Abstract
Background:
Optimization of workflow for breast cancer samples with equivocal human epidermal growth factor receptors 2 (HER2)/neu score 2+ results in routine practice, remains to be a central focus of the on-going efforts to assess HER2 status. According to the College of American Pathologists/American Society of Clinical Oncology guidelines equivocal HER2/neu score 2+ cases are subject for further testing, usually by fluorescence in situ hybridization (FISH) investigations. It still remains on open question, whether quantitative digital image analysis of HER2 immunohistochemistry (IHC) stained slides can assist in further refining the HER2 score 2+.
Aim of this Work:
To assess utility of quantitative digital analysis of IHC stained slides and compare its performance to FISH in cases of breast cancer with equivocal HER2 score 2+.
Materials and Methods:
Fifteen specimens (previously diagnosed as breast cancer and was evaluated as HER 2- score 2+) represented the study population. Contemporary new cuts were prepared for re-evaluation of HER2 immunohistochemical studies and FISH examination. All the cases were digitally scanned by iScan (Produced by BioImagene [Now Roche-Ventana]). The IHC signals of HER2 were measured using an automated image analyzing system (MECES, www.Diagnomx.eu/meces). Finally, a comparative study was done between the results of the FISH and the quantitative analysis of the virtual slides.
Results:
Three out of the 15 cases with equivocal HER2 score 2+, turned out to be positive (3+) by quantitative digital analysis, and 12 were found to be negative in FISH too. Two of these three positive cases proved to be positive with FISH, and only one was negative.
Conclusions:
Quantitative digital analysis is highly sensitive and relatively specific when compared to FISH in detecting HER2/neu overexpression. Therefore, it represents a potential reliable substitute for FISH in breast cancer cases, which desire further refinement of equivocal IHC results.
Keywords: Breast cancer, fluorescence in situ hybridization, human epidermal growth factor receptors 2, quantitative digital image analysis
INTRODUCTION
Breast carcinoma is the most common malignant tumor and the leading cause of carcinoma death in women, with more than 1,000,000 cases occurring worldwide annually.[1] Hormone receptor and human epidermal growth factor receptors 2 (HER2) co-expression are not uncommon in breast cancer; approximately half of breast cancers with HER2 overexpression also co-express hormone receptors.[2]
Human epidermal growth factor receptor 2 testing is routinely performed in patients with a new diagnosis of invasive breast cancer. Accurate testing to identify HER2 status for patients with breast cancer who can benefit from anti-HER2 treatment (e.g., trastuzumab, lapatinib) is a clinical and economic necessity. As a consequence, issues relating to accurate and reliable laboratory assessment of HER2 status in patients with breast cancer are a matter of significant concern to patients, pathologists, and oncologists (De P et al., 2010).[3]
According to the American Society of Clinical Oncology/College of American Pathologists Guidelines, a negative HER2 test is defined as no staining (score 0) or weak, incomplete membrane staining in any proportion of tumor cells (score 1+). Positive HER2 test (score 3+) is defined as uniform intense complete membrane staining of >10% of invasive tumor cells. An equivocal result (2+) is circumferential membrane staining that is incomplete and/or weak/moderate and within >10% of tumor cells OR complete and circumferential membrane staining is intense and within ≤10% of tumor cells (Wolff AC et al., 2013).[4]
Optimization of workflow for breast cancer samples with equivocal HER2/neu score 2+ results in routine practice, remains to be a central focus of the ongoing efforts to assess HER2 status. According to the College of American Pathologists/American Society of Clinical Oncology guidelines equivocal HER2/neu score 2+ cases are subject for further testing, usually by fluorescence in situ hybridization (FISH) investigations (Wolff AC et al., 2007).[5]
It still remains on open question, whether quantitative digital image analysis of HER2 immunohistochemistry (IHC) stained slides can assist in further refining the HER2 score 2+. The College of American Pathologists/American Society of Clinical Oncology guidelines also recommend digital analysis as an effective tool for achieving consistent interpretation of IHC HER2/neu staining, provided that a pathologist confirms the result. Reducing inter- and intra-observer variability is critical toward improving reproducibility in IHC, along with efforts for improving and standardizing procedures for preanalytic specimen handling, antibody selection, and staining and scoring methods.[6]
The aim of this work is to assess utility of quantitative digital analysis of IHC stained slides and compare its performance to FISH in cases of breast cancer with equivocal HER2 score 2+.
MATERIALS AND METHODS
This study is done on histological sections selectively obtained from archival paraffin blocks for 15 lumpectomy/excisional/core biopsy specimens from 15 patients (collected from the Pathology Department, Cairo University labs archives; 2011–2012) diagnosed as invasive breast cancer (ductal, lobular, tubular and medullary carcinomas). The 15 specimens included were diagnosed to have a HER2 immunohistochemical stain score 2+ [Figure 1].
Figure 1.
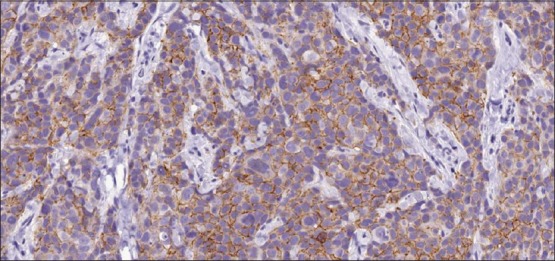
Invasive breast cancer, score 2+ for human epidermal growth factor receptors 2/neu (immunohistochemistry ×200)
Contemporary new cuts from the paraffin blocks were prepared for immunohistochemical stains and FISH test.
The new glass slides (pretreated for antigen retrieval, then with the universal immunostaining supersensitive monoclonal antibodies against HER2) were revised by two separate senior pathologists to re-evaluate the HER2 immunohistochemical stain score and proved to be score 2+.
Immunohistochemical staining for HER2: Paraffin sections on charged slides were pretreated for antigen retrieval, then with the universal immunostainingsupersensitive monoclonal antibodies against HER2 (ultravision detection kit, antipolyvalent, HRP/DAP) with blockage of internal peroxidase activity. Streptavidin biotin peroxidase detection system was used, utilizing DAP as a chromogen and hematoxylin as a counter stain. IHC scoring and Evaluation: According to the American Society of Clinical Oncology/College of American Pathologists Guidelines in 2013, an equivocal result (2+) is circumferential membrane staining that is incomplete and/or weak/moderate and within >10% of tumor cells OR complete and circumferential membrane staining is intense and within ≤10% of tumor cells.
All cases with HER2/new score of 2+ were digitally scanned by iScan (Produced by BioImagene [Now Roche-Ventana]) with Scanning Resolution 0.46 um/pixel at ×20.
Fifteen regions of interest of 1660 × 929 pixel were captured per each virtual slide using the iScan image viewer and selected by a pathologist. Those images were submitted to the automated image analysis system (MECES, www.Diagnomx.eu/meces (website is unavailable currently). Those 15 Images selected from each case were subjected to quantitative measures, which include: The membrane staining program is an appropriate program to measure antigens that are present in the membranes. Appropriate markers are those for receptor analysis. The measurement data includes:
- Score membrane: 0,1,2,3, computed as follows:
- 3+: Total length of intensively stained membrane/total length of detected membranes >0.50
- 2: Total length of stained membrane/total length of detected membranes >0.10
- 1+: Total length of stained membrane/total length of detected membranes >002
- 0+: Elsewhere;
- Score L/V cells: =, 1, 2, 3, computed as follows:
- 3+: Number of cells with ([length of stained membrane/cellular area] >0.50)
- 2+: Number of cells with ([length of stained membrane/cellular area] >0.10)
- 1+: Number of cells with ([length of stained membrane/cellular area] >0.02)
- 0+: Elsewhere.
Number nuclei, total nuclear area (Vv), mean nuclear area, total cellular surface (Lv), mean cell surface, total cell area (Vv), total cytoplasm area (Vv), total length virtual membrane (Sv), mean length Voronoi, membrane, rel length stained membrane (Sv), mean length stained membrane, ratio stained/cellular membrane (%), ratio stained/virtual membranes (%), ratio intensive/total membrane, percent no volume signal-cells (L/V), percent small volume signal cells (L/V), percent moderate volume signal cells (L/V), percent large volume signal cells (L/V) and percent intensively stained cells.
DNA FISH involves the precise annealing of a single-stranded, fluorescently-labeled DNA probe to complementary target sequences. The hybridization of the probe with the cellular DNA site is visible by direct detection using fluorescence microscopy. The locus-specific identifier (LSI) HER-2/neu DNA probe is a 190 Kb Spectrum Orange directly-labeled, fluorescent DNA probe specific for the HER-2/neu gene locus (17q11.2-q12). The chromosome enumeration probe (CEP) 17 DNA probe is a 5.4 Kb SpectrumGreen directly-labeled, fluorescent DNA probe specific for the alpha satellite DNA sequence at the centromeric region of chromosome 17 (17p11.1-q11.1). The probes are pre-mixed and pre-denatured in hybridization buffer for ease of use. Unlabeled blocking DNA is also included with the probes to suppress sequences contained within the target loci that are common to other chromosomes. This PathVysion Kit is designed for the detection of HER-2/neu gene amplification in formalin-fixed, paraffin-embedded human breast tissue specimens by FISH. The assay is rapid, non-radioactive, requires little tumor material, and is capable of detecting as few as 2–8 copies of the oncogene. Formalin-fixed, paraffin-embedded tissue specimens are placed on slides. The DNA is denatured to single-stranded form and subsequently allowed to hybridize with the PathVysion probes. Following hybridization, the unbound probe is removed by a series of washes and the nuclei are counterstained with DAPI (4,6 diamidino-2-phenylindole), a DNA-specific stain that fluoresces blue. Hybridization of the PathVysion probes is viewed using a fluorescence microscope equipped with appropriate excitation and emission filters allowing visualization of the intense orange and green fluorescent signals. Enumeration of the LSI HER-2/neu and CEP 17 signals is conducted by microscopic examination of the nucleus, which yields a ratio of the HER-2/neu gene to chromosome 17 copy number [Figure 2].
Figure 2.
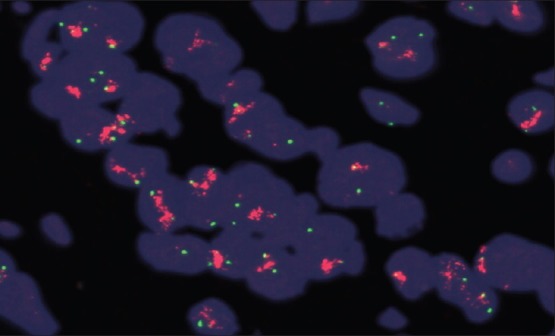
Fluorescence in situ hybridization (FISH), amplified human epidermal growth factor receptors 2/chromosome enumeration probe 17 ratio (red signals, arrows) (FISH ×1000)
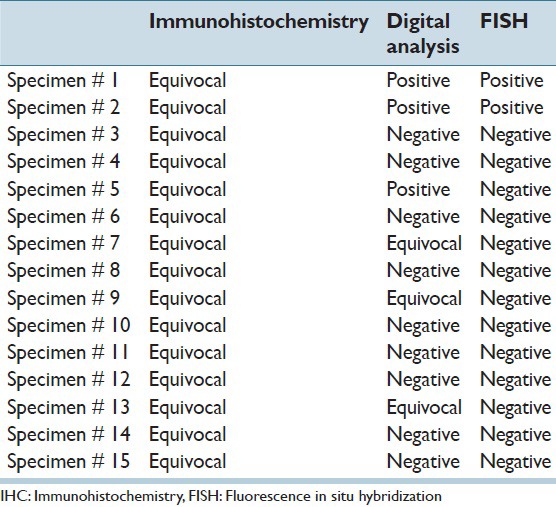
Then lastly, a comparative study was done between the results of the FISH [Figure 3] and the quantitative analysis of the virtual slides [Figure 4].
Figure 3.
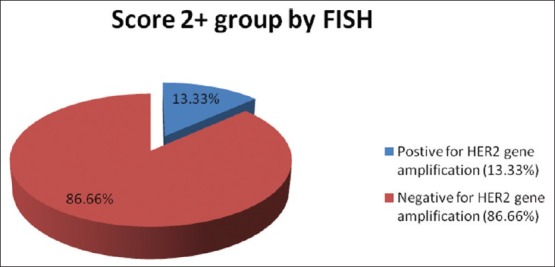
Fluorescence in situ hybridization results for specimens with score 2+
Figure 4.

Example of the running control of formal image (quality quantitative digital analysis) of one image of one of the cases of with score 2+
RESULTS
The Quantitative digital analysis program divided the IHC equivocal cases into positive (3 cases), equivocal (3 cases) and negative (9 cases) [Figure 5]. So three out of the 15 cases with equivocal HER2 score 2+, turned out to be positive (3+) by quantitative digital analysis, and 12 were found to be negative in FISH too. Two of these three positive cases proved to be positive with FISH, and only one was negative [Tables 1 and 2].
Figure 5.
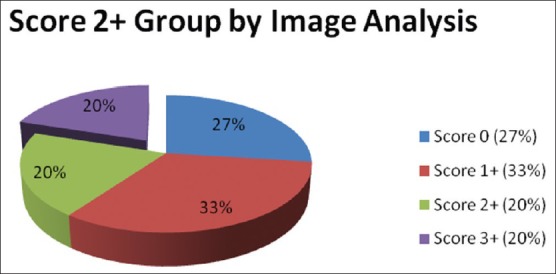
Quantitative digital analysis results for specimens with score 2+
Table 1.
Fluorescence in-situ hybridization results for specimens with score 2+
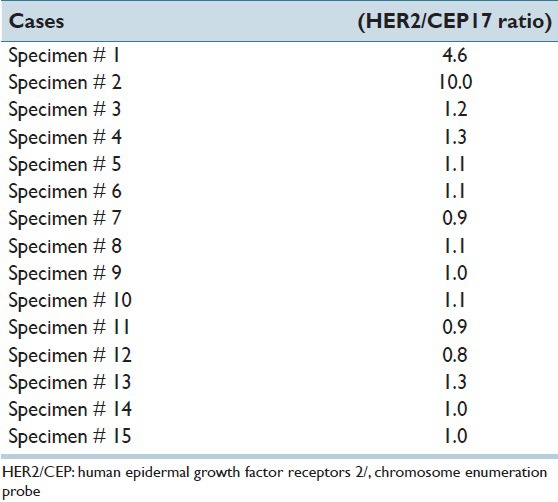
Table 2.
Comparison between IHC, quantitative digital analysis and FISH for specimens with score 2+
DISCUSSION
Human epidermal growth factor receptors 2 immunohistochemical analysis are typically performed using one of the two FDA-approved kits, HercepTest and pathway. An important and somewhat controversial issue is that the recommended ASCO/CAP guidelines are significantly different from the HercepTest scoring algorithm. This panel recommends that more than 10% of tumor cells must show circumferential membrane staining to consider a tumor as strongly positive (3+). All cases that showed circumferential membrane staining that is incomplete and/or weak/moderate and within >10% of tumor cells OR complete and circumferential membrane staining is intense and within ≤10% of tumor cells now being called equivocal (2+) under the ASCO/CAP guidelines.[7] Theoretically, adoption of these criteria could lead to more equivocal immunohistochemical cases, which thus would be subjected to additional FISH testing.
Much has been written on the various inadequacies of current HER2 immunohistochemical modalities. Minot et al.[8] found that by using quantitative digital analysis, the number of equivocal HER2 immunohistochemical results can be reduced, thus reducing the number of cases reflexed to FISH analysis. However, the optimal HER2 testing algorithm is still under debate.[9]
Dendukuri et al.[10] performed a meta-analysis and found that the strategy with the lowest cost-effectiveness ratio involved screening all newly diagnosed cases of breast cancer with immunohistochemical analysis and confirming scores of 2+ or 3+ with FISH. The current ASCO/CAP guidelines recommend that clinicians first perform immunohistochemical analysis, with equivocal results (2+) reflexed to FISH. The published guidelines identify a potential role for quantitative digital analysis as a means to achieve consistent interpretation. Image analysis is being used by some institutions, as shown by a recent survey distributed to participants in the HER2 immunohistochemical analysis proficiency program that indicated that 33% of laboratories are currently using quantitative computer image analysis.[11]
Quantitative digital analysis studies showed positive correlations with FISH that meet or exceed the guideline goal of 95% concordance for negative and positive immunohistochemical results.[12,13,14] Dobson et al.[15] achieved a 95% concordance rate between their quantitative digital analysis technique and FISH by determining the extent of circumferential HER2 staining along with other tissue staining features.
In our study, we found that three out of 15 (20%) of the score + 2 cases are positive by quantitative digital analysis while by using the FISH technique we found two cases [13.33%] to be positive. Both the two cases detected by FISH cases were among the three cases detected by the quantitative digital analysis. These findings are in concordance with those reported by Wang et al.[16] who were the first to show that the quantitative digital analysis had a higher concordance rate and improved sensitivity over manual interpretation compared with FISH testing. Subsequently, Bloom and Harrington[17] also demonstrated that the quantitative digital analysis had higher interobserver agreement than manual interpretation, further suggesting that quantitative digital analysis improves the accuracy and reliability of HER2 immunohistochemical analysis. An additional study assessing quantitative digital analysis-immunohistochemical studies concluded that quantitative digital analysis HER2 assessment is reliable, rapid, and inexpensive and similar to the previous 2 studies, correlated highly with the FISH results.[18]
The sensitivity and specificity of the quantitative digital analysis were calculated using FISH evaluation as the gold standard. In our study, the IA showed high sensitivity (100%) and lower specificity (90%). This finding is in accordance with Dobson et al.[15] Our concordance with FISH ranged between 93%-100%. This finding goes with reports by Minot et al. 2012.[8]
Another significant finding in our study was the decrease in the number of cases being interpreted as equivocal (2+) using our validated IA technique. This observation, coupled with the identification of more IHC negative (0, 1+) cases and a fewer number of cases in the positive (3+) category for quantitative digital analysis-assisted assessments, suggests that the quantitative digital analysis is appropriately downgrading a proportion of these equivocal cases without missing FISH + cases.
CONCLUSIONS
The quantitative digital analysis is highly sensitive and relatively specific when compared to FISH in detecting HER2/neu overexpression. Therefore, it represents a potential reliable substitute for FISH (which is an expensive technique that costs >300 US $ per cases and takes much more time than the simple quantitative analysis) in breast cancer cases which desire further refinement of equivocal IHC results. Digital quantitative analysis technique can also decrease the need of the expensive FISH technique (which is an expensive technique that costs >300 US $ per cases and takes much more time than the simple quantitative analysis) for the cases being interpreted as equivocal (2+) especially in countries with limited resources. Accordingly, we recommend using the quantitative digital analysis technique first to all score 2+ cases and restrict only FISH testing to those cases that appear equivocal with quantitative digital analysis. This will not only save cost and time in laboratory diagnosis, but also will improve patient outcome by introducing appropriate cases to trastuzumab therapy.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2015/6/1/31/158066
REFERENCES
- 1.Rosai J. 10th ed. Ch. 20. USA: Elsevier; 2011. Breast. Rosai and Ackerman's Surgical Pathology; p. 1681. [Google Scholar]
- 2.Cortes J, Baselga J. How to treat hormone receptor-positive, human epidermal growth factor receptor 2-amplified breast cancer. J Clin Oncol. 2009;27:5492–4. doi: 10.1200/JCO.2009.23.8089. [DOI] [PubMed] [Google Scholar]
- 3.De P, Smith BR, Leyland-Jones B. Human epidermal growth factor receptor 2 testing: Where are we? J Clin Oncol. 2010;28:4289–92. doi: 10.1200/JCO.2010.29.5071. [DOI] [PubMed] [Google Scholar]
- 4.Wolff AC, Hammond ME, Hayes DF. Re: Predictability of adjuvant trastuzumab benefit in N9831 patients using the ASCO/CAP HER2-positivity criteria. J Natl Cancer Inst. 2012;104:957–8. doi: 10.1093/jnci/djs243. [DOI] [PubMed] [Google Scholar]
- 5.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–45. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 6.Keay T, Conway CM, O’Flaherty N, Hewitt SM, Shea K, Gavrielides MA. Reproducibility in the automated quantitative assessment of HER2/neu for breast cancer. J Pathol Inform. 2013;4:19. doi: 10.4103/2153-3539.115879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31:3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 8.Minot DM, Voss J, Rademacher S, Lwin T, Orsulak J, Caron B, et al. Image analysis of HER2 immunohistochemical staining. Reproducibility and concordance with fluorescence in situ hybridization of a laboratory-validated scoring technique. Am J Clin Pathol. 2012;137:270–6. doi: 10.1309/AJCP9MKNLHQNK2ZX. [DOI] [PubMed] [Google Scholar]
- 9.Phillips KA, Marshall DA, Haas JS, Elkin EB, Liang SY, Hassett MJ, et al. Clinical practice patterns and cost effectiveness of human epidermal growth receptor 2 testing strategies in breast cancer patients. Cancer. 2009;115:5166–74. doi: 10.1002/cncr.24574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dendukuri N, Khetani K, McIsaac M, Brophy J. Testing for HER2-positive breast cancer: A systematic review and cost-effectiveness analysis. CMAJ. 2007;176:1429–34. doi: 10.1503/cmaj.061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakhleh RE, Grimm EE, Idowu MO, Souers RJ, Fitzgibbons PL. Laboratory compliance with the American Society of Clinical Oncology/College of American Pathologists guidelines for human epidermal growth factor receptor 2 testing: A College of American Pathologists survey of 757 laboratories. Arch Pathol Lab Med. 2010;134:728–34. doi: 10.5858/134.5.728. [DOI] [PubMed] [Google Scholar]
- 12.Masmoudi H, Hewitt SM, Petrick N, Myers KJ, Gavrielides MA. Automated quantitative assessment of HER-2/neu immunohistochemical expression in breast cancer. IEEE Trans Med Imaging. 2009;28:916–25. doi: 10.1109/TMI.2009.2012901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gustavson MD, Bourke-Martin B, Reilly D, Cregger M, Williams C, Mayotte J, et al. Standardization of HER2 immunohistochemistry in breast cancer by automated quantitative analysis. Arch Pathol Lab Med. 2009;133:1413–9. doi: 10.5858/133.9.1413. [DOI] [PubMed] [Google Scholar]
- 14.Turashvili G, Leung S, Turbin D, Montgomery K, Gilks B, West R, et al. Inter-observer reproducibility of HER2 immunohistochemical assessment and concordance with fluorescent in situ hybridization (FISH): Pathologist assessment compared to quantitative image analysis. BMC Cancer. 2009;9:165. doi: 10.1186/1471-2407-9-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobson L, Conway C, Hanley A, Johnson A, Costello S, O’Grady A, et al. Image analysis as an adjunct to manual HER-2 immunohistochemical review: A diagnostic tool to standardize interpretation. Histopathology. 2010;57:27–38. doi: 10.1111/j.1365-2559.2010.03577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S, Saboorian MH, Frenkel EP, Haley BB, Siddiqui MT, Gokaslan S, et al. Assessment of HER-2/neu status in breast cancer. Automated Cellular Imaging System (ACIS)-assisted quantitation of immunohistochemical assay achieves high accuracy in comparison with fluorescence in situ hybridization assay as the standard. Am J Clin Pathol. 2001;116:495–503. doi: 10.1309/TMUW-G4WB-LXJ2-FUDN. [DOI] [PubMed] [Google Scholar]
- 17.Bloom K, Harrington D. Enhanced accuracy and reliability of HER-2/neu immunohistochemical scoring using digital microscopy. Am J Clin Pathol. 2004;121:620–30. doi: 10.1309/Y73U-8X72-B68T-MGH5. [DOI] [PubMed] [Google Scholar]
- 18.Tawfik OW, Kimler BF, Davis M, Donahue JK, Persons DL, Fan F, et al. Comparison of immunohistochemistry by automated cellular imaging system (ACIS) versus fluorescence in-situ hybridization in the evaluation of HER-2/neu expression in primary breast carcinoma. Histopathology. 2006;48:258–67. doi: 10.1111/j.1365-2559.2005.02322.x. [DOI] [PubMed] [Google Scholar]


