014).
Most of the upregulated genes are not significantly expressed during normal retinal development, consistent with a role for Ezh2 in repressing non-relevant gene programs to preserve lineage integrity, as proposed for skeletal muscle stem cells (Conerly et al., 2011; Juan et al., 2011). Comparing RNAseq and previous gene profiling studies from embryonic and adult tissues of diverse developmental origin, we find that loss of Ezh2 in retina leads to changes in expression of many of the same genes. While there are lineage-specific changes in gene expression after loss of Ezh2, many of these genes represent families of developmental regulators originally identified as PRC2 targets in embryonic stem cells (Lee et al., 2006). Promoters of many developmental control genes are poised for expression (Zeitlinger and Stark, 2010), and bivalent domains in their promoter regions containing both permissive H3K4me3 and repressive H3K27me3 modifications have been proposed to silence expression while keeping the genes poised for activation, which can be triggered by loss of H3K27me3 (Voigt et al., 2013). While bivalent domains can be dynamically modified, it is intriguing to consider that for a core set of genes this poised state may be maintained across lineages and into adult tissues (Weishaupt et al., 2010). Since H3K4me3 was not assessed here, it remains to be determined whether such bivalent domains are present in the developing mouse retina, and whether there is variation among the heterogeneous cell populations that are present in this tissue.
H3K27me3 levels do not strictly correspond to gene expression changes after loss of Ezh2
Not all genes with high levels of H3K27me3 were upregulated in the Ezh2 CKO retina. For example, RNAseq analysis showed that no Hoxd genes were upregulated, although Hoxd genes have high levels of H3K27me3 in the newborn mouse retina (Popova et al., 2012), and are known targets for PRC2-mediated repression. For example, in human cells the Hoxd locus is specifically targeted for PRC2-mediated repression via the long non-coding RNA HOTAIR (Rinn et al., 2007). Other genes, including developmental regulators such as Pax3, En1 and Olig2, are also highly marked with H3K27me3 in the P1 mouse retina (Popova et al., 2012), but were not upregulated with loss of Ezh2 in the retina. These data suggest that either other repressive histone modifications are present at these loci, or that the transcription factors necessary for their activation are not present in the retina. In contrast to retina, after loss of Ezh2 from adult mouse neural stem cells, Olig2 is upregulated and prevents neuronal differentiation (Hwang et al., 2014), suggesting context-dependent regulation of gene repression at certain loci.
Some genes that increase in the Ezh2 CKO do not appear to have significant levels of H3K27me3, although we cannot rule out cell type-specific epigenetic regulation (Popova et al., 2014), which can result in different levels of these modifications. Alternatively, these genes may not be directly regulated by Ezh2, but may be regulated by one of the many transcription factors that are derepressed directly.
Chromatin modifications and regulation of retina development
Together with previous studies, our results show that different sets of developmental genes are repressed by distinct complementary epigenetic regulators. Two other well-established repressive chromatin modifications, H3K9me2 and DNA methylation, are also required for retinal progenitor proliferation and/or differentiation, involving genes that are different from those regulated by Ezh2 (Katoh et al., 2012). Many long non-coding RNAs (lncRNAs) interact with PRC2 to coordinate gene silencing (Brockdorff, 2013), and several lncRNAs are expressed in retina and regulate retinal neurogenesis (Meola et al., 2012; Rapicavoli et al., 2010; Rapicavoli et al., 2011). In addition, we found significant upregulation of the PRC1 component Bmi1 in Ezh2 CKO retina. Bmi1 has also been implicated in the regulation of proliferation and differentiation in the developing nervous system (Fasano et al., 2009; Zencak et al., 2005). Thus, multiple repressive complexes are required to coordinate retinal neurogenesis, and there is the potential for cross-talk and complementary interactions in the regulation of different repressive complexes during development.
Supplementary Material
Highlights.
Dynamic patterns of Ezh2 expression and histone H3K27me3 modification during retinal development.
Reduction in postnatal progenitor proliferation after Ezh2 conditional knockout.
Enhanced differentiation of several late born cell types in the early postnatal retina.
RNA-seq shows many non-retinal genes upregulated with Ezh2 loss, including Cdkn2a.
ChIP analysis confirms loss of the H3K27me3 modification at these loci.
Acknowledgments
We thank Joy Corley for technical support, Kathy Moore for advice and for comments on the manuscript, Tim Mosbruger for bioinformatics support, and Ed Levine and Wolfgang Baehr for sharing antibodies. This work was supported by NIH grant EY012274 to MLV, and 1R01EY021482 to TAR, the University of Utah Vision Core 5P30EY014800 and the University of Washington Vision Core P30EY01730. Genomics analysis was through the University of Utah Huntsman Cancer Institute High-Throughput Genomics and Bioinformatic Analysis Shared Resource supported by Award Number P30CA042014 from the National Cancer Institute.
References
- Aldiri I, Moore KB, Hutcheson DA, Zhang J, Vetter ML. Polycomb repressive complex PRC2 regulates Xenopus retina development downstream of Wnt/beta-catenin signaling. Development. 2013;140:2867–2878. doi: 10.1242/dev.088096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldiri I, Vetter ML. PRC2 during vertebrate organogenesis: a complex in transition. Developmental biology. 2012;367:91–99. doi: 10.1016/j.ydbio.2012.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett EA, Wallace VA. Cell fate determination in the vertebrate retina. Trends in neurosciences. 2012;35:565–573. doi: 10.1016/j.tins.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Brockdorff N. Noncoding RNA and Polycomb recruitment. Rna. 2013;19:429–442. doi: 10.1261/rna.037598.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conerly ML, MacQuarrie KL, Fong AP, Yao Z, Tapscott SJ. Polycomb-mediated repression during terminal differentiation: what don’t you want to be when you grow up? Genes & development. 2011;25:997–1003. doi: 10.1101/gad.2054311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Olguin P, Huang Y, Li X, Christodoulou D, Seidman CE, Seidman JG, Tarakhovsky A, Bruneau BG. Epigenetic repression of cardiac progenitor gene expression by Ezh2 is required for postnatal cardiac homeostasis. Nature genetics. 2012;44:343–347. doi: 10.1038/ng.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano CA, Phoenix TN, Kokovay E, Lowry N, Elkabetz Y, Dimos JT, Lemischka IR, Studer L, Temple S. Bmi-1 cooperates with Foxg1 to maintain neural stem cell self-renewal in the forebrain. Genes & development. 2009;23:561–574. doi: 10.1101/gad.1743709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta Y, Lagutin O, Hogan BL, Oliver GC. Retina- and ventral forebrain-specific Cre recombinase activity in transgenic mice. Genesis. 2000;26:130–132. [PubMed] [Google Scholar]
- He A, Ma Q, Cao J, von Gise A, Zhou P, Xie H, Zhang B, Hsing M, Christodoulou DC, Cahan P, Daley GQ, Kong SW, Orkin SH, Seidman CE, Seidman JG, Pu WT. Polycomb repressive complex 2 regulates normal development of the mouse heart. Circulation research. 2012;110:406–415. doi: 10.1161/CIRCRESAHA.111.252205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi Y, Gotoh Y. Epigenetic control of neural precursor cell fate during development. Nat Rev Neurosci. 2010;11:377–388. doi: 10.1038/nrn2810. [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y, Suzki N, Tsuboi M, Endo TA, Toyoda T, Shinga J, Koseki H, Vidal M, Gotoh Y. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron. 2009;63:600–613. doi: 10.1016/j.neuron.2009.08.021. [DOI] [PubMed] [Google Scholar]
- Hwang WW, Salinas RD, Siu JJ, Kelley KW, Delgado RN, Paredes MF, Alvarez-Buylla A, Oldham MC, Lim DA. Distinct and separable roles for EZH2 in neurogenic astroglia. eLife. 2014;3:e02439. doi: 10.7554/eLife.02439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida A, Iwagawa T, Baba Y, Satoh S, Mochizuki Y, Nakauchi H, Furukawa T, Koseki H, Murakami A, Watanabe S. Roles of histone H3K27 trimethylase Ezh2 in retinal proliferation and differentiation. Developmental neurobiology. 2014 doi: 10.1002/dneu.22261. [DOI] [PubMed] [Google Scholar]
- Juan AH, Derfoul A, Feng X, Ryall JG, Dell’Orso S, Pasut A, Zare H, Simone JM, Rudnicki MA, Sartorelli V. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes & development. 2011;25:789–794. doi: 10.1101/gad.2027911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Yamazaki R, Onishi A, Sanuki R, Furukawa T. G9a histone methyltransferase activity in retinal progenitors is essential for proper differentiation and survival of mouse retinal cells. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:17658–17670. doi: 10.1523/JNEUROSCI.1869-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Torre A, Georgi S, Reh TA. Conserved microRNA pathway regulates developmental timing of retinal neurogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E2362–2370. doi: 10.1073/pnas.1301837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugesen A, Helin K. Chromatin repressive complexes in stem cells, development, and cancer. Cell stem cell. 2014;14:735–751. doi: 10.1016/j.stem.2014.05.006. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Molecular cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Meola N, Pizzo M, Alfano G, Surace EM, Banfi S. The long noncoding RNA Vax2os1 controls the cell cycle progression of photoreceptor progenitors in the mouse retina. Rna. 2012;18:111–123. doi: 10.1261/rna.029454.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Molecular and cellular biology. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira JD, Sansom SN, Smith J, Dobenecker MW, Tarakhovsky A, Livesey FJ. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15957–15962. doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov N, Gil J. Epigenetic regulation of the INK4b-ARF-INK4a locus: in sickness and in health. Epigenetics : official journal of the DNA Methylation Society. 2010;5:685–690. doi: 10.4161/epi.5.8.12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova EY, Barnstable CJ, Zhang SS. Cell type-specific epigenetic signatures accompany late stages of mouse retina development. Advances in experimental medicine and biology. 2014;801:3–8. doi: 10.1007/978-1-4614-3209-8_1. [DOI] [PubMed] [Google Scholar]
- Popova EY, Xu X, DeWan AT, Salzberg AC, Berg A, Hoh J, Zhang SS, Barnstable CJ. Stage and gene specific signatures defined by histones H3K4me2 and H3K27me3 accompany mammalian retina maturation in vivo. PloS one. 2012;7:e46867. doi: 10.1371/journal.pone.0046867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JD, Park KY, Chen J, Salinas RD, Cho MJ, Kriegstein AR, Lim DA. The Ink4a/Arf locus is a barrier to direct neuronal transdifferentiation. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2014;34:12560–12567. doi: 10.1523/JNEUROSCI.3159-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli NA, Poth EM, Blackshaw S. The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC developmental biology. 2010;10:49. doi: 10.1186/1471-213X-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli NA, Poth EM, Zhu H, Blackshaw S. The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural development. 2011;6:32. doi: 10.1186/1749-8104-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee KD, Yu J, Zhao CY, Fan G, Yang XJ. Dnmt1-dependent DNA methylation is essential for photoreceptor terminal differentiation and retinal neuron survival. Cell death & disease. 2012;3:e427. doi: 10.1038/cddis.2012.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Schwarz D, Varum S, Zemke M, Scholer A, Baggiolini A, Draganova K, Koseki H, Schubeler D, Sommer L. Ezh2 is required for neural crest-derived cartilage and bone formation. Development. 2014;141:867–877. doi: 10.1242/dev.094342. [DOI] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Molecular cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher F, Rossler R, Brouwer N, Balasubramaniyan V, Boddeke E, Copray S. Differentiation of neural stem cells into oligodendrocytes: involvement of the polycomb group protein Ezh2. Stem cells. 2008;26:2875–2883. doi: 10.1634/stemcells.2008-0121. [DOI] [PubMed] [Google Scholar]
- Snitow ME, Li S, Morley MP, Rathi K, Lu MM, Kadzik RS, Stewart KM, Morrisey EE. Ezh2 represses the basal cell lineage during lung endoderm development. Development. 2015;142:108–117. doi: 10.1242/dev.116947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nature reviews. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC developmental biology. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, Tarakhovsky A. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nature immunology. 2003;4:124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- Testa G. The time of timing: how Polycomb proteins regulate neurogenesis. BioEssays: news and reviews in molecular, cellular and developmental biology. 2011;33:519–528. doi: 10.1002/bies.201100021. [DOI] [PubMed] [Google Scholar]
- Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes & development. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weishaupt H, Sigvardsson M, Attema JL. Epigenetic chromatin states uniquely define the developmental plasticity of murine hematopoietic stem cells. Blood. 2010;115:247–256. doi: 10.1182/blood-2009-07-235176. [DOI] [PubMed] [Google Scholar]
- Wilczynska KM, Singh SK, Adams B, Bryan L, Rao RR, Valerie K, Wright S, Griswold-Prenner I, Kordula T. Nuclear factor I isoforms regulate gene expression during the differentiation of human neural progenitors to astrocytes. Stem cells. 2009;27:1173–1181. doi: 10.1002/stem.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse S, Pugazhendhi D, Brien P, Pell JM. Ezh2 maintains a key phase of muscle satellite cell expansion but does not regulate terminal differentiation. Journal of cell science. 2013;126:565–579. doi: 10.1242/jcs.114843. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A. Developmental gene regulation in the era of genomics. Developmental biology. 2010;339:230–239. doi: 10.1016/j.ydbio.2009.12.039. [DOI] [PubMed] [Google Scholar]
- Zencak D, Lingbeek M, Kostic C, Tekaya M, Tanger E, Hornfeld D, Jaquet M, Munier FL, Schorderet DF, van Lohuizen M, Arsenijevic Y. Bmi1 loss produces an increase in astroglial cells and a decrease in neural stem cell population and proliferation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005;25:5774–5783. doi: 10.1523/JNEUROSCI.3452-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Fuhrmann S, Vetter ML. A nonautonomous role for retinal frizzled-5 in regulating hyaloid vitreous vasculature development. Investigative ophthalmology & visual science. 2008;49:5561–5567. doi: 10.1167/iovs.08-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.