Abstract
The deer mouse (genus Peromyscus) is the most abundant mammal in North America, and it occupies almost every type of terrestrial habitat. It is not surprising therefore that the natural history of Peromyscus is among the best studied of any small mammal. For decades, the deer mouse has contributed to our understanding of population genetics, disease ecology, longevity, endocrinology and behavior. Over a century's worth of detailed descriptive studies of Peromyscus in the wild, coupled with emerging genetic and genomic techniques, have now positioned these mice as model organisms for the study of natural variation and adaptation. Recent work, combining field observations and laboratory experiments, has lead to exciting advances in a number of fields—from evolution and genetics, to physiology and neurobiology.
DOI: http://dx.doi.org/10.7554/eLife.06813.001
Research organism: mouse
Introduction
Peromyscus is a genus of small North American rodents known colloquially as deer mice (Emmons, 1840). When the first Peromyscus specimens were shipped to European systematicists in the late 18th century, their resemblance to the local wood mouse prompted the designation Mus sylvaticus (Hooper, 1968). At the time, little was known of the diversity of rodents worldwide and most were assigned the generic term Mus (Linnaeus, 1758). The name Peromyscus (Gloger, 1841) was first employed, albeit narrowly, in the middle of the 19th century. Quadrupeds of North America (Audubon and Bachman, 1854) recognized only three species now known to belong to Peromyscus, and Mammals of North America (Baird, 1859) included a mere 12. But by the turn of the 20th century, Peromyscus included 143 forms, 42 of which represented monotypic or good biological species (Osgood, 1909). The genus saw several additional revisions throughout the 20th century as North American mammalogy matured and natural history collections expanded. Today 56 species are recognized, the most widespread and diverse being Peromyscus maniculatus (Musser and Carleton, 2005).
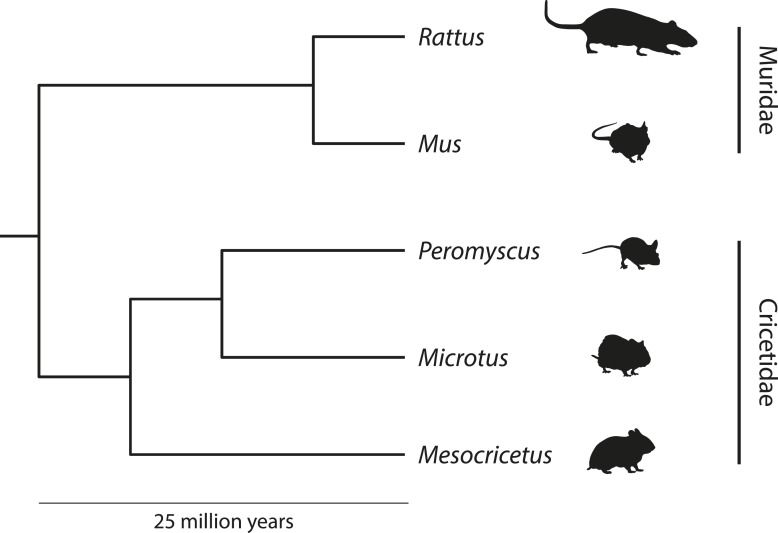
Thus, although not immediately appreciated, Peromyscus includes more species than any other North American mammalian genus and, apart from Mus and Rattus, more is known concerning its biology in the laboratory than any other group of small mammals (Figure 1; King, 1968; Kirkland and Layne, 1989). Several disciplines including ecology, evolution, physiology, reproductive biology and behavioral neuroscience have all employed Peromyscus, inspiring its label as ‘the Drosophila of North American mammalogy’ (Dewey and Dawson, 2001). Arguably, the emergence of Peromyscus as a model system was propelled by our cumulative knowledge of its fascinating and varied natural history.
Figure 1. Simplified phylogeny depicting the relationships among muroid rodent model organisms.
Peromyscus belong to the Cricetidae family, which includes voles (Microtus), hamsters (Mesocricetus), and New World rats and mice. Old World rats and mice belong to the Muridae family, which include the familiar laboratory rat (Rattus norvegicus) and mouse (Mus musculus). Muridae and Cricetidae diverged roughly 25 million years ago. Schematic based on based on phylogeny data from Steppan et al. (2004). Image credit, Nicole Bedford and Hopi Hoekstra.
Distribution and habitat
‘Within the range of one species (maniculatus) it is probable that a line, or several lines, could be drawn from Labrador to Alaska and thence to southern Mexico throughout which not a single square mile is not inhabited by some form of this species’ (Osgood, 1909).
Wilfred H Osgood asserted that some form of Peromyscus had been trapped in nearly every patch of North America ever visited by a mammal collector. Members of the genus are distributed from the southern edge of the Canadian Arctic to the Colombian border of Panama (Figure 2). Various demographic and biogeographic factors (e.g., Pleistocene glacial and pluvial cycles, population expansions, mountain range elevations and sea-level changes) have influenced the diversity and distribution of deer mice (Sullivan et al., 1997; Riddle et al., 2000; Dragoo et al., 2006; Kalkvik et al., 2012; López-González et al., 2014). The result is a mosaic of widespread and restricted species ranges shaped by both dispersal and vicariance events. Our knowledge of the distributions, home ranges and habitat preferences of deer mice comes primarily from the trapping data and field notes of early natural historians (e.g., Sumner, 1917; Dice, 1931; Blair, 1940, 1951). Osgood's influential 1909 taxonomic revision was built on examinations of more than 27,000 specimens from diverse locales that were collected primarily by the US Biological Survey. Today, more than 120,000 Peromyscus specimens are accessioned in Natural History museums across North America and the United Kingdom (Table 1). These invaluable collections document more than a century of dynamic relationships between deer mice and their environment. For example, by comparing past and present-day collecting locales, shifts in the distributions of deer mice have been linked to climate change (Moritz et al., 2008; Yang et al., 2011; Rowe et al., 2014), and morphological analyses of these museum specimens reveal how deer mice respond to changing environments (Grieco and Rizk, 2010).
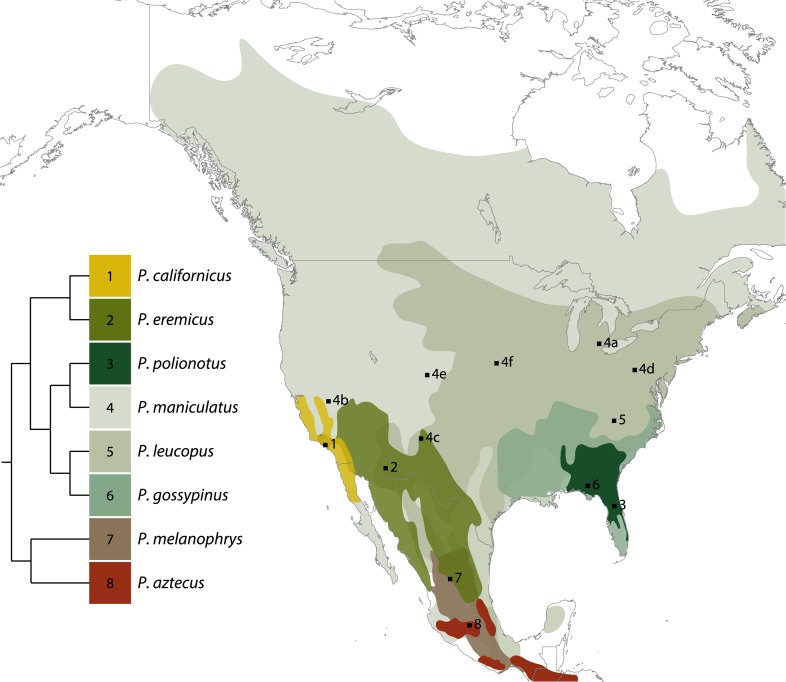
Figure 2. North American distributions of eight Peromyscus species currently maintained as outbred laboratory stocks (based on data from Hall, 1981).
Some ranges are narrow and others are extensive, with many overlapping to a large extent. Simplified tree indicating phylogenetic relationships among taxa is shown; branch lengths are arbitrary (based on data from Bradley et al., 2007). The most widespread and ecologically diverse group is also the best represented in the laboratory: six P. maniculatus subspecies are maintained in laboratories across the United States. Collecting localities of colony founders are indicated by numbered squares (see also Table 2). Image credit, Nicole Bedford and Hopi Hoekstra.
Table 1.
Museums with the largest collections of Peromyscus specimens
| Collection | Location | No. specimens |
|---|---|---|
| Smithsonian National Museum of Natural History | Washington, DC | 38,406 |
| Museum of Vertebrate Zoology | Berkeley, CA | 34,131 |
| American Museum of Natural History | New York, NY | 19,234 |
| Field Museum | Chicago, IL | 8939 |
| Museum of Comparative Zoology | Cambridge, MA | 7754 |
| Canadian Museum of Nature | Ottawa, ON | 6315 |
| Academy of Natural Science | Philadelphia, PA | 2425 |
| Natural History Museum | London, UK | 2238 |
Although not strictly commensal, deer mice (particularly in New England) do enter human households and partake of their larders. According to legend, Walt Disney drew inspiration for Mickey Mouse from the ‘tame field mice’ (most likely Peromyscus leucopus) that would wander into his old Kansas City animation studio (Updike, 1991). Nevertheless, Peromyscus are most commonly trapped in woodlands and brushlands and are also found in tropical and temperate rainforests, grasslands, savannas, swamps, deserts and alpine habitats (Figure 3; Baker, 1968). Local adaptation to these various environments has been the subject of much recent inquiry (e.g., Linnen et al., 2013; Natarajan et al., 2013; MacManes and Eisen, 2014), and the detailed cataloguing of phenotypic diversity by early naturalists inspired much of this work. However, we still require a more complete understanding of ecological diversity across the entire genus, as well as an enlightened view of phylogenetic relationships informed by whole-genome sequences (see Box 1).
Figure 3. The ecology of Peromyscus varies considerably both within and among species.
(A) The forest-dwelling deer mouse, P. maniculatus nubiterrae, perches high on a tree branch in Southwestern Pennsylvania. (B) The beach mouse, P. polionotus phasma, takes shelter among the dune grasses on Florida's Atlantic coast. (C) Its mainland counterpart, the oldfield mouse, P. polionotus sumneri, is typically found in fallow fields and is sympatric with the cotton mouse, P. gossypinus (D), which occupies adjacent stands of long leaf pine. Image credits: A, Evan P Kingsley; B, JB Miller; C, D, Nicole Bedford.
Box 1. Priorities for Peromyscus research.
Discovering as yet untapped ecological diversity
Much of our understanding of Peromyscus biology comes from studies of two ubiquitous species that have proven amenable to laboratory life—P. maniculatus and P. leucopus. However, most Peromyscus species remain comparatively understudied, particularly in Central America and Mexico where taxonomic diversity and endemism (i.e., where species are unique to a given geographic location) is greatest.
Sequencing more Peromyscus genomes and revising their phylogeny
A comprehensive phylogeny based on genome-wide DNA sequences would greatly facilitate the comparative approaches that are the unique advantage of the Peromyscus system. An annotated genome assembly is currently available for P. maniculatus bairdii (Pman_1.0, GenBank assembly accession GCA_000500345.1) and draft sequences are available for P. californicus, P. leucopus and P. polionotus (Baylor College of Medicine, www.hgsc.bcm.edu/peromyscus-genome-project). Several more Peromyscus genomes are being sequenced, but still more are needed.
Identifying where Peromyscus can complement biomedical studies of other laboratory species
The genetically diverse Peromyscus could be used more widely in biomedical research than previously thought. Indeed, certain aspects of human biology—including aging, epigenetics, retinal development and hematology—have been suitably modeled in Peromyscus (e.g., Ungvari et al., 2008; Shorter et al., 2012; Arbogast et al., 2013; Sun et al., 2014).
Adaptation to mountains, cities and deserts
Among North American mammals, the deer mouse is unparalleled in its ability to colonize an impressive array of habitats. The remarkable elevational range of one subspecies (P. m. sonoriensis) stretches from below sea level in Death Valley to above 4300 meters in the adjacent White and Sierra Nevada mountain ranges (Hock, 1964). The ability of deer mice to colonize and thrive in low-oxygen environments is due, in part, to standing genetic variation in globin genes (Snyder, 1981; Natarajan et al., 2015). Storz and colleagues (2007, 2009) pinpointed several amino acid substitutions that confer high hemoglobin-O2 affinity and better aerobic performance at high altitudes. Functional analyses have since identified how precise mutations, and interactions among mutations, affect hemoglobin-O2 affinity, demonstrating that the adaptive value of a given biochemical substitution depends both on the local environment and the genetic background in which it arises (Natarajan et al., 2013).
The process of adapting to urban environments also leaves its mark on the genome (Pergams and Lacy, 2008; Munshi-South and Kharchenko, 2010; Munshi-South and Nagy, 2014). By comparing the brain, liver and gonad transcriptomes of urban and rural populations of P. leucopus, Harris et al. (2013) identified several genes associated with metabolism and immune function exhibiting signatures of selection in New York City's parklands. Similarly, MacManes and Eisen (2014) identified renal transcripts related to solute and water balance experiencing purifying selection in the desert-adapted species, Peromyscus eremicus. Further study of these candidate genes will determine their role in adaptation to new or extreme environments.
Diet and predators
Generally deer mice are granivores, feeding primarily on seeds, but fruits, fungi, green vegetation and insects have been found among their stomach contents and in the nest cavities of their burrows (Gentry and Smith, 1968; Wolff, 1985). However, some species have evolved seasonally specialized diets. In the winter, Peromyscus melanotis prey almost exclusively on monarch butterflies that roost in Mexico's central highlands (Brower et al., 1985). Moreover, on a remote island in British Columbia, Peromyscus keeni feast on auklet eggs during the seabird breeding season (Drever et al., 2000). Deer mice are themselves common prey, contributing to the diets of many predators such as weasels, skunks, lynx, bobcats, foxes, coyotes, hawks and owls (Luttich et al., 1970; Bowen, 1981; Montgomery, 1989; Van Zant and Wooten, 2003). Indeed, avian predation imposes strong selective pressure for cryptic coloration in Peromyscus—a classic example of local adaptation (Vignieri et al., 2010; Linnen et al., 2013).
Parasites and disease
The diversity of parasites is documented for only a few Peromyscus species, and very little is known of the ecological factors that influence infection dynamics. Common internal parasites include pentastomid larvae, cestode tapeworms, nematodes and trematodes (Whitaker, 1968; Pedersen and Antonovics, 2013). External parasites include lice, mites, fleas and ticks (Whitaker, 1968), the latter two being vectors of plague and Lyme disease, respectively (Allred, 1952; Burgdorfer et al., 1982; Gage and Kosoy, 2005).
As a natural reservoir for Borrelia burgdorferi—the bacterial agent of Lyme disease—Peromyscus is the subject of much research on the pathogenesis and transmission of the disease (Bunikis et al., 2004; Ramamoorthi et al., 2005; Schwanz et al., 2011; Baum et al., 2012). Peromyscus also features in ecological modeling efforts to determine how the diversity of the tick host community impacts disease risk (LoGiudice et al., 2003, 2008). One hypothesis for the alarming recent expansion of Lyme disease is that habitat fragmentation associated with human development favors deer mouse populations at the expense of other tick hosts (e.g., squirrels and shrews) that are poor reservoirs for the disease (LoGiudice et al., 2003; Schwanz et al., 2011). Peromyscus is also a notorious carrier of the Sin Nombre hantavirus, responsible for the deaths of 12 people in the Four Corners area of the southwestern United States in 1993.
Longevity
Mortality in natural populations is incredibly high and driven by a combination of factors including limited food supply, competition for territories and predation (Bendell, 1959). As such, most Peromyscus are thought to live less than a year in the wild (Terman, 1968). However, early investigators noted substantially longer natural lifespans in their laboratory colonies (Sumner, 1922; Dice, 1933). With a twofold difference in life expectancy, Sacher and Hart (1978) proposed P. leucopus and Mus musculus as a longevity contrast pair. P. leucopus—which lives up to 8 years and may remain fertile for 5—produces fewer reactive oxygen species, exhibits enhanced antioxidant enzyme activity and less oxidative damage to lipids relative to the short-lived (~3.5 years) laboratory mouse (Sohal et al., 1993; Shi et al., 2013). Measuring the biochemical correlates of longevity in Peromyscus has been integral to providing support for the oxidative stress theory of aging (Ungvari et al., 2008).
Life history
The timing of life history events in Peromyscus—well documented from field and laboratory studies alike—is highly variable both within and among species. Yet studies contrasting the reproductive and developmental patterns of wild and domesticated deer mice have found few significant differences (Millar, 1989; Botten et al., 2000). Here, we highlight life history traits in P. maniculatus, the most commonly used laboratory species. Gestation ranges from 21 to 27 days (average 23.6) and average litter size is 4.6 pups (Millar, 1989). Juveniles first leave the nest between 14 and 16 days of age (Vestal et al., 1980) and become independent of their mother between 18 and 25 days (Millar, 1989). Captive females give birth to their first litter, on average, at 84 days (Haigh, 1983), but males are capable of siring offspring several weeks earlier.
The actual timing of sexual maturation in the wild, however, is often dictated by population density, food availability and season. In response to short day length, many species exhibit seasonal gonadal regression (Trainor et al., 2006), increased aggression (Trainor et al., 2007), impaired spatial memory (Workman et al., 2009) and enhanced immune function (Prendergast and Nelson, 2001). As such, Peromyscus has emerged as a model system for the study of photoperiodism (i.e., the ability to seasonally modulate energetic demands by tracking day length changes). Such studies have been particularly fruitful for understanding the mechanistic basis of gene by environment interactions. For example, day length can reverse the behavioral action of the hormone estradiol by determining which estrogen receptor pathway is expressed and consequently activated (Trainor et al., 2007). While life history traits are strongly affected by environmental cues, substantial genetic variation in the neuroendocrine pathways that control reproductive timing also exists, as demonstrated by selection line experiments with photoperiod responsive and nonresponsive P. leucopus (Heideman et al., 1999; Heideman and Pittman, 2009).
Mating system and parental care
While the majority of Peromyscus species are promiscuous, monogamy has independently evolved at least twice in the genus (Turner et al., 2010). Both Peromyscus californicus (Gubernick and Alberts, 1987; Ribble, 1991) and Peromyscus polionotus (Smith, 1966; Foltz, 1981) are socially and genetically monogamous, and both males and females contribute to the care of offspring. P. californicus, in particular, has become an important neurobiological model for the study of male parental care (Bester-Meredith et al., 1999; Trainor et al., 2003; Lee and Brown, 2007; de Jong et al., 2009, 2010). As a complement, the ability of monogamous P. polionotus to hybridize with promiscuous P. maniculatus allows geneticists to identify the genetic basis of alternate mating systems and their associated phenotypes, from genomic imprinting (Vrana et al., 2000) to parental investment and reproductive traits (e.g., Fisher and Hoekstra, 2010).
Rosenfeld (2015) argues that parental and social behaviors are particularly vulnerable to endocrine disruption, as these traits are dependent upon the organizational and activational effects of androgens and estrogens. Mating system variation between closely related species of deer mice provides an opportunity to test this hypothesis. P. maniculatus males exposed to the endocrine disrupting compound bisphenol A (BPA) during development displayed reduced spatial learning and exploratory behavior—traits known to be associated with male–male competition for mates (Galea et al., 1996; Jašarević et al., 2011). However, these behaviors—which are not subject to sexual selection in females—were unaffected in BPA-exposed females. By contrast, sexual selection favors the evolution of mate guarding and territorial behavior in monogamous males, and it is these traits (rather than spatial learning or exploratory behavior) that are compromised by endocrine disruption in P. californicus (Williams et al., 2013).
Home building
Behavioral genetics studies have historically been restricted to a handful of genetic model organisms that display behaviors of unclear ecological relevance (Fitzpatrick et al., 2005). Sufficient resources are now available—from a medium-density genetic linkage map (Kenney-Hunt et al., 2014) to draft genome sequences (Baylor College of Medicine, Peromyscus Genome Project)—that we can attribute natural variation in Peromyscus behavior to specific genetic variants. For instance, P. maniculatus and P. polionotus display considerable differences in stereotyped burrowing behavior. P. maniculatus digs short, simple burrows in contrast to the long, complex burrows constructed by P. polionotus that consist of an entrance tunnel, nest chamber and escape tunnel (Dawson et al., 1988; Weber et al., 2013). Remarkably, mice raised in the laboratory for several generations recapitulate the species-specific burrow architectures observed in nature (Video 1). Furthermore, the complex burrows of P. polionotus are derived (Weber and Hoekstra, 2009) and likely evolved through changes at only a handful of genetic loci, each affecting distinct behavioral modules (i.e., entrance tunnel length and escape tunnel presence; Weber et al., 2013). Next steps include isolating genetic variants, understanding their effects on the neural circuitry underlying burrowing behavior and quantifying the adaptive value of burrowing in the wild.
Video 1. Innate burrowing behavior in Peromyscus can be directly observed in a laboratory setting. Here, P. polionotus is busy constructing the long entrance tunnel of its complex burrow. Video credit, Nicole Bedford and Hopi Hoekstra.
Pigmentation
Among the several cases of adaptive phenotypic variation in Peromyscus, perhaps the most obvious is coat coloration. Recent advances have identified not only the genes, but also the specific mutations, leading to local variation in coat color. Beach mice (P. polionotus leucocephalus) living on the coastal sand dunes and barrier islands of Florida are considerably paler than their inland counterparts (P. p. subgriseus) that inhabit dark, loamy soils (Figure 4; Howell, 1920; Sumner, 1929). For beach mice on Florida's Gulf Coast, light coloration is due, in part, to a fixed single nucleotide polymorphism (SNP) in the melanocortin-1 receptor (Mc1r) coding region (Hoekstra et al., 2006). However, this Mc1r allele does not contribute to light pelage in Florida's Atlantic coast mice, suggesting that the two populations converged on light coloration independently (Steiner et al., 2007).
Figure 4. Genetic crosses between the pale beach mouse P. polionotus leucocephalus (top row left) and the darker mainland mouse P. p. polionotus (top row right) result in first-generation F1 hybrids, all with intermediate coloration (second row).
Intercrosses between F1 hybrids produce a variable F2 generation, showing a continuous distribution of pigmentation phenotypes ranging from light to dark (third and fourth rows; Steiner et al., 2007). This segregation pattern—initially described by Francis Sumner—is among the earliest empirical evidence that several discrete loci may collectively contribute to a quantitative trait (Dobzhansky, 1937; see also Box 1). Image credit, Nicole Bedford and Hopi Hoekstra.
Similarly, background matching in P. maniculatus of the Nebraska Sand Hills affords a strong selective advantage against avian predators (Linnen et al., 2013). Yet, cryptic coloration is a complex phenotype composed of multiple component traits (i.e., tail stripe, dorsal-ventral boundary, ventral color, dorsal brightness and hue). Linnen and colleagues (2013) identified multiple distinct mutations within the Agouti locus, each associated with a different color trait that independently affected fitness. Thus, parallel studies of Peromyscus pigmentation nicely illustrate the marriage between classical natural history studies and modern molecular techniques, thereby providing new insights into the molecular basis of adaptation.
Peromyscus in the laboratory
Francis Sumner, considered the grandfather of Peromyscus biology (see Box 2), first demonstrated the feasibility of the deer mouse as a laboratory organism in the 1910s and 20s. He famously built the first Peromyscus ‘mouse house’ in what is now referred to as Sumner Canyon at the Scripps Institution in La Jolla, California. When his Peromyscus work at Scripps was discontinued, Sumner bequeathed his stocks to Lee R Dice at the University of Michigan who honed the methods for generating and maintaining Peromyscus colonies in the 1930s and 40s. During this time, Dice began to catalogue single factor genetic mutations in his stocks (e.g., gray, dilute, epilepsy). These mice served as the founding strains for the Peromyscus Genetic Stock Center (PGSC) established in 1985 by Wallace Dawson at the University of South Carolina, which currently maintains wild-derived stocks of six species, as well as 13 coat-color mutants and four additional mutants on P. maniculatus genetic backgrounds. Additional wild-derived stocks are kept in individual laboratories (Table 2) and still more mutants have been cryopreserved. The PGSC also maintains an extensive online reference library (http://stkctr.biol.sc.edu) with more than 3000 citations.
Box 2. Peromyscus and the history of evolutionary thought.
The work of early Peromyscus biologists (particularly Francis B Sumner) informed influential thinkers in population genetics and evolutionary biology, such as Sewall Wright, Theodosius Dobzhansky and JBS Haldane. Since most early 20th century geneticists came from experimentalist backgrounds, many turned to naturalists for data from wild populations (Provine, 1986). At the time, Sumner's work on geographic variation in Peromyscus represented one of the few major studies of evolution in natural populations. As such, Wright closely followed Sumner's analysis of phenotypic intergradation between geographically contiguous P. maniculatus subspecies in California (Sumner, 1918). Wright concluded that the observed quantitative differences in coat color were determined by the accumulation of several discrete (i.e., Mendelian) factors (Wright, 1932). The question of whether continuous (or quantitative) traits are subject to the same rules of inheritance as discrete characters was central to the Modern Evolutionary Synthesis.
Between 1914 and 1930, Sumner carefully measured several quantitative traits—most notably coat color—that varied among geographically distinct subspecies of Peromyscus, which he then crossed in the laboratory (Figure 4; Sumner, 1930). Dobzhansky (1937) highlighted these data as empirical support for the multiple gene hypothesis for the inheritance of quantitative traits. Later, Haldane (1948) applied a theoretical model to the gradient of increasing pigmentation observed in P. polionotus populations from coastal to inland Florida (Sumner, 1929). From these data, he estimated the local strength of selection acting on a putative pigmentation locus in the wild—the dominant white-cheek character (Wc) identified by Blair (1944).
Peromyscus also featured in Dobzhansky's studies of reproductive isolation. Certain P. maniculatus subspecies with overlapping geographic distributions are nevertheless separated by habitat, often with one subspecies inhabiting prairie, open fields or sandy lake beaches, and the other being exclusively forest-dwelling (Dice, 1931). These sub-specific forms readily produce viable and fertile offspring in the laboratory yet remain reproductively isolated in the wild—a prime example of ecological isolation (Dobzhansky, 1937). Peromyscus has thus been a cornerstone of evolutionary biology for nearly a century. These and other studies drew the attention of biologists in many fields, launching the many, varied Peromyscus research programs we see today.
Table 2.
Current laboratory colonies of Peromyscus
| Species | Year | Source population | Location | |
|---|---|---|---|---|
| 1 | P. californicus insignis | 1979–1987 | Santa Monica Mts., CA | PGSC |
| 2 | P. eremicus sp. | 1993 | Tucson, AZ | PGSC |
| 3 | P. polionotus subgriseus | 1952 | Ocala National Forest, FL | PGSC |
| 4a | P. maniculatus bairdii | 1946–1948 | Ann Arbor, MI | PGSC |
| 4b | P. m. sonoriensis | 1995 | White Mtn. Research Station, CA | PGSC |
| 4c | P. m. rufinus | 1998 | Manzano Mtn., NM | UNM |
| 4d | P. m. nubiterrae | 2010 | Powder Mill Nature Reserve, PA | HU |
| 4e | P. m. rufinus | 2014 | Mt. Evans, CO | UIUC |
| 4f | P. m. nebrascensis | 2014 | Lincoln, NE | UIUC |
| 5 | P. leucopus sp. | 1982–1985 | Linville, NC | PGSC |
| 6 | P. gossypinus gossypinus | 2009 | Jackson County, FL | HU |
| 7 | P. melanophrys xenerus | 1970–1978 | Zacatecas, Mexico | UIUC |
| 8 | P. aztecus hylocetes | 1986 | Sierra Chincua, Mexico | UIUC |
The year and population from which the founders were collected are noted. Numbers refer to collecting localities shown in Figure 2. PGSC: Peromyscus Genetic Stock Center; UNM: University of New Mexico; HU: Harvard University; UIUC: University of Illinois at Urbana-Champaign.
While the genetic causes and phenotypic consequences differ among strains, Peromyscus colonies are invariably susceptible to inbreeding depression, which necessitates their maintenance as relatively outbred stocks (Lacy et al., 1996; Joyner et al., 1998). Thus, although the deer mouse is amenable to laboratory life, its biology has not been purposely altered by generations of inbreeding or artificial selection. Life history traits and even behaviors such as burrow construction or ultrasonic vocalization are generally preserved in laboratory strains (Dawson et al., 1988; Millar, 1989; Kalcounis-Rueppell et al., 2010). Thus, the traits we scrutinize in the laboratory (e.g., aerobic performance, photoperiodism, mating and parental behavior) are arguably faithful representations of phenotypes in nature. The ability to study genetically diverse, wild-derived mice under controlled laboratory conditions has opened up several constructive research programs centered on understanding the phenotypic consequences of natural genetic variation.
Conclusions
The tradition of dissecting the genetic basis of ecologically relevant traits in the laboratory began in the early 20th century; in Peromyscus, this effort was lead by Francis Sumner and continues today. In an era of high-throughput sequencing and expanding transgenic technologies, our concept of the genetic model organism is rapidly changing. We can now widen our focus to include the diverse and naturally evolving species that may further our understanding of life outside the laboratory. The emergence of Peromyscus as a model system has been largely driven by the wealth of natural history information available for the genus. Indeed, deer mice form the foundation of much of our understanding of the biology of small mammals. The multitude of ecological conditions to which deer mice have adapted has contributed to an impressive array of biological diversity within a single, ubiquitous genus. While this radiation is fascinating in its own right, Peromyscus is arguably foremost among nascent model systems that may aptly model the genetic complexity of the human condition, which too has long been shaped by natural selection in the wild. We hope that the continued development—primarily through the growth of genetic and genomic resources—of this model system will galvanize research in all corners of biology.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Competing interests:
The authors declare that no competing interests exist.
Author contributions
NLB, conceived and wrote the paper.
HEH, conceived and wrote the paper.
Funding Information
This paper was supported by the following grants:
Howard Hughes Medical Institute to Hopi E Hoekstra.
Natural Sciences and Engineering Research Council of Canada (Conseil de Recherches en Sciences Naturelles et en Génie du Canada) 421595-2012 to Nicole L Bedford.
References
- Allred DM. Plague important fleas and mammals in Utah and the western United States. The Great Basin Naturalist. 1952;12:67–75. [Google Scholar]
- Arbogast P, Glösmann M, Peichl L. Retinal cone photoreceptors of the deer mouse Peromyscus maniculatus: development, topography, opsin expression and spectral tuning. PLOS ONE. 2013;8:1–12. doi: 10.1371/journal.pone.0080910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audubon JJ, Bachman J. The Quadrupeds of North America. Volume 3. New York: V. G. Audubon; 1854. [Google Scholar]
- Baird SF. Mammals of North America. Philadelphia: J. B. Lippincott & Co; 1859. [Google Scholar]
- Baker RH. Habitats and distribution. In: King JA, editor. Biology of Peromyscus (Rodentia) American Society of Mammalogists; 1968. [Google Scholar]
- Baum E, Hue F, Barbour AG. Experimental infections of the reservoir species Peromyscus leucopus with diverse strains of Borrelia burgdorferi, a Lyme disease agent. mBio. 2012;3:1–11. doi: 10.1128/mBio.00434-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendell JF. Food as a control of a population of white-footed mice, Peromyscus leucopus noveboracensis (Fischer) Canadian Journal of Zoology. 1959;37:173–209. doi: 10.1139/z59-021. [DOI] [Google Scholar]
- Bester-Meredith JK, Young LJ, Marler CA. Species differences in paternal behavior and aggression in Peromyscus and their associations with vasopressin immunoreactivity and receptors. Hormones and Behavior. 1999;36:25–38. doi: 10.1006/hbeh.1999.1522. [DOI] [PubMed] [Google Scholar]
- Blair WF. A study of prairie deer-mouse populations in Southern Michigan. American Midland Naturalist. 1940;24:273–305. doi: 10.2307/2420931. [DOI] [Google Scholar]
- Blair WF. Inheritance of the white-cheek character in mice of the genus Peromyscus. Contributions from the Laboratory of Vertebrate Biology. 1944;25:1–7. [Google Scholar]
- Blair WF. Population structure, social behavior, and environmental relations in a natural population of the beach mouse (Peromyscus polionotus leucocephalus) Contributions from the Laboratory of Vertebrate Biology. 1951;48:1–47. [Google Scholar]
- Botten J, Mirowsky K, Kusewitt D, Bharadwaj M, Yee J, Ricci R, Feddersen RM, Hjelle B. Experimental infection model for Sin Nombre hantavirus in the deer mouse (Peromyscus maniculatus) Proceedings of the National Academy of Sciences of USA. 2000;97:10578–10583. doi: 10.1073/pnas.180197197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen WD. Variation in coyote social organization: the influence of prey size. Canadian Journal of Zoology. 1981;59:639–652. doi: 10.1139/z81-094. [DOI] [Google Scholar]
- Bradley RD, Rogers DS, Kilpatrick CW. Toward a molecular phylogeny for Peromyscus: evidence from mitochondrial cytochrome-b sequences. Journal of Mammalogy. 2007;88:1146–1159. doi: 10.1644/06-MAMM-A-342R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower LP, Horner BE, Marty MA, Moffitt CM, Villa-R B. Mice (Peromyscus maniculatus, P. spicilegus, and Microtus mexicanus) as predators of overwintering Monarch butterflies (Danaus plexippus) in Mexico. Biotropica. 1985;17:89–99. doi: 10.2307/2388500. [DOI] [Google Scholar]
- Bunikis J, Tsao J, Luke CJ, Luna MG, Fish D, Barbour AG. Borrelia burgdorferi infection in a natural population of Peromyscus leucopus mice: a longitudinal study in an area where Lyme Borreliosis is highly endemic. Journal of Infectious Diseases. 2004;189:1515–1523. doi: 10.1086/382594. [DOI] [PubMed] [Google Scholar]
- Burgdorfer W, Barbour AG, Hayes SF, Benach JL, Davis JP. Lyme disease—a tick-borne spirochetosis? Science. 1982;216:1317–1319. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- Dawson WD, Lake CE, Schumpert SS. Inheritance of burrow building in Peromyscus. Behavior Genetics. 1988;18:371–382. doi: 10.1007/BF01260937. [DOI] [PubMed] [Google Scholar]
- de Jong TR, Chauke M, Harris BN, Saltzman W. From here to paternity: neural correlates of the onset of paternal behavior in California mice (Peromyscus californicus) Hormones and Behavior. 2009;56:220–231. doi: 10.1016/j.yhbeh.2009.05.001. [DOI] [PubMed] [Google Scholar]
- de Jong TR, Measor KR, Chauke M, Harris BN, Saltzman W. Brief pup exposure induces Fos expression in the lateral habenula and serotonergic caudal dorsal raphe nucleus of paternally experienced male California mice (Peromyscus californicus) Neuroscience. 2010;169:1094–1104. doi: 10.1016/j.neuroscience.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Dewey MJ, Dawson WD. Deer mice: “the Drosophila of North American Mammalogy”. Genesis. 2001;29:105–109. doi: 10.1002/gene.1011. [DOI] [PubMed] [Google Scholar]
- Dice LR. The occurrence of two subspecies of the same species in the same area. Journal of Mammalogy. 1931;12:210–213. doi: 10.2307/1373867. [DOI] [Google Scholar]
- Dice LR. Longevity in Peromyscus maniculatus gracilis. Journal of Mammalogy. 1933;14:147–148. doi: 10.2307/1374020. [DOI] [Google Scholar]
- Dobzhansky T. Genetics and the Origin of Species. New York: Columbia University Press; 1937. [Google Scholar]
- Dragoo JW, Lackey JA, Moore KE, Lessa EP, Cook JA, Yates TL. Phylogeography of the deer mouse (Peromyscus maniculatus) provides a predictive framework for research on hantaviruses. Journal of General Virology. 2006;87:1997–2003. doi: 10.1099/vir.0.81576-0. [DOI] [PubMed] [Google Scholar]
- Drever MC, Blight LK, Hobson KA, Bertram DF. Predation on seabird eggs by Keen's mice (Peromyscus keeni): using stable isotopes to decipher the diet of a terrestrial omnivore on a remote offshore island. Canadian Journal of Zoology. 2000;78:2010–2018. doi: 10.1139/z00-131. [DOI] [Google Scholar]
- Emmons E. Report on the Quadrupeds of Massachusetts. Cambridge: Folsom, Wells, and Thurston; 1840. [Google Scholar]
- Fisher HS, Hoekstra HE. Competition drives cooperation among closely related sperm of deer mice. Nature. 2010;463:801–803. doi: 10.1038/nature08736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick MJ, Ben-Shahar Y, Smid HM, Vet LE, Robinson GE, Sokolowski MB. Candidate genes for behavioural ecology. Trends in Ecology & Evolution. 2005;20:96–104. doi: 10.1016/j.tree.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Foltz DW. Genetic evidence for long-term monogamy in a small rodent, Peromyscus polionotus. American Naturalist. 1981;117:665–675. doi: 10.1086/283751. [DOI] [Google Scholar]
- Gage KL, Kosoy MY. Natural history of plague: perspectives from more than a century of research. Annual Review of Entomology. 2005;50:505–528. doi: 10.1146/annurev.ento.50.071803.130337. [DOI] [PubMed] [Google Scholar]
- Galea LA, Kavaliers M, Ossenkopp KP. Sexually dimorphic spatial learning in meadow voles Microtus pennsylvanicus and deer mice Peromyscus maniculatus. Journal of Experimental Biology. 1996;199:195–200. doi: 10.1242/jeb.199.1.195. [DOI] [PubMed] [Google Scholar]
- Gentry JB, Smith MH. Food habits and burrow associates of Peromyscus polionotus. Journal of Mammalogy. 1968;49:562–565. doi: 10.2307/1378235. [DOI] [Google Scholar]
- Gloger CW. Gemeinnütziges Hand-und Hilfsbuch der Naturgeschichte 1841 [Google Scholar]
- Grieco TM, Rizk OT. Cranial shape varies along an elevation gradient in Gambel's white-footed mouse (Peromyscus maniculatus gambelii) in the Grinnell Resurvey Yosemite transect. Journal of Morphology. 2010;271:897–909. doi: 10.1002/jmor.10839. [DOI] [PubMed] [Google Scholar]
- Gubernick DJ, Alberts JR. The biparental care system of the California mouse, Peromyscus californicus. Journal of Comparative Psychology. 1987;101:169–177. doi: 10.1037/0735-7036.101.2.169. [DOI] [PubMed] [Google Scholar]
- Haldane JB. The theory of a cline. Journal of Genetics. 1948;48:277–284. doi: 10.1007/BF02986626. [DOI] [PubMed] [Google Scholar]
- Hall ER. The Mammals of North America. Volume 2. New York: Wiley; 1981. [Google Scholar]
- Haigh GR. Effects of inbreeding and social factors on the reproduction of young female Peromyscus maniculatus bairdii. Journal of Mammalogy. 1983;64:48–54. doi: 10.2307/1380749. [DOI] [Google Scholar]
- Harris SE, Munshi-South J, Obergfell C, O'Neill R. Signatures of rapid evolution in urban and rural transcriptomes of white-footed mice (Peromyscus leucopus) in the New York metropolitan area. PLOS ONE. 2013;8:1–19. doi: 10.1371/journal.pone.0074938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heideman PD, Bruno TA, Singley JW, Smedley JV. Genetic variation in photoperiodism in Peromyscus leucopus: geographic variation in an alternative life-history strategy. Journal of Mammalogy. 1999;80:1232–1242. doi: 10.2307/1383173. [DOI] [Google Scholar]
- Heideman PD, Pittman JT. Microevolution of neuroendocrine mechanisms regulating reproductive timing in Peromyscus leucopus. Integrative and Comparative Biology. 2009;49:550–562. doi: 10.1093/icb/icp014. [DOI] [PubMed] [Google Scholar]
- Hock RJ. Physiological responses of deer mice to various native altitudes. In: Wiehe WH, editor. The Physiological Effects of High Altitude. New York: Macmillan; 1964. [Google Scholar]
- Hoekstra HE, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science. 2006;313:101–104. doi: 10.1126/science.1126121. [DOI] [PubMed] [Google Scholar]
- Hooper ET. Classification. In: King JA, editor. Biology of Peromyscus (Rodentia) American Society of Mammalogists; 1968. [Google Scholar]
- Howell AH. Description of a new species of beach mouse from Florida. Journal of Mammalogy. 1920;1:237–240. doi: 10.2307/1373248. [DOI] [Google Scholar]
- Jašarević E, Sieli PT, Twellman EE, Welsh TH, Schachtman TR, Roberts RM, Geary DC, Rosenfeld CS. Disruption of adult expression of sexually selected traits by developmental exposure to bisphenol A. Proceedings of the National Academy of Sciences of USA. 2011;108:11715–11720. doi: 10.1073/pnas.1107958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner CP, Myrick LC, Crossland JP, Dawson WD. Deer mice as laboratory animals. ILAR Journal. 1998;39:322–330. doi: 10.1093/ilar.39.4.322. [DOI] [PubMed] [Google Scholar]
- Kalcounis-Rueppell MC, Petric R, Briggs JR, Carney C, Marshall MM, Willse JT, Rueppell O, Ribble DO, Crossland JP. Differences in ultrasonic vocalizations between wild and laboratory California mice (Peromyscus californicus) PLOS ONE. 2010;5:e9705. doi: 10.1371/journal.pone.0009705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkvik HM, Stout IJ, Doonan TJ, Parkinson CL. Investigating niche and lineage diversification in widely distributed taxa: phylogeography and ecological niche modeling of the Peromyscus maniculatus species group. Ecography. 2012;35:54–64. doi: 10.1111/j.1600-0587.2011.06994.x. [DOI] [Google Scholar]
- Kenney-Hunt J, Lewandowski A, Glenn TC, Glenn JL, Tsyusko OV, O'Neill RJ, Brown J, Ramsdell CM, Nguyen Q, Phan T, Shorter KR, Dewey MJ, Szalai G, Vrana PB, Felder MR. A genetic map of Peromyscus with chromosomal assignment of linkage groups (a Peromyscus genetic map) Mammalian Genome. 2014;25:160–179. doi: 10.1007/s00335-014-9500-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JA, editor. Biology of Peromyscus (Rodentia) American Society of Mammalogists; 1968. [Google Scholar]
- Kirkland GL, Layne JN, editors. Advances in the Study of Peromyscus (Rodentia) Lubbock: Texas Tech University Press; 1989. [Google Scholar]
- Lacy RC, Alaks G, Walsh A. Hierarchical analysis of inbreeding depression in Peromyscus polionotus. Evolution. 1996;50:2187–2200. doi: 10.2307/2410690. [DOI] [PubMed] [Google Scholar]
- Lee AW, Brown RE. Comparison of medial preoptic, amygdala, and nucleus accumbens lesions on parental behavior in California mice (Peromyscus californicus) Physiology & Behavior. 2007;92:617–628. doi: 10.1016/j.physbeh.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Linnaeus C. Systema Naturae. 10th edition. Stockholm: Laurentius Salvius; 1758. [Google Scholar]
- Linnen CR, Poh YP, Peterson BK, Barrett RD, Larson JG, Jensen JD, Hoekstra HE. Adaptive evolution of multiple traits through multiple mutations at a single gene. Science. 2013;339:1312–1316. doi: 10.1126/science.1233213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoGiudice K, Duerr ST, Newhouse MJ, Schmidt KA, Killilea ME, Ostfeld RS. Impact of host community composition on Lyme disease risk. Ecology. 2008;89:2841–2849. doi: 10.1890/07-1047.1. [DOI] [PubMed] [Google Scholar]
- LoGiudice K, Ostfeld RS, Schmidt KA, Keesing F. The ecology of infectious disease: effects of host diversity and community composition on Lyme disease risk. Proceedings of the National Academy of Sciences of USA. 2003;100:567–571. doi: 10.1073/pnas.0233733100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-González C, Correa-Ramírez MM, García-Mendoza DF. Phylogeography of Peromyscus schmidlyi: an endemic of the Sierra Madre Occidental, Mexico. Journal of Mammalogy. 2014;95:254–268. doi: 10.1644/13-MAMM-A-166. [DOI] [Google Scholar]
- Luttich S, Rusch DH, Meslow EC, Keith LB. Ecology of red-tailed hawk predation in Alberta. Ecology. 1970;51:190–203. doi: 10.2307/1933655. [DOI] [Google Scholar]
- MacManes MD, Eisen MB. Characterization of the transcriptome, nucleotide sequence polymorphism, and natural selection in the desert adapted mouse Peromyscus eremicus. PeerJ. 2014;2:e642. doi: 10.7717/peerj.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JS. Reproduction and development. In: Kirkland GL, Layne JN, editors. Advances in the Study of Peromyscus (Rodentia) Lubbock: Texas Tech University Press; 1989. [Google Scholar]
- Montgomery WI. Peromyscus and Apodemus: patterns of similarity in ecological equivalents. In: Kirkland GL, Layne JN, editors. Advances in the Study of Peromyscus (Rodentia) Lubbock: Texas Tech University Press; 1989. [Google Scholar]
- Moritz C, Patton JL, Conroy CJ, Parra JL, White GC, Beissinger SR. Impact of a century of climate change on small-mammal communities in Yosemite National Park, USA. Science. 2008;322:261–264. doi: 10.1126/science.1163428. [DOI] [PubMed] [Google Scholar]
- Munshi-South J, Kharchenko K. Rapid, pervasive genetic differentiation of urban white-footed mouse (Peromyscus leucopus) populations in New York City. Molecular Ecology. 2010;19:4242–4254. doi: 10.1111/j.1365-294X.2010.04816.x. [DOI] [PubMed] [Google Scholar]
- Munshi-South J, Nagy C. Urban park characteristics, genetic variation, and historical demography of white-footed mouse (Peromyscus leucopus) populations in New York City. PeerJ. 2014;2:e310. doi: 10.7717/peerj.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser GG, Carleton MD. Peromyscus. In: Wilson DE, Reeder DM, editors. Mammal Species of the World: a Taxonomic and Geographic Reference. 3rd edition. Baltimore: Johns Hopkins University Press; 2005. [Google Scholar]
- Natarajan C, Hoffmann FG, Lanier HC, Wolf CJ, Zachary A, Spangler ML, Weber RE, Fago A, Storz JF. Intraspecific polymorphism, interspecific divergence, and the origins of function-altering mutations in deer mouse hemoglobin. Molecular Biology and Evolution. 2015;32:978–997. doi: 10.1093/molbev/msu403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan C, Inoguchi N, Weber RE, Fago A, Moriyama H, Storz JF. Epistasis among adaptive mutations in deer mouse hemoglobin. Science. 2013;340:1324–1327. doi: 10.1126/science.1236862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osgood WH. Revision of the mice of the American genus Peromyscus. North American Fauna. 1909;28:1–285. doi: 10.3996/nafa.28.0001. [DOI] [Google Scholar]
- Pedersen AB, Antonovics J. Anthelmintic treatment alters the parasite community in a wild mouse host. Biology Letters. 2013;9:20130205. doi: 10.1098/rsbl.2013.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pergams OR, Lacy RC. Rapid morphological and genetic change in Chicago-area Peromyscus. Molecular Ecology. 2008;17:450–463. doi: 10.1111/j.1365-294X.2007.03517.x. [DOI] [PubMed] [Google Scholar]
- Prendergast BJ, Nelson RJ. Spontaneous ‘regression’ of enhanced immune function in a photoperiodic rodent Peromyscus maniculatus. Proceedings of the Royal Society B. 2001;268:2221–2228. doi: 10.1098/rspb.2001.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provine WB. Sewall Wright and Evolutionary Biology. Chicago: The University of Chicago Press; 1986. [Google Scholar]
- Ramamoorthi N, Narasimhan S, Pal U, Bao F, Yang XF, Fish D, Anguita J, Norgard MV, Kantor FS, Anderson JF, Koski RA, Fikrig E. The Lyme disease agent exploits a tick protein to infect the mammalian host. Nature. 2005;436:573–577. doi: 10.1038/nature03812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribble DO. The monogamous mating system of Peromyscus californicus as revealed by DNA fingerprinting. Behavioral Ecology & Sociobiology. 1991;29:161–166. doi: 10.1007/BF00166397. [DOI] [Google Scholar]
- Riddle BR, Hafner DJ, Alexander LF. Phylogeography and systematics of the Peromyscus eremicus species group and the historical biogeography of North American warm regional deserts. Molecular Phylogenetics and Evolution. 2000;17:145–160. doi: 10.1006/mpev.2000.0841. [DOI] [PubMed] [Google Scholar]
- Rosenfeld CS. Bisphenol A and phthalate endocrine disruption of parental and social behaviors. Frontiers in Neuroscience. 2015;9:1–15. doi: 10.3389/fnins.2015.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe KC, Rowe KM, Tingley MW, Koo MS, Patton JL, Conroy CJ, Perrine JD, Beissinger SR, Moritz C. Spatially heterogeneous impact of climate change on small mammals of montane California. Proceedings of the Royal Society B. 2014;282:20141857. doi: 10.1098/rspb.2014.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher GA, Hart RW. Longevity, aging and comparative cellular and molecular biology of the house mouse, Mus musculus, and the white-footed mouse, Peromyscus leucopus. Birth Defects Original Article Series. 1978;14:71–96. [PubMed] [Google Scholar]
- Schwanz LE, Voordouw MJ, Brisson D, Ostfeld RS. Borrelia burgdorferi has minimal impact on the Lyme disease reservoir host Peromyscus leucopus. Vector Borne and Zoonotic Diseases. 2011;11:117–124. doi: 10.1089/vbz.2009.0215. [DOI] [PubMed] [Google Scholar]
- Shi Y, Pulliam DA, Liu Y, Hamilton RT, Jernigan AL, Bhattacharya A, Sloane LB, Qi W, Chaudhuri A, Buffenstein R, Ungvari Z, Austad SN, Van Remmen H. Reduced mitochondrial ROS, enhanced antioxidant defense, and distinct age-related changes in oxidative damage in muscles of long-lived Peromyscus leucopus. American Journal of Physiology. Regulatory, Integrative and Comparative Physiology. 2013;304:R343–R355. doi: 10.1152/ajpregu.00139.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter KR, Crossland JP, Webb D, Szalai G, Felder MR, Vrana PB. Peromyscus as a mammalian epigenetic model. Genetics Research International. 2012:179159. doi: 10.1155/2012/179159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MH. University of Florida; 1966. The evolutionary significance of certain behavioral, physiological, and morphological adaptations of the old-field moue, Peromyscus polionotus. (Unpublished doctoral dissertation) [Google Scholar]
- Snyder LR. Deer mouse hemoglobins: is there genetic adaptation to high altitude? BioScience. 1981;31:299–304. doi: 10.2307/1308147. [DOI] [Google Scholar]
- Sohal RS, Ku HH, Agarwal S. Biochemical correlates of longevity in two closely related rodent species. Biochemical and Biophysical Research Communications. 1993;196:7–11. doi: 10.1006/bbrc.1993.2208. [DOI] [PubMed] [Google Scholar]
- Steiner CC, Weber JN, Hoekstra HE. Adaptive variation in beach mice produced by two interacting pigmentation genes. PLOS Biology. 2007;5:e219. doi: 10.1371/journal.pbio.0050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steppan S, Adkins R, Anderson J. Phylogeny and divergence-date estimates of rapid radiations in muroid rodents based on multiple nuclear genes. Systematic Biology. 2004;53:533–553. doi: 10.1080/10635150490468701. [DOI] [PubMed] [Google Scholar]
- Storz JF. Hemoglobin function and physiological adaptation to hypoxia in high-altitude mammals. Journal of Mammalogy. 2007;88:24–31. doi: 10.1644/06-MAMM-S-199R1.1. [DOI] [Google Scholar]
- Storz JF, Runck AM, Sabatino SJ, Kelly JK, Ferrand N, Moriyama H, Weber RE, Fago A. Evolutionary and functional insights into the mechanism underlying high-altitude adaptation of deer mouse hemoglobin. Proceedings of the National Academy of Sciences of USA. 2009;106:14450–14455. doi: 10.1073/pnas.0905224106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan J, Markert JA, Kilpatrick CW. Phylogeography and molecular systematics of the Peromyscus aztecus species group (Rodentia: Muridae) inferred using parsimony and likelihood. Systematic Biology. 1997;46:426–440. doi: 10.1093/sysbio/46.3.426. [DOI] [PubMed] [Google Scholar]
- Sumner FB. The role of isolation in the formation of a narrowly localized race of deer-mice (Peromyscus) American Naturalist. 1917;51:173–185. doi: 10.1086/279595. [DOI] [Google Scholar]
- Sumner FB. Continuous and discontinuous variations and their inheritance in Peromyscus. American Naturalist. 1918;52:177–208. doi: 10.1086/279662. [DOI] [Google Scholar]
- Sumner FB. Longevity in Peromyscus. Journal of Mammalogy. 1922;3:79–81. doi: 10.2307/1373298. [DOI] [Google Scholar]
- Sumner FB. The analysis of a concrete case of intergradation between two subspecies. Proceedings of the National Academy of Sciences of USA. 1929;15:110–120. doi: 10.1073/pnas.15.2.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner FB. Genetic and distributional studies of three sub-species of Peromyscus. Journal of Genetics. 1930;23:275–376. doi: 10.1007/BF03052609. [DOI] [Google Scholar]
- Sun Y, Desierto MJ, Ueda Y, Kajigaya S, Chen J, Young NS. Peromyscus leucopus mice: a potential animal model for haematological studies. International Journal of Experimental Pathology. 2014;95:342–350. doi: 10.1111/iep.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terman RC. Population dynamics. In: King JA, editor. Biology of Peromyscus (Rodentia) American Society of Mammalogists; 1968. [Google Scholar]
- Trainor BC, Bird IM, Alday NA, Schlinger BA, Marler CA. Variation in aromatase activity in the medial preoptic area and plasma progesterone is associated with the onset of paternal behavior. Neuroendocrinology. 2003;78:36–44. doi: 10.1159/000071704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor BC, Lin S, Finy MS, Rowland MR, Nelson RJ. Photoperiod reverses the effects of estrogens on male aggression via genomic and nongenomic pathways. Proceedings of the National Academy of Sciences of USA. 2007;104:9840–9845. doi: 10.1073/pnas.0701819104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor BC, Martin LB, Greiwe KM, Kuhlman JR, Nelson RJ. Social and photoperiod effects on reproduction in five species of Peromyscus. General and Comparative Endocrinology. 2006;148:252–259. doi: 10.1016/j.ygcen.2006.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner LM, Young AR, Römpler H, Schöneberg T, Phelps SM, Hoekstra HE. Monogamy evolves through multiple mechanisms: evidence from V1aR in deer mice. Molecular Biology and Evolution. 2010;27:1269–1278. doi: 10.1093/molbev/msq013. [DOI] [PubMed] [Google Scholar]
- Updike J. Introduction. In: Yoe C, Morra-Yoe J, editors. The Art of Mickey Mouse. New York: Hyperion; 1991. [Google Scholar]
- Ungvari Z, Krasnikov BF, Csiszar A, Labinskyy N, Mukhopadhyay P, Pacher P, Cooper AJ, Podlutskaya N, Austad SN, Podlutsky A. Testing hypotheses of aging in long-lived mice of the genus Peromyscus: association between longevity and mitochondrial stress resistance, ROS detoxification pathways, and DNA repair efficiency. Age. 2008;30:121–133. doi: 10.1007/s11357-008-9059-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Zant JL, Wooten MC. Translocation of Choctawhatchee beach mice (Peromyscus polionotus allophrys): hard lessons learned. Biological Conservation. 2003;112:405–413. doi: 10.1016/S0006-3207(02)00338-5. [DOI] [Google Scholar]
- Vestal BM, Coleman WC, Chu PR. Age of first leaving the nest in two species of deer mice (Peromyscus) Journal of Mammalogy. 1980;61:143–146. doi: 10.2307/1379974. [DOI] [Google Scholar]
- Vignieri SN, Larson JG, Hoekstra HE. The selective advantage of crypsis in mice. Evolution. 2010;64:2153–2158. doi: 10.1111/j.1558-5646.2010.00976.x. [DOI] [PubMed] [Google Scholar]
- Vrana PB, Fossella JA, Matteson P, del Rio T, O'Neill MJ, Tilghman SM. Genetic and epigenetic incompatibilities underlie hybrid dysgenesis in Peromyscus. Nature Genetics. 2000;25:120–124. doi: 10.1038/75518. [DOI] [PubMed] [Google Scholar]
- Weber JN, Hoekstra HE. The evolution of burrowing behaviour in deer mice (genus Peromyscus) Animal Behaviour. 2009;77:603–609. doi: 10.1016/j.anbehav.2008.10.031. [DOI] [Google Scholar]
- Weber JN, Peterson BK, Hoekstra HE. Discrete genetic modules are responsible for complex burrow evolution in Peromyscus mice. Nature. 2013;493:402–405. doi: 10.1038/nature11816. [DOI] [PubMed] [Google Scholar]
- Whitaker JO. Parasites. In: King JA, editor. Biology of Peromyscus (Rodentia) American Society of Mammalogists; 1968. [Google Scholar]
- Williams SA, Jasarevic E, Vandas GM, Warzak DA, Geary DC, Ellersieck MR, Roberts RM, Rosenfeld CS. Effects of developmental bisphenol A exposure on reproductive-related behaviors in California mice (Peromyscus californicus): a monogamous animal model. PLOS ONE. 2013;8:17–19. doi: 10.1371/journal.pone.0055698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff JO. The effects of density, food, and interspecific interference on home range size in Peromyscus leucopus and Peromyscus maniculatus. Canadian Journal of Zoology. 1985;63:2657–2662. doi: 10.1139/z85-397. [DOI] [Google Scholar]
- Workman JL, Bowers SL, Nelson RJ. Enrichment and photoperiod interact to affect spatial learning and hippocampal dendritic morphology in white-footed mice (Peromyscus leucopus) European Journal of Neuroscience. 2009;29:161–170. doi: 10.1111/j.1460-9568.2008.06570.x. [DOI] [PubMed] [Google Scholar]
- Wright S. The roles of mutation, inbreeding, crossbreeding, and selection in evolution. Proceedings of the Sixth International Congress of Genetics. 1932;1:356–366. [Google Scholar]
- Yang DS, Conroy CJ, Moritz C. Contrasting responses of Peromyscus mice of Yosemite National Park to recent climate change. Global Change Biology. 2011;17:2559–2566. doi: 10.1111/j.1365-2486.2011.02394.x. [DOI] [Google Scholar]