Abstract
Traditionally, human disorders were studied using animal models or somatic cells taken from patients. Such studies enabled the analysis of the molecular mechanisms of numerous disorders, and led to the discovery of new treatments. Yet, these systems are limited or even irrelevant in modeling multiple genetic diseases. The isolation of human embryonic stem cells (ESCs) from diseased blastocysts, the derivation of induced pluripotent stem cells (iPSCs) from patients’ somatic cells, and the new technologies for genome editing of pluripotent stem cells have opened a new window of opportunities in the field of disease modeling, and enabled studying diseases that couldn’t be modeled in the past. Importantly, despite the high similarity between ESCs and iPSCs, there are several fundamental differences between these cells, which have important implications regarding disease modeling. In this review we compare ESC-based models to iPSC-based models, and highlight the advantages and disadvantages of each system. We further suggest a roadmap for how to choose the optimal strategy to model each specific disorder.
Keywords: embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs), disease modeling
1. Introduction
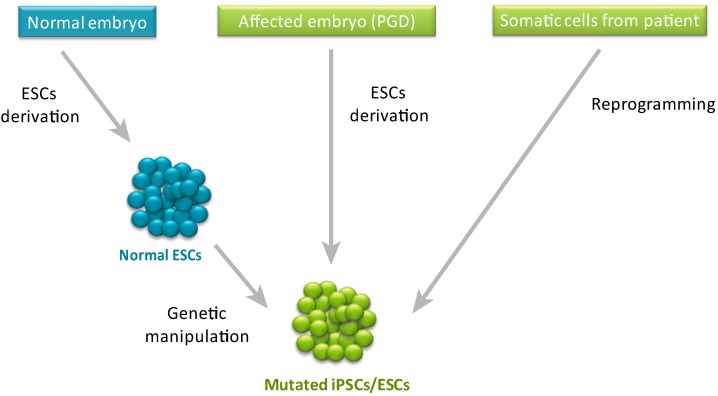
Pluripotent stem cells have an unlimited self-renewal capacity and can differentiate into virtually any adult cell type [1] and even some extra-embryonic tissues [2,3]. These features make human pluripotent stem cells (hPSCs) a useful tool for disease modeling, which overcomes limitations observed in animal and adult human cellular models. While the use of animal models proved to be extremely valuable and successful in many cases [4], there are numerous diseases, such as Lesch-Nyhan syndrome [5], Turner syndrome [6] and Fragile X syndrome [7], that cannot be studied using animal models due to species-specific differences. The use of mature cells from patients can solve the species-specificity issue but this strategy is limited by the fact that it enables studying only a few types of cells at a specific developmental stage, and in many cases requires also transformation of the cells to enable their proliferation in culture. By contrast, due to their unique properties, hPSCs enable exploration of different types of cells, to study the effect of a specific mutation on differentiation or development and can proliferate in vitro without additional transformation. Indeed, since the generation of the first human embryonic stem cells (ESCs) based model (a model for Lesch-Nyhan syndrome by targeting of the HPRT gene in human ESCs) [5] dozens of disease models were generated by reprogramming of somatic cells from patients [1], by derivation of mutant ESCs from affected embryos diagnosed by pre-implantation genetic diagnosis (PGD) or by genetic manipulation of normal ESCs [8] (see Figure 1). While some models were used as a “proof of concept” to demonstrate that hPSCs can be derived from a wide range of disorders [9,10,11] or to show the feasibility of the mutant pluripotent cells to be used as a disease model [12], other models were further used to obtain novel mechanistic or physiological insights regarding the disorders. One example is a model for Amyotrophic Lateral Sclerosis (ALS) by Kiskinis et al. [13].
Figure 1.
Human pluripotent stem cell-based models for genetic disorders can be generated by different techniques. Mutated human pluripotent stem cells can be derived by genetic manipulation of normal pluripotent stem cells, from affected embryos (identified by PGD), or from adult patients (by reprogramming of somatic cells).
The general differences between ESCs and induced pluripotent stem cells (iPSCs) and the utilization of hPSCs for disease modeling has been discussed extensively in the literature [1,14,15,16]. In this review we will focus on the differences between ESC-based models and iPSC-based models, and discuss the effect of genome editing technologies on the field of disease modeling.
2. ESCs vs. iPSCs in Disease Modeling
Theoretically, a given disorder can be equally modeled by iPSCs and by ESCs, as both are pluripotent stem cells. However, several reasons have made iPSCs derived from patients the system of choice:
-
(1)
The use of normal human ESCs to model a genetic disorder requires genetic manipulation to induce the specific mutation that one would like to study. The way to obtain a mutation that will be identical to the natural occurring mutation, seen in patients, is by genome editing. However, the efficiency of genome editing in human ESCs, before the establishment of gene targeting technologies as discussed below, was extremely low (especially in cases where a homozygous mutation was required) [17] and derivation of iPSCs that already contain the specific mutation obviates the needs for this inefficient process.
-
(2)
While the above mentioned limitation can be overcome by derivation of mutant ESCs from affected embryos identified by Preimplantation Genetic Diagnosis (PGD), this procedure is limited to a small number of diseases in which PGD is normally preformed, and can be done only in labs that are associated with in vitro fertilization (IVF) units.
-
(3)
By contrast to iPSCs from affected individuals, in the case of ESCs based models, the correlation between the genotype and the phenotype is not obvious, and the penetrance of the mutation might be low as a results of specific “protective” genetic background [18].
-
(4)
Lastly, in some countries the use of human ESCs is limited or banned due to ethical and religious concerns regarding the use of human embryos for research purposes as was discussed by others [19,20].
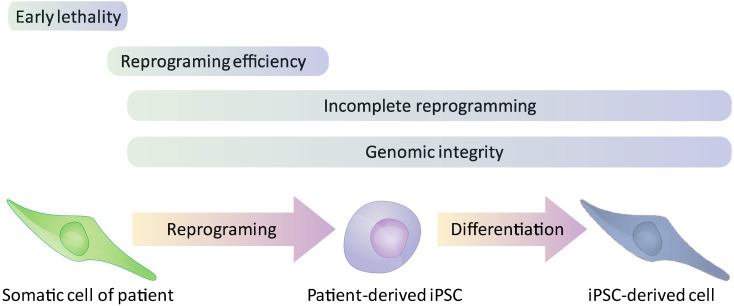
Nevertheless, possible drawbacks in modeling genetic disorders by iPSCs suggest that some disorders or specific aspects within a given disease might be better modeled in ESCs than iPSCs. The generation of a faithful iPSC-based model might be disrupted due to the following reasons (see Figure 2): (1) Incomplete reprogramming as a result of “Epigenetic memory” of the original somatic cells [21,22,23]; (2) Mutations accumulated during the reprogramming process [24] and deleterious effects (such as chromosomal instability, [25]) of the reprogramming process on the genome integrity of iPSCs; (3) Genetic aberrations that significantly decrease the reprogramming efficiency [26]; (4) The absence of appropriate sources of somatic cells such as in the cases of genetic aberration and aneuploidies that lead to very early embryonic lethality [6].
Figure 2.
Limitations in the generation of iPSC-based disease models. The “X-axis” in this scheme depicts the specific stages during the formation of iPSC-based models that might be affected by the different factors that discussed in the main text.
To demonstrate the commonalities and differences between ESC- and iPSC-based models, we compared models for X-linked, autosomal recessive and autosomal dominant disorders in which disease-related phenotypes were observed in both models (Table 1). As expected, in some cases the ESC-based models and the iPSC-based models were similar (spinal muscular atrophy [27,28], Shwachman-Dimond syndrome [29], long QT syndrome [30], and some aspects of myotonic dystrophy [31,32]). However, in other cases the iPSCs were limited in their capacity to model the disorder or specific aspects within the disorders. To demonstrate some of these cases, and to discuss the principles behind them, we will focus on the following disorders: Turner syndrome, Fanconi Anemia, fragile X syndrome and Huntington’s disease.
Table 1.
Comparison between ESCs and iPSC models for human genetic disorders.
| ESCs | iPSCs | ||||
|---|---|---|---|---|---|
| Reference (ref #) | Method of Derivation of Mutate ESCs | Reference (ref #) | Reprogramming Method | ||
| Fragile X | Eiges 2007 [7] | PGD | Urbach 2009 [36] | Retroviruses | |
| Rett syndrome | Cheung 2011 [37] | Retroviruses | |||
| Li 2013 [38] | Gene targeting by TALEN | Marchetto 2010 [39] | Retroviruses | ||
| Turner syndrome | Urbach 2011 [6] | Screen for XO colonies | Li 2012 [35] | Retroviruses and Lentiviruses | |
| Fanconi Anemia | Yung 2013 [25]; Tulpule 2010 [40] | Knockdown of FANCA and FANCD2 | Yung 2013 [25] | Lentiviruses | |
| Liu 2014 [41] | Gene targeting by TALEN | Liu 2014 [41] | Reprogramming | ||
| Spinal muscular atrophy | Wang 2013 [27] | Knockdown of SMN | Ebert 2009 [28] | Lentiviruses | |
| Shwachman-Diamond syndrome | Tulpule 2013 [29] | Knockdown of SBDS | Tulpule 2013 [29] | Retroviruses | |
| Long QT | Bellin 2013 [30] | Gene targeting | Bellin 2013 [30] | Retroviruses | |
| Huntington’s disease | Lu 2013 [42] | Over-expression of HTTexon1 with 23, 73 or 145 glutamine repeats in HESCs | HD iPSC Consortium 2012 [43] | Lentiviruses (OSKM + Nanog + Lin28) |
ESCs > iPSCs (Based on Lu et al.) mHTT aggregated appeared only in ESCs based model |
| Myotonic Dystrophy | Seriola 2011 [31] | Du 2013 [32] | Retroviruses |
ESCs ~ iPSCs TNR becomes stable upon differentiation in ES and iPS. Down-regulation of MMR genes upon differentiation was observed only in ESCs |
|
3. Turner Syndrome
X chromosome monosomy (XO) is one of the most common chromosomal abnormalities, as 3% of all pregnancies start with XO embryos [33]. Yet, approximately 99% of the XO embryos undergo miscarriage during the first trimester [33,34]. The 1% that survive to term are born with Turner syndrome which is characterized by several phenotypes; the most common among them are growth failure, gonadal dysgensis and webbed neck [34]. While Turner syndrome derived iPSCs can be used in order to study the phenotypes of the patient (pending the availability of the required differentiation protocols), they might be problematic in modeling the early lethality of XO embryos, as they represent the exceptional 1% of the cases that survived to term.
In agreement with this notion, gene expression analysis of XO ESCs (derived by screening for ESCs with normal karyotype that lost one of their X chromosomes) revealed a significant effect of X chromosome monosomy on the expression of placental genes and suggests that the reason for the early lethality is abnormal placental development [6]. By contrast, there was almost no effect of X chromosome monosomy on placental gene expression in iPSCs derived from Turner syndrome patients, and even from amniotes of a 20 weeks old embryo [35]. The results suggest that Turner syndrome iPSCs represent the rare cases in which the embryo survived despite the XO karyotype.
Representative examples of ESC and iPSC models for X linked, autosomal recessive and autosomal dominant disorders. As the primary intention of this review is to discuss the differences between the two model systems and to highlight cases in which the ESCs model is a better choice than the iPSCs model, only the characteristics or phenotypes that are relevance for the direct comparison between the two models were mentioned in the table. We refer the readers to the original papers to learn more details about each model. When several studies for a specific disorder were relevant for the comparison between the systems we cited all relevant studies.
4. Fanconi Anemia
Fanconi Anemia (FA) is an autosomal recessive disorder caused by a mutation in any of the 16 FANC genes and characterized by congenital abnormalities, cancer predisposition and progressive bone marrow failure [44]. Initial attempts to reprogram somatic cells from FA patients into iPSCs failed unless using fibroblasts that were first genetically corrected [45]. These results suggested that the FA pathway is essential for the reprogramming pathway (probably due to defective DNA repair and genomic instability of FA cells) and therefore that FA can’t be easily modeled by iPSCs. However, further attempts to reprogram “uncorrected” somatic cells from FA patients under hypoxic conditions [26], and even under normoxic conditions [25], showed that iPSCs can be derived from FA somatic cells, albeit in a very low efficiency and revealed that “…somatic cells harboring mutations that render the FA pathway defective are resistant but not refractory to reprogramming” [26]. Nevertheless, significant chromosomal aberration in uncorrected FA-iPSCs [25], but not in FA-iPSCs derived from “corrected” somatic cells [26] or in human ESCs with stable knockdown of FANCC [25] suggests that the FA pathway is required to prevent DNA damage and chromosomal instabilities associated with the reprogramming process. The severe aneuploidy in the uncorrected FA-iPSCs but not in the ESC-based model for Fanconi anemia suggests that ESCs and not iPSCs should be used to study FA. Surprisingly though, it has been recently shown [41] that FA-iPSCs with a normal karyotype can be derived from FA somatic cells upon episomal reprogramming. Moreover, the FA-iPSCs were very similar to FA-ESCs that were generated by gene targeting of the FANCA gene using the TALEN mediated gene targeting [37]. The FA-iPSCs and the FA-ESCs were extensively studied and compared to isogenic control cells (the original ESCs and target corrected FA-iPSCs) and proved to be a very useful model for different aspects of Fanconi anemia. While the reasons for the differences in the chromosomal stability between the viruses mediated reprogramming and the integration-free episomal mediated reprogramming are still not clear, these results indicate that in some cases, the reprogramming method itself might have a dramatic effect on the quality of the iPSCs and thus, should be taken under consideration when choosing to generate a disease model by reprogramming of somatic cells from patients.
5. Fragile X Syndrome
Fragile X syndrome (FXS) is a trinucleotide repeat disorder and is the leading cause of inherited intellectual disability in males, affecting approximately one in every four thousand boys and one in eight thousand girls worldwide [46,47,48,49]. The mutation leading to the syndrome is a trinucleotide CGG expansion at the 5′ untranslated region of the fragile X mental retardation 1 (FMR1) gene, which is accompanied by epigenetic changes, resulting in the silencing of the gene [49,50]. The product of the FMR1 gene is the fragile X mental retardation protein (FMRP) which is most abundant in the brain and testis and plays a major role in synaptic plasticity [51].
In 2007 human ESCs from FXS affected embryos (FXS-ESCs) were derived for the first time through PGD and enabled the study of the development of the disease [7]. Interestingly, although carrying the full mutation, FXS-ESCs showed both FMR1 mRNA expression and the presence of FMRP. This finding showed that the transcriptional silencing of FMR1 is a developmentally regulated process. Moreover, the study indicated that FMR1 is silenced in FXS embryos only during development and that the inactivation is initiated by chromatin modifications prior to DNA methylation [7]. Other studies on FXS-ESCs supported the finding that FMR1 is expressed in full mutation embryos and is silenced only during differentiation and further demonstrated that FMR1 plays an important role in early stages of neurogenesis and synaptic function [52,53]. Therefore, FX-ESCs are invaluable to study many aspects of FXS, first and foremost the epigenetic silencing mechanism. However, there are also limitations in the FX-ESCs model: FXS is represented by profound variability in patients, ranging from the varying length of the repeats, through the methylation levels, and to the neurological phenotype itself. The degree of intellectual impairment also varies between different individuals, as only about 30% of full mutation carriers display autistic behavior [54,55]. Additionally, some carriers of the full mutation allele do not display any of the syndrome’s phenotypes [56,57]. As this variability is not inherited from the parents and is detected only after PGD analysis, the probability of acquiring numerous human embryonic stem cells displaying the entire spectrum of genetic and epigenetic differences is quite small and may take several years.
In contrast to the FMR1 expression seen in FXS-ESCs, it seems that in FXS derived iPSCs (FXS-iPSCs), despite successful reprogramming of patients derived fibroblasts, the FMR1 gene is resistant to the process and remains methylated and silent [36,58,59]. Thus, while the FXS model in human ESCs demonstrated the temporal silencing of FMR1, in FXS-iPSCs FMR1 was already inactive in the undifferentiated state. This fundamental difference between FXS-ESCs and FXS-iPSCs controls the choice of model according to the question being asked. In order to better understand the different aspects of the initiating steps of the FMR1 silencing such as CGG methylation and the epigenetic silencing, one should use the FXS-ESC model. On the other hand, if one wishes to model neural development, screen for new drugs or understand the CGG expansion mechanism it is preferential to use the FXS-iPSC model to understand the effects of lack of FMRP on developing neurons, as we do not fully understand at which time point during the differentiation process FMR1 is silenced in ESCs in vitro. One example using FX-iPSCs to model Fragile X syndrome is a study aimed to evaluate the reactivation of FMR1 in FXS-iPSCs and their neuronal derivatives through epigenetic modulation drugs. This study showed not only that reactivation is possible but also uncovered additional layers of epigenetic control on FMR1 [60].
6. Huntington’s Disease
Huntington’s disease (HD) is an autosomal dominant neurological disorder caused by a trinucleotide repeat expansion and characterized by a late onset progressive neurodegeneration ending with death [61,62]. In HD, an expansion of a CAG repeat in the first exon of the Huntingtin (HTT) gene leads to a toxic gain of function activity of the mutant Huntingtin protein (mHTT), containing an increased number of polyglutamines at the N terminus [62]. These polyglutamine tails are then cleaved and accumulate as aggregates in the nuclei of neurons [63].
During the past few years, several groups have successfully created iPSC models for HD (HD-iPSCs) [11,43,64,65]. Some have further differentiated HD-iPSCs to neurons and showed increased caspase activity of neural precursors upon growth factor deprivation [64] or increased lysosomal activity in both HD-iPSCs and derived neurons [65]. The most comprehensive work done with the HD-iPSCs model system was performed by the HD-iPSC Consortium, in which several HD-iPSC lines were created and analyzed by a group of different labs [43]. In this work, HD-iPSCs were also differentiated into neural stem cells (NSCs) and neurons. HD-derived NSCs showed differential gene expression accompanied with changes at the protein level as well. Other changes observed were compromised energy metabolism, inability to fire action potential and increased cell death. Neurons also display increased death under different stress conditions most notably in lines containing longer repeats.
HD-iPSCs provide a useful model, however, it was never shown that they accumulate any insoluble aggregates, and thus cannot be used to study the formation and pathological contribution of this aggregates to the development of the disease. In order to study this aspect of the syndrome, normal ESCs were genetically engineered to express the polyglutamine repeats [42]. Neurons derived from these HD-ESCs matured over a period of several months and showed the polyglutamine aggregates. Similar to HD-iPSCs, HD-ESCs derived neurons exhibited progressive death under stress conditions. It was also shown using this model that reduction of mHTT by just 10% is sufficient to prevent toxicity and lowering the expression levels of HTT by up to 90% had no effect on neurons, opening the possibility to screen for new drugs to control the levels of mHTT. Thus, HD-ESCs may provide a stronger tool than HD-iPSCs in our understanding of the initiation and progression of the pathology of HD. However, work done on ESCs derived directly from an embryo with HD did not show the formation of polyglutamine aggregates [66]. Furthermore, HD embryos from PGD are not readily available, and due to the fact that HD is a late onset disease, we do not know the ultimate phenotype of these never developed embryos. In this case, more work should be done on both HD-ESCs and HD-iPSCs in an attempt to obtain more of the molecular phenotypes characteristic of the disease to create a better model system.
7. Disease Modeling by Gene Targeting of hPSCs
As mentioned above, hPSCs based models can be generated by the derivation of ESCs from affected embryos diagnosed by PGD or by genetic manipulation of normal hPSC cells. Down-regulation or over-expression of specific genes can be easily achieved by RNAi technologies (for down regulation) or by introduction of exogenous genes into the genome (for over-expression). While these methods proved to be very informative in some cases, they can’t mimic the natural occurring mutation in the patients and therefore the relevance of the finding to the disease might be questionable in other cases. To overcome this problem, one has to induce a specific mutation that is identical to the mutation occurring in patients. However, until lately, genome editing in mammalian cells was an extremely inefficient process [17], and therefore it was challenging to generate homozygous mutations in human cells using the traditional methods for gene targeting. The development of new technologies for gene targeting, (reviewed in details in [17]) especially the TALEN technology and the Cas/CRISPER technology have dramatically increased the efficiency of gene targeting in mammalian cells and enabled to correct specific mutations or to obtained homozygous mutations in reasonable efficiency in human pluripotent stem cells.
These methods enable, for the first time, the comparison between isogenic cells that differ only in the specific mutation under investigation. This can be achieved by induction of a specific mutation in otherwise normal ESCs, by correction of a specific mutation in iPSCs or by combination of both methods [30,41] One possible drawback in these methods is the possible off-target effect that might result in additional unplanned genetic aberrations [17]. To overcome this possibility it is important to design the targeting sequence in a way that will decrease the off-target effects and to target different sequences of the same gene. Among these methods, the CRISPR technique will probably become the first choice for most labs due to the combination of accuracy, efficiency and accessibility.
8. “Guidelines” for Choosing the Optimal Model System for a Given Disease
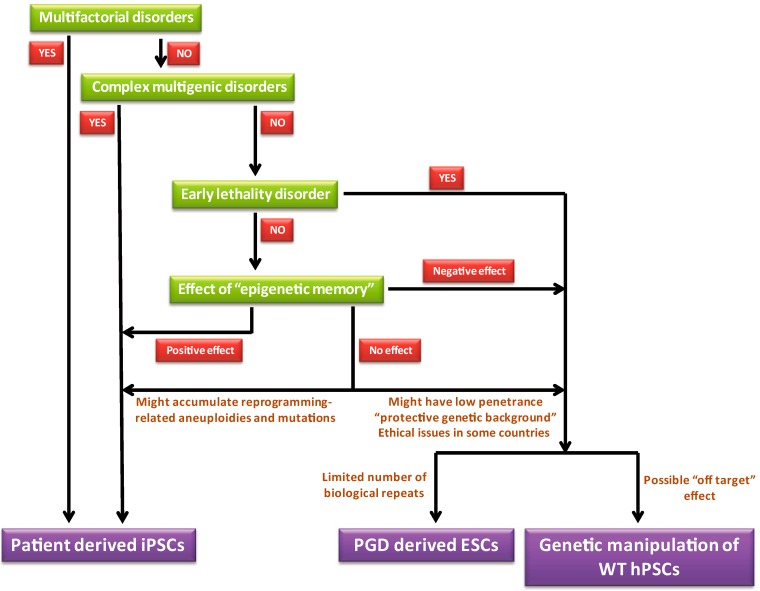
The choice between modeling disorders with ESCs or iPSCs is dependent on several factors. We propose that the optimal model that probably overcomes most if not all the drawbacks mentioned above, is a model that combines both ESCs (that were genetically modified to carry a specific mutation) and iPSCs from patients (with an isogenic control of iPSCs from the same patient in which the mutation was corrected by genomic engineering). Such “combined methods” have been recently generated for long QT syndrome [30] and for Fanconi anemia [41]. However, as was discussed above, in some cases only one of the two methods is doable/informative. In Figure 3 we suggest general guidelines that should assist in choosing the right system for a given disorder. We generated this scheme based on the following assumptions:
Figure 3.
Scheme depicting the steps in choosing the appropriate system for disease modeling. While in some cases there is only one possible option (either ESCs or iPSCs), in other cases both ESCs or iPSCs can be used and the decision between the two methods should be done after the consideration of the advantageous and disadvantageous of each one of the options (some of them are described in the scheme).
Multifactorial disorders in which there is a major contribution to factors other than genetic factors on the disease etiology can be modeled exclusively by iPSCs from patients that already manifested the disease and can’t be modeled by ESCs. Similarly, iPSCs but not ESCs should be used to model multigenic disorders in which the genetic factor cannot be narrowed down into a single gene, (the discussion regarding the use of hPSCs to model these type of disorders is out of the scope of this review). Other than these two groups of disorders, “monogeneic” disorders can be modeled theoretically using both systems but under the following notions: (1) Genetic aberrations that lead to early lethality should not be modeled by iPSCs that derived from individuals that survived to term as they represent the exceptional cases (these rare cases however, can be used to study the effect of the genetic aberration on the phenotype of the exceptional embryos that survived to term and the genetic backgrounds that enable them to escape the early lethality); (2) Possible epigenetic memory in iPSCs has to be taken under consideration. While in general the epigenetic memory is considered to have a negative effect on iPSCs (as the pluripotent cells retain some of their previous identity of adult cells and therefore might not be equivalent to normal pluripotent cells), one can also utilize these phenomena in a positive manner. For example, in cases of hematopoietic disorders, iPSCs that were derived from blood cells might undergo hematopoietic differentiation in a greater efficiency than ESCs or iPSCs derived from other somatic cells [16]; (3) In cases in which no epigenetic effect is predicted, the best choice is to combine both model systems. When only one of the two systems will be used it is important to keep in mind the following limitations of each one of the methods: (a) The reprogramming process itself might results in accumulation of genetic aberrations that under some circumstances might affect the reliability of the model; (b) The penetrance of the mutation in the ESCs based model might not be completed (as a result of “protective” genetic background). By contrast, iPSCs are derived from patients that already manifested the phenotype and therefore one should not be concerned about “protective” genetic background; (c) In the case of PGD-based models the number of available samples (affected embryos) might be limited; (d) Gene targeting by the TALEN or CRISPER systems might lead to off-target effects.
9. Conclusions
Reprogramming of somatic cells from patients is a relatively easy procedure that doesn’t involve the usage of human embryos, nor ethical issues, and results in the formation of iPSCs with the naturally occurring mutation. Therefore, since the first derivation of human iPSCs from normal donors [67,68,69] and from patients [11], this method was considered by many to be the optimal methodology for disease modeling by human pluripotent cells (due to scientific reasons as well as other reasons). Indeed, during the last several years numerous models for genetic disorders were generated by reprogramming of somatic cells from patients. Yet, in many cases the mutant cell lines were not further analyzed to study their relevance to the actual disorders. In this review we focused on iPSCs based models and ESCs based models that have been shown to have a phenotype related to the disease.
To demonstrate that iPSCs can’t always replace ESCs in disease modeling, we focused on four models, each one emphasizes a specific aspect of the differences between ESC-based models and iPSC-based models. In addition to these specific examples, the reprogramming process itself might result in the generation of de-novo mutations [24] that might add “noise” to the system. On the other hand one general advantage of iPSC-based models compared to ESCs-based models is the fact that the patient chosen already manifested the phenotype associated with the mutation. This assures that there is no effect to the specific genetic background on the penetrance of the mutation. Based on the comparison between ESC-based models and iPSC-based models we suggested in Figure 3 a general guidelines to assist in choosing the appropriate model for a given disorder.
Lastly, the development of the “iPSCs technology” by Takahashi and Yamanaka some eight years ago [70], dramatically changed the entire field of pluripotent stem cells biology. While the most desirable application of this technology is probably for cell therapy, there is no doubt that currently the most common application of iPSCs is for disease modeling. In this review we highlighted some of the pros and cons of iPSCs compared to ESCs in regards to disease modeling and discussed the effect of advanced technologies for genome editing on the field. We believe that the field of disease modeling by hPSCs has reached a point wherein the challenge is not to derive pluripotent cells (ESCs or iPSCs) with a specific mutation but rather to better understand the pathophysiology of the disease and finding effective therapies.
Acknowledgments
The Authors would like to acknowledge Nissim Benvenisty for helpful discussions and critical comments on the manuscript.
Author Contributions
Tomer Halevy and Achia Urbach wrote the manuscript and designed the figures.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Robinton D.A., Daley G.Q. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roberts R.M., Loh K.M., Amita M., Bernardo A.S., Adachi K., Alexenko A.P., Schust D.J., Schulz L.C., Telugu B.P.V.L., Ezashi T., et al. Differentiation of trophoblast cells from human embryonic stem cells: To be or not to be? Reproduction. 2014;147 doi: 10.1530/REP-14-0080. [DOI] [PubMed] [Google Scholar]
- 3.Xu R.-H., Chen X., Li D.S., Li R., Addicks G.C., Glennon C., Zwaka T.P., Thomson J.A. BMP4 Initiates Human Embryonic Stem Cell Differentiation to Trophoblast. Nat. Biotechnol. 2002;20:1261–1264. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- 4.Vandamme T.F. Use of rodents as models of human diseases. J. Pharm. Bioallied Sci. 2014;6:2–9. doi: 10.4103/0975-7406.124301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Urbach A., Schuldiner M., Benvenisty N. Modeling for Lesch-Nyhan disease by gene targeting in human embryonic stem cells. Stem Cells. 2004;22:635–641. doi: 10.1634/stemcells.22-4-635. [DOI] [PubMed] [Google Scholar]
- 6.Urbach A., Benvenisty N. Studying early lethality of 45,XO (Turner’s syndrome) embryos using human embryonic stem cells. PLoS One. 2009;4 doi: 10.1371/journal.pone.0004175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eiges R., Urbach A., Malcov M., Frumkin T., Schwartz T., Amit A., Yaron Y., Eden A., Yanuka O., Benvenisty N., et al. Developmental study of fragile X syndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos. Cell Stem Cell. 2007;1:568–577. doi: 10.1016/j.stem.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Maury Y., Gauthier M., Peschanski M., Martinat C. Human pluripotent stem cells for disease modelling and drug screening. Bioessays. 2012;34:61–71. doi: 10.1002/bies.201100071. [DOI] [PubMed] [Google Scholar]
- 9.Verlinsky Y., Strelchenko N., Kukharenko V., Rechitsky S., Verlinsky O., Galat V., Kuliev A. Human embryonic stem cell lines with genetic disorders. Reprod. Biomed. Online. 2005;10:105–110. doi: 10.1016/S1472-6483(10)60810-3. [DOI] [PubMed] [Google Scholar]
- 10.Mateizel I., Spits C., de Rycke M., Liebaers I., Sermon K. Derivation, culture, and characterization of VUB hESC lines. Vitr. Cell. Dev. Biol. Anim. 2010;46:300–308. doi: 10.1007/s11626-010-9284-4. [DOI] [PubMed] [Google Scholar]
- 11.Park I.-H., Arora N., Huo H., Maherali N., Ahfeldt T., Shimamura A., Lensch M.W., Cowan C., Hochedlinger K., Daley G.Q. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimos J.T., Rodolfa K.T., Niakan K.K., Weisenthal L.M., Mitsumoto H., Chung W., Croft G.F., Saphier G., Leibel R., Goland R., et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 13.Kiskinis E., Sandoe J., Williams L., Boulting G., Moccia R., Wainger B., Han S., Peng T., Thams S., Mikkilineni S., et al. Pathways Disrupted in Human ALS Motor Neurons Identified through Genetic Correction of Mutant SOD1. Cell Stem Cell. 2014;14:781–795. doi: 10.1016/j.stem.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puri M.C., Nagy A. Concise review: Embryonic stem cells versus induced pluripotent stem cells: The game is on. Stem Cells. 2012;30:10–14. doi: 10.1002/stem.788. [DOI] [PubMed] [Google Scholar]
- 15.Bilic J., Izpisua Belmonte J.C. Concise review: Induced pluripotent stem cells versus embryonic stem cells: Close enough or yet too far apart? Stem Cells. 2012;30:33–41. doi: 10.1002/stem.700. [DOI] [PubMed] [Google Scholar]
- 16.Cherry A.B.C., Daley G.Q. Reprogrammed cells for disease modeling and regenerative medicine. Annu. Rev. Med. 2013;64:277–290. doi: 10.1146/annurev-med-050311-163324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim H., Kim J.-S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 18.Sandoe J., Eggan K. Opportunities and challenges of pluripotent stem cell neurodegenerative disease models. Nat. Neurosci. 2013;16:780–789. doi: 10.1038/nn.3425. [DOI] [PubMed] [Google Scholar]
- 19.Robertson J.A. Human embryonic stem cell research: Ethical and legal issues. Nat. Rev. Genet. 2001;2:74–78. doi: 10.1038/35047594. [DOI] [PubMed] [Google Scholar]
- 20.De Wert G., Mummery C. Human embryonic stem cells: Research, ethics and policy. Hum. Reprod. 2003;18:672–682. doi: 10.1093/humrep/deg143. [DOI] [PubMed] [Google Scholar]
- 21.Kim K., Doi A., Wen B., Ng K., Zhao R., Cahan P., Kim J., Aryee M.J., Ji H., Ehrlich L.I., et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bar-Nur O., Russ H.A., Efrat S., Benvenisty N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell. 2011;9:17–23. doi: 10.1016/j.stem.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 23.Ma H., Morey R., O’Neil R.C., He Y., Daughtry B., Schultz M.D., Hariharan M., Nery J.R., Castanon R., Sabatini K., et al. Abnormalities in human pluripotent cells due to reprogramming mechanisms. Nature. 2014;511:177–183. doi: 10.1038/nature13551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gore A., Li Z., Fung H.-L., Young J.E., Agarwal S., Antosiewicz-Bourget J., Canto I., Giorgetti A., Israel M.A., Kiskinis E., et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yung S.K., Tilgner K., Ledran M.H., Habibollah S., Neganova I., Singhapol C., Saretzki G., Stojkovic M., Armstrong L., Przyborski S., et al. Brief report: Human pluripotent stem cell models of fanconi anemia deficiency reveal an important role for fanconi anemia proteins in cellular reprogramming and survival of hematopoietic progenitors. Stem Cells. 2013;31:1022–1029. doi: 10.1002/stem.1308. [DOI] [PubMed] [Google Scholar]
- 26.Muller L.U.W., Milsom M.D., Harris C.E., Vyas R., Brumme K.M., Parmar K., Moreau L.A., Schambach A., Park I.H., London W.B., et al. Overcoming reprogramming resistance of Fanconi anemia cells. Blood. 2012;119:5449–5457. doi: 10.1182/blood-2012-02-408674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z.-B., Zhang X., Li X.-J. Recapitulation of spinal motor neuron-specific disease phenotypes in a human cell model of spinal muscular atrophy. Cell Res. 2013;23:378–393. doi: 10.1038/cr.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ebert A.D., Yu J., Rose F.F., Mattis V.B., Lorson C.L., Thomson J.A., Svendsen C.N. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tulpule A., Kelley J.M., Lensch M.W., McPherson J., Park I.H., Hartung O., Nakamura T., Schlaeger T.M., Shimamura A., Daley G.Q. Pluripotent stem cell models of shwachman-diamond syndrome reveal a common mechanism for pancreatic and hematopoietic dysfunction. Cell Stem Cell. 2013;12:727–736. doi: 10.1016/j.stem.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bellin M., Casini S., Davis R.P., D’Aniello C., Haas J., Ward-van Oostwaard D., Tertoolen L.G.J., Jung C.B., Elliott D.A., Welling A., et al. Isogenic human pluripotent stem cell pairs reveal the role of a KCNH2 mutation in long-QT syndrome. EMBO J. 2013;32:3161–3175. doi: 10.1038/emboj.2013.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seriola A., Spits C., Simard J.P., Hilven P., Haentjens P., Pearson C.E., Sermon K. Huntington’s and myotonic dystrophy hESCs: Down-regulated trinucleotide repeat instability and mismatch repair machinery expression bupon differentiation. Hum. Mol. Genet. 2011;20:176–185. doi: 10.1093/hmg/ddq456. [DOI] [PubMed] [Google Scholar]
- 32.Du J., Campau E., Soragni E., Jespersen C., Gottesfeld J.M. Length-dependent CTG.CAG triplet-repeat expansion in myotonic dystrophy patient-derived induced pluripotent stem cells. Hum. Mol. Genet. 2013;22:5276–5287. doi: 10.1093/hmg/ddt386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saenger P. Turner’s syndrome. N. Engl. J. Med. 1996;335:1749–1754. doi: 10.1056/NEJM199612053352307. [DOI] [PubMed] [Google Scholar]
- 34.Ranke M.B., Saenger P. Turner’s syndrome. Lancet. 2001;358:309–314. doi: 10.1016/S0140-6736(01)05487-3. [DOI] [PubMed] [Google Scholar]
- 35.Li W., Wang X., Fan W., Zhao P., Chan Y.C., Chen S., Zhang S., Guo X., Zhang Y., Li Y., et al. Modeling abnormal early development with induced pluripotent stem cells from aneuploid syndromes. Hum. Mol. Genet. 2012;21:32–45. doi: 10.1093/hmg/ddr435. [DOI] [PubMed] [Google Scholar]
- 36.Urbach A., Bar-Nur O., Daley G.Q., Benvenisty N. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell. 2010;6:407–411. doi: 10.1016/j.stem.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheung A.Y.L., Horvath L.M., Grafodatskaya D., Pasceri P., Weksberg R., Hotta A., Carrel L., Ellis J. Isolation of MECP2-null Rett Syndrome patient hiPS cells and isogenic controls through X-chromosome inactivation. Hum. Mol. Genet. 2011;20:2103–2115. doi: 10.1093/hmg/ddr093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y., Wang H., Muffat J., Cheng A.W., Orlando D.A., Lovén J., Kwok S.M., Feldman D.A., Bateup H.S., Gao Q., et al. Global transcriptional and translational repression in human-embryonic-stem-cell-derived Rett syndrome neurons. Cell Stem Cell. 2013;13:446–458. doi: 10.1016/j.stem.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marchetto M.C.N., Carromeu C., Acab A., Yu D., Yeo G.W., Mu Y., Chen G., Gage F.H., Muotri A.R. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tulpule A., William Lensch M., Miller J.D., Austin K., D’Andrea A., Schlaeger T.M., Shimamura A., Daley G.Q. Knockdown of Fanconi anemia genes in human embryonic stem cells reveals early developmental defects in the hematopoietic lineage. Blood. 2010;115:3453–3462. doi: 10.1182/blood-2009-10-246694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu G.-H., Suzuki K., Li M., Qu J., Montserrat N., Tarantino C., Gu Y., Yi F., Xu X., Zhang W., et al. Modelling Fanconi anemia pathogenesis and therapeutics using integration-free patient-derived iPSCs. Nat. Commun. 2014;5:4330. doi: 10.1038/ncomms5330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu B., Palacino J. A novel human embryonic stem cell-derived Huntington’s disease neuronal model exhibits mutant huntingtin (mHTT) aggregates and soluble mHTT-dependent neurodegeneration. FASEB J. 2013;27:1820–1829. doi: 10.1096/fj.12-219220. [DOI] [PubMed] [Google Scholar]
- 43.HD iPSC Consortium Induced pluripotent stem cells from patients with Huntington’s disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell. 2012;11:264–278. doi: 10.1016/j.stem.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D’Andrea A.D., Grompe M. Molecular biology of Fanconi anemia: Implications for diagnosis and therapy. Blood. 1997;90:1725–1736. [PubMed] [Google Scholar]
- 45.Raya A., Rodríguez-Pizà I., Guenechea G., Vassena R., Navarro S., Barrero M.J., Consiglio A., Castellà M., Río P., Sleep E., et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460:53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boyle L., Kaufmann W.E. The behavioral phenotype of FMR1 mutations. Am. J. Med. Genet. Part C Semin. Med. Genet. 2010;154C:469–476. doi: 10.1002/ajmg.c.30277. [DOI] [PubMed] [Google Scholar]
- 47.Penagarikano O., Mulle J.G., Warren S.T. The pathophysiology of fragile X syndrome. Annu. Rev. Genomics Hum. Genet. 2007;8:109–129. doi: 10.1146/annurev.genom.8.080706.092249. [DOI] [PubMed] [Google Scholar]
- 48.Wang T., Bray S.M., Warren S.T. New perspectives on the biology of fragile X syndrome. Curr. Opin. Genet. Dev. 2012;22:256–263. doi: 10.1016/j.gde.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Callan M.A., Zarnescu D.C. Heads-up: New roles for the fragile X mental retardation protein in neural stem and progenitor cells. Genesis. 2011;49:424–440. doi: 10.1002/dvg.20745. [DOI] [PubMed] [Google Scholar]
- 50.Santoro M.R., Bray S.M., Warren S.T. Molecular mechanisms of fragile X syndrome: A twenty-year perspective. Annu. Rev. Pathol. 2012;7:219–245. doi: 10.1146/annurev-pathol-011811-132457. [DOI] [PubMed] [Google Scholar]
- 51.Devys D., Lutz Y., Rouyer N., Bellocq J.P., Mandel J.L. The FMR-1 protein is cytoplasmic, most abundant in neurons and appears normal in carriers of a fragile X premutation. Nat. Genet. 1993;4:335–340. doi: 10.1038/ng0893-335. [DOI] [PubMed] [Google Scholar]
- 52.Turetsky T., Aizenman E., Gil Y., Weinberg N., Shufaro Y., Revel A., Laufer N., Simon A., Abeliovich D., Reubinoff B.E. Laser-assisted derivation of human embryonic stem cell lines from IVF embryos after preimplantation genetic diagnosis. Hum. Reprod. 2008;23:46–53. doi: 10.1093/humrep/dem351. [DOI] [PubMed] [Google Scholar]
- 53.Telias M., Segal M., Ben-Yosef D. Neural differentiation of Fragile X human Embryonic Stem Cells reveals abnormal patterns of development despite successful neurogenesis. Dev. Biol. 2013;374:32–45. doi: 10.1016/j.ydbio.2012.11.031. [DOI] [PubMed] [Google Scholar]
- 54.Hatton D.D., Sideris J., Skinner M., Mankowski J., Bailey D.B., Roberts J., Mirrett P. Autistic behavior in children with fragile X syndrome: Prevalence, stability, and the impact of FMRP. Am. J. Med. Genet. Part A. 2006;140:1804–1813. doi: 10.1002/ajmg.a.31286. [DOI] [PubMed] [Google Scholar]
- 55.Kaufmann W.E., Cortell R., Kau A.S.M., Bukelis I., Tierney E., Gray R.M., Cox C., Capone G.T., Stanard P. Autism spectrum disorder in fragile X syndrome: Communication, social interaction, and specific behaviors. Am. J. Med. Genet. A. 2004;129A:225–234. doi: 10.1002/ajmg.a.30229. [DOI] [PubMed] [Google Scholar]
- 56.Pietrobono R., Tabolacci E., Zalfa F., Zito I., Terracciano A., Moscato U., Bagni C., Oostra B., Chiurazzi P., Neri G. Molecular dissection of the events leading to inactivation of the FMR1 gene. Hum. Mol. Genet. 2005;14:267–277. doi: 10.1093/hmg/ddi024. [DOI] [PubMed] [Google Scholar]
- 57.Smeets H.J., Smits A.P., Verheij C.E., Theelen J.P., Willemsen R., van de Burgt I., Hoogeveen A.T., Oosterwijk J.C., Oostra B.A. Normal phenotype in two brothers with a full FMR1 mutation. Hum. Mol. Genet. 1995;4:2103–2108. doi: 10.1093/hmg/4.11.2103. [DOI] [PubMed] [Google Scholar]
- 58.Sheridan S.D., Theriault K.M., Reis S.A., Zhou F., Madison J.M., Daheron L., Loring J.F., Haggarty S.J. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS One. 2011;6:e26203. doi: 10.1371/journal.pone.0026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alisch R.S., Wang T., Chopra P., Visootsak J., Conneely K.N., Warren S.T. Genome-wide analysis validates aberrant methylation in fragile X syndrome is specific to the FMR1 locus. BMC Med. Genet. 2013;14:18. doi: 10.1186/1471-2350-14-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bar-Nur O., Caspi I., Benvenisty N. Molecular analysis of FMR1 reactivation in fragile-X induced pluripotent stem cells and their neuronal derivatives. J. Mol. Cell Biol. 2012;4:180–183. doi: 10.1093/jmcb/mjs007. [DOI] [PubMed] [Google Scholar]
- 61.Martin J.B., Gusella J.F. Huntington’s disease. Pathogenesis and management. N. Engl. J. Med. 1986;315:1267–1276. doi: 10.1056/NEJM198611133152006. [DOI] [PubMed] [Google Scholar]
- 62.Huntington T., Macdonald M.E., Ambrose C.M., Duyao M.P., Myers R.H., Lin C., Srinidhi L., Barnes G., Taylor S.A., James M., Groat N., et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-E. [DOI] [PubMed] [Google Scholar]
- 63.Landles C., Sathasivam K., Weiss A., Woodman B., Moffitt H., Finkbeiner S., Sun B., Gafni J., Ellerby L.M., Trottier Y., et al. Proteolysis of mutant huntingtin produces an exon 1 fragment that accumulates as an aggregated protein in neuronal nuclei in Huntington disease. J. Biol. Chem. 2010;285:8808–8823. doi: 10.1074/jbc.M109.075028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang N., An M.C., Montoro D., Ellerby L.M. Characterization of human Huntington’s disease cell model from induced pluripotent stem cells. PLoS Curr. 2010;2:1–11. doi: 10.1371/currents.RRN1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Camnasio S., Carri A.D., Lombardo A., Grad I., Mariotti C., Castucci A., Rozell B., Riso P.L., Castiglioni V., Zuccato C., et al. The first reported generation of several induced pluripotent stem cell lines from homozygous and heterozygous Huntington’s disease patients demonstrates mutation related enhanced lysosomal activity. Neurobiol. Dis. 2012;46:41–51. doi: 10.1016/j.nbd.2011.12.042. [DOI] [PubMed] [Google Scholar]
- 66.Bradley C.K., Scott H.A., Chami O., Peura T.T., Dumevska B., Schmidt U., Stojanov T. Derivation of Huntington’s disease-affected human embryonic stem cell lines. Stem Cells Dev. 2011;20:495–502. doi: 10.1089/scd.2010.0120. [DOI] [PubMed] [Google Scholar]
- 67.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 68.Park I.-H., Zhao R., West J.A., Yabuuchi A., Huo H., Ince T.A., Lerou P.H., Lensch M.W., Daley G.Q. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 69.Yu J.Y., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 70.Takahashi K., Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]