Abstract
Arabidopsis Shaggy-like protein kinases (ASKs) are Arabidopsis thaliana homologs of glycogen synthase kinase 3/SHAGGY-like kinases (GSK3/SGG), which are comprised of 10 genes with diverse functions. To dissect the function of ASKβ (AtSK32), ASKβ antisense transgenic plants were generated, revealing the effects of ASKβ down-regulation in Arabidopsis. Suppression of ASKβ expression specifically interfered with pollen development and fertility without altering the plants’ vegetative phenotypes, which differed from the phenotypes reported for Arabidopsis plants defective in other ASK members. The strength of these phenotypes showed an inverse correlation with the expression levels of ASKβ and its co-expressed genes. In the aborted pollen of ASKβ antisense plants, loss of nuclei and shrunken cytoplasm began to appear at the bicellular stage of microgametogenesis. The in silico analysis of promoter and the expression characteristics implicate ASKβ is associated with the expression of genes known to be involved in sperm cell differentiation. We speculate that ASKβ indirectly affects the transcription of its co-expressed genes through the phosphorylation of its target proteins during late pollen development.
Keywords: aborted pollen; antisense suppression; ASKβ (AtSK32, At3g61160); co-expressed genes; serine/threonine kinase; sperm cell formation
INTRODUCTION
Glycogen synthase kinase 3 (GSK3)/SHAGGY-like kinase (SGG) is a multifunctional non-receptor serine/threonine kinase found in all eukaryotes studied to date (Jope and Johnson, 2004; Kaidanovich-Beilin and Woodgett, 2011; Saidi et al., 2012). GSK3/SGG was originally characterized in the animal insulin signaling pathway and is considered to be a key regulator of many developmental processes, such as cell fate specification, cytoskeleton movements, and programmed cell death (reviewed by Saidi et al., 2012).
GSK3s/SGGs in Arabidopsis (Arabidopsis thaliana) are referred to as ASKs (Arabidopsis Shaggy-like protein kinases) or AtSKs (Arabidopsis thaliana Shaggy-like kinases). Arabidopsis contains ten AtSKs, which are grouped into four clades: clade I contains AtSK11/ASKα, AtSK12/ASKγ and AtSK13/ASKε; clade II contains AtSK21/ASKη/BIN2 (BRASSINOSTERIOD-INSENSITIVE 2), AtSK22/ASKι/BIL1 (BIN 2-like 1) and AtSK23/ASKη/BIL2; clade III contains AtSK31/ASKθ and AtSK32/ASKβ and clade IV contains AtSK41/ASKκ and AtSK42/ASKδ (Jonak and Hirt, 2002; Saidi et al., 2012).
ASKs are involved in growth, development and stress responses, and mounting evidence indicates that they function by integrating multiple hormonal signals (Saidi et al., 2012). All ASKs contain a tyrosine residue, which is located at a comparable position to that of the well-studied tyrosine in the activation loop of mitogen-activated protein kinase (Supplementary Fig. S1) (de la Fuente van Bentem et al., 2008). In ASKα/AtSK11 and ASKη/AtSK21/BIN2, this tyrosine residue is phosphorylated and indispensable for its kinase activity (de la Fuente van Bentem et al., 2008; Kim et al., 2009).
ASKs appear to require “priming” phosphorylation of the substrate for their activity with some exceptions, such as BES1 (BRI1-EMS SUPPRESSOR 1) and BZR1 (Brassinazole resistant 1) (Zhao et al., 2002), as do their counterparts in animals: before being phosphorylated by GSK3/SGG, substrates are often phosphorylated by another kinase (Frame and Cohen, 2001). Once primed, the phosphorylated substrate interacts with a phosphate-binding pocket (arginine 142, arginine 226 and lysine 251 in the case of GSK-3β) and thereby partially substitutes T-loop phosphorylation, which is required for kinase activity (Doble and Woodgett, 2003). Among the ten proteins identified as putative substrates of ASKs, five of these (underlined) are phosphorylated at the potential priming sites: PEP carboxykinase PCK1, RanBP domain-containing protein, thioredoxin-regulated beta-amylase TR-BAMY, At1g72790, remorin family protein, At4g11270, nodulin family protein, RabGAP/TBC domain-containing protein AtGYPC1b, glycine-rich protein and At3g01160 (de la Fuente van Bentem et al., 2008).
All ASKs in clade II, along with other ASKs, are involved in brassinosteroid (BR) signaling pathways (Wang et al., 2013a; also reviewed in Saidi et al., 2012). Of the six ASKs (ASKα/AtSK11, ASKγ/AtSK12, ASKη/AtSK21/BIN2, ASKι/AtSK22/BIL1, ASKζ/AtSK23/BIL2, and ASKθ/AtSK31) implicated in brassinosteroid (BR) signaling pathways for their interaction with the BZR1 transcription factor (Kim et al. 2009), all three clade II ASKs have been functionally confirmed for their redundant roles in BR signaling (Yan et al., 2009). Although ASKη (AtSK21/BIN2) acts as a major negative regulator of BR signaling in Arabidopsis, other ASKs in clade II and III can also phosphorylate BES1. For example, the bin2-1 gain-of-function mutation mimics BR deficiency, and BIN2 co-suppression can rescue BR signaling defects observed in a weak bri1 allele (Li and Nam, 2002; Rozhon et al., 2010; Yan et al., 2009). Recently, it was reported that clade II ASKs positively modulate ABA signaling by phosphorylating Snf1-related kinase 2s (SnRK2s) (Cai et al., 2014). In addition, all clade II ASKs interact with tracheary element differentiation inhibitory factor (TDIF) receptor and repress xylem differentiation through BES1 (Kondo et al., 2014).
The functions of ASKs other than those in clade II are largely unknown. Two ASKs in clade I (ASKα/AtSK11 and ASKγ/AtSK12) are up-regulated, especially in sepals and ovule primordia (Dornenals et al., 2000). While down-regulation of either gene using an antisense approach increased the number of sepals/petals and caused the splitting of styles/stigmas, these phenotypes could not be observed when T-DNA knock-out was used (Dal Santo et al., 2012; Dornenals et al., 2000). Additionally, introduction of a point mutation that reduced the primed activity of ASKθ/AtSK31 caused defective cell expansion (Claisse et al., 2007). ASKθ/AtSK31 was subsequently shown to be an additional ASK that is negatively regulated by the BRI1 receptor and phosphorylates BZR1 and BES1 (Rozhon et al. 2010). Although the induction of some ASKs (ASKε/AtSK13, ASKθ/AtSK31 and ASKδ/AtSK42) by salt or osmotic stress conditions has been reported (Charrier et al., 2002), the role of ASK in the salt stress response has been experimentally determined only for another ASK, ASKι/AtSK22/BIL1 (Piao et al., 2001).
In many crop species, male sterility is exploited for the production of F1 seeds with desirable traits. As a result, much effort has focused on establishing male sterile lines without deleterious side effects. We recently identified BrASKβ, an ASKβ homolog showing differential expression in floral buds of genic male sterile plants, as a putative male fertility-related gene (Dong et al., 2013; “BrASK2” was the name used in the reference). In the current study, to gain insight into the role of BrASKβ, we transgenically suppressed the expression of ASKβ in Arabidopsis, the homolog of BrASKβ, using antisense technology, and examined the resulting phenotypes. The results of expression analysis of ASKβ and its co-expressed genes, together with the pollen phenotypes observed in transgenic plants, suggest that ASKβ is essential for pollen development in Arabidopsis.
MATERIAL AND METHODS
Plant materials and growth conditions
Transgenic plants were generated with the Arabidopsis thaliana ecotype Col-0. Arabidopsis plants were grown under long day (16 h light / 8 h dark) conditions with 140 ± 2 μmol/m 2/s light intensity at 22 ± 0.5°C. Seeds were stratified for 3 days under 4°C before sowing in 60 × 60 mm soil pots. To maintain the humidity high enough for seed germination, the pots were covered under transparent polyethylthene film for the first 5 or 6 days. For in vitro growth, seeds were sterilized with 0.1% Triton X-100 (Sigma, USA) and 30% bleach. After the stratification at 4°C for 3 days, seeds were planted on solid media containing half-strength MS medium (Duchefa Biochemie, The Netherlands), 1% sucrose, and 0.8% phyto agar.
Antisense constructs and plant transformation
The full-length coding sequence of ASKβ was cloned from the first-strand cDNA using primer set AKSβ-P2 (Supplementary Table S1) and inserted into T&A cloning vectors (RBC T&A Cloning kit, Real Biotech Corporation, Taiwan). After confirming its sequence and orientation, the ASKβ fragment was subcloned into the pCambina 3300-35S binary vector for plant transformation. Agrobacterium tumefaciens GV3101 carrying the abovementioned binary plasmid was used for Arabidopsis transformation with floral dipping method (Clough and Bent, 1998). Transgenic plants (T1) were selected on half-strength MS media with 25 mg/ml gulfosinate (Sigma-Aldrich, USA) and further confirmed by gDNA PCR for the transgene. T1 plants were selfed and only the T1 lines, which showed the 3:1 segregation ratio for glufosinate resistant at T2 generation, were selected for the further experiment.
Reverse transcription (RT) PCR and qRT-PCR
The first-stand cDNA were synthesized following the Manufacturer’s instructions for ReverTra Ace-α kit (Toybo, Japan). To be used as PCR template, cDNA was diluted to 12.5 ng/μl using NanoDrop ND-1000 (Thermo Scientific). Semi-quantitative RT-PCRs were carried out using a following protocol: denaturation at 94°C for 5 min, 25-cycle amplification (94°C, 30 s; 54°C, 30 s; 72°C, 1 min), and final extension at 72°C for 7 min. Reaction condition for quantitative RT-PCR (qRT-PCR) as follows: 95°C for 30 s and 40 cycles of three-step reaction (95°C for 5 s, 60°C for 20 s, and 72°C for 15 s). Sequences of primers used in this study are shown in Supplementary Table S1. Products from semi-quantitative PCR were separated on 1.5% agarose gels and visualized with ethidium bromide.
Pollen viability, semi-thin sections, and DAPI (4′,6-diamidino-2-phenylindole) staining
To determine the viability and developmental progression of pollen, flowers collected from wild-type and ASKβS antisense transgenic lines were fixed in Carnoy’s solution (alcohol: chloroform: acetic acid = 6:3:1) for 2 h. Then, the anthers were dissected and stained with a solution containing Malachite green, acid fuchsin, and Orange G for 12 h as previously described (Peterson et al., 2010). Semi-thin sections were prepared as previously described (Javelle et al., 2011). Flower samples were cut and fixed in 4% paraformaldehybe overnight. Then, the samples were dehydrated gradually in an ethanol: Histochoice Clearing Agent (Sigma-Aldrich, USA) series and infiltrated with paraffin. Sections were sliced to an 8-micron thickness and mounted on slides. Paraffin was removed from the slides using Histo-clear and an ethanol series. For observation under a light microscope, samples were stained with 0.05% (W/V) toluidine blue O.
For DAPI staining, disrupted anthers were put on microscope slides with several drops of DAPI-staining solution [100 mm NaPO4, pH 7.5, 1 mM EDTA, 3 μg/ml DAPI (Sigma-Aldrich, high grade)]. The anthers on slides were inspected under a fluorescence microscope (Olympus, BX51) with a DAPI filter set, after incubated at room temperature for 5–10 min.
Phylogenetic tree construction
GSK/SGG-related amino acid sequences from Arabidopsis and Chinese cabbage (Brassica rapa) were identified by BLAST searches with the full-length amino acid sequence of ASKβ as a query and a cutoff value of 100 in the TAIR, and BRAD databases (www.arabidopsis.org and http://brassicadb.org/brad/). The retrieved sequences were aligned with ClustalX2.0 (Larkin et al., 2007). The phylogenetic tree was constructed by the neighbor-joining method in Molecular Evolutionary Genetics Analysis (MEGA) software, using “pairwise deletion” option, “Poisson correction” model (Tamura et al., 2013). Bootstrap value for test was set for 1000 replications.
Co-expression and gene network analysis
“Expression Angler”, which enables genome-wide co-expression analysis, was used to find genes that may have similar functions to that of ASKβ (Toufighi et al., 2005). Genes with a PCC (Pearson Correlation coefficient) value above 0.8 were chose for the additional analysis. The expression patterns of the identified genes were examined using Arabidopsis eFP browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) (Winter et al., 2007). Cluster analysis for functional categorization and GO enrichment analysis were performed with the MeV_4_9 (http://www.tm4.org/mev.html) and agriGO (http://bioinfo.cau.edu.cn/agriGO/index.php), respectively (Du et al., 2010). The gene network was produced by GeneMANIA (Zuberi et al., 2013) and modified with Cytoscape (Version 3.1.0) (Shannon et al., 2003).
Motif analysis
Conserved DNA motifs for transcription factor binding in the promoter sequences of the ASKβ co-expressed genes were identified as follows. For each gene, conserved motifs were searched in the 500 bp upstream regions from the transcription start site (TAIR: www.arabidopsis.org) using MEME suite (version 4.9.1) (Bailey et al., 2009). Motifs with lengths of 6–50 nucleotides were searched on both strands using the “zero or one occurrence per sequence” option. Motifs with an E-value ≤ 1.00E + 02 were assessed for their similarity to known motifs using PLACE (Higo et al., 1999).
RESULTS
Shaggy-like kinase genes are conserved between Arabidopsis and Chinese cabbage
In the previous study, BrASKβ (Bra003440), a homolog of ASKβ, was shown to be specifically expressed in fertile buds after tetrad stage and prosed as a putative GMS gene (Dong et al., 2013). To gain further insight into the potential function(s) of ASKβ (and thus BrASKβ), we performed bioinformatics analyses. Using the Arabidopsis ASKβ full-length protein sequence as a query, we searched for the related genes in the Arabidopsis and B. rapa databases (www.arabidopsis.org and http://brassicadb.org/brad/). The results show that a large number of ASKβ relative genes are present in each plant genome, including 10, and 18 genes in Arabidopsis, and B. rapa respectively. An earlier study showed that the ten shaggy-like kinases in Arabidopsis (AtSKs) can be divided into four clades (I, II, III and IV) based on their sequence similarities (Jonak and Hirt, 2002). Phylogenetic analysis using all 28 sequences identified in our database search revealed that all B. rapa homologs are also clustered into four branches along with Arabidopsis sequences, and one to three B. rapa homologs are closely associated with each ASK sequence (Fig. 1).
Fig. 1.
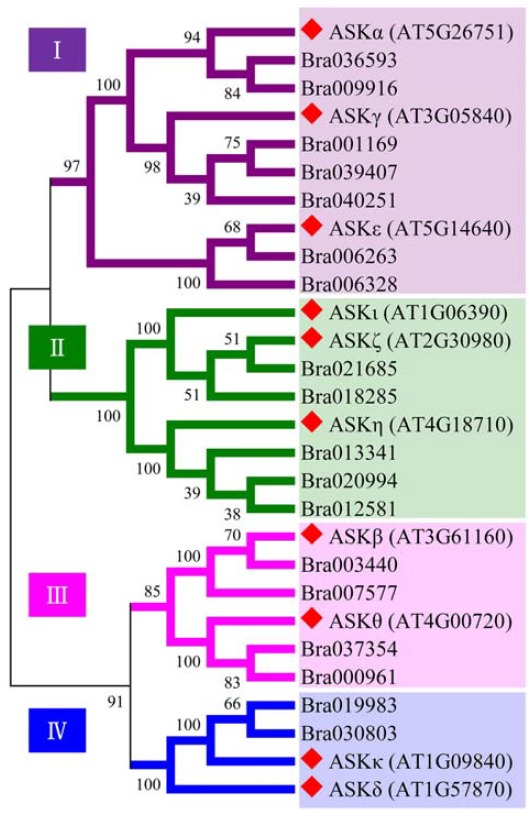
Phylogenetic analysis of ten ASKs and their homologs. Sequences from Arabidopsis thaliana, and Brassica rapa were aligned and used to construct the phylogenetic tree using the Neighbor-joining method with MEGA software (versio 6.0). Diamonds indicate Arabidopsis ASKs. All proteins were clustered into clades I to IV, which is similar to previous results (Jonak and Hirt, 2002).
The orthologue relationships between ASKs and BrSKs have been assinged using InParanoid Version 4.1 (Sonnhammer and Östlund, 2015). The results showed that BrASKβ (Bra003440) is the orthologues of ASKβ (AT3G61160) (Supplementary Table S2). Previously, it was shown that BrASKβ is differentially expressed between male fertile and sterile plants (Dong et al., 2013). Given the differential expression pattern of BrASKβ in sterile or fertile B. rapa floral buds and orthologuous relationship between BrASKβ and ASKβ, we suspected that ASKβ also plays a role during male gametogenesis in Arabidopsis.
ASKβ expression pattern in Arabidopsis suggests its putative function in pollen development
To infer the possible function of ASKβ, the expression patterns were examined in various wild-type Arabidopsis tissues using qRT-PCR and semi-quantitative RT-PCR (Fig. 2 and Supplementary Fig. S2). Relatively low levels of ASKβ expression were detected in roots, rosette leaves, stems, and siliques. By contrast, rather high levels of expression were detected in floral buds, especially later in “floral stage 13–16”. Consistent with these results, two independent publicly available datasets indicate that ASKβ is predominantly expressed in floral tissues (Supplementary Figs. S2B and S2C). Based on the reported expression of group III ASKs, including ASKβ, in developing pollen (Tichtinsky et al., 1998; Wellmer et al., 2004), we speculated that the major function of ASKβ is related to pollen development, with little or no role in other tissues or cell types.
Fig. 2.
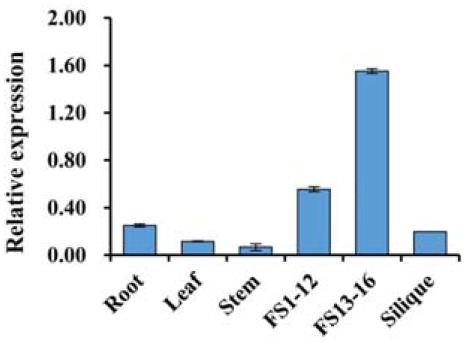
Expression patterns of ASKβ in Arabidopsis. The qRT-PCR was carried out with wild-type Arabidopsis. FS1–12, flower stage 1 to stage 12; FS13–16, flower stage 13 to stage 16 (Smyth et al., 1990). Expression levels were normalized to that of ACTIN7. Data were obtained from two biological replicates and the error bar represents standard deviation.
Knock-down of ASKβ levels causes reduced fertility
We used antisense technology to monitor the phenotypes of plants in which ASKβ expression was reduced. No T-DNA mutant lines harboring insertions in the coding region are available for ASKβ. A line containing an insertion in the promoter region of ASKβ (SAIL_66_D05) did not show any reduction in ASKβ expression compare to WT plants (data not show). Therefore, we adopted antisense approach. The antisense construct included the entire ASKβ coding region driven by constitutive CaMV 35S, since identical nucleotide sequences longer than 21-nucleotide were rarely identified between ASKβ and other members (Fig. 3A). After glufosinate selection, the T1 transformants were further confirmed by genomic DNA PCR using the AKSβ-P2 and 35S primer sets listed in Table S1. In seven independent T2 transgenic lines, variable effects on ASKβ expression were detected (Figs. 3B and 3C): four lines (lines 4, 25, 29 and 33) showed over 70% reductions in ASKβ expression, while two lines (lines 2 and 20) showed 10–30% reductions and one line (line 24) showed no reduction. Unlike the changes in expression observed for ASKβ, the expression levels of ASKθ (another ASK in clade III, which is most closely related to ASKβ) only showed weak or no reductions (Supplementary Fig. S3B). In addition, the expression levels of other ASK genes in Arabidopsis have been determined for WT, antisense transgenic lines 2 and 4, and no significant reduction in gene expression was observed. We assume that the phenotype of ASKβ antisense plants is mainly due to the suppressed expression of ASKβ. During vegetative growth, no phenotypic difference was observed between transgenic and wild-type plants (Fig. 3D). In contrast to vegetative phenotypes, reproductive traits including silique size and seed number per silique exhibited meaningful differences in the antisense transgenic lines. It should be noted that the seed number was greatly affected by the reduced expression of ASKβ but we could not find any correlation between ASKθ expression level and reduced fertility (Figs. 3D and 3E).
Fig. 3.
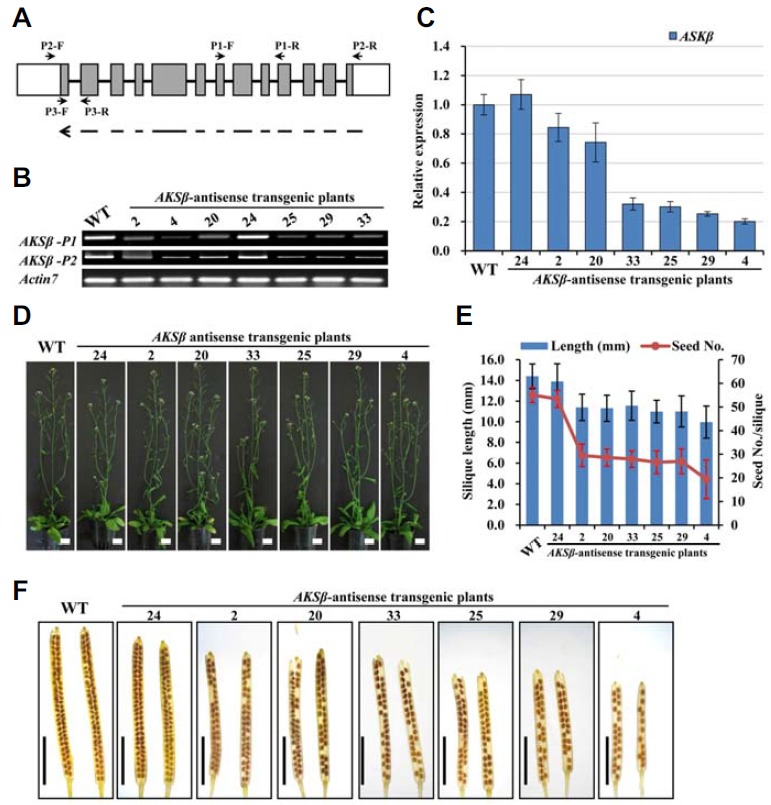
Analysis of ASKβ antisense transgenic Arabidopsis plants. (A) Schematic representation of the AKSβ gene structure and DNA fragments for antisense constructs. Gray boxes indicate exons, while the line and white boxes represent introns and UTRs, respectively. The dotted arrow in the antisense orientation at the bottom shows the DNA region used in the constructs. Arrows with names starting with “P” indicate the positions and directions of primers used: P1 and P2 for Semi-RTPCR, and P3 for qRT-PCR. (B) AKSβ transcript levels in floral buds of wild-type and antisense transgenic lines determined by semi-RT-PCR. ACTIN7 PCR was performed to validate that equal amounts of cDNA were used in each reaction. (C) Transcript levels of AKSβ and ASK measured by qRT-PCR. Expression levels were normalized to that of ACTIN7 and represented relative to wild-type expression levels. Error bars represent standard deviation (SD) of three biologically independent experiments. (D) Whole plant phenotypes showing no obvious differences between wild-type and ASKβ transgenic plants in vegetative growth. Bar = 20 mm. (E) Silique length and seed numbers. Mean values and SD were measured from three experiments using 18 plants per experiment. (F) Silique lengths and seed numbers in ASKβ antisense transgenic lines. Whole-mount images of mature siliques cleared with 0.2 NaOH and 1% SDS solution were used. Bar = 5 mm.
Down-regulation of ASKβ leads to defects in microspore development after the uninucleate stage
Because the predominant expression of ASKβ has been observed in pollen (Tichtinsky et al., 1998; Wellmer et al., 2004), we suspected disrupted pollen development in the antisense transgenic plants resulted in the reduced fertility. To test this, we first investigated the morphology of mature pollen grains with a modified Alexander’s staining (Peterson et al., 2010). We found that significant amounts of pollen grains in the transgenic plants were aborted, even though overall anther morphology seemed to be normal (Supplementary Fig. S3A). The severity of the abortion phenotype showed a clear correlation with the strength of antisense effects on ASKβ expression (Fig. 3 and Supplementary Fig. S3A). As a first step in identifying the function of ASKβ in pollen development, the morphologies of developing male gametophytes at various developmental stages were systematically compared between wild-type and ASKβ antisense plants under light microscopy after Alexander staining (Fig. 4). Before tetrad stage, no differences were observed between wild-type and ASKβ antisense plants (data not show). As shown in Figure 4, the microspores were also very similar in the wild-type and transgenic plants at the tetrad and uninucleate stages. However, clear differences were detected after these stages. From the bicellular stage to the mature stage, wild-type microspores exhibited typical round, gradually enlarging forms. By contrast, some of the ASKβ microspores from the antisense transgenic lines displayed signs of cytosol shrinkage, which appeared to be responsible for the failure to develop intact pollen grains. The defects in ASKβ antisense pollen became more obvious at the tricellular stage. The loss of the inner contents of some mature pollen grains resulted in the production of smaller or shrunken pollen grains. To examine whether ASKβ knock-down also affects tapetal tissue development and differentiation and thereby indirectly causes defects in pollen development, semi-thin transverse sections were produced from anthers at stage 8 to 13 (Supplementary Fig. S4) (Sanders et al., 1999). Following the degradation of the callose wall, microspores are released at anther stage 8. From anther stage 9 to 12 (floral stage 10 to 12), microspores develop into pollen grains. Tapetum degradation occurs during stage 10 and 11, while pollen mitotic divisions are observed during anther stage 12, resulting in the formation of tricellular pollen grains. At stage 13 and 14, anther dehiscence and shrinkage of anther cells occurs, respectively (Ma, 2005). In ASKβ antisense transgenic plants producing defective pollen grains, tapetum differentiation seemed to be identical to that of wild-type plants. The normal anther morphology and tapetum differentiation observed in these lines (Supplementary Figs. S3A and S4) indicate that suppression of ASKβ levels specifically interferes with microspore development without affecting sporophytic tissues in the anther.
Fig. 4.
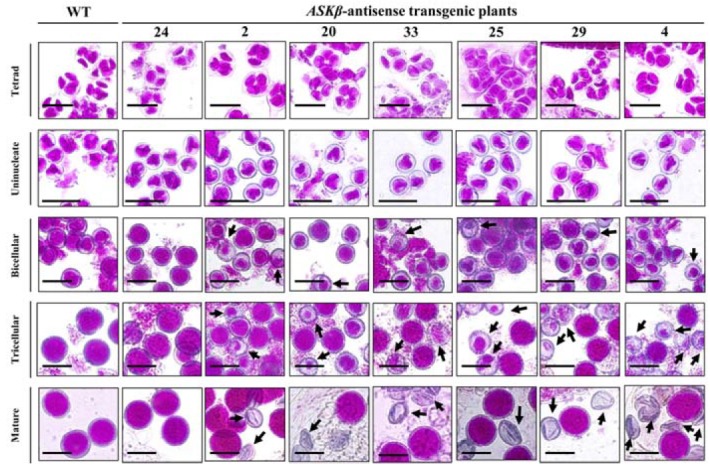
Male gametophyte development in wild-type and ASKβ antisense transgenic lines. Male gametophytes were stained with modified Alexander’s stain (Peterson et al., 2010). Tetrad, uninucleate, bicellular, tricellular and mature indicate the different stages of pollen development. Bar = 20 μm.
Some ASKβ co-expressed genes are involved in the late stage of pollen development
To help elucidate how ASKβ functions during pollen development, we analyzed genes that are co-expressed with ASKβ using ‘Expression Angler’ (Toufighi et al., 2005). A total of 641 co-expressed genes were isolated (Supplementary Table S3) and subjected to gene ontology (GO) annotation enrichment analysis using the singular enrichment analysis tool in agriGO (Du et al., 2010). Genes in the category “pollen development process (GO:0009555)” were highly represented (p value = 1.30E–11), which included 32 genes (Table 1 and Supplementary Table S4). The expression patterns of these 32 genes were reconstructed using Arabidopsis eFP browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) (Winter et al., 2007) (Fig. 5A). All of these genes, as well as ASKβ, were highly expressed in tricellular and mature stage pollen grains, which is in line with the time frame for the ASKβ antisense phenotypes (Fig. 4). The functions of the 32 genes mentioned above are summarized in Table 1; knock-outs of many of these genes have been reported to affect pollen development at later stages of sperm cell differentiation, pollen wall assembly, and pollen tube growth.
Table 1.
List of pollen development-related genes that are co-expressed with ASKβ
| At Locus | R-value to Bait | Gene name | Expression patterns | Phenotype of knock out | References |
|---|---|---|---|---|---|
|
| |||||
| Expression / Phenotype | |||||
| At3g21180 | 0.9830 | ACA9 | Expressed primarily in pollen | Reduction in seed set; defects in pollen tube growth and fertilization | Schiott et al. (2004) |
| At1g24520 | 0.9770 | BCP1 | Diploid tapetum and haploid microspore | Pollen cytoplasmic degeneration and loss of cellular contents | Xu et al. (1995) |
| At4g05330 | 0.9670 | AGD13 | - | ∼5–10% two celled pollen grains | Reňáik et al. (2012) |
| At3g57390 | 0.9650 | AGL18 | Male and female gametophyte, pollen | - | Kofuji et al. (2003) |
| At3g62230 | 0.9640 | DAF1 | Pollen sperm cells | - | Borg et al. (2011) |
| At5g64510 | 0.9580 | TIN1 | Highly expressed in pollen | Abnormal pollen surface morphology | Iwata et al. (2012) |
| At2g32460 | 0.9530 | MYB101 | - | Misarranged male sperm units | Reňáik et al. (2012) |
| At3g01040 | 0.9470 | GAUT13 | Pollen grains and pollen tube | Defected in pollen tube growth | Wang et al. (2013b) |
| At2g02970 | 0.9450 | APY6 | Expressed in mature pollen grains | Minor change in pollen exine pattern | Yang et al. (2013) |
| At5g39400 | 0.9350 | ATPTEN1 | Expressed exclusively in pollen grains | Pollen cell death after mitosis | Gupta et al. (2002) |
| At3g47440 | 0.9340 | TIP5;1 | Pollen sperm cells | Defected in pollen tube in the absence of exogenous nitrogen | Borg et al. (2011) and Soto et al. (2010) |
| At4g35700 | 0.9340 | DAZ3 | Pollen sperm cells | - | Borg et al. (2011) |
| At1g53320 | 0.9160 | ATTLP7 | - | Two celled pollen | Reňáik et al. (2012) |
| At2g35210 | 0.9040 | RPA | - | No sperm cell in mature pollen grains and cytoplasmic degradation | Boavida et al. (2009) |
| At2g03060 | 0.9020 | AGL30 | Inflorescence tissues | Reduced pollen fertility | Adamczyk and Fernandez (2009) |
| At5g22000 | 0.8940 | RHF2A | Stamens and carpels | Defective in the formation of male and female gametophytes | Liu et al. (2008) |
| At3g10470 | 0.8930 | - | - | - | - |
| At1g68610 | 0.8890 | PCR11 | Pollen sperm cells | - | Borg et al. (2011) |
| At1g47270 | 0.8850 | ATTLP6 | - | Two celled pollen | Reňáik et al. (2012) |
| At5g15470 | 0.8750 | GAUT14 | Pollen grains and pollen tube | Defected in pollen tube growth | Wang et al. (2013b) |
| At5g59030 | 0.8680 | COPT1 | Embryos, trichomes, pollen, and root | Displayed an enhanced nutritional induced pollen abnormalities | Sancenon et al. (2004) |
| At5g39650 | 0.8670 | DAU2 | Pollen sperm cells | - | Borg et al. (2011) |
| At5g23670 | 0.8490 | LCB2 | Ubiquitously expressed in Arabidopsis | Pollen contained aberrant endomembrane and lacked an intine layer | Dietrich et al. (2008) |
| At3g50310 | 0.8480 | MKKK20 | Pollen sperm cells | - | Borg et al. (2011) |
| At5g02390 | 0.8380 | DAU1 | Pollen sperm cells | - | Borg et al. (2011) |
| At2g18080 | 0.8340 | EDA2 | - | - | - |
| At5g53520 | 0.8290 | ATOPT8 | Pollen sperm cells | - | Borg et al. (2011) |
| At1g18750 | 0.8290 | AGL65 | Inflorescence tissues | Reduced pollen fertility | Adamczyk and Fernandez (2009) |
| At1g22130 | 0.8270 | AGL104 | Inflorescence tissues | Reduced pollen fertility | Adamczyk and Fernandez (2009) |
| At1g77980 | 0.8240 | AGL66 | Inflorescence tissues | Reduced pollen fertility | Adamczyk and Fernandez (2009) |
| At2g42380 | 0.8180 | ATBZIP34 | Gametophytic and sporophytic tissues | Misshapen and misplaced nuclei; defected in exine shape | Gibalova et al. (2009) |
| At1g19890 | 0.8140 | ATMGH3 | Pollen sperm cells | - | Borg et al. (2011) |
Fig. 5.
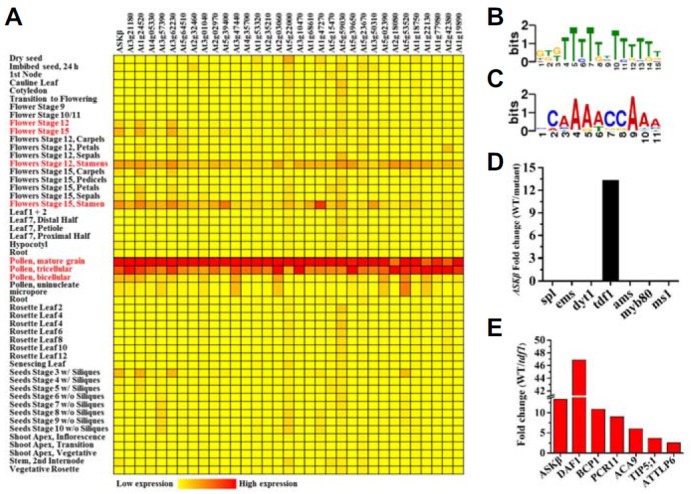
Analysis of putative ASKβ-interacting genes. (A) Expression pattern analysis of ASKβ and co-expressed pollen development-related genes. (B) Sequence Logo of conserved Motif 1, which is annotated as POLLEN1LELAT 52 in PLACE database and is present in ASKβ and its 32 co-expressed genes. (C) Sequence Logo of the conserved Motif 3, which is annotated as MYB1AT in PLACE database and is present in all analyzed genes. (D) ASKβ expression levels in wild-type plants with respect to that in a corresponding mutant background (based on previous reports in Feng et al., 2012; Phan et al., 2011; Wijeratne et al., 2007; Xu et al., 2010; Yang et al., 2007; Zhu et al., 2008). (E) Expression levels of ASKβ and its six co-expressed genes in the tdf1 mutant background, according to data in the above references.
Since co-expressed genes tend to be regulated by the same or similar transcription factor(s) (Coppe et al., 2009), we analyzed the promoter sequences of these genes to gather information about the regulation of ASKβ. Cis-regulatory elements (CREs) are frequently distributed within a region 500 bp upstream of the transcription start site and their distributions largely determine the transcriptional activity of promoters (Wang et al., 2011). Analysis of the promoter regions of ASKβ and 32 co-expressed genes identified six conserved CREs at cutoff values of E ≤ 1.00E + 02 (Supplementary Table S5). In particular, two CRE motifs (motif 1 and 3), which are annotated as POLLEN1LELAT52 and MYB1AT in the PLACE database (Higo et al., 1999), were found in all of the promoter regions (Figs. 5B and 5C; Supplementary Fig. S5; Supplementary Tables S4 and S5). POLLEN1LELAT52 specifies pollen-specific activation and was originally identified in the tomato lat52 gene (Abe et al., 2003; Bate and Twell, 1998), and MYB1AT is a CRE for MYB recognition (Filichkin et al., 2004).
TDF1 (TAPETAL DEVELOPMENT AND FUNCTION 1) and six pollen-expressed genes are a putative upstream regulator and downstream components of ASKβ, respectively
In Arabidopsis, functions of transcription factors controlling anther and pollen development have been well studied using mutant analysis, and many putative downstream genes have been isolated by microarray analysis (Feng et al., 2012; Phan et al., 2011; Wijeratne et al., 2007; Xu et al., 2010; Yang et al., 2007; Zhu et al., 2008). From these previously obtained data sets, we obtained the expression levels of ASKβ and its co-expressed genes, and replotted them as ratios between wild-type and mutants of interest (Figs. 5D and 5E). Among the mutant backgrounds of well-known transcription factors for male gametophyte development, ASKβ expression was strongly reduced only in the tdf1 background (Fig. 5D), while another MYB transcription factor, MYB80 (also known as MYB103), did not show any effects. This result indicates that TDF1 is an upstream transcription factor necessary for ASKβ transcription, but other transcription factors used in the analysis are not. In addition, 6 out of 32 ASKβ co-expressed genes were down-regulated in the tdf1 mutant (Fig. 5E), raising the possibility that they are expressed downstream of TDF1 and are also regulated by ASKβ: At1g24520/BCP1 (Homolog of Brassica campestris pollen protein 1), At3g62230/DAF1 (DUO1-Activated F-box 1), At3g47440/TIP5;1 (putative aquaporin TIP5-1), At1g68610/PCR11 (plant cadmium resistance 11), At3g21180/ACA9 (auto-inhibited calcium ATPase 9), and At1g47270/TLP6 (Tubby-like protein 6).
To test our hypotheses on the positions of upstream or downstream genes in the ASKβ transcriptional cascade, we performed qRT-PCR using floral buds of wild-type and ASKβ antisense transgenic lines (Fig. 6). In this analysis, we also included MYB80, which is a downstream gene of TDF1 (Zhu et al., 2008), to narrow down the position of ASKβ in TDF1- and MYB80-dependent transcriptional regulation. Compared with wild-type plants, the expression levels of TDF1 and MYB80 did not change in the ASKβ antisense transgenic lines (Fig. 6A). Given that ASKβ expression is not affected by the mutation of MYB80 (Fig. 5D), this result indicates that (1) ASKβ is transcriptionally regulated downstream of TDF1, and (2) ASKβ and MYB80 are located in separate branches downstream of TDF1. Six ASKβ co-expressed and TDF1-regulated genes can be categorized by their putative functions or expression characteristic, as summarized in Table 1. Knock-out of BCP1 and ATTLP6 mutations leads to cytoplasmic degradation and defects in late pollen development, respectively (Reňák et al., 2012; Xu et al., 1995). While DAF1, PCR11, and TIP5;1 are involved in sperm cell formation (Borg et al., 2011), ACC9 is important for fertility and mutation in ACC9 results in more than 80% reduction of seed set (Schiott et al., 2004). As expected for the downstream genes of ASKβ, the expression patterns of all six genes examined were quite similar to those of ASKβ in wild-type and antisense transgenic lines with variable ASKβ levels (Figs. 6B–6E).
Fig. 6.
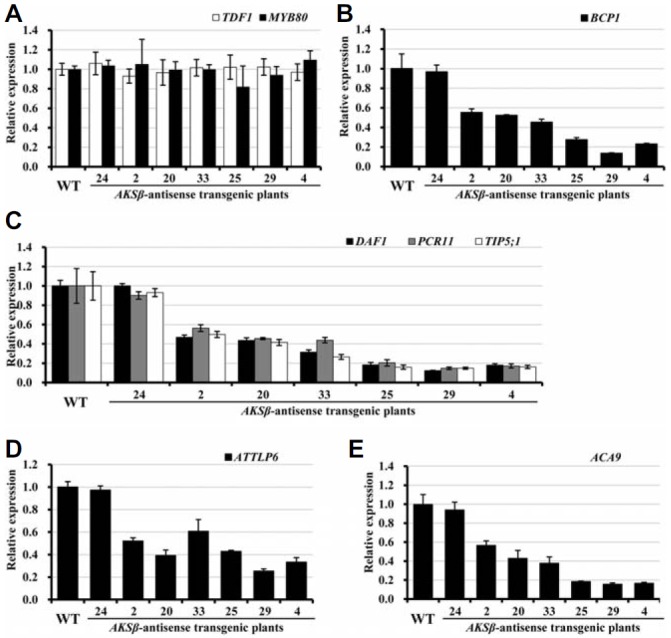
Results of qRT-PCR analyses of putative upstream and downstream genes of ASKβ. Figures are grouped according to their putative functions or expression, as summarized in Table 1: BCP1 for cytoplasm degradation; DAF1, PCR11 and TIP5;1 for sperm cell development; ATTLP7 for two-celled pollen; ACA9 for aborted fertilization. Relative transcript levels in wild-type and ASKβ antisense transgenic lines are shown as follows: (A) TDF1 and MYB80; (B) BCP1; (C) DAF1, PCR11 and TIP5;1; (D) ATTLP6; and (E) ACA9. Expression levels were normalized to that of ACTIN7 and presented as the mean ± SD of three biologically independent experiments.
ASKβ suppression affects sperm cell formation at the bicellular stage
Staining with DAPI allows easy visualization of the number, shapes, and positions of nuclei during late stages of pollen development (Fig. 7). In uninucleate microspores, DAPI-stained pollen from ASKβ antisense plants appeared to be normal. However, loss of cytoplasm and nuclei was observed from the bicellular stage pollen grains, which leads to the productions of slightly smaller and aberrant pollen grains. Defects in pollen grains were more clearly observed in tricellular stage and mature pollen grains (Fig. 7). Together with the results of co-expression and qRT-PCR analysis, these results lead to the conclusion that ASKβ plays an important role in Arabidopsis microgametogenesis through the activation of many important genes required for late pollen development: more specifically for pollen sperm nuclei formation.
Fig. 7.
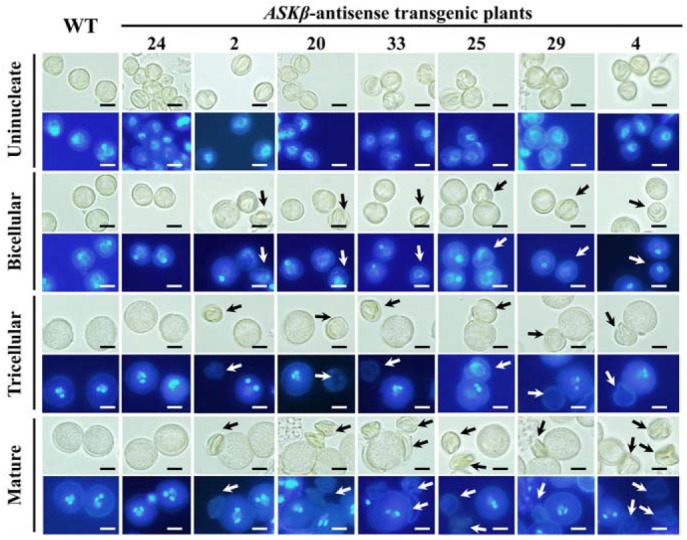
Pollen grains of wild-type and ASKβ antisense transgenic plants stained with DAPI. For each developmental stage and phenotype, the upper and lower panel shows bright-field and fluorescence microscope images, respectively. Aberrant or no staining in pollen grains is indicated with arrows. Bar = 10 μm.
DISCUSSION
Phylogenetic and expression data strongly suggest an orthologous relationship between ASKβ (At3g61160) and BrASKβ (Bra003440)
BrASKβ, which is expressed in fertile floral buds but not in genic male sterile floral buds, was identified as a putative male fertility-related gene and was initially named based on its sequence similarity to ASKβ (Dong et al., 2013). The following observations support the notion that ASKβ is the Arabidopsis ortholog of BrASKβ: (1) these genes display a close association in genome-wide phylogenetic analysis (Fig. 1); (2) both genes are predominantly expressed in developing pollen (Dong et al., 2013; Tichtinsky et al., 1998; Figs. 2 and 5), and (3) their functions are linked to proper pollen development (Dong et al., 2013; Figs. 3 and 4; Supplementary Figs. S3A and S4). In the phylogenetic tree constructed using both Arabidopsis and B. rapa ASK genes, another B. rapa gene, Bra007577, was also found near ASKβ in a clade containing ASKβ and ASKθ (Fig. 1). BrASKβ and Bra007577 may be produced from a common ancestor of Arabidopsis ASKβ by the genome triplication in Brassica lineage (Yang et al., 2005).
Putative function(s) of ASKβ appears to be restricted to pollen development
Most ASKs that have been functionally characterized to date are implicated in BR signaling and are found in clade I, II and III (Saidi et al., 2012). Although the roles of ASKs in flower development and osmotic stress responses have also been determined or proposed, seven of the ten ASKs in Arabidopsis are considered to be involved in BR signaling, including ASKα/AtSK11, ASKγ/AtSK12, ASKε/AtSK13, ASKη/AtSK21/BIN2, ASKι/AtSK22/BIL1, ASKζ/AtSK23/BIL2 and ASKθ/AtSK31. Among these, three ASK members play an additional role in flower development. ASKα/AtSK11 and ASKγ/AtSK12 are preferentially expressed in early floral meristems, and down-regulation of each gene via antisense technology results in similar phenotypes, i.e., an increase in perianth organ number and defects in gynoecium development (Dornelas et al., 2000). Moreover, plants over-expressing the mutated version of ASKθ/AtSK31 (R178A) produce flowers with smaller pedicels, petals, and sepals (Claise et al., 2007). Although Claise and colleagues did not observe any altered phenotypes when they over-expressed a wild-type version of ASKθ/AtSK31 (Claise et al., 2007), a later study reported that over-expression of ASKθ/AtSK31-Myc fusion protein led to pleiotropic phenotypes in BR-deficient transgenic plants (Rozhon et al., 2010). The finding that BR and ABA can act antagonistically, which involves the action of clade II ASKs (ASKη/BIN2, ASKι/BIL1, and ASKζ/BIL2), as well as the observation that over-expression of a clade II ASK (ASKι/BIL1) can induce NaCl tolerance, suggest that ASKs function as integrators of plant hormones and environmental conditions (Cai et al., 2014; Piao et al., 2001; Zhang et al., 2009).
Unlike the phenotypes reported in previous studies of ASKs, our investigation of ASKβ function using antisense transgenic plants revealed that ASKβ is required for fertility in Arabidopsis (Figs. 3 and 4). The phenotypes observed in the current investigation seem to be caused by the selective suppression of the intended target, ASKβ. Most importantly, we found that reduced fertility, which can be explained by the presence of aborted pollen, is not associated with the expression level of ASKθ (Fig. 3). Among ASK gene family members, ASKθ is most closely related to ASKβ (Fig. 1), and these two members (in clade III) are different from the other members, as they contain N-terminal extensions (Supplementary Fig. S1). Although ASKβ and ASKθ show rather high levels of sequence identity across the entire sequences at the nucleotide level, the length of identical regions without any mismatch are less than 21 nucleotides with a few exceptions (data not shown). The notion that antisense effect is specific to ASKβ is further supported by previous studies reporting no morphological defects in pollen development for plants with hypomorphic alleles of ASKθ or reduced expression of ASKα/ASKγ (Claise et al., 2007; Dornelas et al., 2000).
Various phenotypic outcomes reported for plants in which the functions of different ASKs were disrupted may be related to the discrete expression patterns and/or physical interactions of individual ASKs (Supplementary Fig. S6). According to the expression analysis using eFP browser tool, ASK and ASK are widely expressed but are much more highly expressed in senescing leaves, while that of ASKβ is primarily limited to late stage pollen. ASKθ, the most closely related gene to ASKβ, was clearly induced in developing pollen, but only in the tricellular to mature pollen stage. By contrast, ASKβ was clearly expressed beginning at the bicellular stage. In addition, ASK strongly interacts with BR-responsive transcription factors (BEH2, BES1 and BZR1), but ASKβ does not bind to any BR-responsive transcription factor (Rozhon et al., 2010).
ASKβ may regulate pollen development at late microsporogenesis
Previous studies have shown that ASKβ and some of the 32 co-expressed genes, such as BCP1, TIP5;1, COPT1, and AGL65, are expressed in a pollen-specific manner under the control of the MYB transcription factor TDF1 (Adamczyk and Fernandez, 2009; Sancenon et al., 2004; Soto et al., 2008; Wellmer et al., 2004; Xu et al., 1995; Zhu et al., 2008). Our results functionally confirm that ASKβ is expressed during late pollen development downstream of the TDF1 MYB transcription factor (Figs. 4–6; Supplementary Fig. S4). Furthermore, our expression profiling results indicate that ASKβ is an upstream component controlling the expression of BCP1, DAF1, PCR11, TIP5;1, ATTLP6, and ACA9 (Fig. 6). Considering that BCP1 antisense plants also display defects in pollen development and reduced fertility, and the first signs of pollen generation are evident at the bicellular stage (Xu et al., 1995), the phenotypes observed in the ASKβ antisense transgenic plants generated in the current study were at least partly associated with the down-regulation of BCP1 (Fig. 6). Consistent with the current results, the expression of DAF1, PCR11, TIP5;1, ATTLP6, and ACA9 is more pronounced after the tetrad stage when ASKβ expression is strong, and altering the expression of these genes also causes abnormal microgametogenesis (Borg et al., 2011; Reňák et al., 2012; Schiott et al., 2004; Soto et al., 2010). ASKβ may activate regulatory protein(s) that transcriptionally induce the expression of these genes.
ASKβ may function during sperm cell formation. Asymmetric division of the haploid microspore produces the male germline cells for double fertilization in flowering plants (Twell, 2011). In rice, knock-down of a glycosyltransferase, OSGT1, affects pollen intine formation, leading to the disappearance of nuclei and a reduction in pollen size (Moon et al., 2013). In addition, knock-down of Dynamin-related protein 2 (DRP2) results in the production of shrunken mature pollen grains with fewer or no nuclei (Backues et al., 2010). Numerous ASKβ co-expressed genes are involved in sperm cell differentiation (genes and their references are found in Table 1). Among these, mutants of AGD13, ATTLP7, ATTLP6, and MYB101 exhibit two-celled pollen grains or misarranged male sperm units (Reňák et al., 2012), while mutants of PDD6 have no sperm cells in their mature pollen grains and exhibit cytoplasmic degradation (Boavida et al., 2009). Recently, DAF1, PCR11, and TIP5;1 were reported to be target genes of the male germline-specific transcription factor DUO POLLEN 1, which is also required for germline nucleus development (Borg et al., 2014).
The putative roles of ASKβ in pollen development might also involve phosphorylation of co-expressed genes
In addition to dynamic changes in gene expression, post-translational modifications of regulatory proteins are also necessary for proper pollen development (Guan et al., 2014). Therefore, the disturbance of phosphorylation cascades in ASKβ antisense plants should be the primary factor contributing to the disappearance of pollen nuclei and the subsequent appearance of shrunken pollen. To extend our knowledge of the functions of ASKβ during pollen development, we built an expression network involving ASKβ and its 32 co-expressed genes (Fig. 8). For some co-expressed genes, ASKβ kinase is likely to phosphorylate upstream signaling components, resulting in the transcriptional activation of these co-expressed genes. However, it is possible that ASKβ-dependent phosphorylation cascades also directly modify the activities of these co-expressed genes at the protein level. Indeed, analysis using Musite (with a cutoff value of 85%) revealed that all ASKβ co-expressed gene products except DAF1 contain at least one putative serine/threonine-specific phosphorylation site (Supplementary Table S6; Gao et al., 2010). GSK3/SGG kinases can use both priming-phosphorylated proteins and non-priming-phosphorylated proteins as substrates (de la Fuente van Bentem et al., 2008; Doble and Woodgett, 2003; Zhao et al., 2002). Many proteins encoded by ASKβ co-expressed genes may function as substrates of ASKβ and be directly regulated by ASKβ (Supplementary Table S6).
Fig. 8.
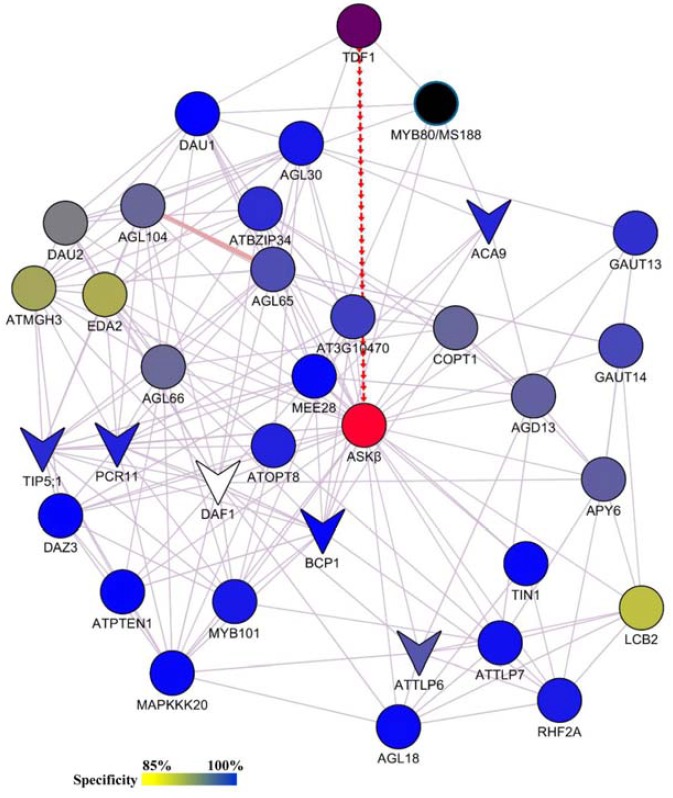
A network composed of ASKβ and its 32 co-expressed genes that have the GO annotation “pollen development process”. ASKβ is indicated in red, while ASKβ co-expressed genes are indicated with different colors (ranging from yellow to blue) based on their maximum specificity value for putative phosphorylation sites predicted by Musite (http://musite.sourceforge.net/). DAF1 does not appear to have any putative phosphorylation sites at a cutoff of 85%. TDF1 and MYB80, the upstream and independent gene of ASKβ, are indicated in dark purple and black, respectively. The genes indicated with a “V” shape are the putative downstream genes of ASKβ identified in this study. Light purple lines show the co-expression interactions, while the thick, light pink line indicates a reported physical interaction (de Folter et al., 2005; Adamczyk and Fernandez, 2009). The regulation of ASKβ by TDF1 confirmed in this study is indicated with red arrows.
In summary, the current results demonstrate that ASKβ function is required for fertility of Arabidopsis plants downstream of TDF1, which is mediated through the coordinated regulation of many pollen development-related genes. Consistent with the expression patterns of ASKβ, we determined that the first signs of deteriorating male gametophytes begin to appear at the bicellular stage. Based on the finding that ASKβ co-expressed and -regulated genes play important roles in sperm cell differentiation, we propose that ASKβ plays a similar role. Identification of the target proteins of ASKβ kinase and investigation of the mechanism defining the distinct roles of ASKβ and ASKθ will further expand our knowledge of the pollen development process and GSK3/SGG-like kinases in plants.
Acknowledgments
This work was supported by a grant from the Next-Generation BioGreen 21 Program (the Next-Generation Genomics Center No. PJ008118), Rural Development Administration, Republic of Korea to Yoonkang Hur. This work was supported by research fund of Chungnam National University to Hankuil Yi.
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamczyk BJ, Fernandez DE. MIKC* MADS domain heterodimers are required for pollen maturation and tube growth in Arabidopsis. Plant Physiol. 2009;149:1713–1723. doi: 10.1104/pp.109.135806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backues SK, Korasick DA, Heese A, Bednarek SY. The Arabidopsis dynamin-related protein2 family is essential for gametophyte development. Plant Cell. 2010;22:3218–3231. doi: 10.1105/tpc.110.077727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bate N, Twell D. Functional architecture of a late pollen promoter: pollen-specific transcription is developmentally regulated by multiple stage-specific and co-dependent activator elements. Plant Mol. Biol. 1998;37:859–869. doi: 10.1023/a:1006095023050. [DOI] [PubMed] [Google Scholar]
- Boavida LC, Shuai B, Yu HJ, Pagnussat GC, Sundaresan V, McCormick S. A collection of Ds insertional mutants associated with defects in male gametophyte development and function in Arabidopsis thaliana. Genetics. 2009;181:1369–1385. doi: 10.1534/genetics.108.090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg M, Brownfield L, Khatab H, Sidorova A, Lingaya M, Twell D. The R2R3 MYB transcription factor DUO1 activates a male germline-specific regulon essential for sperm cell differentiation in Arabidopsis. Plant Cell. 2011;23:534–549. doi: 10.1105/tpc.110.081059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg M, Rutley N, Kagale S, Hamamura Y, Gherghinoiu M, Kumar S, Sari U, Esparza-Franco MA, Sakamoto W, Rozwadowski K, et al. An EAR-dependent regulatory module promotes male germ cell division and sperm fertility in Arabidopsis. Plant Cell. 2014;26:2098–2113. doi: 10.1105/tpc.114.124743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, Liu J, Wang H, Yang C, Chen Y, Li Y, Pan S, Dong R, Tang G, Barajas-Lopez Jde D, et al. GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating clade III SnRK2s in Arabidopsis. Proc. Natl. Acad. Sci. 2014;111:9651–9656. doi: 10.1073/pnas.1316717111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier B, Champion A, Henry Y, Kreis M. Expression profiling of the whole Arabidopsis shaggy-like kinase multigene family by real-time reverse transcriptase-polymerase chain reaction. Plant Physiol. 2002;130:577–590. doi: 10.1104/pp.009175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claisse G, Charrier B, Kreis M. The Arabidopsis thaliana GSK3/Shaggy like kinase AtSK3-2 modulates floral cell expansion. Plant Mol. Biol. 2007;64:113–124. doi: 10.1007/s11103-007-9138-y. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Coppe A, Ferrari F, Bisognin A, Danieli GA, Ferrari S, Bicciato S, Bortoluzzi S. Motif discovery in promoters of genes co-localized and co-expressed during myeloid cells differentiation. Nucleic Acids Res. 2009;37:533–549. doi: 10.1093/nar/gkn948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Santo S, Stampfl H, Krasensky J, Kempa S, Gibon Y, Petutschnig E, Rozhon W, Heuck A, Clausen T, Jonaka C. Stress-induced GSK3 regulates the Redox stress response by phosphorylating glucose-6-phosphate dehydrogenase in Arabidopsis. Plant Cell. 2004;24:3380–3392. doi: 10.1105/tpc.112.101279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Folter S, Immink RG, Kieffer M, Parenicova L, Henz SR, Weigel D, Busscher M, Kooiker M, Colombo L, et al. Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell. 2005;17:1424–1433. doi: 10.1105/tpc.105.031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente van Bentem S, Anrather D, Dohnal I, Roitinger E, Csaszar E, Joore J, Buijnink J, Carreri A, Forzani C, Lorkovic ZJ, et al. Site-specific phosphorylation profiling of Arabidopsis proteins by mass spectrometry and peptide chip analysis. J. Proteome Res. 2008;7:2458–2470. doi: 10.1021/pr8000173. [DOI] [PubMed] [Google Scholar]
- Dietrich CR, Han G, Chen M, Berg RH, Dunn TM, Cahoon EB. Loss-of-function mutations and inducible RNAi suppression of Arabidopsis LCB2 genes reveal the critical role of sphingolipids in gametophytic and sporophytic cell viability. Plant J. 2008;54:284–298. doi: 10.1111/j.1365-313X.2008.03420.x. [DOI] [PubMed] [Google Scholar]
- Doble BW, Woodgett JR. GSK3: tricks of the trade for a multi-tasking kinase. J. Cell. Sci. 2003;116:1175–1186. doi: 10.1242/jcs.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Feng H, Xu M, Lee J, Kim YK, Lim YP, Piao Z, Park YD, Ma H, Hur Y. Comprehensive analysis of genic male sterility-related genes in Brassica rapa using a newly developed Br300K oligomeric chip. PLoS One. 2013;8:e72178. doi: 10.1371/journal.pone.0072178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornelas MC, Van Lammeren AA, Kreis M. Arabidopsis thaliana SHAGGY-related protein kinases (AtSK11 and 12) function in perianth and gynoecium development. Plant J. 2000;21:419–429. doi: 10.1046/j.1365-313x.2000.00691.x. [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38:W64–70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Lu D, Ma X, Peng Y, Sun Y, Ning G, Ma H. Regulation of the Arabidopsis anther transcriptome by DYT1 for pollen development. Plant J. 2012;72:612–624. doi: 10.1111/j.1365-313X.2012.05104.x. [DOI] [PubMed] [Google Scholar]
- Filichkin SA, Leonard JM, Monteros A, Liu PP, Nonogaki H. A novel endo-beta-mannanase gene in tomato LeMAN5 is associated with anther and pollen development. Plant physiol. 2004;134:1080–1087. doi: 10.1104/pp.103.035998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame S, Cohen P. GSK3 takes centre stage more than 20 years after its discovery. Biochem. J. 2001;359:1–16. doi: 10.1042/0264-6021:3590001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Thelen JJ, Dunker AK, Xu D. Musite, a tool for global prediction of general and kinase-specific phosphorylation sites. Mol. Cell. Proteomics. 2010;9:2586–2600. doi: 10.1074/mcp.M110.001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibalova A, Reňák D, Matczuk K, Dupl’akova N, Chab D, Twell D, Honys D. AtbZIP34 is required for Arabidopsis pollen wall patterning and the control of several metabolic pathways in developing pollen. Plant Mol. Biol. 2009;70:581–601. doi: 10.1007/s11103-009-9493-y. [DOI] [PubMed] [Google Scholar]
- Guan Y, Meng X, Khanna R, LaMontagne E, Liu Y, Zhang S. Phosphorylation of a WRKY transcription factor by MAPKs is required for pollen development and function in Arabidopsis. PLoS Genet. 2014;10:e1004384. doi: 10.1371/journal.pgen.1004384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Ting JT, Sokolov LN, Johnson SA, Luan S. A tumor suppressor homolog, AtPTEN1, is essential for pollen development in Arabidopsis. Plant Cell. 2002;14:2495–2507. doi: 10.1105/tpc.005702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis- acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y, Nishino T, Iwano M, Takayama S, Koizumi N. Role of the plant-specific endoplasmic reticulum stress-inducible gene TIN1 in the formation of pollen surface structure in Arabidopsis thaliana. Plant Biotechnol. 2012;29:51–56. [Google Scholar]
- Javelle M, Marco CF, Timmermans M. In situ hybridization for the precise localization of transcripts in plants. J. Vis. Exp. 2011;57:e3328. doi: 10.3791/3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak C, Hirt H. Glycogen synthase kinase 3/SHAGGY-like kinases in plants: an emerging family with novel functions. Trends Plant Sci. 2002;7:457–461. doi: 10.1016/s1360-1385(02)02331-2. [DOI] [PubMed] [Google Scholar]
- Jope RS, Johnson GV. The glamour and gloom of glycogen synthase kinase-3. Trends Biochem. Sci. 2004;29:95–102. doi: 10.1016/j.tibs.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Kaidanovich-Beilin O, Woodgett JR. GSK-3: functional insights from cell biology and animal models. Front. Mol. Neurosci. 2011;4:40. doi: 10.3389/fnmol.2011.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Guan S, Sun Y, Deng Z, Tang W, Shang J, Sun Y, Burlingame AL, Wang Z. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 2009;11:1254–1260. doi: 10.1038/ncb1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofuji R, Sumikawa N, Yamasaki M, Kondo K, Ueda K, Ito M, Hasebe M. Evolution and divergence of the MADS-box gene family based on genome-wide expression analyses. Mol. Biol. Evol. 2003;20:1963–1977. doi: 10.1093/molbev/msg216. [DOI] [PubMed] [Google Scholar]
- Kondo Y, Ito T, Nakagami H, Hirakawa Y, Saito M, Tamaki T, Shirasu K, Fukuda H. Plant GSK3 proteins regulate xylem cell differentiation downstream of TDIF-TDR signalling. Nat. Commun. 2014;5:3504. doi: 10.1038/ncomms4504. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH. Regulation of brassinosteroid signaling by GSK3/SHAGGY-like kinase. Science. 2002;295:1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- Liu J, Zhang Y, Qin G, Tsuge T, Sakaguchi N, Luo G, Sun K, Shi D, Aki S, Zheng N, et al. Targeted degradation of the cyclin-dependent kinase inhibitor ICK4/KRP6 by RING-type E3 ligases is essential for mitotic cell cycle progression during Arabidopsis gametogenesis. Plant Cell. 2008;20:1538–1554. doi: 10.1105/tpc.108.059741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H. Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 2005;56:393–434. doi: 10.1146/annurev.arplant.55.031903.141717. [DOI] [PubMed] [Google Scholar]
- Moon S, Kim SR, Zhao G, Yi J, Yoo Y, Jin P, Lee SW, Jung KH, Zhang D, An G. Rice glycosyltransferase1 encodes a glycosyltransferase essential for pollen wall formation. Plant Physiol. 2013;161:663–675. doi: 10.1104/pp.112.210948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson R, Slovin JP, Chen C. A simplified method for differential staining of aborted and non-aborted pollen grains. Int. J. Plant Biol. 2010;1:e13. [Google Scholar]
- Phan HA, Iacuone S, Li SF, Parish RW. The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell. 2011;23:2209–2224. doi: 10.1105/tpc.110.082651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao HL, Lim JH, Kim SJ, Cheong GW, Hwang I. Constitutive over-expression of AtGSK1 induces NaCL stress response in the absence of NaCl stress and results in enhanced NaCl tolerance in Arabidopsis. Plant J. 2001;27:305–314. doi: 10.1046/j.1365-313x.2001.01099.x. [DOI] [PubMed] [Google Scholar]
- Reňák D, Dupl’áková N, Honys D. Wide-scale screening of T-DNA lines for transcription factor genes affecting male gametophyte development in Arabidopsis. Sex. Plant Reprod. 2012;25:39–60. doi: 10.1007/s00497-011-0178-8. [DOI] [PubMed] [Google Scholar]
- Rozhon W, Mayerhofer J, Petutschnig E, Fujioka S, Jonak C. ASKtheta, a clade-III Arabidopsis GSK3, functions in the brassinosteroid signalling pathway. Plant J. 2010;62:215–223. doi: 10.1111/j.1365-313X.2010.04145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saidi Y, Hearn TJ, Coates JC. Function and evolution of ‘green’ GSK3/Shaggy-like kinases. Trends Plant Sci. 2012;17:39–46. doi: 10.1016/j.tplants.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Sancenon V, Puig S, Mateu-Andres I, Dorcey E, Thiele DJ, Penarrubia L. The Arabidopsis copper transporter COPT1 functions in root elongation and pollen development. J. Biol. Chem. 2004;279:15348–15355. doi: 10.1074/jbc.M313321200. [DOI] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire K, Hsu Y-C, Lee PY, Truong MT, Beals T, Goldberg R. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 1999;11:297–322. [Google Scholar]
- Schiott M, Romanowsky SM, Baekgaard L, Jakobsen MK, Palmgren MG, Harper JF. A plant plasma membrane Ca2+ pump is required for normal pollen tube growth and fertilization. Proc. Natl. Acad. Sci. 2004;101:9502–9507. doi: 10.1073/pnas.0401542101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, Östlund G. InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 2015;43:D234–239. doi: 10.1093/nar/gku1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto G, Alleva K, Mazzella MA, Amodeo G, Muschietti JP. AtTIP1;3 and AtTIP5;1, the only highly expressed Arabidopsis pollen-specific aquaporins, transport water and urea. FEBS Lett. 2008;582:4077–4082. doi: 10.1016/j.febslet.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Soto G, Fox R, Ayub N, Alleva K, Guaimas F, Erijman EJ, Mazzella A, Amodeo G, Muschietti J. TIP5;1 is an aquaporin specifically targeted to pollen mitochondria and is probably involved in nitrogen remobilization in Arabidopsis thaliana. Plant J. 2010;64:1038–1047. doi: 10.1111/j.1365-313X.2010.04395.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichtinsky G, Tavares R, Takvorian A, Schwebel-Dugue N, Twell D, Kreis M. An evolutionary conserved clade of plant GSK-3/shaggy-like protein kinase genes preferentially expressed in developing pollen. Biochim. Biophys. Acta. 1998;1442:261–273. doi: 10.1016/s0167-4781(98)00187-0. [DOI] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. The botany array resource: e-Northerns, expression angling, and promoter analyses. Plant J. 2005;43:153–163. doi: 10.1111/j.1365-313X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- Twell D. Male gametogenesis and germline specification in flowering plants. Sex. Plant Reprod. 2011;24:149–160. doi: 10.1007/s00497-010-0157-5. [DOI] [PubMed] [Google Scholar]
- Wang RS, Pandey S, Li S, Gookin TE, Zhao Z, Albert R, Assmann SM. Common and unique elements of the ABA-regulated transcriptome of Arabidopsis guard cells. BMC Genomics. 2011;12:216. doi: 10.1186/1471-2164-12-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Shang JX, Chen QX, Oses-Prieto JA, Bai MY, Yang Y, Yuan M, Zhang YL, Mu CC, Deng Z, et al. Identification of BZR1-interacting proteins as potential components of the brassinosteroid signaling pathway in Arabidopsis through tandem affinity purification. Mol. Cell. Proteomics. 2013a;12:3653–3665. doi: 10.1074/mcp.M113.029256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang W, Wang YQ, Liu YY, Wang JX, Zhang XQ, Ye D, Chen LQ. Arabidopsis galacturonosyltransferase (GAUT) 13 and GAUT14 have redundant functions in pollen tube growth. Mol. Plant. 2013b;6:1131–1148. doi: 10.1093/mp/sst084. [DOI] [PubMed] [Google Scholar]
- Wellmer F, Riechmann JL, Alves-Ferreira M, Meyerowitz EM. Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell. 2004;16:1314–1326. doi: 10.1105/tpc.021741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijeratne AJ, Zhang W, Sun Y, Liu W, Albert R, Zheng Z, Oppenheimer DG, Zhao D, Ma H. Differential gene expression in Arabidopsis wild-type and mutant anthers: insights into anther cell differentiation and regulatory networks. Plant J. 2007;52:14–29. doi: 10.1111/j.1365-313X.2007.03217.x. [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Knox RB, Taylor PE, Singh MB. Bcp1, a gene required for male sterility in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 1995;92:2106–2110. doi: 10.1073/pnas.92.6.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yang C, Yuan Z, Zhang D, Gondwe MY, Ding Z, Liang W, Zhang D, Wilson ZA. The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell. 2010;22:91–107. doi: 10.1105/tpc.109.071803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Zhao J, Peng P, Chihara RK, Li J. BIN2 functions redundantly with other Arabidopsis GSK3-like kinases to regulate brassicnosteroid signaling. Plant Physiol. 2009;150:710–721. doi: 10.1104/pp.109.138099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TJ, Kim JS, Lim KB, Kwon SJ, Kim JA, Jin M, Park JY, Lim MH, Kim H, Kim SH, et al. The Korea Brassica genome project: a glimpse of the Brassica genome based on comparative genome analysis with Arabidopsis. Comp. Funct. Genomics. 2005;6:138–146. doi: 10.1002/cfg.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Vizcay-Barrena G, Conner K, Wilson ZA. MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell. 2007;19:3530–3548. doi: 10.1105/tpc.107.054981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Wu J, Romanovicz D, Clark G, Roux SJ. Co-regulation of exine wall patterning, pollen fertility and anther dehiscence by Arabidopsis apyrases 6 and 7. Plant Physiol. Biochem. 2013;69:62–73. doi: 10.1016/j.plaphy.2013.04.022. [DOI] [PubMed] [Google Scholar]
- Zhang S, Cai Z, Wang X. The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proc. Natl. Acad. Sci. USA. 2009;106:4543–4548. doi: 10.1073/pnas.0900349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Peng P, Schmitz RJ, Decker AD, Tax FE, Li J. Two putative BIN2 substrates are nuclear components of brassinosteroid signaling. Plant Physiol. 2002;130:1221–1229. doi: 10.1104/pp.102.010918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Chen H, Li H, Gao JF, Jiang H, Wang C, Guan YF, Yang ZN. Defective in tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J. 2008;55:266–277. doi: 10.1111/j.1365-313X.2008.03500.x. [DOI] [PubMed] [Google Scholar]
- Zuberi K, Franz M, Rodriguez H, Montojo J, Lopes CT, Bader GD, Morris Q. GeneMANIA prediction server 2013 update. Nucleic Acids Res. 2013;41:W115–122. doi: 10.1093/nar/gkt533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


