Abstract
Interactions between a pathogen and a vector are plastic and dynamic. Such interactions can be more rapidly accommodated by epigenetic changes than by genetic mutations. Gene expression can be affected by the proximity to the heterochromatin, by local histone modifications, and by the three-dimensional position within the nucleus. Recent studies of disease vectors indicate that gene regulation by these factors can be important for susceptibility to pathogens, reproduction, immunity, development, and longevity. Knowledge about heterochromatin, histone modifications, and nuclear architecture will help our understanding of epigenetic mechanisms that control gene function at traits related to vectorial capacity.
Introduction
Genomic approaches are now the focus for the development of novel vector control strategies [1]. However, vector-pathogen interactions are plastic, and the outcome of these interactions might not be solely explained by genetic variation [2]. Thus, it is critical to expand our current understanding of vector-pathogen-host interactions to a new level that includes epigenetic modifications of the genome. Epigenetic mechanisms include heritable effects on gene expression caused by heterochromatin, nuclear architecture, variations in DNA methylation, post-translational modifications of histones, and non-coding RNAs [3-5]. Epigenomic studies operate with the complete set of epigenetic modifications, and they are predominantly conducted in model organisms (www.modencode.org) [6], as well as in humans (www.epigenome.org) [7] and parasites (http://plasmodb.org) [5]. Vectors are usually the only part in “disease triads” (hosts—pathogens—vectors) that lacks comprehensive epigenomic studies. The aim of this review is to highlight recent research advances and to identify what is still missing in understanding of epigenetic mechanisms in disease vectors. Heterochromatin, histone modifications, and nuclear architecture are well interconnected. Heterochromatin has specific histone modifications, such as H3K9me2, H3K9me3, and H3K27me3 [8], and it can form attachments between chromosomes and the nuclear envelope [9, 10]. Chromatin regions marked with H3K27me3 can mediate intra- and inter-chromosomal interactions in the nucleus [11]. Nuclear architecture provides the positioning of transcriptionally active chromatin marked with H3K4me3 and H3K79me2 away from the nuclear periphery, which is a repressive environment [12]. Therefore, a better understanding of one epigenetic mechanism will help to decipher the role of other mechanisms in controlling vectorial capacity.
Organization of heterochromatin in disease vectors
Chromatin of eukaryotes exists in at least two distinct forms that were originally defined by morphology as darkly stained constitutive heterochromatin, which remains condensed throughout the cell cycle, and as lightly stained euchromatin, which undergoes cycles of condensation and de-condensation [13]. Among arthropods, the most detailed analysis of the heterochromatin has been performed in a nonvector species, Drosophila melanogaster. Heterochromatin in this species plays an important role in the chromosome pairing, gene silencing via position-effect variegation, maintaining genome stability, production of Piwi-interacting RNAs, and organism longevity [3, 4]. Over 77% of the 24 Mb of pericentromeric heterochromatin in Drosophila is represented by transposable elements (TEs) and about 10% is occupied by tandem repeats, which concentrate toward the centromeres [14]. Despite being transcriptionally less active than euchromatin, the fruit fly heterochromatin contains essential protein-coding genes and the ribosomal DNA (rDNA) locus. A genome-wide study of 53 chromatin proteins in Drosophila cells has revealed five principal chromatin types indicated by the colors YELLOW, RED, BLUE, BLACK, and GREEN [15]. GREEN chromatin, marked by histone-lysine methyltransferase SU(VAR)3-9 and heterochromatin protein 1 (HP1), includes the pericentromeric constitutive and facultative heterochromatin. BLUE chromatin, marked by Polycomb group (PcG) proteins and H3K27me3, corresponds to the PcG heterochromatin. A repressive BLACK chromatin, marked by Lamin and Suppressor of Under-Replication (SUUR), corresponds to the intercalary heterochromatin. Finally, YELLOW and RED chromatin types represent housekeeping and tissue specific genes of euchromatin, respectively.
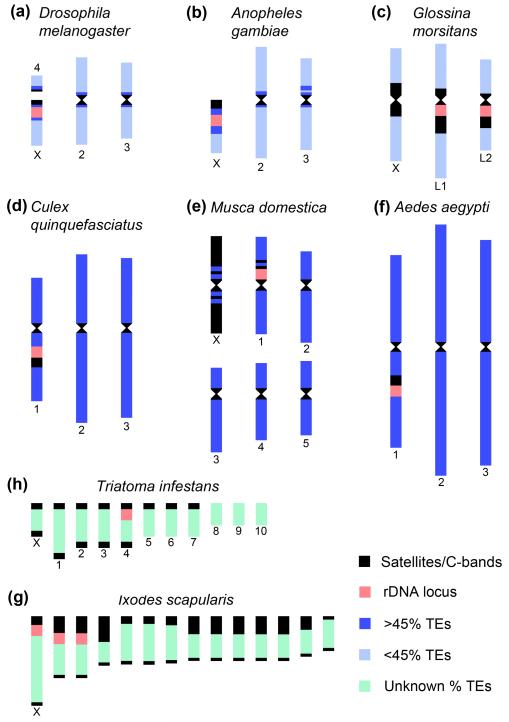
Studies on genomic mapping and characterization of heterochromatin in arthropod vectors are sparse. But even limited numbers of studied cases demonstrate the high diversity in heterochromatin amount and location among species (Figure 1). In the African malaria mosquito Anopheles gambiae, heterochromatin was first described based on Hoechst 33258 staining of mitotic chromosomes [16]. The heterochromatin amount has been found varying among and within species. An early study suggested a possible role of sex chromosome heterochromatin in controlling fertility and mating behavior of An. gambiae [17]. To determine the extent of heterochromatin within the An. gambiae genome assembly, genes have been physically mapped to the euchromatin-heterochromatin transition zone of polytene chromosomes [18]. The study has found that a minimum of 232 genes reside in 16.6 Mb of mapped heterochromatin. Similarly to fruit fly, the heterochromatin in An. gambiae accumulates HP1, includes the rDNA locus, and is enriched with essential protein-coding genes important for establishing, maintaining, and modifying chromatin structure [18]. In the yellow fever mosquito Ae. aegypti, constitutive heterochromatin was first detected by a C-banding technique in pericentromeric areas and in the intercalary region of chromosome 1 [19]. A recent genomic analysis of mapped supercontigs demonstrated a uniformly high coverage of TEs (~52%) across all chromosomal arms with no clear distinction between pericentromeric regions and the rest of chromosomal arms [20]. This organization sharply contrasts with that in An. gambiae, whose genome has a higher TE content in pericentromeric heterochromatin (~54%) and a lower TE content in euchromatic arms (~12%). Thus, increase in the genomic TE content can cause “heterochromatization” of euchromatin seen in Ae. aegypti. In most of the other vector species, heterochromatin has been characterized based on C-banding or fluorescence in situ hybridization (FISH) of repetitive DNA. In the kissing bug Triatoma infestans, a vector of Chagas disease, heterochromatin consists of AT-rich simple tandem repeats and is located in the terminal regions of holokinetic (i.e., with diffuse centromere) chromosomes [21]. In the hard tick Ixodes scapularis, large blocks of pericentromeric heterochromatin of telocentric chromosomes are composed of the few major tandem repeat families [22, 23]. It is likely that satellite DNA repeats rather than TEs mainly contribute to the large genome sizes of the kissing bug (1.7 Gb) [24] and the hard tick (2.1 Gb) [22]. The diversity of the heterochromatin amount and location could potentially result in diverse functions with respect to gene regulation.
Figure 1.
Schematic representation of heterochromatin organization in chromosomes of fruit fly and vector species. Genomes with a low overall TE content (a-c): Drosophila melanogaster [14] (a), Anopheles gambiae [18] (b), Glossina morsitans [54, 59] (c). Genomes with a high overall TE content (d-f): Culex quinquefasciatus [55] (d), Musca domestica [56, 60] (e), Aedes aegypti (f) [20]. Constitutive heterochromatin is marked by the black color representing satellite DNA and C-bands. Chromosome regions with a low (<45%) and a high (>45%) TE contents are indicated by the light blue and dark blue colors, respectively. Genomes with a relatively high tandem repeat (satellite DNA) content (g-h): Triatoma infestans [21] (g), Ixodes scapularis [22, 23] (h). Chromosome lengths reflect the differences in genome sizes of these insects. Heterochromatin is marked by the high satellite DNA and/or TE content (a)-(c) and by the high satellite DNA content in (d)-(g). The rDNA locus is located either within or next to the heterochromatin.
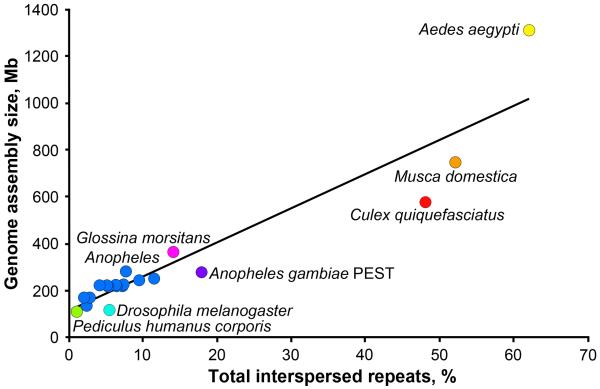
Because heterochromatic regions have many essential and actively transcribed genes [14, 25], mapping, sequencing, and annotation of heterochromatin in vector species are important for genome-wide analyses, such as localization of transcription factors, chromatin modifications, and non-coding RNAs. However, large genome sizes and abundance of repetitive DNA in many vectors remain the major roadblocks toward the assembly of heterochromatic sequences and completion of genome projects. Available genome sequencing data for vector species indicate that the genome sizes can vary more than 10-fold (Table 1). It is clear that the DNA repeat content rather than the gene number correlates well with the genome assembly size (Figure 2). Nevertheless, genome assemblies often miss large portions of heterochromatin. For example, a repetitive satellite DNA Aste72A is completely absent from the genome assembly of An. stephensi mainly obtained by Illumina and 454 sequencing, even though it constitutes a substantial portion of the heterochromatin in the X and Y chromosomes as evident from FISH [26]. Thus, next-generation sequencing technologies have important limitations for providing high-quality finished genome assemblies. Recent studies have demonstrated that a third-generation technology, Pacific Biosciences (PacBio) sequencing, can be a promising alternative approach to assemble structural variants and repeats in complex regions of eukaryotic genomes [27, 28]. A combination of Illumina and PacBio assemblies has given most accurate rDNA operon number predictions and resolution of repetitive regions [29]. Also, recent advances in physical chromosome mapping can facilitate creating reference genome assemblies much needed for investigating heterochromatin in vector species [30]. Elucidating heterochromatin organization will be key to understanding the epigenetic regulation of gene expression in disease vectors.
Table 1.
Sizes and annotation statistics of the sequenced genome assemblies.
| Species name | Common name |
Genome assembly size, Mb* |
Total interspersed repeats, % |
Protein- coding gene number |
Reference |
|---|---|---|---|---|---|
|
Pediculus humanus
corporis |
Human body louse |
110.8 | 1.0 | 10773 | [49] |
|
Drosophila
melanogaster |
Fruit fly | 118.0 | 5.35 | 13733 | [50, 51] |
| Anopheles darlingi | Malaria mosquito |
136.9 | 2.3 | 10481 | [52] |
| Anopheles albimanus | 170.5 | 2.0 | 11911 | [36] | |
| Anopheles christyi | 172.7 | 2.8 | 10738 | ||
| Anopheles dirus | 216.3 | 5.1 | 12781 | ||
| Anopheles sinensis | 220.8 | 6.5 | 16766 | [53] | |
|
Anopheles stephensi (Indian strain) |
221.3 | 7.1 | 11789 | [26] | |
| Anopheles epiroticus | 223.5 | 6.3 | 12078 | [36] | |
| Anopheles funestus | 225.2 | 4.0 | 13344 | ||
|
Anopheles stephensi (SDA strain) |
225.4 | 5.0 | 13113 | ||
| Anopheles melas | 227.4 | 7.3 | 16149 | ||
| Anopheles arabiensis | 246.6 | 9.4 | 13162 | ||
| Anopheles merus | 251.8 | 11.4 | 13887 | ||
|
Anopheles gambiae (PEST strain) |
273.1 | 17.8 | 12810 | ||
|
Anopheles
quadriannulatus |
283.8 | 7.7 | 13349 | [36] | |
| Glossina morsitans | Tsetse fly | 366.2 | 14.0 | 12308 | [54] |
| Culex quinquefasciatus | Southern house mosquito |
579.1 | 48.0 | 18955 | [55] |
| Musca domestica | Common house fly |
750.4 | 52.0 | 14180 | [56] |
| Aedes aegypti | Yellow fever mosquito |
1311.0 | 62.0 | 15784 | [57] |
data for vectors are from VectorBase [58], species are listed in the order of increased
Figure 2.
Correlation between total interspersed repeats and genome assembly sizes for species with sequenced genomes [26, 36, 49, 50, 52-57]. Coefficient of determination, R2=0.88. Color-coded circles represent different species. Blue circles labeled “Anopheles” correspond to 13 Anopheles species from Table 1, except An. gambiae.
Chromatin modifications in disease vectors
Altering chromatin structure by histone modifications, such as acetylation and methylation among others, is the predominant epigenetic regulatory mechanism of transcriptional control in Drosophila. For example, H3K27me3 is associated with Polycomb-mediated silencing, while H3K4me3 and H3K27ac are present in active promoters [8]. A recent work explored the epigenome of An. gambiae by mapping the distribution and levels of two post-translational histone modifications, H3K27ac and H3K27me3 [31]. The enrichment profiles of H3K27ac and H3K27me3 in the mosquito genome have been found to be mutually exclusive and associated with high and low levels of transcription, respectively. The analysis revealed that 6% of the An. gambiae genome is occupied by H3K27ac, whereas 14% is occupied by H3K27me3. Genes marked with H3K27ac display a significant differential enrichment in gene ontology (GO) terms associated with metabolic processes, as well as immune and signaling pathways. In contrast, genes marked with H3K27me3 show differential enrichment in GO terms related to membrane transporters and developmental processes, such as homeobox genes. Another interesting finding of this study is that multiple genes from the Toll, Immune Deficiency (IMD) and Janus kinase/signal transducers and activators of transcription (JAK/STAT) pathways, antimicrobial effectors, and metabolic genes are marked with one of these two histone modification marks. Thus, epigenetic variations in the An. gambiae genome can play a role in the interactions between mosquitoes and pathogens. An important conclusion of the above results is that the mechanisms of regulating chromatin structure and function are largely conserved between Anopheles and Drosophila, and that they are expected to be conserved in many other disease vectors. Thus, the key contribution of the study is that it sets the basis for future epigenetic studies in other mosquito species and insect vectors.
The H3K27me3 marks are produced by E(z), which is the histone methyltransferase component of the Polycomb repressive complex 2 (PRC2) [3]. 3-Deazaneplanocin A (DZNep) is a known histone methylation inhibitor, which is used as an experimental epigenetic drug for cancer therapy [32]. One study has shown that injection of DZNep into pupae of the cotton bollworm Helicoverpa armigera reduced the H3K27me3 mark and the prothoracicotropic hormone gene expression, thereby delaying development and inducing diapause [33]. Another study has provided a simple protocol for examining epigenetic effects of DZNep on the malaria vector An. gambiae [34]. It has demonstrated the negative correlation between the DZNep concentration and the rate of growth and development of immature mosquitoes. Also, adult female mosquitoes fed with blood containing DZNep exhibited a significant reduction in number of viable eggs. Although, the effects of epigenetic drugs are often systemic, modifying the levels of histone marks can be a useful approach to evaluate the global phenotypic changes by chromatin modifications in disease vectors. To identify specific effects on expression of target genes, epigenetic studies must include knockouts of genes encoding for chromatin-modifying enzymes [35].
Sequencing of the genomes of 16 Anopheles species has given the opportunity to explore evolution of epigenetic modifiers in mosquitoes [36]. The study demonstrated that genes enriched for biological process “chromatin organization” and for molecular functions “DNA binding” and “chromatin binding” were among those with the fastest evolutionary rates, indicating the plastic nature of the epigenetic component. At the same time, 75% of epigenetic regulatory genes found in D. melanogaster have also orthologs in An. gambiae [36]. A follow up comparative analysis of epigenetic gene expression between Drosophila and Anopheles revealed distinct tissue-associated expression patterns in fruit fly and mosquito [37]. Almost 85% of the variation in epigenetic gene expression occurred in carcasses, midguts, ovaries, heads, Malpighian tubules, and salivary glands. The results suggest that a subset of epigenetic genes may have different roles in these two species reflecting their differences in development, behavior, and lifestyle. Thus, studies of the genome distribution and function of major chromosomal proteins and histone modifications in vectors will bring new insights into their roles in traits relevant to vectorial capacity (i.e., blood feeding, susceptibility to pathogens, mating behavior). This knowledge will help to design the strategies to manipulate the epigenetic gene function and to fight the diseases.
Nuclear architecture: the role in gene expression and longevity
Evidence for non-random organization of chromosomes into territories gave rise to the idea that the relative positioning of chromosomes and genes with respect to the nuclear periphery is important for gene regulation [38]. In Drosophila, down-regulated genes of interphase chromosomes [12] and gene-poor heterochromatic regions of polytene chromosomes predominantly occupy the nuclear periphery [9, 10]. However, transcriptionally active genes can also be found at the periphery around the nuclear pore complexes in yeast, fruit fly, mouse, and malaria parasite [39]. Tissue-specific transcriptional processes can be regulated by a nuclear architectural protein, CCCTC-binding factor (CTCF). CTCF can mediate inter- and intra-chromosomal interactions that regulate recombination, enhancer-promoter interactions, transcriptional pausing, and alternative mRNA splicing [11].
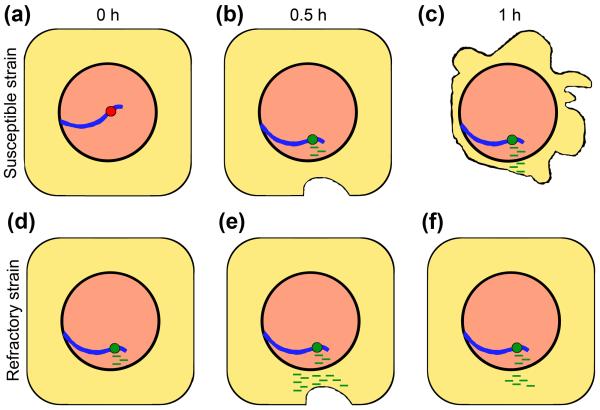
A recent study investigated reorganization in the three-dimensional (3D) position and expression of three genes, actin, ferritin, and Hsp 70, in live intact Biomphalaria glabrata snails upon exposure to Schistosoma mansoni miracidia, a parasitic worm that causes intestinal schistosomiasis (Bilharzia) in humans and other mammals [40]. In the susceptible snails, the actin gene loci moved from the nuclear interior towards a more intermediate position within the nuclei at 30 minutes after infection. Interestingly, the movement preceded the 16-fold increase in gene expression at 1 hour after infection. In the resistant snails, a 22-fold increase in gene expression without major repositioning was observed for the actin gene loci at 30 minutes after infection (Figure 3). The authors hypothesize that the rapid increase in actin expression observed in the resistant snails could be due to pre-positioning of the actin gene in place where transcription is optimal. In contrast, susceptible snails failed to respond as efficiently because they needed ~30 minutes to reposition the actin gene to the transcription factory. Expression of actin could be necessary to support rearrangements in the snail cytoskeleton that is damaged by the worm. The delayed transcription of actin in susceptible snails probably failed to elicit a protection of the cytoskeleton. Yet different patterns of the nuclear repositioning and expression have been observed for the ferritin and Hsp 70 genes. Hsp70 moves and is expressed in the susceptible snails but not in the resistant snails. It is likely that this heat-shock gene is important for the parasite in its efforts to trigger an infection by controlling the snail host genome. The ferritin gene moves and is up-regulated in both susceptible and resistant snails, suggesting its response to tissue damage. This study proposes a hypothesis that timely positioning a locus in the transcription factory could be necessary to confer resistance to a pathogen. Due to the lack of the whole-chromosome visualization, the study could not determine if whole chromosome territories or genes on chromatin loops have made the transition through the nuclear space.
Figure 3.
A schematic representation of changes in the 3D nuclear position and gene expression in a snail vector upon infection with miracidia [40]. The reactions of the cells in the susceptible (a-c) and refractory (d-f) strains are shown at 0, 0.5, and 1 hour after the infection. The strains are different in the spatial position of the chromosome (blue curved line) carrying the actin gene, which is located in the middle of the nucleus in the susceptible strain (a) and closer to the nuclear periphery in the refractory strain (d). Hypothetically, cells in the susceptible strain are damaged by the parasite (c), while cells in the refractory strain are repaired by rearrangements in the cytoskeleton (f). Red circle – inactive gene, green circle – active gene. Green lines – transcripts. Miracidia are not shown.
A detailed study of the 3D genome reorganization in response to infection has not been performed in any arthropod vector species. The chromosome territories in snails have a similar morphology and structure to those of mammalian cells. In contrast, chromosomes of fruit flies and mosquitoes often have so-called Rabl organization, with centromeres and telomeres located at the opposite poles of the interphase nucleus [9, 10]. To study chromosome territories, chromosome paints can be produced for organisms with sequenced genomes by generating DNA FISH probes from oligonucleotide libraries [41]. Such probes can also be developed by using a laser capture microdissection and whole-genome amplification. Microdissected chromosome paints have been used to demonstrate chromosome territories in polytene and nonpolytene interphase nuclei of An. gambiae [42]. A very recent study applied chromosome region-specific microdissected paints to investigate tissue-specific features of chromosome architecture in An. messeae [43]. It found significant differences in the 3D interposition of the nuclear envelope attachment regions of the chromosomes X and 3R between somatic (salivary glands, follicle epithelium) and germ-line (ovarian of nurse cells) tissues, suggesting the link between the nuclear architecture and cell-type differentiation. The chromosome painting method can be applied to detailed analysis of the 3D dynamics of chromosome territories in vectors infected with pathogens.
Sufficient longevity of disease vectors is crucial for a disease transmission, because only older adults are potentially infective, as it takes a specific time for a pathogen to reach an appropriate stage of development inside a vector [44]. It is well known that the nuclear architecture in aged cells significantly differ from that in normal cells. In aging adult fruit flies, nuclei grow in size and assume irregular morphology. Moreover, aberrant nuclear shapes and reduced lifespan can be induced by overexpression of genes encoding for the nuclear envelope proteins Lamin and Kugelkern of adult flies [45, 46]. A new study of polytene nuclei from D. melanogaster salivary glands revealed forces that regulate the nuclear envelope morphology and function. Condensin-II-mediated chromatin condensation can alter the 3D structure and shape of the nucleus via nuclear envelope-chromosome attachments [47]. If the organization of the nuclear architecture is similar among arthropods, than the induced structural changes in the nuclear envelope should cause the reduction in lifespan in the majority of disease vectors.
Conclusions and perspectives
Recent advances in vector genomics created an opportunity to improve our understanding of epigenetic inheritance of species involved in disease transmission. The works reviewed here show that heterochromatin organization, histone modifications, and nuclear architecture can play important roles in phenotypes relevant to vectorial capacity. Several immediate steps have to be taken to make vector species fully accessible to epigenomic studies. First, instead of fragmented and unordered genomic scaffolds, chromosomally mapped high-quality-finished genome assemblies, which include heterochromatin, must be available. An important challenge is to construct chromosome-scale scaffolds from short sequencing reads. Third-generation-sequencing long-read technologies, such as those from Pacific Biosciences and Oxford Nanopore Technologies, could offer the solution of how to create longer scaffolds and to deal with repeat-rich heterochromatic regions [27, 28, 48]. Second, epigenetic regulatory gene sets must be identified in vector species. Although many genes involved in heterochromatin structure, chromatin remodeling, and transcriptional control are conserved among species, other genes with epigenetic functions can evolve rapidly [36]. Third, principles of the 3D genome organization must be studied in vectors to understand its role in gene expression and possible effects on infection and aging. The global patterns of intra- and inter-chromosomal interactions can be studied in 3D using the Hi-C approach, which crosslinks physical contacts between distant genomic regions [11]. The knowledge about epigenetic factors in vectors of human and animal infectious diseases will provide a rich basis for fundamental and applied research aimed at deciphering the mechanisms controlling vectorial capacity.
Highlights.
Heterochromatin contains essential genes and performs important biological functions.
Finished genome assemblies must include heterochromatic sequences.
Histone modifications regulate immunity, development, and reproduction.
Nuclear architecture affects gene expression and longevity.
Acknowledgements
We thank Joanna Bridger, Matty Knight, and the reviewer, for helpful comments. We also thank Philip George for calculating TE content in An. gambiae euchromatin and heterochromatin, Melissa Wade for editing the manuscript, and Nora J. Besansky for the invitation to write a review on this topic. We apologize to all our colleagues whose studies could not be covered because of space limitations. IVS was supported by grants from National Institutes of Health R21AI094289 and R21AI099528. MVS was supported by grants from National Institutes of Health R21AI101345 and R21AI088035.
Abbreviations
- 3D
three-dimensional
- CTCF
CCCTC-binding factor
- DZNep
3-Deazaneplanocin A
- FISH
fluorescence in situ hybridization
- GO
gene ontology
- HP1
heterochromatin protein 1
- Hsp
heat shock protein
- IMD
Immune Deficiency
- JAK/STAT
Janus kinase/signal transducers and activators of transcription
- PacBio
Pacific Biosciences
- PcG
Polycomb group
- PRC2
Polycomb repressive complex 2
- rDNA
ribosomal DNA
- SUUR
Suppressor of Under Replication
- TEs
transposable elements
Footnotes
Conflict of interest
The authors have no conflict of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
(•) of special interest
(••) of outstanding interest
- 1.Gabrieli P, Smidler A, Catteruccia F. Engineering the control of mosquito-borne infectious diseases. Genome Biol. 2014;15:535. doi: 10.1186/s13059-014-0535-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gómez-Díaz E, Jorda M, Peinado MA, Rivero A. Epigenetics of host-pathogen interactions: the road ahead and the road behind. PLoS Pathog. 2012;8:e1003007. doi: 10.1371/journal.ppat.1003007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molla-Herman A, Matias NR, Huynh J-R. Chromatin modifications regulate germ cell development and transgenerational information relay. Current Opinion in Insect Science. 2014;1:10–18. doi: 10.1016/j.cois.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Mtreirek R, Gueguen N, Jensen S, Brasset E, Vaury C. Drosophila heterochromatin: structure and function. Current Opinion in Insect Science. 2014;1:19–24. doi: 10.1016/j.cois.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Hoeijmakers WA, Stunnenberg HG, Bartfai R. Placing the Plasmodium falciparum epigenome on the map. Trends in parasitology. 2012;28:486–495. doi: 10.1016/j.pt.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Contrino S, Smith RN, Butano D, Carr A, Hu F, Lyne R, Rutherford K, Kalderimis A, Sullivan J, Carbon S, et al. modMine: flexible access to modENCODE data. Nucleic Acids Res. 2012;40:D1082–1088. doi: 10.1093/nar/gkr921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rakyan VK, Hildmann T, Novik KL, Lewin J, Tost J, Cox AV, Andrews TD, Howe KL, Otto T, Olek A, et al. DNA methylation profiling of the human major histocompatibility complex: a pilot study for the human epigenome project. PLoS Biol. 2004;2:e405. doi: 10.1371/journal.pbio.0020405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hochstrasser M, Mathog D, Gruenbaum Y, Saumweber H, Sedat JW. Spatial organization of chromosomes in the salivary gland nuclei of Drosophila melanogaster. J Cell Biol. 1986;102:112–123. doi: 10.1083/jcb.102.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinney NA, Sharakhov IV, Onufriev AV. Investigation of the chromosome regions with significant affinity for the nuclear envelope in fruit fly--a model based approach. PLoS ONE. 2014;9:e91943. doi: 10.1371/journal.pone.0091943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ong CT, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet. 2014;15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet. 2006;38:1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- 13.Heitz E. Uber alpha- und beta-Heterochromatin sowie Konstanz und Bau der Chromomeren bei Drosophila. Biol. Zbl. 1934;54:588–609. [Google Scholar]
- 14.Smith CD, Shu S, Mungall CJ, Karpen GH. The Release 5.1 annotation of Drosophila melanogaster heterochromatin. Science. 2007;316:1586–1591. doi: 10.1126/science.1139815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gatti M, Santini G, Pimpinelli S, Coluzz M. Fluorescence banding techniques in the identification of sibling species of the Anopheles gambiae complex. Heredity. 1977;38:105–108. doi: 10.1038/hdy.1977.11. [DOI] [PubMed] [Google Scholar]
- 17.Bonaccorsi S, Santini G, Gatti M, Pimpinelli S, Colluzzi M. Intraspecific polymorphism of sex chromosome heterochromatin in two species of the Anopheles gambiae complex. Chromosoma. 1980;76:57–64. doi: 10.1007/BF00292226. [DOI] [PubMed] [Google Scholar]
- 18.Sharakhova MV, George P, Brusentsova IV, Leman SC, Bailey JA, Smith CD, Sharakhov IV. Genome mapping and characterization of the Anopheles gambiae heterochromatin. BMC Genomics. 2010;11:459. doi: 10.1186/1471-2164-11-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newton ME, Southern DI, Wood RJ. X and Y chromosomes of Aedes aegypti (L.) distinguished by Giemsa C-banding. Chromosoma. 1974;49:41–49. doi: 10.1007/BF00284986. [DOI] [PubMed] [Google Scholar]
- 20 •.Timoshevskiy VA, Kinney NA, deBruyn BS, Mao C, Tu Z, Severson DW, Sharakhov IV, Sharakhova MV. Genomic composition and evolution of Aedes aegypti chromosomes revealed by the analysis of physically mapped supercontigs. BMC Biol. 2014;12:27. doi: 10.1186/1741-7007-12-27. The comparative genomic analysis of Ae. aegypti and An. gambiae reveals vastly different genome structures. The study shows that one Mb of the Ae. aegypti genome has ~12 protein-coding genes and ~1600 TEs, while one Mb of the An. gambiae genome has ~40 protein-coding genes and ~200 TEs.
- 21.Bardella VB, da Rosa JA, Vanzela AL. Origin and distribution of AT-rich repetitive DNA families in Triatoma infestans (Heteroptera) Infect Genet Evol. 2014;23:106–114. doi: 10.1016/j.meegid.2014.01.035. [DOI] [PubMed] [Google Scholar]
- 22.Meyer JM, Kurtti TJ, Van Zee JP, Hill CA. Genome organization of major tandem repeats in the hard tick, Ixodes scapularis. Chromosome Res. 2010;18:357–370. doi: 10.1007/s10577-010-9120-4. [DOI] [PubMed] [Google Scholar]
- 23.Chen C, Munderloh UG, Kurtti TJ. Cytogenetic characteristics of cell lines from Ixodes scapularis (Acari: Ixodidae) J Med Entomol. 1994;31:425–434. doi: 10.1093/jmedent/31.3.425. [DOI] [PubMed] [Google Scholar]
- 24.Panzera F, Ferrandis I, Ramsey J, Salazar-Schettino PM, Cabrera M, Monroy C, Bargues MD, Mas-Coma S, O’Connor JE, Angulo VM, et al. Genome size determination in chagas disease transmitting bugs (hemiptera-triatominae) by flow cytometry. Am J Trop Med Hyg. 2007;76:516–521. [PubMed] [Google Scholar]
- 25.van Steensel B. Chromatin: constructing the big picture. EMBO J. 2011;30:1885–1895. doi: 10.1038/emboj.2011.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang X, Peery A, Hall A, Sharma A, Chen XG, Waterhouse RM, Komissarov A, Riehl MM, Shouche Y, Sharakhova MV, et al. Genome analysis of a major urban malaria vector mosquito, Anopheles stephensi. Genome Biol. 2014;15:459. doi: 10.1186/s13059-014-0459-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huddleston J, Ranade S, Malig M, Antonacci F, Chaisson M, Hon L, Sudmant PH, Graves TA, Alkan C, Dennis MY, et al. Reconstructing complex regions of genomes using long-read sequencing technology. Genome Research. 2014;24:688–696. doi: 10.1101/gr.168450.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogers RL, Cridland JM, Shao L, Hu TT, Andolfatto P, Thornton KR. Landscape of standing variation for tandem duplications in Drosophila yakuba and Drosophila simulans. Mol Biol Evol. 2014;31:1750–1766. doi: 10.1093/molbev/msu124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Utturkar SM, Klingeman DM, Land ML, Schadt CW, Doktycz MJ, Pelletier DA, Brown SD. Evaluation and validation of de novo and hybrid assembly techniques to derive high-quality genome sequences. Bioinformatics. 2014;30:2709–2716. doi: 10.1093/bioinformatics/btu391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharakhov IV. Protocols for cytogenetic mapping of arthropod genomes. Taylor & Francis Group, LLC; Boca Raton, FL.: 2015. [Google Scholar]
- 31 ••.Gómez-Díaz E, Rivero A, Chandre F, Corces VG. Insights into the epigenomic landscape of the human malaria vector Anopheles gambiae. Front Genet. 2014;5:277. doi: 10.3389/fgene.2014.00277. The study has developed the first platform for mosquito comparative epigenomics that can serve as a basis for future studies on the biology of vectors. The analysis linked enrichment or depletion of active (H3K27ac) and repressive (H3K27me3) histone modifications to their target genes in An. gambiae.
- 32.Nakagawa S, Sakamoto Y, Okabe H, Hayashi H, Hashimoto D, Yokoyama N, Tokunaga R, Sakamoto K, Kuroki H, Mima K, et al. Epigenetic therapy with the histone methyltransferase EZH2 inhibitor 3-deazaneplanocin A inhibits the growth of cholangiocarcinoma cells. Oncol Rep. 2014;31:983–988. doi: 10.3892/or.2013.2922. [DOI] [PubMed] [Google Scholar]
- 33.Lu YX, Denlinger DL, Xu WH. Polycomb repressive complex 2 (PRC2) protein ESC regulates insect developmental timing by mediating H3K27me3 and activating prothoracicotropic hormone gene expression. J Biol Chem. 2013;288:23554–23564. doi: 10.1074/jbc.M113.482497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma A, Anderson TD, Sharakhov IV. Toxicological assays for testing effects of an epigenetic drug on development, fecundity and survivorship of malaria mosquitoes. J Vis Exp. 2015:e52041. doi: 10.3791/52041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang L, Mu J, Zhang Q, Ni T, Srinivasan P, Rayavara K, Yang W, Turner L, Lavstsen T, Theander TG, et al. PfSETvs methylation of histone H3K36 represses virulence genes in Plasmodium falciparum. Nature. 2013;499:223–227. doi: 10.1038/nature12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36 ••.Neafsey DE, Waterhouse RM, Abai MR, Aganezov SS, Alekseyev MA, Allen JE, Amon J, Arca B, Arensburger P, Artemov G, et al. Highly evolvable malaria vectors: The genomes of 16 Anopheles mosquitoes. Science. 2015;347:43. doi: 10.1126/science.1258522. The study presents the genomes of 16 anophelines from Africa, Asia, Europe, and Latin America. The genomic analysis of species spanning ~100 million years of evolution revealed a fast rate of evolution of genes important to epigenetic modification.
- 37 •.Jenkins AM, Muskavitch MA. Evolution of an epigenetic gene ensemble within the genus Anopheles. Genome biology and evolution. 2015;7:901–915. doi: 10.1093/gbe/evv041. The study identified seven epigenetic gene family expansion/contraction events within the 12 Anopheles species. Comparative analyses demonstrated tissue-specific expression patterns of epigenetic genes between mosquito and fruit fly.
- 38.Cremer T, Cremer C, Lichter P. Recollections of a scientific journey published in human genetics: from chromosome territories to interphase cytogenetics and comparative genome hybridization. Hum Genet. 2014;133:403–416. doi: 10.1007/s00439-014-1425-5. [DOI] [PubMed] [Google Scholar]
- 39.Brown CR, Silver PA. Transcriptional regulation at the nuclear pore complex. Current Opinion in Genetics & Development. 2007;17:100–106. doi: 10.1016/j.gde.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 40 ••.Arican-Goktas HD, Ittiprasert W, Bridger JM, Knight M. Differential spatial repositioning of activated genes in Biomphalaria glabrata snails infected with Schistosoma mansoni. PLoS Negl Trop Dis. 2014;8:e3013. doi: 10.1371/journal.pntd.0003013. This is the first study of spatial genome reorganization in a disease vector caused by a pathogen. It has uncovered a novel mechanism whereby the nuclear architecture of a snail, B. glabrata, is manipulated by a parasite, S. mansoni, causing a change in gene expression of the vector.
- 41.Beliveau BJ, Joyce EF, Apostolopoulos N, Yilmaz F, Fonseka CY, McCole RB, Chang Y, Li JB, Senaratne TN, Williams BR, et al. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc Natl Acad Sci U S A. 2012;109:21301–21306. doi: 10.1073/pnas.1213818110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.George P, Sharma A, Sharakhov IV. 2D and 3D chromosome painting in malaria mosquitoes. J Vis Exp. 2014:e51173. doi: 10.3791/51173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Artemov G, Bondarenko S, Sapunov G, Stegniy V. Tissue-specific differences in the spatial interposition of X-chromosome and 3R chromosome regions in the malaria mosquito Anopheles messeae Fall. PLoS ONE. 2015;10:e0115281. doi: 10.1371/journal.pone.0115281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cook PE, McMeniman CJ, O’Neill SL. Modifying insect population age structure to control vector-borne disease. Advances in experimental medicine and biology. 2008;627:126–140. doi: 10.1007/978-0-387-78225-6_11. [DOI] [PubMed] [Google Scholar]
- 45.Brandt A, Vilcinskas A. The fruit fly Drosophila melanogaster as a model for aging research. Yellow Biotechnology I: Insect Biotechnologie in Drug Discovery and Preclinical Research. 2013;135:63–77. doi: 10.1007/10_2013_193. [DOI] [PubMed] [Google Scholar]
- 46.Brandt A, Krohne G, Grosshans J. The farnesylated nuclear proteins KUGELKERN and LAMIN B promote aging-like phenotypes in Drosophila flies. Aging cell. 2008;7:541–551. doi: 10.1111/j.1474-9726.2008.00406.x. [DOI] [PubMed] [Google Scholar]
- 47 •.Bozler J, Nguyen HQ, Rogers GC, Bosco G. Condensins exert force on chromatin-nuclear envelope tethers to mediate nucleoplasmic reticulum formation in Drosophila melanogaster. G3 (Bethesda) 2014;5:341–352. doi: 10.1534/g3.114.015685. Experiments with Drosophila salivary gland nuclei led to a model, in which nuclear envelope-chromosome attachments allow chromatin movements to cause aberrant nuclear envelope morphology. This mechanism could be responsible for pathological conditions, such as laminopathies, where distortions in nuclear architecture are associates with premature aging.
- 48.Goodwin S, Gurtowski J, Ethe-Sayers S, Deshpande P, Schatz MC, McCombie WR. Oxford nanopore sequencing and de novo assembly of a eukaryotic genome. BioRxiv pre-print server. 2015 doi: 10.1101/gr.191395.115. http://dx.doi.org/10.1101/013490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kirkness EF, Haas BJ, Sun W, Braig HR, Perotti MA, Clark JM, Lee SH, Robertson HM, Kennedy RC, Elhaik E, et al. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc Natl Acad Sci U S A. 2010;107:12168–12173. doi: 10.1073/pnas.1003379107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 51.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 52.Marinotti O, Cerqueira GC, de Almeida LG, Ferro MI, Loreto EL, Zaha A, Teixeira SM, Wespiser AR, Almeida ESA, Schlindwein AD, et al. The genome of Anopheles darlingi, the main neotropical malaria vector. Nucleic Acids Res. 2013;41:7387–7400. doi: 10.1093/nar/gkt484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou D, Zhang D, Ding G, Shi L, Hou Q, Ye Y, Xu Y, Zhou H, Xiong C, Li S, et al. Genome sequence of Anopheles sinensis provides insight into genetics basis of mosquito competence for malaria parasites. BMC Genomics. 2014;15:42. doi: 10.1186/1471-2164-15-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.International Glossina Genome, I Genome sequence of the tsetse fly (Glossina morsitans): vector of African trypanosomiasis. Science. 2014;344:380–386. doi: 10.1126/science.1249656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arensburger P, Megy K, Waterhouse RM, Abrudan J, Amedeo P, Antelo B, Bartholomay L, Bidwell S, Caler E, Camara F, et al. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science. 2010;330:86–88. doi: 10.1126/science.1191864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scott JG, Warren WC, Beukeboom LW, Bopp D, Clark AG, Giers SD, Hediger M, Jones AK, Kasai S, Leichter CA, et al. Genome of the house fly, Musca domestica L., a global vector of diseases with adaptations to a septic environment. Genome Biol. 2014;15:466. doi: 10.1186/s13059-014-0466-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi Z, Megy K, Grabherr M, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Giraldo-Calderon GI, Emrich SJ, MacCallum RM, Maslen G, Dialynas E, Topalis P, Ho N, Gesing S, VectorBase C, Madey G, et al. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015;43:D707–713. doi: 10.1093/nar/gku1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Willhoeft U. Fluorescence in situ hybridization of ribosomal DNA to mitotic chromosomes of tsetse flies (Diptera: Glossinidae: Glossina) Chromosome Res. 1997;5:262–267. doi: 10.1023/a:1018471620542. [DOI] [PubMed] [Google Scholar]
- 60.Hediger M, Niessen M, Muller-Navia J, Nothiger R, Dubendorfer A. Distribution of heterochromatin on the mitotic chromosomes of Musca domestica L. in relation to the activity of male-determining factors. Chromosoma. 1998;107:267–271. doi: 10.1007/s004120050307. [DOI] [PubMed] [Google Scholar]