Abstract
To assess the public’s propensity for allergic contact dermatitis (ACD), many alternatives to in vivo chemical screening have been developed which generally incorporate a small panel of cell surface and secreted dendritic cell biomarkers. However, given the underlying complexity of ACD, one cell type and limited cellular metrics may be insufficient to predict contact sensitizers accurately. To identify a molecular signature that can further characterize sensitization, we developed a novel system using RealSkin, a full thickness skin equivalent, in co-culture with MUTZ-3 derived Langerhan’s cells. This system was used to distinguish a model moderate pro-hapten isoeugenol (IE) and a model strong pre-hapten p-phenylenediamine (PPD) from irritant, salicylic acid (SA). Commonly evaluated metrics such as CD86, CD54, and IL-8 secretion were assessed, in concert with a 27-cytokine multi-plex screen and a functional chemotaxis assay. Data were analyzed with feature selection methods using ANOVA, hierarchical cluster analysis, and a support vector machine to identify the best molecular signature for sensitization. A panel consisting of IL-12, IL-9, VEGF, and IFN-γ predicted sensitization with over 90% accuracy using this co-culture system analysis. Thus, a multi-metric approach that has the potential to identify a molecular signature may be more predictive of contact sensitization.
Keywords: MUTZ-3, Skin equivalent, Co-culture, In vitro, Skin sensitization, Support vector machine
1. Introduction
Allergic contact dermatitis (ACD) is a common inflammatory skin disease that is mediated by adaptive immunity. It is reported that up to 19.5% of the general population is sensitive to at least one allergen (Thyssen et al., 2007). Traditional in vivo screening assays for sensitizers include the guinea pig maximization assay and a more quantitative assay known as the murine local lymph node assay (LLNA). However, there are still several limitations with these animal models that include their low throughput nature, high cost, variability, and ethical concerns. Furthermore, compared to human clinical data, these in vivo assays are only 73% accurate at predicting the sensitization potential of chemicals (Anderson et al., 2011). For these reasons, the European Union has placed a ban on animal testing of cosmetic ingredients (EU, 2003). Alternatives to animal testing should allow for high throughput screening of chemicals using sensitive metrics that can reliably predict human immunotoxicity.
There are many novel alternatives to animal assays in development that seek to model aspects of the in vivo pathway of allergic contact dermatitis and predict the sensitization potential of chemicals (dos Santos et al., 2009). Sensitizers are commonly haptens, which are molecules that can easily penetrate through the epidermal barrier and bind to nucleophilic regions of proteins in the skin. This conjugated complex serves as the immunogen that initiates the allergic contact dermatitis response (Landsteiner and Jacobs, 1935). A high through-put in chemico assay was developed that measures this peptide binding with a high degree of accuracy (Gerberick et al., 2007). Low through-put in vitro assays that utilize dendritic cells have also been explored to predict the sensitization potential of chemicals using maturation metrics such as surface molecules, cytokine production, chemotaxis, and ability to stimulate a T-cell proliferation response.
A major limitation of many of these systems is that they do not contain a metabolic component to accurately identify a class of haptens known as pro-haptens (Johansson et al., 2011; Gerberick et al., 2007). Pro-haptens are innately non-electrophilic and require chemical activation or biotransformation to form reactive intermediates or products that subsequently bind to peptides to form the immunogen (Hagvall et al., 2008). A metabolic component integrated with dendritic cells would be a valuable addition for the identification of pro-hapten sensitizers. Furthermore, due to the complexity of the in vivo mechanisms that trigger allergic contact dermatitis, a tiered approach that tests for a more comprehensive panel of cellular metrics may be needed. To aid in the analysis of these in vitro metrics, high throughput computational tools can be utilized to identify the best predictors of sensitization and streamline the low through-put collection of cellular data.
To develop a culture platform that is physiologically relevant to humans and capable of metabolizing pro-haptens, a re-constructed full thickness skin model known as RealSkin was used either as a stand-alone assay or co-cultured with Langerhan’s cells derived from the MUTZ-3 cell line. This allows for key in vivo events such as permeation through the skin barrier, metabolic activation of pro-haptens, and a dynamic signaling environment from a variety of cell types in vitro. Post-sensitized Langerhan’s cells were subsequently isolated and characterized for the presence of maturation surface molecules, cytokine secretion, and chemotaxis towards CCL19. The secretome from the post-sensitized cultures was evaluated using hierarchical cluster analysis and support vector machine (SVM) classification tools.
SVM is a powerful machine learning tool that is commonly used in pattern recognition and was previously used to classify skin sensitizers based on molecular structures (Ren et al., 2006). We utilized the support vector machine as a feature selection tool to compare different cytokine secretion profiles as potential predictors and using a small sensitizer panel identified the best molecular signature for sensitization, as a proof of concept for the utility of our approach. This analysis tool can also potentially aid in the understanding of key events in allergic contact dermatitis associated with sensitizer potency differences.
2. Materials and methods
2.1. RealSkin and cell lines
RealSkin, a full thickness skin model from EpiSkin™, consists of a dermal equivalent with a lattice of acido-soluble collagen and normal human adult fibroblasts overlaid by a stratified, well differentiated epidermis layer derived from normal human adult. The RealSkin kit including the tissue model and its respective culture medium was provided by EPISKIN™ (Lyon, France) as a donation from L’Oreal (Paris, France). MUTZ-3 cells were a donation from L’Oreal (Paris, France) and are available for purchase from DSMZ (Braunschweig, Germany). 5637 urinary bladder carcinoma cell line was purchased from ATCC (Manassas, Va). RealSkin was cultured for up to a two week period and the medium was changed every other day. A 1 cm2 biopsy punch was applied to the RealSkin prior to experimental use and the excised piece was placed on top of sterile inserts supplied by L’Oreal (Paris, France). The 5637 cell line was maintained at 37 °C and 5% CO2 in RPMI medium supplemented with 10% FBS, 2% L-glutamine, and 1% penicillin–streptomycin. Media was changed every other day and conditioned medium was collected when the cells were 90% confluent. This conditioned medium was supplemented into the MUTZ-3 culture medium as per the guidelines from DSMZ. MUTZ-3 cell line was maintained at 37 °C and 5% CO2 in alpha-MEM medium with Glutamax, ribonucleosides, and deoxyribonucleosides (Invitrogen) supplemented with 20% heat inactivated FBS, 10% 5637 conditioned medium, 1% penicillin–streptomycin, and 50 µM 2-mercaptoethanol. Media was changed every other day and the cells were split on day five of culture.
2.2. Chemicals
The chemical panel selected to perform the initial proof-ofconcept studies were irritant, salicylic acid (SA), a moderate pro-hapten sensitizer, isoeugenol (IE), and a strong pre-hapten, p-phenylenediamine (PPD). These chemicals were selected as representative pre- and pro-haptens that are currently on the list provided by Sens-It-Iv initiative (Newsletter number 02-2007, January 26. http://www.sens-it-iv.eu/files/newsletter/Sens-it-iv_Newsletter_0.8.html). All chemicals were purchased from Sigma–Aldrich. The stock solutions were prepared in dimethylsulfoxide (DMSO) and then subsequently diluted to their final working concentrations using RPMI cell culture medium (Invitrogen). The final concentration of DMSO was 0.4% across all conditions. These working concentrations were determined from dose–response studies where at least 90% of the skin cells were viable as determined by MTT conversion assay (data not shown). These concentrations are 180 µM and 360 µM for SA, 112 µM and 224 µM for IE, and 185 µM and 92.5 µM for PPD. The chemicals were topically applied on either RealSkin alone or on RealSkin as a co-culture with MUTZ-3 derived Langerhan’s cells for 48 hours (Fig. 1).
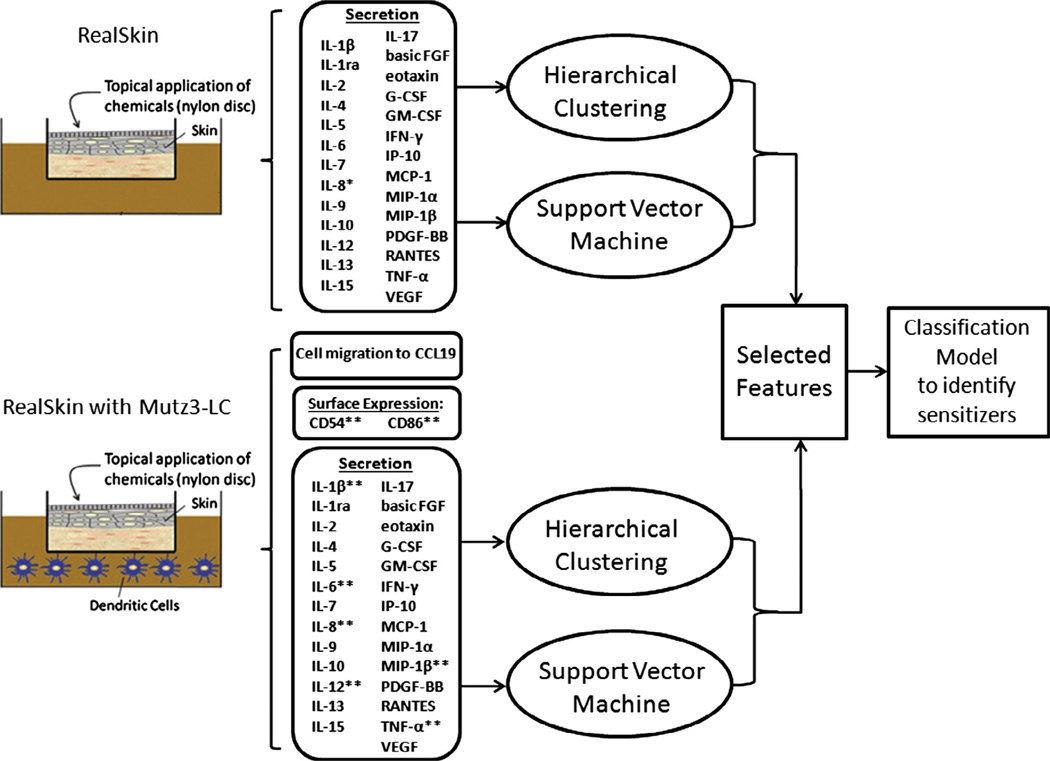
Fig. 1.
Schematic of our experimental system, comparing sensitized RealSkin with Realskin + MUTZ co-cultures. RealSkin was cultured at the air–liquid interface (ALI) and topically dosed with either non-sensitizers (vehicle and salicylic acid (SA)) or sensitizers (isoeugenol (IE) or p-phenylenediamine (PPD)) for 48 hours. For the co-culture configuration, the RealSkin was also cultured at the ALI with the MUTZ-3 derived Langerhan’s cells, cultured in the medium below. The supernatant from each treatment condition was collected and analyzed using the Bioplex assay to screen up to 27 cytokines. Of these cytokines, the * indicates factors that have been previously explored by other groups in literature for skin equivalent sensitization studies and ** indicates factors that have been commonly utilized in dendritic cell based assays (McKim et al., 2012; dos Santos et al., 2009). The secretome data was further analyzed in silico using feature selection tools such as hierarchical cluster analysis and support vector machine to identify potential biomarkers for sensitization. The MUTZ-3 derived LC’s from the co-culture was also analyzed in vitro for surface marker expression changes and chemotaxis towards CCL19.
2.3. Flow cytometry
The phenotype of sensitized MUTZ-LC was determined by flow cytometry. Cells were stained using mouse anti-human monoclonal antibodies to CD54 (IgG1-FITC, R&D Systems) and CD86 (IgG1-FITC, R&D Systems). IgG1-FITC (R&D Systems) was used as an isotype control to assess non-specific binding. Cells were incubated with antibodies for 30 min on ice with 1% mouse serum for blocking, washed three times with PBS, fixed in 4% paraformaldehyde for 15 min, and then re-suspended in the PBS for FACS analysis with a FACScan flow cytometer (Beckton Dickinson, San Jose, Ca.). The data was analyzed using CellQuest Software. Comparisons between different treatment conditions were performed by measuring the stimulation index (SI) which was determined by taking into account the fluorescent intensity and % positive population of cells from the treatment conditions relative to the fluorescent intensity and% positive population of cells treated with the vehicle.
2.4. Trans-well migration assay with CCL19
Sensitized MUTZ-3 LCs co-cultured with RealSkin were harvested and placed into the upper chamber of a 24-well transwell insert (8 µm pore size) at a density of 5 × 104 cells in the presence and absence of 250 ng/mL of chemokine, CCL19. (R&D Systems) Four hours after the cells were exposed to the chemokine, the MUTZ-LCs that migrated were centrifuged and re-suspended in 100 µL of fresh media prior to counting by hemocytometer. The net migration of cells was determined by the following equation using conditions where cells were in the presence and absence of CCL19.
The data was further processed to find the fold difference between all the sensitizer treated conditions with respect to the irritant treated condition. The net migration of LCs for SA treated conditions across a concentration range was averaged since the values were approximately the same. This average net migration for irritant treated cells was then used to determine the fold increase in migration for different sensitizers using the formula below.
2.5. Cytokine multiplex analysis
Supernatant was collected after treating RealSkin alone and RealSkin co-cultured with MUTZ-LCs with salicylic acid, isoeugenol, and p-paraphenylenediamine for 48 hours. Basal alpha-MEM media fortified with 20% heat inactivated FBS and 1% P/S was used as control. The supernatants were then analyzed for 27 human cytokines (IL-1β, IL-1ra, IL-2, IL-4, IL-5, IL-6, IL-8, IL-7, IL-9, IL-10, IL-12, IL-13, IL-15, IL-17, Eotaxin, Basic FGF, G-CSF, GM-CSF, IFNγ, IP-10, MCP-1 (MCAF), MIP-1α, MIP-1β, PDGF-BB, RANTES, TNFα, and VEGF) using a Bioplex Assay following the manufacturer’s instructions. (Bio-Plex Human Cytokine 27-plex panel; Bio-Rad Laboratories, Hercules, CA, USA).
2.6. Hierarchical cluster analysis
Unsupervised agglomerative hierarchical clustering was performed as a feature selection method in Matlab on secretome data from the Bioplex that was normalized to the vehicle treated cultures.
2.7. Support vector machine
The supervised classification approach for distinguishing chemical potency utilizes support vector machine (SVM) as a feature selection method, which determines the best discriminate metrics between the two classes of chemicals (sensitizing vs. non-sensitizing). For each cytokine, the training data is given by {xi, yi}, - i = 1…l, ε − 1, 1, where x denotes the cytokine concentration and y corresponds to the label of the chemical (−1 for non-sensitizers and 1 for sensitizers). Using a linear kernel, the points that lie on the hyperplane that separates the positives and negatives is w · x + b = 0, which leads to the following constraints to the training data:
| (1) |
| (2) |
where w is the norm to the hyperplane and |b|/‖w‖ is the perpendicular distance from the hyperplane to the origin, and ‖w‖ is the Eucledian norm of w. From inequality (1), the perpendicular distance of the hyperplane to the origin is found to be |1 − b|/‖w‖. Similarly, the distance of the hyperplane in (2) to the origin is |−1 −b|/‖w‖. Therefore the margin distance between these two hyperplanes is 2/‖w‖. Thus we can find a pair of hyperplanes that best separates the two classes by minimizing ‖w‖ subject to the constraints (1) and (2). This problem is solved using quadratic programming provided by the bioinformatics toolbox in Matlab. We used the aforementioned method to compute the margin distance 2/||w| for each cytokine. These margin distances and classification accuracy values were subsequently ranked to identify metrics that had the greatest distance of separation between the non-sensitizer (untreated, vehicle, and salicylic acid) and sensitizer treated classes (isoeugenol and paraphenylenediamine).
The support vector machine was also used as a classification model to predict sensitization. Model performance (accuracy, sensitivity, specificity) was assessed using 10-fold cross validation for each individual cytokine metric and molecular signatures identified through various feature selection methods described above.
2.8. Statistical analysis
All data is presented as mean ± standard error. All data in the paper was based on N≥ 3 independent biological replicates. Specifically, the data for the CCL19 migration assay was N = 4 independent biological replicates and the data for all other evaluated metrics (IL-8, CD54, CD86, 27-cytokines) were N = 3 independent biological replicates. To compare the data from the different chemical treatments, we used ANOVA followed by Fisher’s least significant difference post hoc analysis. Statistical significance was determined at p ≤ 0.05 and these values were ranked to perform feature selection on the multi-plex data.
3. Results
3.1. IL-8 secretion
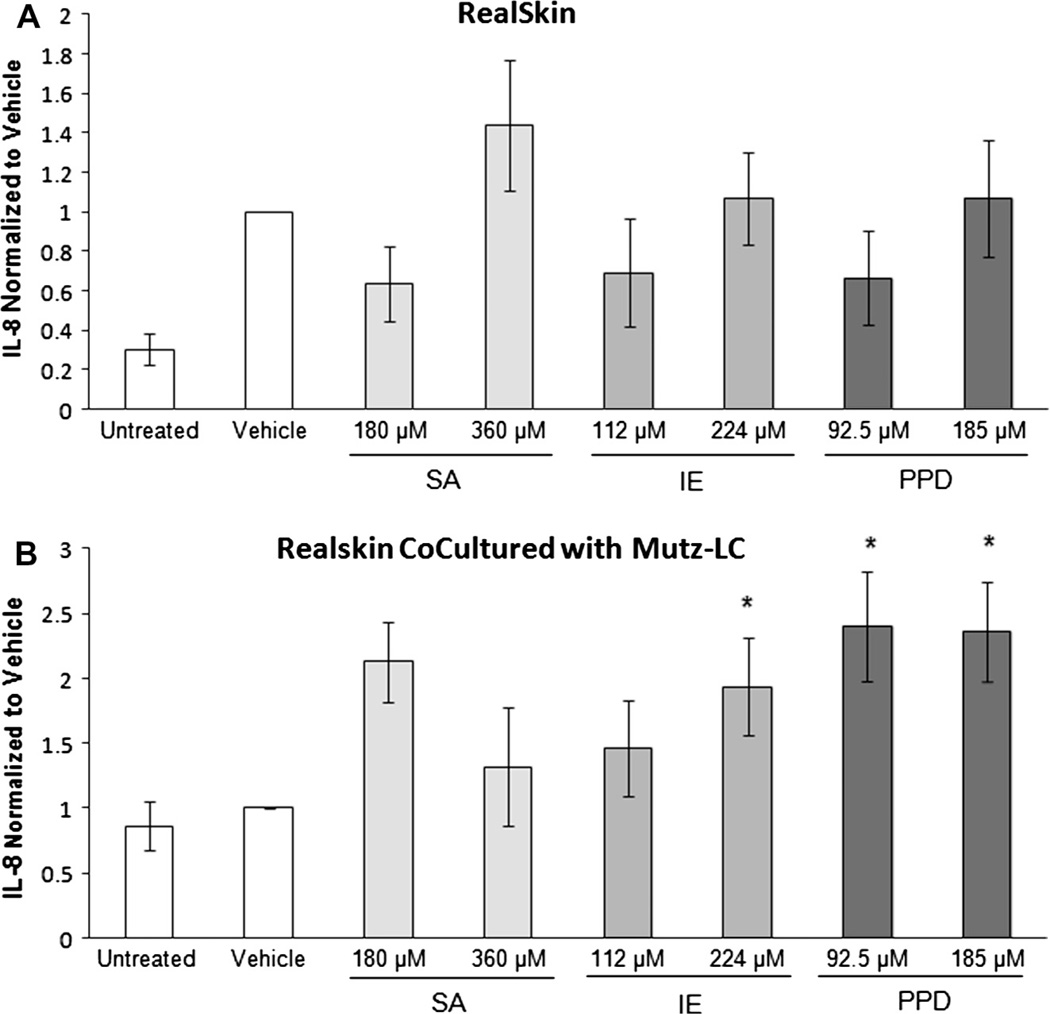
Secretion of IL-8 is a commonly utilized metric for sensitization of several dendritic cell lines and skin cultures (dos Santos et al., 2009). Thus, we assessed IL-8 levels in our co-culture system to see if elevated levels were present in sensitized conditions when normalized to vehicle. IL-8 secretion was evaluated in the supernatant collected from the co-culture configuration with RealSkin and MUTZ-LC 48 hours post-chemical treatment. IL-8 secretion from RealSkin alone indicated that chemical treatments with salicylic acid (SA), isoeugenol (IE), and p-phenylenediamine (PPD) did not significantly alter relative to vehicle treatment (Fig. 2A). Differential secretion between non-sensitized groups (untreated and vehicle) compared to sensitized groups (IE and PPD) were observed (Fig. 2B). However, only PPD induced a significant elevation in secretion levels as compared to the vehicle.
Fig. 2.
Secretion of IL-8 from (A) RealSkin and (B) RealSkin co-cultured with MUTZ-3 LCs. After 48 hours of chemical treatment the secretion level of IL-8 was normalized to the baseline vehicle level. Data is represented as Means ± S.E. for N = 3 independent replicates. (A) RealSkin alone did not induce any significant changes in IL-8 secretion following treatment with the chemicals salicylic acid (SA), isoeugenol (IE) or p-phenylenediamine (PPD). (B) RealSkin Co-Cultured with MUTZ-LCs showed significant IL-8 increases with 224 µM IE and PPD treatments with respect to the untreated and vehicle where p ≤ 0.05. However, these conditions were not significant when compared with the 360 µM dose of SA.
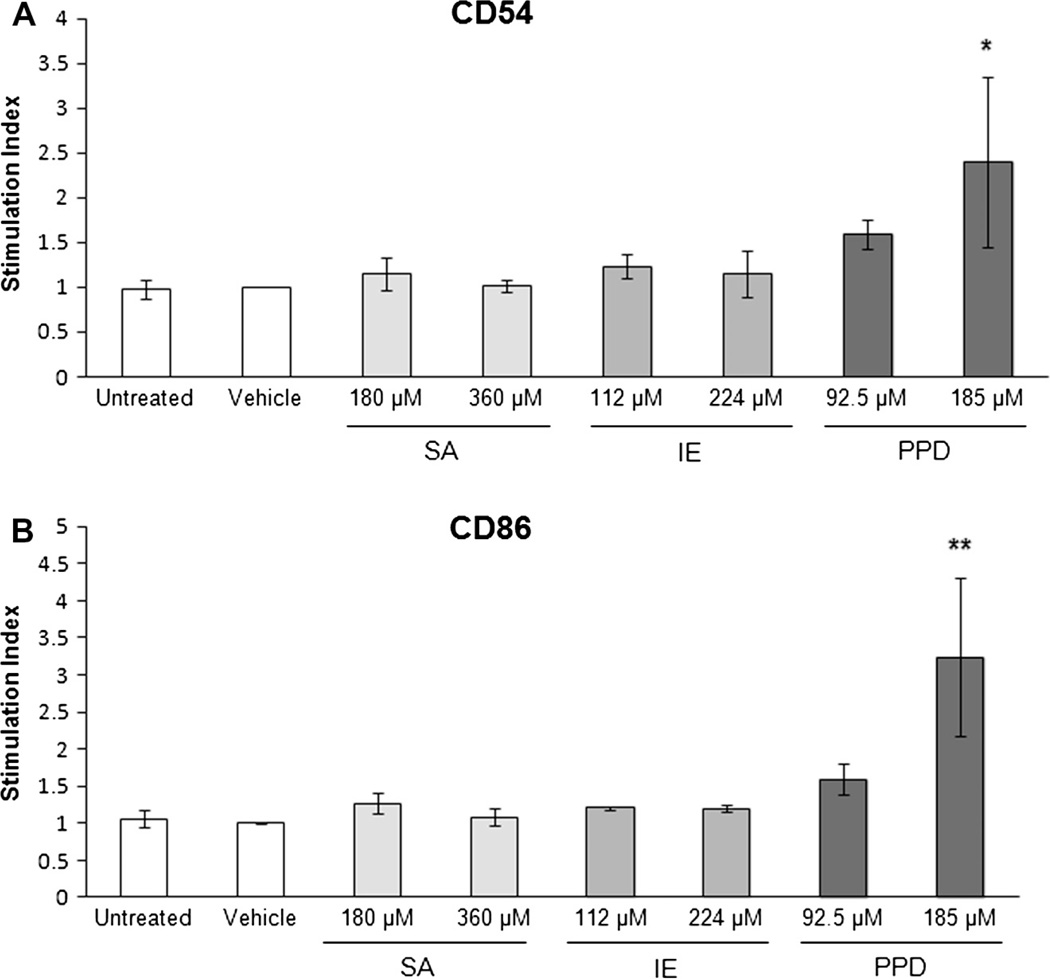
3.2. Evaluation of CD54 and CD86 on MUTZ-LC
Studies were also designed to evaluate the RealSkin full thickness skin co-culture sensitization system using commonly evaluated surface expression metrics for dendritic cell maturation, CD54 and CD86. Sensitizers, isoeugenol (IE) and p-phenylenediamine (PPD) or controls, vehicle (0.4% DMSO in RPMI medium) and irritant salicylic acid (SA) were topically applied to the Real- Skin and DC sensitization was evaluated via immunofluorescence labeling after 48 hours. The flow cytometric results (Fig. 3) of IE and PPD sensitization indicated that exposure to a strong pre-hapten 185 µM PPD, yielded the greatest stimulation for CD86 and CD54. However, isoeugenol, a moderate sensitizer, induced a very mild increase in CD86 and CD54 that was not significant.
Fig. 3.
Expression of CD54 and CD86 following sensitization. The relative percent positive in the sensitized MUTZ-3 LC’s populations was compared to vehicle treatment for (A) CD54 and (B) CD86 expression. The expression level of each corresponding surface molecule treated with vehicle is indicated by the black dotted line. CD54 surface expression did not significantly change with treatments salicylic acid (SA), isoeugenol (IE), or p-phenylenediamine (PPD). CD86 expression increased significantly following treatment with the strong sensitizer PPD. * indicates significance at p ≤ 0.05. Although there is an increased CD86 trend following moderate sensitizer IE treatment, the difference was not significant. Data is represented as Means ± S.E. for N = 3 independent replicates.
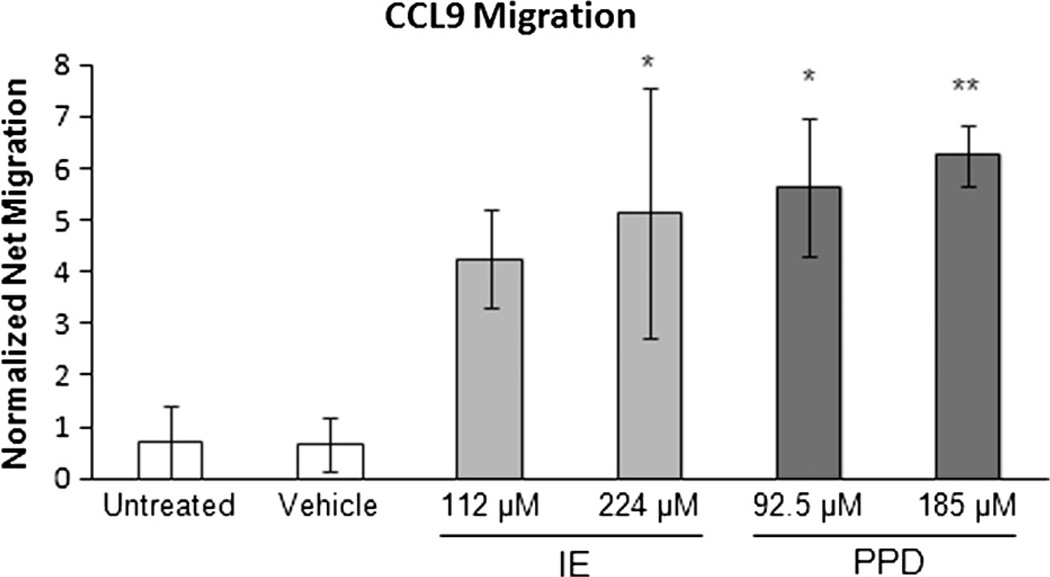
3.3. Evaluation of MUTZ-LC Migration to CCL19
To functionally assess the MUTZ-3 derived LC sensitization mediated chemotaxis towards the chemokine CCL19, a 4 hour trans-well migration assay was performed. After 48 hour sensitizer exposure, an enhanced dose responsive migration towards CCL19 was observed relative to SA for IE and PPD, respectively (Fig. 4).
Fig. 4.
Migration of MUTZ3-LCs co-cultured with RealSkin toward CCL19. Cultures were established and sensitized as described above. Migration was quantified post-exposure to chemical treatments. The data was not normalized to vehicle for the migration assay due to zero net migration when treated with vehicle alone. Instead, the data was normalized to irritant salicylic acid (SA) instead as represented by the dashed line to indicate mean level of migration for SA treated MUTZ3-LCs. Chemotaxis in response to CCL19 exposure was greatest for the strong sensitizer treatment p-phenylenediamine (PPD) followed by the moderate sensitizer isoeugenol (IE). This difference was significant for both sensitizer treatments, where ** indicates p ≤ 0.01. The non-sensitizer treatments (untreated, vehicle, and salicylic acid (SA)) showed minimal cell migration. Data is represented as Means ± S.E. for N = 4 independent replicates.
3.4. Hierarchical cluster analysis of secretome data
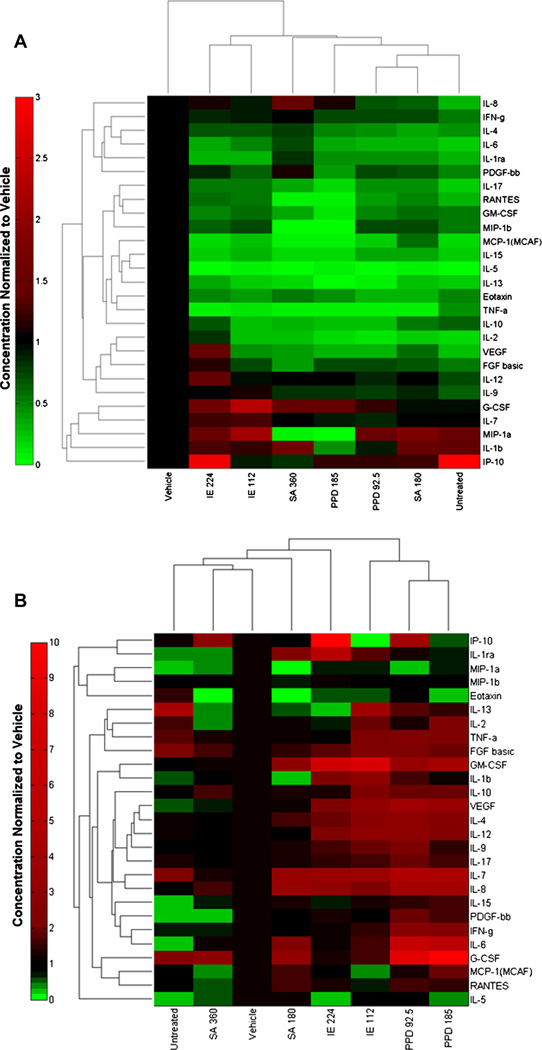
In order to expand our quantitative analysis of RealSkin mediated sensitization, we examined the secreted molecular patterns post-chemical treatment using a 27-cytokine Bioplex screen. The data from this secretome screen was analyzed using high throughput classification tools to identify molecular patterns related to sensitization. An unsupervised agglomerative hierarchical clustering analysis was performed to identify potential cytokines that are elevated post-sensitization and for the two culture conditions (RealSkin treated alone or RealSkin co-cultured with MUTZ-LCs).
The heat map representation of the data shows increased relative levels of secretion in shades of red and decreased levels of secretion in shades of green. The cluster analysis for the RealSkin alone treated with chemicals shows a dendrogram that clusters a node with sensitizer PPD and irritant SA. This demonstrates that these two treatment conditions share similar secretion patterns (Fig. 5A). In contrast, the cluster analysis for the co-culture secretome from RealSkin with MUTZ-LC shows a dendrogram that clusters non-sensitizer treatments (untreated, vehicle, and irritant SA) together. However, the moderate sensitizer, isoeugenol, branches out from this node, indicating a change in the baseline secretion. Moreover, the strong sensitizer PPD, branches off from the isoeugenol treatment conditions and farthest away from the non-sensitizer treatment conditions. Therefore, the non-sensitizer treatment conditions share similar patterns in secretion post-treatment with low levels of inflammatory cytokine secretion and sensitizer treatment conditions show increased levels of inflammatory cytokine secretion. A panel of secreted metrics that is clustered together to reflect these patterns include VEGF, IL-4, IL-9, and IL-12 (Fig. 5B) and are selected as potential candidates to predict skin sensitization.
Fig. 5.
Hierarchical cluster analysis. (A) RealSkin secretome normalized relative to vehicle and (B) RealSkin co-cultured with MUTZ-LCs secretome normalized relative to vehicle after treatment with sensitizers isoeugenol (IE), p-phenylenediamine (PPD), and irritant, salicylic acid (SA) for 48 hours. No discriminant patterns emerged in the RealSkin mono-culture with sensitizer treatment and therefore the cluster analysis does not arrange the treatments according to their actual potency. However, for the co-culture condition, a panel of cytokines (VEGF, IL-4, IL-12, IL-9) show elevated levels of secretion relative to both irritant and vehicle for sensitizer treatments of moderate and strong potency. Heat map data is represented as mean for N = 3 independent replicates.
3.5. Support vector machine analysis on secretome data
Although the hierarchical cluster analysis gave some insight in parsing the potencies of each chemical and identifying potential biomarkers for sensitization based on the multi-plex data, it does not systematically rank the metrics to identify the best possible biomarker(s) quantitatively. Thus, alternative feature selection methods were explored such as the utility of p-values from ANOVA and a support vector machine (SVM) classifier. The SVM calculated the margin distance of separation between two classes of chemicals: non-sensitizer control treatments and sensitizer treatments (IE and PPD). The greater margin distances indicated a greater degree of separation between the two classes for any given metric. Based on this information, we were able to rank each cytokine based on its ability to distinguish between the controls and the sensitizer treatments and select the key features necessary to produce an accurate prediction. The margin distances for the cytokine data from the co-culture assay are greater than the margin distances from the skin equivalent secretome indicating a greater degree of separation from non-sensitized treatment groups and sensitized treatment groups (data not shown). Furthermore, for RealSkin alone, no cytokines were found to be statistically significant and all cytokine metrics showed low accuracies (<75%) for correctly classifying sensitized treatments.
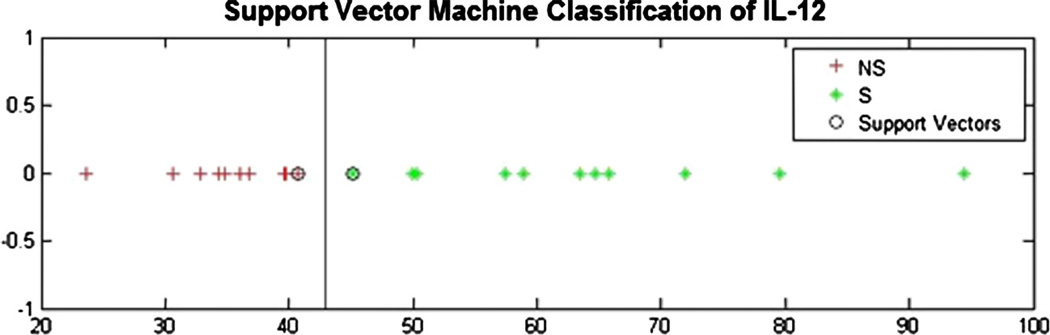
The margin distances of the top ten secretome cytokines collected through the co-culture system with RealSkin and MUTZ-LCs in ranked order are IL-12, IL-9, VEGF, IFN-γ, PDGF, IL-7, IL- 8, GM-CSF and IL-6 (Table 1). A representative scatter plot of the top metric, IL-12 shows the separation of the sensitizer (IE and PPD) data points in contrast to the non-sensitizer (Vehicle, SA) data points that are separated by the margin distance (Fig. 6). The top 3 cytokines, IL-12, IL-9, and VEGF all have accuracy, sensitivity, and specificity values that exceed 80% as individual biomarkers. Using the support vector machine to rank the cytokines by their accuracy instead of by margin distances yields the same panel of cytokines. However, the ranking order is slightly altered to reflect the following hierarchy in descending order of IL-12, IL-9, VEGF, PDGF, IL-4, GM-CSF, IL-6, IL-8, IFN-γ, and IL-7. Ranking the cytokines by p-values determined through ANOVA results in the same cytokines listed in the top 10 as the panel identified by ranking margin distances. However, the order of the top ten cytokine ranking is slightly shuffled to reflect the following order of IL-12, VEGF, IL-9, IFN-γ, IL-4, PDGF, IL-8, IL-7, IL-6, and GM-CSF.
Table 1.
Analysis of margin distances identified by support vector machine analysis. The margin distances quantified from the support vector machine ranked, from greatest to least distance of separation, between non-sensitizer treatment groups and sensitized groups for metrics from a Bioplex screen using supernatant from co-culture of RealSkin with MUTZ-3 derived Langerhan’s cells (RSLC). The accuracy, sensitivity, and specificity for each cytokine metric were determined using a support vector machine classification model with 10-fold cross. The top ten secretion metrics (IL-12, IL-9, VEGF, IFN-γ, IL-4, PDGF, IL-8, IL-7, GM-CSF, and IL-6) that can accurately classify non-sensitizers (vehicle and salicylic acid (SA)) from sensitizers (isoeugenol (IE) and p-phenylenediamine (PPD)) identified by SVM all have p values ≤ 0.05 as determined by ANOVA. IL-12 is the only metric that has an accuracy, sensitivity, and specificity value that exceeds 90%. Data analyzed by the SVM included all treatment conditions (untreated, vehicle, SA, IE, and PPD) and all of their respective concentrations for N = 3 independent replicates.
| Rank | Metrics | Margin distance | Accuracy (%) | Sensitivity (%) | Specificity (%) | P-value |
|---|---|---|---|---|---|---|
| 1 | IL-12 | 0.5564 | 91.67 | 91.67 | 91.67 | <.0001 |
| 2 | IL-9 | 0.3390 | 83.33 | 83.33 | 83.33 | 0.00011 |
| 3 | VEGF | 0.3176 | 83.33 | 83.33 | 83.33 | 0.00022 |
| 4 | IFN-γ | 0.2645 | 66.67 | 66.67 | 66.67 | 0.00146 |
| 5 | IL-4 | 0.2564 | 75.00 | 66.67 | 83.33 | 0.00287 |
| 6 | PDGF-bb | 0.2473 | 79.17 | 83.33 | 75.00 | 0.00732 |
| 7 | IL-8 | 0.2438 | 70.83 | 66.67 | 75.00 | 0.01131 |
| 8 | IL-7 | 0.2403 | 66.67 | 50.00 | 83.33 | 0.01559 |
| 9 | GM-CSF | 0.2391 | 75.00 | 58.33 | 91.67 | 0.02965 |
| 10 | IL-6 | 0.2336 | 70.83 | 58.33 | 83.33 | 0.02993 |
| 11 | IL-17 | 0.2278 | 66.67 | 75.00 | 58.33 | 0.05489 |
| 12 | IL-15 | 0.2233 | 58.33 | 66.67 | 0.50 | 0.083 |
| 13 | IL-10 | 0.2219 | 66.67 | 58.33 | 75.00 | 0.09419 |
| 14 | IL-1β | 0.2196 | 62.50 | 50.00 | 75.00 | 0.09501 |
| 15 | FGF basic | 0.2189 | 66.67 | 66.67 | 66.67 | 0.13044 |
| 16 | TNF-α | 0.2181 | 66.67 | 75.00 | 58.33 | 0.15035 |
| 17 | IL-1ra | 0.2160 | 50.00 | 25.00 | 75.00 | 0.16244 |
| 18 | G-CSF | 0.2147 | 50.00 | 25.00 | 75.00 | 0.22958 |
| 19 | IL-2 | 0.2137 | 54.00 | 50.00 | 58.33 | 0.2565 |
| 20 | IP-10 | 0.2117 | 50.00 | 16.67 | 83.33 | 0.2815 |
| 21 | Rantes | 0.2104 | 50.00 | 50.00 | 50.00 | 0.42441 |
| 22 | Eotaxin | 0.2090 | 33.33 | 33.33 | 33.33 | 0.53499 |
| 23 | MIP-1α | 0.2089 | 33.33 | 25.00 | 41.67 | 0.79776 |
| 24 | MIP-1β | 0.2087 | 25.00 | 33.36 | 16.67 | 0.80659 |
| 25 | IL-13 | 0.2087 | 45.83 | 8.33 | 83.33 | 0.86399 |
| 26 | MCP-1 | 0.2087 | 33.33 | 16.67 | 50.00 | 0.89127 |
| 27 | IL-5 | 0.2085 | 45.83 | 50.00 | 41.67 | 0.90373 |
Fig. 6.
Representative scatter plot that results from the support vector machine analysis for top ranked cytokine IL-12. Red + indicates non-sensitizer (NS) treatments (untreated, vehicle, and salicylic acid (SA)) and green + indicates sensitizer (isoeugenol (IE) and p-phenylenediamine (PPD)) treated conditions. The line separating the two classes in this plot indicates the center of the margin distance boundary. The two encircled points near the margin distance center boundary indicates the critical points that are used as support vectors. IL-12 data analyzed by the SVM classifier included all treatment conditions (untreated, vehicle, SA, IE, and PPD) and all of their respective concentrations for N = 3 independent replicates. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.6. The effect of feature selection on classification performance
We explored whether using cytokines together as a panel identified through different feature selection methods led to greater accuracies in identifying sensitizers than using each secreted metric alone. Ten-fold cross-validation was performed to determine the sensitivity and specificity of the SVM classifier using all of the cytokines in the multi-plex from the co-culture RealSkin and MUTZ-LC supernatant to classify sensitizers, yielding only 75% accuracy with low sensitivity of 67% and specificity of 83% (Table 2).
Table 2.
A classification model that is predictive of skin sensitization was built using a support vector machine (SVM) and metrics identified through various feature selection methods. Classification performance in terms of accuracy, sensitivity, and specificity was determined by 10-fold cross validation. By utilizing all of the 27 cytokines from the Bioplex without performing any feature selection to build a predictive model, the accuracy, sensitivity and specificity of the classifier was very poor. Performing feature selection by ranking the margin distances from the SVM and p-values determined from ANOVA identified IL-12, IL-9, VEGF, IFN-γ as a molecular signature to build the classification model. This classification model performed superiorly as compared to a model built using features selected using hierarchical cluster analysis or by ranking the accuracies computed from the SVM. Data analyzed by all feature selection methods included all treatment conditions (untreated, vehicle, SA, IE, and PPD) and all of their respective concentrations for N = 3 independent replicates.
| Feature selection method | Metrics | Accuracy (%) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|
| No feature selection | All 27 cytokines from bioplex | 75.00 | 67.00 | 83.00 |
| P-values | IL-12, IL-9, VEGF, IFN-γ | 92.00 | 92.00 | 92.00 |
| Hierarchical cluster analysis | IL-4, IL-9, IL-12, VEGF | 91.67 | 91.67 | 91.67 |
| SVM margin distance | IL-12, IL-9, VEGF, IFN-γ | 92.00 | 92.00 | 92.00 |
| SVM accuracy | IL-12, IL-9, VEGF, PDGF | 91.30 | 90.91 | 91.67 |
When hierarchical cluster analysis was used to perform feature selection, a panel of cytokines (IL-12, VEGF, IL-4, IL-9) was identified and used to build the SVM classification model. This feature selection method resulted in a model that performed with accuracy of 91.67% and with a sensitivity and specificity score of 91.67%. When feature selection was performed by ranking the margin distances and p-values, the top 4 metrics (IL-12. IL-9, VEGF, and IFN-γ) were identified and utilized to build a SVM classification model. Both feature selection methods led to the best classification performance where the accuracy, sensitivity, and specificity were all 92% (Table 2). The accuracy value for sensitizer classification was greater when a panel of metrics was used instead of individual cytokine metrics alone (Table 1). Interestingly, the cytokine panel identified using feature selection also performed better than all other cellular metrics including CD54, CD86, and chemotaxis to CCL19 which had respective accuracy scores of 72.73%, 81.83%, and 87.5%. Thus, feature selection methods are useful tools to identify potential molecular signatures to build predictive classification models of skin sensitization.
4. Discussion
Predictive in vitro assays that can accurately identify skin sensitizers and fully replace animal testing are in demand. Although high through-put alternatives such as the peptide binding assays accurately predicted 89% of the 82 sensitizers tested, it failed to accurately identify a class of haptens known as pro-haptens (Gerberick et al., 2007). Pro-haptens require auxiliary chemical reactions to transform the innately inert molecule into a hapten that will bind to peptides in the skin and serve as the antigen that triggers the allergy response. Furthermore, there is evidence that pro-hapten transformation involves cytochrome p450 enzymes present in the epidermis and dermis (Gotz et al., 2012; Bergstrom et al., 2007). Although peptide binding provides a helpful initial tool for evaluating the sensitization potential of chemicals, other cellular factors such as skin permeation, metabolism, cytokine signaling environment, and dendritic cell activation should also be considered.
To address the need for a metabolic component that is more in vivo-like, we developed a co-culture assay system where a full thickness skin model with keratinocytes and fibroblasts known as RealSkin was cultured together with MUTZ-3 derived Langerhan’s cells. RealSkin’s™ mRNA levels for phase I and phase II enzymes were found to be more similar to excised skin than the epidermal skin model, EpiSkin™. This is thought to occur due to fibroblasts in RealSkin which modulate the enzymes’ expression levels (Luu-The et al., 2009). Dermal fibroblasts also have immunomodulatory properties that can facilitate dendritic cell maturation though soluble signals such as TNF-alpha and direct cell–cell contact (Saalbach et al., 2007). Thus, a full thickness skin model that contains both epidermal keratinocytes and dermal fibroblasts co-cultured with Langerhan’s cells can ensure that the dynamic signaling environment across different cell types during sensitization is preserved in vitro. The MUTZ-3 cell line is derived from myeloid leukemia and is established as a viable cell source for differentiation into Langerhan’s cells or dendritic cells upon exposure to different growth factors and cytokines (Azam et al., 2006; Nelissen et al., 2009).
In a previous study by Oewehand et al., a full thickness skin equivalent model with MUTZ-3 derived Langerhan’s included within the epidermis was developed and showed dendritic cell maturation responses upon exposure to skin sensitizers. Here, the LCs were stained for CD83 via immunohistochemistry, IL-1β and CCR7 mRNA were measured, and migration into the dermis of the skin equivalent was observed (Ouwehand et al., 2011). This system is uniquely poised to evaluate the induction of LC migration out of sensitized skin and may also be useful in identifying important sensitization metrics which can be ranked in importance using our analysis tools. Thus far secretome analysis was not performed so we are unable to compare the results of their and our systems in identifying important sensitization metrics. In our co-culture assay the LC are not incorporated directly into the skin equivalent. Nevertheless, we are able to take advantage of the many benefits of skin equivalents. These advantages include the ability to use commercially available products to topically apply chemicals, preservation of the cellular cross talk between keratinocytes, fibroblasts, and LCs, and a source of xenobiotic metabolism for pre/pro-haptens to sensitize the MUTZ-LCs placed below the skin. Thus, with the MUTZ-LCs cultured below the skin compartment, the two components are easily separated and we were able to assess multiple dendritic cell maturation metrics. A small panel of chemicals that include representative pre-/pro-haptens isoeugenol and p-phenylenediamine and irritant salicylic acid were selected in this initial study to show proof-of-concept. Others have also reported small chemical panels for initial system characterization (Tietze and Blomeke 2008; dos Santos et al., 2009; Nelissen et al., 2009; Python et al., 2009). Future studies that incorporate additional chemicals will be necessary to further validate our approach.
Based on the results from our co-culture sensitization platform, we were able to distinguish PPD as a sensitizer relative to vehicle and irritant salicylic acid controls, through CD54 and CD86 surface expression. However, both surface molecules were not elevated for the moderately potent isoeugenol. CD86 and CD54 have been reported as promising surface molecule biomarkers for sensitization with several cell lines including THP-1, U-937, KG-1 and MUTZ-3 (dos Santos et al., 2009). However, an evaluation of CD86 on MUTZ-3 cells using a panel of 20 sensitizers (where PPD and IE were included) and 20 non-sensitizers (where DMSO and SA were included) corroborated our findings where CD86 was upregulated for PPD verses SA and DMSO, but IE was not (Johansson et al., 2011). Thus, CD86 and CD54 may not be sufficient to accurately predict sensitization as standalone metrics.
In addition to surface molecule expression, the level of IL-8 mRNA or secretion is also a commonly evaluated metric of sensitization among several different cell-based assays. Secreted IL-8 was measured from RealSkin alone and from the co-culture of RealSkin with the MUTZ-LCs. RealSkin alone did not secrete IL-8 in a discriminate fashion for sensitizers as compared to the control treatments. IL-8 secretion from the co-culture of RealSkin and MUTZ-LC was able to correctly identify only PPD as a sensitizer. While reports in literature are generally supportive of IL-8 as a distinguisher of sensitization, there are still several reports with false negatives where a contact sensitizer was mis-classified (Toebak et al., 2006). Similar to CD54 and CD86, IL-8 expression may be better suited as a metric that is evaluated within a panel of additional cellular markers. In addition to molecular metrics, a trans-well migration assay was used to functionally assess the MUTZ-LC’s post-sensitization. A dose response with respect to concentrations of the sensitizer was observed for both IE and PPD. These results indicate that a functional assessment of the sensitized LCs to migrate in response to chemokine CCL19 can be used to distinguish pro-hapten IE and pre-hapten PPD from irritant SA and vehicle control (0.4% DMSO in RPMI medium) in our co-culture system.
Several prominent cellular metrics such as CD54, CD86, and IL-8 performed poorly on correctly identifying the less potent sensitizer, isoeugenol. Although these metrics are useful as an initial screening tool, the need to find more sensitive metrics and assays still persist. Thus, a 27-cytokine screen was performed to assess the secretome from the topical application of sensitizers on Real-Skin alone and on RealSkin co-cultured with MUTZ-LCs. An unsupervised classification tool known as hierarchical cluster analysis was used to analyze the secretome data and identify molecular patterns of sensitization. Based on the results of hierarchical cluster analysis, RealSkin secretome did not show any distinguishing cytokine metrics that could be used to cluster sensitizer IE and PPD relative to non-sensitizers SA and vehicle. However, hierarchical clustering of the co-culture system grouped the vehicle and SA treatments with moderate sensitizer IE branching off and with strong PPD treatments clustering adjacent to the IE treatments. Thus, the cluster pattern for this culture configuration also separates treatments by the potency of sensitizers. This suggests that cytokines secreted through co-culture can discriminate between non-sensitizers and sensitizers more accurately than cytokines secreted from the skin alone. Secreted metrics that were identified as prospective predictors in the co-culture setup for the identification of sensitizers include VEGF, IL-4, IL-9, and IL-12. Interestingly, several of these metrics include cytokines currently implicated during skin sensitization and are involved in the immune regulation of T-cells (Kimber et al., 1995; Muller et al., 1995; Watanabe et al., 2002).
To further aid in the discovery of sensitization molecular signatures, a supervised classifier known as a support vector machine was used to classify and identify the best secreted metrics that were capable of distinguishing between sensitizers and non-sensitizers. A support vector machine is a powerful machine learning tool that has been extensively applied in the area of pattern recognition and classification. SVM analysis was compared with linear discriminate analysis (LVA) in screening 130 organic compounds based on 6 molecular descriptors associated with quantitative structure activity relationships (QSAR) based on electrophile– nucleophile reactions that occur during skin sensitization (Ren et al., 2006). Based on this study, SVM methods proved to be superior in classifying compounds as non-strong/moderate or strong/ moderate sensitizers according to their QSAR properties when these in silico results were compared to the Gerberick LLNA database. Additional physicochemical property descriptors were utilized such as hydrophobicity and polarity of the test molecules. However, limited descriptors derived from in vitro biological experiments were applied in this study. Since then, genomic data collected post-sensitization from MUTZ-3 cells has also been used to built a classification model with a support vector machine where 89% accuracy was achieved using a 200 gene signature (GARD assay) (Johansson et al., 2011, 2014). Here, we report the novel use of a SVM for sensitizer classification applied to biological activity at the protein level to build a classification model and develop a feature selection tool to compare different biomarkers. Furthermore, the molecular signature that we’ve identified utilizes four secreted soluble proteins that are easily detectable by ELISA or multi-plex and do not require the more labor intensive nucleic acid based protocols.
By comparing the computed margin distances from the secreted metrics of the RealSkin alone and the co-culture of RealSkin with MUTZ-LCs, the co-culture system showed superior assay performance in the correct prediction of both the moderate pro-hapten isoeugenol and strong pre-hapten PPD. This is likely due to the more in vivo-like nature of the co-culture system where the dynamic cellular interactions between keratinocytes, fibroblasts and Langerhan’s cells are preserved. By systematically ranking the margin distances and accuracy values, a panel of cytokines capable of distinguishing sensitizers was identified. This panel includes IL-12, IL-9, VEGF, IFN-γ, IL-4, PDGF, IL-8, IL-7, GM-CSF, IL-6, and IL-17 with the top three metrics with accuracies, sensitivities, and specificities all exceeding 83.33%. By ranking the statistical p-values from ANOVA, we found the same panel of potential cytokine markers.
After identifying cytokines of interest, we explored whether using them as a panel led to greater accuracies in identifying sensitizers than using each secreted metric alone. Using 10-fold cross-validation, it was determined that using a panel of metrics identified independently through cluster analysis, SVM, and ANOVA improved the accuracy, specificity, and sensitivity in correctly identifying PPD and IE as sensitizers better than using a single cellular metric alone. The results indicate that traditional feature selection methods such as ANOVA and hierarchical cluster analysis validate our support vector machine feature selection approach. However, additional benefits of using SVM as a feature selection tool over traditional statistical methods based on filtering significant p-values or cluster analysis are that SVM accounts for dependencies amongst the features, enables quantitative ranking of individual metrics, interacts with the classifier, and affords superior computational complexity (Saeys et al., 2007). The ability to interact with the classifier is especially important as the classification model is progressively trained with more biological information provided by expanding the panel of chemicals and cellular metrics.
The classifier accuracy, sensitivity, and specificity were all greater than 90% when the best secreted features were selected based on our small panel of sensitizers. This supports the recent notion that a molecular signature or a battery of assays needs to be implemented together to correctly identify potential contact allergens (Bauch et al., 2012; Guyard-Nicodeme, 2014). The true predictive power of our specific molecular signature will need to undergo validation studies with an expanded panel of chemicals. However, the accuracy of our classification model suggests that this identified signature validates our feature selection method by SVM and the need to use a panel of several metrics. Thus, as more chemicals are evaluated, it is feasible that the number of predictive metrics may expand as was the case in the GARD study where initial studies utilized 10 genes and was later expanded to 200 genes (Johansson et al., 2011). Furthermore, additional sensitizers or varying potencies will need to be evaluated to determine whether the system can accurately predict potencies of chemicals as well as their sensitizing potential.
Our molecular panel includes secreted products that are implicated during antigen presentation such as cytokines IL-12, IFN-γ, and IL-9. VEGF is a potent vasodilator that facilitates lymphocyte infiltration into the skin during an allergy response (Brown et al., 1995; Watanabe et al., 2004). Thus, all proteins identified are physiologically relevant to allergic contact dermatitis. Recent literature also indicates IL-18 as an effective predictor of skin sensitization using epidermal equivalents and the NCTC2544 keratinocyte cell line (Corsini et al., 2009; Gibbs et al., 2013). Although IL-18 was not measured as part of our 27-cytokine Bioplex screen, we did identify IFN-γ as a predictive marker of skin sensitization. Since IL-18 is a potent inducer of IFN-γ production and promoter of a Th-1 response, it is feasible that there could be some IL-18 release in our co-culture model (Okamura et al., 1995). However, previous studies with skin equivalents indicate that the accuracy of IL-18 as a biomarker depends on cytotoxicity levels that yield <50% (Gibbs et al., 2013). None the less, future studies could benefit from characterizing IL-18 release and this cellular metric could be compared to the others evaluated in this paper using our SVM selection approach. Also, these studies should include more cytotoxic concentrations as part of the dose response studies to allow for the full predictive potential of IL-18 release as a biomarker.
Although the use of skin equivalents may be more time and resource intensive than submerged cell cultures if they are developed in-house, these inconveniences could be significantly reduced if the skin equivalents are purchased directly through a vendor. Although RealSkin is currently not available for purchase through EpiSkin, there are several full thickness skin equivalents available on market such as MatTek’s EpidermFT, and CellSystems’ AST2000 (Koeper et al., 2007; Varnum et al., 2012). Commercially available epidermal equivalents such as MatTek EpiDerm™, SkinEthic ™ EpiSkin, and SkinEthic™ RHE may also be used in a co-culture model (Gibbs et al., 2013). There is evidence that assays that use of epidermal equivalents may be more amenable to inter-laboratory transfers to generate reproducible results than submerged cell lines that are more sensitive to specific culture conditions (Gibbs et al., 2013; Teunis et al., 2013). Ultimately, it is clear that a single assay and cellular metric of sensitization is insufficient as a stand-alone predictor and tiered strategies using a battery of assays improved the overall accuracy of identifying known skin sensitizers (Bauch et al., 2012; Guyard-Nicodeme, 2014). Thus, we envision this type of co-culture system to be utilized as a second tier assay following in silico or peptide binding screening methods. Unlike studies where a battery of assays with different cell sources, metrics, and specifications are evaluated separately, we envision a more streamlined approach where several key in vivo sensitization steps such as permeation, metabolism, keratinocyte activation, and dendritic cell maturation can all be measured within a single assay system.
In conclusion, we established an assay system that utilizes an organotypic skin model co-cultured with differentiated Langerhan’s cells from the MUTZ-3 cell line as a potential alternative to animal testing. Furthermore, we describe a novel feature selection method to identify key biomarkers of sensitization using a support vector machine to rank the margin distances from a panel of 27 secreted cellular metrics. Unlike standard dendritic cell based assays, chemicals can be topically applied to the skin model and permeate through the skin to activate the MUTZ-LCs below. Additional benefits of a co-culture system include the metabolic capabilities of the skin model to convert pro- and pre-hapten sensitizers into electrophilic products and the preserved cellular interactions between keratinocytes, fibroblasts, and Langerhan’s cells. Based on our comprehensive analysis of multiple cellular metrics, we found that CD54, CD86, and IL-8 may be unreliable markers to identify potential skin sensitizers in our system. However, using a multi-plex screen combined with unsupervised and supervised classification tools, we identified a molecular pattern of cytokines that were expressed after sensitization. Furthermore, the support vector machine was utilized as a tool to systematically rank these cytokines of interest and selected the best panel of sensitization biomarkers in our system. This panel showed greater accuracy, sensitivity, and specificity than using a single secreted metric. Future studies will focus on evaluating an expanded panel of sensitizers with wider ranges of dosages to further optimize and validate our culture approach and molecular signature. The support vector machine feature selection method and classification model we have developed can be continually expanded to compare cellular metrics obtained by our laboratory and by others in training the classification model to predict sensitizers and their respective sensitizer potencies.
Acknowledgements
The authors would like to thank L’Oreal for their generous gift of RealSkin and the NIH Biotechnology Training Grant T32GM8339-21.
Abbreviations
- ACD
allergic contact dermatitis
- ALI
air liquid interface
- CD
cluster of differentiation
- DC
dendritic cell
- DMSO
dimethylsulfoxide
- IL
interleukin
- IE
isoeugenol
- IFN
interferon
- LC
Langerhan’s cell
- LLNA
local lymph node assay
- MUTZ-LC
MUTZ-3 differentiated Langerhan’s cell
- NS
non-sensitizer
- PPD
p-phenylenediamine
- QSAR
quantitative, structure activity relationship
- SA
salicylic acid
- S
sensitizer
- SVM
support vector machine.
Footnotes
Conflict of Interest
The authors declare that there are no conflicts of interest.
Transparency Document
The Transparency document associated with this article can be found in the online version.
References
- Anderson SE, Siegel PD, et al. The LLNA: a brief review of recent advances and limitations. J. Allergy (Cairo) 2011;2011:424203. doi: 10.1155/2011/424203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam P, Peiffer JL, et al. The cytokine-dependent MUTZ-3 cell line as an in vitro model for the screening of contact sensitizers. Toxicol. Appl. Pharmacol. 2006;212(1):14–23. doi: 10.1016/j.taap.2005.06.018. [DOI] [PubMed] [Google Scholar]
- Bauch C, Kolle SN, et al. Putting the parts together: combining in vitro methods to test for skin sensitizing potentials. Regul. Toxicol. Pharmacol. 2012;63(3):489–504. doi: 10.1016/j.yrtph.2012.05.013. [DOI] [PubMed] [Google Scholar]
- Bergstrom MA, Ott H, et al. A skin-like cytochrome P450 cocktail activates prohaptens to contact allergenic metabolites. J. Invest. Dermatol. 2007;127(5):1145–1153. doi: 10.1038/sj.jid.5700638. [DOI] [PubMed] [Google Scholar]
- Brown LF, Olbricht SM, et al. Overexpression of vascular permeability factor (VPF/VEGF) and its endothelial cell receptors in delayed hypersensitivity skin reactions. J. Immunol. 1995;154(6):2801–2807. [PubMed] [Google Scholar]
- Corsini E, Mitjans M, et al. Use of IL-18 production in a human keratinocyte cell line to discriminate contact sensitizers from irritants and low molecular weight respiratory allergens. Toxicol. In Vitro. 2009;23(5):789–796. doi: 10.1016/j.tiv.2009.04.005. [DOI] [PubMed] [Google Scholar]
- dos Santos GG, Reinders J, et al. Progress on the development of human in vitro dendritic cell based assays for assessment of the sensitizing potential of a compound. Toxicol. Appl. Pharmacol. 2009;236(3):372–382. doi: 10.1016/j.taap.2009.02.004. [DOI] [PubMed] [Google Scholar]
- EU. Directive 2003/15/EC of the European parliament and of the council of 27 February 2003 amending council directive 76/768/EEC on the approximation of the laws of the member states relating to cosmetic products. Off. J. Eur. Union. 2003;L66:26–35. [Google Scholar]
- Gerberick GF, Vassallo JD, et al. Quantification of chemical peptide reactivity for screening contact allergens: a classification tree model approach. Toxicol. Sci. 2007;97(2):417–427. doi: 10.1093/toxsci/kfm064. [DOI] [PubMed] [Google Scholar]
- Gibbs S, Corsini E, et al. An epidermal equivalent assay for identification and ranking potency of contact sensitizers. Toxicol. Appl. Pharmacol. 2013;272(2):529–541. doi: 10.1016/j.taap.2013.07.003. [DOI] [PubMed] [Google Scholar]
- Gotz C, Pfeiffer R, et al. Xenobiotic metabolism capacities of human skin in comparison with a 3D epidermis model and keratinocyte-based cell culture as in vitro alternatives for chemical testing: activating enzymes (Phase I) Exp. Dermatol. 2012;21(5):358–363. doi: 10.1111/j.1600-0625.2012.01486.x. [DOI] [PubMed] [Google Scholar]
- Guyard-Nicodeme M, Gerault E, et al. Development of a multiparametric in vitro model of skin sensitization. J. Appl. Toxicol. 2014 doi: 10.1002/jat.2986. [DOI] [PubMed] [Google Scholar]
- Hagvall L, Baron JM, et al. Cytochrome P450-mediated activation of the fragrance compound geraniol forms potent contact allergens. Toxicol. Appl. Pharmacol. 2008;233(2):308–313. doi: 10.1016/j.taap.2008.08.014. [DOI] [PubMed] [Google Scholar]
- Johansson H, Lindstedt M, et al. A genomic biomarker signature can predict skin sensitizers using a cell-based in vitro alternative to animal tests. BMC Genomics. 2011;12:399. doi: 10.1186/1471-2164-12-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson H, Rydnert F, et al. Genomic allergen rapid detection in-house validation – a proof of concept. Toxicol. Sci. 2014;139(2):362–370. doi: 10.1093/toxsci/kfu046. [DOI] [PubMed] [Google Scholar]
- Kimber I, Holliday MR, et al. Cytokine regulation of chemical sensitization. Toxicol. Lett. 1995;82–83:491–496. doi: 10.1016/0378-4274(95)03497-8. [DOI] [PubMed] [Google Scholar]
- Koeper LM, Schulz A, et al. In vitro differentiation of skin sensitizers by cell signaling pathways. Toxicology. 2007;242(1–3):144–152. doi: 10.1016/j.tox.2007.09.019. [DOI] [PubMed] [Google Scholar]
- Landsteiner K, Jacobs J. Studies on the sensitization of animals with simple chemical compounds. J. Exp. Med. 1935;61(5):643–656. doi: 10.1084/jem.61.5.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu-The V, Duche D, et al. Expression profiles of phases 1 and 2 metabolizing enzymes in human skin and the reconstructed skin models Episkin and full thickness model from Episkin. J. Steroid Biochem. Mol. Biol. 2009;116(3–5):178–186. doi: 10.1016/j.jsbmb.2009.05.011. [DOI] [PubMed] [Google Scholar]
- McKim JM, Jr, Keller DJ, 3rd, et al. An in vitro method for detecting chemical sensitization using human reconstructed skin models and its applicability to cosmetic, pharmaceutical, and medical device safety testing. Cutan. Ocul. Toxicol. 2012;31(4):292–305. doi: 10.3109/15569527.2012.667031. [DOI] [PubMed] [Google Scholar]
- Muller G, Saloga J, et al. IL-12 as mediator and adjuvant for the induction of contact sensitivity in vivo. J. Immunol. 1995;155(10):4661–4668. [PubMed] [Google Scholar]
- Nelissen I, Selderslaghs I, et al. MUTZ-3-derived dendritic cells as an in vitro alternative model to CD34+ progenitor-derived dendritic cells for testing of chemical sensitizers. Toxicol. In Vitro. 2009;23(8):1477–1481. doi: 10.1016/j.tiv.2009.08.022. [DOI] [PubMed] [Google Scholar]
- Okamura H, Tsutsi H, et al. Cloning of a new cytokine that induces IFN-gamma production by T cells. Nature. 1995;378(6552):88–91. doi: 10.1038/378088a0. [DOI] [PubMed] [Google Scholar]
- Ouwehand K, Spiekstra SW, et al. Technical advance. Langerhans cells derived from a human cell line in a full-thickness skin equivalent undergo allergen-induced maturation and migration. J. Leukoc Biol. 2011;90(5):1027–1033. doi: 10.1189/jlb.0610374. [DOI] [PubMed] [Google Scholar]
- Python F, Goebel C, et al. Comparative DNA microarray analysis of human monocyte derived dendritic cells and MUTZ-3 cells exposed to the moderate skin sensitizer cinnamaldehyde. Toxicol. Appl. Pharmacol. 2009;239(3):273–283. doi: 10.1016/j.taap.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Ren Y, Liu H, et al. Classification study of skin sensitizers based on support vector machine and linear discriminant analysis. Anal. Chim. Acta. 2006;572(2):272–282. doi: 10.1016/j.aca.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Saalbach A, Klein C, et al. Dermal fibroblasts induce maturation of dendritic cells. J. Immunol. 2007;178(8):4966–4974. doi: 10.4049/jimmunol.178.8.4966. [DOI] [PubMed] [Google Scholar]
- Saeys Y, Inza I, et al. A review of feature selection techniques in bioinformatics. Bioinformatics. 2007;23(19):2507–2517. doi: 10.1093/bioinformatics/btm344. [DOI] [PubMed] [Google Scholar]
- Teunis M, Corsini E, et al. Transfer of a two-tiered keratinocyte assay: IL-18 production by NCTC2544 to determine the skin sensitizing capacity and epidermal equivalent assay to determine sensitizer potency. Toxicol. In Vitro. 2013;27(3):1135–1150. doi: 10.1016/j.tiv.2012.06.004. [DOI] [PubMed] [Google Scholar]
- Thyssen JP, Linneberg A, et al. The epidemiology of contact allergy in the general population–prevalence and main findings. Contact Dermatitis. 2007;57(5):287–299. doi: 10.1111/j.1600-0536.2007.01220.x. [DOI] [PubMed] [Google Scholar]
- Tietze C, Blomeke B. Sensitization assays: monocyte-derived dendritic cells versus a monocytic cell line (THP-1) J. Toxicol. Environ. Health A. 2008;71(13–14):965–968. doi: 10.1080/15287390801989168. [DOI] [PubMed] [Google Scholar]
- Toebak MJ, Pohlmann PR, et al. CXCL8 secretion by dendritic cells predicts contact allergens from irritants. Toxicol. In Vitro. 2006;20(1):117–124. doi: 10.1016/j.tiv.2005.06.039. [DOI] [PubMed] [Google Scholar]
- Varnum SM, Springer DL, et al. The effects of low-dose irradiation on inflammatory response proteins in a 3D reconstituted human skin tissue model. Radiat. Res. 2012;178(6):591–599. doi: 10.1667/RR2976.1. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Mamelak AJ, et al. Anti-vascular endothelial growth factor receptor-2 (Flk-1/KDR) antibody suppresses contact hypersensitivity. Exp. Dermatol. 2004;13(11):671–681. doi: 10.1111/j.0906-6705.2004.00240.x. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Unger M, et al. Contact hypersensitivity: the mechanism of immune responses and T cell balance. J. Interferon Cytokine Res. 2002;22(4):407–412. doi: 10.1089/10799900252952181. [DOI] [PubMed] [Google Scholar]