Abstract
Human parvovirus B19 (B19V) is a human pathogen that belongs to genus Erythroparvovirus of the Parvoviridae family, which is composed of a group of small DNA viruses with a linear single-stranded DNA genome. B19V mainly infects human erythroid progenitor cells and causes mild to severe hematological disorders in patients. However, recent clinical studies indicate that B19V also infects nonerythroid lineage cells, such as myocardial endothelial cells, and may be associated with other disease outcomes. Several cell culture systems, including permissive and semipermissive erythroid lineage cells, nonpermissive human embryonic kidney 293 cells and recently reported myocardial endothelial cells, have been used to study the mechanisms underlying B19V infection and B19V DNA replication. This review aims to summarize recent advances in B19V studies with a focus on the mechanisms of B19V tropism specific to different cell types and the cellular pathways involved in B19V DNA replication including cellular signaling transduction and cell cycle arrest.
Keywords: B19V, cell cycle, DDR, DNA damage response, DNA replication, Epo/EpoR signaling, human parvovirus B19, hypoxia, tropism
Human parvovirus B19 (B19V) was discovered in 1975 by Cossart and colleagues when screening for hepatitis B virus in a panel of human serum samples [1]. The virus was described as 23 nm in diameter, a typical capsid size of a parvovirus. The virus came from the serum sample coded as panel B number 19, and thereafter was named ‘Parvovirus B19.’ Most commonly, B19V infection causes erythema infectiosum or fifth disease (also named ‘slapped cheek syndrome’), which was first identified by Anderson et al. in the early 1980s [2]. In addition to infections in children, B19V is also highly infectious and spreads easily among adults with a seropositive rate ranging from 60 to 90% [3]. It causes a spectrum of clinical complications in vulnerable populations, including arthropathy in healthy adults (particularly in middle-aged women), persistent anemia in immunosuppressed patients, transient aplastic crisis in patients with increased erythropoiesis (such as sickle-cell disease patients), and hydrops fetalis and congenital anemia in pregnant women [4–6]. Noteworthy, in addition to these common manifestations, accumulating clinical reports indicate that B19V infection is associated with cardiovascular diseases, for example, myocarditis, in both adults and children [7–12]. The association of B19V infection to several liver diseases, for example, acute and chronic hepatitis, acute fulminant liver failure and autoimmune hepatitis, has been also reported [13–16]; however, only erythroid progenitor cells from fetal liver have been shown permissive to B19V replication [17]. And, therefore, the mechanism for B19V infection of liver cells will not be discussed in this review.
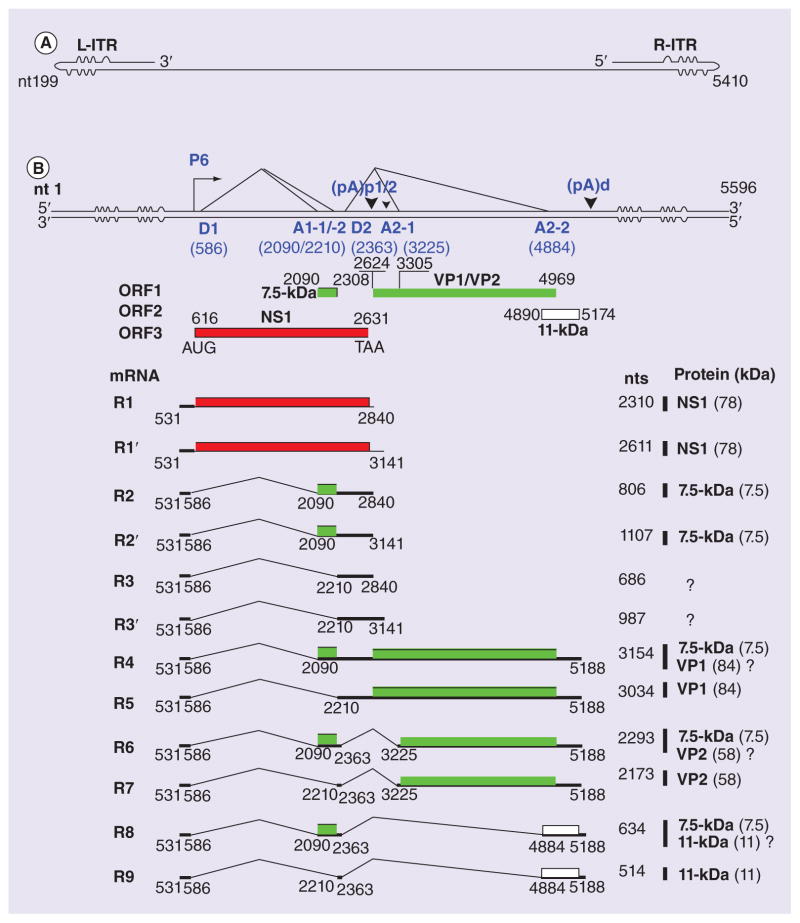
B19V is a member of the Erythroparvovirus genus within the Parvovirinae subfamily of the Parvoviridae family [18]. It has a linear ssDNA genome of 5596 nucleotides. It is flanked by two identical inverted terminal repeats (ITRs) that form an imperfect palindrome at each end (Figure 1A) [19–21], a feature also shared by adeno-associated virus 2 (AAV2) and human parvovirus 4 (PARV4) [22]. By contrast, all other members of the Parvovirinae subfamily have asymmetric terminal repeats [23]. Under the P6 promoter located at map unit 6, the replicative form (RF) of the B19V genome transcribes nine major viral mRNAs (R1-9) that encode capsid proteins (VP1 and VP2) and nonstructural proteins (NS1, 11 and 7.5 kDa) (Figure 1B) [24–26]. A B19V DNA infectious clone (B19V RF DNA M20; Figure 1B) has been constructed [27], based on which, mutagenesis studies have confirmed that NS1 is the only protein essential for B19V DNA replication; whereas VP1/2 and 11-kDa proteins are required for progeny virus production [28].
Figure 1. Structure of the human parvovirus B19 genome and the human parvovirus B19 genetic map.
(A) Schematic diagram of the minus strand of the B19V ssDNA genome. Identical ITRs are present at each end of the genome, and these are depicted with unpaired or mismatched bases in the palindromes represented by ‘bulges’ or ‘bubbles,’ respectively. (B) Schematic diagrams of the duplex RF of the B19V genome (B19V J35 strain; GenBank accession number: AY386330), which has the capability to express viral genes, replicate and produce progeny virions. The left- and right-hand ITRs (L-ITR and R-ITR), P6 promoter, RNA initiation site, splice donors (D1 and D2), splice acceptors (A1-1, A1-2, A2-1 and A2-2) and proximal and distal polyadenylation sites ((pA)p1/2 and (pA)d) are indicated, along with the nine major mRNAs (R1-9) and three minor mRNAs (R1′-3′). The numbers of nucleotides (nt) are indicated in each case. The proteins encoded by each mRNA are shown on the right, while the question mark (?) denotes that the protein translated from the mRNA is currently unknown. The size of the ITRs and the NS1- and VP1-encoding regions diagrammed are not to scale.
B19V: Human parvovirus B19; ITR: Inverted terminal repeat; NS1: Nonstructural proteins; RF: Replicative form; VP1: Capsid proteins.
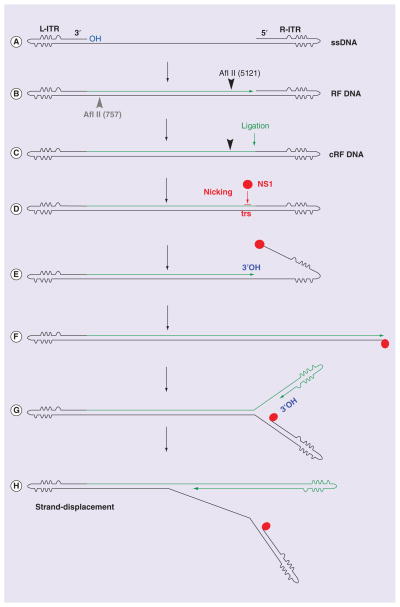
B19V is an autonomous parvovirus, representing a majority of the Parvoviridae family members that can replicate by themselves in host cells, in contrast to AAV that requires coinfection of a helper virus, such as adenovirus, for replication [29]. Generally, DNA replication of autonomous parvoviruses happens in the S phase of the host cell cycle [30–33] and follows a ‘rolling hairpin’ model of DNA replication [34,35]. Because an in vitro reconstitution B19V DNA replication system has not been established, current knowledge on B19V DNA replication is largely derived from AAV, whose genome has two ITRs, and the autonomous parvovirus minute virus of mice. A hairpin-primed ssDNA replication model of B19V is proposed here (Figure 2). In principle, the B19V ssDNA genome uses 3′-end hairpin as a self-primer (3′OH) to extend viral ssDNA into the dsDNA genome by cellular replication proteins, a step in so-called first-strand DNA synthesis (Figure 2B). The extended 3′ end is presumably ligated to the 5′ end of the genome by an unknown cellular ligase to form a partial circular DNA structure (Figure 2C) [36–38]. The dsDNA form of the virus transcribes viral mRNAs for the expression of viral proteins. The largest non-structural protein (NS1), a multiple-functional protein with site-specific endonuclease activity and DNA helicase activity, nicks the junction at the so-called terminal resolution site (trs) between the 5′-end hairpin (Figure 2D) and the newly synthesized viral DNA to form a novel 3′ primer that initiates the strand displacement and rolling hairpin-dependent DNA replication (Figure 2E–H). The elongated viral genomes (both replicative and double-replicative intermediates) are resolved by NS1 nicking and release ssDNA that is finally packaged into the capsid.
Figure 2. Rolling-hairpin model of human parvovirus B19 DNA replication.
(A & B) The B19V ssDNA genome as shown is first converted into RF DNA that is primed by the 3′OH of the L-ITR, a process that may not require viral proteins [39]. (C) The 3′ end of the newly synthesized complementary strand is likely ligated to the R-ITR, resulting in the formation of cRF DNA as the major conversion product cRF DNA.
(D) Further replication of cRF DNA requires NS1 to specifically bind the Ori and nicks the top strand at the trs. (E) This event creates a new 3′OH to lead DNA synthesis following melting of the hairpinned ITR, which subsequently repairs the ITR and results in an open-ended duplex replication intermediate. (G) The repaired ITR is then denatured, which likely requires the helicase activity of the NS1, and reannealed, in a process termed reinitiation, to form a double-hairpinned intermediate, which creates a new 3′ primer (3′OH) (H) to initiate a round of strand displacement synthesis.
B19V: Human parvovirus B19; cRF: Closed replicative form; ITR: Inverted terminal repeat; L-ITR: Left-hand inverted terminal repeat; NS1: Nonstructural proteins; RF: Replicative form; R-ITR: Right-hand inverted terminal repeat.
Understanding the mechanism underlying B19V DNA replication, a critical step to developing antivirus strategies has been impeded by the difficulty of efficiently propagating B19V in an in vitro cell culture system. Several breakthroughs have been made recently to improve and expand B19V infection in in vitro cultures of cells, including permissive, semipermissive and nonpermissive cells, which have greatly advanced the understanding of B19V infection and DNA replication. This article will summarize these studies and discuss the cellular requirements for B19V tropism and B19V DNA replication in these different cell systems.
B19V tropism for erythroid lineage cells
B19V infection and viral DNA replication are restricted by the narrow tropism of the virus. In nature, B19V mainly infects human erythroid progenitor cells (hEPCs) from bone marrow and liver [17,40–41], although restricted infection of other tissues has been frequently reported. B19V DNA replication was first observed in suspension cultures of human erythroid bone marrow from patients with hemolytic anemia [41], and its replication efficiency was greatly enhanced during infection of isolated hematopoietic progenitor cells from normal human bone marrow [40]. Identification of hEPCs as B19V target cells was first observed by Mortimer et al. [42] and Young and colleagues [43]. Further studies showed that B19V infection causes reduction of in vitro-cultured colony-forming unit erythroid and burst-forming unit erythroid cells, but only in the first few days in culture [44], indicating that efficient B19V DNA replication requires certain differentiation stages of the hEPCs. In particular, B19V only infects hEPCs with surface marker CD36+ differentiated from CD34+ human hematopoietic stem cells (HSCs), but not the HSCs and myeloid lineage cells [45–47]. The establishment of ex vivo-expanded CD36+ hEPCs from HSCs by Wong et al. has greatly advanced studies of B19V infection [47]. CD36+ hEPCs are highly permissive to B19V infection with productive B19V DNA replication occurring on a few interval days during the period of post-differentiation from CD34+ HSCs to CD36+ hEPCs [47]. Therefore, productive B19V infection only occurs for a short time during hEPC differentiation. It is not clear whether the differentiation status of CD36+ hEPCs affects B19V entry and virus trafficking and what are the cellular factors fluctuating during the differentiation to affect productive B19V infection.
A few megakaryocyte–erythroid lineage cell lines were documented to support B19V infection, including erythroid leukemic cell lines (KU812Ep6 and JK-11) [48–50], and human megakaryocytic leukemia cell lines (MB-02, UT7/Epo and UT7/Epo-S1) [51–53]. A comparison study evaluating their differences in permissiveness to B19V infection showed that the UT7/Epo-S1 cell line was the most sensitive cell line to B19V infection, based on detection of the viral NS1 protein and increased viral DNA production [53]. Although B19V infection in UT7/Epo-S1 cells is less efficient than that in hEPCs [54], it is particularly useful for the transfection of the B19V infectious clone and subsequent mutagenesis studies, since transfection is extremely difficult in hEPCs due to the nature of this type of primary cells.
At least two major factors have been identified to account for the tropism of B19V for hEPCs and these megakaryocyte–erythroid lineage leukemic cell lines. First, all these types of cells express globoside (erythrocyte P antigen), which is the primary receptor for B19V [55]. B19V capsid directly interacts with the globoside on erythroid cells, and pre-incubation antigloboside antibody or purified globoside prevents B19V infection of human bone marrow cells [55]. Individuals whose erythrocytes do not have globoside are naturally resistant to B19V infection [56]. Globoside is also present in red blood cells and some nonerythroid cells, such as fetal myocytes, placenta, megakaryocytes and some endothelial cells [57–60], which might explain the diverse clinical manifestations in different tissues. Besides the primary receptor globoside, integrin α5β1 and Ku80 were proposed to be coreceptors for B19V [61,62]; however, cell surface expression of Ku80 was shown to be very low in ex vivo-expanded CD36+ hEPCs (<5%) and other B19V-permissive cells [45,63]. Therefore, the erythroid tropism of B19V cannot be simply explained by the presence of the receptor/coreceptors. The second explanation is that all these megakaryocyte–erythroid lineage cells require erythropoietin (Epo) for growth, and Epo receptor (EpoR) signaling is required for B19V DNA replication [45]. In hEPCs, Epo binds to EpoR and activates the Janus kinase 2 (Jak2) by autophosphorylation [64]. Activated Jak2 further phosphorylates EpoR and initiates a kinase cascade with three major pathways, including the signal transducer and activator of transcription 5A (STAT5A), mitogen-activated protein kinase (ERK/MAPK) kinase (MEK) and phosphatidylinositol-3 kinase (PI3K). B19V infection of hEPCs is Epo concentration-dependent [45]. Unlike globoside, EpoR is not required for B19V entry because Epo exposure after virus entry still enables B19V DNA replication. In fact, the EpoR signaling of Jak2 phosphorylation and the MEK/ERK activation play a key role in facilitating B19V DNA replication in hEPCs [45].
In addition to these findings, a recent study showed that the VP1 unique region (VP1u) of the B19V capsid protein VP1 is essential for B19V binding and internalization during B19V infection of in UT7/Epo-S1 cells [65]. Purified-recombinant VP1u can also be internalized in UT7/Epo-S1 cells. The N-terminal 29 amino acids of the VP1u have been shown to be essential for binding and internalization of the VP1u, which is independent of the PLA2 activity that was thought to be critical to B19V infectivity [66]. Interestingly, the VP1u-interacting cellular partner was uniquely expressed on UT7/Epo-S1 and KU812Ep6 cells, but not on nonerythroid lineage cells, such as HeLa, HEK 293 and HepG2 cells [65], indicating the unique role of VP1u in facilitating B19V binding and internalization in erythroid lineage cells. Although the specific VP1u-interatcing molecule (receptor) has not been identified, this study presents a novel parvovirus internalization mechanism.
In conclusion, in in vitro cultures, B19V is mainly permissive to hEPCs and a few megakaryocyte–erythroid lineage leukemic cell lines. The tropism of B19V for erythroid lineage cells is largely due to the B19V receptor globoside and Epo/EpoR signaling, as well as VP1u that facilitates B19V internalization.
B19V infection of myocardial endothelial cells & monocytic cells
A few other nonerythroid lineage cells have been reported to support B19V infection [63,67–68]. B19V infection has been suggested to be associated with myocarditis and acute and chronic inflammatory cardiomyopathies, since B19V DNA was frequently detected in patients who have these symptoms [67,69–71]. B19V was also shown to be the most frequent pathogen detected in patients with normal coronary anatomy that clinically mimics acute myocardial infarction [69]. In addition, in vivo studies have demonstrated that B19V can also productively infect endothelial cells from heart [72] and placental tissues [73], indicating endothelial cells could be a natural target for B19V infection.
A recent study by Kietzell et al. identified a new route for the B19V infection of myocardial endothelial cells [63]. There are no major differences in surface expression of B19V receptor/coreceptors among UT7/Epo-S1 and primary endothelial cells, which are isolated from the pulmonary arteria (human pulmonary artery cells [HPAEC]), the umbilical vein (human umbilical vein endothelial cells [HUVEC]) and the aorta (human aortic endothelial cells [HAoEC]). B19V binds primary endothelial cells at a similar level as that of UT7/Epo-S1 cells. However, B19V internalization is deficient in these endothelial cells, a deficiency which is significantly enhanced by pre-incubation of the virus with anti-B19V antibodies. Mechanistically, the B19V-antibody complex might enter the cells through endocytosis mediated by the direct interaction of antibody-bound complement factor C1q with its receptor CD93 on the cell surface. Kietzell et al.’s study explains that the B19V genome and virus were detected in myocardial samples from patients with cardiac diseases [67], and provides an explanation for the frequent prevalence of B19V in endothelial cells from a variety of tissues, which may be related to the spread of B19V to other cell types [63]. However, the study does not provide evidence of B19V DNA replication in endothelial cells.
An early study has shown an antibody-dependent enhancement of B19V infection in monocytic cell line U937 cells [68]. B19V DNA was detected in infected U937 cells but with abortive B19V DNA replication. By addition of anti-B19V IgG, B19V DNA replication was significantly increased. However, the antibody-enhancement pathway appears to be different between endothelial and monocytic cells, as monocytic cells are speculated to be Fc receptor-mediated enhancement of B19V internalization [68].
In conclusion, collective evidence indicates that B19V enters myocardial endothelial cells, but lacks sufficient support to replicate in them. There is a long road ahead to prove B19V is a causative agent of myocardial diseases. It is attractive to speculate that the endocytosed B19V genome could be sensed by cytosol nor nuclear innate immunity DNA sensors, which induce proinflammatory cytokine secretion [74], and subsequently inflammatory cardiomyopathies. Additionally, the fact that B19V infects both the monocytic cell line and myocardial endothelial cells through an antibody-enhancement pathway suggests that the antibody-mediated B19V entry might be a common mechanism for B19V infection of nonerythroid lineage cells.
Identification of the B19V minimum DNA replication origin
Abortion of B19V infection was thought previously to be due to a block in full-length transcription maturation, as well as the in the conversion of viral ssDNA to double-stranded replicative intermediates [75–77]. With the available infectious DNA of B19V [27], recent studies have suggested that the abortive B19V infection in nonpermissive cells is largely due to the inefficient replication or nonreplication of B19V DNA in these cells. With the help of adenovirus, B19V DNA replicates in nonpermissive human embryonic kidney 293 cells (293 cells) [35,78]. 293 cells either infected with adenovirus or transfected with the pHelper plasmid, which contains the adenovirus genes E2A, E4orf6 and VA RNA, support B19V DNA replication [35]. In line with this, adenovirus infection also enhances B19V DNA replication in UT7/Epo-S1 cells. One explanation for this phenomenon is that adenovirus E1A protein transactivates the B19V promoter in nonpermissive cells [79]. Also, both B19V and AAV have symmetric ITRs at each end of the viral genome; thus, helped by adenovirus, B19V DNA replication in 293 cells may share the same mechanism as that in adenovirus-helped AAV DNA replication. A detailed examination of the function of the adenovirus gene products in B19V DNA replication confirmed that the E4orf6 protein and VA RNA functioned similarly to help B19V DNA replication as they do during adeno-associated virus 2 replication, while E2A had no stimulatory effect on B19V DNA replication or gene expression [78]. More specifically, the E4orf6 protein serves as a scaffold to form a cullin 5-based E3 ubiquitin ligase complex that targets cellular proteins, such as p53 and Mre11, for degradation, while the VA RNA binds and inactivates protein kinase PKR, a (ds)RNA-dependent protein kinase [78]. Notably, adenovirus gene products are not required for the DNA replication of human bocavirus and the canine virus analogue, minute virus of canines (MVC) [80,81]. Both human bocaviruses and MVC have asymmetric terminal hairpin structures, suggesting the differences of terminal hairpin structures may account for the different replication mechanisms of parvoviruses.
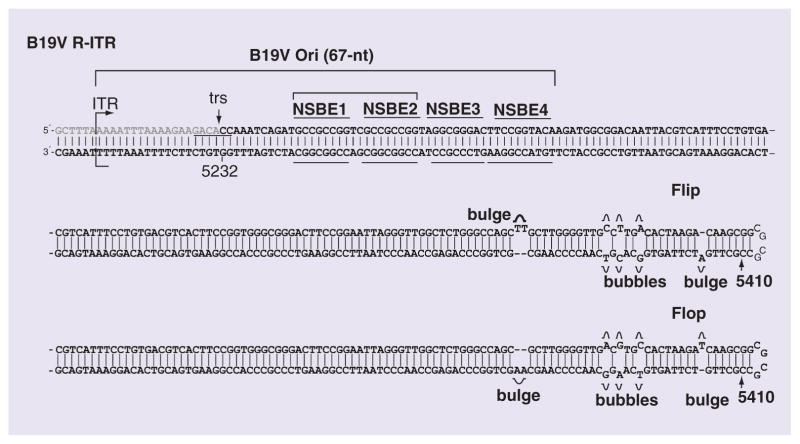
By using the 293 cell transfection system, a B19V DNA replication origin has been identified [35]. The minimum origin of B19V DNA replication is only 67 nucleotides (nucleotide 5214 to 5280 of GenBank: AY386330), which covers a NS1 trs and four repeated NS1 binding elements (NSBEs) (Figure 3). B19V NS1 specifically binds to the NSBE1–NSBE2 region in an in vitro binding assay, while NSBE3 and NSBE4 may provide binding sites for potential cellular factors [82], which should assemble a nucleoprotein complex involving cellular factors to separate the dsDNA strand and enable NS1 to nick the top strand at the trs (Figure 2D). Surprisingly, the mutant B19V infectious DNA with deletion of either the left-hand or the right-hand ITR still replicates in 293 cells with adenovirus infection or pHelper transfection. In addition, transfection of a B19V DNA fragment containing the NS1 expression cassette and only the Ori replicates in 293 cells in the presence of the three adenovirus gene products, as well as in UT7/Epo-S1 cells [35]. Based on these results, a hairpin-independent replication model has been proposed for B19V DNA replication [35]. It was expected that NS1 has the ability to reverse the direction of the replication (Figure 2F & G), regardless of the repairing/annealing of the ITR structure [35], for second-strand replacement synthesis (Figure 3H). However, the in-depth mechanism is not understood yet.
Figure 3. Structure of the human parvovirus B19 right-hand-inverted terminal repeat and the human parvovirus B19 minimal DNA replication origin (Ori).
The B19V right-hand ITR (R-ITR) at 365 nt is depicted in both the ‘Flip’ and the ‘Flop’ orientation, and the trs and NSBEs thought to comprise the NS1 binding site are indicated. The ITR is a nearly perfect palindromic structure; the exceptions are the three unpaired bases at two sites (shown as ‘bulges’) and three mismatched bases at three sites (shown as ‘bubbles’). The nucleotide sequences of the minimal B19V DNA replication origin (Ori) are indicated. The ITR sequence refers to the B19V J35 strain (GenBank accession number: AY386330).
B19V: Human parvovirus B19; ITR: Inverted terminal repeat; NSBE: NS1-binding element; trs: Terminal resolution site.
Despite of these interesting observations in B19V-permissive UT7/Epo-S1 cells and B19V nonpermissive 293 cells, it is not clear whether the hairpin-independent replication is employed during B19V infection of hEPCs. Also, little is known about its relevance to natural infection. The fact that B19V DNA replicates in nonpermissive 293 cells with the helper function of adenovirus provides a possibility that B19V coinfection with other viruses in nonerythroid lineage cells, for example, myocardial endothelial cells, facilitates a high level of B19V DNA replication, which may result in some disease outcomes and awaits more in vivo evidence.
Hypoxic conditions promote productive B19V infection of erythroid lineage cells
Although the ex vivo-expanded EPCs enable B19V DNA replication at a high efficiency, there is still a huge discrepancy in the production of progeny virions from infected hEPCs and during viremia of B19V-infected patients (1013 genomic copies per milliliter of plasma), indicating that other factors remain unidentified to recapture the in vivo B19V infection of human bone marrow in patients in vitro. One of these factors could be the oxygen level in human bone marrow, which is much lower than that in in vitro cell culture conditions [83].
In fact, a significant enhancement of B19V DNA replication as well as progeny virus production has been observed during B19V infection of hEPCs when they were cultured under 1% O2 (hypoxia) [54,84–85]. Interestingly, hypoxia actually reduces the differentiation potential and the proliferation rate of the hEPCs [54]. The productive B19V infection under hypoxia is facilitated neither through an increase in virus entry or intracellular trafficking nor through the network regulated by hypoxia-inducible factor, a common signaling sensor for hypoxia-induced signaling transduction [54]. Strikingly, hypoxia regulates Epo/EpoR receptor signaling, which is essential for B19V DNA replication [45], and thereby, enhances B19V DNA replication. Two pathways mediated by the Epo/EpoR/JAK2 pathways, including upregulation of STAT5 signaling and downregulation of MEK/ERK signaling, boost B19V DNA replication in both hEPCs and UT7/Epo-S1 cells [54].
A promising application of this finding is to study B19V DNA replication by transfecting the B19V infectious clone in UT7/Epo-S1 cells precultured under hypoxia. The inoculated infectious clone replicated efficiently and produced progeny virions that were highly infectious in EPCs [54], which holds promise to perform mutagenesis study of B19V DNA replication.
DNA damage response-facilitated B19V DNA replication
A large number of DNA viruses have been shown to induce a DNA damage response (DDR), and some of them, including parvovirus [86], require the DDR for efficient viral DNA replication [87,88]. DDR is triggered by damaged DNA structures, such as ssDNA breaks, dsDNA breaks and stalled replication forks. The signaling transduction is conducted by a set of host defense machinery that is composed of a number of signaling sensors, transducers and effectors. Three major DDR sensors have been identified, including ataxia telangiectasia-mutated kinase (ATM), ATM- and Rad3-related kinase (ATR) and DNA-dependent protein kinase catalytic subunit (DNA-PKcs). Each of these DDR sensors recognizes specific types of damaged (cellular) DNA structures and transduces a kinase cascade to numerous downstream mediators/effectors, which result in either cell cycle arrest for DNA repairing or apoptosis [89].
B19V infection of hEPCs activates the DDR signaling for the facilitation of viral DNA genome replication [90]. The B19V genome has two identical ITRs separated by a large ssDNA gap (Figure 2A), which is a perfect trigger to activate the ATR signaling [91]. In addition, B19V infection activates ATM and DNA-PKcs, possibly due to the intermediate replicative viral genome (Figure 2G&H) during B19V replication or the cross-interaction between DDR signaling. Notably, B19V hijacks ATR and DNA-PKcs, but not ATM, for viral DNA replication [90]. The DDR activation is associated with viral DNA replication status but not individual B19V structural or nonstructural proteins [92], suggesting that the DDR-involving proteins directly interact with the replicating B19V genome.
It is not clear how B19V replication is facilitated by the DDR signaling. Possibly, the ATR signaling plays a role in the first-strand DNA synthesis, which could be a DNA repair-associated DNA replication, and the DNA-PKcs activation recruits DNA ligase IV to ligate the RF DNA (Figure 2C).
Late S-phase-dependent B19V DNA replication
Cell-cycle arrest is required for a number of DNA viruses to modulate host microenvironment in such a way that favors viral DNA replication. Early studies have shown that B19V infections of both EPCs and UT7/Epo-S1 cells induced G2/M arrest, a status of 4N DNA content [53,92–94]. However, a more careful examination of B19V-infected cells using 5-bromo-2′-deoxyuridine (BrdU) incorporation combined with 4′,6′-diamidino-2-phenylindole hydrochloride (DAPI) staining demonstrated that B19V infection actually induces a cell-cycle status with 4N DNA content as well as with BrdU-incorporation, suggesting a late S phase arrest [30]. Expression of NS1 alone induces a true G2/M arrest, a status with 4N DNA content and without BrdU incorporation [30]. The NS1-induced G2/M arrest has been reported to be caused by the deregulation of E2F family transcription [93], and is not caused by B19V infection-induced DDR [92]. B19V infection-induced late S-phase arrest suggests that replication of the B19V DNA genome, as with other autonomous parvoviruses whose DNA replication depends on host cells arrested at S phase [31–33], requires cellular replication factors expressed in S phase. Similarly, during early infection of parvovirus MVC, MVC DNA replication induces a DDR, which in turn arrests the cells at S phase [95,96]. The S phase arrest further facilitates MVC DNA replication. We speculate that B19V DNA replication-induced DDR causes cells arrested at S phase, while expression of the NS1 solely arrests cells at a status with 4N DNA content, and that the compromising of these two arrests confers the infected cells at late S phase.
Specific cellular replication factors, such as the DNA polymerase, have not been identified for B19V DNA replication, largely because of the essential role of these factors for proliferation and survival, and the lack of an in vitro DNA replication system of B19V. Nevertheless, several S phase factors, including DNA polymerase delta, proliferating cell nuclear antigen (PCNA), replication factor complex 1 (RFC1), Cyclin A and mini-chromosome maintenance complex 2 (MCM2) are found to appear in the B19V DNA replication center [30]. Knockdown of MCM2 and MCM5 to a level that does not affect cell proliferation blocks B19V DNA replication, confirming that the S-phase cellular DNA replication factors are recruited by an unknown mechanism for B19V DNA replication. It would be important to know whether these factors are recruited through a DDR-dependent pathway [97].
Conclusion
B19V is the only member of the Erythroparvovirus genus of the Parvoviridae family, which infects humans. Although initially B19V was identified to infect erythroid lineage cells in bone marrow or fetal liver and to cause several mild to severe human hemalogical disorders, accumulating clinical reports indicate that B19V also infects nonerythroid lineage cells and may be associated with other diseases, such as myocarditis.
The selective B19V infection of erythroid lineage cells is due to the primary receptor globoside and Epo/EpoR signaling. B19V also infects myocardial endothelia cells and a monocytic cell line through an antibody-mediated enhancement pathway. In the presence of adenovirus gene products, B19V replicates in nonpermissive 293 cells and produces infectious progeny virions.
Mechanistic study of B19V DNA replication in erythroid lineage cells is greatly facilitated by the establishment of ex vivo-expanded CD36+ hEPCs and hypoxic conditions for B19V infection. B19V replicates in late S phase of infected cells by recruiting both cellular DNA replication/repairing factors (possibly through DDR signaling) to facilitate first strand synthesis. Thereafter, the dsDNA genome becomes competent for gene expression, NS1 binding and nicking at the replication origin, which initiates strand displacement of viral DNA synthesis, likely, through a hairpin-independent mechanism. Meanwhile, EpoR signaling, which is further enhanced under hypoxia, is crucial for B19V DNA replication and progeny virion production.
Future perspective
Studies of B19V infection and DNA replication have greatly increased our understanding of the B19V life cycle, which will shed light on identifying anti-B19V strategies and eventually a therapeutic approach to B19V infection-caused severe hematological disorders.
Although an in vitro cell culture system of B19V has been improved recently, the system still does not recapitulate B19V infection in vivo, which produces progeny virions at a high yield. A number of questions about the detailed mechanisms involved in B19V infection await exploration. For example, it is necessary to understand how EpoR signaling (STAT5 and MEK) affects B19V replication, what minimal cellular replication factors are involved in B19V DNA synthesis and whether these replication factors are recruited through a DNA repair- and S phase-dependent pathway. B19V infection arrests erythroid lineage cells at late S phase. Whether this represents a compromised condition forced by the NS1 protein and DDR signaling requires further study. Also, the transmission for transmission of B19V from the respiratory tract to human bone marrow, along with its infection of the myocardial system, liver system and possibly even more unidentified organs, warrants further investigation.
EXECUTIVE SUMMARY.
B19V infection of erythroid lineage cells
The primary receptor globoside and likely the coreceptor integrin α5β1 have been indicated for the Human parvovirus B19 (B19V) tropism for erythroid lineage cells.
Erythropoietin/erythropoietin receptor signaling is essential for B19V tropism and productive DNA replication.
Hypoxia boosts productive B19V infection through enhancement of erythropoietin receptor signaling.
B19V infection hijacks DNA damage response signaling to facilitate virus DNA replication.
B19V infection induces late S phase, which favors virus DNA replication.
B19V infection of nonpermissive cells
In the presence of adenovirus gene products, B19V DNA replicates in 293 cells.
A 67-nucleotide region of the B19V minimum DNA replication origin has been identified and a hairpin-independent model of B19V DNA replication has been proposed.
B19V infects myocardial cells and monocytic cell line U937 through an antibody-mediated entry pathway.
Footnotes
Financial & competing interests disclosure
The study was supported by NIH R01 AI070723. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
For reprint orders, please contact: reprints@futuremedicine.com
References
Papers of special note have been highlighted as:
• of interest;
•• of considerable interest
- 1.Cossart YE, Field AM, Cant B, Widdows D. Parvovirus-like particles in human sera. Lancet. 1975;1(7898):72–73. doi: 10.1016/s0140-6736(75)91074-0. [DOI] [PubMed] [Google Scholar]
- 2.Anderson MJ, Jones SE, Fisher-Hoch SP, et al. Human parvovirus, the cause of erythema infectiosum (fifth disease)? Lancet. 1983;1(8338):1378. doi: 10.1016/s0140-6736(83)92152-9. [DOI] [PubMed] [Google Scholar]
- 3.Cohen BJ, Buckley MM. The prevalence of antibody to human parvovirus B19 in England and Wales. J Med Microbiol. 1988;25(2):151–153. doi: 10.1099/00222615-25-2-151. [DOI] [PubMed] [Google Scholar]
- 4.Young NS, Brown KE. Parvovirus B19. N Engl J Med. 2004;350(6):586–597. doi: 10.1056/NEJMra030840. [DOI] [PubMed] [Google Scholar]
- 5.Brown KE. The expanding range of parvoviruses which infect humans. Rev Med Virol. 2010;20(4):231–244. doi: 10.1002/rmv.648. [DOI] [PubMed] [Google Scholar]
- 6.Heegaard ED, Brown KE. Human parvovirus B19. Clin Microbiol Rev. 2002;15(3):485–505. doi: 10.1128/CMR.15.3.485-505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molina KM, Garcia X, Denfield SW, et al. Parvovirus B19 myocarditis causes significant morbidity and mortality in children. Pediatr Cardiol. 2013;34(2):390–397. doi: 10.1007/s00246-012-0468-4. [DOI] [PubMed] [Google Scholar]
- 8.Koepsell SA, Anderson DR, Radio SJ. Parvovirus B19 is a bystander in adult myocarditis. Cardiovasc Pathol. 2012;21(6):476–481. doi: 10.1016/j.carpath.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Dina J, Villedieu F, Labombarda F, et al. Childhood myocarditis and parvovirus B19 genotypes. J Clin Virol. 2011;50(1):61–64. doi: 10.1016/j.jcv.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 10.Bock CT, Klingel K, Kandolf R. Human parvovirus B19-associated myocarditis. N Engl J Med. 2010;362(13):1248–1249. doi: 10.1056/NEJMc0911362. [DOI] [PubMed] [Google Scholar]
- 11.Adda J, Machado S, Eberst E, Roubille C, Macia JC, Roubille F. Parvovirus B19 infection and acute myocarditis. Intern Med. 2010;49(1):79. doi: 10.2169/internalmedicine.49.2606. [DOI] [PubMed] [Google Scholar]
- 12.Papadogiannakis N, Tolfvenstam T, Fischler B, Norbeck O, Broliden K. Active, fulminant, lethal myocarditis associated with parvovirus B19 infection in an infant. Clin Infect Dis. 2002;35(9):1027–1031. doi: 10.1086/342574. [DOI] [PubMed] [Google Scholar]
- 13.Mogensen TH, Jensen JM, Hamilton-Dutoit S, Larsen CS. Chronic hepatitis caused by persistent parvovirus B19 infection. BMC Infect Dis. 2010;10:246. doi: 10.1186/1471-2334-10-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bihari C, Rastogi A, Saxena P, et al. Parvovirus B19 associated hepatitis. Hepat Res Treat. 2013;2013:472027. doi: 10.1155/2013/472027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bihari C, Rastogi A, Rangegowda D, et al. Parvovirus B19 associated acute hepatitis and hepatosplenomegaly. Clin Res Hepatol Gastroenterol. 2014;38(1):e9–e10. doi: 10.1016/j.clinre.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 16.Rauff B, Idrees M, Shah SA, et al. Hepatitis associated aplastic anemia: a review. Virol J. 2011;8:87–88. doi: 10.1186/1743-422X-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morey AL, Fleming KA. Immunophenotyping of fetal haemopoietic cells permissive for human parvovirus B19 replication in vitro. Br J Haematol. 1992;82(2):302–309. doi: 10.1111/j.1365-2141.1992.tb06422.x. [DOI] [PubMed] [Google Scholar]
- 18.Cotmore SF, Agbandje-McKenna M, Chiorini JA, et al. The family Parvoviridae. Arch Virol. 2014;159(5):1239–1247. doi: 10.1007/s00705-013-1914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deiss V, Tratschin JD, Weitz M, Siegl G. Cloning of the human parvovirus B19 genome and structural analysis of its palindromic termini. Virology. 1990;175(1):247–254. doi: 10.1016/0042-6822(90)90205-6. [DOI] [PubMed] [Google Scholar]
- 20.Astell CR, Blundell MC. Sequence of the right hand terminal palindrome of the human B19 parvovirus genome has the potential to form a ‘stem plus arms’ structure. Nucleic Acids Res. 1989;17(14):5857. doi: 10.1093/nar/17.14.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shade RO, Blundell MC, Cotmore SF, Tattersall P, Astell CR. Nucleotide sequence and genome organization of human parvovirus B19 isolated from the serum of a child during aplastic crisis. J Virol. 1986;58(3):921–936. doi: 10.1128/jvi.58.3.921-936.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lou S, Xu B, Huang Q, et al. Molecular characterization of the newly identified human parvovirus 4 in the family Parvoviridae. Virology. 2012;422(1):59–69. doi: 10.1016/j.virol.2011.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cotmore SF, Tattersall P. Structure and organization of the viral genome. In: Kerr J, Cotmore SF, Bloom ME, Linden RM, Parrish CR, editors. Parvoviruses. Hodder Arnold; London, UK: 2005. pp. 73–94. [Google Scholar]
- ••24.Ozawa K, Ayub J, Hao YS, Kurtzman G, Shimada T, Young N. Novel transcription map for the B19 (human) pathogenic parvovirus. J Virol. 1987;61(8):2395–2406. doi: 10.1128/jvi.61.8.2395-2406.1987. The B19V transcription map was revealed during infection of human bone marrow cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo W, Astell CR. A novel protein encoded by small RNAs of parvovirus B19. Virology. 1993;195(2):448–455. doi: 10.1006/viro.1993.1395. [DOI] [PubMed] [Google Scholar]
- 26.St AJ, Astell CR. Identification and characterization of a family of 11-kDa proteins encoded by the human parvovirus B19. Virology. 1993;192(1):121–131. doi: 10.1006/viro.1993.1014. [DOI] [PubMed] [Google Scholar]
- ••27.Zhi N, Zadori Z, Brown KE, Tijssen P. Construction and sequencing of an infectious clone of the human parvovirus B19. Virology. 2004;318(1):142–152. doi: 10.1016/j.virol.2003.09.011. The first B19V infectious clone was constructed. [DOI] [PubMed] [Google Scholar]
- 28.Zhi N, Mills IP, Lu J, Wong S, Filippone C, Brown KE. Molecular and functional analyses of a human parvovirus B19 infectious clone demonstrates essential roles for NS1, VP1, and the 11-kilodalton protein in virus replication and infectivity. J Virol. 2006;80(12):5941–5950. doi: 10.1128/JVI.02430-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berns KI. Parvovirus replication. Microbiol Rev. 1990;54(3):316–329. doi: 10.1128/mr.54.3.316-329.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo Y, Kleiboeker S, Deng X, Qiu J. Human parvovirus B19 infection causes cell cycle arrest of human erythroid progenitors at late S phase that favors viral DNA replication. J Virol. 2013;87(23):12766–12775. doi: 10.1128/JVI.02333-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deleu L, Pujol A, Faisst S, Rommelaere J. Activation of promoter P4 of the autonomous parvovirus minute virus of mice at early S phase is required for productive infection. J Virol. 1999;73(5):3877–3885. doi: 10.1128/jvi.73.5.3877-3885.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oleksiewicz MB, Alexandersen S. S-phase-dependent cell cycle disturbances caused by Aleutian mink disease parvovirus. J Virol. 1997;71(2):1386–1396. doi: 10.1128/jvi.71.2.1386-1396.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parris DS, Bates RC. Effect of bovine parvovirus replication on DNA, RNA, and protein synthesis in S phase cells. Virology. 1976;73(1):72–78. doi: 10.1016/0042-6822(76)90061-1. [DOI] [PubMed] [Google Scholar]
- 34.Cotmore SF, Tattersall P. A rolling-hairpin strategy: basic mechanisms of DNA replication in the parvoviruses. In: Kerr J, Cotmore SF, Bloom ME, Linden RM, Parrish CR, editors. Parvoviruses. Hoddler Arond; London, UK: 2005. pp. 171–181. [Google Scholar]
- ••35.Guan W, Wong S, Zhi N, Qiu J. The genome of human parvovirus B19 virus can replicate in non-permissive cells with the help of adenovirus genes and produces infectious virus. J Virol. 2009;83(18):9541–9553. doi: 10.1128/JVI.00702-09. B19V genome replicates in nonpermissive 293 cells with the help of adenovirus gene products, and the B19V minimum replication origin was identified. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bratosin S, Laub O, Tal J, Aloni Y. Mechanism for circularization of linear DNAs: circular parvovirus MVM DNA is formed by a “noose” sliding in a “lasso”-like DNA structure. Proc Natl Acad Sci USA. 1979;76(9):4289–4293. doi: 10.1073/pnas.76.9.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lochelt M, Delius H, Kaaden OR. A novel replicative form DNA of Aleutian disease virus: the covalently closed linear DNA of the parvoviruses. J Gen Virol. 1989;70(Pt 5):1105–1116. doi: 10.1099/0022-1317-70-5-1105. [DOI] [PubMed] [Google Scholar]
- 38.Cotmore SF, Tattersall P. In vivo resolution of circular plasmids containing concatemer junction fragments from minute virus of mice DNA and their subsequent replication as linear molecules. J Virol. 1992;66(1):420–431. doi: 10.1128/jvi.66.1.420-431.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baldauf AQ, Willwand K, Mumtsidu E, Nuesch JP, Rommelaere J. Specific initiation of replication at the right-end telomere of the closed species of minute virus of mice replicative-form DNA. J Virol. 1997;71(2):971–980. doi: 10.1128/jvi.71.2.971-980.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Srivastava A, Lu L. Replication of B19 parvovirus in highly enriched hematopoietic progenitor cells from normal human bone marrow. J Virol. 1988;62(8):3059–3063. doi: 10.1128/jvi.62.8.3059-3063.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••41.Ozawa K, Kurtzman G, Young N. Replication of the B19 parvovirus in human bone marrow cell cultures. Science. 1986;233(4766):883–886. doi: 10.1126/science.3738514. The first study to establish B19V infection in in vitro-cultured cells. [DOI] [PubMed] [Google Scholar]
- 42.Mortimer PP, Humphries RK, Moore JG, Purcell RH, Young NS. A human parvovirus-like virus inhibits haematopoietic colony formation in vitro. Nature. 1983;302(5907):426–429. doi: 10.1038/302426a0. [DOI] [PubMed] [Google Scholar]
- 43.Young N, Harrison M, Moore J, Mortimer P, Humphries RK. Direct demonstration of the human parvovirus in erythroid progenitor cells infected in vitro. J Clin Invest. 1984;74(6):2024–2032. doi: 10.1172/JCI111625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi T, Ozawa K, Takahashi K, Asano S, Takaku F. Susceptibility of human erythropoietic cells to B19 parvovirus in vitro increases with differentiation. Blood. 1990;75(3):603–610. [PubMed] [Google Scholar]
- ••45.Chen AY, Guan W, Lou S, Liu Z, Kleiboeker S, Qiu J. Role of erythropoietin receptor signaling in parvovirus B19 replication in human erythroid progenitor cells. J Virol. 2010;84(23):12385–12396. doi: 10.1128/JVI.01229-10. The Epo/EpoR signaling is required for B19V infection of human erythroid progenitor cells (hEPCs) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Filippone C, Franssila R, Kumar A, et al. Erythroid progenitor cells expanded from peripheral blood without mobilization or preselection: molecular characteristics and functional competence. PLoS ONE. 2010;5(3):e9496. doi: 10.1371/journal.pone.0009496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••47.Wong S, Zhi N, Filippone C, et al. Ex vivo-generated CD36+ erythroid progenitors are highly permissive to human parvovirus B19 replication. J Virol. 2008;82(5):2470–2476. doi: 10.1128/JVI.02247-07. Ex vivo-expanded CD36+ hEPCs are highly permissive for B19V infection during interval differentiation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blumel J, Eis-Hubinger AM, Stuhler A, Bonsch C, Gessner M, Lower J. Characterization of parvovirus B19 genotype 2 in KU812Ep6 cells. J Virol. 2005;79(22):14197–14206. doi: 10.1128/JVI.79.22.14197-14206.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miyagawa E, Yoshida T, Takahashi H, et al. Infection of the erythroid cell line, KU812Ep6 with human parvovirus B19 and its application to titration of B19 infectivity. J Virol Methods. 1999;83(1–2):45–54. doi: 10.1016/s0166-0934(99)00105-6. [DOI] [PubMed] [Google Scholar]
- 50.Takahashi T, Ozawa K, Takahashi K, et al. DNA replication of parvovirus B 19 in a human erythroid leukemia cell line (JK-1) in vitro. Arch Virol. 1993;131(1–2):201–208. doi: 10.1007/BF01379092. [DOI] [PubMed] [Google Scholar]
- 51.Munshi NC, Zhou S, Woody MJ, Morgan DA, Srivastava A. Successful replication of parvovirus B19 in the human megakaryocytic leukemia cell line MB-02. J Virol. 1993;67(1):562–566. doi: 10.1128/jvi.67.1.562-566.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong S, Brown KE. Development of an improved method of detection of infectious parvovirus B19. J Clin Virol. 2006;35(4):407–413. doi: 10.1016/j.jcv.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 53.Morita E, Tada K, Chisaka H, et al. Human parvovirus B19 induces cell cycle arrest at G(2) phase with accumulation of mitotic cyclins. J Virol. 2001;75(16):7555–7563. doi: 10.1128/JVI.75.16.7555-7563.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••54.Chen AY, Kleiboeker S, Qiu J. Productive parvovirus B19 infection of primary human erythroid progenitor cells at hypoxia is regulated by STAT5A and MEK signaling but not HIF alpha. PLoS Pathog. 2011;7(6):e1002088. doi: 10.1371/journal.ppat.1002088. Hypoxia conditions significantly enhance B19V DNA replication in hEPCs through STAT5A and MEK signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown KE, Anderson SM, Young NS. Erythrocyte P antigen: cellular receptor for B19 parvovirus. Science. 1993;262(5130):114–117. doi: 10.1126/science.8211117. [DOI] [PubMed] [Google Scholar]
- 56.Brown KE, Hibbs JR, Gallinella G, et al. Resistance to parvovirus B19 infection due to lack of virus receptor (erythrocyte P antigen) N Engl J Med. 1994;330(17):1192–1196. doi: 10.1056/NEJM199404283301704. [DOI] [PubMed] [Google Scholar]
- 57.Marcus DM, Kundu SK, Suzuki A. The P blood group system: recent progress in immunochemistry and genetics. Semin Hematol. 1981;18(1):63–71. [PubMed] [Google Scholar]
- 58.Fellous M, Couillin P, Neauport-Sautes C, Frezal J, Billardon C, Dausset J. Studies of human alloantigens on man-mouse hybrids: possible syntheny between HL-A and P systems. Eur J Immunol. 1973;3(9):543–548. doi: 10.1002/eji.1830030904. [DOI] [PubMed] [Google Scholar]
- 59.Jordan JA, DeLoia JA. Globoside expression within the human placenta. Placenta. 1999;20(1):103–108. doi: 10.1053/plac.1998.0353. [DOI] [PubMed] [Google Scholar]
- 60.Rouger P, Gane P, Salmon C. Tissue distribution of H, Lewis and P antigens as shown by a panel of 18 monoclonal antibodies. Rev Fr Transfus Immunohematol. 1987;30(5):699–708. doi: 10.1016/s0338-4535(87)80138-1. [DOI] [PubMed] [Google Scholar]
- 61.Munakata Y, Saito-Ito T, Kumura-Ishii K, et al. Ku80 autoantigen as a cellular coreceptor for human parvovirus B19 infection. Blood. 2005;106(10):3449–3456. doi: 10.1182/blood-2005-02-0536. [DOI] [PubMed] [Google Scholar]
- 62.Weigel-Kelley KA, Yoder MC, Srivastava A. Alpha5beta1 integrin as a cellular coreceptor for human parvovirus B19: requirement of functional activation of beta1 integrin for viral entry. Blood. 2003;102(12):3927–3933. doi: 10.1182/blood-2003-05-1522. [DOI] [PubMed] [Google Scholar]
- ••63.von Kietzell K, Pozzuto T, Heilbronn R, Grossl T, Fechner H, Weger S. Antibody-mediated enhancement of parvovirus b19 uptake into endothelial cells mediated by a receptor for complement factor c1q. J Virol. 2014;88(14):8102–8115. doi: 10.1128/JVI.00649-14. B19V infection of myocardial endothelial cells is mediated through an antibody-dependent route. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oda A, Sawada K, Druker BJ, et al. Erythropoietin induces tyrosine phosphorylation of Jak2, STAT5A, and STAT5B in primary cultured human erythroid precursors. Blood. 1998;92(2):443–451. [PubMed] [Google Scholar]
- •65.Leisi R, Ruprecht N, Kempf C, Ros C. Parvovirus B19 uptake is a highly selective process controlled by VP1u, a novel determinant of viral tropism. J Virol. 2013;87(24):13161–13167. doi: 10.1128/JVI.02548-13. The N-terminus of VP1u is essential for B19V internalization of UT7/Epo-S1 cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Filippone C, Zhi N, Wong S, et al. VP1u phospholipase activity is critical for infectivity of full-length parvovirus B19 genomic clones. Virology. 2008;374(2):444–452. doi: 10.1016/j.virol.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schenk T, Enders M, Pollak S, Hahn R, Huzly D. High prevalence of human parvovirus B19 DNA in myocardial autopsy samples from subjects without myocarditis or dilative cardiomyopathy. J Clin Microbiol. 2009;47(1):106–110. doi: 10.1128/JCM.01672-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Munakata Y, Kato I, Saito T, Kodera T, Ishii KK, Sasaki T. Human parvovirus B19 infection of monocytic cell line U937 and antibody-dependent enhancement. Virology. 2006;345(1):251–257. doi: 10.1016/j.virol.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 69.Kuhl U, Pauschinger M, Bock T, et al. Parvovirus B19 infection mimicking acute myocardial infarction. Circulation. 2003;108(8):945–950. doi: 10.1161/01.CIR.0000085168.02782.2C. [DOI] [PubMed] [Google Scholar]
- 70.Lower FE, Menon S, Sanchez JA. Association of parvovirus B19 with plasma cell-rich myocardial infiltrates after heart transplantation. J Heart Lung Transplant. 2001;20(7):755–758. doi: 10.1016/s1053-2498(01)00233-9. [DOI] [PubMed] [Google Scholar]
- 71.Lambot MA, Noel JC, Peny MO, Rodesch F, Haot J. Fetal parvovirus B19 infection associated with myocardial necrosis. Prenat Diagn. 1999;19(4):389. [PubMed] [Google Scholar]
- 72.Bultmann BD, Klingel K, Sotlar K, et al. Fatal parvovirus B19-associated myocarditis clinically mimicking ischemic heart disease: an endothelial cell-mediated disease. Hum Pathol. 2003;34(1):92–95. doi: 10.1053/hupa.2003.48. [DOI] [PubMed] [Google Scholar]
- 73.Pasquinelli G, Bonvicini F, Foroni L, Salfi N, Gallinella G. Placental endothelial cells can be productively infected by Parvovirus B19. J Clin Virol. 2009;44(1):33–38. doi: 10.1016/j.jcv.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 74.Tam JC, Bidgood SR, McEwan WA, James LC. Intracellular sensing of complement C3 activates cell autonomous immunity. Science. 2014;345(6201):1256070. doi: 10.1126/science.1256070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu JM, Green SW, Shimada T, Young NS. A block in full-length transcript maturation in cells nonpermissive for B19 parvovirus. J Virol. 1992;66(8):4686–4692. doi: 10.1128/jvi.66.8.4686-4692.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gallinella G, Manaresi E, Zuffi E, et al. Different patterns of restriction to B19 parvovirus replication in human blast cell lines. Virology. 2000;278(2):361–367. doi: 10.1006/viro.2000.0673. [DOI] [PubMed] [Google Scholar]
- 77.Leruez M, Pallier C, Vassias I, Elouet JF, Romeo P, Morinet F. Differential transcription, without replication, of non-structural and structural genes of human parvovirus B19 in the UT7/EPO cell as demonstrated by in situ hybridization. J Gen Virol. 1994;75(Pt 6):1475–1478. doi: 10.1099/0022-1317-75-6-1475. [DOI] [PubMed] [Google Scholar]
- 78.Winter K, von Kietzell K, Heilbronn R, Pozzuto T, Fechner H, Weger S. Roles of E4orf6 and VA I RNA in adenovirus-mediated stimulation of human parvovirus B19 DNA replication and structural gene expression. J Virol. 2012;86(9):5099–5109. doi: 10.1128/JVI.06991-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ponnazhagan S, Woody MJ, Wang XS, Zhou SZ, Srivastava A. Transcriptional transactivation of parvovirus B19 promoters in nonpermissive human cells by adenovirus type 2. J Virol. 1995;69(12):8096–8101. doi: 10.1128/jvi.69.12.8096-8101.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang Q, Deng X, Yan Z, et al. Establishment of a reverse genetics system for studying human bocavirus in human airway epithelia. PLoS Pathog. 2012;8(8):e1002899. doi: 10.1371/journal.ppat.1002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sun Y, Chen AY, Cheng F, Guan W, Johnson FB, Qiu J. Molecular characterization of infectious clones of the minute virus of canines reveals unique features of bocaviruses. J Virol. 2009;83(8):3956–3967. doi: 10.1128/JVI.02569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tewary SK, Zhao H, Deng X, Qiu J, Tang L. The human parvovirus B19 non-structural protein 1 N-terminal domain specifically binds to the origin of replication in the viral DNA. Virology. 2014;449:297–303. doi: 10.1016/j.virol.2013.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chow DC, Wenning LA, Miller WM, Papoutsakis ET. Modeling pO(2) distributions in the bone marrow hematopoietic compartment. II. Modified Kroghian models. Biophys J. 2001;81(2):685–696. doi: 10.1016/S0006-3495(01)75733-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Caillet-Fauquet P, Draps ML, Di GM, de LY, Laub R. Hypoxia enables B19 erythrovirus to yield abundant infectious progeny in a pluripotent erythroid cell line. J Virol Methods. 2004;121(2):145–153. doi: 10.1016/j.jviromet.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 85.Pillet S, Le GN, Hofer T, et al. Hypoxia enhances human B19 erythrovirus gene expression in primary erythroid cells. Virology. 2004;327(1):1–7. doi: 10.1016/j.virol.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 86.Luo Y, Qiu J. Parvovirus infection induced DNA damage response. Future Virol. 2013;8:245–257. doi: 10.2217/fvl.13.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Turnell AS, Grand RJ. DNA viruses and the cellular DNA-damage response. J Gen Virol. 2012;93(Pt 10):2076–2097. doi: 10.1099/vir.0.044412-0. [DOI] [PubMed] [Google Scholar]
- 88.Lilley CE, Schwartz RA, Weitzman MD. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 2007;15(3):119–126. doi: 10.1016/j.tim.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 89.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••90.Luo Y, Lou S, Deng X, et al. Parvovirus B19 infection of human primary erythroid progenitor cells triggers ATR-Chk1 signaling, which promotes B19 virus replication. J Virol. 2011;85(16):8046–8055. doi: 10.1128/JVI.00831-11. B19V infection of hEPCs induces a DNA damage response that facilitates viral DNA replication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9(8):616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lou S, Luo Y, Cheng F, et al. Human parvovirus B19 DNA replication induces a DNA damage response that is dispensable for cell cycle arrest at G2/M phase. J Virol. 2012;86(19):10748–10758. doi: 10.1128/JVI.01007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wan Z, Zhi N, Wong S, et al. Human parvovirus B19 causes cell cycle arrest of human erythroid progenitors via deregulation of the E2F family of transcription factors. J Clin Invest. 2010;120(10):3530–3544. doi: 10.1172/JCI41805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Morita E, Sugamura K. Human parvovirus B19-induced cell cycle arrest and apoptosis. Springer Semin Immunopathol. 2002;24(2):187–199. doi: 10.1007/s00281-002-0099-6. [DOI] [PubMed] [Google Scholar]
- 95.Luo Y, Chen AY, Qiu J. Bocavirus infection induces a DNA damage response that facilitates viral DNA replication and mediates cell death. J Virol. 2011;85(1):133–145. doi: 10.1128/JVI.01534-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Luo Y, Deng X, Cheng F, Li Y, Qiu J. SMC1-mediated intra-S phase arrest facilitates Bocavirus DNA replication. J Virol. 2013;87(7):4017–4032. doi: 10.1128/JVI.03396-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shechter D, Gautier J. MCM proteins and checkpoint kinases get together at the fork. Proc Natl Acad Sci USA. 2004;101(30):10845–10846. doi: 10.1073/pnas.0404143101. [DOI] [PMC free article] [PubMed] [Google Scholar]