Protein-mediated “chromosome kissing” between two DNA sites in trans (or in cis) is known to facilitate three-dimensional control of gene expression and DNA replication. Choudhury et al. show that the replication terminator protein Fob1 of Saccharomyces cerevisiae promoted chromosome kissing that initiated rDNA recombination and controlled the replicative life span.
Keywords: replication terminator protein, protein oligomerization, replication termination, autoinhibitory control, recombination, chromosome kissing, replicative life span
Abstract
Protein-mediated “chromosome kissing” between two DNA sites in trans (or in cis) is known to facilitate three-dimensional control of gene expression and DNA replication. However, the mechanisms of regulation of the long-range interactions are unknown. Here, we show that the replication terminator protein Fob1 of Saccharomyces cerevisiae promoted chromosome kissing that initiated rDNA recombination and controlled the replicative life span (RLS). Oligomerization of Fob1 caused synaptic (kissing) interactions between pairs of terminator (Ter) sites that initiated recombination in rDNA. Fob1 oligomerization and Ter–Ter kissing were regulated by intramolecular inhibitory interactions between the C-terminal domain (C-Fob1) and the N-terminal domain (N-Fob1). Phosphomimetic substitutions of specific residues of C-Fob1 counteracted the inhibitory interaction. A mutation in either N-Fob1 that blocked Fob1 oligomerization or C-Fob1 that blocked its phosphorylation antagonized chromosome kissing and recombination and enhanced the RLS. The results provide novel insights into a mechanism of regulation of Fob1-mediated chromosome kissing.
“Chromosome kissing” refers to protein-mediated and physiologically relevant interactions between two sites located on either different chromosomes or the same chromosome. Intrachromosomal kissing interactions are also called chromosome looping (Schleif 1992). Chromosome kissing facilitates three-dimensional mechanisms of control of different DNA transactions. For example, Reb1-mediated termination of DNA replication in fission yeast (Singh et al. 2010) and forkhead protein-dependent control of the timing of replication initiation (Knott et al. 2012) are modulated by chromosome kissing. Differentiation of naïve T helper cells into the Th1 and Th2 forms (Spilianakis et al. 2005) and X-chromosome inactivation (Xu et al. 2006) are some of the other examples of biological activities controlled by kissing interactions. There are several examples of biological regulation by chromosome looping in prokaryotes (Hochschild and Ptashne 1988; Mukherjee et al. 1988a,b; Lobell and Schleif 1990; Miron et al. 1992; Schleif 1992) and eukaryotes (Tolhuis et al. 2002). The principal objective of this study was to unravel a mechanism of regulation of “chromosome kissing” and investigate the DNA transactions regulated by it.
Recent methods designed to detect interchromosomal and intrachromosomal interactions have made it possible to map interacting sites in eukaryotic nuclei on a genome-wide scale (Dekker et al. 2002; Splinter et al. 2004; Simonis et al. 2007; de Laat et al. 2008). However, the proteins responsible for promoting most of these interactions and their physiological significance are known in only very few cases. There is also little or no information available on the mechanisms of regulation of kissing interactions. In mammalian cells, a complex of cohesin and CTCF (CCCTC-binding factor) promotes formation of topological domains of potentially interacting chromosomal loops (Zuin et al. 2014). Cohesin has also been implicated in chromosomal loop stabilization by interaction with a transcription mediator presumably at the stem of a DNA stem–loop structure (Kagey et al. 2010). Within the topological domains, specific loop-to-loop interaction by chromosome kissing probably involves sequence-specific DNA-binding proteins that form protein bridges between the interacting sites by oligomerization (Singh et al. 2010; this study).
Recombination (e.g., translocation) at a distance promoted by long-range protein–DNA interactions is potentially significant because, among other things, it can give rise to fused oncogenes or place a quiescent oncogene under the control of a strong promoter, resulting in its pathogenic overexpression. Chromosome translocations are caused not just by random DNA breakage and stochastic rejoining of the broken ends. Rather, breaks located in close proximity to each other in the nucleoplasm tend to undergo preferential rejoining (Roix et al. 2003; Roukos and Misteli 2014).
The Fob1 protein binds to two tandem terminator (Ter) sites called Ter1 and Ter2 (also called replication fork barriers [RFBs]) (Brewer 1988; Brewer et al. 1992), located in the nontranscribed spacer 1 (NTS1) between the end of the DNA encoding the 35S precursor rRNA and that of the 5S RNA-encoding DNA in each of the rDNA repeats. Fob1–Ter interaction causes polar replication fork arrest (Kobayashi et al. 2004; Mohanty and Bastia 2004; for reviews of replication termination, see Bastia and Mohanty 1996, 2006; Dalgaard et al. 2011; Bastia and Zaman 2014). The fork arrest at Ter sites in rDNA prevents head on collision between the leftward moving fork and the rightward moving 35S transcript (Brewer 1988; Takeuchi et al. 2003). Transcription replication interference can cause genetic instability (Liu and Alberts 1995; Mirkin and Mirkin 2007; Aguilera and Garcia-Muse 2012; Bermejo et al. 2012; Lin and Pasero 2012) that significantly increases the “genetic load,” which can presage induction of cancer.
In the absence Sir2 (a NAD-dependent histone deacetylase), Fob1-mediated fork arrest is known to promote recombination between rDNA repeats (Kobayashi et al. 2004; Kobayashi and Ganley 2005). The presence of Sir2 in the nucleolar milieu suppresses recombination and transcription by RNA polymerase II (Pol II) but not by RNA Pol I or Pol III. Intrachromatid recombination generates rDNA circles, and their excision reduces the number of rDNA repeats. Although it is known that intrachromatid recombination in mother cells causes a decrease in their replicative life span (RLS) (Sinclair and Guarente 1997; Defossez et al. 1999), little is known about the molecular mechanisms that regulate the process.
Sir2 is loaded onto rDNA as a part of the complex called RENT1 that consists of Net1, Cdc14, Tof2, Lrs4, and Csm1 proteins (Straight et al. 1999; Moazed 2001; Huang and Moazed 2003; Huang et al. 2006). There are two foci at which RENT1 gets loaded onto rDNA. Loading at the Ter sites requires Fob1–Net1 interaction, whereas that near the promoter region of 35S RNA requires RENT1–RNA Pol I interaction (Huang et al. 2006). We reported previously that silencing by RENT1 and fork arrest are two independent functions of Fob1 (Bairwa et al. 2010). Sir2 is known to suppress intrachromatid but not interchromatid recombination in the rDNA array (Kobayashi et al. 2004; Kobayashi and Ganley 2005).
Using Saccharomyces cerevisiae as a model system, the principal focus of this study was to determine whether and how chromosome kissing is regulated and whether it promoted recombination between two geographically separated sites in rDNA. Incidentally, we also studied the effect of the control mechanism on RLS that was used as an additional biological readout. Here, we show that oligomerization of Fob1 promotes chromosome kissing at Ter sites of the NTS1 of rDNA. The kissing interaction triggers intrachromatid recombination that is known to control the RLS. We show further that the C-terminal domain of Fob1 (C-Fob1) is inhibitory and interacts with its N-terminal domain (N-Fob1) to antagonize not only Fob1 oligomerization but also its interaction with other proteins. Consistent with the proposition that phosphorylation of C-Fob1 is the trigger that breaks its intramolecular interaction with N-Fob1, we discovered that certain Thr/Ser residues of C-Fob1, when replaced by Ala, caused constitutive C-Fob1–N-Fob1 interaction, whereas phosphomimetic Asp substitutions at the same locations counteracted this effect, thereby illuminating the mechanism of control of chromosome kissing, recombination, and RLS.
Results
Isolation of mutant forms of Fob1 defective in Fob1 oligomerization and chromosome kissing
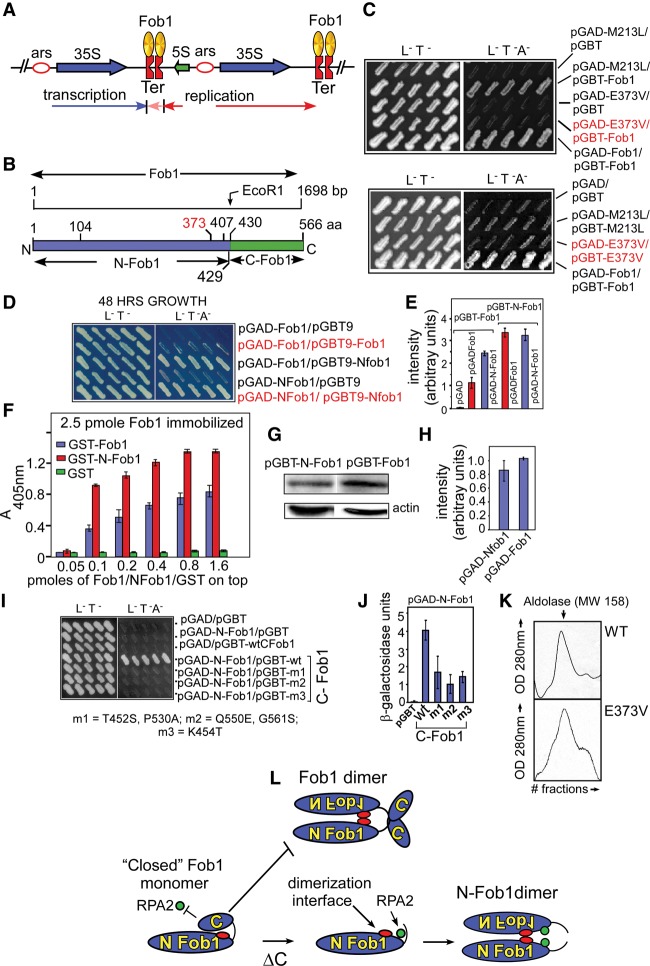
Fob1 interacts with itself (Mohanty and Bastia 2004). Schematic diagrams of the rDNA repeats (Fig. 1A) and the Fob1 ORF are shown in Figure 1B. The C-terminal EcoR1 site provided us with a convenient landmark at which to split the ORF into its N-terminal (N-Fob1) and C-terminal (C-Fob1) domains (Fig. 1B). The N-Fob1 protein was biologically active, as shown by its ability to arrest replication forks and silence rDNA in vivo in comparison with wild-type Fob1 (Supplemental Fig. S1A,B). The C-Fob1 peptide did not seem to be misfolded because, as shown later, it retained specific protein–protein interaction with N-Fob1 and biochemically did not behave like a globally misfolded, sticky protein.
Figure 1.
Mutant forms of Fob1 defective in Fob1 oligomerization and the presence of an autoinhibitory domain at the C-terminal end of Fob1. (A) Physical map of the rDNA repeat unit. Each rDNA unit has 35S and 5S rRNA-encoding sequences separated by two nontranscribed spacers. (B) Schematic representation of the primary structure of Fob1. (C) Yeast two-hybrid (Y2H) analyses of Fob1 mutants defective in protein oligomerization. (D) Y2H analysis of interaction between Fob1 and itself or N-Fob1 and between N-Fob1 and N-Fob1. (E) β-Galactosidase reporter assay showing that the C-terminal deletion of Fob1 enhances Fob1–Fob1 interaction by approximately twofold. (F) Biochemical confirmation by ELISA of the data shown in D and E. (G) Western blots showing the relative levels of Fob1 and N-Fob1 present in pGBT9 vector; the blots were developed using anti-Gal4 DNA-binding domain antibodies with actin used as a loading control. (H) Quantification of the data (ratios of the intensities of GBT9-Fob1/actin and GBT9-N-Fob1/actin) with standard error bars (from three experiments). (I) Y2H analyses and isolation of mutant forms of C-Fob1 by reverse Y2H (YR2H) selection; interaction of the wild-type and three mutant forms of C-Fob1 with N-fob1 is shown (the nucleotide substitution of the mutants of C-Fob1 is shown below). (J) Confirmation of the Y2H analysis (shown in I) by quantifications of the β-galactosidase reporter activities. (K) Gel filtration profiles of wild-type (WT) and E373V mutant forms of yeast from which GST tags were removed by digestion with PreScission protease. (L) A model showing Fob1 monomers in the closed form in which the C-terminal domain of Fob1 is in association with the N-terminal region of the protein-blocking interactions with RPA2 and retarding Fob1 oligomerization. Deletion of the C termini allows the N-Fob1 to interact with not only itself but also RPA2.
Our stratagem to isolate mutants defective in Fob1-mediated chromosome kissing was based on the expectation that the long-range interactions are likely to involve oligomerization of the protein, and therefore mutants defective in the latter are also likely to be kissing-defective (Singh et al. 2010). We amplified the Fob1 ORF by low-fidelity PCR and selected mutants defective in Fob1–Fob1 interaction using a reverse yeast two-hybrid (YR2H) selection as described in the Materials and Methods. We recovered two mutants—namely, M213L and E373V—the former of which did not interact with itself but did interact with wild-type Fob1. In contrast, the latter failed to interact not only with itself but also wild-type Fob1 even after 10 d of growth on Ade dropout indicator plates (Fig. 1C). The E373V mutant caused the strongest defect in Fob1–Fob1 interaction and therefore was selected for further analysis.
In principle, heterodimerization of a pGAD-Fob1 with a pGBT-Fob1 should activate the reporter genes present in the yeast two-hybrid (Y2H) indicator strain, and the E373V mutation is expected to interrupt this dimeric interaction. Alternatively, it is possible that the E373V mutation might be interrupting oligomerization of a dimer. The monomeric molecular mass of wild-type Fob1 is 62.8 kDa. We purified both the wild-type and the E373V forms of the proteins after expression as a GST fusion in yeast and purification after digestion with “PreScission” protease (GE Life Sciences) to remove the GST tag and performed gel filtrations to determine the relative molecular mass. The protein mobility on the gel filtration column with respect to that of the markers of known molecular mass revealed that both forms of Fob1 appeared to migrate as a dimer (Fig. 1K). This observation suggested that the E373V mutation might be inactivating a dimer to oligomer interface. However, this interpretation is subject to the caveat that the protein could still be a monomer because its molecular conformation (e.g., possibly a flexible rod) might cause it to mimic the hydrodynamic behavior of a dimer on a gel filtration column. Indeed, the Reb1 protein of Schizosaccharomyces pombe, which is an ortholog of Fob1, migrates as a dimer on a gel filtration column, but subsequent determination of its crystal structure showed that the protein is a monomer that forms dimers only after binding to DNA (our unpublished results). Therefore, pending more definitive confirmatory evidence, we provisionally interpreted the data as oligomerization of a monomer.
C-Fob1 contains an inhibitory domain
Does C-Fob1 regulate the biological activities of N-Fob1? We addressed this question by performing Y2H analysis of interaction of Fob1 with itself, Fob1 with N-Fob1, and N-Fob1 with itself (Fig. 1D). The quantitative interaction data were generated by the measurement of the specific activities of the lacZ reporter (Fig. 1E). The data revealed that full-length Fob1 had an approximately twofold lower level of interaction with itself in comparison with that between Fob1 and N-Fob1 and between N-Fob1 and itself. Additional confirmation of the data was provided by ELISA experiments in which Fob1 without a GST tag was immobilized on a plastic surface and challenged separately with GST-tagged Fob1 and GST-N-Fob1. The data showed that N-Fob1 had a higher affinity for Fob1 as contrasted to Fob1 binding to itself (Fig. 1F). This interpretation could be subject to the caveat that a significantly higher level of N-Fob1 expressed by the two-hybrid vector, in comparison with that of Fob1, might mimic the data and give the false impression of a higher affinity of interaction between N-Fob1 and itself or between N-Fob1 and Fob1 in comparison with that of Fob1–Fob1 interaction. To address this question, we performed Western blots of both forms of Fob1 expressed from a pGBT9 vector using antibodies against the DNA-binding domain of Gal4 (Fig. 1G,H). The results showed that the consistent increase in the Fob1 oligomerization by deletion of C-Fob1 could not be attributed to the relative intracellular levels of N-Fob1 and Fob1, with actin used as an internal control.
If C-Fob1 inhibited the activities of N-Fob1 by protein–protein interaction, one would expect the two separated domains to physically interact with each other in trans. Furthermore, the interaction should be abolished by missense mutations in C-Fob1 (or N-Fob1) that perturbed the interaction interface. Y2H analysis provided clear evidence supporting physical interaction between the two domains (Fig. 1I). Quantification of the β-galactosidase produced in each case by the LacZ reporter further authenticated the interaction (Fig. 1J). We examined the interactions between N-Fob1 and C-Fob1 further by isolating mutant forms of C-Fob1 that failed to interact with N-Fob1. The amino acid alterations that disrupted N-Fob1–C-Fob1 interactions are shown (Fig. 1I, bottom). All three mutants—namely, m1–m3—were defective in interaction with N-Fob1, as shown by both lack of growth on Ade dropout plates and the magnitude of β-galactosidase activities of the lacZ reporter in comparison with that of wild-type C-Fob1 (Fig. 1J). We also biochemically confirmed the interaction data as shown in Supplemental Figure S2B. The data have been summarized schematically in Figure 1L.
Does the C-terminal truncation of Fob1 enhance not only Fob1–Fob1 interaction but also its interaction with other proteins? In order to address this question, guided by our unpublished results, we examined the interaction between Fob1 and RPA2. Y2H analysis showed that RPA2 either did not interact with full-length Fob1 or did so very poorly. However, the interaction was dramatically enhanced by deleting the C terminus of Fob1 (amino acid residues 430–566) (Supplemental Fig. S2A,C). Taken together, these results led us to conclude that C-Fob1 was inhibitory and antagonized not only Fob1 oligomerization but also its interaction with RPA2.
We constructed the full-length Fob1 ORF with the m3 (K454T) mutation located in the C-terminal domain and observed that it had a modestly lower level of interactions with both wild-type Fob1 and N-Fob1 as contrasted to the same interactions by Fob1 lacking the m3 mutation (Supplemental Fig. S3A,B). Therefore, the m3 mutation located in trans reduced interaction between m3 C-Fob1 and wild-type N-Fob1 to a greater extent than when it was presented in cis. We suggest that this difference can be attributed to a lowering of the entropic component of the interaction brought by physical tethering of the two domains of Fob1 when m3 was present in intact Fob1.
Phosphorylation of C-Fob1 relieved its inhibitory activity
Is there a regulatory mechanism that converts the “closed” Fob1 to an “open” active form in vivo? In this context, at least two alternative possibilities come to mind: (1) a post-translational modification of C-Fob1 by either acetylation, phosphorylation, or some other covalent modification or (2) noncovalent interaction with a ligand that competes out C-Fob1–N-Fob1 interactions. Because of the known association of Fob1 with the histone deacetylase Sir2, we first looked for acetylation as a possible post-translational modification, but the results were negative (data not shown). We then looked for phosphorylation of Fob1 as a possible mechanism to break N-Fob1–C-Fob1 interaction. The residues of Fob1 that are known to be phosphorylated are shown in Figure 2A (based on phosphoGRID [http://www.phosphogrid.org] data; S. Humphrey and M. Mann, pers. comm.). All phosphorylation sites determined so far are located in C-Fob1. We substituted A residues at the T504 and S519 separately with phosphomimetic D at the same locations and investigated whether a double mutation, T504A–S519A, would keep the Fob1 in a constitutively “closed” state and whether the corresponding phosphomimetic D substitutions might induce a constitutively “open” conformation that would permit Fob1 oligomerization.
Figure 2.
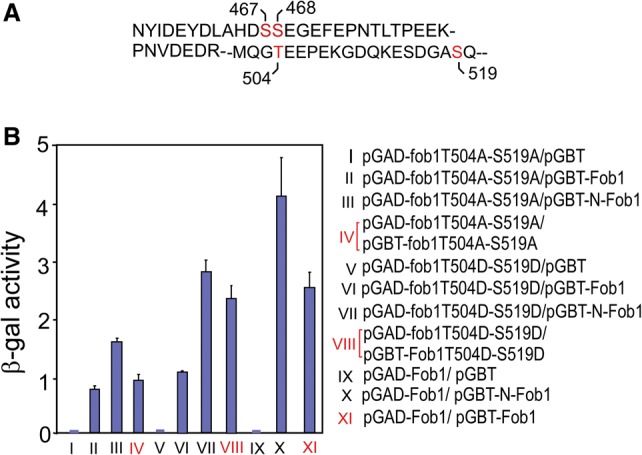
Phosphorylation of some residues of the C terminus of Fob1 relieved its inhibitory activity. (A) The residues of Fob1 that are known to be phosphorylated are shown in red. (B) Y2H analyses of T504A–S519A and T504D–S519D double mutants of Fob1. The T504A–S519A double mutant showed significantly reduced interaction with wild-type Fob1, N-Fob1, and itself in comparison with that of the T504D–S519D mutant.
The effects of double A and double D substitutions in C-Fob1 were compared with and contrasted to each other, wild-type Fob1, and wild-type N-Fob1 by Y2H analyses. The data were obtained from three sets of experiments, with each set measured in triplicate. In each case, the interactions of AA with AA, DD with DD, each of the double mutants with wild-type Fob1, and each of the double mutants with N-Fob1 as measured by the reporter lacZ-specific activities are shown (Fig. 2B). Inspection of the data showed that the T504A–S519A double mutant had significantly reduced interaction with wild-type Fob1, N-Fob1, and itself in comparison with the corresponding interactions with the T504D–S519D phosphomimetic form. For example, Figure 2B shows that the reporter activity induced by the double A-substituted fob1 interacting with itself (Fig. 2B, IV) was 38% of that elicited by the interaction of wild-type Fob1 with itself (Fig. 2B, VIII) and that the self-interaction of the double D-substituted Fob1 restored the reporter activity close to that of the wild-type level (taken as 100%) (Fig. 2B, XI). Both the double A and double D mutants were hypomorphic in comparison with similar interactions by the wild-type protein. This is not unusual because, despite the phosphomimetic attributes of an aspartic acid substitution, it is not identical in structure to a phosphoserine and therefore does not completely recapitulate the properties of a phosphorylated wild-type protein.
It should be noted that the aforementioned mutations similarly regulated the interaction of Fob1 with RPA2 (Supplemental Fig. S2D).
Since residues 430–566 of the protein were dispensable for biological activity of Fob1 (Supplemental Fig. S1), it is unlikely that two residues, 504 and 519, of C-Fob1 are directly involved in protein–protein interactions with N-Fob1. Rather, it is more likely that the C terminus plays a regulatory role by “masking” the oligomerization interface by either imposing a steric block or the induction of an allosteric change in Fob1 that blocks its oligomerization and interactions with other proteins.
The oligomerization-defective mutant E373V of N-Fob1 failed to support integrative recombination
We investigated the possible biological consequences of the aforementioned mutations by investigating their impact on integrative recombination in rDNA. The rDNA units undergo at least two kinds of recombination; namely, intrachromatid and interchromatid. The former but not the latter is suppressed by Sir2, which also represses transcription from the Epro promoter (Kobayashi and Ganley 2005) that makes the region recombinogenic by displacing cohesin (Fig. 3A, panels i,ii). The intrachromatid recombination was mimicked in trans by the integrative recombination between a plasmid DNA that contained NTS1, including two tandem Ter sites, Epro, and the associated sequences. The plasmid readily integrated into the chromosomal rDNA array in a Δsir2 strain (LPY11) in which the Epro-initiated transcription displaced the cohesin rings from the Ter region (Fig. 3A, panels i,ii). We reported previously that this recombination does not require Rad51 but is totally dependent on Rad52 as well as Tof1 and Csm3, components of the fork protection complex, but not Mrc1 (Mohanty et al. 2009; for a general review on recombination, see Paques and Haber 1999). It also requires fork arrest at Ter. The plasmid integration process described below (Fig. 3) is identical in its requirements to that of intrachromatid recombination in the rDNA repeats (Mohanty et al. 2009). Therefore, it is a convenient tool with which to investigate the latter.
Figure 3.
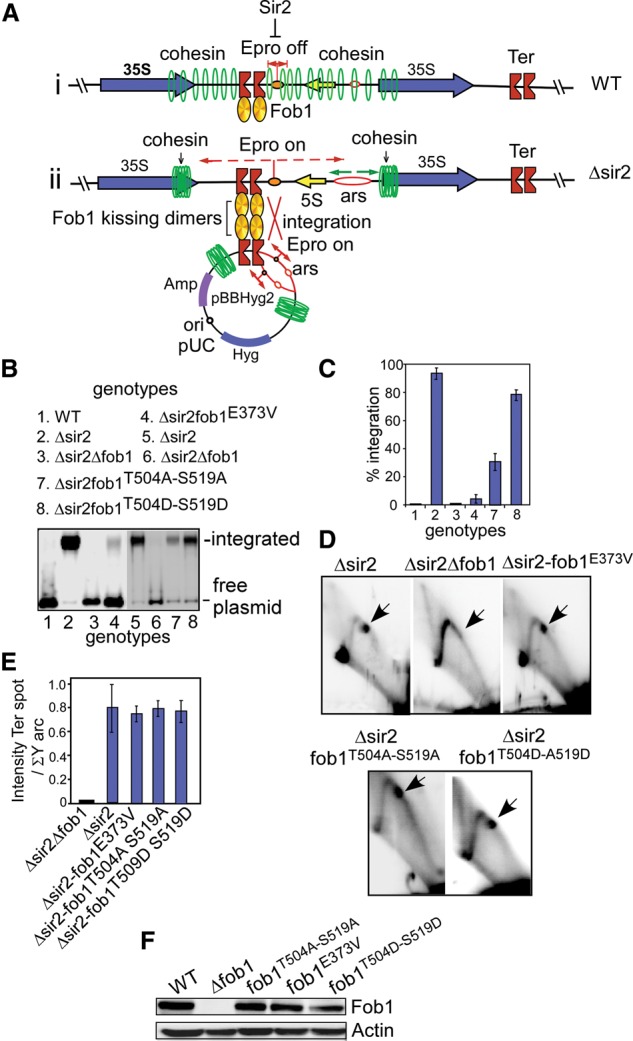
An oligomerization-defective mutant of Fob1 fails to promote integrative recombination and is controlled by phosphorylation of C-Fob1. (A, panel i) Diagram showing that, in wild-type (WT) cells, Sir2 keeps the bidirectional promoter Epro off, and cohesin rings are covering the NTS1 spacer that includes the Ter sites to prevent intrachromatid recombination. (Panel ii) Removal of Sir2 turns on Epro, and the cohesin rings are swept away by transcription emanating from Epro from the spacer region, making it potentially recombinogenic. The presence of a plasmid that has the Ter site, Epro, and the NTS1 region pairs with the corresponding Ter sites of the chromosomal rDNA by chromosome kissing promoted by tetramerization of dimeric Fob1. (B) Southern blots of total DNA from cells of the genotypes shown at the top plus the resident plasmid pBBHyg2 probed with labeled hygromycin (Hyg) sequence probe. Consistent with A, no plasmid integration occurs in a wild-type strain (lane 1), vigorous integration occurs in Δsir2 (lanes 2,5), no detectable recombination occurs in Δsir2 Δfob1 cells (lanes 3,6), and greatly reduced (>90%) integration occurs in the Δsir2fob1E373V mutant form (lane 4). Lane 7 shows reduced recombination in fob1504A–519A in comparison with lane 8, which shows that, in phosphomimetic fob1 504D–519D, recombination was enhanced. (C) Quantification of the recombination data shown in B. (D) Two-dimensional gel analyses of fork arrest in the cells of the designated genotypes. (E) Quantification of the data shown in D. (F) Western blots of wild-type Fob1, Fob1E373V, Fob1T504A–S519A, and Fob1T504D–S519D showed approximately equal intracellular levels of protein with respect to actin.
We constructed the yeast strains Δsir2, Δsir2Δfob1, and Δsir2 fob1E373V, with the integrated Fob1 being transcribed from its natural promoter in chromosome XII. We transformed the reporter plasmid pBBHyg2 that carried a single copy of the NTS1 into each of the strains (Fig. 3A), and, after multiple cycles of growth (approximately the same number for each strain), total DNA was prepared from each culture and resolved in 0.8% agarose gels. The DNA was blotted onto Nytran membranes and probed with a labeled DNA probe containing the hygromycin (Hyg) sequence. The experimental scheme and a representative blot of a one-dimensional agarose gel are shown in Figure 3, A and B, respectively. As expected, the data showed that there was no plasmid integration in the wild-type or Δfob1 cells, but extensive integration occurred in a Δsir2 strain (Fig. 3B, lanes 1–3). In contrast, a Δsir2 fob1E373V congenic strain showed very low levels of integration of the plasmid DNA into the chromosomal rDNA array (Fig. 3B). The results from three independent replicates of the experiment were in agreement with each other (see Fig. 3C). We concluded from the data that the absence of Sir2 and replication fork arrest by Fob1 are both necessary but not sufficient for promoting plasmid integration. Fob1 oligomerization, which presumably promotes synaptic (kissing) interactions between the plasmid and the rDNA at the Ter sites, is a critical factor in promoting recombination.
The aforementioned interpretation would be valid if and only if the fob1E373V mutation did not cause a significant reduction in replication fork arrest, which is essential for integrative recombination (Benguria et al. 2003; Mohanty et al. 2009). We addressed this issue by preparing replication intermediates from each of the strains used in the experiments and resolving these in two-dimensional (2D) Brewer-Fangman gels. The results (Fig. 3D), as expected, did not reveal detectable fork arrest at the Ter site in the Δsir2Δfob1 strain but showed approximately similar magnitude (intensity of termination spot divided by the integrated intensity of the Y arc) of fork arrest in the Δsir2 and Δsir2fob1E373V cells (Fig. 3E). We conclude from the data that (1) presumptive chromosome kissing by Fob1 between pairs of Ter sites in trans initiated recombination by promoting Ter synapsis, and (2) the presumptive kissing activity of the protein can be dissociated from its ability to arrest replication forks by the E373V mutation. Direct evidence for Fob1-mediated kissing is presented later. Recruitment of RPA2 by Fob1 (Supplemental Fig. S2A,C) might be promoting a postsynaptic step in recombination catalyzed by Rad52 (Mohanty et al. 2009).
If C-Fob1–N-Fob1 interaction blocked Fob1 oligomerization and thereby integrative recombination, a deletion of the C-Fob1 domain (as in Δsir2fob1E373ΔC) should partially suppress the phenotypes of E373V point mutations as manifested in a reduction of integrative recombination frequency. This was expected to occur by increased Fob1 oligomerization caused by the unmasking of the oligomerization interface located in the N-Fob. We tested this prediction by constructing a E373VΔC strain and performed plasmid integration analysis as described above. We discovered that the mutation caused a partial enhancement of integration of the plasmid in the fob1E373VΔC strain (35% of the wild-type level) over that of fob1E373V (<5% of the wild-type level). Y2H analysis showed that, whereas fob1E373V failed to interact with itself, the C-terminally deleted form of the point mutant showed enhanced (∼40% of the wild-type level) interaction (Supplemental Fig. S4A,B).
We then examined the effect of C-Fob1 phosphorylation on integrative recombination by comparing and contrasting the relative levels of fob1T504A–S519A and fob1T504D–S519D. The frequency of integrative recombination was diminished in the double A mutant. It was partially restored in the double D mutant (Fig. 3B,C). The fob1 mutant strains promoted equivalent levels of fork arrest at Ter (Fig. 3D,E). Furthermore, the enhancement or attenuation of recombination could not be attributed to a higher or lower intracellular levels of Fob1 protein because Western blots showed that the FOB1, fob1E373V, fob1A-substituted, and fob1D-substituted mutant forms all showed approximately equal intracellular levels of the different forms of the protein with respect to actin, which was used as an internal control (Fig. 3F). Taken together, the data are consistent with the interpretation that the probable “freezing” of Fob1 in the closed state (caused by loss of phosphorylation at the sites by the T504A–S519A mutations) blocked recombination, and the phosphomimetic mutant partially relieved the block by inducing a constitutively open conformation.
Direct demonstration of Fob1-mediated chromosome kissing by circular chromosome conformation capture (4C)
Although the preceding data were consistent with the interpretation that Fob1-mediated chromosome kissing was indispensable for integrative recombination, we wished to obtain direct evidence for such interactions. We performed 4C analysis (Dekker 2006; Simonis et al. 2007) with some modifications (see the extended protocols in the Supplemental Material for details; Singh et al. 2010). The sequence around the Ter sites between the two natural Afl III sites located in the NTS1 regions of rDNA array in chromosome XII (Fig. 4A,B, shown in blue) was used as the bait, and the presumptive prey was the same sequence present in the plasmid pBBHyg2 (Fig. 4A,B, shown in red) excepting that the NTS1 sequence was tagged at the 3′ end (right end) with a 149-base-pair (bp)-long marker sequence (Fig. 4A,B, shown in green) to help distinguish it from the otherwise identical chromosomally located NTS1 sequences (Fig. 4A,B). The F1–F3 primer pairs were used to make sure that the DNA samples were recovered after the multiple manipulations constituting the 4C protocol (Fig. 4C). Successful capture of chromosome kissing between the plasmid and chromosomal rDNA was expected to generate a diagnostic PCR product of 525 bp + 149 bp = 674 bp (Fig. 4C, red–green). As expected, it was recovered from both Δsir2 (LPY11) and Δsir2Δrad52 cells (Fig. 4C, red arrows) but not from the Δsir2Δfob1 or Δsir2fob1E373V cells. The data showed that the kissing interaction (1) was Fob1-dependent, (2) was dependent on Fob1 oligomerization, and (3) preceded the Rad52-catalyzed step of recombination. As expected, control experiments showed that no signal (the 674-bp product) was detectable in “no cross-link” and “no ligation” controls.
Figure 4.
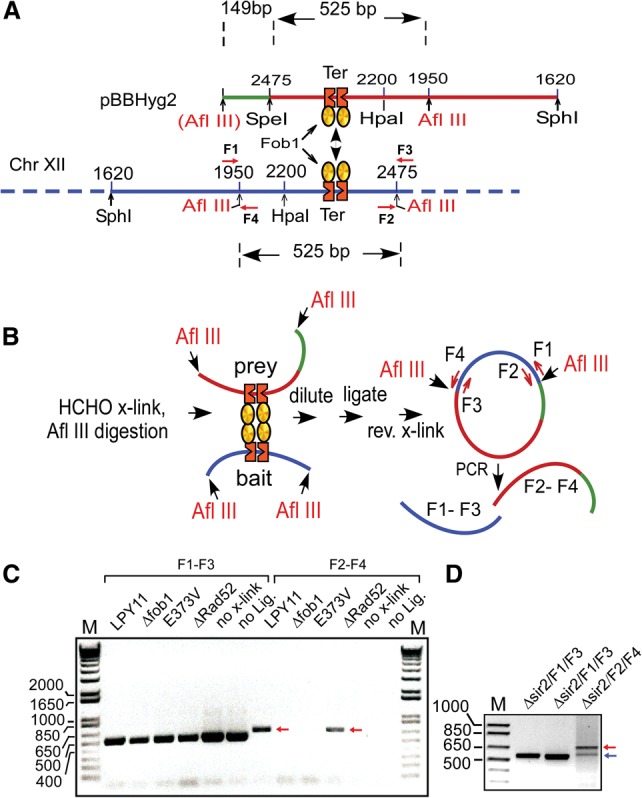
A direct demonstration of Fob1-mediated chromosome kissing by 4C analyses. (A) Restriction maps of the plasmid-borne NTS1 region of rDNA showing the Ter sites and the naturally present left Afl III site (in red), the sequence tag (green) engineered to distinguish it from chromosomal NTS1 site (blue), and the engineered right Afl III site immediately to the right of the green tag. (B) Schematic representation of the 4C reaction and the primer pairs used for PCR amplification. (C) Agarose gel electrophoresis of the diagnostic PCR products indicative of chromosome kissing (red arrows) LPY11 contains Δsir2. (D) Agarose gel electrophoretic pattern showing both the PCR product caused by chromosomal rDNA Ter site interaction in trans with the plasmid-borne Ter site in trans (red arrow) and another chromosomal Ter site in cis (blue arrow).
It has not escaped our attention that the 4C approach is also expected to capture interaction between pairs of chromosomal rDNA repeats. In two out of three sets of experiments, we observed that such interactions are detectable, albeit at a lower level than that between plasmid-borne and chromosomal rDNA interactions (Fig. 4D, blue arrow). It should also be kept in mind that HCHO is a virtual zero-length cross-linker that is not known to cross-link two complexes unless these are in direct physical contact with each other. Taken together, the data provide direct and compelling evidence for Fob1-mediated Ter–Ter kissing.
Ectopic transcription-induced Fob1-mediated HOT1 recombination was not affected by abolition of chromosome kissing and was not regulated by Fob1 phosphorylation
Passage of transcription through a pair of DNA repeats is known to cause recombination (Huang and Keil 1995; Prado et al. 1997). It was reported that when the Ter sites present in the highly recombinogenic HOT1 sequence of yeast are inserted into ectopic locations, transcription catalyzed by RNA Pol I causes high-frequency recombination between flanking repeated sequences. The HOT1 sequence inserted into one of a pair of his3 alleles flanking an ADE5 reporter (the yeast strain is also marked by ade2). Fob1 causes high-frequency recombination between the his3 repeats, resulting in the excision of the ADE5 marker as nonreplicating circular DNA (Fig. 5A) that causes red–white sectoring. In contrast, the strain carrying a deletion of fob1 produces smooth red colonies (Fig. 5A,B; Keil and Roeder 1984; Voelkel-Meiman et al. 1987).
Figure 5.
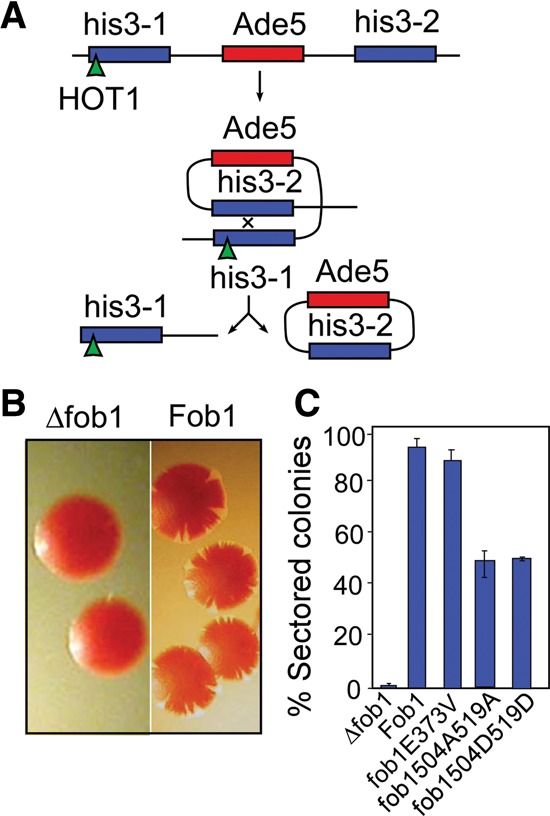
An oligomerization-defective mutation in Fob1 promotes transcription-induced recombination at an ectopic Fob1-binding site located in the His3 ORF (HOT1 recombination). (A) Schematic representation of the recombination process showing the elimination of the Ade5 marker that produces red–white sectoring. (B) Representative photographs showing sectoring and nonsectoring cells. (C) Quantification of the recombination frequency (plasmid sectoring).
We wished to investigate whether chromosome kissing promoted HOT1 recombination. The reporter strain shown in Figure 5A containing a fob1 deletion was separately complemented with a plasmid-borne wild-type FOB1 or the “nonkissing” mutant fob1E373V, the colonies were allowed to grow for a few days on low-adenine plates, and the percentage of sectored colonies was scored. The data showed that both wild-type FOB1 and fob1E373V showed no significant difference in the recombination frequency (Fig. 5C), thereby supporting the conclusion that Fob1-mediated chromosome kissing was not necessary for HOT1 recombination. We further investigated the possible impact of fob1T504A–S519A and fob1T504D–S519D mutations on HOT1 activity. The data showed that the recombination frequencies in the A-substituted mutant were not significantly different from that of the D-substituted form, which was close to that of the wild-type cells (Fig. 5C). Why are the substitution mutations hypomorphic to that of the wild type if all of these strains bind to Ter sites with approximately equivalent affinity? Keeping in mind that Fob1 binding to Ter is necessary but not sufficient for HOT1 activity, it is possible that the two mutant forms do not completely recapitulate the ability of the wild-type protein to promote Fob1-mediated activation of transcription at a HOT1 site. The apparent independence of HOT1 activity on Fob1-mediated long-range interactions, as discussed later, helps to distinguish between two alternative models of transcription-dependent recombination.
The nonkissing mutant form of Fob1 (E373V) extended the RLS of yeast cells
We integrated the E373V mutation into the Fob1 locus in our standard yeast strain, YPK9, previously used for RLS measurements (Jazwinski 1993). Several independent colonies of each strain were used for measurement of RLS by micromanipulation, separation of the daughter cells from the mother cells, and counting the number of generations after which the mother cells ceased to bud. The percentage of survival as a function of the number of generations was plotted, and the data (Fig. 6A; Supplemental Table 3) showed that the 50% survival for the wild-type and fob1E373V strains was 18.7 and 29 generations, respectively. The corresponding maxima were 27 and 47 generations, respectively. Thus, the data (Supplemental Table 3) showed a significant enhancement of the life span of the kissing-defective fob1E373V mutant over that of the wild-type FOB1 strain. Although cells containing the fob1 E373VΔC mutation partially suppressed the integrative recombination defect (Supplemental Fig. S4C), apparently the magnitude of the suppression was not enough to be reflected in the RLS in comparison with that of wild-type FOB1 (Supplemental Fig. S4D). Why did the deletion of C-Fob1 from a Fob1 E373V fail to enhance the RLS of the double mutant (Supplemental Fig. S4D)? This could have been caused by possible pleiotropic, secondary, and tertiary effects caused by the relatively large fob1ΔC deletion and the point mutation contained in the fob1E371VΔC strain on Fob1 structure and function.
Figure 6.
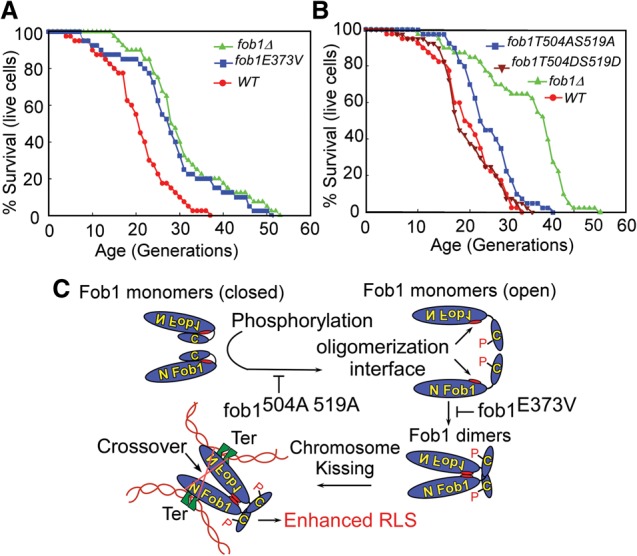
The RLSs of the wild-type (WT) and mutant forms of Fob1. (A) Life span analyses showing that the fob1-E373V mutation significantly enhances the RLS in comparison with the wild-type Fob1. (B) RLS data showing that the T504A–S519A mutations that appear to lock Fob1 dimers in a closed conformation enhance the life span, whereas the phosphomimetic T504D–S519D mutants reduce the life span in comparison with the former mutant. (C) A model depicting the sequence of events that regulate integrative recombination and RLS. Enhanced RLS as shown in the model is caused by the fob1 E3733V and the phosphomimetic fob1 S467D S468D S519D triple mutants.
RLS is controlled by phosphorylation of C-Fob1
Since our data showed that phosphorylation of T504 and S519 in C-Fob1 controlled protein–protein interactions involving Fob1, including its oligomerization, we wished to investigate the possible impact of the fob1T504A–S519A mutant and its phosphomimetic variant, fob1T504D–S519D, on RLS. We performed RLS analysis of the mutants in comparison with the wild-type strain, and the data showed that the A-substituted double mutant had a significantly higher level of RLS in comparison with the D-substituted variant and the wild-type cells (Fig. 6B; see Supplemental Table 3 for statistical analysis of the RLS data.). We conclude from the data that phosphorylation of C-Fob1 regulated the RLS presumably by its impact on the intrachromatid recombination frequency. A summary of the regulatory pathway that controls chromosome kissing, involving Fob1 oligomerization and the phosphorylation of C-Fob1, is shown schematically in Figure 6C.
Discussion
Contact between homologous chromosomes as a potential regulatory mechanism was first suggested by Muller (1941) on the basis of his observations that homologous chromosomes of Diptera show somatic pairing. He postulated that a regulatory protein located on one chromatid could diffuse across and bind to the cognate site on the paired homolog and modulate gene activity in trans, a phenomenon subsequently called “transvection” (Lewis 1985). Recent works using chromosome conformation capture (3C) techniques have revealed that interactions between nonhomologous chromosomes called “chromosome kissing” also occur, and the process controls biological activities such as differentiation and replication (Spilianakis et al. 2005; Singh et al. 2010; Knott et al. 2012) by “nonhomologous transvection.” The present work shows that chromosome kissing can also promote recombination at rDNA and control RLS.
The first new insight provided by this work is that Fob1 promotes the critical synaptic contacts between pairs of Ter sites by chromosome kissing and was obligatory for intrachromatid recombination and life span control. Previously, it was suggested that recombination in rDNA was triggered by recruitment of Top1 to the Ter region generating chromosome breaks (Di Felice et al. 2005) that somehow got together, perhaps by association of Fob1 with inner nuclear membrane proteins, to promote rejoining (Mekhail et al. 2008; Chan et al. 2011). However, the biochemical attributes of the E373V mutations of Fob1 reported here unequivocally establish that chromosome kissing at Ter is the major mechanism that brings Ter sites into close proximity to promote recombination. The second novel insight provided by this work is that phosphorylation of C-Fob1 (and probably also its dephosphorylation) allows Fob1 to be in reversible, closed, and open states that control many of the physiological activities of the protein, including its oligomerization, regulation of chromosome kissing, and intrachromatid recombination in rDNA and RLS. Although Fob1-mediated RLS was previously reported (Sinclair and Guarente 1997; Sinclair et al. 1998; Defossez et al. 1999) and the cyclic degradation of Fob1 during the cell cycle by the anaphase promoting complex is known (Menzel et al. 2014), the molecular mechanism of regulation of various DNA transactions catalyzed by Fob1 was unknown. A summary of the mechanism of control of Fob1-mediated chromosome kissing is presented in Figure 6C. The model suggests that Fob1 monomers exist as a closed structure brought about by the intramolecular interaction between N-Fob1 and C-Fob1, with the latter serving as an inhibitory module. Phosphorylation of C-Fob1 breaks the interaction helping to convert the protein to an open form. A specific domain gets unmasked in the process, leading to Fob1 oligomerization, Ter–Ter kissing, intramolecular recombination, and reduction of RLS.
It was reported that in nondividing cells of dogs and humans, there is progressive loss of rDNA as a function of aging (Strehler and Chang 1979; Strehler et al. 1979). However, the molecular mechanism driving this process in mammalian cells is presently unknown. Although Fob1-induced and Sir2-antagonized recombination in an rDNA array appears to be the dominant mechanism of aging in yeast, it has been suggested to be a yeast-specific mechanism (Rine 2005). However, the aforementioned connection between rDNA instability and aging in mammalian cells tends to caution one against jumping to such a conclusion.
There are other mechanisms of aging that appear to be conserved across species boundaries. For example, caloric restriction (Jiang et al. 2000) mediated by the TOR and Sch9 kinases as an aging mechanism appears to be conserved from yeast to mammals (Kaeberlein et al. 2005; Rine 2005; Steinkraus et al. 2008). Damaged nucleosomes that trigger unwarranted transcriptional noise (Feser et al. 2013) and H4K16 acetylation necessary for maintenance of telomeric heterochromatin (Dang et al. 2009) are other pathways of aging that might be evolutionarily conserved (Kaeberlein et al. 2007). Another conserved pathway involves the yeast longevity genes RAS2 and LAG1, the first longevity gene cloned as such that encodes ceramide synthase (D'Mello et al. 1994; Sun et al. 1994). The Neurospora orthologs of these genes interact to regulate circadian rhythm as well as chronological life span and RLS (Case et al. 2014). An interaction between these genes is associated with human longevity and healthy aging. Not surprisingly, circadian disruption curtails life span in several species and elicits aging-related phenotypes in humans. Mitochondria to nucleus signaling is yet another conserved mechanism operating in yeast, C. elegans, Drosophila, mouse, and in human cells (Jazwinski 2014). Interestingly, this mechanism, which is normally induced during the yeast RLS, suppresses the deleterious effects on longevity of rDNA circle formation (Borghouts et al. 2004), raising the possibility that rDNA circles may not be a major contributor to aging in yeast. Furthermore, circle formation does not appear to be a mechanism of aging of yeast strains isolated from the wild (Stumpferl et al. 2012). Indeed, there are many inconsistencies with the notion that rDNA circles are an important factor in yeast aging, and there is significantly more support for the instability of rDNA itself as the critical factor (Ganley and Kobayashi 2014). There is also support for the notion that excessive transcription of rDNA during aging may play a role (Kim et al. 1999; Jazwinski 2000; Jazwinski et al. 2010; Stumpferl et al. 2012), and this may be related to the regulation of protein translation in the cytoplasm (Steinkraus et al. 2008). Thus, there remains much to learn about the role of different DNA transactions at the rDNA locus in yeast replicative aging.
As contrasted to integrative recombination, HOT1-induced recombination did not require Fob1-mediated chromosome kissing. Two alternative mechanisms were proposed to explain Fob1-provoked and transcription-induced HOT1 recombination: (1) Fob1 promotes it by acting as an enhancer of Pol I transcription at the ectopic HOT1 locus (Keil and Roeder 1984; Zehfus et al. 1990; Huang and Keil 1995), or (2) Fob1 does not directly enhance Pol I transcription but promotes long-range interaction between the HOT1 locus placed in chromosome III and the rDNA array present in chromosome XII in the nucleolus. It was suggested that the more abundant transcription factors, including RNA Pol I, present in the nucleolus promoted vigorous transcription-mediated HOT1 activity (Wai et al. 2000). Our observation that HOT1 recombination was undiminished even in the absence of Fob1-mediated long-range interactions tends to support the first model. We suggest that the E373V mutation did not impact HOT1 activity because chromosome kissing is not required for the process. Consistent with this observation, phosphorylation of C-Fob1 or lack of it also did not seem to affect the frequency of HOT1 recombination.
Genome-wide 4C analysis of the yeast nucleus has revealed that rDNA is present at a separate location at the nuclear periphery, whereas all of the other chromosomes associated with each other at their centromeres in the center of the nucleus, with the two arms of each radiating out from this center to the nuclear periphery (Duan et al. 2010). It is possible that this placement of the rDNA in the periphery of the yeast nucleus acts as a topological barrier to interaction between rDNA and binding sites possibly present or ectopically placed on other chromosomes. This might inhibit kissing interactions between the HOT1 locus present on chromosome III and the rDNA array in chromosome XII.
Is it phosphorylation-mediated chromosome kissing or Fob1–RPA2 and/or Fob1-mediated Top1 recruitment to the regions of the Ter sites rather than Fob1-induced chromosome kissing that triggers recombination of rDNA and RLS? Although it is known that an RPA complex is needed for Rad51/Rad52-mediated homologous recombination (Sung et al. 2003), the recombination event requires RPA2-catalyzed steps that follow rather than precede the Fob1-mediated synaptic interaction. RPA is also involved in a checkpoint response by and loading of Mec1 at a single-stranded region using the adapter protein Ddc2 (Bandhu et al. 2014). However, the latter is provoked by fork-stalling caused by genotoxic stress. Interestingly, fork arrest at natural Ter sites of rDNA does not provoke a checkpoint response despite fork-stalling (Bentsen et al. 2013).
It should also be noted that although Top1 was postulated to induce the DNA breaks that are processed by homologous recombination, Fob1 does not directly interact with Top1, but the latter is believed to be recruited by preferential binding to bent DNA induced by Fob1 (Di Felice et al. 2005). It has been shown that Top1 actually inhibits recombination at rDNA (Kim and Wang 1989; Christman et al. 1993). Therefore, Top1 recruitment is expected to be undiminished in the E373V and the double A- and double D-substituted mutants in comparison with wild-type Fob1 because the Fob1 mutants used in this work retain normal Ter-binding activity. In summary, neither RPA2–Fob1 interaction nor Top1 recruitment indirectly by Fob1 provide alternative mechanisms that can supersede or bypass the critical role of the Fob1–Fob1 interaction in the control of recombination and RLS.
Although the rDNA repeats of budding yeast are present in a single cluster in chromosome XII, those of fission yeast are present in two separate clusters at either end of chromosome III. In humans, rDNA clusters are present in five different chromosomes; namely, XIII, XIV, XV, XXI, and XXII (Henderson et al. 1972). It is therefore tempting to speculate whether the Reb1 protein of fission yeast and its homolog, TTF1, of higher eukaryotes also promote interchromosomal long-range interactions that would promote recombination to correct the rDNA sequences against mutational drift and also coordinate regulation of their functions. One would expect that the process must be tightly controlled so as not to generate frequent translocations among the five nonhomologous chromosomes and consequent loss of genome stability. In this context, perhaps it is worth noting that although the Reb1 protein of S. pombe promotes specific interaction between a Ter site located on chromosome II and a major and a minor site located on chromosome I to generate cooperativity at a distance and robust fork arrest (Singh et al. 2010), frequent recombination between the two chromosomes at the interacting sites has not been reported, suggesting suppression of recombination between the nonhomologous chromosomes despite the homology at the rDNA arrays. These studies provide us with additional reasons to favor rDNA instability rather than circle formation as the probable cause for aging in yeast.
It is perhaps interesting to speculate why yeast cells have evolved such a phosphorylation-mediated kissing mechanism for controlling recombination. It is known that rDNA arrays undergo expansion and contraction of repeats in response to physiological cues such as availability of nutrients. It is likely that a Fob1 phosphorylation- dependent mechanism evolved to provide the molecular basis of rDNA copy number homoeostasis.
Future work will be directed at the identification of the kinase that promotes Fob1 phosphorylation and the hypothetical phosphatase that would reverse the process. Identification and functional analyses of other players anticipated to be involved in chromosome kissing is also a major goal. In conclusion, our work provides novel mechanistic information about regulation of chromosome kissing and replication Ter protein-mediated recombination in rDNA and its impact on the RLS of yeast cells.
Materials and methods
Strains and plasmids
A complete list of strains and plasmids is shown in Supplemental Table 1.
Oligonucleotides
Oligonucleotides are listed in Supplemental Table 2.
Y2H and reverse two-hybrid analysis
Y2H interactions were carried out using the yeast strain PJ69-4A as described previously (James et al. 1996; Mohanty and Bastia 2004). Wild-type genes and their mutant versions were cloned in pGAD424 and pGBT9 vectors. The plasmids were transformed in pairs into PJ69-4A, and colonies containing the plasmid pair were patched on SD/Leu−Trp− and SD/Leu−Trp− Ade− plates. Colonies growing on SD/Leu−Trp− Ade− were considered putative positive interactors, and their counterparts from SD/Leu−Trp− plates were analyzed further by β-galactosidase assays as described in the Clontech manual. YR2H selections were carried out by mutagenizing one of the ORFs (e.g., FOB1) of an interacting pair and selecting for colonies that grew in the SD/Leu−Trp− plates but not SD/Leu−Trp− Ade− plates.
Random and site-directed mutagenesis
The FOB1 ORF was mutagenized by low-fidelity PCR. Each mutagenesis reaction was carried out by vent exo− DNA polymerase in four reaction tubes containing three dNTPS at 0.25 mM each and one dNTP at 0.0625 mM. After PCR amplification, four PCR products were mixed to generate a pool of mutants. Competent yeast cells containing pGBT9-based plasmids were transformed with the PCR products along with linearized pGAD424 vector for in vivo recombination and circularization of pGAD424 with mutagenized Fob1 PCR products. The transformed cells were diluted and plated on SD/Leu−Trp− plates to select cells that contained pGBT9-based plasmid and circularized pGAD424 + Fob1 PCR products. Colonies that grew on the SD/Leu−Trp− but not the SD/Leu−Trp− Ade− were expected to contain either a putative noninteracting mutant of Fob1 or a blank vector that circularized without an insert. Extraction of plasmid followed by PCR amplification and sequencing was done to confirm whether the pGAD424 plasmid contained the correct Fob1 ORF with a potential mutant.
HOT1 assay
The colony color-sectoring assay was carried out as described (Keil and Roeder 1984; Voelkel-Meiman et al. 1987; Mohanty and Bastia 2004).
Plasmid integration assay
The plasmid integration assay was performed as described (Mohanty et al. 2009).
2D agarose gel electrophoresis
Preparation and separation of replication intermediates by 2D gel electrophoresis were performed according to modifications of published procedures (Brewer and Fangman 1987) as modified (Mohanty et al. 2006).
Protein–protein interaction in vitro
Wild-type Fob1, its E373V mutant, and N-terminal Fob1 were expressed and purified from yeast as GST fusions. Whenever necessary, the GST moiety was cleaved off with PreScission protease (GE Life Sciences), and the Fob1 was purified by passage through a glutathione agarose column followed by a gel filtration step. C-terminal Fob1 and its mutants with a kinase tag were cloned into a PMAL vector that included a TEV protease site that enabled us to cleave off the kinase-tagged protein from the MBP (maltose-binding protein). Fob1 fusion proteins expressed in yeast and Escherichia coli were immobilized on glutathione sepharose or amylose resin, depending on the affinity tag, and purified. One picomole of each protein was immobilized in the respective affinity beads that were allowed to interact with an increasing range of 0.1–0.3 pmol of 32P-labeled putative interacting proteins (as in Fig. 1G) or peptides, eluted, and resolved in SDS–polyacrylamide gels. The images from the PhosphorImager were quantified with ImageJ software from the National Institutes of Health.
ELISA
ELISA was performed as described (Mulugu et al. 2001). Fob1-GST was expressed in yeast and purified. Part of the fusion protein was digested with PreScission protease to cleave the linker between the Fob1 and the GST moiety, and Fob1 was repurified through a glutathione agarose affinity column. N-Fob1-GST was purified by the same procedure from yeast excepting that the GST moiety was left in the fusion protein. Fob1 (without GST; 2.5 pmol) was adsorbed onto each well of a plastic microtitre plate; washed, blocked; overlaid separately with a range (0.05–1.6 pmol) of GST, Fob1-GST, or N-Fob1-GST; and allowed to interact with the immobilized fob1. The binding was quantified by adding goat anti-GST IgG and washing, and interaction with anti-goat IgG-alkaline phosphatase antibody and the absorbance were measured at 405 nm with a plate reader.
Western blot analysis
Logarithmic phase cultures were harvested, washed, and lysed in RIPA buffer (50 mM Tri-Cl at pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate with 1 mM PMSF, complete protease inhibitor mix) by mixing with an equal amount of glass beads for 5 min. One percent SDS was added and incubated for 5 min at 95°C. The beads were washed with equal volumes of RIPA buffer and centrifuged at 13,000 rpm for 25 min, and supernatant was collected. The lysates were quantified by the Bradford assay, and equal amounts of each sample were vigorously mixed with 2× SDS loading buffer, boiled for 5 min, and resolved by 10% SDS-PAGE. The resolved protein samples were transferred onto Hybond enhanced chemiluminescence nitrocellulose membrane (GE Healthcare), and the blots were incubated in the presence of anti-DB Gal4 antibody (1:2500 dilution), anti-Fob1 antibody (1:250 dilution), and anti-β-actin antibody (1:500 dilution, used as loading control; Santa Cruz Biotechnology) for 1 h at room temperature. Secondary antibody treatment, washing, and detection by chemiluminescence were performed as directed by the supplier's manual (ECL, Western blotting detection reagents, and analysis system) (GE Healthcare). Quantification was done by either ImageQuant or ImageJ software.
RLS determination
RLS determination has been described previously (Egilmez and Jazwinski 1989). Briefly, cells growing logarithmically on YPG (YP with 3% glycerol instead of glucose) to suppress growth of petites were spotted (1–2 μL) on YPD 2% agar plates. Individual unbudded cells were micromanipulated to isolated spots on the plate and allowed to bud. The mother cells were removed, and the RLS determination was initiated with these newborn daughters (virgin cells) that had never budded. Forty virgin cells of each strain were followed at 30°C. After each division, the new bud was removed from the cell, which was now counted as one generation older. RLS determinations were performed on at least two separately isolated clones of each strain. Statistical differences in the RLSs of strains in any given experiment were evaluated using the Mann-Whitney test, with two-tailed P-values reported.
Statistical analysis of RLS data
Statistical analysis of RLS data is presented in Supplemental Table 3. Additional information is available in the Supplemental Material.
Supplementary Material
Acknowledgments
We thank Bruce Stillman, Stephen Brill, Oscar Aparicio, Ralph Keil, and Bonita Brewer for strains and plasmids. We thank Toni Carr and Steve Brill for a critical reading of the manuscript. We are grateful to Sean Humphrey and Matthias Mann for sharing unpublished data. This work was supported by grant GM-098013 and South Carolina Clinical and Translational Research grant UL1RR029882 to D.B., and grant AG006168 to S.M.J. DNA sequencing services were provided to S.M.J. by the Genomics and Biostatistics Core supported by grant P20 GM103629. This article is dedicated to Professor Sherman Weissman of Yale University.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.260844.115.
References
- Aguilera A, Garcia-Muse T. 2012. Causes of genome instability. Annu Rev Genet 47: 1–32. [DOI] [PubMed] [Google Scholar]
- Bairwa NK, Zzaman S, Mohanty BK, Bastia D. 2010. Replication fork arrest and rDNA silencing are two independent and separable functions of the replication terminator protein Fob1 of Saccharomyces cerevisiae. J Biol Chem 285: 12612–12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandhu A, Kang J, Fukunaga K, Goto G, Sugimoto K. 2014. Ddc2 mediates Mec1 activation through a Ddc1- or Dpb11-independent mechanism. PLoS Genet 10: e1004136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastia D, Mohanty BK. 1996. Mechanisms for completing DNA replication. In DNA replication in eukaryotic cells (ed. DePamphilis M), pp. 177–215. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Bastia D, Mohanty BK. 2006. Termination of DNA Replication. In DNA replication and human disease (ed. DePamphilis M), pp. 155–174. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Bastia D, Zaman S. 2014. Mechanism and physiological significance of programmed replication termination. Semin Cell Dev Biol 30: 165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benguria A, Hernandez P, Krimer DB, Schvartzman JB. 2003. Sir2p suppresses recombination of replication forks stalled at the replication fork barrier of ribosomal DNA in Saccharomyces cerevisiae. Nucleic Acids Res 31: 893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsen IB, Nielsen I, Lisby M, Nielsen HB, Gupta SS, Mundbjerg K, Andersen AH, Bjergbaek L. 2013. MRX protects fork integrity at protein–DNA barriers, and its absence causes checkpoint activation dependent on chromatin context. Nucleic Acids Res 41: 3173–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R, Lai MS, Foiani M. 2012. Preventing replication stress to maintain genome stability: resolving conflicts between replication and transcription. Mol Cell 45: 710–718. [DOI] [PubMed] [Google Scholar]
- Borghouts C, Benguria A, Wawryn J, Jazwinski SM. 2004. Rtg2 protein links metabolism and genome stability in yeast longevity. Genetics 166: 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BJ. 1988. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell 53: 679–686. [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Fangman W. 1987. Localization of replication origins in ars plasmids in Saccharomyces cerevisiae. Cell 51: 463–471. [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Lockshon D, Fangman W. 1992. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell 71: 267–271. [DOI] [PubMed] [Google Scholar]
- Case ME, Griffith J, Dong W, Tigner IL, Gaines K, Jiang JC, Jazwinski SM, Arnold J, Georgia Centenarian S. 2014. The aging biological clock in Neurospora crassa. Ecol Evol 4: 3494–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JN, Poon BP, Salvi J, Olsen JB, Emili A, Mekhail K. 2011. Perinuclear cohibin complexes maintain replicative life span via roles at distinct silent chromatin domains. Dev Cell 20: 867–879. [DOI] [PubMed] [Google Scholar]
- Christman MF, Dietrich FS, Levin NA, Sadoff BU, Fink GR. 1993. The rRNA-encoding DNA array has an altered structure in topoisomerase I mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci 90: 7637–7641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgaard JZ, Godfrey EL, MacFarlane RJ. 2011. Eukaryotic replication barriers: how, why and where forks stall. In Current advances in DNA replication (ed. Seligmann H). InTech, Rijeka, Croatia. [Google Scholar]
- Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL. 2009. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature 459: 802–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defossez PA, Prusty R, Kaeberlein M, Lin SJ, Ferrigno P, Silver PA, Keil RL, Guarente L. 1999. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell 3: 447–455. [DOI] [PubMed] [Google Scholar]
- Dekker J. 2006. The three ‘C’s of chromosome conformation capture: controls, controls, controls. Nat Methods 3: 17–21. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. 2002. Capturing chromosome conformation. Science 295: 1306–1311. [DOI] [PubMed] [Google Scholar]
- de Laat W, Klous P, Kooren J, Noordermeer D, Palstra RJ, Simonis M, Splinter E, Grosveld F. 2008. Three-dimensional organization of gene expression in erythroid cells. Curr Top Dev Biol 82: 117–139. [DOI] [PubMed] [Google Scholar]
- Di Felice F, Cioci F, Camilloni G. 2005. FOB1 affects DNA topoisomerase I in vivo cleavages in the enhancer region of the Saccharomyces cerevisiae ribosomal DNA locus. Nucleic Acids Res 33: 6327–6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Mello NP, Childress AM, Franklin DS, Kale SP, Pinswasdi C, Jazwinski SM. 1994. Cloning and characterization of LAG1, a longevity-assurance gene in yeast. J Biol Chem 269: 15451–15459. [PubMed] [Google Scholar]
- Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS. 2010. A three-dimensional model of the yeast genome. Nature 465: 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egilmez NK, Jazwinski SM. 1989. Evidence for the involvement of a cytoplasmic factor in the aging of the yeast Saccharomyces cerevisiae. J Bacteriol 171: 37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, Tyler JK. 2013. Elevated histone expression promotes life span extension. Mol Cell 39: 724–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley AR, Kobayashi T. 2014. Ribosomal DNA and cellular senescence: new evidence supporting the connection between rDNA and aging. FEMS Yeast Res 14: 49–59. [DOI] [PubMed] [Google Scholar]
- Henderson AS, Warburton D, Atwood KC. 1972. Location of ribosomal DNA in the human chromosome complement. Proc Natl Acad Sci 69: 3394–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochschild A, Ptashne M. 1988. Interaction at a distance between λ repressors disrupts gene activation. Nature 336: 353–357. [DOI] [PubMed] [Google Scholar]
- Huang GS, Keil RL. 1995. Requirements for activity of the yeast mitotic recombination hotspot HOT1: RNA polymerase I and multiple cis-acting sequences. Genetics 141: 845–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Moazed D. 2003. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev 17: 2162–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Brito IL, Villen J, Gygi SP, Amon A, Moazed D. 2006. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev 20: 2887–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craige CA. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinski SM. 1993. The genetics of aging in the yeast Saccharomyces cerevisiae. Genetica 91: 35–51. [DOI] [PubMed] [Google Scholar]
- Jazwinski SM. 2000. Metabolic control and ageing. Trends Genet 16: 506–511. [DOI] [PubMed] [Google Scholar]
- Jazwinski SM. 2014. The retrograde response: when mitochondrial quality control is not enough. Biochim Biophys Acta 1833: 400–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinski SM, Kim S, Dai J, Li L, Bi X, Jiang JC, Arnold J, Batzer MA, Walker JA, Welsh DA, et al. 2010. HRAS1 and LASS1 with APOE are associated with human longevity and healthy aging. Aging Cell 9: 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang JC, Jaruga E, Repnevskaya MV, Jazwinski SM. 2000. An intervention resembling caloric restriction prolongs life span and retards aging in yeast. FASEB J 14: 2135–2137. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. 2005. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310: 1193–1196. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Burtner CR, Kennedy BK. 2007. Recent developments in yeast aging. PLoS Genet 3: e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil RL, Roeder GS. 1984. Cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell 39: 377–386. [DOI] [PubMed] [Google Scholar]
- Kim RA, Wang JC. 1989. A subthreshold level of DNA topoisomerases leads to the excision of yeast rDNA as extrachromosomal rings. Cell 57: 975–985. [DOI] [PubMed] [Google Scholar]
- Kim S, Benguria A, Lai CY, Jazwinski SM. 1999. Modulation of life-span by histone deacetylase genes in Saccharomyces cerevisiae. Mol Biol Cell 10: 3125–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott SR, Peace JM, Ostrow AZ, Gan Y, Rex AE, Viggiani CJ, Tavare S, Aparicio OM. 2012. Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae. Cell 148: 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Ganley AR. 2005. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 309: 1581–1584. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Horiuchi T, Tongaonkar P, Vu L, Nomura M. 2004. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell 117: 441–453. [DOI] [PubMed] [Google Scholar]
- Lewis EB. 1985. Regulation of the genes of the bithorax complex in Drosophila. Cold Spring Harb Symp Quant Biol 50: 155–164. [DOI] [PubMed] [Google Scholar]
- Lin YL, Pasero P. 2012. Interference between DNA replication and transcription as a cause of genomic instability. Curr Genomics 13: 65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Alberts BM. 1995. Head-on collision between a DNA replication apparatus and RNA polymerase transcription complex. Science 267: 1131–1137. [DOI] [PubMed] [Google Scholar]
- Lobell RB, Schleif RF. 1990. DNA looping and unlooping by AraC protein. Science 250: 528–532. [DOI] [PubMed] [Google Scholar]
- Mekhail K, Seebacher J, Gygi SP, Moazed D. 2008. Role for perinuclear chromosome tethering in maintenance of genome stability. Nature 456: 667–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel J, Malo ME, Chan C, Prusinkiewicz M, Arnason TG, Harkness TA. 2014. The anaphase promoting complex regulates yeast lifespan and rDNA stability by targeting Fob1 for degradation. Genetics 196: 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin EV, Mirkin SM. 2007. Replication fork stalling at natural impediments. Microbiol Mol Biol Rev 71: 13–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron A, Mukherjee S, Bastia D. 1992. Activation of distant replication origins in vivo by DNA looping as revealed by a novel mutant form of an initiator protein defective in cooperativity at a distance. EMBO J 11: 1205–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D. 2001. Common themes in mechanisms of gene silencing. Mol Cell 8: 489–498. [DOI] [PubMed] [Google Scholar]
- Mohanty BK, Bastia D. 2004. Binding of the replication terminator protein Fob1p to the Ter sites of yeast causes polar fork arrest. J Biol Chem 279: 1932–1941. [DOI] [PubMed] [Google Scholar]
- Mohanty BK, Bairwa NK, Bastia D. 2006. The Tof1p–Csm3p protein complex counteracts the Rrm3p helicase to control replication termination of Saccharomyces cerevisiae. Proc Natl Acad Sci 103: 897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty BK, Bairwa NK, Bastia D. 2009. Contrasting roles of checkpoint proteins as recombination modulators at Fob1-Ter complexes with or without fork arrest. Eukaryot Cell 8: 487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Erickson H, Bastia D. 1988a. Detection of DNA looping due to simultaneous interaction of a DNA-binding protein with two spatially separated binding sites on DNA. Proc Natl Acad Sci 85: 6287–6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Erickson H, Bastia D. 1988b. Enhancer–origin interaction in plasmid R6K involves a DNA loop mediated by initiator protein. Cell 52: 375–383. [DOI] [PubMed] [Google Scholar]
- Muller HJ. 1941. Induced mutations in Drosophila. Cold Spring Harb Symp Quant Biol 9: 151–167. [Google Scholar]
- Mulugu S, Potnis A, Shamsuzzaman, Taylor J, Alexander K, Bastia D. 2001. Mechanism of termination of DNA replication of Escherichia coli involves helicase–contrahelicase interaction. Proc Natl Acad Sci 98: 9569–9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques P, Haber J. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 63: 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F, Piruat JI, Aguilera A. 1997. Recombination between DNA repeats in yeast hpr1Δ cells is linked to transcription elongation. EMBO J 16: 2826–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine J. 2005. Twists in the tale of the aging yeast. Science 310: 1124–1125. [DOI] [PubMed] [Google Scholar]
- Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T. 2003. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat Genet 34: 287–291. [DOI] [PubMed] [Google Scholar]
- Roukos V, Misteli T. 2014. The biogenesis of chromosome translocations. Nat Cell Biol 16: 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleif R. 1992. DNA looping. Annu Rev Biochem 61: 199–223. [DOI] [PubMed] [Google Scholar]
- Simonis M, Kooren J, de Laat W. 2007. An evaluation of 3C-based methods to capture DNA interactions. Nat Methods 4: 895–901. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. 1997. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell 91: 1033–1042. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Mills K, Guarente L. 1998. Molecular mechanisms of yeast aging. Trends Biochem Sci 23: 131–134. [DOI] [PubMed] [Google Scholar]
- Singh S, Sabatinos S, Forsburg SL, Bastia D. 2010. Regulation of replication termination by Reb1 protein-mediated action at a distance. Cell 142: 868–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. 2005. Interchromosomal associations between alternatively expressed loci. Nature 435: 637–645. [DOI] [PubMed] [Google Scholar]
- Splinter E, Grosveld F, de Laat W. 2004. 3C technology: analyzing the spatial organization of genomic loci in vivo. Methods Enzymol 375: 493–507. [DOI] [PubMed] [Google Scholar]
- Steinkraus KA, Kaeberlein M, Kennedy BK. 2008. Replicative aging in yeast: the means to the end. Annu Rev Cell Dev Biol 24: 29–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Shou W, Dowd GJ, Turck CW, Deshaies RJ, Johnson AD, Moazed D. 1999. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell 97: 245–256. [DOI] [PubMed] [Google Scholar]
- Strehler BL, Chang MP. 1979. Loss of hybridizable ribosomal DNA from human post-mitotic tissues during aging: II. Age-dependent loss in human cerebral cortex—hippocampal and somatosensory cortex comparison. Mech Ageing Dev 11: 379–382. [DOI] [PubMed] [Google Scholar]
- Strehler BL, Chang MP, Johnson LK. 1979. Loss of hybridizable ribosomal DNA from human post-mitotic tissues during aging: I. Age-dependent loss in human myocardium. Mech Ageing Dev 11: 371–378. [DOI] [PubMed] [Google Scholar]
- Stumpferl SW, Brand SE, Jiang JC, Korona B, Tiwari A, Dai J, Seo JG, Jazwinski SM. 2012. Natural genetic variation in yeast longevity. Genome Res 22: 1963–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Kale SP, Childress AM, Pinswasdi C, Jazwinski SM. 1994. Divergent roles of RAS1 and RAS2 in yeast longevity. J Biol Chem 269: 18638–18645. [PubMed] [Google Scholar]
- Sung P, Krejci L, Van Komen S, Sehorn MG. 2003. Rad51 recombinase and recombination mediators. J Biol Chem 278: 42729–42732. [DOI] [PubMed] [Google Scholar]
- Takeuchi Y, Horiuchi T, Kobayashi T. 2003. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev 17: 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. 2002. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol Cell 20: 1453–1465. [DOI] [PubMed] [Google Scholar]
- Voelkel-Meiman K, Keil RL, Roeder GS. 1987. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell 48: 1071–1079. [DOI] [PubMed] [Google Scholar]
- Wai HH, Vu L, Oakes M, Nomura M. 2000. Complete deletion of yeast chromosomal rDNA repeats and integration of a new rDNA repeat: use of rDNA deletion strains for functional analysis of rDNA promoter elements in vivo. Nucleic Acids Res 28: 3524–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Tsai CL, Lee JT. 2006. Transient homologous chromosome pairing marks the onset of X inactivation. Science 311: 1149–1152. [DOI] [PubMed] [Google Scholar]
- Zehfus BR, McWilliams AD, Lin YH, Hoekstra MF, Keil RL. 1990. Genetic control of RNA polymerase I-stimulated recombination in yeast. Genetics 126: 41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuin J, Dixon JR, van der Reijden MI, Ye Z, Kolovos P, Brouwer RW, van de Corput MP, van de Werken HJ, Knoch TA, van IWF, et al. 2014. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci 111: 996–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.