Abstract
Direct mass spectrometry (MS) analysis of biofluids with simple procedures represents a key step in translation of MS technologies to the clinical and point-of-care applications. The current study reports the development of a single-step method using slug flow microextraction and nanoESI (electrospray ionization) for MS analysis of organic compounds in blood and urine. High sensitivity and quantitation precision have been achieved for analysis of therapeutic and illicit drugs in 5 μL samples. Real-time chemical derivatization has been incorporated for analyzing anabolic steroids. The monitoring of enzymatic functions has also been demonstrated with the cholinesterase in wet blood.
Keywords: analytical methods, online reaction, ambient ionization, mass spectrometry, microextraction
Mass spectrometry (MS) has been demonstrated as a powerful tool for chemical and biological analysis. The high specificity, high sensitivity and high precision in quantitation are achieved traditionally in laboratory by eliminating the matrix effect through sample extraction and chromatographic separation prior to the MS analysis. The development of ambient ionization,[1] especially with the recent demonstration using the paper spray,[2] has indicated a promising future for direct MS analysis of high quantitation performance but using highly simplified protocols consuming ultra-small amounts of samples. This would be extremely important for the translation of the MS analysis to out-of-lab applications, especially point-of-care (POC) diagnosis. The underlying principle for a successful development along this direction is to minimize the sample consumption and to achieve high efficiency in an integrated process for the analyte extraction and ionization. In this study, we developed a new method using slug flow microextraction (SFME) and nanoESI (electrospray ionization) to perform a one-step analysis of biofluid samples. Excellent sensitivity and high quantitation precision have been obtained with blood and urine samples of only 5 μL. More importantly, we demonstrated how to incorporate a variety of different processes using a simple device, including liquid-liquid extraction, internal standard (IS) incorporation, chemical derivatization or even enzymatic reactions, which are necessary for a high performance mass analysis.
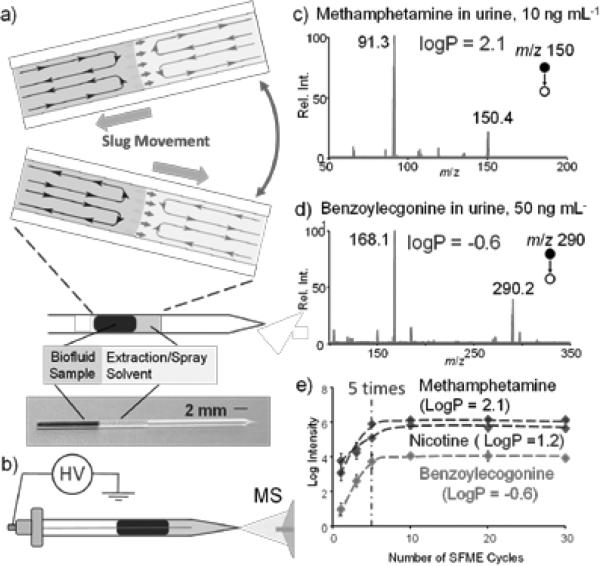
A disposable glass capillary of 0.8 mm i.d. (Figure 1a) with a pulled tip for nanoESI was used to perform the entire sampling ionization process. Two adjacent liquid plugs were formed by sequentially injecting 5 μL organic solvent and 5 μL urine or blood sample into the capillary. The liquid-liquid extraction of the analytes from the biofluid into the organic solvent is expected, but at a fairly low efficiency due to the small interfacing area. However, the extraction speed could be significantly improved with the slug flows induced by the movements of the two liquid plugs, which can be facilitated by tilting the capillary (Figure 1a) or by applying a push-and-pull force through air pressure (Figure S1). The slug flows is formed due to the friction with the capillary wall,[3] and the flows inside each plug (Figure 1a) transfer the analytes to and away from the liquid-liquid interface, therefore significantly improving the extraction efficiency. After the extraction process, the organic solvent plug can be simply pushed to the tip of the capillary; a stainless steel wire was then inserted through the biofluid sample to reach the organic solvent plug; a high voltage was applied to generate the nanoESI for MS analysis (Figure 1b). The selection of the organic solvent is critical. It needs to be immiscible with the biofluid samples, have good solubility for the target analytes and be suitable for nanoESI. Several organic solvents have been tested (see supporting information) and ethyl acetate of a weak polarity was found to provide the optimal performance for analyzing a broad range of chemical compounds in urine (Figure 1c, d) and blood samples (Figure S3).
Figure 1.
a) In-capillary sample extraction using the slug flow micro-extraction and b) subsequent MS analysis with nanoESI. MS/MS spectra for c) 10 ng mL−1 methamphetamine in 5 μL urine and d) 50 ng mL−1 benzoylecgonine in 5 μL urine. e) Impact of the number of SFME cycles on the extraction of the analytes, intensities of the MS/MS product ions monitored for methamphetamine (m/z 150 → 91) nicotine (m/z 163 → 130) and benzoylecgonine (m/z 290 → 168), each at 50 ng/mL in urine samples 2 kV used for nanoESI.
The extraction process with the slug flows have been shown to be very efficient, as tested for extracting methamphetamine, nicotine and benzoylecgonine (a main metabolite of cocaine) from urine samples. The equilibrium was reached after tilting the capillary 5 times (Figure 1e). Limits of detection (LODs) as good as 0.05 ng/mL for verapamil, have been obtained for the whole blood samples using SFME-nanoESI (Table 1). Fewer extraction cycles were needed for reaching equilibrium if the blood samples were diluted to reduce the viscosity. The distribution of the analyte between the sample and extraction phase can be relatively estimated by the partitioning coefficient (logP, see Equation S1 and derivation in supporting information). For methamphetamine with a logP of 2.1, its concentration in the organic extraction solvent can be 100 times higher than in the urine sample after SFME, which certainly explains the good LOD of 0.03 ng/mL achieved with urine samples (Table 1). The logP value for benzoylecgonine is −0.6, which means it has higher solubility in urine than in organic solvents and the extraction into ethyl acetate was a dilution process; however, an LOD of 0.08 ng/mL was achieved regardless. This indicates that the limiting factor in the detection of the benzoylecgonine in raw urine samples might not be the absolute amount or concentration of the benzoylecgonine, but the interference by the matrix effects, such as the ionization suppression due to the high concentrations of salts in the urine sample.[4] An efficient separation of the benzoylecgonine from the salts was achieved in the SFME process. Even with a lower benzoylecgonine concentration in the extraction phase, the ionization efficiency and the overall sensitivity of the analysis were improved significantly.
Table 1.
Limits of detection (LODs) for analytes in urine and/or whole blood samples using SFME-nanoESI.
| Analyte | Sample | Derivatization | Sample volume (μL) | LOD (ng/mL) |
|---|---|---|---|---|
| Methamphetamine | Urine | NA | 5 | 0.03 |
| Blood | NA | 5 | 0.1 | |
| Benzoylecgonine LOD | Urine | NA | 5 | 0.1 |
| Blood | NA | 5 | 1 | |
| Verapamil | Blood | NA | 5 | 0.05 |
| Amitriptyline | Blood | NA | 5 | 0.08 |
| Epitestosterone | Urine | Hydroxylamine | 5 | 0.7 |
| 6-Dehydrocholestenone | Urine | Hydroxylamine | 5 | 0.6 |
| 5α –Androstan-3β, 17β –Diol-16-one | Urine | Hydroxylamine | 5 | 0.2 |
| Stigmastadienone | Urine | Hydroxylamine | 5 | 0.8 |
In addition to the sensitivity, adequate precision in quantitation is often mandatory for clinical and POC applications.[5] Simple means for accurate incorporation of internal standard are important[6] but can be challenging for samples of small volumes taken by minimally invasive methods. Using the SFME-nanoESI, the IS compounds could be spiked in the extraction phase (Figure 2 inset) and subsequently mixed with the analyte during the slug flow extraction process. This method was tested for quantitation of methamphetamine in bovine blood samples with methamphetamined8 as the IS spiked in ethyl acetate at 2 ng/mL. The blood samples were diluted 10 times and then analyzed using the SFME-nanoESI and MRM analysis (transitions m/z 150 to 91 and m/z 158 to 94 for the analyte and IS, respectively) (Figure 2 inset). The measured analyte-to-IS ratios (A/IS) are plotted as a function of the original analyte concentration in blood as shown in Figure 2. A good linearity was obtained, which is governed by the partitioning process (see derivation in Supporting Information). RSDs better than 10% were obtained for samples of concentrations higher than 10 ng/mL.
Figure 2.
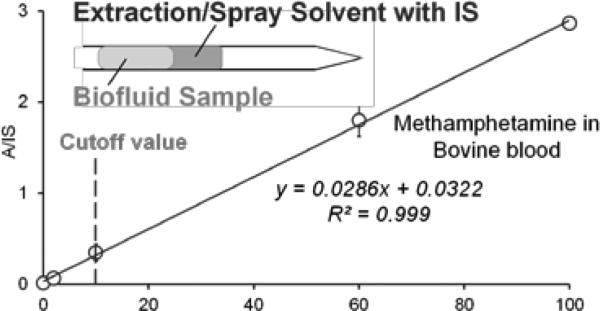
Quantitative analysis of whole blood spiked with methamphetamine (1–100 ng mL−1). The blood samples were diluted 10 times for decrease in viscosity. Methamphetamine-d8 (2 ng mL−1) in extraction solvent ethyl acetate.
Chemical derivatization is an effective way of altering the properties of the target analytes to improve the efficiency of separation or ionization for MS analysis.[7] For example, the steroids in urine or blood samples are expected to be well extracted into an organic phase using the SFME; however, the efficiency for the subsequent ionization by nanoESI would be low due to the low proton affinity of the steroid molecules. The reaction with hydroxyl amine has previously been proved to be effective in improving the ionization efficiency of the steroids,[8] and thereby was used in this study as an example. An additional liquid plug of 5 μL water containing 50 mM hydroxyl amine was injected between the 5 μL ethyl acetate and 5 μL urine sample spiked with 200 ng ml−1 epitestosterone (Figure 3). With 5 SFME cycles, the hydroxyl amine solution mixed well with the urine sample. The MS/MS analysis of the reaction product m/z 304 produced spectra of significantly improved signal-to-noise ratios (S/Ns) (Figure 3b,c). The reactive SFME-nanoESI was applied for analysis of a series of anabolic steroids in 5 μL urine samples, including epitestosterone, 6-Dehydrocholestenone, 5α –Androstan-3β, 17β –Diol-16-one and stigmastadienone, with LODs of 0.7, 0.6, 0.2 and 0.8 ng/mL obtained, respectively (Table 1).
Figure 3.
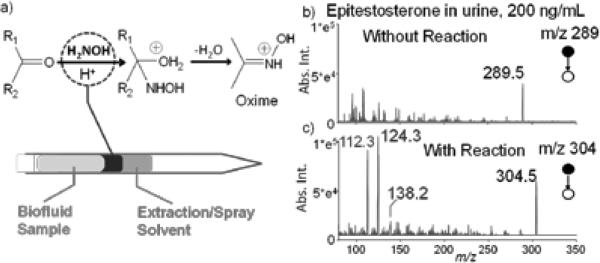
a) Reactive SFME-nanoESI with a reagent plug injected between the biofluid sample and the extraction solvent. MS/MS spectra of b) direct and c) reactive SFME-nanoESI analysis of 200 ng mL−1 epitestosterone in synthetic urine. 5 μL water containing 50 mM hydroxyl amine was used for the reagent liquid plug.
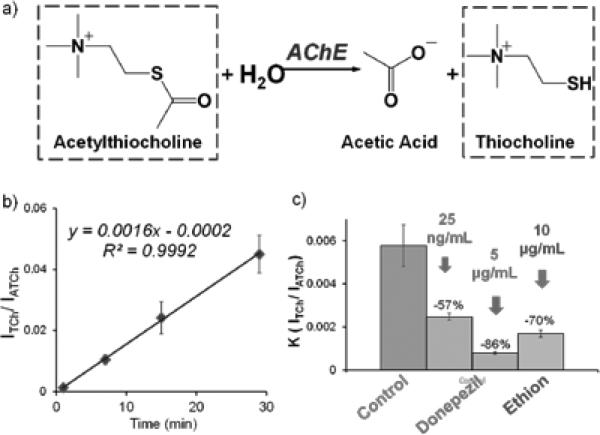
Using the liquid-liquid extraction process with SFME, the analysis can now be performed directly with wet blood samples. This provides an opportunity for probing the chemical and biological properties that only exist with the original liquid samples. For instance, the enzymatic functions of the proteins are typically quenched in the dried blood spots or after the traditional lab procedure for sample extraction. In this study, we applied the SFME-nanoESI for monitoring the enzymatic activity of cholinesterase (ChE) in whole blood samples. The ChE facilitates the enzymatic conversion of acetylthiocholine (ATCh) to thiocholine (TCh) (Figure 4a). The blood sample was diluted 10 times to slow down the reaction rate as well as to facilitate the slug flows for SFME. The substrate acetylthiocholine iodine was added into the diluted blood sample at a concentration of 1.8 mg/mL, and then 5 μL sample was taken immediately and injected into the capillary with 5 μL extraction phase. The capillary with the sample and the extraction solvent was left in room temperature 25 °C for incubation. The SFME-nanoESI could be performed repeatedly on the same sample and the ratio of the substrate ATCh and the reaction product TCh (Figure S7) could be monitored as a function of time to characterize the enzymatic activity of the ChE. A potential problem in this approach would be the damage to the enzyme function by the organic solvent. The impact by organic extraction phase was investigated for ethyl acetate and other solvents such as chloroform with a 5 min incubation. It was found that the reduction of ChE activity due to the contact with ethyl acetate was minimal but much more severe (more than 60% decrease) with chloroform (see Supporting Information). A weakly polar solvent like ethyl acetate can better preserve the enzyme structures.[9]
Figure 4.
a) The reaction scheme of the enzymatic conversion of acetylthiocholine (ATCh) to thiocholine (TCh) catalyzed by cholinesterase (ChE). b) Progression curve of ATCh digestion determined by SFME-nanoESI. The incubation was over 30 minute and catalyzed by blood cholinesterase (ChE) in room temperature. Acetylthiocholine iodide standard was added into human whole blood with a final concentration of 1.8 mg/mL before incubation. The intensity ratios of product ions from thiocholine (m/z 102 → 61) and enzyme substrate (m/z 162 → 103) were monitored using MRM; c) Evaluation of blood ChE with different levels of enzyme inhibition. The ChE activity in blood sample was determined by SFME-nanoESI after 5 min incubation.
Using ethyl acetate as the extraction solvent, the SFME-nanoESI was performed repeatedly over 30 min, with 5 cycles for SFME and 5s nanoESI at 1500 V for each analysis. The TCh/ACTh ratio is plotted as a function of time in Figure 4b, which is characteristic for the enzymatic activity of the ChE. An enzyme inihibition study was then carried out as a validation of this method. Two ChE inhibitors, donepezil (a therapeutic drug for Alzhimer's disease) and ethion (a neuron toxicant), were spiked seperately into blood samples, simulating the enzyme inhibitions at different degrees. The compromized enzyme activities were then determined uisng the SFME-nanoESI method with 5 min incubation. In comparion with the blood samples without adding the inhibitors, the defficiencies measured are resported in Figure 4c for blood samples treated with donpezil at 25 ng/mL and 5 μg/mL, and with ethion at 10 μg/mL. The percent decreases observed are consistent with the findings reported for previouse studies.[10]
In summary, the combination of the slug flow microextraction with nanoESI enabled a high-sensitivity direct analysis of the organic compounds in biofluids. Multiple types of processes for sample treatments, which traditionally require complex setups in lab, can now be incorporated into a one-step analysis with extremely simplified operation procedure. Since the biofluid samples are directly analyzed without being made into dried spots, an efficient liquid-liquid extraction can be designed based on the partitioning properties. The chemical and biological properties of the wet biofluids can also be retained and characterized thereby. The extraction process can be turned on and off by controlling the movements of the sample and extraction plugs. This allows an online monitoring of the chemical and biological reactions in a biofluid sample of only 5 μL. With the increasing interest in the translation of MS technologies to the clinical applications, this development has a profound implication on designing disposable sample cartridges with adequate function for direct analysis. This could ultimately lead to an elimination of the traditional lab procedures requiring complex setups and expertise. Its implementation with miniature mass spectrometers[11] would produce a powerful solution for POC diagnosis.
Experimental Section
All the experiments were carried out with a TSQ Quantum Access Max (Thermo Fisher Scientific, San Jose, CA, USA).The bovine blood was purchased from Innovative Research Inv. (Novi, MI, USA). The human pooled blood for enzymatic reaction study was purchased from BioreclamationIVT (Baltimore, MD, USA). The synthetic urine was purchased from CST Technologies (Great Neck, NY, USA). The steroids were purchased from Steraloids Inc. (Newport, RI, USA). All other chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Supplementary Material
Footnotes
This research was supported by Nation Institutes of Health (1R01GM106016)
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.201xxxxxx.((Please delete if not appropriate))
Contributor Information
Yue Ren, Weldon School of Biomedical Engineering, Purdue University West Lafayette, IN 47907 (USA).
Morgan N. McLuckey, Weldon School of Biomedical Engineering, Purdue University West Lafayette, IN 47907 (USA)
Jiangjiang Liu, Weldon School of Biomedical Engineering, Purdue University West Lafayette, IN 47907 (USA).
Zheng Ouyang, Weldon School of Biomedical Engineering, Purdue University West Lafayette, IN 47907 (USA).
References
- 1.a Cooks RG, Ouyang Z, Takats Z, Wiseman JM. Science. 2006;311:1566–1570. doi: 10.1126/science.1119426. [DOI] [PubMed] [Google Scholar]; b Monge ME, Harris GA, Dwivedi P, Fernández FM. Chemical Reviews. 2013;113:2269–2308. doi: 10.1021/cr300309q. [DOI] [PubMed] [Google Scholar]
- 2.Wang H, Liu JJ, Cooks RG, Ouyang Z. Angewandte Chemie-International Edition. 2010;49:877–880. doi: 10.1002/anie.200906314. [DOI] [PubMed] [Google Scholar]
- 3.a Burns JR, Ramshaw C. Lab on a Chip. 2001;1:10–15. doi: 10.1039/b102818a. [DOI] [PubMed] [Google Scholar]; b Kashid MN, Harshe YM, Agar DW. Industrial & Engineering Chemistry Research. 2007;46:8420–8430. [Google Scholar]
- 4.a Silber RH, Porter CC. Journal of Biological Chemistry. 1954;210:923–932. [PubMed] [Google Scholar]; b Casari C, Andrews ARJ. Forensic Science International. 2001;120:165–171. doi: 10.1016/s0379-0738(00)00466-7. [DOI] [PubMed] [Google Scholar]
- 5.Chace DH. Chemical Reviews. 2001;101:445–478. doi: 10.1021/cr990077+. [DOI] [PubMed] [Google Scholar]
- 6.Liu J, Cooks RG, Ouyang Z. Analytical Chemistry. 2013;85:5632–5636. doi: 10.1021/ac401056q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yates Iii JR. Nat Meth. 2011;8:633–637. [Google Scholar]
- 8.Huang G, Chen H, Zhang X, Cooks RG, Ouyang Z. Analytical Chemistry. 2007;79:8327–8332. doi: 10.1021/ac0711079. [DOI] [PubMed] [Google Scholar]
- 9.a Nakanishi K, Kamikubo T, Matsuno R. Bio-Technology. 1985;3:459–464. [Google Scholar]; b Ilett KF, Reid WD, Sipes IG, Krishna G. Experimental and Molecular Pathology. 1973;19:215–229. doi: 10.1016/0014-4800(73)90080-4. [DOI] [PubMed] [Google Scholar]
- 10.a Rogers SL, Friedhoff LT, Apter JT, Richter RW, Hartford JT, Walshe TM, Baumel B, Linden RD, Kinney FC, Doody RS, Borison RL, Ahem GL. Dementia. 1996;7:293–303. doi: 10.1159/000106895. [DOI] [PubMed] [Google Scholar]; b Dewan A, Patel AB, Pal RR, Jani UJ, Singel VC, Panchal MD. Clinical Toxicology. 2008;46:85–88. doi: 10.1080/15563650701517251. [DOI] [PubMed] [Google Scholar]
- 11.Li L, Chen T-C, Ren Y, Hendricks PI, Cooks RG, Ouyang Z. Analytical Chemistry. 2014;86:2909–2916. doi: 10.1021/ac403766c. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.