In this review, Schmidt and Cech cover human telomerase biogenesis, trafficking, and activation, comparing key aspects with the analogous events in other species.
Keywords: biogenesis, cancer, recruitment, telomerase, telomere, trafficking
Abstract
Telomerase is the ribonucleoprotein enzyme that catalyzes the extension of telomeric DNA in eukaryotes. Recent work has begun to reveal key aspects of the assembly of the human telomerase complex, its intracellular trafficking involving Cajal bodies, and its recruitment to telomeres. Once telomerase has been recruited to the telomere, it appears to undergo a separate activation step, which may include an increase in its repeat addition processivity. This review covers human telomerase biogenesis, trafficking, and activation, comparing key aspects with the analogous events in other species.
Human chromosomes are capped by telomeres, repetitive DNA tracts bound by the six-protein shelterin complex. The shelterin complex prevents telomeres from being recognized as sites of DNA damage and recruits telomerase (Palm and de Lange 2008; Nandakumar and Cech 2013). Telomerase is an RNA-containing reverse transcriptase that adds telomeric repeat DNA to chromosome ends (Blackburn and Collins 2011). This prevents successive shortening of telomeres caused by the failure of the DNA replication machinery to duplicate the very end of each chromosome, termed the end replication problem (Levy et al. 1992). Once telomeres shrink to a critical length, cells enter replicative senescence or alternatively undergo programmed cell death, a major tumor-suppressive mechanism. Thus, continuously dividing cells such as germ cells, stem cells, and, importantly, most cancer cells require telomerase activity for survival (Stewart and Weinberg 2006; Shay and Wright 2011). The importance of telomerase in oncogenesis is highlighted by the recent identification of recurrent mutations in the promoter of the gene for the telomerase protein component hTERT (human telomerase reverse transcriptase) (Horn et al. 2013; Huang et al. 2013), the most frequent mutation in some cancer types (Heidenreich et al. 2014). These promoter mutations are associated with increased hTERT expression, telomerase activity, and telomere length (Borah et al. 2015). In the other direction, deficiencies in telomerase activity, its maturation, or recruitment to telomeres can lead to human diseases such as aplastic anemia and dyskeratosis congenita (Armanios and Blackburn 2012).
Chromosome ends in human cells consist of 5–15 kb of telomeric TTAGGG repeats. The majority of these repeats are double-stranded, but the very end of each chromosome is made up of a single-stranded 3′ overhang (Blackburn 2005). This single-stranded region is the substrate or primer for telomerase. This DNA anneals with the template region in the telomerase RNA (TR) component, allowing the TERT to synthesize a telomeric repeat (Cech 2004). Once a telomeric repeat has been added, the 3′ end of the chromosome is repositioned, and telomerase can synthesize additional repeats, a process called repeat addition processivity (RAP) (Greider 1991).
The double-stranded region of the human telomere is bound by TRF1 and TRF2, which specifically recognize the telomeric repeat sequence. TRF1 and TRF2 play critical roles in suppressing DNA damage signaling at the telomere and recruit the rest of the shelterin complex (Palm and de Lange 2008). TIN2 binds to TRF1 and TRF2 and serves as a linker between the double-stranded and single-stranded binding complexes of the telomere (Ye et al. 2004; Abreu et al. 2010). The shelterin component POT1 (Baumann and Cech 2001) specifically associates with the single-stranded 3′ overhang of the telomere, preventing it from being recognized by the DNA damage machinery (Denchi and de Lange 2007). POT1 is linked to the shelterin complex by TPP1, which in turn binds to TIN2 (Liu et al. 2004; Ye et al. 2004). In addition to promoting POT1 binding, TPP1 is required to recruit telomerase to the telomere (Xin et al. 2007; Abreu et al. 2010; Tejera et al. 2010). In summary, the six shelterin components integrate chromosome end protection and telomerase recruitment to allow continuous cell division in many cell types.
Telomere maintenance requires proper assembly of the protein and RNA components of telomerase into a ribonucleoprotein (RNP) as well as a number of cofactors involved in the maturation, stability, and subcellular localization of telomerase. This review is focused on the biogenesis, trafficking, and activation of human telomerase. We compare and contrast human telomerase with telomerases in other species, highlight recent findings, identify open questions, and describe new technologies that promise to advance our understanding of telomere biology.
Biogenesis of human telomerase
The human telomerase RNP is composed of the TR component (hTR), the TERT (hTERT), and the accessory proteins dyskerin, NOP10, NHP2, and GAR1. The formation of the telomerase RNP is a multistep process that requires additional cofactors. Mutations in many of the core telomerase RNP subunits as well as the maturation factors lead to telomerase deficiency diseases such as dyskeratosis congenita or aplastic anemia, and the resultant genome instability can be oncogenic (Armanios and Blackburn 2012).
TR synthesis and maturation
TRs are divergent in size and sequence between species, ranging from ∼150 nucleotides (nt) in ciliates to ∼1150 nt in Saccharomyces cerevisiae and >2000 nt in Neurospora crassa. While TR size and sequence are not highly conserved, four functional features are present in all TRs: the template for reverse transcription, the pseudoknot domain, a stem–loop that interacts with TERT, and a 3′ element required for RNA stability (Fig. 1A; Theimer and Feigon 2006).
Figure 1.
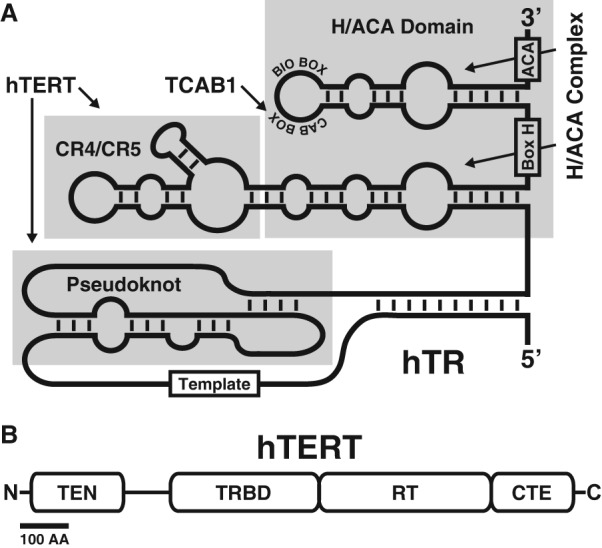
Functional domains of hTR and hTERT. (A) Secondary structure of the hTR. The pseudoknot and CR4/CR5 domains interact with the hTERT. The template for telomeric repeat synthesis is indicated. Two copies of the H/ACA complex, each composed of dyskerin, NHP2, NOP10, and GAR1, associate with the H/ACA domain of hTR to stabilize the RNA in the nucleus. TCAB1 interacts with the CAB box to facilitate telomerase localization to Cajal bodies. (B) hTERT contains four functional domains. The telomerase N-terminal (TEN) domain participates in catalysis and drives telomerase localization to telomeres. The TR-binding domain (TRBD) interacts with hTR. The reverse transcriptase (RT) and C-terminal extension (CTE) form the catalytic core of telomerase. Bar, 100 amino acids.
Fully mature hTR (451 nt) contains all of the functional features mentioned above (Zhang et al. 2011). Like other vertebrate TRs, the 3′ stabilizing element is an H/ACA motif consisting of two hairpins connected by a short single-stranded stretch, the H-box, and a terminal ACA region (Fig. 1A; Mitchell et al. 1999; Egan and Collins 2012a). Vertebrate TRs share this H/ACA motif with small nucleolar and small Cajal body (CB) RNAs (snoRNAs and scaRNAs) that associate with the cofactors dyskerin, NOP10, NHP2, and GAR1 (Kiss et al. 2010). H/ACA scaRNAs and snoRNAs function as guides, specifying the targets for the pseudouridylase dyskerin (Kiss et al. 2010). In contrast, the only known functions of the association between hTR and dyskerin are RNA stabilization and nuclear retention, and no pseudouridylation targets for hTR have been identified.
Unlike scaRNAs and snoRNAs, whose precursors are contained in intronic regions of mRNAs, hTR is transcribed from its own promoter by RNA polymerase II (Feng et al. 1995). The mature form of hTR is not polyadenylated; instead, its 3′ end is formed by exonucleolytic cleavage up to the boundary formed by the H/ACA domain, where further cleavage is prevented by dyskerin associated with the RNA (Fu and Collins 2003). The tetrameric complex of dyskerin, NOP10, NHP2, and the chaperone NAF1 binds to hTR cotranscriptionally (Darzacq et al. 2006). Rapid association of this complex with hTR is crucial for hTR accumulation. The importance of the formation of the complex between hTR and dyskerin, NOP10, NHP2, and NAF1 is highlighted by a number of telomerase deficiency diseases associated with mutations in these factors (Armanios and Blackburn 2012). These mutations are thought to affect either binding of these proteins to hTR or the formation of a dyskerin–NOP10–NHP2–NAF1 complex capable of binding to hTR, either of which would cause reduced levels of hTR and thus of telomerase activity.
In addition to the canonical functional features described above, hTR contains more specialized sequence elements in the terminal loop of its 3′ hairpin (Fig. 1A). The BIO-box stimulates hTR stability by promoting H/ACA RNP formation (Egan and Collins 2012b). The CAB-box, a motif that hTR shares with the scaRNAs, is required for trafficking hTR to the CB, where it receives its TMG cap, and NAF1 is replaced by GAR1 (Jády et al. 2004; Girard et al. 2008; Kiss et al. 2010).
hTERT
In most organisms, the TERT protein contains four functional domains: the telomerase N-terminal (TEN) domain, TR-binding domain (TRBD), reverse transcriptase domain, and C-terminal extension (CTE) (Fig. 1B; Blackburn and Collins 2011). In the red flour beetle Tribolium castaneum, for which the only high-resolution structure of TERT has been solved, the TEN domain appears to be missing (Gillis et al. 2008). In hTERT, the TEN domain plays a key role in telomerase recruitment to telomeres and participates in the catalysis of telomeric repeat synthesis (Jurczyluk et al. 2011; Robart and Collins 2011; Schmidt et al. 2014; Wu and Collins 2014). The TRBD and reverse transcriptase domain provide the key interaction sites with hTR (Lai et al. 2001). The reverse transcriptase domain contains the active site of the telomerase enzyme, with clear homology with the reverse transcriptase domains of retrotransposon and retroviral reverse transcriptases (Lingner et al. 1997; Nakamura et al. 1997). The CTE is part of the reverse transcriptase, and previous proposals of an additional role in telomerase localization to telomeres require re-evaluation as discussed below.
Telomerase assembly
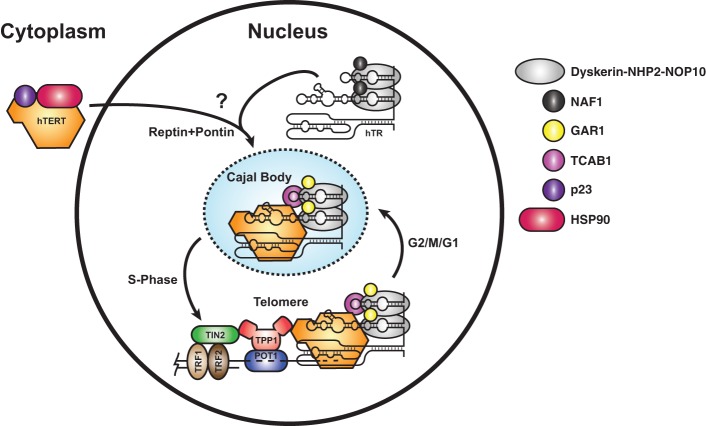
Several reports have indicated that hTERT associates with the chaperones HSP90 and p23 as well as the AAA+ ATPases Pontin and Reptin (Fig. 2; Holt et al. 1999; Venteicher et al. 2008). Chemical inhibition of HSP90 reduces telomerase activity. Interestingly, immunoprecipitation of both HSP90 and p23 enrich for active telomerase, indicating that they interact with mature telomerase RNPs (Holt et al. 1999). Knockdown of Pontin reduces telomerase activity as well as hTR levels (Venteicher et al. 2008). In contrast to HSP90 and p23, Reptin and Pontin seem to associate with a pool of hTERT that is not incorporated into an enzymatically active telomerase RNP, hinting at a potential role in telomerase assembly (Venteicher et al. 2008). In addition, Reptin and Pontin have also been shown to interact with dyskerin and are necessary for H/ACA RNP assembly, a crucial process required for hTR stability (King et al. 2001; Venteicher et al. 2008). It is therefore not clear whether the loss of telomerase activity after Pontin depletion is caused by reduced hTR levels, disrupted telomerase assembly, or a combination of both. Similarly, interpreting the requirement of HSP90 for telomerase maturation is complicated by the fact that chemical inhibition of a key chaperone in human cells potentially has pleiotropic and indirect effects.
Figure 2.
Telomerase assembly, maturation, and recruitment to telomeres. hTERT is synthesized in the cytoplasm and associates with the chaperones HSP90 and p23. hTR cotranscriptionally binds dyskerin, NOP10, NHP2, and NAF1, with NAF1 subsequently being replaced by GAR1. Assembly of hTR and hTERT into catalytically active telomerase is aided by the AAA+ ATPases Reptin and Pontin. A question mark indicates that the subcellular location of assembly is still under investigation. Telomerase is recruited to CBs by its interaction with TCAB1. In S phase of the cell cycle, telomerase is recruited to telomeres by the interaction of the TEN domain of hTERT with the TEL (TPP1 glutamate [E] and leucine [L]-rich) patch of the OB-fold domain of the shelterin component TPP1.
hTERT contains putative nuclear localization signal (NLS) and nuclear export signal (NES), suggesting that nuclear–cytoplasmic shuttling of hTERT could be involved in telomerase assembly or activity regulation (Fig. 2; Chung et al. 2012). It is noteworthy that mutation of the hTERT NLS has only minimal effects on telomerase activity and cellular immortalization by hTERT, while the loss of the NES on telomerase function has yet to be tested. Future studies are needed to address the importance of nuclear–cytoplasmic trafficking of hTERT for telomerase maturation and activity.
In S. cerevisiae, telomerase assembly is thought to occur in the cytoplasm, thereby requiring export of the TR out of the nucleus (Gallardo et al. 2008). Import of the assembled telomerase RNP into the nucleus requires the association of the Ku70/80 heterodimer and presumably the Sm heteroheptamer with TLC1, the budding yeast TR (Seto et al. 1999; Gallardo et al. 2008). Each of these protein complexes has a specific binding site on the TLC1 RNA (Seto et al. 1999, 2002; Stellwagen et al. 2003). Furthermore, a recent study has demonstrated that the assembly state of the budding yeast telomerase holoenzyme is regulated in a cell cycle-dependent manner (Tucey and Lundblad 2014). Est3, a telomerase subunit required for full enzymatic activity, associates with a preformed Est2/TERT–Est1–TLC1 complex at the G2/M phase of the cell cycle, after the completion of DNA replication (Morris and Lundblad 1997; Tucey and Lundblad 2014). The active telomerase complex then disassembles by dissociation of Est2, the yeast TERT (Tucey and Lundblad 2014). Curiously, yeast Est3 is a structural homolog of the human TPP1 telomere protein, but how they might perform homologous functions is difficult to understand at this point (Lee et al. 2008; Yu et al. 2008; Rao et al. 2014). Human proteins related in sequence to Est1 have been identified and shown to associate with telomerase activity (Reichenbach et al. 2003; Snow et al. 2003). It must be noted that these proteins (hEST1A–C) are also involved in the nonsense-mediated decay pathway, and how they may promote telomerase activity is unclear (Redon et al. 2007).
In contrast to yeast, the site of telomerase assembly, the sequential steps, and its cell cycle-dependent regulation are only beginning to be investigated in human cells. In the absence of hTERT, hTR has a diffuse nuclear localization, presumably stabilized by the dyskerin complex associated with its 3′ domain (Tomlinson et al. 2008). However, whether hTR shuttles out of the nucleus to assemble with hTERT or hTERT is imported into the nucleus for assembly is unknown. hTERT transcription and activity peak in S phase (Zhu et al. 1996; Xi and Cech 2014), and telomerase recruitment to telomeres is restricted to S phase (Jády et al. 2006; Tomlinson et al. 2006). Thus, similar to yeast, telomerase action occurs after completion of DNA replication, but the regulatory paradigms underlying these observations remain to be determined.
A major hurdle in analyzing the assembly of human telomerase and its spatiotemporal regulation has been the inability to detect endogenous hTERT by Western blot or microscopy approaches due to its low abundance (∼500 molecules per cell in cancer cells) (Xi and Cech 2014). Overexpression of telomerase components has been shown to disregulate its subcellular localization and is therefore not the ideal approach for studying telomerase assembly and localization (Zhong et al. 2012). With the advent of CRISPR-mediated genome editing and superresolution microscopy techniques with single-molecule sensitivity, it is only a matter of time until endogenous hTERT can be reliably detected to study telomerase assembly and its cell cycle-dependent regulation in human cells.
Structural organization of the telomerase RNP
hTERT interacts with two structural elements of hTR. The TRBD associates with the CR4/5 region in the penultimate hairpin of hTR (Fig. 1; Mitchell and Collins 2000). In addition, the reverse transcriptase domain has been shown to associate with the pseudoknot region of hTR, but the affinity is thought to be substantially weaker than the TRBD–CR4/5 interaction (Robart and Collins 2011). Recently, the structure of the TRBD bound to the CR4/5 region of TR from the teleost fish Oryzias latipes was determined by X-ray crystallography, demonstrating that the TRBD associates with the P6 and P6.1 stems as well a highly conserved adenosine at the junction between the two loops (Huang et al. 2014).
High-resolution structures are available for several structural elements of TRs as well as individual domains of TERT from diverse organisms (Zhang et al. 2011; Nandakumar and Cech 2013). The TEN domain and TRBD of Tetrahymena thermophila TERT have been solved by X-ray crystallography (Jacobs et al. 2006; Rouda and Skordalakes 2007). Additional TRBD structures are available from Takifugu rubripes (the Japanese pufferfish), O. latipes (see above), and T. castaneum, the last of which was solved in the context of the full reverse transcriptase subunit (Gillis et al. 2008; Harkisheimer et al. 2013; Huang et al. 2014). As a cautionary note, the T. castaneum TR subunit has yet to be discovered; thus, it is not proven that the putative T. castaneum TERT is a bona fide telomerase reverse transcriptase.
Lower-resolution information about the full holoenzyme is available from electron microscopy structures of the human and T. thermophila telomerase RNPs. The human telomerase RNP purified from HEK293T cells overexpressing hTERT and hTR forms a dimeric structure ∼28 nm in length (Sauerwald et al. 2013). The dimerization is proposed to be mediated by the 3′ end of hTR and possibly the H/ACA motif-associated proteins dyskerin, NOP10, NHP2, and GAR1. While other studies agree that multiple copies of dyskerin, NOP10, NHP2, and GAR1 associate with the human telomerase RNP, the presence of two catalytically active hTERT subunits has been controversial (Egan and Collins 2010). Additionally, the biological significance of a dimeric telomerase RNP is unclear. It could perhaps facilitate telomerase recruitment to telomeres by providing multiple binding sites, thus increasing the affinity for its telomeric receptor. Furthermore, it has been proposed that an enzymatically inactive hTERT subunit can exert an inhibitory effect on the activity of the second hTERT molecule in the dimer (Sauerwald et al. 2013). Such allosteric regulation could have substantial implications for disease-associated hTERT mutations that reduce its catalytic activity, potentially causing a dominant-negative loss of function in a heterozygous setting.
The structure of T. thermophila telomerase was solved by electron microscopy to a resolution of ∼25 Å in the presence of the accessory subunits p65, p50, Teb1 and the 7-1-4 complex, composed of p75, p45, and p19 (Jiang et al. 2013). In contrast to human telomerase, the T. thermophila RNP is monomeric (Bryan et al. 2003). Mapping of high-resolution structures of holoenzyme components into the electron microscopy density revealed several key features about the organization of T. thermophila telomerase. The cofactors p50 and Teb1, which stimulate both RAP and telomerase activity, are closely associated with the presumed position of the TEN domain of tTERT (Min and Collins 2009; Hong et al. 2013; Jiang et al. 2013). Teb1 contains three OB-fold domains, A, B, and C, which are necessary to stimulate telomerase activity (Min and Collins 2010). Teb1A and Teb1B have been shown to specifically associate with telomeric ssDNA, an activity comparable with that of POT1 in the human system (Min and Collins 2009; Zeng et al. 2011). The most C-terminal OB-fold, Teb1C, does not associate with DNA but seems to mediate the interaction of Teb1 with the TEN domain of telomerase, comparable with the OB-fold of TPP1 (Min and Collins 2010; Jiang et al. 2013; Schmidt et al. 2014). Importantly, p50 stabilizes the interaction of Teb1 with telomerase and also facilitates the interaction of the 7-1-4 complex with the T. thermophila RNP (Jiang et al. 2013).
Telomerase localization to CBs
Telomerase localizes to CBs for most of the cell cycle. While the factors and molecular determinants that drive telomerase localization to CBs in human cells are well defined, the physiological relevance of the process is only starting to be understood.
Telomerase is targeted to CBs by the CAB-box in the terminal hairpin of hTR and requires its association with hTERT (Fig. 1A; Richard et al. 2003; Tomlinson et al. 2008). Like all scaRNAs, hTR localization to CBs is driven by its interaction with the protein TCAB1/WDR-79, which resides in CBs and associates with the CAB-box (Fig. 2; Venteicher et al. 2009). The physiological importance of the TCAB1 for telomerase function is highlighted by autosomal recessive TCAB1 mutations that are associated with telomerase deficiencies in human patients (Zhong et al. 2011). Interestingly, missense mutations in TCAB1 that cause dyskeratosis congenita do not disrupt its interaction with hTR but rather prevent proper TCAB1 folding by the chaperonin TRiC (Freund et al. 2014). While these observations clearly demonstrate the importance of TCAB1 for telomere maintenance, TCAB1 is not required for the enzymatic activity of telomerase, ruling out an obligatory role of CBs in telomerase assembly (Venteicher et al. 2009). Furthermore, even though several steps of hTR maturation are thought to occur in CBs (see above), loss of TCAB1 function has no effect on steady-state hTR levels (Venteicher et al. 2009). To add more intrigue to the puzzle, telomerase localization to telomeres has a differential requirement for CBs and TCAB1. Eliminating CBs by depletion of coilin prevents telomerase recruitment to telomeres, but this phenotype can be rescued by overexpression of telomerase (Stern et al. 2012). In contrast, in the absence of TCAB1, not even telomerase overexpression can force telomerase localization to telomeres (Stern et al. 2012). In total, these observations suggest that CBs in general and specifically TCAB1 make a major contribution to telomerase trafficking and recruitment to telomeres but are not obligatory for telomerase assembly and enzymatic activity.
Different models for the role of CBs in telomerase trafficking have been proposed. Live-cell microscopy has shown that telomeres transiently associate with CBs during S phase of the cell cycle, potentially to deliver telomerase to telomeres by a handoff mechanism (Jády et al. 2006). Such a mechanism would restrict the number of telomeres that can be elongated in each cell cycle and would require some sort of active transport machinery that could deliver telomeres to CBs or vice versa. It is worth noting that hTR is frequently found at telomeres, which are clearly distinct and far apart from CBs (Jády et al. 2006). Alternatively, telomerase could be released from CBs in S phase to allow it to traffic to telomeres. Localization of telomerase to telomeres and CBs is not mutually exclusive. Overexpression of telomerase triggers its localization, along with that of most CB components, to virtually every telomere in the cell, most likely overriding the regulatory processes that restrict telomerase–telomere recruitment to S phase (Zhong et al. 2012). Curiously, TCAB1 is required for overexpressed telomerase to localize to telomeres (Stern et al. 2012). TCAB1 shares this phenotype with the TEL patch of TPP1 and the TEN domain of hTERT, which directly mediate the interaction of telomerase with the telomere (Nandakumar et al. 2012; Zhong et al. 2012; Schmidt et al. 2014; see below). It is therefore tempting to speculate that TCAB1 is also involved in mediating the interaction between telomerase and the telomere. Alternatively, TCAB1 might be required to facilitate the post-translational modification of telomerase to allow it to localize to telomeres. Future studies using superresolution or live-cell imaging to detect hTERT molecules at endogenous expression levels will be necessary to address these fundamental questions regarding the role of CBs in telomerase trafficking.
Telomerase recruitment to telomeres
Although most enzymes encounter their substrates by simple diffusion, both telomerase and its chromosome end substrate have very low abundance: ∼30 telomerases/64 telomeres in a yeast haploid cell in late S phase and ∼250 telomerases/184 telomeres in a human cell in late S phase (Mozdy and Cech 2006; Xi and Cech 2014). Thus, it is perhaps not surprising that the telomerase enzyme is recruited to telomeres rather than simply encountering them by diffusion. The yeast pathway in which the telomerase accessory subunit Est1 binds to the telomere protein Cdc13 was worked out by Lundblad and colleagues (Pennock et al. 2001). In retrospect, the first evidence for human telomerase recruitment was DAT (dissociates activities of telomerase) mutants of TERT identified in the Counter laboratory (Armbruster et al. 2001, 2004) that were catalytically active in vitro but unable to act in vivo. However, the molecular understanding of DAT mutants had to await the discovery of the human telomeric end-capping proteins.
The shelterin component TPP1 is the key telomeric component necessary for telomerase recruitment to telomeres (Fig. 2; Xin et al. 2007). In addition, TPP1 in complex with POT1 stimulates telomerase RAP in vitro and has therefore been proposed to be a processivity factor for telomerase action at telomeres (Wang et al. 2007). TPP1 is composed of an N-terminal OB-fold domain required for telomerase recruitment, a central domain that directly binds to POT1, and a C-terminal domain necessary for its association with TIN2, which integrates both TPP1 and POT1 into the shelterin complex (Houghtaling et al. 2004; Liu et al. 2004; Ye et al. 2004). Mutational analysis of the TPP1 OB-fold domain has revealed a patch of amino acids (termed the TEL patch) that is essential for telomerase recruitment to telomeres in vivo and telomerase RAP stimulation in vitro (Nandakumar et al. 2012; Sexton et al. 2012; Zhong et al. 2012). The TEL patch is thought to directly interact with telomerase (Schmidt et al. 2014). In addition, a recent study has described a single amino acid deletion in the TEL patch (TPP1K170Δ) associated with Hoyeraal-Hreidarsson syndrome, a severe form of dyskeratosis congenita (Kocak et al. 2014). Similar to other TEL patch mutations, TPP1K170Δ fails to recruit telomerase to telomeres in vivo and displays reduced stimulation of telomerase RAP in vitro (Kocak et al. 2014).
Several DAT mutations were described in the TEN domain of hTERT (N-DAT), which had enzymatic activity but failed to immortalize human fibroblasts (Armbruster et al. 2001). Because fusion of one of the N-DAT mutants to POT1 rescued immortalization, it was proposed that the DAT mutants were defective in telomere maintenance because they were unable to localize to telomeres (Armbruster et al. 2004). Additional mutants in the TEN domain were later identified that affected telomerase localization to telomeres in vivo (V144M, a dyskeratosis congenita-associated allele) or were defective in RAP stimulation by TPP1 in vitro (G100V) (Zaug et al. 2010; Zhong et al. 2012; Schmidt et al. 2014).
The identification of N-DAT mutants did not rule out the presence of a bridging factor between the TEL patch of TPP1 and the TEN domain of telomerase. This hurdle was overcome in a recent study that identified mutations in the TEN domain of hTERT (K78E) and the TEL patch of TPP1 (E215K) that were individually detrimental to telomerase recruitment in vivo and telomerase RAP stimulation by TPP1–POT1 in vitro but rescued both phenotypes when combined (Schmidt et al. 2014). These mutations invert the charges at individual positions in the TEN domain and the TEL patch; therefore, the likely explanation for these observations is that K78 in the TEN domain and E215 in the TEL patch are involved in a direct charge–charge interaction. Inverting that charge at one of the two residues destabilizes the interaction between the TEL patch and the TEN domain, but reversing the charge at both positions re-establishes the interaction and suppresses the loss-of-function phenotypes of the individual mutations. While these experiments demonstrated that a direct interaction between TPP1 and hTERT is necessary for telomerase recruitment to telomeres, they did not demonstrate that it is sufficient. It is therefore possible that additional factors facilitate the interaction between the telomere and telomerase, such as TCAB1 (see above).
The original proposal of a second set of DAT mutants in the CTE (C-DAT) has not been tested in the same detail as applied to the N-DAT region. Direct telomerase enzyme assays, instead of the PCR-based TRAP assay, confirmed that mutations in the C-DAT region result in substantial loss of activity. In addition, disease-associated mutations in this region (F1127L and E1117X) also suffer from decreased activity or do not display a DAT phenotype (Zhong et al. 2012; Zaug et al. 2013). Thus, the CTE of TERT deserves further study to confirm or deny its role in recruitment.
While the molecular interactions underlying human telomerase recruitment to telomeres are fairly well understood, its restriction to the end of S phase remains an unanswered question. Several studies have observed telomerase localization to telomeres specifically in S phase (Jády et al. 2006; Tomlinson et al. 2006), which also coincides with an increase in hTERT mRNA levels (Xi and Cech 2014). In addition, a phosphorylation site in the OB-fold domain of TPP1 (S111) could potentially aid activation of telomerase at telomeres in S, G2, and M phase (Zhang et al. 2013). Introduction of mutations of S111 in different cell types has led to conflicting results. In HeLa cells, it triggered telomere shortening, while expression in embryonic stem cells had no discernible effect when compared with wild-type TPP1 (Zhang et al. 2013; Sexton et al. 2014). Therefore, the timing of telomerase action at telomeres requires further study. Likely regulatory events include cell cycle-specific post-translational modifications of telomere or telomerase components or potentially the restriction of the assembly of telomerase into an active RNP in a cell cycle-dependent manner. A precedent for post-translational modification regulating telomerase recruitment to telomeres exists in fission yeast. Phosphorylation of the telomere component Ccq1 by the DNA damage kinase Rad3 (ATR) is required to drive its association with the Est1 subunit of the fission yeast telomerase holoenzyme (Moser et al. 2011; Yamazaki et al. 2012). In addition, as described above, telomerase assembly is regulated in a cell cycle-dependent manner in budding yeast, where an active telomerase holoenzyme is formed only after the completion of DNA replication (Tucey and Lundblad 2014).
Telomerase activation and telomere length regulation
Telomere length in telomerase-positive tissues is determined by a number of variables, among which are the absolute telomerase levels, the frequency of telomerase–telomere interactions, and the number of telomeric repeats added to each telomere upon recruitment. The number of repeats that telomerase adds to the chromosome ends depends on how fast it can catalyze nucleotide addition (activity) and how many consecutive repeats it can add before dissociating (RAP). Both activity and RAP are subject to regulation by cofactors. It is worth noting that “telomerase recruitment” as assessed by cytolocalization or telomere chromatin immunoprecipitation simply indicates that telomerase is somewhere at the telomere, not necessarily at the very end of the DNA; telomerase “activation” could perhaps involve the relocation of telomerase from an internal portion of the telomere to engagement with the DNA 3′ terminus.
In vitro, the POT1–TPP1 complex substantially increases the RAP of telomerase while only having a small impact on activity (Wang et al. 2007). POT1–TPP1 is proposed to decrease the dissociation rate of telomerase from its telomeric substrate and aid the translocation step (Fig. 3; Latrick and Cech 2010). TEL patch mutants of TPP1 are defective in these RAP stimulatory events, thereby identifying a specific region of TPP1 that enhances the telomerase translocation step of the catalytic cycle and decreases telomeric DNA dissociation (Dalby et al. 2015). A recent single-molecule study has provided data conflicting with these observations. Activity measurements of single telomerase molecules in the presence and absence of POT1–TPP1 demonstrated that POT1–TPP1 increased the rate of product formation and processivity of telomerase and triggered a more rapid dissociation from the telomeric primer (Hwang et al. 2014). An important assumption underlying this study is that POT1–TPP1 is capable of sliding on ssDNA, an interesting hypothesis that awaits conclusive experimental verification (Hwang et al. 2012). The importance of processivity stimulation by POT1–TPP1 in vivo is difficult to test because telomerase fails to localize to telomeres if the interaction between TPP1 and telomerase is lost.
Figure 3.
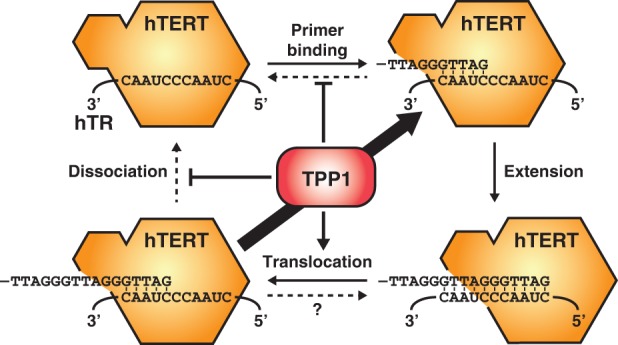
The telomerase catalytic cycle and its modulation by TPP1. Telomere extension requires multiple steps: primer DNA binding; extension of the primer using dGTP, TTP, and dATP as substrates; translocation of the RNA template relative to the active site and translocation of the DNA primer relative to the template; and recycling (thick arrow) for another round of extension. TPP1 aids translocation of the telomeric primer and reduces the rate of product dissociation from cycling telomerase to increase RAP.
In addition to the TEL patch amino acids in TPP1, a residue on the opposite face of the same OB-fold domain (L104) has been implicated in regulating the activity of telomerase. Mutations in L104 cause a short telomere phenotype even though they bind similar amounts of telomerase when compared with other OB-fold mutants that support longer telomeres (Sexton et al. 2014). To confirm the role of L104 in the TPP1 OB-fold in telomerase activity stimulation, more quantitative information about this mutant TPP1's biochemical effect on telomerase enzymatic activity, its localization to telomeres, and its effect on telomerase recruitment to telomeres is necessary. If, for instance, a lower amount of L104 mutant TPP1 localized to telomeres than wild-type TPP1, the expectation would be a shorter telomere length set point.
In other species, telomerase recruitment to telomeres and activation of its enzymatic activity have been found to be separable steps. In budding yeast, recruitment is facilitated by the interaction of Cdc13, a telomeric protein, with the Est1 subunit of the telomerase holoenzyme (Pennock et al. 2001). Est3 associates with telomerase in a cell cycle-dependent manner (see above), activates telomerase activity in vitro, and is required for telomere maintenance in vivo but it is not thought to be required for recruitment (Morris and Lundblad 1997; Talley et al. 2011). In addition to a TEL patch-like surface required for its interaction with telomerase, Est3 also contains surface residues that are required for telomerase activation but are not involved in the interaction with telomerase, potentially comparable with L104 in TPP1 (Rao et al. 2014). Similarly, in fission yeast, two factors—Ccq1 (fission yeast-specific) and Tpz1 (the TPP1 homolog)—are required for telomerase recruitment to telomeres and telomerase activation (Miyoshi et al. 2008; Tomita and Cooper 2008; Jun et al. 2013; Armstrong et al. 2014). Intriguingly, just as described for Est3 and TPP1, one allele of Tpz1 (K75A) is fully capable of recruiting telomerase to telomeres but fails to maintain telomere length, most likely due to its inability to activate telomerase (Armstrong et al. 2014). While there is a large amount of evidence supporting residues in TPP1 and its homologs being involved in telomerase activation, the structural biology and biochemistry underlying this process are unknown. It is tempting to speculate that TPP1 transiently interacts with the DNA substrate or, potentially, the TR to facilitate the translocation step of the telomerase catalytic cycle, but further mechanistic studies will be necessary to test this hypothesis (Fig. 3).
In addition to the positive regulators of telomerase activity mentioned above, there are also factors that counteract telomerase activity at telomeres. Each telomerase RNP is thought to add 50–60 nt to most telomeres after a single initiation event (Zhao et al. 2009). The CST complex, composed of CTC1, STN1, and TEN1, has been proposed to set the upper limit of telomere elongation by binding to the telomeric ssDNA and displacing telomerase once the telomeric overhang has reached a certain length (Chen et al. 2012). In addition, the CST complex is required to promote C-strand fill-in by recruiting the primase Polα to the telomeric overhang (Chen et al. 2012; Wu et al. 2012). Another negative regulator of telomere length in human cells is TERRA RNA, whose transcription initiates in subtelomeric regions of telomeres and which contains UUAGGG repeats; TERRA may inhibit telomerase activity in cis by directly binding telomerase (Schoeftner and Blasco 2008; Redon et al. 2010). Finally, specific conformations of the telomeric DNA, such as T loops and G quadruplexes, could prevent telomerase from accessing the telomere (Zaug et al. 2005; Doksani et al. 2013; Jun et al. 2013). While there is evidence that T loops and G quadruplexes exist at telomeres, the degree to which they regulate telomerase activity in vivo remains an unanswered question.
Summary and conclusions
Telomere length maintenance is a key process underlying normal human development and is defective in various disease settings. Inappropriate telomere lengthening, circumventing tumor-suppressive telomere shortening during cell division, is a hallmark of cancer, while defects in telomere maintenance lead to human diseases such as dyskeratosis congenita. Telomerase is the key factor responsible for telomere lengthening in most human cells. Its production is up-regulated in many cancers, and its function is impaired in telomeropathies. Great strides have been made in recent years to understand how telomerase is assembled and recruited to the telomere and how it synthesizes telomeric repeats. However, many key questions remain unanswered. With the advent of CRISPR–Cas9-mediated genome editing and sensitive superresolution microscopy approaches, many of these questions will be answered in the coming years. Such studies will certainly lead to a greater understanding of telomere biology and hopefully to novel therapeutic approaches for treating cancer and other telomere maintenance-related diseases.
Acknowledgments
We thank members of the Cech laboratory for useful discussion and critical reading of the manuscript. J.C.S. is a Merck fellow of the Damon Runyon Cancer Research Foundation (DRG-2169-13). T.R.C. is an investigator of the Howard Hughes Medical Institute. This work was supported by National Institutes of Health grant R01 GM099705 to T.R.C.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.263863.115.
References
- Abreu E, Aritonovska E, Reichenbach P, Cristofari G, Culp B, Terns RM, Lingner J, Terns MP. 2010. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol Cell Biol 30: 2971–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios M, Blackburn EH. 2012. The telomere syndromes. Nat Rev Genet 13: 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster BN, Banik SS, Guo C, Smith AC, Counter CM. 2001. N-terminal domains of the human telomerase catalytic subunit required for enzyme activity in vivo. Mol Cell Biol 21: 7775–7786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster BN, Linardic CM, Veldman T, Bansal NP, Downie DL, Counter CM. 2004. Rescue of an hTERT mutant defective in telomere elongation by fusion with hPot1. Mol Cell Biol 24: 3552–3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong CA, Pearson SR, Amelina H, Moiseeva V, Tomita K. 2014. Telomerase activation after recruitment in fission yeast. Curr Biol 24: 2006–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, Cech TR. 2001. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292: 1171–1175. [DOI] [PubMed] [Google Scholar]
- Blackburn EH. 2005. Telomeres and telomerase: their mechanisms of action and the effects of altering their functions. FEBS Lett 579: 859–862. [DOI] [PubMed] [Google Scholar]
- Blackburn EH, Collins K. 2011. Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb Perspect Biol 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borah S, Xi L, Zaug AJ, Powell NM, Dancik GM, Cohen SB, Costello JC, Theodorescu D, Cech TR. 2015. Cancer. TERT promoter mutations and telomerase reactivation in urothelial cancer. Science 347: 1006–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan TM, Goodrich KJ, Cech TR. 2003. Tetrahymena telomerase is active as a monomer. Mol Biol Cell 14: 4794–4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR. 2004. Beginning to understand the end of the chromosome. Cell 116: 273–279. [DOI] [PubMed] [Google Scholar]
- Chen L-Y, Redon S, Lingner J. 2012. The human CST complex is a terminator of telomerase activity. Nature 488: 540–544. [DOI] [PubMed] [Google Scholar]
- Chung J, Khadka P, Chung IK. 2012. Nuclear import of hTERT requires a bipartite nuclear localization signal and Akt-mediated phosphorylation. J Cell Sci 125: 2684–2697. [DOI] [PubMed] [Google Scholar]
- Dalby AB, Hofr C, Cech TR. 2015. Contributions of the TEL-patch amino acid cluster on TPP1 to telomeric DNA synthesis by human telomerase. J Mol Biol 427: 1291–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Kittur N, Roy S, Shav-Tal Y, Singer RH, Meier UT. 2006. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J Cell Biol 173: 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denchi EL, de Lange T. 2007. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448: 1068–1071. [DOI] [PubMed] [Google Scholar]
- Doksani Y, Wu JY, de Lange T, Zhuang X. 2013. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell 155: 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan ED, Collins K. 2010. Specificity and stoichiometry of subunit interactions in the human telomerase holoenzyme assembled in vivo. Mol Cell Biol 30: 2775–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan ED, Collins K. 2012a. Biogenesis of telomerase ribonucleoproteins. RNA 18: 1747–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan ED, Collins K. 2012b. An enhanced H/ACA RNP assembly mechanism for human telomerase RNA. Mol Cell Biol 32: 2428–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, Adams RR, Chang E, Allsopp RC, Yu J. 1995. The RNA component of human telomerase. Science 269: 1236–1241. [DOI] [PubMed] [Google Scholar]
- Freund A, Zhong FL, Venteicher AS, Meng Z, Veenstra TD, Frydman J, Artandi SE. 2014. Proteostatic control of telomerase function through TRiC-mediated folding of TCAB1. Cell 159: 1389–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Collins K. 2003. Distinct biogenesis pathways for human telomerase RNA and H/ACA small nucleolar RNAs. Mol Cell 11: 1361–1372. [DOI] [PubMed] [Google Scholar]
- Gallardo F, Olivier C, Dandjinou AT, Wellinger RJ, Chartrand P. 2008. TLC1 RNA nucleo–cytoplasmic trafficking links telomerase biogenesis to its recruitment to telomeres. EMBO J 27: 748–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis AJ, Schuller AP, Skordalakes E. 2008. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature 455: 633–637. [DOI] [PubMed] [Google Scholar]
- Girard C, Verheggen C, Neel H, Cammas A, Vagner S, Soret J, Bertrand E, Bordonné R. 2008. Characterization of a short isoform of human Tgs1 hypermethylase associating with small nucleolar ribonucleoprotein core proteins and produced by limited proteolytic processing. J Biol Chem 283: 2060–2069. [DOI] [PubMed] [Google Scholar]
- Greider CW. 1991. Telomerase is processive. Mol Cell Biol 11: 4572–4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkisheimer M, Mason M, Shuvaeva E, Skordalakes E. 2013. A motif in the vertebrate telomerase N-terminal linker of TERT contributes to RNA binding and telomerase activity and processivity. Structure 21: 1870–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidenreich B, Rachakonda PS, Hemminki K, Kumar R. 2014. TERT promoter mutations in cancer development. Curr Opin Genet Dev 24: 30–37. [DOI] [PubMed] [Google Scholar]
- Holt SE, Aisner DL, Baur J, Tesmer VM, Dy M, Ouellette M, Trager JB, Morin GB, Toft DO, Shay JW, et al. 1999. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev 13: 817–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K, Upton H, Miracco EJ, Jiang J, Zhou ZH, Feigon J, Collins K. 2013. Tetrahymena telomerase holoenzyme assembly, activation, and inhibition by domains of the p50 central hub. Mol Cell Biol 33: 3962–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, et al. 2013. TERT promoter mutations in familial and sporadic melanoma. Science 339: 959–961. [DOI] [PubMed] [Google Scholar]
- Houghtaling BR, Cuttonaro L, Chang W, Smith S. 2004. A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr Biol 14: 1621–1631. [DOI] [PubMed] [Google Scholar]
- Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. 2013. Highly recurrent TERT promoter mutations in human melanoma. Science 339: 957–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Brown AF, Wu J, Xue J, Bley CJ, Rand DP, Wu L, Zhang R, Chen JJ-L, Lei M. 2014. Structural basis for protein-RNA recognition in telomerase. Nat Struct Mol Biol 21: 507–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H, Buncher N, Opresko PL, Myong S. 2012. POT1–TPP1 regulates telomeric overhang structural dynamics. Structure 20: 1872–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H, Opresko P, Myong S. 2014. Single-molecule real-time detection of telomerase extension activity. Sci Rep 4: 6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SA, Podell ER, Cech TR. 2006. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat Struct Mol Biol 13: 218–225. [DOI] [PubMed] [Google Scholar]
- Jády BE, Bertrand E, Kiss T. 2004. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J Cell Biol 164: 647–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády BE, Richard P, Bertrand E, Kiss T. 2006. Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol Biol Cell 17: 944–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Miracco EJ, Hong K, Eckert B, Chan H, Cash DD, Min B, Zhou ZH, Collins K, Feigon J. 2013. The architecture of Tetrahymena telomerase holoenzyme. Nature 496: 187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun H-I, Liu J, Jeong H, Kim J-K, Qiao F. 2013. Tpz1 controls a telomerase-nonextendible telomeric state and coordinates switching to an extendible state via Ccq1. Genes Dev 27: 1917–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurczyluk J, Nouwens AS, Holien JK, Adams TE, Lovrecz GO, Parker MW, Cohen SB, Bryan TM. 2011. Direct involvement of the TEN domain at the active site of human telomerase. Nucleic Acids Res 39: 1774–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King TH, Decatur WA, Bertrand E, Maxwell ES, Fournier MJ. 2001. A well-connected and conserved nucleoplasmic helicase is required for production of box C/D and H/ACA snoRNAs and localization of snoRNP proteins. Mol Cell Biol 21: 7731–7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T, Fayet-Lebaron E, Jády BE. 2010. Box H/ACA small ribonucleoproteins. Mol Cell 37: 597–606. [DOI] [PubMed] [Google Scholar]
- Kocak H, Ballew BJ, Bisht K, Eggebeen R, Hicks BD, Suman S, O'Neil A, Giri N, NCI DCEG Cancer Genomics Research Laboratory, NCI DCEG Cancer Sequencing Working Group, et al. 2014. Hoyeraal-Hreidarsson syndrome caused by a germline mutation in the TEL patch of the telomere protein TPP1. Genes Dev 28: 2090–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CK, Mitchell JR, Collins K. 2001. RNA binding domain of telomerase reverse transcriptase. Mol Cell Biol 21: 990–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latrick CM, Cech TR. 2010. POT1–TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J 29: 924–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Mandell EK, Tucey TM, Morris DK, Lundblad V. 2008. The Est3 protein associates with yeast telomerase through an OB-fold domain. Nat Struct Mol Biol 15: 990–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy MZ, Allsopp RC, Futcher AB, Greider CW, Harley CB. 1992. Telomere end-replication problem and cell aging. J Mol Biol 225: 951–960. [DOI] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. 1997. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276: 561–567. [DOI] [PubMed] [Google Scholar]
- Liu D, Safari A, O'Connor MS, Chan DW, Laegeler A, Qin J, Songyang Z. 2004. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol 6: 673–680. [DOI] [PubMed] [Google Scholar]
- Min B, Collins K. 2009. An RPA-related sequence-specific DNA-binding subunit of telomerase holoenzyme is required for elongation processivity and telomere maintenance. Mol Cell 36: 609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min B, Collins K. 2010. Multiple mechanisms for elongation processivity within the reconstituted Tetrahymena telomerase holoenzyme. J Biol Chem 285: 16434–16443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Collins K. 2000. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol Cell 6: 361–371. [DOI] [PubMed] [Google Scholar]
- Mitchell JR, Cheng J, Collins K. 1999. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol 19: 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Kanoh J, Saito M, Ishikawa F. 2008. Fission yeast Pot1–Tpp1 protects telomeres and regulates telomere length. Science 320: 1341–1344. [DOI] [PubMed] [Google Scholar]
- Morris DK, Lundblad V. 1997. Programmed translational frameshifting in a gene required for yeast telomere replication. Curr Biol 7: 969–976. [DOI] [PubMed] [Google Scholar]
- Moser BA, Chang Y-T, Kosti J, Nakamura TM. 2011. Tel1ATM and Rad3ATR kinases promote Ccq1–Est1 interaction to maintain telomeres in fission yeast. Nat Struct Mol Biol 18: 1408–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy AD, Cech TR. 2006. Low abundance of telomerase in yeast: implications for telomerase haploinsufficiency. RNA 12: 1721–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR. 1997. Telomerase catalytic subunit homologs from fission yeast and human. Science 277: 955–959. [DOI] [PubMed] [Google Scholar]
- Nandakumar J, Cech TR. 2013. Finding the end: recruitment of telomerase to telomeres. Nat Rev Mol Cell Biol 14: 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J, Bell CF, Weidenfeld I, Zaug AJ, Leinwand LA, Cech TR. 2012. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature 492: 285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T. 2008. How shelterin protects mammalian telomeres. Annu Rev Genet 42: 301–334. [DOI] [PubMed] [Google Scholar]
- Pennock E, Buckley K, Lundblad V. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104: 387–396. [DOI] [PubMed] [Google Scholar]
- Rao T, Lubin JW, Armstrong GS, Tucey TM, Lundblad V, Wuttke DS. 2014. Structure of Est3 reveals a bimodal surface with differential roles in telomere replication. Proc Natl Acad Sci 111: 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon S, Reichenbach P, Lingner J. 2007. Protein–RNA and protein–protein interactions mediate association of human EST1A/SMG6 with telomerase. Nucleic Acids Res 35: 7011–7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon S, Reichenbach P, Lingner J. 2010. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res 38: 5797–5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbach P, Höss M, Azzalin CM, Nabholz M, Bucher P, Lingner J. 2003. A human homolog of yeast Est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr Biol 13: 568–574. [DOI] [PubMed] [Google Scholar]
- Richard P, Darzacq X, Bertrand E, Jády BE, Verheggen C, Kiss T. 2003. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. EMBO J 22: 4283–4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robart AR, Collins K. 2011. Human telomerase domain interactions capture DNA for TEN domain-dependent processive elongation. Mol Cell 42: 308–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouda S, Skordalakes E. 2007. Structure of the RNA-binding domain of telomerase: implications for RNA recognition and binding. Structure 15: 1403–1412. [DOI] [PubMed] [Google Scholar]
- Sauerwald A, Sandin S, Cristofari G, Scheres SHW, Lingner J, Rhodes D. 2013. Structure of active dimeric human telomerase. Nat Struct Mol Biol 20: 454–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JC, Dalby AB, Cech TR. 2014. Identification of human TERT elements necessary for telomerase recruitment to telomeres. Elife 3: e03563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA. 2008. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol 10: 228–236. [DOI] [PubMed] [Google Scholar]
- Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR. 1999. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401: 177–180. [DOI] [PubMed] [Google Scholar]
- Seto AG, Livengood AJ, Tzfati Y, Blackburn EH, Cech TR. 2002. A bulged stem tethers Est1p to telomerase RNA in budding yeast. Genes Dev 16: 2800–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton AN, Youmans DT, Collins K. 2012. Specificity requirements for human telomere protein interaction with telomerase holoenzyme. J Biol Chem 287: 34455–34464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton AN, Regalado SG, Lai CS, Cost GJ, O'Neil CM, Urnov FD, Gregory PD, Jaenisch R, Collins K, Hockemeyer D. 2014. Genetic and molecular identification of three human TPP1 functions in telomerase action: recruitment, activation, and homeostasis set point regulation. Genes Dev 28: 1885–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay JW, Wright WE. 2011. Role of telomeres and telomerase in cancer. Semin Cancer Biol 21: 349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow BE, Erdmann N, Cruickshank J, Goldman H, Gill RM, Robinson MO, Harrington L. 2003. Functional conservation of the telomerase protein Est1p in humans. Curr Biol 13: 698–704. [DOI] [PubMed] [Google Scholar]
- Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE. 2003. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes Dev 17: 2384–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern JL, Zyner KG, Pickett HA, Cohen SB, Bryan TM. 2012. Telomerase recruitment requires both TCAB1 and Cajal bodies independently. Mol Cell Biol 32: 2384–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SA, Weinberg RA. 2006. Telomeres: cancer to human aging. Annu Rev Cell Dev Biol 22: 531–557. [DOI] [PubMed] [Google Scholar]
- Talley JM, DeZwaan DC, Maness LD, Freeman BC, Friedman KL. 2011. Stimulation of yeast telomerase activity by the ever shorter telomere 3 (Est3) subunit is dependent on direct interaction with the catalytic protein Est2. J Biol Chem 286: 26431–26439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejera AM, Stagno d'Alcontres M, Thanasoula M, Marion RM, Martinez P, Liao C, Flores JM, Tarsounas M, Blasco MA. 2010. TPP1 is required for TERT recruitment, telomere elongation during nuclear reprogramming, and normal skin development in mice. Dev Cell 18: 775–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer CA, Feigon J. 2006. Structure and function of telomerase RNA. Curr Opin Struct Biol 16: 307–318. [DOI] [PubMed] [Google Scholar]
- Tomita K, Cooper JP. 2008. Fission yeast Ccq1 is telomerase recruiter and local checkpoint controller. Genes Dev 22: 3461–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, Terns MP. 2006. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol Biol Cell 17: 955–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson RL, Abreu EB, Ziegler T, Ly H, Counter CM, Terns RM, Terns MP. 2008. Telomerase reverse transcriptase is required for the localization of telomerase RNA to cajal bodies and telomeres in human cancer cells. Mol Biol Cell 19: 3793–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucey TM, Lundblad V. 2014. Regulated assembly and disassembly of the yeast telomerase quaternary complex. Genes Dev 28: 2077–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. 2008. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell 132: 945–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE. 2009. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 323: 644–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. 2007. The POT1–TPP1 telomere complex is a telomerase processivity factor. Nature 445: 506–510. [DOI] [PubMed] [Google Scholar]
- Wu RA, Collins K. 2014. Human telomerase specialization for repeat synthesis by unique handling of primer-template duplex. EMBO J 33: 921–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Takai H, de Lange T. 2012. Telomeric 3′ overhangs derive from resection by Exo1 and Apollo and fill-in by POT1b-associated CST. Cell 150: 39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi L, Cech TR. 2014. Inventory of telomerase components in human cells reveals multiple subpopulations of hTR and hTERT. Nucleic Acids Res 42: 8565–8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O'Connor MS, Songyang Z. 2007. TPP1 is a homologue of ciliate TEBP-β and interacts with POT1 to recruit telomerase. Nature 445: 559–562. [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Tarumoto Y, Ishikawa F. 2012. Tel1(ATM) and Rad3(ATR) phosphorylate the telomere protein Ccq1 to recruit telomerase and elongate telomeres in fission yeast. Genes Dev 26: 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye JZ-S, Donigian JR, van Overbeek M, Loayza D, Luo Y, Krutchinsky AN, Chait BT, de Lange T. 2004. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J Biol Chem 279: 47264–47271. [DOI] [PubMed] [Google Scholar]
- Yu EY, Wang F, Lei M, Lue NF. 2008. A proposed OB-fold with a protein-interaction surface in Candida albicans telomerase protein Est3. Nat Struct Mol Biol 15: 985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug AJ, Podell ER, Cech TR. 2005. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc Natl Acad Sci 102: 10864–10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug AJ, Podell ER, Nandakumar J, Cech TR. 2010. Functional interaction between telomere protein TPP1 and telomerase. Genes Dev 24: 613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug AJ, Crary SM, Jesse Fioravanti M, Campbell K, Cech TR. 2013. Many disease-associated variants of hTERT retain high telomerase enzymatic activity. Nucleic Acids Res 41: 8969–8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z, Min B, Huang J, Hong K, Yang Y, Collins K, Lei M. 2011. Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA. Proc Natl Acad Sci 108: 20357–20361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Kim N-K, Feigon J. 2011. Architecture of human telomerase RNA. Proc Natl Acad Sci 108: 20325–20332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen L-Y, Han X, Xie W, Kim H, Yang D, Liu D, Songyang Z. 2013. Phosphorylation of TPP1 regulates cell cycle-dependent telomerase recruitment. Proc Natl Acad Sci 110: 5457–5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Sfeir AJ, Zou Y, Buseman CM, Chow TT, Shay JW, Wright WE. 2009. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell 138: 463–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong F, Savage SA, Shkreli M, Giri N, Jessop L, Myers T, Chen R, Alter BP, Artandi SE. 2011. Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenita. Genes Dev 25: 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong FL, Batista LFZ, Freund A, Pech MF, Venteicher AS, Artandi SE. 2012. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell 150: 481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Kumar R, Mandal M, Sharma N, Sharma HW, Dhingra U, Sokoloski JA, Hsiao R, Narayanan R. 1996. Cell cycle-dependent modulation of telomerase activity in tumor cells. Proc Natl Acad Sci 93: 6091–6095. [DOI] [PMC free article] [PubMed] [Google Scholar]