Abstract
Acylation of lysine is an important protein modification regulating diverse biological processes. It was recently demonstrated that members of the human Sirtuin family are capable of catalyzing long-chain deacylation, in addition to the well-known NAD+-dependent deacetylation activity.1 Here we provide a detailed kinetic and structural analysis that describes the interdependence of NAD+ and acyl-group length for a diverse series of human Sirtuins, SIRT1, SIRT2, SIRT3 and SIRT6. Steady-state and rapid-quench kinetic analyses indicated that differences in NAD+ saturation and susceptibility to nicotinamide inhibition reflect unique kinetic behavior displayed by each Sirtuin and depend on acyl-substrate chain length. Though the rate of nucleophilic attack of the 2′-hydroxyl on the C1′-O-alkylimidate intermediate varies with acyl substrate chain length, this step remains rate-determining for SIRT2 and SIRT3; however for SIRT6, this step is no longer rate-limiting for long-chain substrates. Co-crystallization of SIRT2 with myristoylated peptide and NAD+ yielded a co-complex structure with reaction product 2′-O-myristoyl-ADP-ribose, revealing a latent hydrophobic cavity to accommodate the long chain acyl group, and suggesting a general mechanism for long chain deacylation. Comparing two separately solved co-complex structures containing either a myristoylated peptide or 2′-O-myristoyl-ADP-ribose indicate there are conformational changes at the myristoyl-ribose linkage with minimal structural differences in the enzyme active site. During the deacylation reaction, the fatty acyl group is held in a relatively fixed position. We describe a kinetic and structural model to explain how various Sirtuins display unique acyl-substrate preferences and how different reaction kinetics influence NAD+ dependence. The biological implications are discussed.
Mammalian Sirtuins (SIRT1-7) are a family of nicotinamide adenine dinucleotide (NAD+)-dependent protein deacylases implicated in a wide range of cellular processes. The physiological importance of Sirtuins has spurred interest in elucidating their catalytic properties, substrates and molecular functions. Beyond ε-amino deacetylation of lysine residues, which was the first established activity attributed to these enzymes, recent in vitro evidence indicate that many Sirtuins catalyze longer chain deacylations, including the removal of hexanoyl, octanoyl, decanoyl, dodecanoyl, myristoyl, and palmitoyl groups.1, 2 The results broaden the acylation landscape targeted by Sirtuins and might explain the broad diversity in biological functions. However, a detailed kinetic and structural understanding of catalytic deacylation activities is lacking.
Protein acylation is emerging as a potential cellular control mechanism and Sirtuins play a major role in regulating acylation status.3 In addition to acetyl-CoA, other abundant cellular acyl-CoAs serve as acyl donor molecules for the modification of lysine residues. Acyl-CoAs are derived from carbohydrate, protein and fatty acid metabolism, therefore their abundance is dictated by the metabolic status of the cell.4 Increased concentrations of reactive acyl-CoAs may drive protein acylation, as previously indicated with acetyl-CoA in yeast and acetyl-phosphate in E. coli.5-7 Recent in vivo studies identified a series of short and medium chain acyl groups – propionyl, butyryl, succinyl, glutaryl, malonyl, and crotonyl – as post-translational modifications of lysine residues in histone and non-histone proteins located in multiple cellular compartments, including the nucleus and mitochondria.8-15 Furthermore, these studies found that mitochondrial localized SIRT5 could catalyze desuccinylation, demalonylation and deglutarylation in vitro, and mice lacking SIRT5 displayed global increases in these modifications.9, 14, 16 SIRT6, a nuclear localized Sirtuin with poor in vitro deacetylase activity, was recently established as a lysine demyristoylase in vitro and in vivo, acting to demyristoylate TNFα.1, 2 Providing further in vivo evidence for the prevalence of longer (>C6) chain acylations is difficult as traditional methods for identifying and localizing these modifications, including immunoenrichment using modification specific antibodies and mass spectrometry, have yet to be optimized for this purpose. However, Jiang et al. identified a number of cellular acylated proteins using a fluorescent reporter based assay, providing additional evidence that these modifications exist.2
Previously, we showed that SIRT1, SIRT2, SIRT3, SIRT4, SIRT5, and SIRT6 could all catalyze long-chain deacylations, but with varying degrees of specificity and efficiency.1 The mechanistic basis underlying these unique deacylation profiles was not investigated. In particular, the links between NAD+ dependence and the nature of the acyl-group is unclear. NAD+ metabolism is known to affect the cellular functions of some Sirtuins,17 however if alterations in NAD+ binding are dependent on acyl substrate and how diverse acyl-groups affect the various catalytic steps remain unknown.
Here, we performed a series of kinetic and structural studies to explain the unique deacylation signatures for human Sirtuins SIRT1, SIRT2, SIRT3 and SIRT6. These human Sirtuins exist in distinct sub-cellular compartments and represent two phyla of Sirtuin enzymes.18 Using acetylated, hexanoylated, deconylated and myristoylated peptides as substrates, we find the Km for NAD+ and the sensitivity to nicotinamide inhibition are dependent on the Sirtuin, as well as the chain length of the acylated substrate. Our results show that SIRT1, SIRT2, SIRT3 and SIRT6 exhibit varying catalytic efficiencies and substrate preferences among the various acyl modifications. Pre-steady-state kinetic analysis provides insight into the microscopic rate constants that contribute to the kcat, kcat/Km, NAD and Km, NAD and reveal differences between Sirtuins that are not observed in the macroscopic rate constants. The rate of nucleophilic attack of the 2′-hydroxyl on the C1′-O-alkylimidate intermediate varies with acyl substrate chain length and remains the rate-determining step for SIRT2 and SIRT3. However, with long-chain substrates, this step is no longer rate-limiting for SIRT6. Furthermore, we co-crystallized SIRT2 with myristoylated peptide, and with myristoylated peptide and NAD+, which allowed direct observation of the product. Through an induced fit mechanism, conformational changes in the zinc-binding domain and helix bundle region create a cavity to accommodate the acyl-chain. Comparison of the crystal structures of SIRT2 in complex with myristoylated peptide and with 2′-O-myristoyl-ADP-ribose reveal the conformational changes in the myristoyl group occur near the myristoyl-ribose linkage, while the fatty acyl group is held in a fixed position during the deacylation reaction in the hydrophobic cavity suited for binding the acyl chain. The kinetic and structural characterization provides a more complete understanding of the mechanisms that control Sirtuin deacylase activity. Further, these results may aid in the interpretation of cellular studies and allow for the design of rational small molecule regulators of Sirtuin activity.
Experimental Procedures
Expression and purification of recombinant human SIRT1, SIRT2, SIRT3 and SIRT6
His-tagged SIRT1, SIRT2, SIRT3 and SIRT6 were overexpressed in BL21 DE3 E. coli strain. Overexpression was initiated by growing cells to an OD600 of 0.6-0.8 at 37 °C. To induce expression, 0.5 mM isopropyl-1-thio-D-galactopyranoside (IPTG) was added and cells were grown at room temperature for 6 h (SIRT1 and SIRT2) or 18 hours. Cells were harvested by centrifugation and stored at −80 °C. SIRT1,19 SIRT2,20 SIRT321 and SIRT622 were purified as reported previously. Protein concentrations were determined by the Bradford assay.
Synthesis and analysis of the acyl H3K9 peptides
Peptides corresponding to residues 4-17 of histone H3 (Acetyl: AcQTARKacSTGGKAPR-WW-NH2, Hexanoyl: QTARKhexSTGGKAPR-WW-NH2, Decanoyl: QTARKdecSTGGKAPR-WW-NH2, and Myristoyl: Ac-QTARKmyrSTGGKAPRWW-NH2) were synthesized by standard solid phase peptide synthesis on a Prelude instrument (Protein Technologies). The side chain of lysine 9 was protected with 1-(4,4-dimethyl-2,6-dioxocyclohex-1-ylidene)-3-methylbutyl (ivDde) group. Following synthesis, the amino terminus was acetylated using acetic anhydride. The ivDde group was then removed by incubating the peptide-resin with 4% hydrazine in dimethylformamide (DMF) for 10 minutes. The liquid was extracted and the incubation was repeated for a total of seven times. The peptide-resin was then incubated with various acyl anhydrides for 2 × 20 minutes. The acylation solutions contained 528 mM acyl anhydride (acetic, hexanoic, decanoic, and myristic) in toluene. The resin was then washed with DMF followed by dichloromethane (DCM). The peptides were cleaved with a mixture of trifluoroacetic acid (TFA), 5% thioanisole, and 2.5% ethanedithiol. The cleaved peptides were then precipitated in ice-cold ether and lyophilized. The peptides were purified over a preparative C18 HPLC column. The chromatographic purity of the peptide was determined to be ≥95% and mass spectrometric analysis on a Bruker REFLEX II: MALDI-TOF (matrix-assisted laser desorption ionization time-of-flight) instrument confirmed the identification of the peptide. The acetylated peptide was dissolved in H2O and the myristoylated and deconylated peptide were dissolved in 50% DMSO. Two tryptophan residues were added to the C-terminus for quantification at 280 nm.
Determination of kinetic parameters for Sirtuin-dependent deacylation
Deacylation reactions were analyzed by reversed phase high-performance liquid chromatography on a Kinetex C18 column (100 Å, 100 × 4.6 mm, 2.6 μm, Phenomenex) by monitoring the formation of the deacylated product at 214 nm. Sirtuin (0.2 μM) or 0.5 μM SIRT6 were incubated with 2.5-1000 μM NAD+, saturating acetyl-lysine, hexanoyl-lysine, decanoyl-lysine, or myristoyl-lysine in 20 mM sodium phosphate, pH 7.5, at 37 °C. To ensure saturating concentrations of acyl-peptide substrates, approximate Km values for acyl-peptide and the concentration required to saturate the reaction were experimentally determined. The approximate Km, acyl-peptide values for the Sirtuins studied were in the low μM (<1-20 μM) range, and below the detection limit of the HPLC where accurate quantitation is possible. Reactions were quenched with TFA at various time points and centrifuged at 21,000xg for 5 min. Time points were chosen such that all reactions remained within steady-state initial velocity during the course of the reaction. Deacetylation reactions were analyzed using a gradient of 33-100% B (30% acetonitrile with 0.02% TFA) in 20 min at 1.4 mL min−1. Dehexanoylation, dedecanoylation and demyristoylation reactions were analyzed using a gradient of 3 to 100% B (acetonitrile with 0.02% TFA) in 10 min at 1.6 mL min−1. The product and substrate peaks were quantified, rates of deacylation were determined, and the data were fitted to the Michaelis-Menten equation. Three independent experiments were performed for each Sirtuin, acyl-substrate pair. The average of the three experiments were plotted and fitted to obtain Km,NAD, kcat/Km,NAD, and kcat. Error bars represent the standard deviation of the mean.
Steady-state kinetic data analysis
To demonstrate the effects of reaction kinetics on the acyl-substrate-dependent Sirtuin activities in terms of the microscopic rate constants, we used the net rate constant method of Cleland23 to derive the initial velocity equation for the proposed kinetic mechanism under saturating acyl-peptide substrate concentration (Figure 1A). The kinetic analysis was performed under saturating acyl-peptide substrate concentration, forcing all free enzyme into an enzyme-acyl peptide substrate complex. Thus, k1 and k2 do not contribute to the initial velocity measurements. Based on the steady-state approximation, k8 (rebinding of nicotinamide) will be equal to zero and assuming a random product release of OAADPr and deacylated peptide,24 we defined a single kinetic constant, k11, to encompass the rate of product release, allowing the equation to be simplified to (Eq. 2).
| (Eq. 2) |
The rate of nicotinamide release (k7) is believed to be fast relative to all other steps. Using Figure 1A as the kinetic model, as well as the previous assumptions, we can derive the equation for kcat (Eq. 3) and kcat/Km, NAD (Eq. 4), which are equal to:
| (Eq. 3) |
| (Eq. 4) |
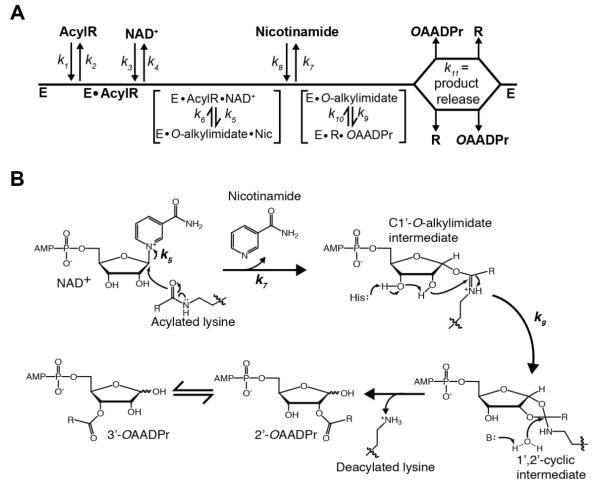
Figure 1. Proposed Sirtuin deacylation mechanism and kinetic scheme.
A, Sirtuin enzymes follow a sequential mechanism in which both acylated substrate (AcylR) (k1) and NAD+ (k3) bind prior to any catalytic step. SIRT1, SIRT2 and SIRT3 must bind AcylR prior to NAD+, while SIRT6 is the only mammalian Sirtuin capable of binding AcylR or NAD+ in random order. A ternary complex is formed, followed by nicotinamide formation (k5) and release (k7), and transfer of the acyl group from AcylR to ADP-ribose (k9). Deacylated substrate (R) and O-acyl-ADPr (OAADPr) are randomly released (k11). B, Proposed deacylation mechanism. Nucleophilic addition of the acyl oxygen on the 1′-carbon of the nicotinamide ribose forms the C1′-O-alkylimidate intermediate. A conserved histidine residue in the active site activates the 2′-hydroxyl group of NAD+ ribose. The activated hydroxyl attacks the O-alkylimidate carbon to afford the 1′2′-cyclic intermediate. A base activated water molecule attacks the cyclic intermediate resulting in the formation of deacylated lysine and OAADPr. Rate constants are indicated.
The kcat value reflects chemical catalysis (k5, k9, k10) as well as deacylated substrate and OAADPr release (k11). However, because turnover is often controlled by a single slow step, the kcat expression will likely simplify to a single rate constant (either k5, k9, k10, or k11). The kcat/Km value for NAD+ encompasses both NAD+ binding (k3, k4) and NAD+ cleavage, k5. The kcat/Km measures the apparent rate of enzyme capture of the substrate to form a productive complex destined to form products and reflects enzyme activity at low substrate levels.25
The Km value for NAD+ is the ratio of kcat and kcat/Km, NAD and is the steady-state Michaelis constant for the Sirtuin-NAD+-acyl-substrate ternary complex. The Km reflects both binding and catalysis and is equal to (Eq. 5):
| (Eq. 5) |
A change in Km for NAD+ can be due to a change in kcat, k5 or NAD+ binding (change in k3 or in k4) (Figure 1A).
Rapid quench analysis for determining the rates of product formation
The rates of nicotinamide and deacylated peptide formation were determined under single-turnover conditions using a Hi-Tech RQF-63 rapid-quench-flow device (Hi-Tech Scientific, Bradford-on-Avon, U.K.) as previously described.24 Single turnover reactions for SIRT2 contained 400 μM NAD+, 2.5 μM acyl peptide, and 12 μM SIRT2 in 20 mM potassium phosphate, pH 7.5 at 37 °C. Reactions were monitored from 100 ms up to 30,000 ms and were quenched with TFA to a final concentration of 1% on the rapid quench flow system. Single turnover reactions for SIRT3 contained 2.5 μM acyl peptide and varying NAD+ depending on acyl-substrate as follows; 1 mM NAD+ (acetyl peptide), 2 mM NAD+ (hexanoyl peptide), or 600 μM NAD+ (decanoyl and myristoyl peptides) and 12 μM SIRT3 in 20 mM potassium phosphate, pH 7.5 at 37 °C. Reactions were monitored from 100 ms up to 30,000 ms and were quenched with TFA to a final concentration of 1% on the rapid quench flow system. Single turnover reactions for SIRT6 contained 300 μM NAD+, 5 μM acyl peptide, and 18 μM SIRT6 in 20 mM potassium phosphate, pH 7.5 at 37 °C. Reactions were monitored from 5 s to 180 s and were manually quenched with TFA to a final concentration of 1%. For reactions containing hexanoylated, decanoylated and myristoylated substrates, nicotinamide, NAD+, deacylated product and acylated substrate were analyzed by reverse-phase HPLC using a gradient of 0-4% B (acetonitrile with 0.02% TFA) over 12.5 minutes followed by a gradient of 20-100% B in 30 minutes on a Vydac 201SP104 C18 small pore column. For reactions containing the acetylated substrate, solvent B was 30% acetonitrile with 0.02% TFA, and the same gradient was used for separation. Product and reactant peaks were quantified. The concentration of nicotinamide was determined based on a standard curve measured at 260 nm. The concentration of deacylated peptide was determined as described above for steady-state measurements. To obtain the first-order rate constant (k), the plot of product concentration formed over time was fit to a single-exponential equation, P = [S]0(1 – e−kt), where P is the concentration of product formed, [S]0 is the initial concentration of the limiting substrate, and t is the reaction time.26 For all acyl substrates, complete conversion of the peptide was not attainable. Therefore, to fit these analogs [S]0 was set to 2.5 μM.
Nicotinamide inhibition assay
Sirtuin (0.2 μM) or 0.5 μM SIRT6 was incubated with saturating NAD+ (SIRT1: Acetyl-500 μM, Long chain-300 μM; SIRT2: Acetyl-500 μM, Long chain-300 μM; SIRT3: Acetyl-500 μM, Long chain: 600 μM; SIRT6: Hexanoyl-800 μM, Dec-300 μM, Myr-300 μM) and saturating acyl-lysine peptides (SIRT1: Acetyl-50 μM, Hex-40 μM, Dec-20 μM, Myr-15 μM; SIRT2: Acetyl-50 μM, Hex-15 μM, Dec-15 μM, Myr-15 μM; SIRT3: Acetyl-70 μM, Hex-70 μM, Dec-20 μM, Myr-15 μM; SIRT6: Hex-200 μM, Dec-50 μM, Myr-50 μM) in the presence of 0 – 1.5 mM nicotinamide in 20 mM sodium phosphate, pH 7.5, at 37 °C. Reactions were quenched and analyzed as described above. The data were fitted to the Langmuir isotherm equation to determine the half maximal inhibitory concentration (IC50)27, where [I] is the concentration of the inhibitor and x is the fractional activity minima. Each reaction was performed 2 or 3 times and the average rate was plotted. Error bars represent standard deviation of mean.
| (Eq. 1) |
Crystallization and Structure Determination of SIRT2
GST-tagged SIRT2 was overexpressed in BL21 (DE3) E. coli strain using the same method as that used for His-SIRT2 described above, and purified using Glutathione Sepharose 4B resin (GE Healthcare) according to the manufacturer’s protocol. After thrombin digestion, SIRT2 was further purified by ion exchange chromatography (Resource Q column (GE Healthcare), 20 mM Tris-HCl, pH 8.0, 0 – 1 M NaCl). SDS-PAGE was used to pool SIRT2-containing fractions. Myristoylated peptides (TNF-αK20myr (LPKKmyrTGG) and H3K9myr (TARKmyrSTG)) were used for crystallization (purchased from Toray Research Center, Japan). SIRT2 (15 - 20 mg/ml) and myristoylated peptide (final concentration 1 mM) were mixed and crystallization screening using commercially available kits (Hampton Research) was carried out. Each drop consisting of 0.4 μL of protein/peptide mixture and 0.4 μL of reservoir solution were equilibrated against 80 μL of reservoir solution at 293K by the sitting-drop vapor-diffusion method. Crystals for X-ray crystallography were obtained using 20-25% PEG3350 and 0.2 M potassium formate for H3K9myr peptide complex, or 0.2 M ammonium acetate for TNF-αK20myr peptide complex. We also crystallized SIRT2 with myristoylated peptide and NAD+ to obtain a 2′-O-acyl ADP ribose structure, and crystals were obtained in a condition using 20-25% PEG2000 monomethylether and 0.1 M BisTris pH 6.0-6.5. Crystals were grown in a week. Crystals were frozen by liquid nitrogen, using 10% 2-methyl-2,4-pentanediol as a cryoprotectant. X-ray diffraction data were collected at 100 K in a nitrogen gas stream at the synchrotron beamlines PF-BL5A, PF-BL17A, PF-AR NW12A at Photon Factory, KEK (Proposal No. 2013G674). Data were processed with the programs denzo and scalepack from the HKL package28 (HKL Research, Charlottesville, NC). The structures were solved by molecular replacement by using the program Molrep.29 The SIRT2 structure in the apo-form (PDB IDs: 1J8F and 3ZGO) was used as a search model. Refinement and model building were performed with REFMAC530 and Coot.31 The geometric quality of the model was assessed with MolProbity.32 Data collection and refinement statistics are summarized in Table 2. Atomic coordinates and structure factors have been deposited in the PDB (entries 4Y6L for Human SIRT2 in complex with myristoylated peptide (H3K9myr), 4Y6O Human SIRT2 in complex with myristoylated peptide (TNF-αK20myr) and 4Y6Q Human SIRT2 in complex with 2-O-myristoyl-ADP-ribose). We also re-refined the SIRT6 coordinates in complex with a myristoylated peptide (PDB ID: 3ZG6)2 and used the re-refined coordinates throughout this paper.
Table 2.
Data collection and refinement statistics.
| Crystallization | ||||
| co-crystallized with | TNF-αK20myr | H3K9myr | TNF-αK20myr NAD+ |
|
|
| ||||
|
Data collection statistics
| ||||
| X-ray source | PF-BL5A | PF-BL5A | PF-BL17A | |
| Space group | P1 | P1 | P21 | |
| Unit cell | a (Å) | 37.1 | 37.1 | 61.2 |
| b (Å) | 48.7 | 48.8 | 142.3 | |
| c (Å) | 96.8 | 97.1 | 62.6 | |
| α (°) | 101.2 | 101.0 | 90.0 | |
| β (°) | 91.3 | 91.5 | 94.3 | |
| γ (°) | 112.3 | 112.0 | 90.0 | |
| Wavelength (Å) | 1.000 | 1.000 | 0.980 | |
| Resolution (Å) | 50.0 - 1.6 (1.63 - 1.60) |
50.0 - 1.6 (1.63 - 1.60) |
50 - 1.85 (1.88 - 1.85) |
|
| Unique reflections | 77514 | 79168 | 90675 | |
| Completeness (%)a | 95.7 (92.2) | 95.6 (93.6) | 98.3 (96.6) | |
| Rmerge(%) a | 4.9 (20.8) | 2.3 (14.0) | 6.3 (64.3) | |
| I/σ(I)a | 14.3 (3.2) | 38.5 (14.5) | 17.0 (1.8) | |
| Wilson B (Å2) | 10.3 | 9.8 | 41.9 | |
| Refinement statistics | |||
|---|---|---|---|
| Resolution range (Å) | 20.0 - 1.6 | 20.0 - 1.6 | 20.0 - 1.9 |
| No. of reflections | 73362 | 74836 | 74580 |
| No. of non-hydrogen atoms | 5030 | 4988 | 9471 |
| R work (%) | 21.3 | 21.7 | 25.1 |
| R free (%) | 24.0 | 23.7 | 27.1 |
| R.m.s. deviations | |||
| Bond length (Å) | 0.007 | 0.007 | 0.009 |
| Bond angle (degree) | 1.291 | 1.252 | 1.445 |
| B-factors (Å2) | |||
| Protein | 16.5 | 15.2 | 22.3 |
| Substrate/Product | 26.4 | 21.5 | 20.3 |
| Waters | 20.6 | 20.7 | 19.3 |
| Ramachandran plot (%) | |||
| Favoured region | 97.5 | 97.6 | 98.1 |
| Allowed region | 2.0 | 2.4 | 1.8 |
| Outlier region | 0.5 | 0.0 | 0.1 |
Values in parentheses are for the highest resolution shell
Results
Catalytic efficiency and NAD+ binding kinetics of human Sirtuin enzymes with various acyl-substrates
Our previous results indicate that each Sirtuin has a unique acyl substrate specificity pattern,1 however the molecular basis for these patterns was not investigated. The initial characterization of the deacylation activity was performed at saturating peptide concentration (5 mM) and limiting NAD+, suggesting differences in acyl-substrate reactivity could be due to alterations in NAD+ binding and/or catalysis. To investigate a kinetic model that can explain isoform specific human Sirtuin deacylation activity, we determined the steady-state kinetic parameters kcat (Eq. 3), kcat/Km, NAD (Eq. 4) and Km, NAD (Eq. 5) for SIRT1, SIRT2, SIRT3 and SIRT6, using four different acyl-group substrates representing a range of short, medium and long acyl chain lengths. Analyzed together, these kinetic parameters provide insight into the Sirtuin kinetic mechanism (Figure 1A and B) and an understanding of the relationship between the deacylation activities of diverse human Sirtuins and NAD+ binding.
Human Sirtuins were overexpressed in E. coli and purified as previously described.19-22 Sirtuin enzyme (0.2 μM or 0.5 μM SIRT6) was incubated with a histone H3 lysine 9 (H3K9) acetylated, hexanoylated (C6), decanoylated (C10) or myristoylated (C14) peptide at saturating concentrations, with varied NAD+ levels. To interrogate only the effect of the acyl group on catalysis, the same peptide sequence, representing amino acids 4-17 of histone H3 with two tryptophan residues added for peptide quantitation (QTARKacylSTGGKAPR-WW), was used in all assays. Steady-state deacylation rates were determined using an HPLC-based assay that monitors the formation of deacylated product.1
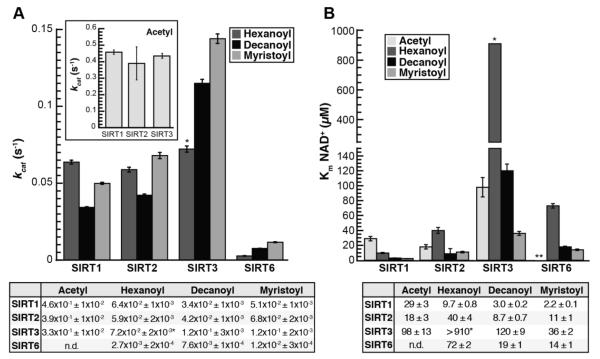
Data from three independent experiments were fitted to the Michaelis-Menten equation (Figure S1), and the resulting average kcat, Km, NAD, and kcat/Km, NAD values were compared (Figure 2 and 3). A Km, NAD for SIRT6 with the acetylated substrate was not determined due to the prohibitively high concentration of acetylated peptide needed to perform the assay at saturating peptide conditions (estimated at nearly 4.5 mM1). Additionally, the hexanoylated peptide was a poor substrate for SIRT3 and we were unable to saturate the reaction with NAD+. Therefore, only the kcat/Km, NAD value was accurately obtained for this reaction, resulting in estimates for Km, NAD and kcat.
Figure 2. kcat and Km, NAD for SIRT1, SIRT2, SIRT3, and SIRT6 deacylation reactions.
A, kcat, inset: kcat for acetylated substrate plotted separately for clarity; Calculated kcat values determined from non-linear regression fits to Michaelis-Menten shown below (n ≥ 3, ± standard deviation). *Estimate for SIRT3 resulting from inability to saturate reaction with NAD+. B, Km for NAD+, Calculated Km, NAD values determined from non-linear regression fits to Michaelis-Menten shown below. *Estimate for SIRT3, **Km, NAD with acetylated peptide for SIRT6 was not measured due to prohibitively high peptide substrate necessary to saturate the reaction (n ≥ 3, ± standard deviation of mean).
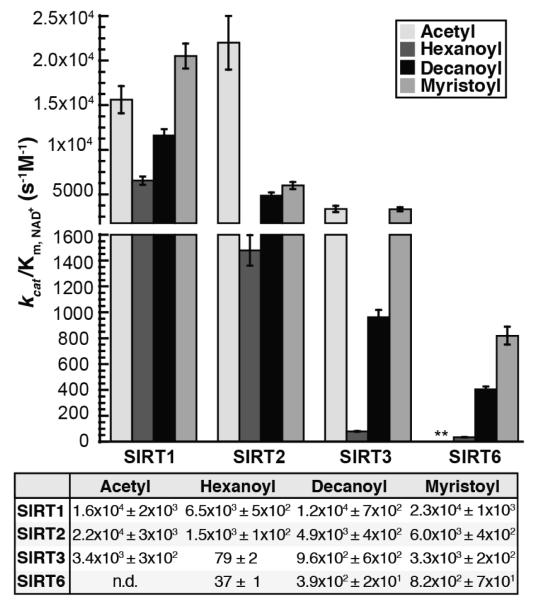
Figure 3. kcat/Km,NAD for SIRT1, SIRT2, SIRT3, and SIRT6 deacylation reactions.
kcat/Km,NAD determined for SIRT1, SIRT2, SIRT3, and SIRT6 in the presence of acetyl-, hexanoyl-, decanoyl-, and myristoyl-lysine H3K9 peptides. Calculated parameters determined from non-linear regression fits to Michaelis-Menten shown below. **kcat/Km,NAD with acetylated peptide for SIRT6 was not measured due to prohibitively high peptide substrate necessary to saturate the reaction (n ≥ 3, ± standard deviation of mean)
SIRT1, SIRT2 and SIRT3 displayed the highest kcat with the acetylated substrate (Figure 2A, inset), while having similar kcat values between the hexanoylated, decanoylated and myristoylated substrates (Figure 2A). The kcat values for SIRT6 increase nearly 4-fold as the chain length increases from hexanoyl to myristoyl (Figure 2A, Table S1). The Km, NAD values for SIRT1 decrease as the length of the acyl group increases from acetyl to myristoyl (Figure 2B, Table S1). The Km, NAD values for SIRT2 and SIRT3 are more varied, however the Km, NAD does decrease as the chain length increases from hexanoyl to myristoyl, similar to the decrease in Km, NAD observed with SIRT6 (Figure 2B, Table S1). For all Sirtuins tested the kcat/Km, NAD increased as acyl chain length increases from hexanoyl (C6) to myristoyl (C14) (Figure 3). With respect to kcat/Km, NAD values for SIRT2, there was a significant preference for the acetylated substrate, where the rates were 4 – 15 times higher than that of other acyl-substrates. The highest kcat/Km, NAD values for SIRT3 were observed with the acetylated and myristoylated substrates and were 42 and 3.5 times higher than the rates with the hexanoylated and decanoylated substrates, respectively (Table S1). SIRT6 preferred myristoylated and decanoylated substrates relative to a hexanoyl substrate, with kcat/Km, NAD values 22 and 10.5 times higher, respectively. Although kcat/Km, NAD values with SIRT1 are similar with all acyl-substrates analyzed, SIRT1 displayed a decreased rate with the hexanoylated substrate relative to acetylated, decanoylated and myristoylated substrates. Together, the results indicate that while each Sirtuin displays varied specificity and catalytic efficiency for deacylation, there exists interdependence between the acylated substrate and NAD+ capture (kcat/Km, NAD).
Nicotinamide inhibition analysis
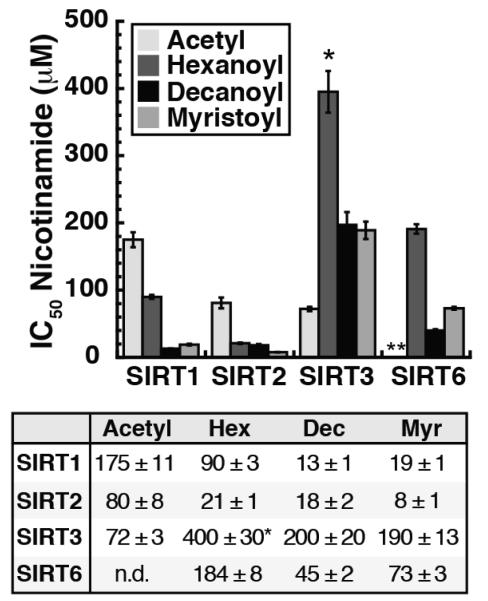
Nicotinamide is the first product released during the deacylation reaction and exhibits noncompetitive inhibition with respect to NAD+ and acylated substrate.24 Previous studies demonstrated that free radiolabeled nicotinamide is capable of re-binding the active site and intercepting the O-alkylimidate intermediate to reform NAD+ through nicotinamide exchange.33 The slower the second chemical step of the reaction (k9) or rate of association and disassociation of nicotinamide (k7, k8), the more susceptible the enzyme is to nicotinamide inhibition (Figure 1A and B). To investigate the effect of the acyl substrate on the ability of nicotinamide to inhibit deacylation, we determined the IC50 of nicotinamide with SIRT1, SIRT2, SIRT3 and SIRT6 in the presence of acetylated, hexanoylated, decanoylated and myristoylated substrates. We found that the IC50 of nicotinamide varied with the Sirtuin and acyl-substrate analyzed (Figure 4).
Figure 4. Nicotinamide IC50 values.
SIRT1, SIRT2, SIRT3 and SIRT6 display varied susceptibilities to nicotinamide inhibition dependent upon acyl-substrate (n ≥ 2, ± standard deviation). *Due to the high Km for NAD+ in the presence of the hexanoylated peptide (>910 μM) the assay was performed in the presence of 800 μM NAD+. **Nicotinamide IC50 for SIRT6 in the presence of acetylated peptide was not measured due to prohibitively high peptide substrate concentration necessary to saturate the reaction.
Steady-state nicotinamide inhibition assays were carried out at fixed, saturating concentrations of acetylated, hexanoylated, decanoylated or myristoylated peptides, saturating NAD+ and Sirtuin (0.2 μM or 0.5 μM SIRT6) with varied nicotinamide (0 to 1500 μM). Reaction rates were quantified, fitted to the Langmuir equation (Eq. 1) to determine the half maximal inhibitory concentration (IC50) (Figure S2), and average IC50 of 2-3 independent experiments were compared (Figure 4). We were unable to measure nicotinamide inhibition for SIRT6 with the acetylated substrate due to the prohibitively high concentration of acetyl-peptide needed for saturation and the IC50,nicotinamide for SIRT3 with the hexanoylated peptide, a poor SIRT3 substrate, is estimated due to the inability to saturate the reaction with NAD+. For SIRT1 and SIRT2, the IC50 for nicotinamide is the highest for the acetylated substrate and decreases as the acyl chain length increases, following the trends observed in kcat (Figure 4 and 3A). Interestingly, SIRT3 and SIRT6 display the greatest susceptibility to nicotinamide inhibition in the presence of the substrate that had the highest kcat (Figure 4 and 3A). SIRT3 is more inhibited by nicotinamide with the acetylated substrate than with the longer chain acyl substrates and SIRT6 is more inhibited by nicotinamide with the myristoylated and decanoylated peptides relative to the hexanoylated substrate (Figure 4). The results indicate that in addition to the interdependence between acylated substrate and NAD+ binding, susceptibility to nicotinamide inhibition is also dependent on the acylated substrate for each Sirtuin.
Rapid quench analysis: resolving individual chemical steps in catalysis
The steady-state reaction parameters and nicotinamide inhibition assays suggest differences in catalytic efficiency that result from the use of diverse acyl-substrates are due to both catalysis and NAD+ binding. Changes in kcat/Km, NAD reflect the rate of the reaction through the first committed step and include both NAD+ binding and cleavage (Figure 1A, Eq. 4). In addition, the differences in kcat measured with the various acylated peptides for a given Sirtuin can be due to differences in the rate of slowest reaction step or can be caused by a change in the rate-limiting step for the entire reaction, for example, from chemistry to product release. To provide mechanistic details about the individual steps involved in catalysis and product formation, and to assess whether the distinct kinetic profiles in our initial velocity analysis could be explained by a change in the rate limiting step of the reaction, a pre-steady-state, rapid-quench kinetic analysis was performed. This analysis was performed for SIRT2 and SIRT3 with acetylated, hexanoylated, decanoylated and myristoylated substrates and for SIRT6 with hexanoylated, decanoylated and myristoylated substrates. SIRT1 was excluded from the analysis due to the co-elution of the high levels of SIRT1 needed for single turnover analysis and the acylated peptides during HPLC analysis, limiting our ability to accurately quantify deacylated product formation. Assays were performed with limiting acyl-substrate, allowing only a single turnover of the enzyme. Sirtuin enzyme (12 μM SIRT2, 12 μM SIRT3, 18 μM SIRT6) was incubated with saturating concentrations of NAD+ and 2.5 μM acyl-peptide or 5 μM acyl-peptide for SIRT6. Under these conditions, all acyl-peptide is assumed to be bound to Sirtuin.34 At six to eight time points, reactions were rapidly quenched with TFA and nicotinamide, deacylated peptide and acylated peptide were resolved and quantified by HPLC. To determine rates of nicotinamide (k5) and deacylated product formation, data from two to three separate experiments per Sirtuin enzyme were fitted to a single exponential (Figure S3). The rate of deacylated product formation represents the rate of the nucleophilic attack of the 2′-hydroxyl on the C1′-O-alkylimidate intermediate in the second chemical step of the deacylation reaction (k9). The experiments were in excellent agreement and the average rates and standard deviations are presented in Table 1. For all enzymes and substrates analyzed the rate of nicotinamide formation (k5) exceeded the rate of the nucleophilic attack of the 2′-hydroxyl (k9) (Figure 1B).
Table 1. Rapid-quench rates of nicotinamide and deacylated product formation.
Average rate and standard deviation of two to three independent experiments.
| Nicotinamide formeition rate, k5 (s−1) | ||||
|---|---|---|---|---|
| Acetyl | Hexanoyl | Decanoyl | Myristoyl | |
| SIRT2 | 1.6 ± 3×10−1 | 2.9×10−1 ± 3×10−2 | 4.3×10−1 ± 6×10−2 | 9×10−1 ± 2×10−1 |
| SIRT3 | 3.8 ± 6×10−1 | 2.8×10−1 ± 5×10−2 | 4.0×10−1 ± 4×10−2 | 7.6×10−1 ± 6×10−2 |
| SIRT6 | N.D. | 4.4×10−3 ± 6×10−4 | 7.3×10−2 ± 8×10−3 | 1.4×10−1 ± 2×10−2 |
| Deacylated peptide formation rate, k9 (s−1) | ||||
|---|---|---|---|---|
| Acetyl | Hexanoyl | Decanoyl | Myristoyl | |
| SIRT2 | 5.3×10−1 ± 6×10−2 | 1.1×10−1 ± 1×102 | 9.0×10−2 ± 8×10−3 | 5.3×10−2 ± 5×10−3 |
| SIRT3 | 6.5×10−1 ± 2×10−2 | 8×10−2 ± 1×10−2 | 1.0×10−1 ± 1×10−2 | 6.5×10−2 ± 6×10−3 |
| SIRT6 | N.D. | 2.3×10−3 ± 1×10−4 | 3.3×10−2 ± 3×10−3 | 4.2×10−2 ± 4×10−3 |
Structural insight into catalysis of long-chain deacylation
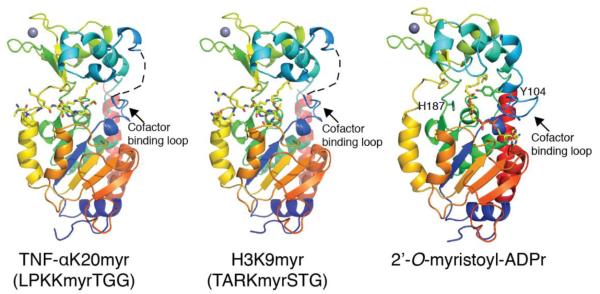
To investigate the molecular basis for varied sensitivity to acyl chain length observed in the rate constants and provide insight into the deacylation mechanism as well as the conformational changes the protein, substrates and products undergo during catalysis, we solved crystal structures of SIRT2 in complex with myristoylated TNF-αK20 or H3K9 substrates at 1.6 Å resolution. To obtain the product complex, we crystallized SIRT2 with myristoylated TNF-αK20 and NAD+, and solved the crystal structure of SIRT2 in complex with 2′-O-myristoyl-ADP-ribose, at 1.9 Å resolution (Figure 5, Table 2). The structures are similar to published SIRT2 structures35-37, including a crystal structure published by Teng et al. of SIRT2 in complex with an H3K9myr peptide, which appeared while this manuscript was being reviewed.38 The overall folds are similar between the substrate and product structures (RMS deviation of 0.7 Å), with the exception of the highly flexible cofactor-binding loop39, which only shows continuous electron density in the SIRT2 – 2′-O-myristoyl-ADPr product structure (Figure 5).
Figure 5. Crystal structure of SIRT2 in complex with TNFα-K20myr, H3K9myr or 2′-O-myristoyl-ADP-ribose.
Overall structural features of SIRT2 in the presence of myristoylated substrates or product. The cofactor binding loop was not visible in the SIRT2 – TNFα-K20myr or SIRT2 – H3K9myr structures and is shown as a dashed line for clarity. Also highlighted in the 2′-O-myristoyl-ADPr structure are the locations and orientations of Tyr104 and the catalytic histidine, His187.
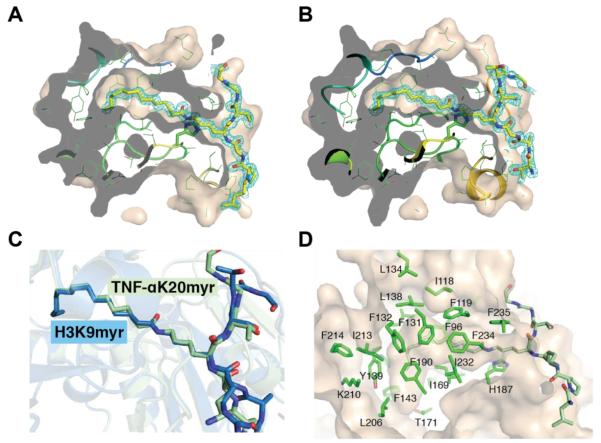
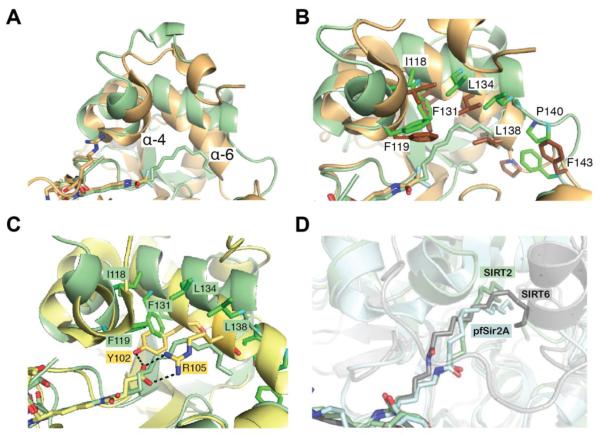
Continuous electron density was observed for the myristoyl-lysine residue of the TNF-αK20myr and H3K9myr peptides (Figure 6A and B). Superimposition of the TNF-αK20myr and H3K9myr structures indicates that while there are changes in the orientation of side chain residues in the peptide substrate, the myristoylated lysine residue adopts a nearly identical conformation in the two structures with an RMS deviation of less than 0.2 Å (Figure 6C). Similar to previously published SIRT6 – H3K9myr and pfSir2A – H3K9myr structures2, 40, the aliphatic chain of the myristoylated lysine is located in a hydrophobic pocket (Figure 6D). To examine the conformational differences in SIRT2 that are needed to accommodate the myristoyl chain in the active site compared to a smaller acetyl-like moiety, the SIRT2 – H3K9myr structure was superimposed with a crystal structure of SIRT2 bound to an ε-trifluoroacetyl lysine peptide inhibitor.35 The structures align well (RMS deviation of 0.4 Å) with the exception of portions of the zinc-binding domain and the helix bundle region (Figure 7A). The greatest differences are observed in α-helix 4 and 6. Amino acids Ile118, Phe119, Phe131, Leu134 and Leu138 shift away from the active site (Figure 7B), suggestive of an induced fit mechanism to accommodate the longer acyl chains. Further, previous structural analysis of SIRT5 in complex with a succinylated substrate identified Tyr-102 and Arg-105, located in α-helix 6, as the amino acids responsible for binding the carboxyl group (Figure 7C),41 indicating there is likely a common mechanism for substrate binding among the Sirtuins that contain a helix bundle region, including human SIRT1 – SIRT5. Superimposition of the SIRT2, SIRT6 and pfSir2A structures reveals the myristoyl chain can bind in a number of different conformations, depending on the hydrophobic pocket and organization of the active site for a given Sirtuin (Figure 7D). As observed in the SIRT6 coordinate file from Jiang et al.2 (PDB ID: 3ZG6), one of the water molecules (A2146 in PDB ID: 3ZG6) is in close contact to the myristoylated peptide (less than 2.2 Å, Figure S4A). We re-refined the SIRT6 coordinate without the water molecule, and concluded the myristoylated peptide should be in a trans conformation (Figure S4B). The newly refined structure was used in the comparison presented in Figure 7D.
Figure 6. Analysis of myristoyl-lysine binding pocket.
A, 2Fo-Fc omit electron density map (cyan mesh, 1σ) of the TNF-αK20 myristoylated peptide. Molecular surface of SIRT2 cut at the level of the hydrophobic cavity are shown. The myristoylated peptides are drawn as sticks, in which yellow, blue and red represent C, N and O atoms, respectively. B, 2Fo-Fc omit electron density map (cyan mesh, 1σ) of the H3K9 myristoylated peptide represented as shown in A. C, superimposition of SIRT2 – TNFα-K20myr (light green) or SIRT2 – H3K9myr (blue) highlighting the orientation of the myristoylated lysine chain. D, hydrophobic pocket in SIRT2 that accommodates the myristoyl chain.
Figure 7. SIRT2 – TNFα-K20myr structural comparison.
A, superimposition of SIRT2 - TNFα-K20myr (light green) with SIRT2 - ε-trifluoroacetyl lysine peptide inhibitor (PDB ID: 4L3O, brown), highlighting the differences in the helix bundle region. B, same structures as in A, highlighting the orientations of specific amino acids. C, superimposition of SIRT2 – TNFα-K20myr (light green) with SIRT5- H3K9succinyl (PDB: 4F4U, yellow). The amino acids in α-helix 6 that interact with the peptide substrates are shown in stick representation. D, superimposition of SIRT2 – TNFα-K20myr (light green) with SIRT6 – H3K9myr (PDB ID: 3ZG6, light blue) and pfSir2A – H3K9myr (PDB ID: 3U3D, grey), highlighting the various orientations of the myristoyl group.
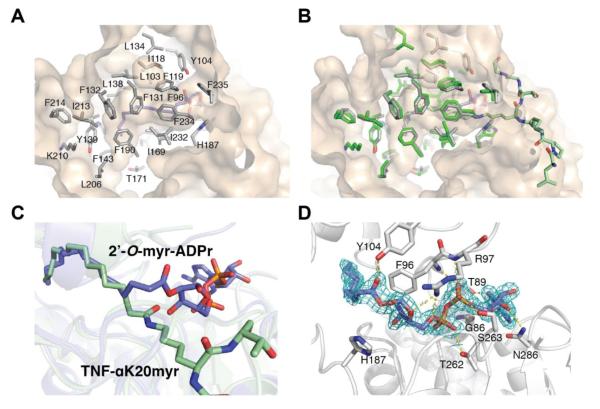
Similar to the H3K9myr and TNF-αK20myr structures, the myristoyl chain of 2′-O-myristoyl-ADPr is bound in the hydrophobic pocket (Figure 8A) and there is minimal movement of the side chains (Figure 8B), indicating the hydrophobic pocket remains relatively unchanged during the course of the reaction. In support of this observation, superimposition of the TNF-αK20myr and 2′-O-myristoyl-ADPr structures reveals the majority of the myristoyl chain adopts a similar conformation between the substrate and product structures (Figure 8C) (RMS deviation of 1.2 Å ). The difference between the acyl chains is found in C3, which is flipped and rotated in the myristoyl chain of 2′-O-myristoyl-ADPr. The movement is similar to what was previously observed in a co-crystal structure of SIRT5 in complex with the 1′,2′-cylic intermediate. In the structure, the lysine side chain is rotated 18° and C3 of the succinyl group is flipped, while the carboxylate of the succinyl lysine remains in contact with Tyr-102 and Arg-105.41 These observations suggest a general mechanism to facilitate deacylation of longer acyl substrates may be to hold the chain beyond C3 in a relatively fixed position during the reaction.
Figure 8. Comparison of SIRT2 – 2′-O-myristoyl-ADPr with SIRT2 – TNFα-K20myr structure.
A, hydrophobic pocket in SIRT2 that accommodates the myristoyl chain of 2′-O-myristoyl-ADPr. B, superimposition of SIRT2 –2′-O-myristoyl-ADPr (grey) and SIRT2 – TNFα-K20myr (green) structures highlighting the hydrophobic residues in the myristoyl chain binding site. C, overlay as in B highlighting differences in myristoyl chain between substrate and product. D, 2Fo-Fc omit electron density map (blue mesh, 1σ) of 2′-O-myristoyl-ADPr molecule and hydrogen bonding network surrounding 2′-O-myristoyl-ADPr. Also shown is the catalytic histidine residue.
In addition to the rotation of C3 in 2′-O-myristoyl-ADPr, there is a corresponding rotation of C1′ and C2′ (Figure 8C) in the ribose. The orientation is stabilized by a hydrogen bond between the carbonyl oxygen and the hydroxyl group of Tyr104 located in the cofactor-binding loop of SIRT2 (Figure 8D). The ADP-ribose interacts with SIRT2 through a number of side chain and backbone hydrogen bonds which are shown in Figure 8D. Interestingly, the catalytic histidine residue (His187) does not hydrogen bond to the 3′-hydroxyl of the ribose as is observed in the SIRT5 - cyclic intermediate structure (Figure 8D).41
Discussion
Kinetic mechanism demonstrates the interdependence of NAD+ binding and nicotinamide inhibition on specific deacylation reactions
We combined our NAD+ steady-state kinetic, pre-steady-state and inhibition analyses to develop a model for each Sirtuin that explains the relationship between NAD+ binding and the acyl chain length of the peptide substrate. The established Sirtuin kinetic mechanism describes ordered binding of acyl-substrate followed by NAD+ (Figure 1A).24 Supra-physiological levels of NAD+ can induce NAD+ binding in non-productive complexes in non-mammalian Sirtuins42, 43 and near millimolar binding constants for NAD+ for SIRT1 and SIRT3 have been measured in vitro.44 However, to date only SIRT6 has been shown to bind NAD+ tightly (KD = 27 μM) in the absence of an acylated substrate22, suggesting SIRT6 may be capable of random substrate binding. In the case of random substrate binding to SIRT6, saturating with peptide will cause a random mechanism to behave as an ordered mechanism45, allowing us to utilize the same kinetic model to describe the behavior of all Sirtuins studied (Figure 1A).
The kcat/Km, NAD values for SIRT1 are similar with all acyl-substrates analyzed (Figure 3). However, the kcat value increased with the acetylated substrate compared to the longer acylated substrates (Figure 2A). The results suggest that as the acyl chain length increases, the rate of turnover is more adversely affected compared to the rate of NAD+ capture by SIRT1 to form productive catalytic complexes. SIRT2, a cytosolic Sirtuin with distinct protein substrates from SIRT1, displays similar trends in kcat and kcat/Km, NAD as SIRT1 (Figure 2A and 3). However, the catalytic efficiency is lower for SIRT2 in the presence of the longer acyl chains (Figure 3), indicating the decrease in kcat/Km, NAD for SIRT2, particularly with the hexanoylated substrate, is driven by a weaker affinity of SIRT2 for NAD+ or decreased rate of NAD+ cleavage (k5) in the presence of the longer chain acyl groups compared to SIRT1 (Figure 3). The mitochondrial Sirtuin, SIRT3, displays different trends to those observed with SIRT1 and SIRT2. SIRT3 has the highest kcat/Km, NAD for acetylated and myristoylated peptides (Figure 3) and the lowest for the hexanoylated substrate. The kcat is similar between the hexanoylated, deconylated and myristoylated substrates (Figure 2A) while the Km for NAD+ is significantly higher in the presence of the hexanoylated substrate (Km, NAD > 910 μM versus <75 μM for other substrates) (Figure 2B), indicating the rate of NAD+ binding or cleavage is decreased. While use of the same peptide sequence for all Sirtuin isoforms allows for a dissection of the role of acyl chain length on catalysis, different amino acid sequence could impart additional kinetic differences.
The rate of partitioning the C1′-O-alkylimidate intermediate between continuation to products and reversal to acylated substrate and NAD+ dictates the sensitivity to nicotinamide inhibition, and the results indicate this partitioning is dependent on the acyl substrate. The slower conversion of the long-chain O-alkylimidate intermediates (k9) relative to deacetylation likely increases the sensitivity of SIRT2 to inhibition by nicotinamide, leading to decreases observed in nicotinamide IC50 with long chain acyl groups (Figure 3). Interestingly, nicotinamide is a more potent inhibitor of SIRT3 in the presence of the acetylated substrate. The result is similar to previous observations made for SIRT5,44 a mitochondrial Sirtuin whose preferred desuccinylase activity is strongly inhibited by nicotinamide while the deacetylase activity displays very poor inhibition. Differences in nicotinamide inhibition with SIRT5 were proposed to be caused by differences in the organization of the active site in the presence of an acetylated peptide compared to a succinylated peptide.44 Along with the data presented here, it is reasonable to suggest that the ability of nicotinamide to inhibit depends on the efficiency of binding the intermediate form of the enzyme, as well as the relative partitioning of the O-alkylimidate intermediates toward products or reactants.
To investigate in greater detail the individual steps in the SIRT2 and SIRT3 reactions that contribute to the observed patterns in the kinetic constants, pre-steady-state kinetics monitoring nicotinamide and deacylated peptide formation were performed with acetylated, hexanoylated, decanoylated and myristoylated substrates. For all peptides analyzed, SIRT2 cleaved NAD+ at a rate greater than kcat (Table 1, Figure 2A) and followed a similar trend to that observed in kcat/Km, NAD (Figure 3, Table 1). Therefore, nicotinamide formation (k5) is not the rate-limiting step. The fold increases from hexanoylated to myristoylated substrates are similar to the changes in kcat/Km, NAD (Figure 2B, Table S1), indicating the increase in kcat/Km, NAD is driven by a faster rate of NAD+ cleavage (k5), not a change in the rate of NAD+ binding (k3, k4) (Figure 1A). Similarly, SIRT3 cleaved NAD+ at a rate greater than kcat with all acylated substrates analyzed. However, as the chain length increases from acetyl to hexanoyl, the change in the rate of nicotinamide formation is significantly less than the fold change in kcat/Km, NAD (Table S1). Therefore, weaker binding of NAD+ (k3, k4) likely contributes to the decrease in kcat/Km, NAD in the presence of the hexanoylated substrate as opposed simply a decrease in the rate of NAD+ cleavage (k5).
A step after nicotinamide formation, either the second chemical step of the reaction (attack of 2′-OH on the imidate intermediate), hydrolysis of the cyclic intermediate or product release (Figure 1A and B), is rate limiting. Because the rapid quench assay detects the formation of the deacylated product upon rapid quenching of the enzyme, the rate represents deacylated peptide formation of the labile 1′,2′-cyclic intermediate, which upon the acid quench yields deacylated product. Therefore, if all reaction steps prior to deacylated product and O-Acyl-ADPr (OAADPr) release are faster than kcat, we conclude that hydrolysis of the cyclic intermediate or the release of deacylated product or OAADPr must be rate limiting. With acetylated, hexanoylated, decanoylated and myristoylated substrates, SIRT2 and SIRT3 form deacylated peptide at rates similar to kcat (≤ 2-fold) (Table 1, Figure 2A), indicating the rate limiting step of deacylation is the nucleophilic attack of the 2′-hydroxyl on the C1′-O-alkylimidate intermediate (k9).
SIRT6, a Sirtuin with unique catalytic properties1, has a distinct kinetic profile from its paralogs, SIRT1, SIRT2 and SIRT3. As acyl chain length increases, kcat and kcat/Km increase (Figure 2A and 3). SIRT6 lacks the conserved helix bundle region, which we hypothesize, is responsible for accommodating the acylated substrate in SIRT1 – SIRT5 (Figure 7A and B). Instead, SIRT6 contains an extended N-terminal loop which covers over the NAD+ and acyl-substrate binding sites.2, 22 Continuous electron density for the N-terminal twelve amino acids is only observed in the co-crystal structure of SIRT6 in complex with a myristoylated peptide and ADP-ribose, indicating the loop is highly flexible.2 The ordering of the loop in the presence of the longer acyl chains might allow for synergistic binding of NAD+ and secure the catalytically competent conformation, leading to increased turnover (Figure 2A).
To determine whether the differing trend in kcat from SIRT2 and SIRT3 could be explained by differences in the rate limiting step of the reaction, we performed pre-steady-state kinetic analysis, and similar to SIRT2 and SIRT3 the rate of nicotinamide formation proceeds faster than kcat with all peptides tested (Table 1, Figure 2A). The increases in the rate of nicotinamide formation (k5) are similar to the increases in kcat/Km, NAD (Table 1 and S1, Figure 3), indicating the increase in kcat/Km, NAD is driven by faster NAD+ cleavage (k5) as opposed to differences in NAD+ binding (k3, k4).
We found that unlike SIRT2 and SIRT3, the rate-limiting step of the SIRT6 deacylation reaction appears to be dependent on acyl-chain length. The rate of dehexanoyl product formation is equal to kcat, while the rate of dodecanoyl and demyristoyl product formation is faster than kcat (~3-4-fold) (Table 1, Figure 2A). The results are consistent with the attack of the 2′-OH in the second chemical step (k9) being rate limiting in the dehexanoylation reaction, while hydrolysis of the 1′,2′-cyclic intermediate or the release of deacylated product or OAADPr appears to be the rate limiting step with decanoylated and myristoylated substrates (Figure 1B). The rate of k9 with the myristoylated substrate begins to approach the values observed for the more efficient SIRT2 and SIRT3 enzymes (Table 1), further highlighting the ability of SIRT6 to function as an efficient demyristoylase.
Taken together, the results indicate SIRT2 and SIRT3 may operate under the same kinetic mechanism where the same rate-limiting step is maintained among the diverse acyl substrates, while the nature of rate-limiting step of SIRT6 depends on the length of the acyl chain. With the exception of SIRT2 and SIRT3 with the myristoylated substrate, while the individual rates of k5 and k9 differ between Sirtuins and between peptides, the difference between the rates of k5 and k9 for a given Sirtuin and substrate are within an order of magnitude of each other (Table 1). This suggests the rate of NAD+ cleavage is coupled to the rate of attack of the 2′-hydroxyl on the C1′-O-alkylimidate intermediate, such that the rate of NAD+ cleavage does not far exceed the rate of the second chemical step.
Structural insight into long chain deacylation
Long-chain deacylation was initially believed to be limited to SIRT6 and pfSir2A, due to the identification of a hydrophobic binding cavity for the longer acyl groups. However, our previous1 and current analyses demonstrate human Sirtuins SIRT1, SIRT2 and SIRT3 function as efficient long-chain deacylases. The crystal structures of SIRT2 bound myristoylated substrates identified a latent hydrophobic binding pocket, which is able to accommodate the longer myristoyl chain (Figure 6D). SIRT1 and SIRT3 have similarly positioned hydrophobic amino acids (Figure S5), indicating these Sirtuins have a latent hydrophobic binding pocket to accommodate the long acyl chains, as evidenced by long-chain deacylation activity. Additionally, structural alignment of SIRT2 in complex with a myristoyl-lysine peptide or an ε-trifluoroacetyl-lysine peptide identified α-helix 4 and 6 in the helix bundle region as having the largest conformational change in the presence of the myristoyl chain compared to the smaller ε-trifluoroacetyl ligand (Figure 7A). The movement places several hydrophobic amino acids in a position to bind the myristoyl chain. The amino acids are similar in SIRT1 and SIRT3, however Phe312 and Ile316 in SIRT1 are larger than the respective leucine residues in SIRT2 and SIRT3 (Figure S5). The increased van der Waals forces and size of the amino acids might help lock the longer acyl chains and NAD+ in a suitable conformation for catalysis, leading to the observed increase in kcat/Km, NAD with the longer acyl chains. Despite having very similar hydrophobic pockets, SIRT2 and SIRT3 display varying catalytic efficiencies that depend on the nature of the acylated substrate (Figure 3), indicating there are additional factors that affect NAD+ capture (kcat/Km, NAD) that would not be evident from the crystal structure alone. These differences include the dynamics of the acyl-substrate binding pocket for each Sirtuin as well as the highly flexible cofactor-binding loop39, which adopts different conformations depending on the ligand(s) bound. However, the rates of k9 for SIRT2 and SIRT3 are similar with the hexanoylated, decanoylated and myristoylated intermediates (Table S1), suggesting once the O-alkylimidate intermediate is formed, the chain is held in a similar conformation allowing for attack of the 2′-hydroxyl.
In vivo implications
Genetic, metabolic and pharmacological-induced alterations in NAD+ levels have been linked to changes in Sirtuin activity.17 However, inconsistent observations have confounded a clear picture of the connection between NAD+ and each Sirtuin. Here, we provide a model that describes the interrelationships between the different deacylation specificities, NAD+ dependence and chemical mechanism of individual human Sirtuin members. These results have broad implications and provide molecular insight into the cellular behavior of Sirtuin activity. NAD+ is an essential co-substrate in metabolism and its biosynthesis is compartmentalized.46 Studies estimate cellular levels of NAD+ to be 365 μM with 70% of this pool located in the mitochondria and 10-100 μM in the nucleus and cytoplasm.8, 47 Consistent with adaption to compartmentalized NAD+, mitochondrial SIRT3 has a higher Km for NAD+ with all substrates tested when compared to the nuclear and cytoplasmic Sirtuins (Figure 2B).
Taken together, our observations support the idea that individual Sirtuin activity has evolved to operate in a sub-cellular compartment where changes in NAD+ levels will directly affect Sirtuin activity and substrate preference. Moreover, the generally low Km values for NAD+ with long-chain acyl substrates combined with the low Km values for long-chain acyl substrates suggests that these deacylations are not controlled appreciably by cellular NAD+ fluctuations, and might reflect a housekeeping function to limit the adverse effects of spurious long-chain protein acylation from acyl-CoAs. The low Km values are indicative of a strong commitment to catalysis when Sirtuins encounter these modifications in the cell. Recent literature suggests many acyl-lysine modifications exist in vivo and much remains to be explored about the potential Sirtuin substrates that contain these modifications.2, 3, 9, 13-16
Concluding remarks
We present an integrated kinetic and structural analysis providing insight into potential regulation of in vivo Sirtuin activity. We find the Km for NAD+ and susceptibility to nicotinamide inhibition varies with Sirtuin and is dependent upon the acyl substrate. Nicotinamide is considered a general Sirtuin product inhibitor and widely used in in vivo and in vitro studies. Our results reveal careful consideration should be taken when using this commonly employed Sirtuin inhibitor in cell-based and in vitro studies and suggest targeted nicotinamide inhibition is possible. We provide the first rapid-quench analysis of human Sirtuins, showing nicotinamide formation is rapid when compared to product formation or release for SIRT2, SIRT3 and SIRT6 for all acylated substrates tested. Rapid-quench analysis establishes the nucleophilic attack of the 2′-hydroxyl on the C1′-O-alkylimidate intermediate as the rate limiting step of catalysis for SIRT2 and SIRT3 and it is independent of the acylated substrate, while the rate limiting step for SIRT6 depends on the length of the acyl chain and appears to change from attack of the 2′-hydroxyl group with the hexanoylated substrate to the hydrolysis of the cyclic intermediate or the release of deacylated product or OAADPr with decanoylated or myristoylated substrates. Our structural studies indicate the myristoyl chain is held in a relatively fixed position during catalysis and reveals a general mechanism for long chain deacylation. Taken together, this study establishes the interdependence of NAD+ binding, acyl-substrate and nicotinamide inhibition on the Sirtuin chemical mechanism. As diverse deacylation activities for Sirtuins are further characterized in cells, this compilation of kinetic parameters will support the development of targeted small molecule inhibitors and activators.
Supplementary Material
Acknowledgments
Funding Information
This work was supported by NIH Traineeship 5T32GM08349 (K.E.D.), NIH Grant GM06538 (J.M.D.), and the Project for Development of Innovative Research on Cancer Therapeutics (P-DIRECT) from The Ministry of Education, Culture, Sports, Science and Technology of Japan (A.I.). The purchase of the Bruker REFLEX®II in 1996 was partially funded by NSF Award #9520868 to the University of Wisconsin – Madison Department of Chemistry.
Abbreviations
- NAD+
Nicotinamide adenine dinucleotide
- NAM
Nicotinamide
- TNFα
Tumor necrosis factor alpha
- OAADPR
O-acyl-ADP-ribose
- Myr
Myristoyl
- DMF
dimethylformamide
- DCM
dichloromethane
- TFA
trifluoroacetic acid
Footnotes
Supporting information available: NAD+ saturation curves (Figure S1), nicotinamide inhibition curves (Figure S2), single turnover kinetics (Figure S3), re-refinement of the SIRT6 coordinates in complex with a myristoylated peptide (Figure S4), sequence alignment (Figure S5), rate constant comparisons (Table S1). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- [1].Feldman JL, Baeza J, Denu JM. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem. 2013;288:31350–31356. doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, Du J, Kim R, Ge E, Mostoslavsky R, Hang HC, Hao Q, Lin H. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013;496:110–113. doi: 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wagner GR, Hirschey MD. Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol Cell. 2014;54:5–16. doi: 10.1016/j.molcel.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].He W, Newman JC, Wang MZ, Ho L, Verdin E. Mitochondrial sirtuins: regulators of protein acylation and metabolism. Trends Endocrinol Metab. 2012;23:467–476. doi: 10.1016/j.tem.2012.07.004. [DOI] [PubMed] [Google Scholar]
- [5].Cai L, Sutter BM, Li B, Tu BP. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell. 2011;42:426–437. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Weinert BT, Iesmantavicius V, Moustafa T, Scholz C, Wagner SA, Magnes C, Zechner R, Choudhary C. Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol Syst Biol. 2014;10:716. doi: 10.1002/msb.134766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Weinert BT, Iesmantavicius V, Wagner SA, Scholz C, Gummesson B, Beli P, Nystrom T, Choudhary C. Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol Cell. 2013;51:265–272. doi: 10.1016/j.molcel.2013.06.003. [DOI] [PubMed] [Google Scholar]
- [8].Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr., Weissman S, Verdin E, Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, He B, Chen W, Zhang S, Cerione RA, Auwerx J, Hao Q, Lin H. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Garrity J, Gardner JG, Hawse W, Wolberger C, Escalante-Semerena JC. N-lysine propionylation controls the activity of propionyl-CoA synthetase. J Biol Chem. 2007;282:30239–30245. doi: 10.1074/jbc.M704409200. [DOI] [PubMed] [Google Scholar]
- [11].Liu B, Lin Y, Darwanto A, Song X, Xu G, Zhang K. Identification and characterization of propionylation at histone H3 lysine 23 in mammalian cells. J Biol Chem. 2009;284:32288–32295. doi: 10.1074/jbc.M109.045856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhang K, Chen Y, Zhang Z, Zhao Y. Identification and verification of lysine propionylation and butyrylation in yeast core histones using PTMap software. Journal of proteome research. 2009;8:900–906. doi: 10.1021/pr8005155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhang Z, Tan M, Xie Z, Dai L, Chen Y, Zhao Y. Identification of lysine succinylation as a new post-translational modification. Nature chemical biology. 2011;7:58–63. doi: 10.1038/nchembio.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, Luo H, Zhang Y, He W, Yang K, Zwaans BM, Tishkoff D, Ho L, Lombard D, He TC, Dai J, Verdin E, Ye Y, Zhao Y. The first identification of lysine malonylation substrates and its regulatory enzyme. Molecular & cellular proteomics : MCP. 2011;10:M111. doi: 10.1074/mcp.M111.012658. 012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Xie Z, Dai J, Dai L, Tan M, Cheng Z, Wu Y, Boeke JD, Zhao Y. Lysine succinylation and lysine malonylation in histones. Molecular & cellular proteomics : MCP. 2012 doi: 10.1074/mcp.M111.015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tan M, Peng C, Anderson KA, Chhoy P, Xie Z, Dai L, Park J, Chen Y, Huang H, Zhang Y, Ro J, Wagner GR, Green MF, Madsen AS, Schmiesing J, Peterson BS, Xu G, Ilkayeva OR, Muehlbauer MJ, Braulke T, Muhlhausen C, Backos DS, Olsen CA, McGuire PJ, Pletcher SD, Lombard DB, Hirschey MD, Zhao Y. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 2014;19:605–617. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Imai SI, Guarente L. NAD and sirtuins in aging and disease. Trends Cell Biol. 2014 doi: 10.1016/j.tcb.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- [19].Borra MT, Smith BC, Denu JM. Mechanism of human SIRT1 activation by resveratrol. J Biol Chem. 2005;280:17187–17195. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- [20].Borra MT, O’Neill FJ, Jackson MD, Marshall B, Verdin E, Foltz KR, Denu JM. Conserved enzymatic production and biological effect of O-acetyl-ADP-ribose by silent information regulator 2-like NAD+-dependent deacetylases. J Biol Chem. 2002;277:12632–12641. doi: 10.1074/jbc.M111830200. [DOI] [PubMed] [Google Scholar]
- [21].Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pan PW, Feldman JL, Devries MK, Dong A, Edwards AM, Denu JM. Structure and biochemical functions of SIRT6. J Biol Chem. 2011;286:14575–14587. doi: 10.1074/jbc.M111.218990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cleland WW. Partition analysis and the concept of net rate constants as tools in enzyme kinetics. Biochemistry. 1975;14:3220–3224. doi: 10.1021/bi00685a029. [DOI] [PubMed] [Google Scholar]
- [24].Borra MT, Langer MR, Slama JT, Denu JM. Substrate specificity and kinetic mechanism of the Sir2 family of NAD+-dependent histone/protein deacetylases. Biochemistry. 2004;43:9877–9887. doi: 10.1021/bi049592e. [DOI] [PubMed] [Google Scholar]
- [25].Northrop DB. On the Meaning of Km and V/K in Enzyme Kinetics. Journal of Chemical Education. 1998;75:1153. [Google Scholar]
- [26].Smith BC, Denu JM. Sir2 deacetylases exhibit nucleophilic participation of acetyl-lysine in NAD+ cleavage. J Am Chem Soc. 2007;129:5802–5803. doi: 10.1021/ja070162w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wu J, Xie N, Wu Z, Zhang Y, Zheng YG. Bisubstrate Inhibitors of the MYST HATs Esa1 and Tip60. Bioorganic & medicinal chemistry. 2009;17:1381–1386. doi: 10.1016/j.bmc.2008.12.014. [DOI] [PubMed] [Google Scholar]
- [28].Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- [29].Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta crystallographica. Section D, Biological crystallography. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta crystallographica. Section D, Biological crystallography. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- [31].Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica. Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica. Section D, Biological crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Jackson MD, Schmidt MT, Oppenheimer NJ, Denu JM. Mechanism of nicotinamide inhibition and transglycosidation by Sir2 histone/protein deacetylases. J Biol Chem. 2003;278:50985–50998. doi: 10.1074/jbc.M306552200. [DOI] [PubMed] [Google Scholar]
- [34].Smith BC, Denu JM. Sir2 deacetylases exhibit nucleophilic participation of acetyl-lysine in NAD+ cleavage. Journal of the American Chemical Society. 2007;129:5802–5803. doi: 10.1021/ja070162w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yamagata K, Goto Y, Nishimasu H, Morimoto J, Ishitani R, Dohmae N, Takeda N, Nagai R, Komuro I, Suga H, Nureki O. Structural basis for potent inhibition of SIRT2 deacetylase by a macrocyclic peptide inducing dynamic structural change. Structure. 2014;22:345–352. doi: 10.1016/j.str.2013.12.001. [DOI] [PubMed] [Google Scholar]
- [36].Moniot S, Schutkowski M, Steegborn C. Crystal structure analysis of human Sirt2 and its ADP-ribose complex. Journal of structural biology. 2013;182:136–143. doi: 10.1016/j.jsb.2013.02.012. [DOI] [PubMed] [Google Scholar]
- [37].Finnin MS, Donigian JR, Pavletich NP. Structure of the histone deacetylase SIRT2. Nature structural biology. 2001;8:621–625. doi: 10.1038/89668. [DOI] [PubMed] [Google Scholar]
- [38].Teng YB, Jing H, Aramsangtienchai P, He B, Khan S, Hu J, Lin H, Hao Q. Efficient Demyristoylase Activity of SIRT2 Revealed by Kinetic and Structural Studies. Scientific reports. 2015;5:8529. doi: 10.1038/srep08529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sanders BD, Jackson B, Marmorstein R. Structural basis for sirtuin function: what we know and what we don’t. Biochimica et biophysica acta. 2010;1804:1604–1616. doi: 10.1016/j.bbapap.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhu AY, Zhou Y, Khan S, Deitsch KW, Hao Q, Lin H. Plasmodium falciparum Sir2A preferentially hydrolyzes medium and long chain fatty acyl lysine. ACS chemical biology. 2012;7:155–159. doi: 10.1021/cb200230x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhou Y, Zhang H, He B, Du J, Lin H, Cerione RA, Hao Q. The bicyclic intermediate structure provides insights into the desuccinylation mechanism of human sirtuin 5 (SIRT5) J Biol Chem. 2012;287:28307–28314. doi: 10.1074/jbc.M112.384511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Avalos JL, Boeke JD, Wolberger C. Structural basis for the mechanism and regulation of Sir2 enzymes. Mol Cell. 2004;13:639–648. doi: 10.1016/s1097-2765(04)00082-6. [DOI] [PubMed] [Google Scholar]
- [43].Min J, Landry J, Sternglanz R, Xu RM. Crystal structure of a SIR2 homolog-NAD complex. Cell. 2001;105:269–279. doi: 10.1016/s0092-8674(01)00317-8. [DOI] [PubMed] [Google Scholar]
- [44].Fischer F, Gertz M, Suenkel B, Lakshminarasimhan M, Schutkowski M, Steegborn C. Sirt5 deacylation activities show differential sensitivities to nicotinamide inhibition. PLoS One. 2012;7:e45098. doi: 10.1371/journal.pone.0045098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Cook PF, Cleland WW. Enzyme kinetics and mechanism. Garland Science; London ; New York: 2007. [Google Scholar]
- [46].Nikiforov A, Dolle C, Niere M, Ziegler M. Pathways and subcellular compartmentation of NAD biosynthesis in human cells: from entry of extracellular precursors to mitochondrial NAD generation. J Biol Chem. 2011;286:21767–21778. doi: 10.1074/jbc.M110.213298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Zhang Q, Piston DW, Goodman RH. Regulation of corepressor function by nuclear NADH. Science. 2002;295:1895–1897. doi: 10.1126/science.1069300. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.