Abstract
While increasing evidence indicates the important function of histone methylation during development, how this process influences cardiac development in vertebrates has not been explored. Here, we elucidate the functions of two histone H3 lysine 4 (H3K4) methylation enzymes, SMYD3 and SETD7, during zebrafish heart morphogenesis using gene expression profiling by whole mount in situ hybridization and antisense morpholino oligonucleotide (MO)-based gene knockdown. We find both smyd3 and setd7 are highly expressed within developing zebrafish heart and knock-down of these genes led to severe defects in cardiac morphogenesis without altering the expressions pattern of heart markers, including cmlc2, vmhc, and amhc. Furthermore, double knock-down by coinjection of smyd3 and setd7 MOs caused the synergistic defects in heart development. As similar to knock-down effect, overexpression of these genes also caused the heart morphogenesis defect in zebrafish. These results indicate that histone modifying enzymes, SMYD3 and SETD7, appear to function synergistically during heart development and their proper functioning is essential for normal heart morphogenesis during development.
Keywords: heart morphogenesis, histone methyltransferase, SETD7, SMYD3, zebrafish
INTRODUCTION
During vertebrate development, heart is formed by a highly stereotypic and organized process, and the expression of genes in these processes need be tightly regulated in a spatiotemporal manner (Barnett et al., 2012; Wang, 2012). Recent reports suggest that epigenetic regulation modulates genetic program during differentiation and development of various organs including heart and somitic lineages (Wamstad et al., 2012). For instance, modifications of histone substantially alter the accessibility and structure of chromatin, therefore modifying transcriptional activity of specific gene clusters. Histones are known to undergo specific modification such as methylation and acetylation: while methylation and demethylation of histone are coordinated by histone methyltransferases (HMTs) and demethylase (HDMs), acetylation of histone is modulated by acetyltransferases (HAT) and deacetylases (HDAC) (Bhaumik et al., 2007; Helin and Dhanak, 2013). Due to their significance in regulating genetic program during development and diseases, recent efforts were directed to identify specific factors that mediate this process (Arrowsmith et al., 2012; Cayuso Mas et al., 2011; Kim et al., 2012a; 2012b). In case of methylation, histone H3 lysine 4 residue (H3K4) is sequentially modified through mono-, di-, and tri-methylation status, each of which step is mediated by specific enzymes. MLLs (Mll1–5), SETDB1, and SETDB2 have broad roles for H3K4 methylation, and can methylate all type of H3K4, such as non-, mono-, di-methylated H3K4 (Black et al., 2012; Sims and Reinberg, 2004). In contrast, SMYD3 methylate mono- and di-methylated H3K4, and SETD7 can only catalase the non-methylated H3K4 to transit to mono-methylated H3K4 (Hamamoto et al., 2004; Wang et al., 2001).
SET and MYND domain-containing proteins (SMYDs), involving SMYD1–5, are high conserved protein family across plant, fungi, and animal as well as some protozoa, and have two functional protein domains, SET and MYND domains (Del Rizzo and Trievel, 2011; Dillon et al., 2005). Especially, SET domain is important for histone lysine methylation activity and MYND domain can mediate the protein-protein interaction and bind to DNA motifs. SMYD3 was originally reported as histone lysine methyltransferase (HMT), which methlylates the mono-(H3K4me1) and di-methylated lysine 4 residue (H3K4me2) to generate H3K4me3, of histone H3, but SMYD3 also can bind to 5′-CCCTCC-3′ motif on promoter region of Nkx2.8 gene to induce its expression in cancer cells (Hamamoto et al., 2004). Recent researches showed the global decrease of histone H4 lysine 5 (H4K5) methylation in SMYD3 knock-down condition and structural preference for histone H4 lysine 20 (H4K20) of SMYD3, suggesting the substrate diversity of SMYD3 (Foreman et al., 2011; Van Aller et al., 2012). It is known that SMYD3 is highly expressed in a various cancers, including liver, prostate, rectal, and breast carcinomas, and stimulates the oncogenic activities, such as cell proliferation, adhesion and migration, of tumor cells (Frank et al., 2006; Hamamoto et al., 2006; Silva et al., 2008). In addition, SMYD3 is essential for myogenic differentiation through MyoD regulation (Proserpio et al., 2013). Actually, the deficient zebrafish embryos displayed the developmental cardiac and somite abnormalities, illuminating its functions in cardiac and somite muscle formation (Fujii et al., 2011).
SET domain containing protein 7 (SETD7), also termed as SET7/9, is another type of histone lysine methyltransferase (HMT) and only have SET domain for methyltransferase activity, but not MYND domain (Wang et al., 2001). It was initially discovered as a specific methyltransferase for only non-methylated histone H3 lysine 4 (H3K4) that converted to mono-methylated histone H3 lysine 4 (H3K4me1) (Wang et al., 2001). Subsequently, recent accumulating evidence suggests that non-histone proteins, such as Yap, DNMT1, E2F1, and STAT3, are also the substrates for SETD7 (Estève et al., 2009; Oudhoff et al., 2013; Pradhan et al., 2009; Yang et al., 2010). The mouse knock out allele of SETD7 is a viable and did not show any developmental and gross defect (Campaner et al., 2011; Lehnertz et al., 2011). In zebrafish developmental model, knock-down of setd7 showed the defects in skeletal muscle formation and myofibril structures (Tao et al., 2011).
Here, we examined the function of smyd3 and setd7 in heart development in zebrafish. Collectively, our results demonstrate that SMYD3 and SETD7 function synergistically for skeletal muscle development and non-redundantly for heart morphogenesis.
METHODS AND MATERIALS
Zebrafish husbandry
Zebrafish AB strain as wild-type control for all experiments, expect the experiments using Tg(cmlc2:EGFP) line. Zebrafish strains were maintained as previously described (Westerfield, 1993). To visualize the heart morphology in live embryos, Tg(cmlc2:EGFP) embryos were subjected to several experiments (Huang et al., 2003).
Injection of anti-sense morpholino oligonucleotides (MOs)
Control, smyd3, and setd7 MOs were designed by author and ordered to Genetools Company. MOs were injected with 0.1% phenol-red solution into zebrafish embryos at 1–2 cell stages as desired concentrations. The sequences of MOs used are Control MO, 5′-CCTCTTACCTCAGTTACAATTTATA-3′; smyd3 MO, 5′-AAGAGCTCTTACCTGACACTGAACG-3′; and setd7 MO, 5′-AGCTTCTCGTACCTTCCACCACTTC-3′. Embryos injected MOs were observed or collected at desired developmental stages for further experiment.
Whole mount in situ hybridization
To make digoxigenin-labelled antisense ribo-probe, zebrafish smyd1a (NM_205540.1), smyd2 (NM_001013550.1), smyd3 (NM_001037400.1) and setd7 (NM_001002456.1) full ORFs were cloned into pGEM-T vector (Promega) after amplification of 24 hpf cDNA. Cloned genes were validated by sequencing. Following primers were used for PCR reaction: smyd1a forward, 5′-AGCATGACCGTGGAGAAGAC-3′; smyd1a reverse, 5′-TGTTCATGCTTTGATCTGCAC-3′; smyd2 forward, 5′-AGCA GCAGGTCAATAGCGATG-3′; smyd2 reverse, 5′-TTCCTCCCATTATGTCTGCTC-3′; smyd3 forward, 5′-ATGGACGGTCACTG CAGATGA-3′; smyd3 reverse, 5′-CTAGAGCGTGTTGAGCTC AGC-3′; setd7 forward, 5′-AACATGGACAGCGATGATGAC-3′, and setd7 reverse, 5′-CAGGTGTTCAGAGGAGTC-3′. The reaction producing ribo-probes were carried out according to manufacture protocol (Roche). Whole mount in situ hybridization were routinely performed as previously described (Thisse and Thisse, 2008). As brief, zebrfish embryos were fixed by 4% paraformaldehyde (PFA) and stored in methanol. To experiment, fixed embryos washed and digested with proteinase K solution as desired concentration and times. Hybridization reaction was carried out at 65°C, and the detection of hybridized RNA probe was achieved by anti-digoxigenin Fab fragment conjugated to alkaline phosphatase and BCIP/NBT substrate (Roche). Staining were stopped according to desired detection status and imaged in 75% glycerol solution for proper orientation.
Over-expression of smyd3 and setd7 by the injection of synthetic mRNA
To generate the synthetic smyd3 and setd7 mRNAs, we first cloned the full open reading frames (ORF) of smyd3 and setd7 into pCS2+ expression vector, which have SP6 promoter sequence and SV40 poly-A tail sequence for in vitro transcription. These plasmids were cut by NotI digestion to linearize, and capping mRNA were produced using m7G(5′)PPP(5′)G (Roche, Germany) and SP6 RNA polymerase (Roche, Germany). Synthetic mRNA was diluted as 100 μg/ml and phenol red was added prior to injection as 0.1% solution. Injection was achieved at 1–2 cell stages as optimal concentrations (about 100–200 pg/embryo).
RESULTS
To delineate functions of HMTs, such as smyd1a, smyd2, smyd3, and setd7, during heart development, we have first analyzed the expression pattern of these genes by whole mount in situ hybridization analyses and conventional reverse-transcriptase (RT) PCR. Transcripts of all HMTs examined appear to be maternally deposited, and widely detected at early stages. At later stages, the expression becomes gradually restricted to several organs, including somite, brain and eye (Figs. 1 and 2). While smyd1a and smyd2 showed higher expressions in developing somite region from 10 somite stage (ss) than smyd3 and setd7, hinting their roles in somitogenesis (Figs. 1 and 2) (Sesé et al., 2013), the developmental expression regions of smyd3 and setd7 were highly expressed within the heart forming region at 36 h post-fertilization (hpf) and 48 hpf, respectively (red arrow heads in Fig. 2). These gene expression profiling data suggest functions of zygotic SMYD3 and SETD7 in heart development.
Fig. 1.
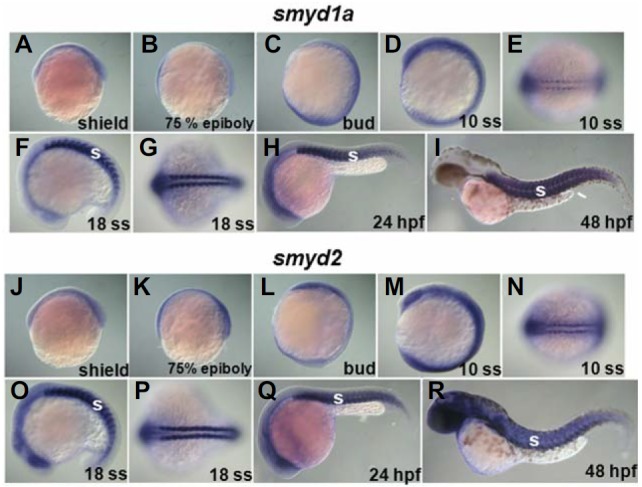
The expressions of smyd1a and smyd2 are ubiquitous in early developmental stages and restricted into somite during late development in zebrafish. (A–I) Images by whole mount in situ hybridization experiment using smyd1a ribo-probe. (J–R) Developmental expressions of smyd2. Lateral views (A–D, F, H–M, O, Q, and R) and dorsal views (E, G, N, and, P). Anterior is left (E–F and N–R). S; somite.
Fig. 2.
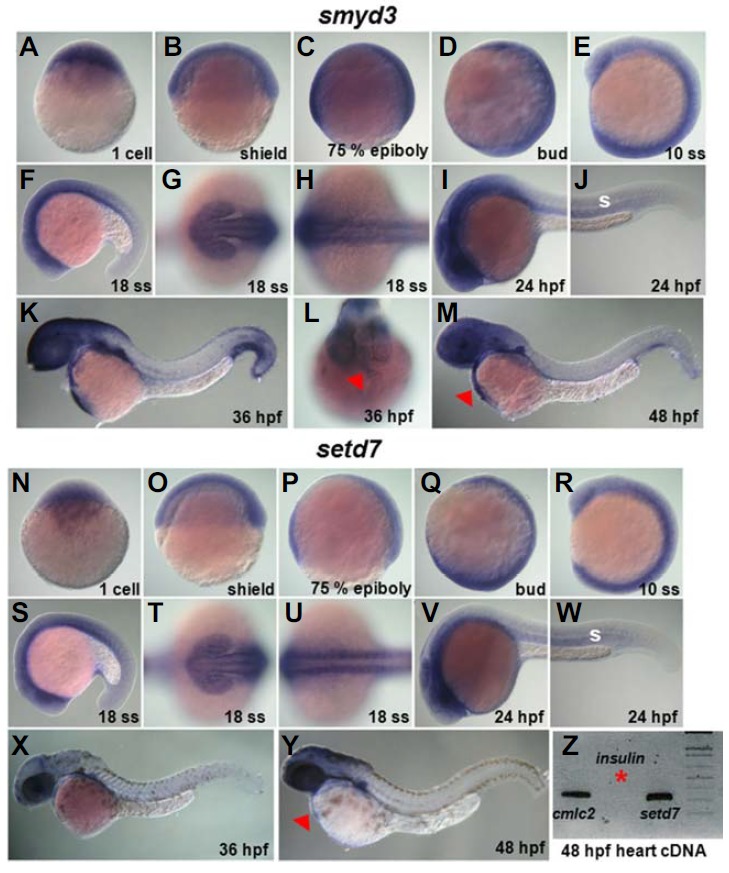
smyd3 and setd7 express in developing heart in zebrafish. (A–M) Developmental expressions of smyd3 by whole mount in situ hybridization experiment. (N–Y) The detection of setd7 expressions by whole mount in situ hybridization. Lateral views (AF, I–K, M–S, and V–Y), anterior views (G, L, and T), and dorsal views (H, U). Anterior is left (F–K, M, and S–Y). Red arrowheads point to heart regions. S; somite. (Z) setd7 expression in heart was determined by conventional reverse-transcriptase (RT) PCR method using 48 hpf zebrafish cDNA. insulin (red asterisk), which not expressed in heart sample prepared here, was used as control.
To further delineate the roles of HMTs in heart development, we carried out knock-down experiments of SMYD3 and SETD7. First, to determine the functional requirement of SMYD3 during zebrafish heart development, we knocked SMYD3 down with a morpholino (MO)-based gene targeting system. For this purpose, splicing blocking MO that binds to the junction of exon2 and intron2 of smyd3 pre-matured mRNA was designed (Fig. 3A). To validate the MO efficacy, PCR primers were designed to detect normal and aberrant smyd3 transcripts in wild-type embryos and smyd3 morphants, and carried out conventional RT-PCR (Fig. 3B). In smyd3 morphant embryo, mature transcript that amplified by F+R2 primer was not presented, while aberrant transcript that produced by F+R1 primer was displayed (Fig. 3B), validating the efficacy of smyd3 MO. In addition, smyd3 expression was drastically reduced in smyd3 morphants at 22 ss, suggesting non-sense mediated decay of aberrant transcript caused by MO injection (Fig. 3C). At optimal concentration (approximately 3.0 ng/embryo), gross morphology of smyd3 morphants was comparable to control embryos (Fig. 3D and data not shown). However, pronounced heart edema (red arrow) was apparent in smyd3 morphants at 48 hpf, suggesting that SMYD3 function may be essential for cardiac development (Fig. 3D).
Fig. 3.
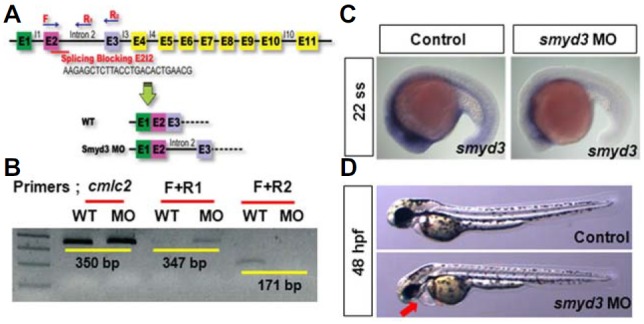
The knock-down of smyd3 causes the developmental cardiac edema. (A) Schematic showing the design of splicing blocking morpholino oligonucleotide (MO) targeting to smyd3. (B) Efficacy validation of smyd3 MO by conventional RT-PCR using 24 hpf cDNA. Normal PCR product by F and R2 primers was largely decreased, while aberrant PCR product was presented by F and R1 primers in smyd3 morphant embryos. (C) Images by whole mount in situ hybridization experiment at 22 somite stage (ss). Endogenous smyd3 expression was reduced by smyd3 MO injection. (D) Phenotypes of embryos injected control and smyd3 MO. Red arrow points to cardiac edema at 48 hpf. Anterior is left.
Similarly, we examined the function of SETD7 during development. MOs targeting setd7 was designed in a way to target the junction of exon 1 and intron 1 of setd7 pre-mature mRNA (Fig. 4A). The efficacy of the MO was validated by conventional RT-PCR (Fig. 4B). In the setd7 MO-injected embryos, normal transcript was largely abolished, detecting by PCR primer F+R1 (Fig. 4B). In addition, expression of setd7 was noticeably decreased by MO injection at 22 ss, suggest that setd7 MO is a functional to block the normal transcription of setd7 gene (Fig. 4C). Similar to SMYD3 deficiency, the knock-down embryos of setd7 at optimal concentration (1.0 ng/embryo) displayed cardiac edema (red arrow) without causing any obvious morphological defects (Fig. 4D and data not shown). Collectively, our results illustrate that the proper activities of H3K4 methyltransferases, specifically SMYD3 and SETD7, are important for zebrafish heart formation.
Fig. 4.
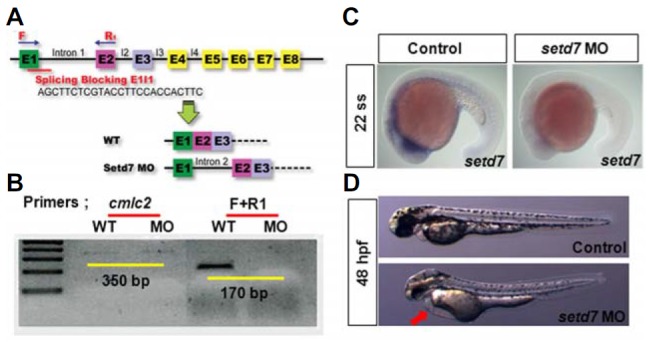
SETD7-deficent embryo shows the developmental heart edema. (A) Design of the splicing blocking morpholino oligo (MO) targeting to setd7. (B) Validation of setd7 MO efficacy. Normal transcript of setd7 presenting RT- PCR by F and R1 primers was largely reduced. (C) The endogenous expression of setd7 was determined by whole mount in situ hybridization experiment in control and setd7 knock-down embryos. (D) Phenotypes of embryos injected control and setd7 MO. Red arrow points to cardiac edema at 48 hpf. Anterior is left.
During histone H3 lysine 4 (H3K4) methylation, SETD7 can only methylate non-methylated H3K4 to produce mono-methylated H3K4 (H3K4me1), which then is further methylated by SMYD3 to become H3K4me2 and H3K4me4 (Hamamoto et al., 2004; Wang et al., 2001). Individual roles of SMYD3 and SETD7 during cardiac and skeletal muscle development in zebrafish have been reported (Fujii et al., 2011; Tao et al., 2011). Given that SMYD3 and SETD7 successively methylate H3K4, we can assume that these two enzymes function synergistically during zebrafish development. To test this possibility, we examined whether co-injection of smyd3 and setd7 MOs at a suboptimal concentration can give rise a synergistic effect for induction of developmental defects (Fig. 5). The smyd3 and setd7 MOs, by themselves at optimal concentrations (3.0 ng/embryo for smyd3 and 1.0 ng/embryo for setd7), produced no discernible gross morphological defects except cardiac edema at 48 hpf (Figs. 3 and 4). However, co-injection of these MOs at optimal concentrations caused severe somitogenesis defects as indicated by the phenotype of shortened and curled body morphology (Fig. 5A). In addition, the severity of the abnormalities was sharpened by co-injection of increased concentrations of these MOs (Fig. 5B). As expected, these results suggest that SMYD3 and SETD7 have a synergistic function in zebrafish development.
Fig. 5.
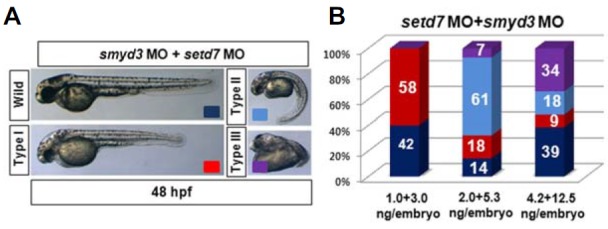
Synergistic defect of body formation by double knock-down of smyd3 and setd7. (A) Developmental phenotypes of embryos injected both smyd3 and setd7 MOs. Embryos were injected both MOs at 1–2 cell stages and imaged at 48 hpf. The embryos in type I group showed shortened body axis. Type II group was sorted by appearance of curved body formation. Monster phenotypes were grouped into type III. Anterior is left. (B) Phenotypic contributions were determined by titrated-MO injections. Ratios were indicated in graph (N > 100).
Next, we delineated the synergistic roles of SMYD3 and SETD7 in zebrafish heart development. For this purpose, we used the heart-specific transgenic fish Tg(cmlc2:EGFP) line to allow easy visualization of the heart morphology in live embryos (Fig. 6A). The injections of control, smyd3, setd7, and smyd3 + setd7 MOs were carried out at the optimal concentrations (3.0 ng/embryo for smyd3, 1.0 ng/embryo for setd7). Morphants without body morphological defects were selected for further analyses of heart development. At 48 hpf, heart tubes in the smyd3 and setd7 morphants displayed defects in the looping process and remained linear in shape (Fig. 6A). Single knockdown of these genes caused a defect in heart morphogenesis, suggesting that each gene is indispensable for heart morphogenesis. Interestingly, double knock-down of SMYD3 and SETD7 presented a more severe defect than single knockdown of either of those genes (Fig. 6A). However, the global expression level of clmc2 detected by whole mount in situ hybridization (Kim et al., 2014) was not altered in these smyd3, setd7, and smyd3 + setd7 morphant embryos at 48 hpf (data not shown), suggesting that specification and/or maintenance of cardiomyocytes were not compromised in those embryos. Collectively, our results show that SMYD3 and SETD7 likely function synergistically in zebrafish development. Moreover, our data suggest that heart development is more sensitive than somitogenesis to the decrease of SMYD3 and SETD7 levels.
Fig. 6.
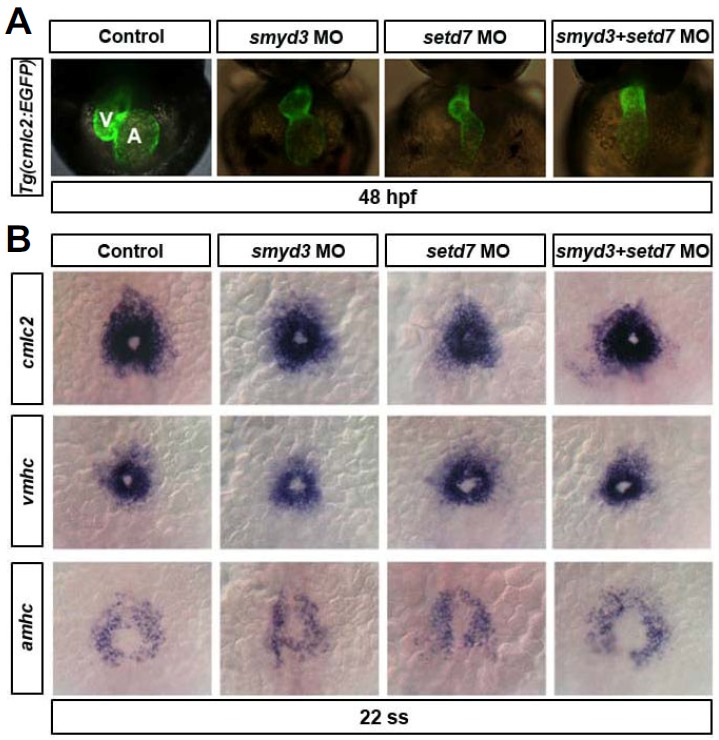
Heart morphogenesis was affected by smyd3 and/or setd7 knock-down in Tg(cmlc2:EGFP) fish, but not the expressions of early heart markers. (A) Developmental heart morphology showing the GFP expression by cmlc2 promoter in Tg(cmlc2:EGFP) transgenic fish. MOs were injected at 1–2 cell stages and imaged by 48 hpf. smyd3 and setd7 knock-down cause the looping defect in zebraifsh developing heart. (B) Early heart marker expressions by whole mount in situ hybridization. cmlc2, vmhc, and amhc expressions were not changed in smyd3 and setd7 knock-down embryos. MOs were injected at 1–2 cell stage and embryos were collected at 22 somite stage (ss). Dorsal views and anterior is up.
To test whether the developing heart defects in the SMYD3-and SETD7-deficient embryos are caused by the expressional changes of early cardiac muscle genes, we determined the early expressions of cmlc2, vmhc, and amhc (Kim et al., 2014) by whole mount in situ hybridization experiment. The expression level of these genes was not altered by the knock-down of SMYD3 and/or SETD7 and the gathered shape of cardiac precursor cells in these morphant embryos was also comparable to wild-type embryos (Fig. 6B). This finding suggests that SMYD3 and SETD7 are dispensable for proper expression of early cardiac muscle genes and differentiation of cardiac precursor cells.
To investigate whether enforced expression of smyd3 and setd7 causes the cardiac defect in zebrafish, synthetic mRNA encoding SMYD3 or SETD7 was injected (Fig. 7A). The co-injection of smyd3 and setd7 mRNA as optimal dose (each 100–200 pg/embryo) caused similar developmental defects as in embryos lack either smyd3 or setd7, such as defective body axis formation. Moreover, these embryos showed pronounced cardiac edema, indicating the defects in heart formation (red arrow in Fig. 7B). To better examine the defects caused by overexpression of these smyd3 and setd7, we analyzed the expression of known cardiac markers, including cmlc2, vmhc, and amhc. Despite the obvious cardiac edema, the expression of these genes was unaltered in embryos overexpressing smyd3 and setd7 (Fig. 7C). Therefore, it appears that excessive H3K4 methyltransferase activity can lead to defects in heart and skeletal muscle development in zebrafish.
Fig. 7.
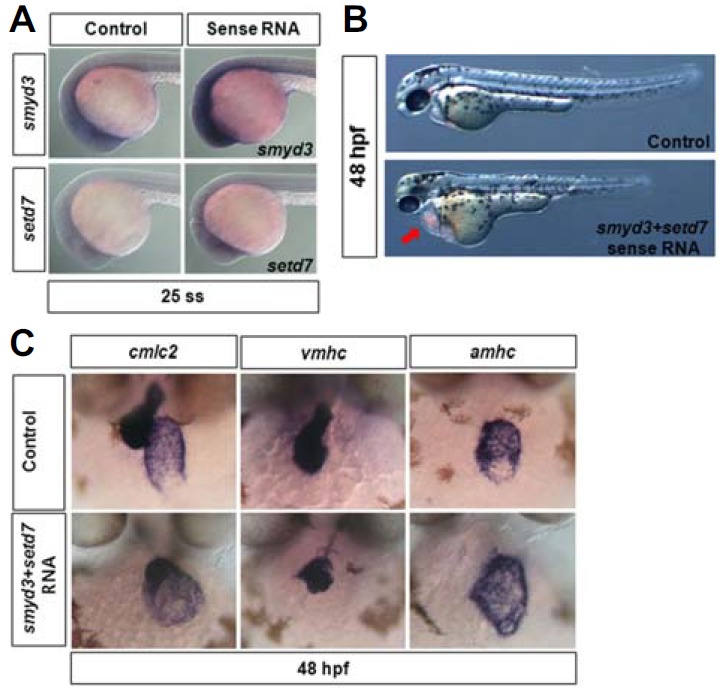
Augmented activities of SMYD3 and SETD7 cause the defect in heart morphogenesis. (A) Endogenous expressions of smyd3 and setd7 were determined by whole mount in situ hybridization experiment in control and synthetic smyd3- or setd7 mRNA-injected embryos. Higher expression was shown in embryos injected sense RNA compared with control at 25 ss. Developmental phenotypes of control and both smyd3 and setd7 sense RNA-injected embryos at 48 hpf. Embryos injected sense RNA of these genes displayed the heart edema (red arrow). (C) Heart morphology was addressed by expression shapes of heart marker genes, such as cmlc2, vmhc, and amhc, in control and sense RNA injected embryos at 48 hpf. Expression levels of heart marker genes were not changed, but expression shapes were largely altered in embryos injected sense RNA. MOs were injected at 1–2 cell stage and embryos were collected at 48 hpf. Frontal views and anterior is up.
Taken together, our results indicate that methylation can provide essential regulatory input during organ development in zebrafish. Since similar developmental defects were observed in embryos with either excessive or reduced level of SMDY3 and/or SETD7 activity, it appears that appropriate level of histone methylation is critical to ensure the formation of cardiac and skeletal muscle in zebrafish.
DISCUSSION
Heart morphogenesis in vertebrate development is a highly complicated event that achieved by correct cell specification, migration, proliferation, apoptosis, and transition as well as controlled beating of heart myocytes (Barnett et al., 2012). In this process, proper gene expressions for structure of cardiac chamber at early developmental stages are critical for late heart forming event. Our data presented here indicates that proper level of histone methylation, which is mediated by a series of enzymes commonly known as HMTs, including smyd3, and setd7 appears to be essential for heart development.
During zebrafish development, the expression pattern of HMTs indicates that transcripts encoding these proteins may be maternally deposited. In addition, considering their ubiquitous expression, these proteins may provide essential functions in development by regulating histone methylation for all types of cells (Figs. 1 and 2). However, knock-down of smyd3 and setd7 in zebrafish embryos did not cause any obvious defects in early development, such as in gastrulation movement, germ layer formation, cell proliferation, or cell death. Therefore, these genes may redundantly function to regulate early vertebrate development (Figs. 1 and 2). In later developmental stages, the expressions of all HMTs tested in this paper were gradually restricted and often increased in specific organs, including somite, brain, eye primordial cells, fin primordial region, and heart. For instance, smyd2 becomes highly expressed within eye primodium, while smyd3 and setd7 are enriched in developing heart, suggest that each HMTs might have a non-redundant and organ-specific roles during late vertebrate developmental periods (Figs. 1 and 2).
It was report that knocked-down smyd3 and setd7 by MO led to developmental defects in heart and/or skeletal muscle (Fujii et al., 2011; Tao et al., 2011). Consistent with previous reports, we also find that MO injections as high concentrations (over 10 ng/embryo for smyd3 MO and 4.2 ng/embryo for setd7 MO) cause the similar somite defects. However, the optimal injection conditions (concentrations under 3.0 ng/embryo for smyd3 and 1.0 ng/embryo for setd7 MO) morphant embryos showed only developmental heart abnormality without any somite defects, indicating that each organ may have different sensitivity to the fluctuation in Smyd3 and Setd7 level during organogenesis.
Interestingly, in skeletal muscle development, SETD7 directly interacts with MyoD to induce myogenin and MEF2 expression to generate differentiated myocytes (Tao et al., 2011). Therefore, it is tempting to speculate that SETD7 manipulation in zebrafish may lead to the alteration in expression of certain structural genes. However, we did not find any discernible change in the expression of cmlc2, vmhc, and amhc in our SMYD3 and SETD7 double-deficient or overexpressed embryos, suggesting the presence of distinct mechanisms underlying SETD7-mediated differentiation of the cardiac muscle and skeletal muscle cells. Moreover, SMYD3 and SETD7 appear to be dispensable for the specification of cardiomyocytes, since the lack of SMYD3 or SETD7 did not abrogate the expression of cardiac markers (Figs. 6 and 7).
Here, we have shown the knock-down and overexpression phenotypes of SMYD3 and SETD7 in zebrafish development. The results suggest that H3K4 methyltransferases are crucial genetic regulators for normal heart development. These findings extend our current knowledge of the roles of HMTs and increase our understanding of the functional mechanism of HMTs in heart development.
Acknowledgments
We thank the Korea Zebrafish Organogenesis Mutant Bank (ZOMB) and Zebrafish Information Network (ZFIN) for providing zebrafish lines. This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF-2013R1A1A2009570) funded by the Ministry.
REFERENCES
- Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat. Rev. Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- Barnett P, van den Boogaard M, Christoffels V. Localized and temporal gene regulation in heart development. Curr. Top. Dev. Biol. 2012;100:171–201. doi: 10.1016/B978-0-12-387786-4.00004-X. [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. Mol. Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol. Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campaner S, Spreafico F, Burgold T, Doni M, Rosato U, Amati B, Testa G. The methyltransferase Set7/9 (Setd7) is dispensable for the p53-mediated DNA damage response in vivo. Mol. Cell. 2011;43:681–688. doi: 10.1016/j.molcel.2011.08.007. [DOI] [PubMed] [Google Scholar]
- Cayuso Mas J, Noël ES, Ober EA. Chromatin modification in zebrafish development. Methods Cell Biol. 2011;104:401–428. doi: 10.1016/B978-0-12-374814-0.00022-7. [DOI] [PubMed] [Google Scholar]
- Del Rizzo PA, Trievel RC. Substrate and product specificities of SET domain methyltransferases. Epigenetics. 2011;6:1059–1067. doi: 10.4161/epi.6.9.16069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6:227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estève PO, Chin HG, Benner J, Feehery GR, Samaranayake M, Horwitz GA, Jacobsen SE, Pradhan S. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc. Natl. Acad. Sci. USA. 2009;106:5076–5081. doi: 10.1073/pnas.0810362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman KW, Brown M, Park F, Emtage S, Harriss J, Das C, Zhu L, Crew A, Arnold L, Shaaban S, Tucker P. Structural and functional profiling of the human histone methyltransferase SMYD3. PLoS One. 2011;6:e22290. doi: 10.1371/journal.pone.0022290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank B, Hemminki K, Wappenschmidt B, Klaes R, Meindl A, Schmutzler RK, Bugert P, Untch M, Bartram CR, Burwinkel B. Variable number of tandem repeats polymorphism in the SMYD3 promoter region and the risk of familial breast cancer. Int. J. Cancer. 2006;118:2917–2918. doi: 10.1002/ijc.21696. [DOI] [PubMed] [Google Scholar]
- Fujii T, Tsunesumi S, Yamaguchi K, Watanabe S, Furukawa Y. Smyd3 is required for the development of cardiac and skeletal muscle in zebrafish. PLoS One. 2011;6:e23491. doi: 10.1371/journal.pone.0023491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, Yagyu R, Nakamura Y. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat. Cell Biol. 2004;6:731–740. doi: 10.1038/ncb1151. [DOI] [PubMed] [Google Scholar]
- Hamamoto R, Silva FP, Tsuge M, Nishidate T, Katagiri T, Nakamura Y, Furukawa Y. Enhanced SMYD3 expression is essential for the growth of breast cancer cells. Cancer Sci. 2006;97:113–118. doi: 10.1111/j.1349-7006.2006.00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helin K, Dhanak D. Chromatin proteins and modifications as drug targets. Nature. 2013;502:480–488. doi: 10.1038/nature12751. [DOI] [PubMed] [Google Scholar]
- Huang CJ, Tu CT, Hsiao CD, Hsieh FJ, Tsai HJ. Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev. Dyn. 2003;228:30–40. doi: 10.1002/dvdy.10356. [DOI] [PubMed] [Google Scholar]
- Kim B, Kang S, Kim SJ. Differential promoter methylation and histone modification contribute to the brain specific expression of the mouse Mbu-1 gene. Mol. Cells. 2012a;34:433–437. doi: 10.1007/s10059-012-0182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Kim MJ, Koo TH, Kim JD, Koun S, Ham HJ, Lee YM, Rhee M, Yeo SY, Huh TL. Histone deacetylase is required for the activation of Wnt/β-catenin signaling crucial for heart valve formation in zebrafish embryos. Biochem. Biophys. Res. Commun. 2012b;423:140–146. doi: 10.1016/j.bbrc.2012.05.098. [DOI] [PubMed] [Google Scholar]
- Kim JD, Kim HJ, Koun S, Ham HJ, Kim MJ, Rhee M, Huh TL. Zebrafish Crip2 plays a critical role in atrioventricular valve development by downregulating the expression of ECM genes in the endocardial cushion. Mol. Cells. 2014;37:406–411. doi: 10.14348/molcells.2014.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnertz B, Rogalski JC, Schulze FM, Yi L, Lin S, Kast J, Rossi FM. p53-dependent transcription and tumor suppression are not affected in Set7/9-deficient mice. Mol. Cell. 2011;43:73–680. doi: 10.1016/j.molcel.2011.08.006. [DOI] [PubMed] [Google Scholar]
- Oudhoff MJ, Freeman SA, Couzens AL, Antignano F, Kuznetsova E, Min PH, Northrop JP, Lehnertz B, Barsyte-Lovejoy D, Vedadi M, et al. Control of the hippo pathway by Set7-dependent methylation of Yap. Dev. Cell. 2013;26:188–194. doi: 10.1016/j.devcel.2013.05.025. [DOI] [PubMed] [Google Scholar]
- Pradhan S, Chin HG, Estève PO, Jacobsen SE. SET7/9 mediated methylation of non-histone proteins in mammalian cells. Epigenetics. 2009;4:383–387. doi: 10.4161/epi.4.6.9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proserpio V, Fittipaldi R, Ryall JG, Sartorelli V, Caretti G. The methyltransferase SMYD3 mediates the recruitment of transcriptional cofactors at the myostatin and c-Met genes and regulates skeletal muscle atrophy. Genes Dev. 2013;27:1299–1312. doi: 10.1101/gad.217240.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesé B, Barrero MJ, Fabregat MC, Sander V, Izpisua Belmonte JC. SMYD2 is induced during cell differentiation and participates in early development. Int. J. Dev. Biol. 2013;57:357–364. doi: 10.1387/ijdb.130051ji. [DOI] [PubMed] [Google Scholar]
- Silva FP, Hamamoto R, Kunizaki M, Tsuge M, Nakamura Y, Furukawa Y. Enhanced methyltransferase activity of SMYD3 by the cleavage of its N-terminal region in human cancer cells. Oncogene. 2008;27:2686–2692. doi: 10.1038/sj.onc.1210929. [DOI] [PubMed] [Google Scholar]
- Sims RJ, Reinberg D. From chromatin to cancer: a new histone lysine methyltransferase enters the mix. Nat. Cell Biol. 2004;6:685–687. doi: 10.1038/ncb0804-685. [DOI] [PubMed] [Google Scholar]
- Tao Y, Neppl RL, Huang ZP, Chen J, Tang RH, Cao R, Zhang Y, Jin SW, Wang DZ. The histone methyltransferase Set7/9 promotes myoblast differentiation and myofibril assembly. J. Cell Biol. 2011;194:551–565. doi: 10.1083/jcb.201010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- Van Aller GS, Reynoird N, Barbash O, Huddleston M, Liu S, Zmoos AF, McDevitt P, Sinnamon R, Le B, Mas G, et al. Smyd3 regulates cancer cell phenotypes and catalyzes histone H4 lysine 5 methylation. Epigenetics. 2012;7:340–343. doi: 10.4161/epi.19506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- Wang QT. Epigenetic regulation of cardiac development and function by polycomb group and trithorax group proteins. Dev. Dyn. 2012;241:1021–1033. doi: 10.1002/dvdy.23796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish book : a guide for the laboratory use of zebrafish (Brachydanio rerio) Eugene, USA: University of Oregon Press; 1993. [Google Scholar]
- Yang J, Huang J, Dasgupta M, Sears N, Miyagi M, Wang B, Chance MR, Chen X, Du Y, Wang Y, et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc. Natl. Acad. Sci. 2010;107:21499–21504. doi: 10.1073/pnas.1016147107. [DOI] [PMC free article] [PubMed] [Google Scholar]


